Abstract
In order to improve the detection specificity of iron oxide nanoparticles (IONPs) delivered to tumors, we embedded saturation pulses into the sweep imaging using Fourier transformation (SWIFT) sequence to suppress long T2 tissues and fat. Simulation of the Bloch equation was first conducted to study behavior of the saturation pulses of various lengths under different T2 and off-resonance conditions. MR experiments were then conducted using in vivo mouse xenografts and a phantom consisting of IONPs, vegetable oil, and explanted tumor specimen, without and with long T2 suppression under a 7 T magnetic field. For the in vivo study, arginine-glycine-aspartate (RGD) coated 10 nm IONPs (RGD-IONPs) were delivered to tumors implanted in nude mice through both intra-tumor and intravenous injections. Histological studies confirmed that RGD-IONPs efficiently homed to tumors through RGD-integrin interaction. Compared to conventional SWIFT, the proposed method resulted in sufficient suppression on long T2 species but less influence on short T2 species. For both the in vivo and ex vivo studies, significantly improved contrast-to-noise ratio (CNR) was achieved between the IONPs and the long T2 species.
Keywords: Long T2 suppression, Sweep imaging with Fourier transformation, SWIFT, Iron oxide nanoparticles, IONPs, Tumor
1. Introduction
Iron oxide nanoparticles (IONPs) have been widely used as a T2/T2*-shortening contrast agent in magnetic resonance imaging (MRI) for various applications, such as cell labeling and tracking, and drug delivery [1–8]. IONPs are usually recognized as a negative contrast agent, inducing hypointensities or signal void on MR images acquired with regular pulse sequences, such as the fast spin echo (FSE) and spoiled gradient echo (SPGR) sequences. A major challenge arises when one attempts to differentiate IONPs from tissues with short T2/T2* values, air cavities, or susceptibility artifacts, especially when the signal-to-noise ratio (CNR) is very low.
These problems can be mitigated with the use of recently developed pulse sequences such as ultra-short echo time (UTE) and sweeping imaging using Fourier transforms (SWIFT) sequences [9–13]. These sequences shorten the time interval between radiofrequency (RF) transmission and reception, and reduce data sampling down to only a few microseconds, detecting mainly the T1-shortening effect of IONPs that are manifested as positive contrast, inducing hyperintensities or signal increase on MR images. This development is important for improving both the detection specificity of IONPs as well as the image SNR. For example, the SWIFT sequence has been successfully applied to detect IONP-labeled stem cells grafted in the myocardium [14], while the UTE has been used to enhance IONP-based tumor detection [5,15]. However, long T2 species (such as fluids) and fat also appear bright in the UTE and SWIFT images, posing a challenge to detect the IONPs among these tissues. Therefore, suppressions of long T2 species and fat are crucial for improving the contrast and detection specificity of IONPs.
Various suppression approaches have been developed, and they can be mainly classified into two categories. The first strategy is focused on distinguishing changes of image contrast, when different scan parameters are applied. For example, one widely used approach is to scan an object using the UTE sequence with dual echo times [15–18]. The signal void of short T2 species is more severe in the second echo acquisition than that in the first one, while neither images show obvious changes for long T2 species. The subtraction of the two acquisitions consequently suppresses the signal from long T2 species to achieve the enhancement of short T2 species.
The second strategy is to implement RF saturation pulses. Because long T2 species have a narrow spectrum in the frequency domain and are sensitive to a long RF pulse (with short bandwidth), hence they can be suppressed by using a long RF pulse, with little influence on short T2 species [16,19]. An early study utilized a long rectangular π/2 pulse to excite long T2 species at the water resonant frequency, followed by a gradient spoiler to dephase the excited long T2 species, while leaving short T2 species minimally affected [19]. With the consideration of fat saturation, Du et al. shifted the central frequency of the suppression pulse between water and fat to suppress both components and improve the detection specificity of short T2 species using the UTE sequence at a 3 T magnetic field strength [20]. Instead of using a single banded pulse, an alternative method implemented either two separated RF pulses or a single dual-banded RF pulse focused at water and fat frequencies [19,21–23]. For example, Luhach et al. have investigated the prostate-to-bone tumor in ex vivo specimens using the SWIFT sequence inserted with a dual band Gaussian pulse [24].
In this work, we propose to suppress long T2 and fatty species in order to improve the detection specificity of the delivered IONPs to tumors in vivo and ex vivo using the SWIFT sequence. In order to achieve the goal with a simple design of the suppression pulses, we proposed to embed two Gaussian pulses into the SWIFT sequence, referred to as SWIFT-2sat. Similar with the UTE based saturation method [20], the SWIFT sequence embedded with only one Gaussian pulse, referred to as SWIFT-1sat, was also conducted to provide a comparison. To increase the amount of delivered IONPs, arginine-glycine-aspartate coated 10 nm IONPs (RGD-IONPs) were injected into 4 T1 tumor xenograft models for the in vivo study. Ex vivo scans were also performed with explanted tumors. Finally, histological studies were performed to verify the results of the MR studies.
2. Materials and methods
2.1. Bloch equation simulation
The Bloch equation simulation was performed to calculate the ratio between z-component of magnetization, Mz, and the initial magnetization, M0, after Gaussian pulses are applied with various pulse durations in the absence of any off-resonance effect [25]. The T2 of the magnetization varied from 10−2 to 102 ms, while the T1 was set to a large value (103 s) to minimize the influence of longitudinal relaxation. The amplitude of the Gaussian pulses was determined by the assumption that a perfect 90° flip angle can be achieved with an infinite long T2.
Then, the off-resonance behavior of the Gaussian pulses was investigated using the Bloch equation simulation [25], assuming T1 and T2 of the magnetization to be 103 s and 50 ms, respectively. The frequency offset varied from − 500 Hz to 500 Hz.
2.2. Arginine-Glycine-Aspartate (RGD) coated IONPs
c(Arg-Gly-Asp-DTyr-Lys) or c(RGDyK) is an RGD derivative. The water-soluble IONPs (Fe3O4) with carboxyl groups on the surface (10 nm, from Ocean Nanotech, Inc.) were mixed with N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-Hydroxysuccinimide (NHS) (100×) in the borate buffer (pH 8.3). After incubating at room temperature for 30 minutes, the activated particles were collected by centrifugation, washed 3 times by phosphate-buffered saline (PBS) (pH = 7.4), and re-dispersed in PBS. c (RGDyK) (200×) in dimethyl sulfoxide (DMSO) was added to the particles solution, and the mixture was incubated at room temperature for 2 hours. The resulting RGD conjugated IONPs were collected by centrifugation, washed, and re-dispersed in PBS.
2.3. Animal preparations
Animal studies were performed according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of University of Georgia. Mouse breast cancer cell line 4 T1 was cultured in 1640 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2 humidified condition. For tumor inoculation, a total of ~106 cells were subcutaneously injected to the right hind leg of three nude mice. The tumor size was inspected every three days, and was computed using the following equation: volume (mm3) = length × (width)2/2. MRI and histological studies were conducted about one week later when the average tumor size reached about 200 mm3. In detail, to first demonstrate the proposed approaches, intra-tumor injection of 0.1 mL aqueous suspensions of non-conjugated IONPs (5 mM Fe3+ in concentration) was conducted to one mouse to induce an area of highly concentrated IONPs inside the tumor. In vivo MR scans were conducted before and immediately after the injection. Secondly, the RGD-modified IONPs (10 mg Fe/Kg) were intravenously injected to the tail veins of two mice, and allowed 24 hours to target and accumulate in the tumors. Then, different approaches (in vivo MRI, ex vivo MRI and histology) were applied to examine the distribution of the RGD-IONPs inside the tumors.
2.4. Tumor specimen phantom
To visualize the detailed distribution of the RGD-IONPs, the tumor was harvested by surgery after the in vivo MR scans, and fixed in formalin for 72 hours before the tumor specimen was immerged into 1% agar gel inside a cylindrical glass container. To provide referential materials of different T2 values and resonant frequencies, two 0.3 mL plastic vials, filled with vegetable oil and aqueous suspensions of 1 mM IONPs, were also inserted into the agar gel to make a phantom.
2.5. MR experiments
All MR experiments were performed on a 7 T Varian Magnex small animal scanner (Agilent Technologies, Santa Clara, CA) with a maximum gradient strength of 440 mT/m. A high-pass birdcage coil (3.8 cm diameter) was used for both RF transmission and signal reception.
2.6. In vivo MR scans
A 2D FSE sequence was utilized to generate T2-weighted anatomical axial images of the tumor-bearing mice with the following parameters: TR/TE = 2 s/30 ms, echo train length = 8, spectral width of acquisition (sw) = 100 kHz, field-of-view (FOV) = 3.5 cm, slice thickness (thk) = 1 mm, image size = 256 × 256, scan time = 1.4 minutes. However, to minimize the image distortion due to intra-tumor injection of IONPs, a shorter TE was set to 12 ms rather than 30 ms.
Both 3D SPGR and SWIFT sequences were implemented to generate T1-weighted images. For the SPGR sequence, we utilized the shortest achievable TE for the scanner, and the scan parameters are listed in the following: flip angle = 6°, TR/TE = 6.5/3.15 ms, sw = 62.5 kHz, FOV = 50 × 50 × 50 mm3, size of image matrix = 256 × 256 × 256, scan time = 7.2 minutes.
For the SWIFT sequence, a gapped hyperbolic secant pulse, with the dimensionless shape factor n = 1 and truncation factor β = 7.6, was implemented to excite a 6° flip angle. The pulse was divided into 256 segments, and each segment consisted of 4 μs pulse element and 12 μs free induction decay for data sampling. The dead time between the end of a pulse element and the beginning of data sampling was set to 3.9 μs, referred to as the effective TE for SWIFT. The TR of SWIFT was set to 6 ms. The pulse bandwidth (bw) and the spectral width were both set to 62.5 kHz in this study. The entire 3D radial k-space consisted of 64,000 spokes, and covered a spherical FOV with the radius equal to 50 mm. The scan time of the SWIFT sequence was about 6.5 minutes. For both of the SPGR and SWIFT sequences, 512 dummy scans were implemented to achieve a steady state at the beginning of the scans.
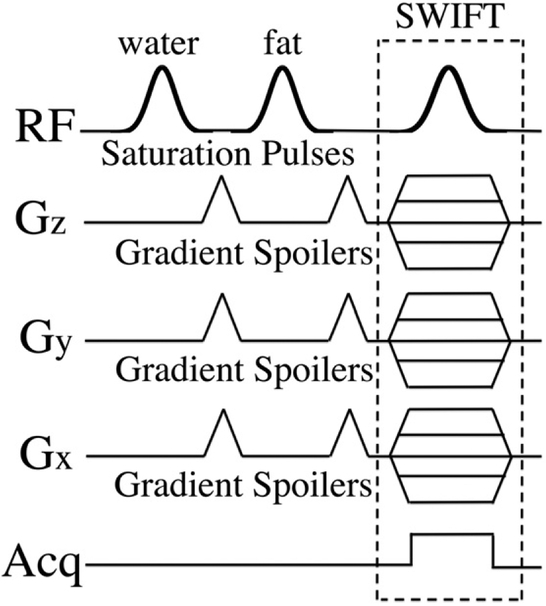
Additionally, the SWIFT-2sat sequence, as illustrated in Fig. 1, was conducted to further enhance contrast and detection specificity of IONPs. Specifically, the magnetization preparation was performed using two 90° Gaussian pulses placed at water and fat frequencies under 7 T magnet. Although the magnetic field was carefully shimmed before the experiment, the in vivo scan might still suffer poor field homogeneity. So in order to improve the tolerance of field inhomogeneity, a relatively short pulse length (8 ms) was selected for the two Gaussian pulses, according to the simulation result (shown in Fig. 2). Two 1 ms gradient spoilers were then presented immediately after each Gaussian pulse to dephase the excited long T2 species and fat, which makes the time interval between the two RF pulses equal to 1 ms. The scan time of the SWIFT-2sat sequence was about 7.7 minutes.
Fig. 1.
This figure illustrates the proposed SWIFT-2sat sequence. Two saturation pulses, placed at the water and fat resonant frequencies, are embedded in the regular SWIFT sequence to suppress long T2 and fat signals. Gradient spoilers follow each saturation pulse to dephase the excited signals.
Fig. 2.
The ratios of the longitudinal magnetization, Mz, and the initial magnetization, M0, were calculated after a 90° Gaussian pulse using the Bloch equation simulation. (a) Without any off-resonance effect, the ratio was plotted as a function of T2 for different pulse lengths. (b) By setting T2 to 50 ms, the ratio was plotted as a function of frequency offset for different pulse lengths. For all the two situations, T1 is set as 1000 s.
As a comparison, the SWIFT-1sat sequence used a single 90° Gaussian pulse, centered at the mid-point between water and fat frequencies, to suppress the long T2 tissues and fat simultaneously, followed by a gradient spoiler. By setting the pulse duration to 2 ms and 4 ms, two situations were investigated to study how the pulse width influences the saturation performance for the SWIFT-1sat sequence. The corresponding scan times were about 6.7 and 6.8 minutes, respectively. Note that for the above two methods, 16 radial spokes in the k-space were sampled using the SWIFT sequence after every saturation procedure.
2.7. Ex vivo MR scans
To further conduct quantitative measurement of IONP contrast improvement, the tumor of the mouse with systematic injection of RGD-IONPs was harvested by surgery. A phantom was made containing a vial of aqueous IONPs suspensions (1 mM Fe3+), a vial of vegetable oil, and the tumor specimen harvested. Both T2 and maps of the specimen were measured using spin echo and spoiled gradient echo sequences with five echo times: TE = 10, 20, 30, 40, and 50 ms for the spin echo sequence, and TE = 2.64, 6.74, 10.84, 14.94, and 19.04 ms for the spoiled gradient echo sequence. The rest scan parameters of both sequences were listed in the following: TR = 2.5 s, sw = 100 kHz, FOV = 352 mm2, thk = 1 ms, image size = 256 × 256, 16 axial slices, scan time = 10.7 minutes.
Additionally, 3D images were acquired by using the SPGR, SWIFT, and SWIFT-1sat sequences with the same scan parameters as that of the in vivo experiment. Compared with the in vivo experiment, a much better field homogeneity could be achieved for the phantom. Therefore, several longer durations (8, 16, 24, and 32 ms) of the Gaussian pulse at the water frequency were implemented to investigate the resulted performance of the SWIFT-2sat sequence on the long T2 suppression, resulting in the scan times approximately equal to 7.7, 8.2, 8.7, and 9.3 minutes. The fat saturation was fulfilled by using a fixed 8 ms Gaussian pulse with the flip angle of 135°.
2.8. Histological studies
Except using the MRI scans, the existence of RGD-IONPs inside the tumor was also verified using double staining of Prussian blue and murine β3. In detail, the snap-frozen tumor tissues were cut into slices of 8 μm thickness and mounted on glass slides. After dried at room temperature for 20 minutes, the slices were fixed in 10% formalin (w/v) for 30 minutes.
For the immunochemistry staining of murine β3, the fixed slices were incubated with 0.3% H2O2 solution in PBS for 10 minutes to block endogenous peroxidase activity and then gently washed by flowing water for 5 minutes 3 times. The slices were incubated with goat serum (5%, w/v) for 1 hour at room temperature and then washed with PBS. Primary antibody (ab75872) was then added and incubated with the slices overnight at 4°C. After gently washing by PBS (1×, pH 7.4) for 5 minutes 3 times, the slices were incubated with the second antibody (ab6721) for another 1 hour at room temperature. The slices were washed by PBS and then developed with 3,3'-diaminobenzidine (DAB) substrate solution until desired color intensity was reached.
Following the staining of murine β3, the resulting slices were immersed in Prussian blue staining working solution (a mixture of equal volume of 20% hydrochloric acid and 10% potassium ferrocyanide solution) for 40 minutes at room temperature. After being gently washed by PBS, the slices were dehydrated in a concentration gradient of ethanol (75%, 90%, and 100%). Subsequently, xylene was applied, and the tissues were mounted with Canada balsam. The images were acquired on a Nikon Eclipse 90i microscope.
2.9. Data processing
Images acquired using Cartesian sampling were obtained directly from fast Fourier transforms of the k-space data using Matlab (MathWorks, Natick, MA). The raw data of the SWIFT scans were first processed using house-developed code written in LabVIEW (National Instruments, Austin, TX), and then interpolated with a Kaiser-Bessel function onto a Cartesian grid utilizing compiled Matlab mex code, resulting in the image size = 256 × 256 × 256 [26]. Fast Fourier transforms of the post-processed raw data were performed to generate the complex SWIFT images at last.
Regarding the phantom images, the agar gel and the two vials containing IONPs and vegetable oil were segmented simply based on the difference of their image intensities [27], so that their mean intensities could then be calculated within the segmented regions. Noise standard deviation was estimated using regions outside the phantom. The SNRs of the IONPs, oil, and agar gel were derived as a ratio of the corresponding mean intensities over the noise standard deviation. The contrast-to-noise ratio (CNR) was calculated as a subtraction between the corresponding SNRs, respectively.
3. Results
Fig. 2 gives results of the Bloch equation simulation, illustrating the ratio of Mz over M0 as functions of T2 (Fig. 2a) and off-resonance frequency (Fig. 2b), respectively. Fig. 2a indicates that during the long Gaussian pulses, the short T2 species are barely affected and a smaller flip angle is resulted due to loss of coherence. For example, a 90° Gaussian pulse with the duration of 32 ms can only excite components with T2 of 1 ms to a flip angle about 30° (Mz/M0 ~ 0.85). However, even though a long Gaussian pulse has less influence on short T2 components, it is more susceptible to field off-resonance effects. As shown in Fig. 2b, the same 32 ms Gaussian pulse only has a full width at half maximum (FWHM) about 100 Hz, but the FWHM of a 2 ms Gaussian pulse is close to 1 kHz, which is more tolerant to field inhomogeneity. The latter was therefore selected for the SWIFT-1sat sequence to cover the frequency gap between water and fat under a 7 T magnetic field.
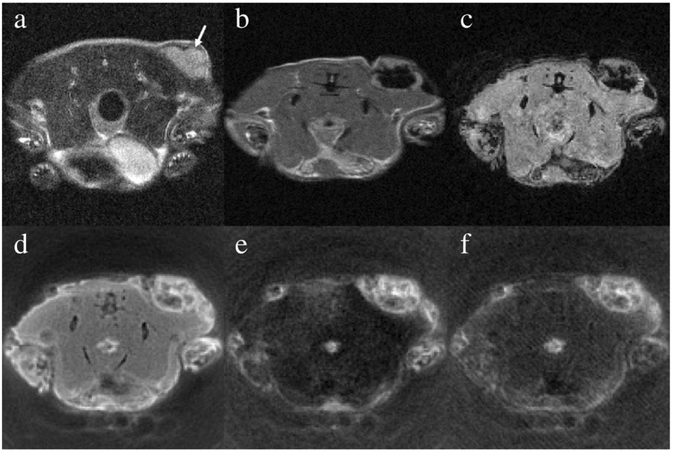
For the mouse, which received the intra-tumor injection of IONPs, T2-weighted pre-injection FSE image was acquired and shown in Fig. 3a, where the tumor appears brighter than the surroundings muscles. Compared to the pre-injection image, two post-injection MR images acquired using FSE and SPGR sequences showed significant signal loss inside the tumors in Fig. 3b and c, which are attributed to IONP-induced T2 shortening. With a much shorter TE of 3.9 μs, almost 3 orders of magnitude smaller than that used in the SPGR, signal void was significantly mitigated inside the tumor in the SWIFT image, as shown in Fig. 3d. By regarding the muscles as a long T2 material in this study, Fig. 3e shows an image taken by using the SWIFT-2sat sequence, for which two 8 ms suppression pulses were selected to improve its tolerance to the off-resonance effect. In Fig. 3e, we observed significantly enhanced tumor-to-tissue contrast, mainly owing to suppressed signals from long T2 species. As a comparison, the SWIFT-1sat sequence was conducted with a single 2 ms Gaussian pulse to suppress both long T2 components and fat, and the result shown inFig. 3f was comparable to Fig. 3e, with similar positive contrast from the IONPs. However, an increase of pulse duration to 4 ms (not shown in the Fig. 3) narrowed the FWHM about half of the frequency shift between water and fat at 7 T, and the single Gaussian pulse failed to suppress both the long T2 species and fat.
Fig. 3.
This figure presents the in vivo MR images before and after intra-tumor injection of the aqueous IONP suspension. (a) The pre-injection image was acquired using the fast spin echo sequence with TE = 30 ms. The tumor is indicated by the arrow. Immediately after the injection, the mouse was scanned using (b) the fast spin echo sequence with TE = 12 ms, (c) the spoiled gradient echo sequence with TE = 3.15 ms, (d) the SWIFT sequence with TE = 3.9 –s, (e) the SWIFT-2sat sequence with two 8 ms Gaussian pulses, and (f) the SWIFT-1sat sequence with one 2 ms Gaussian pulse.
Fig. 4presents the in vivo T2-weighted MR images for the mouse with systematic delivery of RGD-IONPs through tail vein injection. Compared with the surrounding muscles, the tumor shows a positive contrast in Fig. 4a, indicating that the tumor tissue had a longer T2 in the absence of RGD-IONPs. A presumable tumor necrotic core was observed and indicated by an arrow. After 24 hours post-injection, signal void in the T2-weighted FSE image (Fig. 4b) and T1-weighted SPGR image (Fig. 4c) was observed inside the tumor, especially surrounding the necrotic core, which is attributable to the delivered RGD-IONPs. Benefiting from the extremely short TE of the SWIFT sequence, no significant signal void was seen in Fig. 4d, but no significant contrast enhancement for the IONPs was observed either within the tumor region. An image taken by using the SWIFT-2sat method to suppress tissues and fat surrounding the tumor is shown in Fig. 4e, and as a result, it highlighted the delivered RGD-IONPs. Although an 8 ms Gaussian pulse was utilized to improve tolerance of field inhomogeneity, the off-resonance effect was still presented at the left side of Fig. 4e, where mouse tissues were not completely suppressed at the air-tissue boundary. Fig. 4f was acquired using the SWIFT-1sat sequence with a 2 ms Gaussian pulse, which seemed to indicate an over-saturation of the RGD-IONPs while suppressing the long T2 components.
Fig. 4.
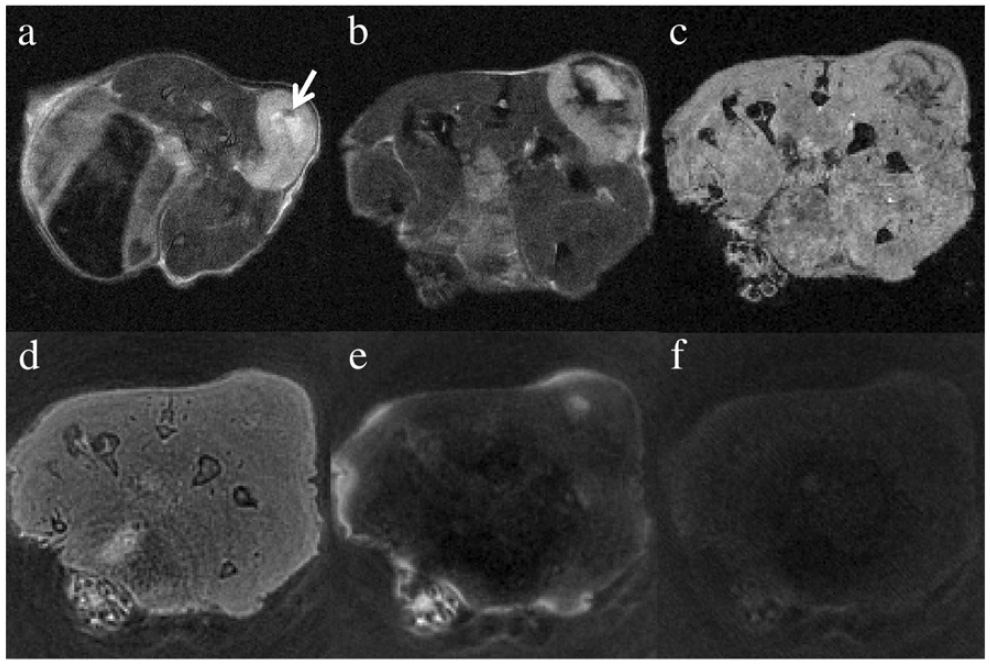
This figure presents in vivo MR images of the mouse before and after the intravenous injection of the RGD-IONPs. (a) The pre-injection was acquired using the fast spin echo sequence with TE = 30 ms. The arrow indicates a presumably necrotic core inside the tumor. After 24 hours of the intravenous injection, the mouse was scanned using (b) the fast spin echo sequence with TE = 30 ms, (c) the spoiled gradient echo sequence with TE = 3.15 ms, and (d) the SWIFT sequence with TE = 3.9 μs. To suppress tissues with long T2, (e) the SWIFT-2sat sequence with two 8 ms Gaussian pulses and (f) the SWIFT-1sat sequence with one 2 ms Gaussian pulse were implemented.
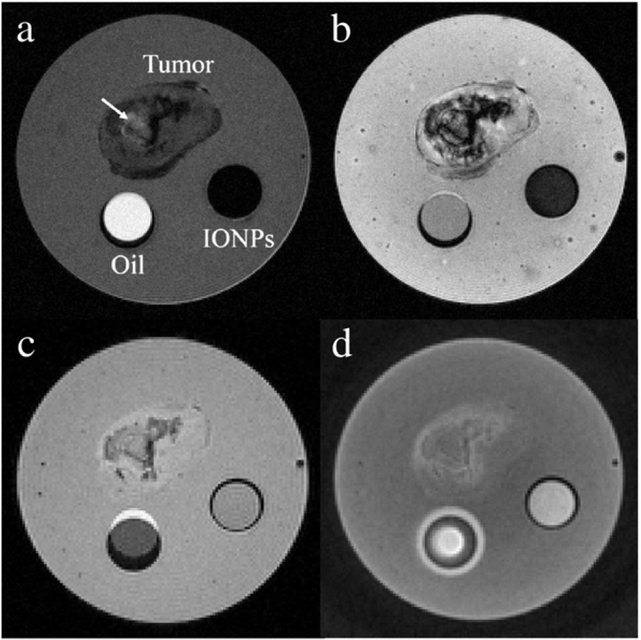
For the ex vivo experiment, Fig. 5 presents MR images of the phantom, consisting of two vials of aqueous IONPs suspension and vegetable oil, as well as the tumor specimen harvested from the mouse shown in Fig. 4. IONP-induced signal loss inside the vial was observed in both T2- and -weighted images (Fig. 5a–b). By using a 3D SPGR sequence with the shortest TE of 3.15 ms, the IONPs showed similar image intensity with the surrounding agar gel (Fig. 5c). Attributed to an extremely short TE, the SWIFT sequence is able to present the IONP suspension as positive contrast relative to the agar gel in Fig. 5d. Based on the negative contrast in Fig. 5a–b, the distribution of the delivered RGD-IONPs could be fairly specified in the tumor, but with long echo times, the signal void led to a poor signal-to-noise ratio (SNR). In Fig. 5a, the presumable necrotic core, firstly founded in Fig. 4, was indicated by the arrow. In Fig. 5c–d of the T1-weighted SPGR and SWIFT images, the tumor region is associated with a better SNR but a poor image contrast, making it hard to specify the RGD-IONPs.
Fig. 5.
This figure shows the MR images of the phantom using different scan protocols. The phantom consists of a vial of aqueous suspensions of 1 mM IONPs, a vial of vegetable oil, and the tumor specimen with the delivered RGD-IONPs. The images were acquired using (a) the spin echo sequence with TE = 30 ms, (b) the spoiled gradient echo sequence with TE = 10.84 ms, (c) the fast spoiled gradient echo sequence with TE = 3.15 ms, and (d) the SWIFT sequence with TE = 3.9 μs.
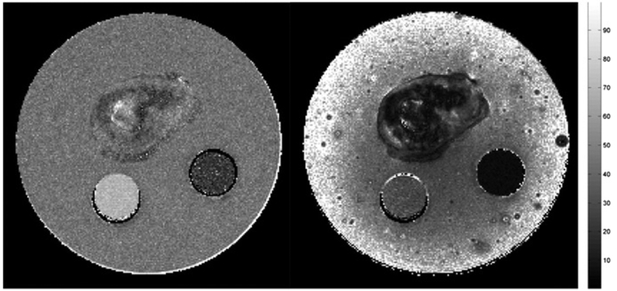
To further investigate the properties of the RGD-IONPs distributed regions, both the T2 and maps of the phantom were derived and presented in Fig. 6a and b, respectively. Due to inhomogeneous distribution of IONPs in the tumor, the T2 values of the area containing RGD-IONPs in the tumor are approximately between 20 ms to 30 ms, while the values are generally shorter than 10 ms. As a reference, the T2 and values of the 1 mM IONP suspension are about 23 ms and 5.5 ms, respectively.
Fig. 6.
This figure shows the (a)T2 and (b) maps of the phantom. The scale bar is in the unit of [ms].
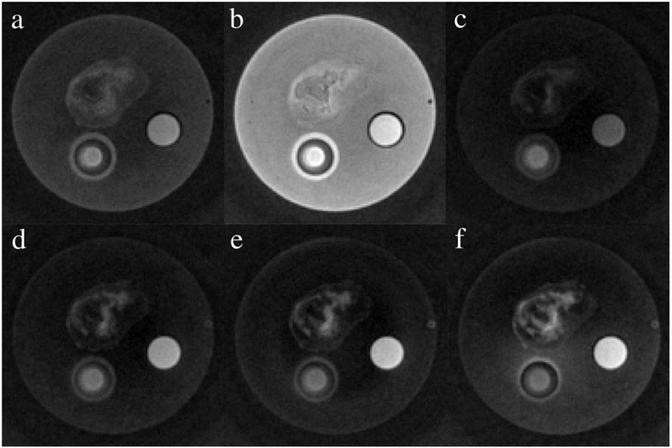
In order to improve detection specificity of IONP, different pulse widths for long T2 and fat suppressions were implemented for the SWIFT-1sat and SWIFT-2sat methods. Fig. 7a was obtained using the SWIFT-1sat sequence with a 2 ms Gaussian pulse. Inhomogeneous distribution of the RGD-IONPs in the tumor is clearly revealed, highlighted as positive contrast. The center area remained dark, likely a necrotic core. Noticeable suppressions of agar gel and oil were observed, but the SWIFT-1 sat also caused undesired suppression of IONPs both in the tumor and the vial. A further increase of the pulse width to 4 ms caused inefficient suppression of the long T2 species, leading to suboptimal contrast of the IONPs (Fig. 7b). This is probably because the FWHM, about 500 Hz for the 4 ms Gaussian pulse, is too narrow to cover the frequency gap between water and fat. According to the Bloch equation simulation in Fig. 2b, for SWIFT-2sat, we compared the performance of different pulse lengths (8, 16, 24, and 32 ms) for long T2 suppression, with a fixed 8 ms pulse length for fat suppression. As shown in Fig. 7c-f, the tumor tissue, agar gel, and oil were better suppressed compared with Fig. 7a, and the RGD-IONPs were clearly delineated inside the tumor, indicating a better performance of the SWIFT-2sat sequence. Consistent with the simulation result in Fig. 2a, these ex vivo results also confirmed that a short pulse might decrease the signal from IONPs. For example, details of the distribution of the delivered RGD-IONPs in the tumor region shown in Fig. 7c were suppressed compared with that in Fig. 7f. But on the other hand, Fig. 7f suffered the field off-resonance effect, when using a 32 ms Gaussian pulse with a narrow FWHM, resulting in that the agar gel located at lower half of the phantom was not fully suppressed.
Fig. 7.
This figure shows the phantom images obtained by using different suppression methods. (a) and (b) were acquired using the SWIFT-1sat sequence with 2 ms and 4 ms Gaussian pulses, respectively. For the images acquired using the SWIFT-2sat sequence, the first Gaussian pulse was set to (c) 8 ms, (d) 16 ms, (e) 24 ms, (f) and 32 ms to suppress long T2 species, respectively. And the second Gaussian pulse was fixed to 8 ms for fat suppression.
To further compare the performance of the SWIFT-1sat and SWIFT-2sat, the SNRs of the aqueous IONP suspension (SNRIONP), the vegetable oil (SNRoil), and the agar gel (SNRgel) were derived using Figs. 5d and 7 and presented in Table 1. For all of the three different imaging methods listed in the table, the IONP suspension has higher SNR values than those of the agar gel. Using the SWIFT-2sat method, the SNRgel was decreased to about one-third (from 21.2 down to 7.4) compared to that using the regular SWIFT method. Similarly, the oil signal was also suppressed about half (from 32 down to 13.6) by the SWIFT-2sat method. With a 2 ms Gaussian pulse, the SWIFT-1sat method also approximately reduced the SNRs of agar gel by 50% and oil by 30%, but not as good as the SWIFT-2sat method. When the Gaussian pulse was increased to 4 ms for the SWIFT-1sat sequence, the SNR values of the three materials were almost the same as that resulted from the regular SWIFT method.
Table 1.
SNR and CNR for the phantom study.
| Regular SWIFT |
SWIFT-1sat |
SWIFT-2sat |
||||||
|---|---|---|---|---|---|---|---|---|
| 2 ms | 4 ms | 8 ms | 16 ms | 24 ms | 32 ms | |||
| SNR | IONPs | 27.5 ± 2.8 | 21.5 ± 2.3 | 27.8 ± 3.2 | 14.0 ± 1.5 | 21.7 ± 2.1 | 24.4 ± 2.4 | 26.8 ± 2.6 |
| Gel | 21.2 ± 2.0 | 11.1 ± 1.2 | 22.0 ± 2.1 | 6.1 ± 0.8 | 6.0 ± 0.8 | 5.8 ± 0.8 | 7.4 ± 1.0 | |
| Oil | 32.0 ± 5.3 | 20.7 ± 3.4 | 28.6 ± 5.2 | 17.1 ± 1.7 | 15.5 ± 1.4 | 14.0 ± 1.3 | 13.6 ± 1.4 | |
| CNR | IONPs-Gel | 6.3 | 10.4 | 5.8 | 7.9 | 15.7 | 18.6 | 19.4 |
| IONPs-Oil | −4.5 | 0.8 | −0.8 | −3.1 | 6.2 | 10.4 | 13.2 | |
Table 1 presents the mean and standard deviation of the SNR as well as the resulted CNR of the IONP suspension, the agar gel, and the vegetable oil in the phantom, acquired by using different scan protocols. For the SWIFT-2sat sequence, the fat saturation was fulfilled by using a fixed 8 ms Gaussian pulse with the flip angle of 135°.
In addition to SNR, the CNR between the IONP suspensions and the agar gel (referred to as CNRIONP-gel) as well as that between the IONP suspensions and the vegetable oil (referred to as CNRIONP-oil) were also derived. Unlike the T2- or -weighted scans, the SWIFT sequence was able to generate positive CNRIONP-gel. About 40% increase of CNRIONP-gel was reached using the SWIFT-1sat method with a 2 ms Gaussian pulse length. When using the SWIFT-2sat sequence, the highest CNRIONP-gel and CNRIONP-oil were about 3times larger than that obtained with the regular SWIFT sequence. Both the CNRIONP-gel and CNRIONP-oil became larger along with the increase of the pulse length, which again implies that a short pulse duration may partially suppress the signal from IONPs.
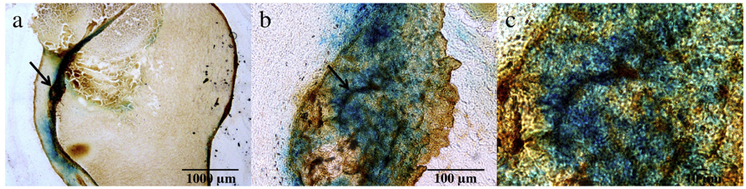
To further investigate the tumor targeting, we performed histological studies with the dissected tumor tissues (Fig. 8). Specifically, Prussian blue and integrin β3 double staining were conducted. Prussian blue staining is a sensitive histochemical test for Fe and has been widely used to study distribution of iron-containing nanoparticles both ex vivo and in vivo. In the current tumor tissues, a high level of iron deposit (blue) was found in the masses, suggesting good accumulation in the tumor. This is consistent to the signal loss shown in the post-injection MR images before suppression, and indicates that the highlighted regions by using SWIFT-2sat and SWIFT-1sat are more likely due to the IONPs in the tumor. An anti-integrin β3 antibody was used to reveal the distribution of integrin αvβ3 in the tissue samples. Integrinαvβ3 is known to be overexpressed on neoplastic vasculature and also, on cancer cells of various types, such as U87MG cells. Indeed, positive β3 staining (brown) was found across the tumor tissues. Overall good correlation was found between the two stainings, indicating that the particle accumulation was mainly mediated by RGD-integrin interaction.
Fig. 8.
This figure presents the histological studies of the tumor tissues with the delivered RGD-IONPs. Figures (b) and (c) show magnified areas of the arrow-pointed regions in (a) and (b), respectively. The results of the Prussian blue staining and the murine (β3 staining indicate that the tumor targeting was mainly mediated by RGD-integrin interactions, and the highlighted regions in the MR images acquired by using SWIFT-2sat and SWIFT-1sat are more likely due to the IONPs in the tumor.
4. Discussion
In this study, both in vivo and ex vivo experiments were conducted using various pulse sequences to acquire MR images for detection of IONPs deposited in the implanted tumors. Generally, T2- and -weighted images are usually associated with a poor SNR, which is attributed to the signal incoherence caused by IONPs due to the T2-shortening effect. Such an effect is manifested by negative contrast of IONPs, which compromises the detection of IONPs as a contrast agent. The SWIFT sequence can improve the SNR of the IONP deposited regions; however, the contrast between IONPs and surrounding tissues is improved only marginally. In order to further improve the detection specificity of IONPs, we introduced herein the SWIFT-2sat sequence, which combines the SWIFT sequence with magnetization preparation The two Gaussian pulses target the frequencies of water and fat, respectively. It is expected that fat and long T2 species, such as tumor tissues, can be efficiently saturated in the acquired SWIFT-2sat images. On the other hand, short T2 species, mainly IONPs in this study, are minimally influenced due to their insensitivity to the long RF pulses.
SWIFT-2sat was demonstrated on a 7 TMR scanner, and compared with the SWIFT-1satmethod. Since the SWIFT-1sat method only used one saturation pulse with its central frequency placed at the mid-point of the water and fat, the FWHM of the implemented pulse has to be set wide enough to cover the frequency gap between water and fat. This corresponds to a 2 ms pulse duration under 7 T according to the Bloch equation simulation (the red curve in Fig. 2b). Because short T2 species are also subjected to RF pulses with wide FWHM, the signals from IONPs were partially saturated by the 2 ms Gaussian pulse as well. Alonger pulse duration of 4 ms, on the other hand, failed to achieve the saturation result, as shown in Fig. 7b.
Highly concentrated IONPs appear bright in the T1-weighted SWIFT images. With an intra-tumor delivery, the IONPs in the tumor could be easily delineated from the surrounding tissues in Fig. 3d, even though the contrast was further improved in Fig. 3e and 3f by using the SWIFT-2sat and SWIFT-1sat sequences. However, it is practically impossible to deliver such a high concentration of IONPs to the areas of interest, such as tumors. At a low iron concentration, the spectral profile of the IONPs could become narrow and close to that of regular tissues [19], so that saturation using a short RF pulse may downgrade the image SNR and reduce the contrast between IONPs and surrounding tissues. As demonstrated in Table 1, the SNR of the IONPs increases from 14 to 26.8, which is about 2 times higher, when the pulse duration increases from8 msto 32 msin the SWIFT-2sat method. Another example can be found in Fig. 4f wherein the utilization of a 2 ms Gaussian pulse led to an over-saturation of all the species, including RGD IONPs.
Both the simulation and the experimental results suggest that the pulse duration is dependent on several factors, such as field homogeneity, the magnetic field strength, the size of the tumor, the amount of IONPs injected, the amount of long T2 components and fat present in the sample, etc. As a result, the pulse duration should be determined with a proper consideration of all or most of these factors. Off resonance effect may also comprise the image quality with the remaining long T2 signal unsuppressed. This is crucial especially for an in vivo scan, since a homogeneous field shimming is usually harder to achieve compared with a phantom scan. For example, with an 8 ms Gaussian pulse, long T2 signals were suppressed around the center area of the mouse body, but they still existed at the peripheral air-tissue boundary in Fig. 4e. However, for the ex vivo scan, insufficient suppression only happened when a 32 ms Gaussian pulse was applied (as shown in Fig. 7f). Therefore, a proper selection of the pulse duration is required to optimize image contrast and improve the detection specificity of IONPs, and the optimal pulse duration may vary on a case-by-case basis. We believe that the SWIFT-2sat sequence has an advantage over the SWIFT-1sat in the presence of a low iron concentration. However, the performance of the two is similar in the presence of a high iron concentration, where such an example can be found in Fig. 3(e) and (f).
In addition to optimization of the pulse length, we also tried to minimize the inhomogeneous signal suppression from possible sources, such as inhomogeneous B0 and B1 fields. In this study, a careful shimming procedure of the B0 magnetic field was conducted before all the scans. And a birdcage coil, instead of a surface coil, was used in order to provide a more homogeneous B1 field. One of potential drawbacks of our approach is that if the concentration of IONPs is too low in the target site to make the local T2 values lower enough than those of the surrounding tissues, the proposed method will be unable to sufficiently suppress the surrounding tissues and consequently detect the accumulated IONPs. Thus, to solve such an issue, future work will be conducted not only on optimizing pulse duration, designing tailored RF pulses using the Shinnar-Le Roux algorithm [28,29], but also on improving the delivery efficiency of IONPs using more robust coating materials. Another drawback is that the number of mice used in this study was not statistically very large, although this work consisted of in vivo, ex vivo, and histological experiments. During the experiments, we did find that the image quality and contrast for different mice were not solely affected by the saturation pulses, but also by many other factors, such as field shimming and anatomical variations among individual mice. Therefore, repetitions of both in vivo and ex vivo experiments will also be included in the further work to make the results more statistically sound. Nevertheless, in addition to improving detection specificity of IONPs, our method is potentially useful in clinical scans to enhance image contrast of short T2 materials, such as cortical bones, teeth, and calcifications in various bio-tissues.
5. Conclusion
In conclusion, the SWIFT sequence in combination with Gaussian pulses for suppression of long T2 components can successfully enhance the contrast of RGD-IONP, and therefore improves its detection specificity. Compared with the original SWIFT and SWIFT-1sat sequences, the SWIFT-2sat sequence effectively suppressed long T2 species while generating minimum influence on short T2 species, including the RGD-IONPs inside the tumor, even in the presence of a low iron concentration.
Acknowledgments
We gratefully acknowledge support from the following grants: Faculty Research Grant from the University of Georgia (Zhao), S10RR023706 from the National Center for Research Resources, and 5R00CA153772 from NIH (J. X.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources. In addition we wish to thank Dr. Michael Garwood, Dr. Curtis Corum, and Dr. Djaudat Idiyatullin from the Center of Magnetic Resonance Research at the University of Minnesota, for providing their supports on the SWIFT sequence. We also thank Dr. Khan Hekmetyar for assisting MR scan at the BioImaging Research Center (BIRC) of the University of Georgia.
References
- [1].Mills PH, Hitchens TK, Foley LM, Link T, Ye Q Weiss CR, et al. Automated detection and characterization of SPIO-labeled cells and capsules using magnetic field perturbations. Magn Reson Med 2012;67(1):278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu W, Frank JA. Detection and quantification of magnetically labeled cells by cellular MRI. EurJ Radiol 2009;70(2):258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Richards JM, Shaw CA, Lang NN, Williams MC, Semple SI, Macgillivray TJ, et al. In vivo mononuclear cell tracking using superparamagnetic particles of iron oxide: feasibility and safety in humans. Circ Cardiovasc Imaging 2012;5(4):509–17. [DOI] [PubMed] [Google Scholar]
- [4].Ramaswamy S, Schornack PA, Smelko AG, Boronyak SM, Ivanova J, Mayer JE Jr, et al. Superparamagnetic iron oxide (SPIO) labeling efficiency and subsequent MRI tracking of native cell populations pertinent to pulmonary heart valve tissue engineering studies. NMR Biomed 2012;25(3):410–7. [DOI] [PubMed] [Google Scholar]
- [5].Zhang LJ, Zhong XD, Wang LY, Chen HW, Wang YA, Yeh JL, et al. T1 weighted ultrashort echo time method for positive contrast imaging of magnetic nanoparticles and cancer cells bound with the targeted nanoparticles. J Magn Reson Imaging 2011;33(1):194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hasenpusch G, Geiger J, Wagner K, Mykhaylyk O, Wiekhorst F, Trahms L, et al. Magnetized aerosols comprising superparamagnetic iron oxide nanoparticles improve targeted drug and gene delivery to the lung. Pharm Res 2012;29(5):1308–18. [DOI] [PubMed] [Google Scholar]
- [7].Singh I, Rana V. Iron oxide induced enhancement of mucoadhesive potential of Eudragit RLPO: formulation, evaluation and optimization of mucoadhesive drug delivery system. Expert Opin Drug Deliv 2013;10(9):1179–91. [DOI] [PubMed] [Google Scholar]
- [8].Ji C, Miller PA, Miller MJ. Iron transport-mediated drug delivery: practical syntheses and in vitro antibacterial studies of tris-catecholate siderophore-aminopenicillin conjugates reveals selectively potent antipseudomonal activity. J Am Chem Soc 2012;134(24):9898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tyler DJ, Robson MD, Henkelman RM, Young IR, Bydder GM. Magnetic resonance imaging with ultrashort TE (UTE) PULSE sequences: technical considerations. J Magn Reson Imaging 2007;25(2):279–89. [DOI] [PubMed] [Google Scholar]
- [10].Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging.J Comput Assist Tomogr 2003;27(6):825–6. [DOI] [PubMed] [Google Scholar]
- [11].Idiyatullin D, Corum C, Park JY, Garwood M. Fast and quiet MRI using a swept radiofrequency. J Magn Reson 2006;181(2):342–9. [DOI] [PubMed] [Google Scholar]
- [12].Idiyatullin D, Corum C, Moeller S, Garwood M. Gapped pulses for frequency-swept MRI.J Magn Reson 2008;193(2):267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garwood M MRI of fast-relaxing spins. J Magn Reson 2013;229:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou R, Idiyatullin D, Moeller S, Corum C, Zhang H, Qiao H, et al. SWIFT detection of SPIO-labeled stem cells grafted in the myocardium. Magn Reson Med 2010;63(5):1154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Girard OM, Du J, Agemy L, Sugahara KN, Kotamraju VR, Ruoslahti E, et al. Optimization of iron oxide nanoparticle detection using ultrashort echo time pulse sequences: comparison of T1, T2*, and synergistic T1–T2* contrast mechanisms. Magn Reson Med 2011;65(6):1649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rahmer J, Blume U, Bornert P. Selective 3D ultrashort TE imaging: comparison of “dual-echo” acquisition and magnetization preparation for improving short-T 2 contrast. Magma 2007;20(2):83–92. [DOI] [PubMed] [Google Scholar]
- [17].Martirosian P, Schraml C, Springer F, Schwenzer NF, Wurslin C, Schick F, et al. Positive contrast MR imaging of tendons, ligaments, and menisci by subtraction of signals from a double echo steady state sequence (Sub-DESS). Magn Reson Med 2014;71(1):294–301. [DOI] [PubMed] [Google Scholar]
- [18].Rahmer J, Bornert P, Groen J, Bos C. Three-dimensional radial ultrashort echo time imaging with T2 adapted sampling. Magn Reson Med 2006;55(5):1075–82. [DOI] [PubMed] [Google Scholar]
- [19].Larson PE, Gurney PT, Nayak K, Gold GE, Pauly JM, Nishimura DG. Designing long T2 suppression pulses for ultrashort echo time imaging. Magn Reson Med 2006;56(1):94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Du J, Bydder M, Takahashi AM, Carl M, Chung CB, Bydder GM. Short T2 contrast with three-dimensional ultrashort echo time imaging. Magn Reson Imaging 2011;29(4):470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Du J, Carl M, Bae WC, Statum S, Chang EY, Bydder GM, et al. Dual inversion recovery ultrashort echo time (DIR-UTE) imaging and quantification of the zone of calcified cartilage (ZCC). Osteoarthritis Cartilage 2013;21(1):77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Du J, Carl M, Bydder M, Takahashi A, Chung CB, Bydder GM. Qualitative and quantitative ultrashort echo time (UTE) imaging of cortical bone. J Magn Reson 2010;207(2):304–11. [DOI] [PubMed] [Google Scholar]
- [23].Springer F, Steidle G, Martirosian P, Grosse U, Syha R, Schabel C, et al. Quick water-selective excitation of fast relaxing tissues with 3D UTE sequences. Magn Reson Med 2014;71:534–43. [DOI] [PubMed] [Google Scholar]
- [24].Luhach I, Idiyatullin D, Lynch C, Corum C, Martinez G, Garwood M, et al. Rapid ex vivo imaging of PAIII prostate to bone tumor with SWIFT-MRI. Magn Reson Med 2013. 10.1002/mrm.24979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Murase K, Tanki N. Numerical solutions to the time-dependent Bloch equations revisited. Magn Reson Imaging 2011;29(1):126–31. [DOI] [PubMed] [Google Scholar]
- [26].Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding. IEEE Trans Med Imaging 1991;10(3):473–8. [DOI] [PubMed] [Google Scholar]
- [27].Gonzalez RC, Woods RE, Eddins SL. Digital Image processing using MATLAB. Upper Saddle River (NJ): Pearson Prentice Hall; 2004. [Google Scholar]
- [28].Lee KJ. General parameter relations for the Shinnar-Le Roux pulse design algorithm. J Magn Reson 2007;186(2):252–8. [DOI] [PubMed] [Google Scholar]
- [29].Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm. IEEE Trans Med Imaging 1991;10(1):53–65. [DOI] [PubMed] [Google Scholar]