Abstract
Heat treatment of colostrum is performed on modern dairy farms to reduce pathogenic contamination before hand-feeding the colostrum to newborn calves; however, limited data are available concerning effects of heat treatment on biologically active proteins in colostrum. The objective of this exploratory study was to investigate effects of heat treatment and length of heat treatment on colostrum protein profile. Colostrum samples were collected from Holstein cows within 12 h after parturition and assigned to the following groups: heat treatment at 60°C for 0 (untreated control), 30, 60, or 90 min. Samples were fractionated using acid precipitation, followed by ultracentrifugation and ProteoMiner (Bio-Rad Laboratories, Hercules, CA) treatment, and tandem-mass tagging was used to comparatively assess the low abundance protein profile. A total of 162 proteins were identified with more than 2 peptides in the low abundance protein enriched fraction. Of these, 62 differed in abundance by more than 2-fold in heat treated samples compared with the unheated control. The majority of proteins affected by heat treatment were involved in immunity, enzyme function, and transport-related processes; affected proteins included lactadherin, chitinase-3-like protein 1, and complement component C9. These results provide a foundation for further research to determine optimum heat treatment practices to ensure newborn calves are fed colostrum-containing proteins with the highest nutritional and biological value.
Keywords: low abundance milk protein, mass spectrometry, casein, whey
INTRODUCTION
Colostrum is a complex mixture of proteins, lipids, simple carbohydrates (primarily lactose), vitamins, and minerals. Several of these factors including immunoglobulins, growth factors, and cytokines can influence immune and gastrointestinal development following colostrum consumption in the first day of life (Ingram et al., 1956; Burrin et al., 1996; Yamanaka et al., 2003). In addition to these known compounds, recent data suggest that even small peptides present at low abundance in colostrum and milk may affect whole organ systems in the neonate. Data supporting this “lactocrine hypothesis” were reviewed by Bartol et al. (2013), detailing the mechanisms by which relaxin, a peptide secreted in sow colostrum and milk, can enhance development of the reproductive tract of female piglets. Such data indicate the vast potential of colostrum components for neonatal development.
On many dairy farms, newborn calves are hand-fed colostrum during the first 2 to 3 d after birth to ensure adequate consumption (Yang et al., 2015). One disadvantage of hand-feeding colostrum is increased risk of bacterial contamination of colostrum before feeding. Heat treatment of colostrum at 60°C for 60 min is becoming a common management tool to limit bacterial exposure in newborn calves (NAHMS, 2016). This protocol sufficiently reduced bacterial contamination, while minimally affecting IgG concentration and colostrum viscosity (Godden et al., 2006; Elizondo-Salazar et al., 2010). Little attention was paid to other potentially important colostrum proteins when heat treatment protocols were developed. A recent study that fed either unheated or heat-treated colostrum to calves observed reduced cytokine absorption in calves that received heat-treated colostrum, implying that heat treatment can affect proteins that would otherwise be absorbed and function in calf development (Gelsinger and Heinrichs, 2017).
Early work on the bovine colostrum proteome used centrifugation and ultracentrifugation to remove the fat layer and isolate whey from the casein pellet (Yamada et al., 2002; Palmer et al., 2006). Immunoabsorption and 2-D gel electrophoresis were used to separate proteins before microsequencing and tandem MS analysis. Differences in the 2-dimensional electrophoretic gel patterns were observed between colostrum and mature bovine milk. At that time, 29 low abundance proteins were identified as unique to colostrum (Yamada et al., 2002). Almost a decade later, an ion-exchange-based protein fractionation method was performed on bovine colostrum and fresh milk and identified over 140 uniquely colostral proteins (Le et al., 2011). Shortly thereafter, Nissen et al. (2012) identified over 400 proteins in bovine colostrum after fractionating colostrum samples into 8 different fluid and solid fractions using a range of fractionating techniques: ultracentrifugation, acid precipitation, and ultrafiltration. The most expansive analysis of the bovine colostrum proteome to date used ProteoMiner enrichment, affinity chromatography depletion, gel separation, and nano-HPLC-MS/ MS techniques to identify over 600 proteins that occur in low abundance in colostrum (Altomare et al., 2016).
Based on gene ontology, the majority of proteins present in colostrum are involved in immune, enzymatic, and transport pathways (Zhang et al., 2015a), but the importance of each protein to neonatal calf development is yet to be determined. Additionally, no global investigation has been done to determine effects of heating on the colostrum proteome. Thus, the objective of this study was to compare the protein profile of colostrum before and after exposure to heat treatment for 30, 60, or 90 min. Durations of 30 and 60 min were chosen because they are currently used on dairy farms. A 90-min duration was chosen to provide an example and to represent any on-farm case when colostrum may be heated for a longer duration than recommended.
MATERIALS AND METHODS
Sample Collection and Heat Treatment
All animal procedures were approved by the Pennsylvania State University Institutional Animal Care and Use Committee (#46062). A detailed description of colostrum collection and preparation of unheated and 60-min heat treatment colostrum treatments is given by Gelsinger and Heinrichs (2017). Briefly, total first-milking colostrum was collected within 12 h after parturition from approximately 20 individual cows and stored at −20°C until treatment preparation. Approximately 114 L of colostrum was thawed at 4°C, pooled, and divided into 2 equal batches. One batch remained unheated and the other was heated to 60°C for 60 min (heat60) before returning to storage at −20°C as described by Gelsinger and Heinrichs (2017). The heat30 and heat90 treatments were created by heating 1-L aliquots of the unheated colostrum treatment to 60°C for 30 or 90 min, respectively. Aliquots were heated using a water bath and constant agitation. Colostrum and water temperatures were monitored constantly in both heating systems, and colostrum temperature never exceeded 60°C.
At the time of treatment preparation, 50 mL of whole colostrum was collected and submerged in a dry ice ethanol bath immediately following heat treatment, or pooling in the case of unheated colostrum, and stored at −80°C for low abundance protein analysis. An additional 50 mL of colostrum was centrifuged at 4,000 × g at 4°C for 10 min. The skim milk fractions were isolated and stored at −20°C for high abundance protein analysis. All samples were packaged in dry ice and shipped to the University of Vermont (Burlington) for proteomics analysis.
High Abundance Protein Analysis
Skimmed colostrum samples (n = 1 sample/treatment) were thawed at 4°C overnight. Thawed samples were mixed thoroughly by vortexing, followed by sonication (Bransonic Model 220 sonicator, Branson Ultrasonics, Danbury, CT) at 33 W for 15 min in an ice water bath to maintain a sample temperature of less than 25°C. Following sonication, a 0.5-mL aliquot of milk was pipetted into a borosilicate test tube. Sample preparation and high abundance protein analysis were performed using methods outlined by Bordin et al. (2001) with modifications outlined by Tacoma et al. (2016). Briefly, 0.5 mL of reducing buffer (6.0 M guanidine hydrochloride, 5.0 mM trisodium citrate dehydrate, 20.0 mM dithiothreitol) was added to each sample, and samples were incubated at room temperature for 1 h, after which buffer without the dithiothreitol was added. The sample was transferred to a syringe and ejected through a 0.45-μm regenerated cellulose membrane syringe filter (Sartorius, Goettingen, Germany) into an HPLC autosampler vial. A Shimadzu (Kyoto, Japan) HPLC system fitted with a C4 reversed-phase microbore analytical column (150 × 2.1 mm, 300 Å pore diameter, and 5 μm particle size, Vydac 214MS, Grace Davison Inc., Columbia, MD) maintained at 40°C was used for protein separation. Samples were maintained in the 15°C autosampler chamber before injection of 4 μL of sample into the HPLC. For protein separation, the following solvent protocol was applied: an increased linear gradient from 26.5 to 28% eluent B (90% acetonitrile and 0.1% trifluoroacetic acid in ultrapure water) in 2.5 min (0.60% B/min), an isocratic elution at 28.0% B for 4 min, then from 28.6 to 30.6% B in 3.4 min (0.70% B/min); and from 30.6 to 33.5% B in 2.9 min (1.0% B/min), followed by an isocratic elution at 33.5% B for 3 min, an increase from 33.5 to 36.1% B in 2.6 min (1.0% B/min), an isocratic elution at 36.1% B for 5 min, an increase from 36.1 to 37% B in 1.5 min (0.6% B/min), an isocratic elution at 37% B for 2 min, and a final increase to 41% B in 6.5 min (0.60% B/min), for a total run time of 42 min at a flow rate of 0.50 mL/ min. Eluent A was composed of 10% (vol/vol) acetonitrile and 0.1% trifluoroacetic acid in ultrapure water. A detection wavelength of 214 nm was used, and LC solution software from Shimadzu (Kyoto, Japan) was used for chromatograph integration. Individual peaks were identified by comparison to injections of standard protein solutions prepared in our laboratory from purchased isolated proteins (Sigma, St. Louis, MO). For quantification of total α-CN, as well as β-CN, κ-CN, α-LA, and the A and B variants of β-LG, standard curves were directly determined by injecting known concentrations of the standard protein solutions. The constituent αS1- and αS2-CN proteins are not readily available as isolates; therefore, quantification of αS1-CN and αS2-CN was performed by interpolating the results from the total α-CN standard curve for semiquantitative comparisons between experimental groups.
Protein Fractionation and Low Abundance Protein Enrichment
Whole colostrum samples (n = 1 sample/treatment) were thawed at 4°C overnight and fractionated and enriched as described by Tacoma et al. (2016). Briefly, mammalian protease inhibitor (Protease Inhibitor Cocktail, Sigma) was added to samples before centrifugation at 4,000 × g for 15 min at 4°C. The skim fraction was isolated and underwent a second centrifugation using the same conditions. The separated skim milk fraction was collected and to this fraction 60 mM of CaCl2 was added for CN depletion as outlined by Kunz and Lonnerdal (1990). The pH of each sample was adjusted to using 30% acetic acid (Fisher Scientific, Fair Lawn, NJ) and centrifuged at 189,000 × g for 70 min at 4°C. The supernatant was collected, frozen to −80°C, then lyophilized. One hundred milligrams of the resulting whey powder was reconstituted in PBS, and the protein concentration of each sample was determined using the bicinchoninic acid assay kit (Pierce Biotechnology, Rockford, IL). The low abundance protein fraction of each sample was enriched using the ProteoMiner kit (Bio-Rad Laboratories, Hercules, CA), where 10 mg of protein was loaded onto each column and enrichment was performed as per kit instructions. The protein concentration of each sample was determined using the bicinchoninic acid assay kit (Pierce Biotechnology).
Protein Digestion, Labeling, and Identification
A known quantity of protein from each sample (99 μg) was combined with 1 μg of Saccharomyces cerevisiae GAPDH (Sigma) for use as an internal control, for subsequent digestion and labeling efficiencies, and samples (100 μg of protein/sample) were subjected to alkylation, protein digestion, and labeling using isobaric tags as per kit instructions (TMT Isobaric Mass Tag ging Kit, Pierce Biotechnology). Labeled samples were combined in equal volume, frozen to −80°C, and submitted to the Vermont Genetics Network Proteomics Facility (University of Vermont, Burlington) for liquid chromatography (LC)-MS. Prior to LC-MS separa tion, 4 μL of the mixture was dried under vacuum and resuspended in 10 μL of solution containing 2.5% acetonitrile and 2.5% formic acid in water. Peptide identification and quantification was performed in triplicate on the Q-Exactive MS coupled to an EASY-nLC (Thermo Fisher Scientific, Waltham, MA) fitted with a 100 μm × 120 mm capillary column packed with Halo C18 (2.7 μm particle size, 90 nm pore size, Michrom Bioresources, Auburn, CA). Peptides within the 5 μL of sample were separated by LC at a flow rate of 300 nL/min using the following solvent gradient: 2.5 to 35% of acetonitrile/0.1% formic acid for 150 min, and increase from 35 to 100% acetonitrile/0.1% formic acid in 1 min, an isocratic elution at 100% acetonitrile/0.1% formic acid for 8 min, an immediate return to 2.5% acetonitrile/0.1% formic acid, and a final hold at 2.5% acetonitrile/0.1% formic acid. The peptides were introduced into the MS via a nanospray ionization source and a laser-pulled ~3-μm orifice with a spray voltage of 2.0 kV, and data were acquired in a data-dependent “top 10” acquisition mode with lock mass function activated (m/z 371.1012; use lock masses: best; lock mass injection: full MS). A survey scan from m/z 350 to 1,600 at 70,000 resolution (AGC target 1e6; maximum injection time 100 ms; profile mode) was followed by 10 higher-energy collisional dissociation tandem MS/ MS scans on the most abundant ions at 35,000 resolution (AGC target 1e5; maximum injection time 100 ms; profile mode). The MS/MS scans were acquired with an isolation width of 1.2 m/z and a normalized collisional energy of 35%. Dynamic exclusion was enabled (peptide match: preferred; exclude isotopes: on; underfill ratio: 1%; exclusion duration: 30 s). The SEQUEST and Mascot search engines on Proteome Discoverer 1.4 (Thermo Fisher Scientific, Waltham, MA) were used to search product ion spectra of the bovine proteins against a curated Bovine Uniprot (Bos taurus database; UP000009136; 24,346 entries; downloaded December 9, 2015; www.uniprot.org) with sequences in forward and reverse orientations. The product ion spectra were re-searched against a Saccharomyces cerevisiae database to verify the digestion and labeling efficiencies across samples. The following search parameters were enabled: full trypsin enzymatic activity, a maximum of 2 missed cleavages, and peptide molecular weight between 350 and 5,000 g/mol; mass tolerance at 20 ppm for precursor ions and 0.02 Da for fragment ions, dynamic modifications on Met (+15.9949 Da: oxidation), dynamic TMT6plex modification (the TMT6plex and TMT10plex have the same isobaric mass) on N-termini and Lys (229.163 Da), as well as static modification on Cys (+57.021 Da). Percolator node was included in the workflow to limit the false positive rates to <1% in the data set. Quantification was performed using Reporter Ion Quantification Node in the Proteome Discoverer 1.4. All protein identification and quantification information (<1% false positives; with protein grouping enabled) was exported to Microsoft Excel 2010 (Microsoft Corp., Redmond, WA) spreadsheets. Gene ontology analysis was performed using the identified accession numbers searched against the Bos taurus database in the PANTHER Classification System (Thomas et al., 2003; Mi et al., 2010). Proteins were clustered on the basis of cellular component and biological process. These results were exported into Microsoft Excel 2011 and figures were generated for comparative interpretation. Proteins that were found to differ in abundance >2-fold from the unheated control were grouped based on whether their abundance increased or decreased in response to heat treatment. Standard error of the technical triplicates and heatmaps were calculated and generated, respectively, using JMP Pro (version 13.0.0, SAS Institute Inc., Cary, NC).
RESULTS
Total CN and whey concentrations were higher in the heat60 and heat90 samples compared with the unheated and heat30 treatments (Table 1). The proteins primarily responsible for this increase in both the heat60 and heat90 were β-CN, αS2-CN, α-LA, and β-LG variant A, which were 9.75, 13.21, 19.10, and 17.34% greater in heat90 compared with unheated treatments, respectively, as determined by HPLC analysis.
Table 1.
High abundance protein content in bovine colostrum at different heat treatments1
| Item | Unheated | Heat30 | Heat60 | Heat90 |
|---|---|---|---|---|
| Whey (mg/mL of skim milk) | ||||
| α-LA | 3.05 | 3.44 (11.34) | 3.67 (16.89) | 3.77 (19.10) |
| β-LG variant A (β-LGA) | 19.11 | 21.03 (9.13) | 22.52 (15.14) | 23.12 (17.34) |
| β-LG variant B (β-LGB) | 4.87 | 5.25 (7.24) | 5.31 (8.29) | 5.30 (8.11) |
| Total α-LA, β-LGA, β-LGB | 27.03 | 29.72 (9.05) | 31.50 (14.19) | 32.20 (16.06) |
| CN (mg/mL of skim milk) | ||||
| β-CN | 31.00 | 30.21 (−2.62) | 33.42 (7.24) | 34.35 (9.75) |
| κ-CN | 14.62 | 14.46 (−1.11) | 15.66 (6.64) | 14.50 (−0.83) |
| Total α-CN | 43.42 | 43.09 (−0.77) | 47.13 (7.87) | 47.14 (7.89) |
| αS1 | 38.56 | 38.29 (−0.71) | 41.82 (7.80) | 41.54 (7.17) |
| αS2 | 4.86 | 4.80 (−1.25) | 5.31 (8.47) | 5.60 (13.21) |
| Total CN | 89.04 | 87.77 (−1.45) | 96.20 (7.44) | 96.00 (7.25) |
Values in parentheses show percentage difference between unheated and treatment. Heat treatment of colostrum at 60°C for 0 (unheated), 30 (heat30), 60 (heat60), or 90 (heat90) min.
A total of 162 colostrum whey proteins with a peptide count ≥2 were identified by LC-MS/MS analysis across the different heat treatment samples (Supplemental Table S1; https://doi.org/10.3168/jds.2017-13211), 62 of which differed in abundance by least 2-fold in heated compared with unheated samples (Supplemental Table S1; https://doi.org/10.3168/jds.2017-13211). Figure 1 depicts the relative differences in ratio abundance of low abundance proteins that differed by at least 2-fold compared with the unheated sample for comparative assessment. Gene ontological functions of these 62 proteins are depicted in Figure 2. The most prominent biological processes clusters were cellular process (GO: 0009987, 16.7%), localization (GO:0051179, 13.7%), metabolic process (GO:0008152, 12.7%), immune system process (GO:0002376, 11.8%), and biological regulation (GO:0065007, 10.8%). Some proteins with potential bioactivity grouped in these clusters include lactadherin, chitinase-3-like protein 1, complement component C9, thrombospondin-1, and zinc-α−2-glycoprotein (Supplemental Table S1, https://doi.org/10.3168/jds.2017-13211; Yamada et al., 2002; Le et al., 2011; Senda et al., 2011). A total of 44 proteins exhibited lower abundance in heat-treated compared with unheated samples. The biological clusters that had the largest depletion in abundance compared with the unheated sample were cellular process (GO: 0009987, 19.10%), localization (GO:0051179, 14.7%), and metabolic process (GO:0008152, 14.7%). Some relatively stable proteins that were not drastically affected by heat treatment included lactotransferrin, cathelicidin-6, and apolipoprotein A-IV (Supplemental Table S1; https://doi.org/10.3168/jds.2017-13211). A total of 18 proteins were identified to be present at higher concentrations in the heated samples (>2-fold increase compared with unheated samples). These proteins included many fragments of higher abundance proteins that were not removed during the fractionation process, but also include zinc-α−2-glycoprotein, a known bioactive protein, as well as 6 proteins known to be involved in immune processes. Immune system process (GO:002376, 17.6%) and biological regulation (GO:0065007, 14.7%) were the biological processes that encompassed the greatest proportion of proteins observed to be higher in heated samples.
Figure 1.
Heat map illustration of proteins that were increased 2-fold (A) or decreased by 50% (B) in abundance in heated samples compared with the unheated control. Heat treatment of colostrum at 60°C for 0 (unheated), 30 (heat30), 60 (heat60), or 90 (heat90) min. Color version available online.
Figure 2.
Gene ontological (GO) categories of low abundance proteins identified in bovine colostrum that increased 2-fold (A) or decreased by 50% (B) in abundance in heated samples compared with the unheated control.
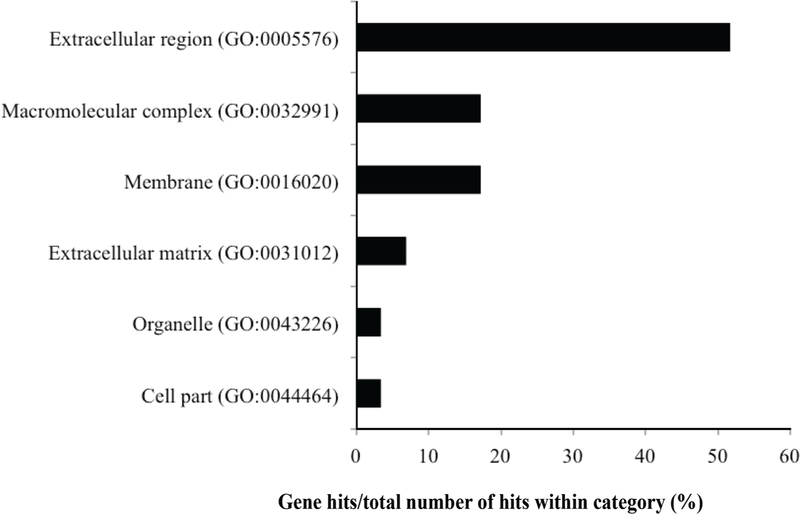
Although most of the affected proteins appeared to followed a linear pattern of change across heat treatments, a select few proteins did not appear to follow either of these patterns (Figures 1A and 1B; Supplemental Table S1; https://doi.org/10.3168/jds.2017-13211). For instance, histone H1.2 and H1.3 (accession numbers P02253 and A7MAZ5) were relatively stable in colostrum samples heated for 30 or 90 min, but were identified at 37% of abundance in the heat60 sample compared with the unheated sample. Serotransferrin, on the other hand, more than doubled in abundance in heat30 and heat60 samples compared with the unheated and heat90 samples. Other proteins, including 3 uncharacterized proteins (accession numbers G1N514, G5E513, and G5E5T5) were relatively stable in samples heat treated for 30 and 60 min and then were approximately 50% lower abundance in samples heat treated for 90 min (Supplemental Table S1; https://doi.org/10.3168/jds.2017-13211). The original cellular locations of the 62 proteins identified in Figure 1 are presented in Figure 3. Of these proteins, 51.7, 17.2, and 17.2% were located in the extracellular region, plasma membrane, and macromolecular complexes, respectively.
Figure 3.
Cellular location of bovine colostrum low abundance proteins that changed in abundance more than 2-fold in heated samples compared with the unheated control. GO = gene ontology.
DISCUSSION
The effect of heat treatment on the proteome of first-milking colostrum was analyzed in the current study. The concentrations of high abundance proteins rapidly change during consecutive milkings after parturition (Parrish et al., 1948; Madsen et al., 2004; Sobczuk-Szul et al., 2013). Content of these high abundance proteins in the unheated colostrum analyzed in the current research was in line with previous research outlining total casein content of first-milking colostrum (Madsen et al., 2004) and was within the reported ranges of first milking α-LA and β-LG concentrations (Marnila and Korhonen, 2002; Sobczuk-Szul et al., 2013; McGrath et al., 2016). Typically, decreased whey protein solubility is observed when milk is heated to 40 to 60°C. Above this range, whey proteins denature due to disruption of noncovalent bonds stabilizing the secondary and tertiary structure (Pelegrine and Gasparetto, 2005). Casein proteins on the other hand, are more resistant to heat treatment due to random coiling of the primary chain held together by strong covalent and noncovalent bonds (Akkerman, 2014) and can withstand heating at 140°C for 15 to 20 min. In this study, the higher total casein and whey content in samples subjected to longer heat treatment time is likely explained by the dissociation of some of the high abundance proteins from micelles and other cellular structures, allowing these proteins to become soluble (Anema, 1998; Akkerman, 2014).
Using a 2-fold cutoff, the majority of low abundance proteins (62%) were unaffected by heat treatment, irrespective of the length of heat treatment time. This heat resistance has been demonstrated previously where multiple bioactive proteins have been shown to be resistant to heat and acid denaturation (Wada and Lönnerdal, 2015; Deglaire et al., 2016; Peila et al., 2016).
Unexpectedly, several low abundance proteins, including zinc-α−2-glycoprotein, apparently increased in concentration as the colostrum was heated. This may be a result of heat stability of these proteins in combination with the methodology, or could be a result of biochemical effects of heating on cell stability. In considering the former possibility, because an equal amount of lyophilized sample (99 μg of protein) was labeled for LC-MS/MS analysis for each sample, any heat-resistant proteins would compose a greater proportion of the protein content of the sample and labeling would reflect that. These proteins would consequently be identified to be in higher amounts in the heated samples relative to the unheated sample. Biochemically, the increase in concentration of some proteins could be a result of sloughed mammary epithelial cells lysing during heat treatment, releasing the cell contents into colostrum. Released cellular proteins would then contribute to the increased abundance of mammary-derived proteins, which may account for the high number of proteins identified to have originated from the plasma membrane. The increase in other proteins may be related to the release of proteins from the milk fat globule membrane (MFGM) during heat treatment. The MFGM-associated proteins found at lower abundance in heated samples include lactadherin, protein disulfide-isomerase, Ser protease HTRA1, and β-galactosyltransferase 1. Gene ontological analysis of the cellular location of the low abundance proteins identified in the colostrum samples shown in Figure 3 indicated that most proteins translocate after synthesis to the plasma membrane and either remain in the phospholipid bilayer or are secreted extracellularly from their tissue origin (Rainard and Riollet, 2006; Stelwagen et al., 2009).
Functional analysis based on gene ontology indicated that the majority of low abundance proteins whose ratios changed due to heat treatment were associated with immune functions, implying that calves that receive heat-treated colostrum may experience a different degree of immune support compared with calves that received unheated colostrum. Several studies reported greater absorption of IgG in calves that received heat-treated colostrum (Johnson et al., 2007; Elizondo-Salazar and Heinrichs, 2009). In contrast, absorption of IGF-1, lactoferrin, and IL-1β was decreased in calves that received heat-treated colostrum (Abd El-Fattah et al., 2014; Gelsinger and Heinrichs, 2017). The study that used the unheated and heat60 treatments described here for feeding calves observed greater circulating IL-1β in calves that received unheated colostrum for up to 2 wk after birth (Gelsinger and Heinrichs, 2017). Growth patterns also tended to differ between treatment groups (Gelsinger and Heinrichs, 2017). Taken together, these data indicate that changes in the colostrum proteome due to heating can alter calf development, potentially through a decreased bioactive protein content regardless of length of heat treatment. Though this study used a composite milk sample including a large number of cattle to decrease individual animal bias, such as somatic score, which could affect the milk proteome as seen in mature milk (Reinhardt et al., 2013; Smolenski et al., 2014; Zhang et al., 2015b), replication of the analytical methods using a larger number of individual biological replicates and technical replicates is needed to confirm the current results. However, the results do highlight that additional studies are warranted to explore specific mechanisms by which the colostrum proteome affects neonatal immune development.
CONCLUSIONS
The effect of heat treatment on the colostrum proteome was investigated in this exploratory study using a range of pre-fractionation and enrichment techniques followed by detection and identification using MS. As heat treatment of colostrum becomes more common on farms for reducing bacterial exposure, examining the colostrum proteome after heat treatment will provide a more realistic review of proteins that are actually consumed and available to the calf. The length of heat treatment altered the composition of both high and low abundance proteins within bovine colostrum. The majority of low abundance proteins affected by heat in this investigation were involved in cellular and immune processes. Further insights into the effect of heat treatment on the colostrum proteome and greater understanding of the roles of low abundance proteins in neonatal growth and development are crucial to ensure provision of high quality colostrum to neonatal ruminants.
Supplementary Material
ACKNOWLEDGMENTS
This research is a component of USDA Regional Research Project NC-2042; Management Systems to Improve the Economic and Environmental Sustainability of Dairy Enterprises and was supported by the USDA-National Institute of Food and Agriculture HATCH Grant number VT-H02009. Equipment use included pieces funded by National Institutes of Health (NIH) Grant Numbers 5 P30 RR032135 from the Center of Biomedical Research Excellence (Bethesda, MD) Program of the National Center for Research Resources and 8 P30 GM 103498 from the National Institute of General Medical Sciences. The Vermont Genetics Network Proteomics Facility is supported through NIH grant P20GM103449 from the IDeA Network for Biomedical Research Excellence (Bethesda, MD) Program of the National Institute of General Medical Sciences.
REFERENCES
- Abd El-Fattah AE, Ragab Abd Rabo FH, El-Dieb SM, and Satar El-Kashef HA 2014. Preservation methods of buffalo and bovine colostrum as a source of bioactive components. Int. Dairy J 39:24–27. 10.1016/j.idairyj.2014.04.008. [DOI] [Google Scholar]
- Akkerman M 2014. The effect of heating processes on milk whey protein denaturation and rennet coagulation properties. MS Thesis. Department of Food Science, Aarhus University, Aarhus, Denmark. [Google Scholar]
- Altomare A, Fasoli E, Colzani M, Parra XMP, Ferrari M, Cilurzo F, Rumio C, Cannizzaro L, Carini M, Righetti PG, and Aldini A 2016. An in depth proteomic analysis based on ProteoMiner, affinity chromatography and nano-HPLC-MS/MS to explain the potential health benefits of bovine colostrum. J. Pharm. Biomed. Anal 121:297–306. [DOI] [PubMed] [Google Scholar]
- Anema SG 1998. Effect of milk concentration on heat-induced, pH dependent dissociation of casein from micelles in reconstituted skim milk at temperatures between 20 and 120°C. J. Agric. Food Chem 46:2299–2305. 10.1021/jf970909+. [DOI] [Google Scholar]
- Bartol FF, Wiley AA, Miller DJ, Silva AJ, Roberts KE, Davolt MLP, Chen JC, Frankshun AL, Camp ME, Rahman KM, Vallet JL, and Bagnell CA 2013. Lactation biology symposium: Lactocrine signaling and developmental programming. J. Anim. Sci 91:696–705. [DOI] [PubMed] [Google Scholar]
- Bordin G, Raposo FC, de la Calle B, and Rodriguez AR 2001. Identification and quantification of major bovine milk proteins by liquid chromatography. J. Chromatogr. A 928:63–76. 10.1016/S0021-9673(01)01097-4. [DOI] [PubMed] [Google Scholar]
- Burrin DG, Wester TJ, Davis TA, Amick S, and Heath JP 1996. Orally administered IGF-I increases intestinal mucosal growth in formula-fed neonatal pigs. Am. J. Physiol 270:R1085–R1091. 10.20.33.4. [DOI] [PubMed] [Google Scholar]
- Deglaire A, De Oliveira SC, Jardin J, Briard-Bion V, Emily M, Ménard O, Bourlieu C, and Dupont D 2016. Impact of human milk pasteurization on the kinetics of peptide release during in vitro dynamic term newborn digestion. Electrophoresis 37:1839–1850. 10.1002/elps.201500573. [DOI] [PubMed] [Google Scholar]
- Elizondo-Salazar JA, and Heinrichs AJ 2009. Feeding heat-treated colostrum or unheated colostrum with two different bacterial concentrations to neonatal dairy calves. J. Dairy Sci 92:4565–4571. 10.3168/jds.2009-2188. [DOI] [PubMed] [Google Scholar]
- Elizondo-Salazar JA, Jayarao BM, and Heinrichs AJ 2010. Effect of heat treatment of bovine colostrum on bacterial counts, viscosity, and immunoglobulin G concentration. J. Dairy Sci 93:961–967. 10.3168/jds.2009-2388. [DOI] [PubMed] [Google Scholar]
- Gelsinger SL, and Heinrichs AJ 2017. Comparison of immune responses in calves fed heat-treated or unheated colostrum. J. Dairy Sci 100:4090–4101. 10.3168/jds.2016-12010. [DOI] [PubMed] [Google Scholar]
- Godden S, McMartin S, Feirtag J, Stabel J, Bey R, Goyal S, Metzger L, Fetrow J, Wells S, and Chester-Jones H 2006. Heat treatment of bovine colostrum. II: Effects of heating duration on pathogen viability and immunoglobulin. J. Dairy Sci 89:3476–3483. 10.3168/jds.S0022-0302(06)72386-4. [DOI] [PubMed] [Google Scholar]
- Ingram PL, Lovell R, Wood PC, Aschaffenburg R, Bartlett S, Kon SK, Palmer J, Roy JHB, and Shillam KWG 1956. Bacterium coli antibodies in colostrum and their relation to calf survival. J. Pathol. Bacteriol 72:561–568. [Google Scholar]
- Johnson JL, Godden SM, Molitor T, Ames T, and Hagman D 2007. Effects of feeding heat-treated colostrum on passive transfer of immune and nutritional parameters in neonatal dairy calves. J. Dairy Sci 90:5189–5198. 10.3168/jds.2007-0219. [DOI] [PubMed] [Google Scholar]
- Kunz C, and Lonnerdal B 1990. Human-milk proteins: Analysis of casein and casein subunits by anion-exchange chromatography, gel electrophoresis, and specific staining methods. Am. Soc. Clin. Nutr 51:37–46. [DOI] [PubMed] [Google Scholar]
- Le A, Barton LD, Sanders JT, and Zhang Q 2011. Exploration of bovine milk proteome in colostral and mature whey using an ion-exchange approach. J. Proteome Res 10:692–704. 10.1021/pr100884z. [DOI] [PubMed] [Google Scholar]
- Madsen BD, Rasmussen MD, Nielsen MO, Wiking L, and Larsen LB 2004. Physical properties of mammary secretions in relation to chemical changes during transition from colostrum to milk. J. Dairy Res 71:263–272. [DOI] [PubMed] [Google Scholar]
- Marnila P, and Korhonen H 2002. Colostrum. Pages 591–597 in Encyclopedia of Dairy Sciences Fuquay JW, Fox PF, and McSweeney PLH, ed. Academic Press, London, UK. [Google Scholar]
- McGrath BA, Fox PF, McSweeney PLH, and Kelly AL 2016. Composition and properties of bovine colostrum: A review. Dairy Sci. Technol 96:133–158. [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, and Thomas PD 2010. PANTHER version 7: Improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 38:D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAHMS (National Animal Health Monitoring System). 2016. Dairy cattle management practices in the United States, 2014 USDA-APHIS-VS, CEAH, Fort Collins, CO. [Google Scholar]
- Nissen A, Bendixen E, Ingvartsen KL, and Røntved CM 2012. In-depth analysis of low abundant proteins in bovine colostrum using different fractionation techniques. Proteomics 12:2866–2878. 10.1002/pmic.201200231. [DOI] [PubMed] [Google Scholar]
- Palmer DJ, Kelly VC, Smit AM, Kuy S, Knight CG, and Cooper GJ 2006. Human colostrum: Identification of minor proteins in the aqueous phase by proteomics. Proteomics 6:2208–2216. [DOI] [PubMed] [Google Scholar]
- Parrish DB, Wise GH, Hughes JS, and Atkeson FW 1948. Properties of the colostrum of the dairy cow. II. Effect of prepartal rations upon the nitrogenous constituents. J. Dairy Sci 33:457–465. [Google Scholar]
- Peila C, Coscia A, Bertino E, Cavaletto M, Spertino S, Icardi S, and Gazzolo D 2016. Effects of Holder pasteurization on the protein profile of human milk. Ital. J. Pediatr 42:36 10.1186/s13052-016-0248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrine DHG, and Gasparetto CA 2005. Whey proteins solubility as function of temperature and pH. LWT Food Sci. Technol (Campinas: ) 38:77–80. 10.1016/j.lwt.2004.03.013. [DOI] [Google Scholar]
- Rainard P, and Riollet C 2006. Innate immunity of the bovine mammary gland. Vet. Res 37:369–400. 10.1051/vetres:2006007. [DOI] [PubMed] [Google Scholar]
- Reinhardt TA, Sacco RE, Nonnecke J, and Lippolis JD 2013. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteomics 82:141–154. [DOI] [PubMed] [Google Scholar]
- Senda A, Fukuda K, Ishii T, and Urashima T 2011. Changes in the bovine whey proteome during the early lactation period. Anim. Sci. J 82:698–706. 10.1111/j.1740-0929.2011.00886.x. [DOI] [PubMed] [Google Scholar]
- Smolenski GA, Broadhurst MK, Stelwagen K, Haigh BJ, and Wheeler TT 2014. Host defence related responses in bovine milk during an experimentally induced Streptococcus uberis infection. Proteome Sci 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczuk-Szul M, Wielgosz-Groth Z, Wroński M, and Rzemieniewski A 2013. Changes in the bioactive protein concentrations in the bovine colostrum of Jersey and Polish Holstein-Friesian cows. Turk. J. Vet. Anim. Sci 37:43–49. [Google Scholar]
- Stelwagen K, Carpenter E, Haigh B, Hodgkinson A, and Wheeler TT 2009. Immune components of bovine colostrum and milk. J. Anim. Sci 87:3–9. 10.2527/jas.2008-1377. [DOI] [PubMed] [Google Scholar]
- Tacoma R, Fields J, Ebenstein DB, Lam Y-W, and Greenwood SL 2016. Characterization of the bovine milk proteome in early lactation Holstein and Jersey breeds of dairy cows. J. Proteomics 130:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, and Narechania A 2003. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 13:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, and Lönnerdal B 2015. Bioactive peptides released from in vitro digestion of human milk with or without pasteurization. Pediatr. Res 77:546–553. 10.1038/pr.2015.10. [DOI] [PubMed] [Google Scholar]
- Yamada M, Murakami K, Wallingford JC, and Yuki Y 2002. Identification of low-abundance proteins of bovine colostral and mature milk using two-dimensional electrophoresis followed by microsequencing and mass spectrometry. Electrophoresis 23:1153–1160. . [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Hagiwara K, Kirisawa R, and Iwai H 2003. Proinflammatory cytokines in bovine colostrum potentiate the mitogenic response of peripheral blood mononuclear cells from newborn calves through IL-2 and CD25 expression. Microbiol. Immunol 47:461–468. 10.1111/j.1348-0421.2003.tb03371.x. [DOI] [PubMed] [Google Scholar]
- Yang M, Zou Y, Wu ZH, Li SL, and Cao ZJ 2015. Colostrum quality affects immune system establishment and intestinal development of neonatal calves. J. Dairy Sci 98:7153–7163. 10.3168/jds.2014-9238. [DOI] [PubMed] [Google Scholar]
- Zhang L, Boeren S, Hageman JA, van Hooijdonk ACM, Vervoort JM, and Hettinga KA 2015a. Perspective on calf and mammary gland development through changes in the bovine milk proteome over a complete lactation. J. Dairy Sci 98:5362–5373. 10.3168/jds.2015-9342. [DOI] [PubMed] [Google Scholar]
- Zhang L, Boeren S, van Hooijdonk ACM, Vervoort JM, and Hettinga K 2015b. A proteomic perspective on the changes in milk proteins due to high somatic cell count. J. Dairy Sci 98:5339–5351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.