SUMMARY
5-hydroxymethylcytosine (5hmC) is enriched in brain and has been recognized as an important DNA modification. However, the roles of 5hmC and its writers, ten-eleven translocation (Tet) proteins, in stress-induced response have yet to be elucidated. Here, we show that chronic restraint stress (CRS) induced depression-like behavior in mice and resulted in a 5hmC reduction in prefrontal cortex (PFC). We found that loss of Tet1 (Tet1 KO) led to resistance to CRS, whereas loss of Tet2 (Tet2 KO) increased the susceptibility of mice to CRS. Genome-wide 5hmC profiling identified the phenotype-associated stress-induced dynamically hydroxymethylated loci (PA-SI-DhMLs), which are strongly enriched with hypoxia-induced factor (HIF) binding motifs. We demonstrated the physical interaction between TET1 and HIF1α induced by CRS and revealed that the increased HIF1α binding under CRS is associated with SI-DhMLs. These results suggest that TET1 could regulate stress-induced response by interacting with HIFIα.
Graphical Abstract
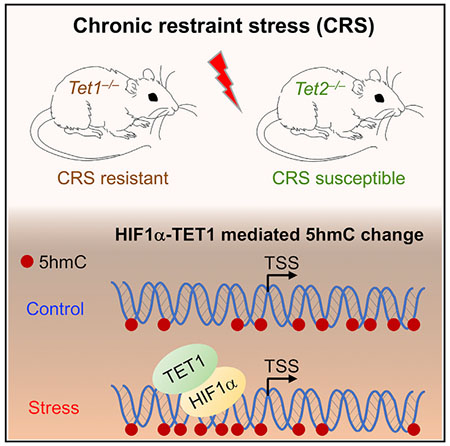
In Brief
The roles of 5-hydroxymethylcytosine (5hmC) and its writers, Tet proteins, in stress-induced response remain unclear. Cheng et al. show that Tet1 knockout mice exhibit resistance, whereas Tet2 knockout mice have increased susceptibility to stress. Biochemical and genome-wide analyses suggest that Tet1 could regulate stress-induced response by interacting with Hif1α.
INTRODUCTION
Depression is a mental disorder characterized by a pervasive and persistent low mood, which is usually accompanied by a loss of interest in activities once pleasurable (Wong and Licinio, 2001). Depression can be triggered by various causes, including genetic, psychological, and environmental factors (Uher and McGuffin, 2008). Among these causes, chronic stress in particular has been shown to result in decreased serotonin and other neurotransmitters in the brain and thus to be a direct link to depression (Caspi et al., 2003; Risch et al., 2009). However, the etiology of depression remains obscure, because neither traditional genetic nor environmental approaches alone can explain this complex neuropsychiatric disorder (Sun et al., 2013).
Several mechanisms have been shown to contribute to the disease pathogenesis of depression; among them, epigenetic regulations were highlighted (Covington et al., 2009, 2011; Hunter et al., 2009; Weaver et al., 2004). Epigenetics is defined as the mitotically and/or meiotically heritable changes in gene expression that are caused by mechanisms without altering the DNA sequence, primarily including post-translational histone modification and DNA methylation (Berger et al., 2009). Histone acetylation is associated with stress-induced depression, suggesting histone acetylation can be an essential substrate for antidepressant action, and histone deacetylase inhibitors were reported to convey antidepressant effects (Schroeder et al., 2007; Tsankova et al., 2006). Moreover, increased histone methylation repressed growth factor BDNF (brain-derived neurotrophic factor) gene expression in chronic defeat-stressed mouse hippocampus, while increased 5-methylcytosine (5mC) in the glucocorticoid receptor gene promoter resulted in early life stress in a rat model (Tsankova et al., 2006; Weaver et al., 2004).
5-hydroxymethylcytosine (5hmC), a core component in the process of active DNA demethylation, can be generated from 5mC oxidized by ten-eleven translocation (Tet) proteins (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). 5hmC can be further oxidized by Tet proteins to produce 5-formylcytosine and 5-carboxylcytosine, which are quickly removed from the genome by thymine-DNA glycosylase (TDG) to initiate base excision repair (BER) (He et al., 2011; Ito et al., 2011; Maiti and Drohat, 2011). Not just a transitory intermediate in DNA demethylation, 5hmC has been reported to be stably enriched in the CNS and bound by specific 5hmC binding proteins (Song et al., 2011; Spruijt et al., 2013; Szulwach et al., 2011). Therefore, 5hmC is recognized as an important epigenetic mark and has a biological function in several developmental and neurological diseases (Cheng et al., 2015). However, it is not yet known whether 5hmC- and Tet-mediated regulation are involved in the stress-induced response.
Here we employed the chronic restraint stress (CRS) paradigm to investigate the role of 5hmC and Tet proteins in stress-induced response. We found that CRS led to depression-like behavior in wild-type (WT) mice and resulted in a significant decrease of 5hmC in prefrontal cortex (PFC). The loss of Tet1 led to resistance to CRS, whereas the loss of Tet2 increased the susceptibility of mice to CRS. Through genome-wide profiling, we identified the stress-induced hydroxymethylated loci. These loci were enriched with hypoxia-induced factor (HIF) 1α binding motifs. Our subsequent biochemical and chromatin immunoprecipitation sequencing (ChIP-seq) analyses suggested that TET1 plays a vital role in stress-induced responses by interacting with HIF1α.
RESULTS
CRS Influences Genome-wide 5hmC Distribution in PFC
It has been shown that environmental exposures can induce epigenetic changes. To understand the dynamics of 5hmC in the mouse brain, we employed a well-established paradigm for chronic stress to model environmental exposure: 7- to 8-week-old WT C57BL/6 adult male mice were exposed to normal or restraint environments (Figure S1A) (O’Mahony et al., 2010; Zhang et al., 2015). We then performed the forced swimming test (FST) and tail suspension test (TST) to evaluate stress responses in the mice. The restraint environment successfully altered mouse behavior; a significant increase in immobility time occurred in the restrained, stressed group for both tests (Figure 1A). Amygdala, hippocampus, and PFC are the three main brain regions linked to stress response (Arnsten, 2009; Roozendaal et al., 2009; Savitz and Drevets, 2009). We dissected these three brain regions, as well as two other depression-associated brain regions, the hypothalamus and the cerebellum, from the stressed and control mice. Dot blot analysis of genomic DNA from these brain regions showed a significant 5hmC reduction in the PFC, suggesting potentially important roles of 5hmC in PFC upon stress (Figure 1B).
Figure 1. Chronic Restraint Stress Causes Depression-like Behavior and Leads to Dynamic Change of 5hmC in Prefrontal Cortex.
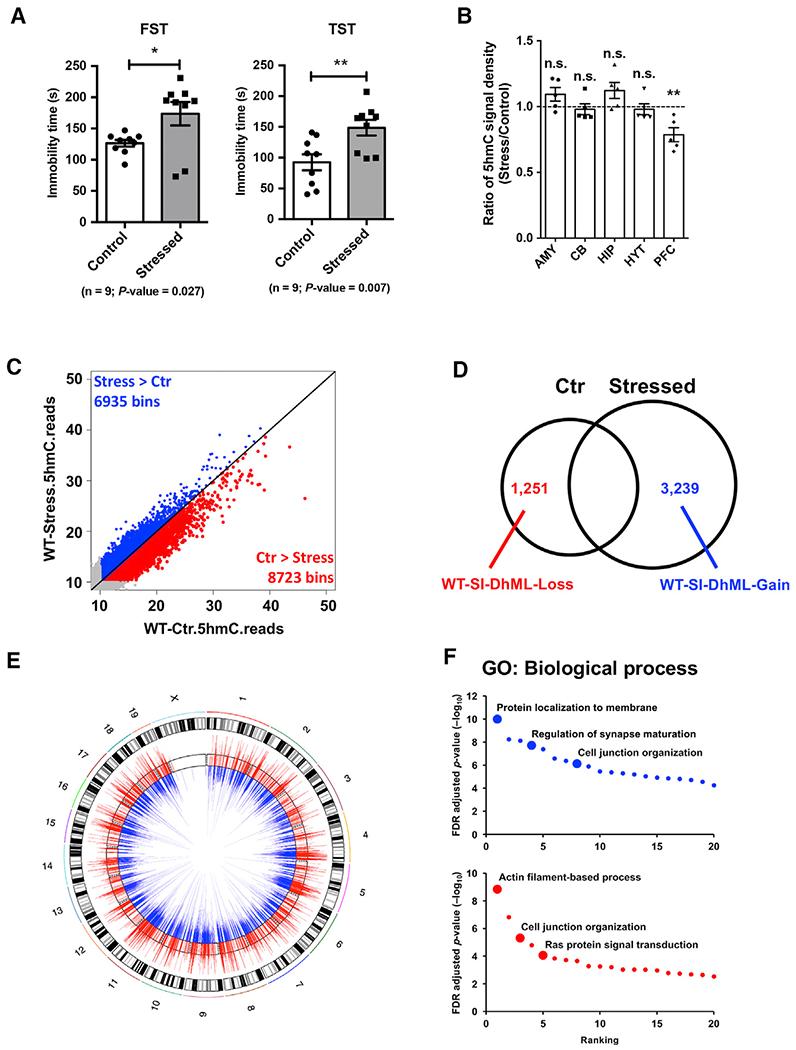
(A) Chronic restraint stress (CRS) resulted in longer immobility times in forced swimming test (FST) and tail suspension test (TST) in wild-type (WT) C57BL/6 adult male mice. n = 9, error bars indicate mean ± SEM; two-tailed unpaired t test; *p < 0.05; **p < 0.01.
(B) 5hmC-specific dot blot (quantified by ImageJ software) revealed a significant decrease of 5hmC (stressed versus control) in prefrontal cortex (PFC). Ratio of 5hmC signal density was calculated by normalizing quantified 5hmC levels in stressed mice to those in littermate control mice. AMY, amygdala; CB, cerebellum; HIP, hippocampus; HYT, hypothalamus; PFC, prefrontal cortex; NS, not significant. n = 5, error bars indicate mean ± SEM; two-tailed unpaired t test; **p < 0.01.
(C) Genome-scale patterns of 5hmC exhibited general reduction upon CRS. Global 5hmC normalized reads in control and stressed mouse PFC were counted in 10-kb binned mouse genome (mm9). Bins with more than 10 reads are colored. 6,935 bins (blue) showed more 5hmC reads in stressed PFC than in control, whereas 8,723 bins (red) contained fewer 5hmC reads in stressed mouse PFC.
(D) Identification of the stress-induced dynamically hydroxymethylated gain-of-5hmC or loss-of-5hmC loci (i.e., WT-stress-induced dynamically hydroxymethylated loci [SI-DhML]-Gain or WT-SI-DhML-Loss) by overlapping 5hmC-seq peaks in control and stressed WT samples.
(E) Circular map view of WT-SI-DhML-Gain (blue) and WT-SI-DhML-Loss (red) on chromosomes. Each mouse chromosome is shown in the black-and-white outer track. 5hmC read ratios between control and stressed PFC were calculated on these loci and are shown in the inner track, with blue bars demonstrating higher 5hmC in stressed mouse PFC and red bars showing lower 5hmC in stressed mouse PFC.
(F) Gene ontology (GO) analysis with WT-SI-DhML-Gain and WT-SI-DhML-Loss revealed enrichment of synapse maturation-, cell junction organization-, and Ras protein signaling-related function.
See also Figure S1.
To better understand the dynamic change of 5hmC in the stressed PFC, we profiled the genome-wide 5hmC distribution by employing a previously established chemical labeling and affinity purification method, coupled with high-throughput sequencing technology (Song et al., 2011). Genome-wide distribution of 5hmC in PFC was evaluated by counting normalized 5hmC mapped reads in 10-kb binned mouse genome (mm9). We found more bins had higher 5hmC reads in control PFC samples compared with those from stressed PFC samples (Figure 1C). This finding is consistent with our observation in the dot blot experiment, confirming that CRS resulted in a global reduction of 5hmC in the PFC of stressed mice.
To investigate the specific stress-induced dynamically hydroxymethylated loci (SI-DhMLs) in WT mice (WT-SI-DhMLs), we identified and characterized the stress-induced gain- or loss-of-5hmC loci (i.e., WT-SI-DhML-Gain or WT-SI-DhML-Loss, respectively) (Figures 1D, 1E, and S1B). In total, 3,239 WT-SI-DhML-Gain and 1,251 WT-SI-DhML-Loss were identified. Genomic features associated with these known WT-SI-DhMLs revealed that most were located in exon, intron, and intergenic regions, as well as enriched in promoter and gene body regions (Figures S1C and S1D). To explore the biological relevance of these identified WT-SI-DhMLs, we performed gene ontology (GO) analyses and found that WT-SI-DhMLs were strongly associated with various important biological processes, including protein localization to membrane, regulation of synapse maturation, actin filament-based process, and Ras protein signal transduction (Figure 1F). Both WT-SI-DhML-Gain and WT-SI-DhML-Loss were involved in pathways significantly associated with major depressive disorder, such as cell junction organization (Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015).
Loss of Tet1 and Tet2 Leads to the Distinct Responses to CRS in Mice
Considering that Tet proteins are responsible for generating 5hmC and the subsequent demethylation process (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009), we assessed the impact of CRS on Tet expression (Figure S1E). PFC RNA was extracted from control and stressed WT mice, respectively, and qRT-PCR determined the expression levels of three Tet genes (Tet1–Tet3). We found that CRS resulted in a significant decrease of Tet1 expression but had the least effect on Tet2 and Tet3.
To explore the epigenetic mechanism involved in stress-induced depression-like behavior, we used the previously generated global Tet1 knockout (Tet1 KO on a C57BL/6 background) and Tet2 knockout (Tet2 KO on a 129 background) mice to perform the same CRS as we did for WT mice (Figure 1). Given the different genetic background of the two mutant lines, we used Tet1 KO and Tet2 KO male mice, along with their respective littermate male WT mice as controls, for these analyses (Figure 2). FST and TST occurred before the mice were exposed to stress to assess the difference between WT and Tet1 KO and Tet2 KO mice under the non-stress condition. Then FST and TST were done at 1- and 2-week time points after stress. Our behavior test results demonstrated the distinct roles of Tet1 and Tet2 in response to restraint stress (Figure 2). Compared with littermate WT control (Tet1+/+) mice, Tet1 KO mice consistently showed less immobility time in FST, irrespective of the stress status (Figures 2A and S2A). In TST, Tet1 KO mice were more active than Tet1+/+ mice under the non-stress condition, and significant less immobility time was found in Tet1 KO mice after CRS exposure (Figures 2B and S2B). These results show that loss of Tet1 could increase stress resistance.
Figure 2. Tet1 KO and Tet2 KO Mice Exhibit Distinct Responses to CRS.
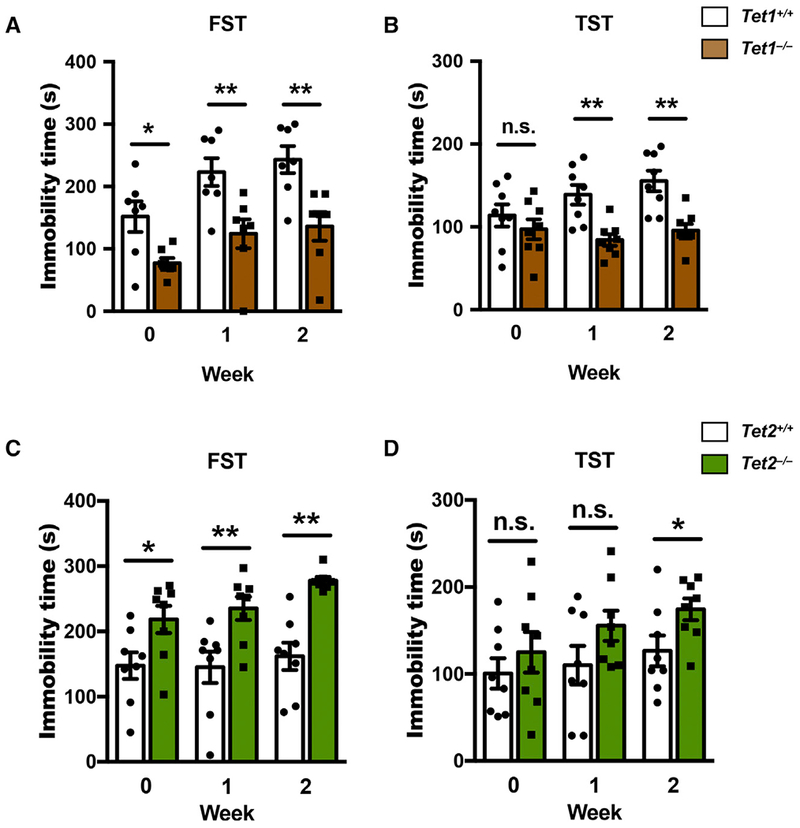
(A–D) FST and TST occurred with Tet1 KO (Tet1−/−, on C57BL/6 background) or Tet2 KO (Tet2−/−, on 129 background) mice and their littermate control (Tet1+/+ or Tet2+/+) mice at a different time point (0, 1, and 2 weeks) after CRS. Compared with respective littermate control mice, Tet1−/− mice showed significant less immobility time in both FST (A) and TST (B), whereas Tet2−/− mice showed significant more immobility time in both FST (C) and TST (D). n = 7–8, error bars indicate mean ± SEM; two-tailed unpaired t test; *p < 0.05; **p < 0.01. See also Figure S2.
Tet2 KO mice showed an opposite response to CRS. In FST, Tet2 KO mice had significantly longer immobility times, irrespective of the stress status (Figure 2C). When compared with littermate WT control (Tet2+/+) mice, Tet2 KO mice exhibited a strong stressed phenotype before exposure to CRS, and their immobility time in FST was still significant increased at week 2, even though the Tet2+/+ mice did not show a significant increase in immobility upon stress, which is likely due to the genetic background of Tet2 KO mice (Figure S2C). In TST, Tet2 KO mice had a significantly longer immobility time 2 weeks after CRS (Figure 2D), while no significant change of immobility time of Tet2 KO mice across weeks was observed (Figure S2D). These data suggest that loss of Tet2 could increase susceptibility to stress stimuli.
To examine the potential compensatory effect of the three Tet enzymes, we performed qRT-PCR analysis using the PFC RNAs from both Tet1 KO and Tet2 KO mice. We did not observe change of the expression of Tet2/3 in Tet1 KO mice or Tet1/3 in Tet2 KO mice (Figure S2E). In addition, upon the stress condition (control versus stressed), we did not find a significant change of Tet2/3 in Tet1 KO mice and Tet1/3 in Tet2 KO mice (Figure S2F). These results suggest that there is little compensatory effect of Tet enzymes in response to stress.
Identification of Phenotype-Associated SI-DhMLs
We further profiled 5hmC using the PFC DNA samples of Tet1 KO and Tet2 KO mice to link the 5hmC status with stress-resistant and stress-susceptible phenotypes. Based on the distinct responses of Tet1 KO and Tet2 KO mice upon the exposure to CRS in FST and TST tests, we next sought to identify the phenotype-associated SI-DhMLs (PA-SI-DhMLs). Because Tet1 KO mice showed resistance to stress and performed like non-stressed WT mice, we defined PA-SI-DhML-Gain as those DhMLs that were found only in stressed WT PFC samples, not in control WT and stressed Tet1 KO PFC samples (Figure 3A). In contrast, we defined PA-SI-DhML-Loss as those DhMLs found only in control WT and stressed Tet1 KO PFC samples, not in stressed WT PFC samples. Overall 2,950 PA-SI-DhML-Gain and 66 PA-SI-DhML-Loss were identified in Tet1 KO mice. Moreover, Tet2 KO mice showed susceptibility to stress and performed like stressed WT mice. Thus, we also found 1,128 PA-SI-DhML-Gain and 630 PA-SI-DhML-Loss in Tet2 KO mice (Figure 3A). The 2,950 PA-SI-DhML-Gain that were identified in Tet1 KO mice contain most of the 3,239 WT-SI-DhML-Gain (91%) in WT mice and highly overlap with 1,128 PA-SI-DhML-Gain (87%) that were identified in Tet2 KO mice, indicating that Tet1 is responsible for generating the stress-induced gain-of-5hmC loci. Similarly, we found that the 630 PA-SI-DhML-Loss that were identified in Tet2 KO mice have half of the 1,251 WT-SI-DhML-Loss, suggesting that Tet2 is more likely to contribute on the stress-induced loss-of-5hmC loci.
Figure 3. Identification and Characterization of Phenotype-Associated SI-DhMLs in PFC.
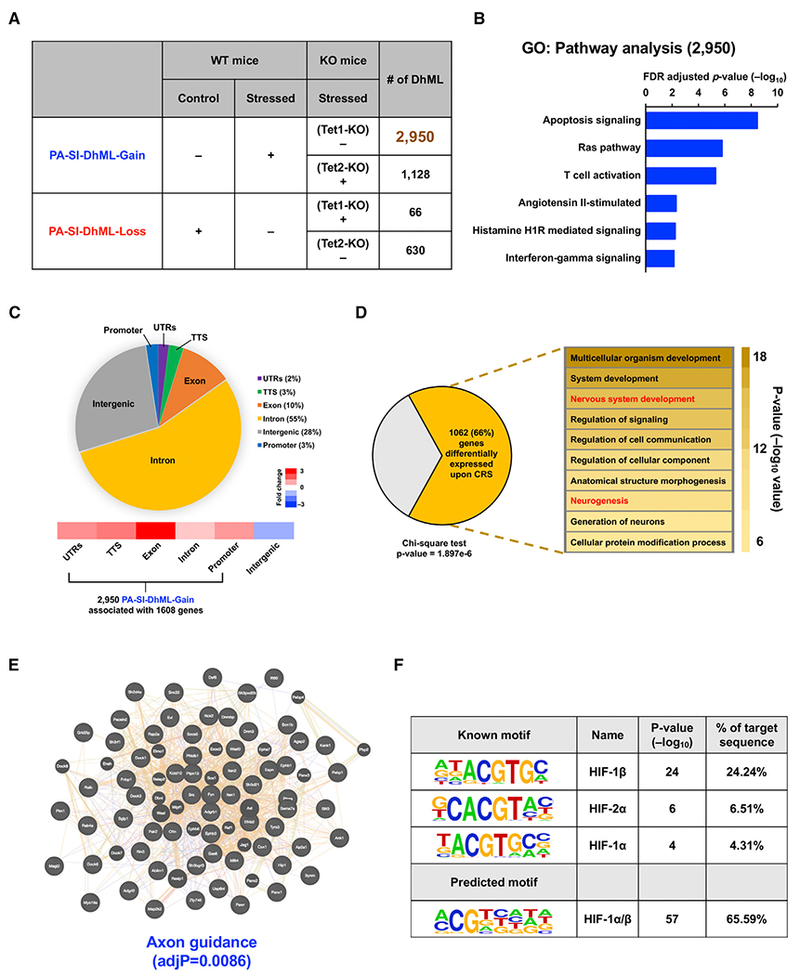
(A) Identification of gain-of-5hmC and loss-of-5hmC loci of phenotype-associated SI-DhMLs (PA-SI-DhMLs), i.e., PA-SI-DhML-Gain and PA-SI-DhML-Loss.
(B) GO analysis on pathways with 2,950 PA-SI-DhML-Gain suggested that these gain-of-5hmC loci were significantly involved in the Ras and T cell activation pathways associated with depression.
(C) Genomic annotation of2,950 PA-SI-DhML-Gain to demonstrate their percentage of each genomic region and enrichment versus expected values. More than 70% of PA-SI-DhML-Gain occurred in the gene body region, which was highly enriched over the expected value.
(D) Significant number of PA-SI-DhML-Gain-associated genes (1,062 of 1,608) were differentially expressed under CRS (p value= 1.897e—6, chi-square test). GO analysis on these 1,062 genes indicated significant enrichment in neurological development-related functions (in red).
(E) Protein coding genes that associated with 2,950 PA-SI-DhML-Gain were selected and then subjected to protein-protein interaction (PPI) analyses using WebGestalt online software. PPI analyses revealed that the proteins associated with PA-SI-DhML-Gain were significantly involved in axon guidance (adjusted p value [adjP] = 0.0086).
(F) Motif search on 2,950 PA-SI-DhML-Gain revealed significant enrichment of the hypoxia-induced factor (HIF) binding motif. The motif alignment score is 0.71.
See also Figure S3.
Stress-Induced 5hmC Alteration and Gene Expression
We performed the GO analysis for the 2,950 PA-SI-DhML-Gain and 630 PA-SI-DhML-Loss to determine whether these loci are associated with any pathways involved in stress response. We found that the 2,950 PA-SI-DhML-Gain significantly associated with specific critical pathways, including Ras and T cell activation, which are involved in depression (Figure 3B). No significant association of any pathway was observed for the 630 PA-SI-DhML-Loss (data not shown). Therefore, we focused only on these 2,950 PA-SI-DhML-Gain for the following analyses.
Genomic annotation of these 2,950 PA-SI-DhML-Gain revealed biased genomic features. For example, 72% of the PA-SI-DhML-Gain were associated with intragenic regions, such as promoter, intron, and exon, while only 28% of the PA-SI-DhML-Gain were located at intergenic regions (Figure 3C). Among them, the PA-SI-DhML-Gain at the exon showed the highest enrichment versus expected value. Additional GO analyses showed that the 2,950 PA-SI-DhML-Gain were significantly associated with the genes involved in neuronal functions, such as regulation of synapse maturation (Figure S3A).
Furthermore, to link PA-SI-DhMLs and altered gene expression, global transcriptomes from control and stressed PFC were assessed by RNA sequencing (RNA-seq) (Table S1). Among the 2,950 PA-SI-DhMLs, we found about 70% of PA-SI-DhMLs were in the intragenic region of 1,608 genes (Figure 3C). A sizable number of genes (492 upregulated and 570 downregulated) were differentially expressed (p value = 1.897e—6, chi-square test) (Figures 3D and S3B). These differentially expressed (DE) genes were significantly enriched in biological functions related to neurological development (Figure 3D). Furthermore, we performed additional analyses using WebGestalt (Web-Based Gene Set Analysis Toolkit), which contains mouse protein-protein interaction modules and identifies hierarchical modules (Wang et al., 2013). The genes associated with PA-SI-DhML-Gain showed significant correlation with the genes involving in axon guidance, ephrin receptor signaling pathway, and endocytosis (Figures 3E, S3C, and S3D).
We next sought to examine the known DNA sequence motifs associated with these 2,950 PA-SI-DhMLs to further explore the possible mechanism. To this end, we used hypergeometric optimization of motif enrichment (HOMER) to search the known motifs in the mm9 genome. We found that the top 10 significant known motifs contained three members of HIF gene family: HIF1α, HIF1β, and HIF2α (Figure 3F). The de novo motif search also found several motifs; the sequence of the top-ranked motif was similar to those of HIF motifs, of which the alignment score is 0.71 (Figure 3F).
Stress-Induced HIF1α Binding Associates with the Dynamic Change of 5hmC
Given that CRS led to the downregulation of Tet1 (Figure 1D), we asked whether CRS also affected Hif1α expression. We examined Hif1α expression in mRNA and protein levels but were not able to find significant changes between control and stressed WT mouse PFC (Figures 4A and 4B). To determine the physical interaction of endogenous TET1 and HIF1α, we performed immunoprecipitation by using an antibody specifically against HIF1α or TET1 using WT PFC lysate (Figure 4C). We observed that HIF1α only interacted with TET1 in the mice exposed to CRS, suggesting HIF1α might recruit TET1 upon CRS.
Figure 4. TET1 Physically Interacts with HIF1α upon CRS and Leads to Dynamic 5hmC Alterations.
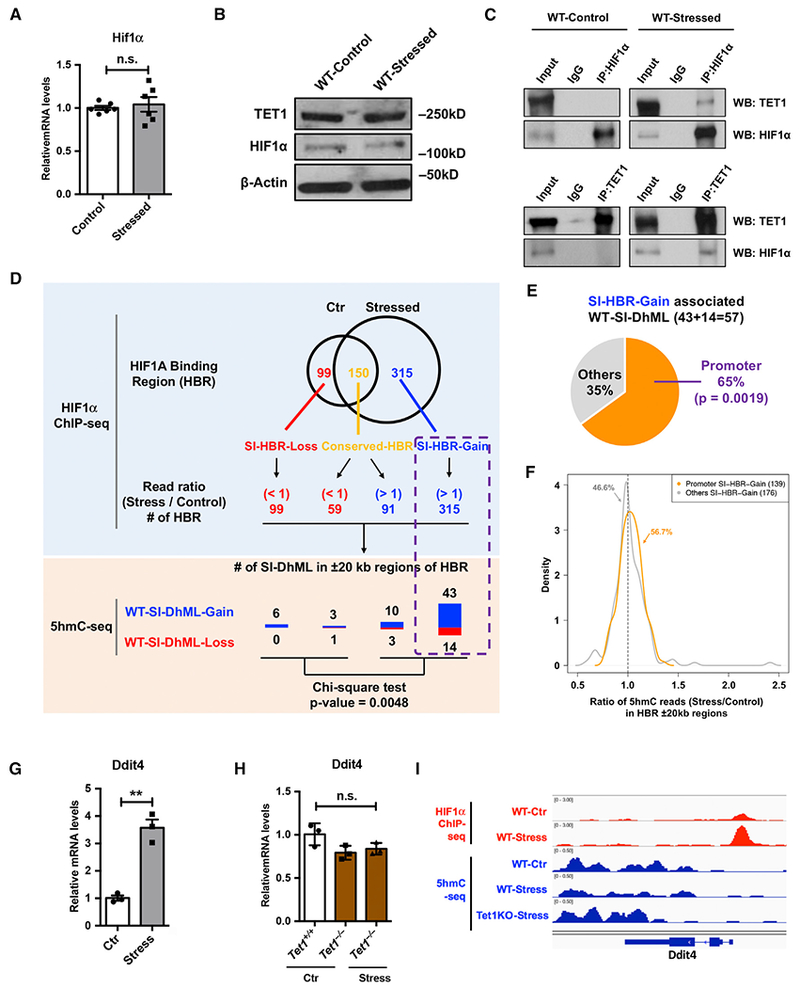
(A and B) qRT-PCR (A) and western blot (B) analysis showed little impact of CRS on TET1 and HIF1α protein in WT mouse PFC. β-actin is used as a loading control.
(C) By using an antibody against HIF1α or TET1, immunoprecipitation followed by western blot analyses revealed that only under the CRS condition could TET1 physically interact with HIF1α.
(D) The HIF1α binding regions (HBRs) with increased ChIP-seq reads (stress and control) were highly associated with WT-SI-DhMLs and PA-SI-DhMLs. Stress-induced differential HBRs were identified by overlapping HIF1α ChIP-seq peaks in WT-control and WT-stress samples, including 99SI-HBR-Loss, 150 conserved HBRs, and 315SI-HBR-Gain. ChIP-seq reads (stress and control) was further examined in conserved HBRs, revealing 59 HBRs with decreased ChIP-seq reads (read ratio < 1) and 91 HBRs with increased ChIP-seq reads (read ratio > 1). WT- and PA-SI-DhMLs in ±20-kb regions of each HBR were investigated, and we found that the HBRs with increased ChIP-seq reads were highly associated with WT-SI-DhMLs (p value = 0.0048, chi-square test).
(E) SI-HBR-Gain in promoter regions associated with significantly more WT-SI-DhMLs. In SI-HBR-Gain-associated WT-SI-DhMLs, 37 of 57 (65%) WT-SI-DhMLs were found to be significantly associated with those promoter-associated SI-HBR-Gain (p value = 0.0019, binomial test).
(F) Ratio density distribution of 5hmC normalized read counts (stress versus control, ±20-kb regions) in two SI-HBR-Gain lists: promoter and non-promoter (other)-associated SI-HBR-Gain. 56.7% of promoter SI-HBR-Gain show enriched 5hmC in stressed WT PFC compared to control, followed by 46.6% of non-promoter SI-HBR-Gain.
(G and H) CRS increased Ddit4 expression in WT mice, but not in Tet1 KO mice.
(G) qRT-PCR analysis of Ddit4 in WT PFC revealed a significant increase of Ddit4 mRNA level upon CRS. n = 3, error bars indicate mean ± SEM; two-tailed unpaired t test; **p < 0.01.
(H) qRT-PCR determined Ddit4 expression in the PFC of control Tet1+/+ and Tet1−/− mice and stressed Tet1−/− mice. Ddit4 exhibited little change upon the loss of Tet1, regardless of the stress condition. n.s., not significant. n = 3, error bars indicate mean ± SEM; two-tailed unpaired t test.
(I) 5hmC dynamic changes in gene body ± a 1.5-kb region of Ddit4 were visualized by an integrated genomics viewer (IGV). HIF1α ChIP-seq data are in red, and 5hmC-seq data are in blue.
See also Figure S4.
To further determine the roles of the TET1-HIF1α interaction, we performed chromatin immunoprecipitation (ChIP) using an anti-HIF1α antibody. Stress-induced HIF1α binding regions (HBRs) were found by overlapping HIF1α ChIP-seq peaks in WT-control and WT-stress samples, revealing 99 SI-HBR-Loss (stress-induced loss of HBRs), 150 conserved HBRs and 315 SI-HBR-Gain (stress-induced gain of HBRs) (Figures 4D and S4A). ChIP-seq reads (stress and control) were further examined from the conserved HBRs, and we identified 59 HBRs with decreased HIF1α binding and 91 HBRs with increased HIF1α binding in stressed PFC (Figure S4B). We hypothesized that TET1 could be recruited by HIF1α under the condition of CRS, resulting in the dynamic changes of 5hmC in specific loci. To test this, WT-SI-DhMLs in ±20-kb regions of each HBR group were investigated. To this end, the HBRs with decreased HIF1α binding were associated with a limited number of WT-SI-DhMLs, while the HBRs with increased HIF1α binding were highly associated with WT-SI-DhMLs (p value = 0.0048, chi-square test) (Figure 4D). Moreover, we found that these WT-SI-DhMLs were significantly associated with the SI-HBR-Gain found in gene promoters (promoter HBRs) (Figure 4E) and that promoter HBRs resulted in more 5hmC upregulation than non-promoter HBRs (Figure 4F).
We further analyzed the correlation between promoter HBRs and gene expression and found that most promoter HBR-associated genes were differentially expressed (Table S2). The most significantly changed genes (with log2 fold change [Log2FC] > 0.5 or Log2FC < −0.5) included 4 upregulated and 19 down-regulated genes associated with dynamically changed 5hmC (Figure S4C; Table S2). Many of these genes have been reported to be associated with major depression disorder (MDD). For example, one of the upregulated genes, Ddit4 (DNA damage-inducible transcript 4), which has been reported to be associated with depression (Ota et al., 2014), showed a significant increase in the PFC of stressed mice. To validate the elevated Ddit4 expression in RNA-seq data, we performed qRT-PCR analysis and observed significant upregulation of Ddit4 in WT PFC upon CRS (Figure 4G). Moreover, we found that in Tet1 KO mice, Ddit4 had limited change in either the normal or the CRS condition (Figure 4H). We then examined whether 5hmC alteration was associated with the change in Ddit4 gene expression and found that 5hmC reduced in WT mice upon stress but increased in stressed Tet1 KO mice (Figures 4I and S4D). These data suggest that Tet1-mediated epigenetic modulation could contribute to the regulation of MDD-linked genes.
DISCUSSION
Epidemiological studies have linked a combined contribution of both genetic and environmental factors, particularly stress, to an increased risk for depression (Hammen, 2005; Kessler, 1997). Epigenetic modifications, including post-translational histone modifications and DNA methylation, could reflect the molecular responses to environmental changes and thus provide a potential mechanism for gene-environment interactions (Klengel and Binder, 2015; Nestler, 2014). Our present study demonstrates the dynamic change of 5hmC in mouse PFC in response to CRS. Genome-wide 5hmC profiling showed the SI-DhMLs were involved with pathways that are significantly associated with MDD. More importantly, we observed that the two core 5hmC writers, Tet1 andTet2, play distinct roles in stress-induced depression-like behaviors, in which Tet1 KO and Tet2 KO mice, respectively, show resistance and susceptibility to CRS. By investigating the PA-SI-DhMLs, we have found that in stressed mice, Tet1 handles generation of most gain-of-5hmC regions (PA-SI-DhML-Gain), in which HIF binding motifs are strongly enriched. We demonstrate the physical interaction between TET1 and HIF1α under CRS and reveal that the increased HIF1α binding is highly associated with increased 5hmC modification. These results suggest that HIF1α may recruit TET1 upon CRS and dynamically influence 5hmC status.
TET family members (TET1, TET2, and TET3) are the writers that regulate 5hmC status and play important roles in various biological processes; however, their expression patterns and functions are different (Wu and Zhang, 2011). Previous studies show that Tet1 and Tet2, despite some overlaps, have distinct roles in mouse embryonic stem cell (mESC) development, re-programing, and neurodevelopment (Koh et al., 2011; Li et al., 2017; Piccolo et al., 2013; Zhang et al., 2013). For example, loss of Tet1 causes a reduction in the self-renewal capability of adult neuronal stem cells, with limited impact on the differentiation potential (Zhang et al., 2013). We have demonstrated opposite results in Tet2−/− mice (Li et al., 2017), indicating that Tet1 and Tet2 have distinct roles in the regulation of neurogenesis. Our present data have demonstrated that Tet1 KO and Tet2 KO mice exhibit strikingly distinct phenotypes in response to stress. By comparing the SI-DhMLs that associated with stress-resistant and stress-susceptible phenotypes, we show that Tet1 is responsible for the stress-induced gain-of-5hmC regions, based on the finding that Tet1-dependent PA-SI-DhML-Gain contains most WT-SI-DhML-Gain (2,950 of 3,239 SI-DhMLs, ~91%). These PA-SI-DhML-Gain are significantly involved in various important pathways that are associated with MDD and influence expression of the genes associated with neurodevelopment. However, the Tet1 KO and Tet2 KO mice that we used in this study were on different genetic backgrounds (C57BL/6 for Tet1 KO versus 129 for Tet2 KO). It is well known that different mouse strains might respond differently to external stimuli, such as environmental stress. In our analyses, Tet1+/+ control mice behaved similarly to WT C57BL/6 mice (Figure 1). When compared with Tet2+/+ control mice, Tet2 KO mice exhibited a stressed phenotype even before they were exposed to stress, and their immobility time in FST significantly increased at week 2 after the exposure to stress; however, unlike Tet1+/+ controls, the Tet2+,+ control mice did not show a significant increase in immobility upon the exposure to stress, which is likely due to a different genetic background. Despite this, we believe our data support that loss of Tet2 would result in increased stress susceptibility.
TET1-mediated 5hmC changes were reported as an essential epigenetic component of the hypoxic response in neuroblastoma (Mariani et al., 2014). Thus, the discovery that PA-SI-DhML-Gain is strongly enriched with HIF binding motifs indicates an exciting possibility of interaction between TET1 and HIF proteins in regulating 5hmC in stress-induced response. Our biochemical results demonstrate that TET1 specifically interacts with HIF1α under CRS conditions in WT mouse PFC, although CRS has no impact on the expression of TET1 and HIF1α (Figures 4A–4C). To validate whether TET1 can interact with HIF1α under CRS and affect 5hmC status, we performed ChIP-seq using an antibody against HIF1α and identified differentially changed HBRs. Only the HBRs with increased ChIP-seq reads (stress and control) are associated with WT-SI-DhMLs (Figure 4D), suggesting that HIF1α may recruit TET1 upon CRS and regulates 5hmC status.
REDD1 (also known as DDIT4 and RTP801) is an inhibitor of mTORC1 (mammalian target of rapamycin complex-1) (Corradetti et al., 2005) and has been shown to be upregulated in stressed rat PFC and postmortem PFC of human subjects with MDD (Ota et al., 2014). In addition, manipulating REDD1 in vivo reveals that REDD1 plays critical roles in neuronal atrophy and depressive behavior caused by chronic stress exposure (Ota et al., 2014). Consistent with these works, our results showed a significant increase of Ddit4 in stressed WT mouse PFC (Figure 4G), which corresponded with the depression-like behavior. In Tet1 KO mouse PFC, however, we observed limited change in Ddit4 expression upon CRS (Figure 4H), which was consistent with stress resistance. Moreover, our ChIP-seq data showed significantly more HIF1α binding to the promoter region of Ddit4 in the PFC of stressed mice, suggesting a potential role of the TET1-HIF1α complex in regulating Ddit4 gene expression. These data suggest that Tet1-mediated modulation may play an important role in regulating depression-linked genes in response to stress.
In summary, our data presented here reveal a Tet-mediated modulation in stress-induced response. The distinct phenotypes of Tet1 KO and Tet2 KO mice in response to CRS enable us to investigate the potential roles of Tet1 andTet2 in stress response. We demonstrate that TET1 can physically interact with HIF1α upon CRS and influence the adjacent 5hmC dynamics, which may further regulate the expression of neurodevelopment-associated genes. These results suggest that TET1, by interacting with HIF1α, plays an important role in stress response.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Peng Jin (peng.jin@emory.edu). There are no restrictions on any data or materials presented in this paper.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
C57BL/6 WT mice were used for the initial CRS experiment (Figures 1 and S1). To further explore the epigenetic mechanism involved in stress-induced depression-like behavior, we used the previously generated global Tet1 knockout (Tet1 KO) and Tet2 knockout (Tet2 KO) mice (Figures 2 and S2). Tet1 KO line was obtained from The Jackson Laboratory (stock number 017358, B6;129S4-Tet1tm1.1Jae/J (Dawlaty et al., 2011), and has been bred with C57BL/6 WT mice. Tet2 KO line was provided by Dr. Mingjiang Xu (Li et al., 2011), which was generated in 129/sv background and has been maintained in 129/sv background. All animal procedures occurred according to protocols approved by Emory University Institutional Animal Care and Use Committee and University Committee on Animal Care of Soochow University.
METHOD DETAILS
Chronic Restraint Stress (CRS)
Mice were individually placed into a modified well-ventilated 50 mL centrifuge tube (Corning Inc., Corning, NY) daily from 10am to 12pm for 2 hours in their home cages for two weeks. The restrained mice could change from supine to prone position but were not able to move forward or backward in tubes. The non-restrained control mice remained undisturbed in their home cages. After restraint stress, mice were released from the tube and returned to their home cages.
Forced Swimming Test
Forced swimming test (FST) was performed as described previously (Porsolt et al., 1977). In brief; each mouse was placed in a glass cylinder (20 cm high, 15 cm in diameter) with warm water (23–25°C, 14 cm in depth) for 6 min. Fresh water of the same temperature was used for each test. Mice were gently dried after removal from the bath and returned to their home cages. Immobility time was recorded when the animal floated in the water or made the little movement for floatation.
Tail Suspension Test
Tail suspension test (TST) was performed according to the method outlined in previous reports with a minor modification (Steru et al., 1985). Each mouse was suspended by adhesive tape placed 1 cm from the tip of the tail. The distance between the tip of tail of each mouse and the desktop was about 45 cm. Each mouse was suspended for 6 min and the immobility time was recorded.
Genomic DNA Preparation
Mouse amygdala, cerebellum, hippocampus, hypothalamus and PFC were dissected from the brain. Genomic DNAs were isolated by different brain tissues in 600 μL digestion buffer (100 mM Tris-HCl, pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl), Proteinase K (Thermo Fisher, cat# EO0491) treatment at 55°C for overnight. The second day, 600 μL of Phenol:Chloroform:Isoamyl Alcohol (25:24:1 Saturated with 10 mM Tris, pH 8.0, 1 mM EDTA) (Sigma-Aldrich, cat# P-3803) was added to samples, mixed completely, and centrifuged for 10 min at 12,000 rpm. The aqueous layer solution was transferred into a new Eppendorf tube and precipitated with 600 μL isopropanol. The pellet was washed with 75% ethanol, air-dried, and eluted with Nuclease-Free Water (Ambion).
Dot Blot
Dot blot was performed on a Bio-Dot Apparatus (cat#170-6545, BIO-RAD) as described previously (Song et al., 2011) using rabbit antibody to 5hmC (1:10,000, cat#39769, Active Motif) as the primary antibody, incubated overnight at 4°C. Horseradish peroxidase–conjugated antibody to rabbit (1:5,000, #A-0545, Sigma) was used as a secondary antibody, and incubated for 45 min at 20–25°C. Standard DNA templates were loaded (cat#D5405, ZYMO) for the quantification and to verify the specificity of antibodies. The density of each dot signal was quantified by ImageJ software (Schneider et al., 2012).
RNA Isolation and qPCR
PFC samples were homogenized in TRIzol (Invitrogen, cat# 15596026) and processed according to the manufacturer’s instructions. RNA was then reverse transcribed using a SuperScript III First-Strand Synthesis System (Thermo Fisher, cat# 18080051). cDNA was quantified by qPCR using TaqMan Universal PCR Master Mix (Thermo Fisher, cat# 4304437) and SYBR Green PCR Master Mix (Thermo Fisher, cat# 4309155). Each reaction was run in triplicate and analyzed following the ΔΔCt method using glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as normalization control. All experiments were repeated at least twice. All primer sequences are listed in Table S3.
Immunoprecipitation (IP) and Western Blot Analysis
Two mice PFC samples was dissected, combined, homogenized and lysed for 30 min on ice in IP lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1mM EDTA, 5% Glycerol) with cOmplete ULTRA Tablets, Mini, EASYpack Protease Inhibitor Cocktail (Sigma-Aldrich, 05892970001), and then centrifuged at 4°C with 12,000 rpm for 15 min. Protein concentration was determined using Pierce BCA Protein Assay Kit (Thermo Fisher, cat# 23225). The supernatants were then incubated with either HIF-1 alpha Antibody (Novus Biologicals, cat# NB100-134) orTET-1 Antibody (GeneTex, cat# GTX124207) overnight. Rabbit IgG-Agarose (Sigma-Aldrich) was used as control IgG. The beads were washed three times. The immunoprecipitates were eluted with 2× Laemmli Sample Buffer (Bio-Rad), boiled at 95°C for 8 min, and the following immuno-blotting assays were carried out to detect the target proteins as indicated. Rabbit anti-HIF1α (1:1,000; Novus Biologicals, cat# NB100-134), -TET1 (1:1,000; GeneTex, cat# GTX124207) and -β-Actin (1:3000; Abcam, cat# ab8227) were used for western blot.
Chromatin Immunoprecipitation (ChIP) and ChIP-seq
PFC from two mice was combined, homogenized gently in cold PBS (with protease inhibitor cocktail), then treated with 1% formaldehyde for 10 min at room temperature with gentle shaking. Fixation was terminated by adding 2M fresh glycine to reach a 0.125 M final concentration, and then shaking continued for an additional 5 min. The cell pellet was collected by spinning down at 3,750 rpm for 10 min at 4°C. The cell pellet was then resuspended in 1 mL Nuclei Swelling Buffer (10 mM HEPES/pH 7.9, 0.5% NP-40,1.5 mM MgCl2,10 mM KCl, 0.5 mM DTT, and protease inhibitor cocktail), incubated on ice for 10 min, and centrifuged at 5,000 rpm for 5 min. Nuclear pellets were further lysed in 200 μl SDS lysis buffer (20 mM HEPES/pH7.9, 25% glycerol, 0.5% NP-40, 0.5% Triton X-100, 0.42 M NaCl, 1.5 mM MgCl2,0.2 mM EDTA), and protease inhibitor cocktail). Cell lysate was sonicated 8 times with 0-3 Power Output for 30 s each (at least 30 s cooling on ice between each 30 s sonication) to obtain DNA fragments between 200-500 bp. After sonication, nuclear lysate was cleared by centrifugation at 13,000 rpm for 10 min at 4°C to collect the supernatant. The nuclear lysate was diluted with 4 volumes of dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl/pH8.0, and protease inhibitor cocktail). Diluted nuclear lysate was pre-cleared by 30 μl pre-washed Protein-G Dynabeads (Thermo Fisher, cat# 10009D) for 2 hours at 4°C. After pre-clear, immunoprecipitation was performed with 5 μg ChIP grade antibody specific to HIF-1 alpha (Novus Biologicals, cat# NB100-134) with overnight rotation at 4°C. 30 μL pre-washed Protein-G Dynabeads (Thermo Fisher, cat# 10009D) were added for an additional 2 h. The beads were then washed with a series of wash buffers for 5 min in the following order: Wash Buffer I (0.1% SDS, 1% Triton X-100,2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl/pH 8.1), Wash Buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl/pH 8.1), Wash Buffer III (0.25 M LiCl, 1% NP-40,1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl/pH 8.1). Then the beads were washed twice with TE buffer before being eluted with 350 μl Elution Buffer (1% SDS and 0.1 M NaHCO3). The elution was incubated at 65°C for at least 6 h to reverse the formaldehyde cross-linking, and then DNA fragments were purified using QIAquick PCR Purification Kit (QIAGEN, cat#28106). ChIPed DNA was subjected for library preparation. 5 ng of ChIPed DNA and input DNA was used for preparation of sequencing libraries following the instructions of Illumina’s ChIP-seq sample prep kit (cat# IP-102-1001). All sequencing libraries were run on Illumina Hi-seq 2000 machines.
5hmC–specific Chemical Labeling, Affinity Purification and Sequencing
5hmC enrichment was performed using a previously described procedure with an improved selective chemical labeling method (Szulwach et al., 2011). DNA libraries were generated following the Illumina protocol for “Preparing Samples for ChIP Sequencing of DNA” (Part# 111257047 Rev. A) using 25–50 ng of input genomic DNA or 5hmC-captured DNA to initiate the protocol. All sequencing libraries were run on Illumina Hi-seq 2000 machines.
RNA-seq
Three biological replicates (both control and stressed mice PFC RNA) were subjected to RNA-seq. 4 μg of total RNA was used for RNA-seq library construction following instructions of Illumina mRNA sample prep kit (cat# RS-100-0801). In brief, the poly-A–containing mRNA was purified using poly-T oligo-attached magnetic beads. The mRNA was then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA Polymerase I and RNaseH. These cDNA fragments went through an end repair process, the addition of a single ‘A’ base, and then ligation of the adapters. These products were gel purified and enriched with PCR to create the final cDNA libraries. The library constructs were run on the bioanalyzer to verify the size and concentration before sequencing on the Illumina HiSeq2000 machines. RNA-seq reads were aligned using TopHat v2.0.1349 (Trapnell et al., 2009), and differential FPKM (Fragments Per Kilobase of transcript per Million mapped reads) expression values were extracted using Cufflinks v2.2.1 (Trapnell et al., 2010). Genes with log2FC > 0.1 or < −0.1 and FPKM > 0.5 are defined as differentially expressed genes.
Sequence Alignment, Binning, and Peak Identification
Processing of sequencing data was performed as previously described (Szulwach et al., 2011). Briefly, FASTQ sequence files from biological replicates were concatenated and aligned to the M. musculus reference genome (NCBI37v1/mm9) using Bowtie 1.1.0 (Langmead et al., 2009), keeping unique non-duplicate genomic matches with no more than 2 mismatches within the first 25 bp. Unique, non-duplicate reads from input genomic DNA and each 5-hmC–enriched sequence set were counted in 100-, 1,000- and 10,000-bp bins genome wide and normalized to the total number of non-duplicate reads in millions. Input normalized values were subtracted from 5-hmC–enriched values per bin to generate normalized 5-hmC signals. Bins from each sample were clustered using uncentered correlation with pairwise complete linkage (Cluster3.0) to broadly assess patterns of 5-hmC–regulated loci. Overall, clustering of different sized bins generated similar clustering profiles. All data presented are those binned at 10,000 bp.
Identification and Annotation of SI-DhMLs
To identify the DhML, we employed a Poisson-based peak identification algorithm (MACS) using unique non-duplicate reads (Zhang et al., 2008). First we used MACS to call peaks from the replicates of each conditions (effective genome size = 1.87 × 109, tag size = 38, bandwidth = 200, P value cutoff = 1.00 × 10~5). By overlapping the peaks in littermate Control and Stressed samples, we identified the stress induced stress-unique peaks (SI-DhML-Gain) and control-unique peaks (SI-DhML-Loss). Only the DhML present in all replicates were used in the following analyses. DhML with genomic features was performed by overlapping defined sets of DhML with known genomic features obtained from UCSC Tables for NCBI37v1/mm9 by using HOMER (Hypergeometric Optimization of Motif EnRichment) software (Heinz et al., 2010). DhML were assigned to a given genomic feature if overlapping ≥ 1 bp. The percentage of total DhML in a defined set was divided by the percentage expected to overlap each genomic feature by chance, based on the percentage of genomic space occupied by that genomic feature, to determine the fold changes from expected values. GREAT (Genomic Regions Enrichment of Annotations Tool, version 3.0) and DAVID (The Database for Annotation, Visualization and Integrated Discovery) was employed to perform GO analyses (Huang et al., 2009; McLean et al., 2010). WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) was used for protein network analysis (Wang et al., 2013). HOMER Motif Analysis software performed Known and De novo Motif searches.
Read Coverage and Visualization
Genomic views of read coverage were generated using Integrated Genomics Viewer tools and browser (IGV 1.4.2, http://www.broadinstitute.org/igv/) (Robinson et al., 2011) with a window size of 25 and a read extend of 200.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed in Prism 7.0 (GraphPad Software). Datasets were analyzed for significance using either unpaired Student’s two-tailed t tests or ANOVA with multiple comparison post hoc tests; all data are presented as mean ± s.e.m. Samples and animal groups with p value < 0.05 were considered statistically. Pearson’s chi-square tests with Yates’ continuity correction were performed in R software (http://www.r-project.org/). Sample sizes and descriptions of sample collection are provided in each figure legend.
DATA AND SOFTWARE AVAILABILITY
The sequencing data are archived at Gene Expression Omnibus under accession number GSE106220. The customized R codes were used to generate Figures 1C, 1E, 4D–4F, S1B, S3B, S4B, and S4D, and will be provided upon request to the Lead Contact.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-5-Hydroxymethylcytosine (5-hmC) | Active Motif | Cat# 39769, RRID:AB_10013602 |
| Rabbit polyclonal anti-HIF1 alpha | Novus Biologicals | Cat# NB 100-134, RRID:AB_350071 |
| Rabbit polyclonal anti-TET1 (clone N3C1) | GeneTex | Cat# GTX124207, RRID:AB_11176491 |
| Rabbit polyclonal anti-beta Actin | Abcam | Cat# ab8227, RRID:AB_2305186 |
| Anti-Rabbit IgG | Sigma-Aldrich | Cat# A0545, RRID:AB_257896 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Proteinase K | Thermo Fisher | Cat# EO0491 |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | Sigma-Aldrich | Cat# P-3803 |
| 5-mC & 5-hmC DNA Standard Set | Zymo Research | Cat# D5405 |
| TRIzol | Invitrogen | Cat# 15596026 |
| cOmplete ULTRA Tablets, Mini, EASYpack Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# 05892970001 |
| Dynabeads Protein G | Thermo Fisher | Cat# 10009D |
| Critical Commercial Assays | ||
| SuperScript III First-Strand Synthesis System | Thermo Fisher | Cat# 18080051 |
| TaqMan Universal PCR Master Mix | Thermo Fisher | Cat# 4304437 |
| SYBR Green PCR Master Mix | Thermo Fisher | Cat# 4309155 |
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat# 23225 |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28106 |
| Illumina’s ChIP-seq sample prep kit | Illumina | Cat# IP-102-1001 |
| Illumina mRNA sample prep kit | Illumina | Cat# RS-100-0801 |
| Deposited Data | ||
| 5hmC-seq | This paper | GEO: GSE106220 |
| ChIP-seq | This paper | GEO: GSE106220 |
| RNA-seq | This paper | GEO: GSE106220 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: B6;129S4-Tet1tm1.1Jae/J | The Jackson Laboratory | Stock No: 017358 |
| Mouse: Tet2:nlacZ/nGFP (129/sv & C57BL/6 mixed genetic background) | Li et al., 2011 | N/A |
| Oligonucleotides | ||
| Primers for qPCR, see Table S3 | This paper | N/A |
| Software and Algorithms | ||
| TopHat | Trapnell et al., 2009 | http://tophat.cbcb.umd.edu/ |
| Cufflinks | Trapnell et al., 2010 | http://cufflinks.cbcb.umd.edu/ |
| Bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| MACS | Zhang et al., 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/index.html |
| GREAT | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| DAVID | Huang et al., 2009 | https://david.ncifcrf.gov/ |
| WebGestalt | Wang et al., 2013 | http://www.webgestalt.org/webgestalt_2013/ |
| IGV | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
| Prism 7 | GraphPad | https://www.graphpad.com/ |
| R | R Project | https://www.r-project.org/ |
Highlights.
Chronic stress induces depression-like behaviors and alters 5hmC distribution in PFC
Tet1 and Tet2 knockout mice exhibit distinct responses to chronic stress
TET1 physically interacts with HIF1α under chronic stress conditions
Stress-induced HIF1α binding associates with the dynamic change of 5hmC
ACKNOWLEDGMENTS
The authors thank Deborah Cook for critical reading of the manuscript. This work was supported by grants from the National Key R&D Program of China (2017YFE0103700) and the National Natural Science Foundation of China (81120108011 and 81771454) to X.X. and in part by NIH grants (NS051630, MH102690, NS097206, and AG052476 to P.J.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.061.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Arnsten AF (2009). Stress signalling pathways that impair prefrontal cortex structure and function. Nat. Rev. Neurosci 10, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, and Shilatifard A (2009). An operational definition of epigenetics. Genes Dev 23, 781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, and Poulton R (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Bernstein A, Chen D, and Jin P (2015). 5-hydroxymethylcytosine: a new player in brain disorders? Exp. Neurol 268, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, and Guan KL (2005). The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem 280, 9769–9772. [DOI] [PubMed] [Google Scholar]
- Covington HE 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ 3rd, Wu EY, et al. (2009). Antidepressant actions of histone deacetylase inhibitors. J. Neurosci 29, 11451–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, et al. (2011). A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, and Jaenisch R (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammen C (2005). Stress and depression. Annu. Rev. Clin. Psychol 1, 293–319. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, and McEwen BS (2009). Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. USA 106, 20912–20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, and Zhang Y (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC. (1997). The effects of stressful life events on depression. Annu. Rev. Psychol 48, 191–214. [DOI] [PubMed] [Google Scholar]
- Klengel T, and Binder EB (2015). Epigenetics of stress-related psychiatric disorders and gene 3 environment interactions. Neuron 86, 1343–1357. [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, and Heintz N (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, and Xu M (2011). Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yao B, Chen L, Kang Y, Li Y, Cheng Y, Li L, Lin L, Wang Z, Wang M, et al. (2017). Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun 8, 15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, and Drohat AC (2011). Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem 286, 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani CJ, Vasanthakumar A, Madzo J, Yesilkanal A, Bhagat T, Yu Y, Bhattacharyya S, Wenger RH, Cohn SL, Nanduri J, et al. (2014). TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep 7, 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ (2014). Epigenetic mechanisms of depression. JAMA Psychiatry 71, 454–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium (2015). Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci 18, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony CM, Sweeney FF, Daly E, Dinan TG, and Cryan JF (2010). Restraint stress-induced brain activation patterns in two strains of mice differing in their anxiety behaviour. Behav. Brain Res 213, 148–154. [DOI] [PubMed] [Google Scholar]
- Ota KT, Liu RJ, Voleti B, Maldonado-Aviles JG, Duric V, Iwata M, Dutheil S, Duman C, Boikess S, Lewis DA, et al. (2014). REDD1 is essential for stress-induced synaptic loss and depressive behavior. Nat. Med 20, 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, et al. (2013). Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell 49, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsolt RD, Le Pichon M, and Jalfre M (1977). Depression: a new animal model sensitive to antidepressant treatments. Nature 266, 730–732. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, and Merikangas KR (2009). Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA 301, 2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdo´ ttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP. (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, McEwen BS, and Chattarji S (2009). Stress, memory and the amygdala. Nat. Rev. Neurosci 10, 423–433. [DOI] [PubMed] [Google Scholar]
- Savitz J, and Drevets WC (2009). Bipolar and major depressive disorder: neuroimaging the developmental-degenerative divide. Neurosci. Biobehav. Rev 33, 699–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder FA, Lin CL, Crusio WE, and Akbarian S (2007). Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol. Psychiatry 62, 55–64. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. (2011). Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol 29, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C,unzel M, Wagner M,M€uller M, Khan F, et al. (2013). Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- Steru L, Chermat R, Thierry B, and Simon P (1985). The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology (Berl.) 85, 367–370. [DOI] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, and Nestler EJ (2013). Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology 38, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. (2011). 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, and Rao A (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, and Nestler EJ (2006). Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci 9, 519–525. [DOI] [PubMed] [Google Scholar]
- Uher R, and McGuffin P (2008). The moderation by the serotonin transporter gene of environmental adversity in the aetiology of mental illness: review and methodological analysis. Mol. Psychiatry 13, 131–146. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, and Zhang B (2013). Web-based gene set analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, and Meaney MJ (2004). Epigenetic programming by maternal behavior. Nat. Neurosci 7, 847–854. [DOI] [PubMed] [Google Scholar]
- Wong ML, and Licinio J (2001). Research and treatment approaches to depression. Nat. Rev. Neurosci 2, 343–351. [DOI] [PubMed] [Google Scholar]
- Wu H, and Zhang Y (2011). Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25, 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen L, Yang L, Hua X, Zhou B, Miao Z, Li J, Hu H, Namaka M, Kong J, and Xu X (2015). Combined use of spatial restraint stress and middle cerebral artery occlusion is a novel model of post-stroke depression in mice. Sci. Rep 5, 16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data are archived at Gene Expression Omnibus under accession number GSE106220. The customized R codes were used to generate Figures 1C, 1E, 4D–4F, S1B, S3B, S4B, and S4D, and will be provided upon request to the Lead Contact.


