SUMMARY
Protein translation in eukaryotes is cell-cycle dependent, with translation rates more robust in G1 phase of the cell cycle than in mitosis. However, whether the fundamental cell-cycle control machinery directly activates protein translation during the G1/S cell-cycle transition remains unknown. Using the early divergent eukaryote Trypanosoma brucei as a model organism, we report that the G1 cyclin-dependent kinase CRK1 phosphorylates two translation initiation factors, eIF4E4 and PABP1, to promote the G1/S cell-cycle transition and global protein translation. Phosphorylation of eIF4E4 by CRK1 enhances binding to the m7G cap structure and interaction with eIF4E4 and eIF4G3, and phosphorylation of PABP1 by CRK1 promotes association with the poly(A) sequence, self-interaction, and interaction with eIF4E4. These findings demonstrate that cyclin-dependent kinase-mediated regulation of translation initiation factors couples global protein translation with the G1/S cell-cycle transition.
Graphical Abstract
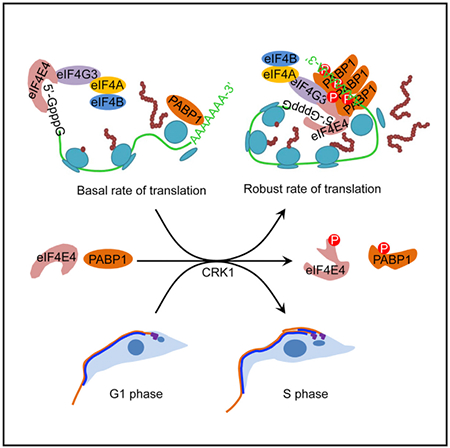
In Brief
Protein translation is cell-cycle dependent, with more robust translation rates in the G1 phase of the cell cycle than in mitosis. An et al. show that the G1 cyclin-dependent kinase CRK1 phosphorylates translation initiation factors eIF4E4 and PABP1 to couple protein translation initiation with the G1/S cell-cycle transition.
INTRODUCTION
All nuclear-encoded mRNAs in eukaryotes contain a modified 5′ end termed cap structure (m7GppN, in which m7G is 7-methylguanylate and N indicates any nucleotides) (Shatkin, 1976) and a 3′ polyadenylate (poly(A)) tail. Cap-dependent protein translation is mediated by eIF4F, a eukaryotic initiation factor complex composed of the cap-binding protein eIF4E; the RNA helicase eIF4A; and the scaffold protein eIF4G, which interacts with eIF4E and eIF4A (Gingras et al., 1999). eIF4G also interacts with eIF3, another initiation factor complex that associates with the 40S small ribosomal subunit (Gingras et al., 1999), and with the poly(A)-binding protein (PABP) (Sachs and Davis, 1989), thereby causing the circulation of the mRNA (Wells et al., 1998). The formation of a closed loop of mRNA facilitates recruitment of the 43S pre-initiation complex, which is composed of the 40S small ribosomal subunit and several initiation factors, to the mRNA, and thus promotes translation initiation (Kozak, 2006).
Protein translation rates fluctuate during the cell cycle in animals (Pyronnet and Sonenberg, 2001). Translation is robust in G1 phase of the cell cycle, but is globally repressed during mitosis (Fan and Penman, 1970; Konrad, 1963; Prescott and Bender, 1962; Tanenbaum et al., 2015). Activation of cap-dependent protein translation requires phosphorylation of eIF4E at Ser209 by the mitogen-activated protein kinase (MAPK)-interacting kinase MnK (Flynn and Proud, 1995; Joshi et al., 1995), which enhances the binding affinity of eIF4E to the cap structure (Minich et al., 1994) and promotes assembly of a stable eIF4F complex (Bu et al., 1993). Suppression of cap-dependent translation in mitosis coincides with eIF4E dephosphorylation (Bonneau and Sonenberg, 1987) and is attributed to the increased level of hypophosphorylated eIF4E-binding protein (BP) (Pyronnet et al., 2001), which competes with eIF4G for the common binding site on eIF4E (Haghighat et al., 1995; Mader et al., 1995) and thus blocks the eIF4F complex assembly (Pyronnet et al., 2001). eIF4E-BP is phosphorylated by the mammalian target of rapamycin (mTOR), an atypical serine/threonine protein kinase (Burnett et al., 1998), thereby releasing eIF4E and activating translation (Gingras et al., 2001). The cyclin-dependent kinase 1 (CDK1) also phosphorylates eIF4E-BP (Heesom et al., 2001; Herbert et al., 2002) and can substitute for mTOR to activate cap-dependent translation in mitosis (Shuda et al., 2015). Other studies found that the translation of some specific mRNAs during mitosis is mediated by a cap-independent mechanism involving the internal ribosomal entry site (IRES) (Cornelis et al., 2000; Pyronnet et al., 2000). However, it was suggested that gene-specific translational regulation in mitosis is mainly to repress but not activate translation (Tanenbaum et al., 2015).
In Trypanosoma brucei, an early divergent protozoan, the genome encodes six eIF4E homologs (eIF4E1–eIF4E6), five eIF4G homologs (eIF4G1–eIF4G5), two eIF4A homologs (eIF4A1 and eIF4A2) (Freire et al., 2017), and two PABP homologs (PABP1 and PABP2) (Kramer et al., 2013). Two distinct eIF4F complexes have been detected in T. brucei, but the dominant eIF4F complex is assembled by eIF4E4, eIF4G3, and eIF4A1, and it associates with PABP1 through eIF4G3 and eIF4E4 (Freire et al., 2011; Moura et al., 2015; Zinoviev et al., 2011). eIF4E4 has the highest binding affinity to the m7G cap among the T. brucei eIF4E homologs (Freire et al., 2011) and is the dominant eIF4E homolog co-purified in the polysomal fraction (Klein et al., 2015). Notably, T. brucei appears to lack the homolog of eIF4E-BP, an inhibitor of the eIF4F complex assembly and a highly conserved protein found in most eukaryotes, except Caenorhabditis elegans (Zinoviev and Shapira, 2012), suggesting that T. brucei likely adopts a cap-dependent translation control mechanism that is distinct from most eukaryotes studied so far.
Initiation of protein translation is essential for the G1/S cell-cycle transition in eukaryotes, as mutation of Cdc33, the yeast eIF4E homolog, arrested cells at G1 phase (Altmann and Trachsel, 1989; Brenner et al., 1988) and loss of the TOR function in yeast and mammals resulted in G1 arrest (Heitman et al., 1991; Wicker et al., 1990). Therefore, robust protein translation during G1 phase depends on the TOR-mediated activation of eIF4F complex assembly (Pyronnet and Sonenberg, 2001). Using T. brucei as a model organism, we report that activation of translation initiation during G1 phase requires the G1 cyclin-dependent kinase CRK1-mediated phosphorylation of translation initiation factors. Phosphorylation of eIF4E4 and PABP1 promotes eIF4E4-eIF4G3 interaction and eIF4E4-PABP1 interaction and enhances eIF4E4 binding to the m7G cap and PABP1 binding to the poly(A) sequence. Our results demonstrate that the fundamental cell-cycle control machinery operating at the G1/S cell-cycle boundary directly activates translation initiation, uncovering the mechanism for cell-cycle-dependent translational control.
RESULTS
The Translation Initiation Factors eIF4E4 and PABP1 Are Substrates of CRK1 in T. brucei
Our recent chemical genetic approach identified several CRK1 substrates, including two translation initiation factors, eIF4E4 and PABP1 (Hu et al., 2016). This approach also identified Ser102 in eIF4E4 and Thr473, Ser477, and Thr484 in PABP1 as CRK1 phosphosites (Hu et al., 2016). Further in vitro kinase assays using purified recombinant proteins followed by mass spectrometry identified six additional phosphosites—Ser90, Ser114, Ser154, Ser175, Ser229, and Thr234—in eIF4E4, but no additional phosphosites in PABP1. The seven phosphosites in eIF4E4 and the three phosphosites in PABP1 were confirmed to be in vivo phosphosites (Urbaniak et al., 2013). Five of the seven phosphosites in eIF4E4 lie in the unusual N-terminal extension, and all three phosphosites in PABP1 are located in the proline-rich region between the fourth RNA recognition motif (RRM) and the PAB C-terminal (PABC) domain (Figure 1A). These CRK1 phosphosites in eIF4E4 and PABP1 are each followed by a proline residue, conforming to the minimal CDK consensus sequence (S/T*-P or S/T*-P-x-K/R, where the asterisk indicates the phosphorylation site and x represents any amino acid) (Holt et al., 2009). A search of potential CDK phosphosites in the yeast and human eIF4E and PABP homologs from the list of phosphosites identified by mass spectrometry (Holt et al., 2009; Rigbolt et al., 2011; Sharma et al., 2014; Swaney et al., 2013; Zhou et al., 2013) suggests one or several putative CDK phosphosites (Figure 1A), but whether these yeast and human homologs are genuine CDK substrates remains to be determined.
Figure 1. eIF4E4 and PABP1 Are Substrates of CRK1.
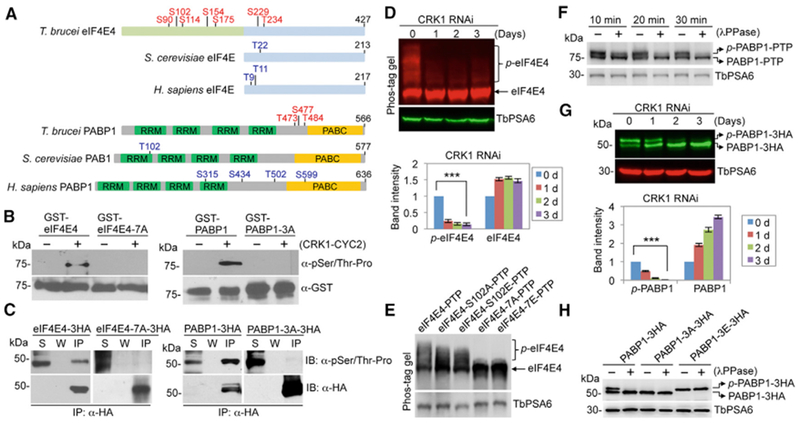
(A) Schematic illustration of the eIF4E and PABP homologs in T. brucei, Saccharomyces cerevisiae, and Homo sapiens. The CRK1 phosphosites in T. brucei eIF4E4 and PABP1 are shown in red, whereas the putative Cdk phosphosites in yeast and human eIF4E and PABP homologs are indicated in blue. RRM, RNA recognition motif; PABC, polyadenylate binding C terminus.
(B) Phosphorylation of eIF4E4 and PABP1 by CRK1 in vitro. An in vitro kinase assay was carried out using purified recombinant proteins. Phosphorylated proteins were detected by anti-pSer/Thr-Pro antibody, and glutathione S-transferase (GST)-fusion proteins were detected by anti-GST antibody.
(C) eIF4E4 and PABP1 are phosphorylated in vivo in T. brucei. eIF4E4, PABP1, and the phospho-deficient mutants (eIF4E4-7A and PABP1-3A) were ectopically expressed in T. brucei, immunoprecipitated, and immunoblotted with anti-pSer/Thr-Pro antibody and anti-HA antibody. IP, immunoprecipitated protein; S, supernatant after immunoprecipitation; W, wash solution.
(D) Effect of CRK1 depletion on eIF4E4 phosphorylation. Phos-tag was included in SDS-PAGE gel to bind to phosphorylated residues of proteins. Non-phospho-eIF4E4 and the slower migrating phosphorylated eIF4E4 (p-eIF4E4) were detected by anti-eIF4E4. The graph below the western blots shows the quantitation of band intensity. Error bars indicate SDs (n = 3). ***p < 0.001.
(E) Effect of phosphosite mutation on the migration of eIF4E4 on SDS-PAGE gel containing Phos-tag solution. PTP-tagged wild-type and mutant eIF4E4 proteins were ectopically expressed and detected with anti-protein A antibody.
(F) Phosphorylated PABP1 (p-PABP1) was detected as a slower migrating band on SDS-PAGE. The lysate of cells expressing protein C-tobacco etch virus-protein A (PTP)-tagged PABP1 was treated with lambda protein phosphatase (λPPase) and detected with anti-Protein A (anti-ProtA) antibody.
(G) Effect of CRK1 RNAi on PABP1 phosphorylation. PABP1 was tagged with a triple HA epitope and detected with anti-HA antibody. The graph shows the quantitation of band intensity. Error bars indicate SDs (n = 3). ***p < 0.001.
(H) Effect of phosphosite mutation on the migration of PABP1 on SDS-PAGE. Triple HA-tagged wild-type and mutant PABP1 were ectopically expressed and detected by with anti-HA antibody. In (D)–(H), TbPSA6 served as the loading control.
Mutation of the seven phosphosites in eIF4E4 and the three phosphosites in PABP1 abolished phosphorylation by CRK1 in vitro (Figure 1B), confirming that these sites are the only CRK1 phosphosites in the two proteins. Immunoprecipitation of eIF4E4, PABP1, and their respective mutants bearing the phosphosite mutations from T. brucei cell lysate, followed by western blotting with the anti-phospho-Ser/Thr-Pro antibody, which specifically detects CDK phosphosites, showed that the seven phosphosites in eIF4E4 and the three phosphosites in PABP1 are phosphorylated in vivo in T. brucei (Figure 1C). Because phosphorylated eIF4E4 could not be separated from non-phosphorylated eIF4E4 by regular SDS-PAGE, we used Phos-tag, which binds to phosphates on phosphorylated proteins and causes slower migration of phosphorylated proteins (Kinoshita et al., 2006), to label phosphorylated eIF4E4. Depletion of CRK1 by RNAi significantly decreased the level of phospho-eIF4E4 (Figure 1D), and mutation of the seven phosphosites to alanine completely abolished eIF4E4 phosphorylation (Figure 1E). These results demonstrated that eIF4E4 is an in vivo substrate of CRK1. Phosphorylated PABP1 (p-PABP1) was detected as a slower migrating band on the western blot, which is sensitive to lambda protein phosphatase treatment (Figure 1F), and its level was significantly decreased upon CRK1 depletion (Figure 1G). Furthermore, mutation of the three phosphosites in PABP1 to alanine (PABP1-3A) completely abolished PABP1 phosphorylation in T. brucei, whereas mutation of these phosphosites to glutamate (PABP1-3E) mimicked the migration of the p-PABP1 (Figure 1H). These results demonstrated that PABP1 is an in vivo substrate of CRK1.
eIF4E4 and PABP1 Are Required for the G1/S Cell-Cycle Transition
Since eIF4E4 and PABP1 are substrates of CRK1, a crucial regulator controlling the G1/S cell-cycle transition in T. brucei (Tu and Wang, 2004), we hypothesized that they are involved in promoting the G1/S cell-cycle transition. To test this hypothesis, we generated an eIF4E4 conditional knockout (cKO) cell line and a PABP1 RNAi cell line. Western blotting confirmed the knockout of native eIF4E4 protein and the gradual depletion of the ectopically protein C-tobacco etch virus-protein A (PTP)-tagged eIF4E4 protein upon removal of tetracycline, and the knock down of the endogenously PTP-tagged PABP1 protein upon tetracycline induction (Figure 2A). Depletion of eIF4E4 caused a moderate growth defect, whereas knock down of PABP1 inhibited cell growth (Figure 2B). Flow cytometry analyses showed that the depletion of eIF4E4 and knock down of PABP1 both caused a significant increase in G1 cells and a corresponding decrease in S phase and G2/M cells (Figure 2C). Furthermore, we quantitated the cells at different cell-cycle stages before and after eIF4E4 depletion and PABP1 knockdown. T. brucei cells at different cell-cycle stages can be distinguished by the numbers of the kinetoplast, the nucleus, and the mature basal body (BB). G1 and S phase cells have one nucleus and one kinetoplast (1N1K), but they have one or two mature BBs, respectively. G2 cells have one nucleus, two kinetoplasts, and two mature BBs (1N2K2BB). Mitotic cells are either 1N2K2BB or 2N2K2BB, and cells at the post-mitosis stage and the cytokinesis stage are 2N2K2BB (Figure 2D). By these criteria, we confirmed that eIF4E4 depletion and PABP1 knockdown resulted in a significant increase in G1 cells, and correspondingly, S phase cells, G2/M cells, and post-mitotic cells decreased (Figure 2D). Finally, we carried out a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay and found that eIF4E4 depletion and PABP1 knockdown impaired EdU incorporation (Figure 2E), indicating that DNA replication was inhibited. These results demonstrated that eIF4E4 and PABP1 are each required for the G1/S cell-cycle transition.
Figure 2. eIF4E4 and PABP1 Are Required for the G1/S Cell-Cycle Transition.
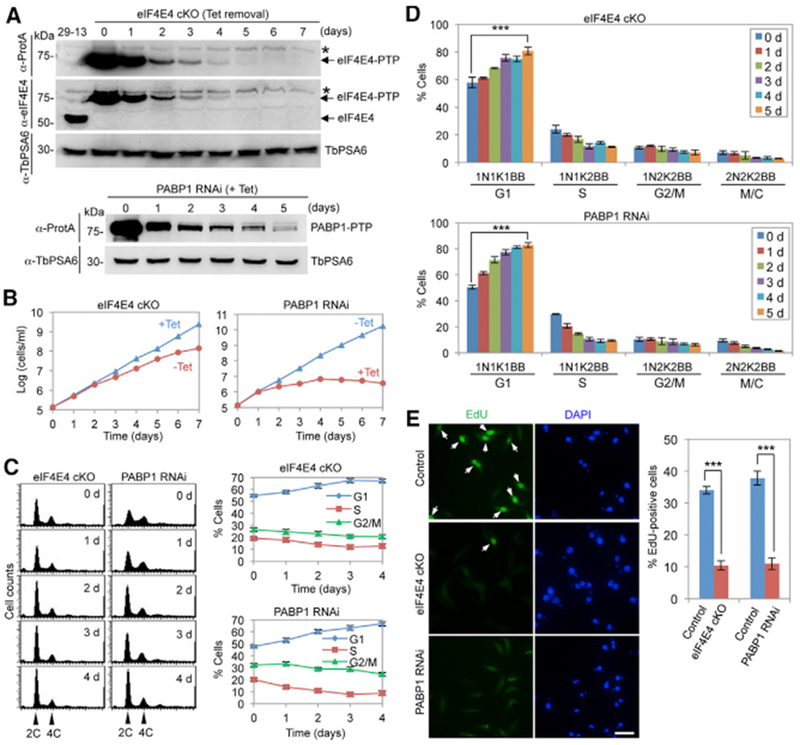
(A) Depletion of eIF4E4 by cKO and of PABP1 by RNAi. eIF4E4-PTP was detected by anti-ProtA antibody, and eIF4E4 was detected by anti-eIF4E4 antibody. PABP1 -PTP was detected by anti-ProtA antibody. TbPSA6 served as the loading control. The asterisk indicates a non-specific band.
(B) Effect of eIF4E4 cKO and PABP1 RNAi on cell proliferation.
(C) Flow cytometry analysis of eIF4E4 cKO and PABP1 RNAi cells. Shown are the flow cytometry histograms (left) and the quantitative data (right). Error bars indicate SDs (n = 3).
(D) Effect of eIF4E4 cKO and PABP1 RNAi on cell-cycle progression. Shown are the percentages of control cells, eIF4E4 cKO cells, and PABP1 RNAi cells at different cell-cycle stages. For each time point, 100 cells were counted. Error bars indicate SDs (n = 3). ***p < 0.001.
(E) Effect of eIF4E4 cKO and PABP1 RNAi on DNA replication. Shown are EdU incorporation of control cells, eIF4E4 cKO (−Tet, 5 days), and PABP1 RNAi (+Tet, 3 days) (left) and quantitative data (right). Error bars indicate SDs (n = 3). ***p < 0.001. Scale bar: 10 μm.
Phosphorylation of eIF4E4 and PABP1 Is Required for the G1/S Cell-Cycle Transition
We next investigated whether CRK1-mediated phosphorylation of eIF4E4 and PABP1 is required for the G1/S cell-cycle transition. We tested the dominant-negative effect of the overexpressed eIF4E4 phospho-deficient mutant. Ectopic expression of PTP-tagged eIF4E4, eIF4E4-7A, and eIF4E4-7E was confirmed by western blotting (Figure 3A). Overexpression of eIF4E4 and eIF4E4-7E did not affect cell proliferation, but overexpression of eIF4E4-7A caused a moderate growth defect (Figure 3B), similar to eIF4E4 cKO (Figure 2B). These results suggest that phosphorylation of eIF4E4 is required for cell proliferation. We then quantitated the cells at different cell-cycle stages using the criteria established above and found that overexpression of eIF4E4-7A, but not eIF4E4 and eIF4E4-7E, resulted in a significant increase in G1 cells (Figure 3C). Overexpression of eIF4E4-7A also decreased EdU incorporation (Figure 3D). These results demonstrated that phosphorylation of eIF4E4 is required for the G1/S cell-cycle transition.
Figure 3. Phosphorylation of eIF4E4 Is Required for the G1/S Cell-Cycle Transition.
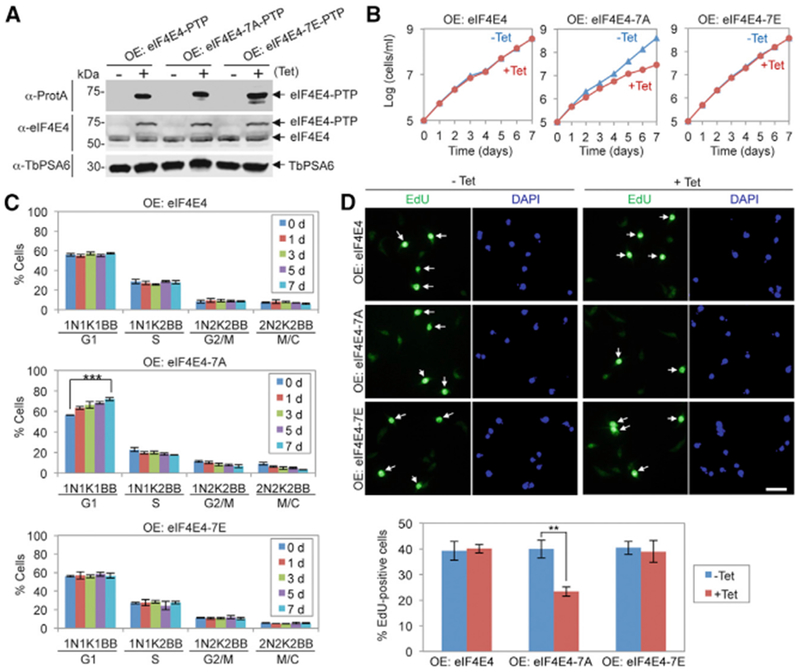
(A) Ectopic overexpression (OE) of PTP-tagged wild-type and mutant eIF4E4 proteins.
(B) Effect of overexpression of eIF4E4, eIF4E4-7A, and eIF4E4-7E on cell proliferation.
(C) Effect of overexpression of eIF4E4, eIF4E4-7A, and eIF4E4-7Eon cell-cycle progression. Shown are the percentages of cells at different cell-cycle stages. For each time point, 100 cells were counted. Error bars indicate SDs (n = 3). ***p < 0.001.
(D) Effect of overexpression of eIF4E4, eIF4E4-7A, and eIF4E4-7E on DNA replication. Shown are the fluorescence micrographs of EdU-stained cells (top) and the percentages of EdU+ cells before (−Tet) and after (+Tet, 5 days) tetracycline induction. Error bars indicate SDs (n = 3). **p < 0.01. Scale bar: 10 μm.
We used the RNAi complementation approach to test the function of PABP1 mutants. We first generated a PABP1-3′ UTR RNAi cell line by targeting the 3′ UTR of PABP1 (Figure 4A), and then ectopically expressed triple hemagglutinin (3HA)-tagged PABP1, PABP1-3A, and PABP1-3E, each of which bears a different 3′ UTR, in the PABP1-3′ UTR RNAi cell line (Figure 4A). Expression of PABP1 or PABP1-3E in the PABP1-3′ UTR RNAi cell line rescued cell proliferation (Figure 4B), but expression of PABP1-3A in the PABP1-3′ UTR RNAi cell line resulted in a moderate growth defect (Figure 4B), indicating that the phosphorylation of PABP1 is important for cell proliferation. Furthermore, quantitation of cells at different cell-cycle stages showed that the expression of PABP1-3A, but not PABP1 and PABP1-3E, caused a significant increase in G1 cells (Figure 4C), and EdU incorporation assays showed that the expression of PABP1-3A impaired DNA replication (Figure 4D). These results demonstrated that the phosphorylation of PABP1 is also important for the G1/S cell-cycle transition.
Figure 4. Phosphorylation of PABP1 Is Required for the G1/S Cell-Cycle Transition.
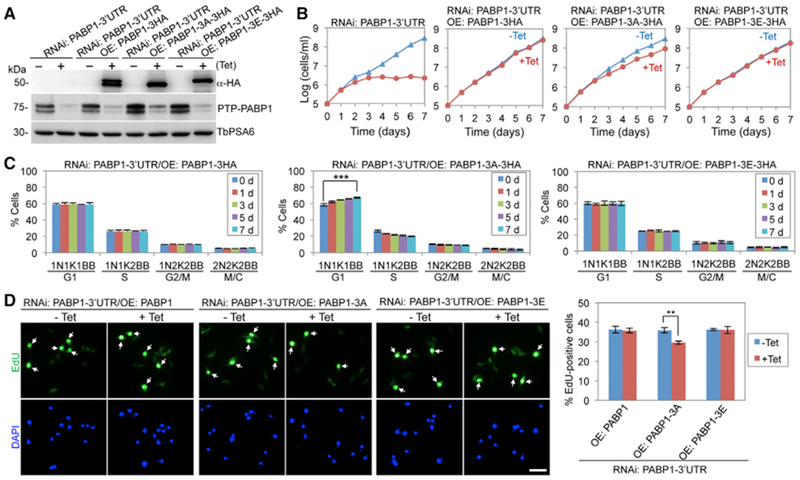
(A) Complementation of PABP1-3′ UTR RNAi by 3HA-tagged wild-type and mutant PABP1 proteins. Ectopic 3HA-tagged PABP1 was detected with anti-HA antibody, and endogenous PTP-tagged PABP1 was detected by anti-ProtA antibody. TbPSA6 served as the loading control.
(B) Effect of PABP1 phosphorylation on cell proliferation.
(C) Effect of PABP1 phosphorylation on cell-cycle progression. Shown are the percentages of cells at different cell-cycle stages. For each time point, 100 cells were counted. Error bars indicate SDs (n = 3). ***p < 0.001.
(D) Effect of PABP1 phosphorylation on DNA replication. Shown are the fluorescence micrographs of EdU-stained cells (top) and the percentages of EdU+ cells before (−Tet) and after (+Tet, 5 days) tetracycline induction. Error bars indicate SDs (n = 3). **p < 0.01. Scale bar: 10 μm. **p < 0.01; ***p < 0.001.
Phosphorylation of eIF4E4 and PABP1 Promotes Interactions among Translation Initiation Factors
We investigated the effect of eIF4E4 and PABP1 phosphorylation by CRK1 on the formation of the translation initiation complex. Previous work demonstrated that eIF4E4 interacts with PABP1 and eIF4G3 (Freire et al., 2011; Moura et al., 2015; Zinoviev et al., 2011). Co-immunoprecipitation confirmed that eIF4E4 interacts with PABP1 (Figure 5A) and eIF4G3 (Figure 5B). While eIE4E4-7A pulled down significantly less PABP1 and eIF4G3 than eIF4E4, eIF4E4-7E restored the interaction with PABP1 and eIF4G3 (Figures 5A and 5B), suggesting that the phosphorylation of eIF4E4 is required for binding to PABP1 and eIF4G3. Conversely, while PABP1-3A pulled down significantly lower amounts of eIF4E4 than PABP1, PABP1-3E restored the interaction with eIF4E4 (Figure 5C), suggesting that the phosphorylation of PABP1 is required for interaction with eIF4E4. However, PABP1-3A, PABP1-3E, and PABP1 all pulled down similar amounts of eIF4G3 (Figure 5D), indicating that the phosphorylation of PABP1 is not required for interaction with eIF4G3. To further corroborate these results, we tested the PABP1-eIF4E4 interaction and the eIF4G3-eIF4E4 interaction in CRK1 RNAi cells. Depletion of CRK1 impaired the interaction between PABP1 and eIF4E (Figure 5E) and the interaction between eIF4G3 and eIF4E4 (Figure 5F). These results demonstrated that the phosphorylation of eIF4E4 and PABP1 promotes the eIF4E4-PABP1 interaction and eIF4E4-eIF4G3 interaction.
Figure 5. Phosphorylation of eIF4E4 and PABP1 by CRK1 Is Required for Interactions among Translation Initiation Factors.
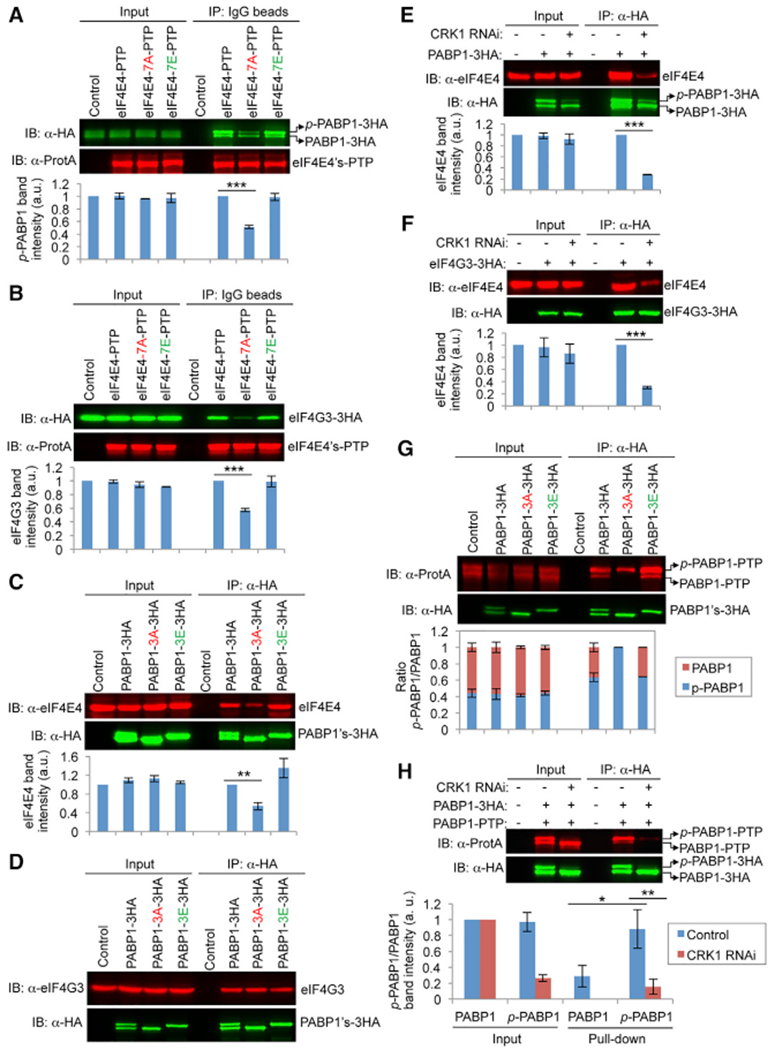
(A) Effect of eIF4E4 phosphorylation on interaction with PABP1. Immunoprecipitation (IP) was carried out with immunoglobulin G (IgG) beads, and immunoblotting (IB) was performed with anti-HA and anti-ProtA antibodies. p-PABP1-3HA, phosphorylated PABP1-3HA.
(B) Effect of eIF4E4 phosphorylation on interaction with eIF4G3.
(C) Effect of PABP1 phosphorylation on interaction with eIF4E4.
(D) Effect of PABP1 phosphorylation on interaction with eIF4G3.
(E) Effect of CRK1 RNAi on the interaction between PABP1 and eIF4E4.
(F) Effect of CRK1 RNAi on the interaction between eIF4E4 and eIF4G3.
(G) Effect of PABP1 phosphorylation on self-interaction. IP was performed with anti-HA beads, and IB was performed with anti-HA and anti-ProtA antibodies. p-PABP1-PTP, phosphorylated PABP1-PTP.
(H) Effect of CRK1 RNAi on PABP1 self-interaction. In all of the panels except (D), the graphs below the western blots show the quantitation of band intensity. Error bars indicate SDs (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001.
Previous work demonstrated that the proline-rich region of human PABP1 is required for self-interaction and for cooperative binding to poly(A) (Melo et al., 2003). We thus hypothesized that the phosphorylation of PABP1 in the proline-rich linker region by CRK1 assists in PABP1 self-interaction, thereby promoting cooperative binding to the poly(A) sequence. Co-immunoprecipitation showed that PABP1 and PABP1-3E pulled down both phospho-PABP1 and non-phospho-PABP1, but the amount of phospho-PABP1 was significantly higher than that of non-phospho-PABP1 (Figure 5G). Moreover, PABP1-3A only pulled down phospho-PABP1 (Figure 5G). These results demonstrated that non-phospho-PABP1 does not interact with itself, but does interact with phospho-PABP1. These results also suggest that PABP1 self-interaction requires phosphorylation by CRK1. Co-immunoprecipitation of PABP1 in CRK1 RNAi cells showed that the depletion of CRK1 impaired PABP1 self-interaction (Figure 5H), further confirming that phosphorylation of PABP1 promotes self-interaction.
Finally, to test whether the interactions between these translation initiation factors are cell-cycle dependent and are mediated by mRNAs, we carried out co-immunoprecipitation experiments using cell-cycle-synchronized cells and treated the cell lysate with RNase (Figures S1 and S2). The results showed that these protein-protein interactions are independent of the cell cycle (Figure S1) and are not mediated by mRNAs (Figure S2).
Phosphorylation Strengthens eIF4E4 Association with the m7G Cap and PABP1 Association with the Poly(A) Sequence
We examined whether phosphorylation of eIF4E4 affects its association with the m7G cap structure. 7-Methylguanosine 5′-triphosphate (m7GTP) pull-down assays showed that a significantly lower amount of eIF4E4 was pulled down from CRK1 RNAi cells than from the non-induced control cells (Figure 6A), indicating that the association of eIF4E4 with the m7G cap requires CRK1 phosphorylation. Using synchronized cells, we showed that the association of eIF4E4 with the cap structure is not cell-cycle dependent (Figures S3A and S3B). In addition, phospho-PABP1 (p-PABP1) and eIF4G3, which were co-precipitated with phospho-eIF4E4, were also significantly decreased in CRK1 RNAi cells (Figure 6A), suggesting that CRK1 depletion disrupted the association of the eIF4E4-PABP1-eIF4G3 complex with the m7G cap. Finally, the m7GTP pull-down assay showed that a significantly lower amount of eIF4E-7A but a significantly higher amount of eIF4E4-7E than wild-type eIF4E4 were pulled down (Figure 6B), further demonstrating that the association of eIF4E4 with the m7G cap is strengthened by CRK1-mediated phosphorylation.
Figure 6. Phosphorylation of eIF4E4 and PABP1 Promotes Their Respective Association with the m7GTP Cap and the Poly(A) Sequence.
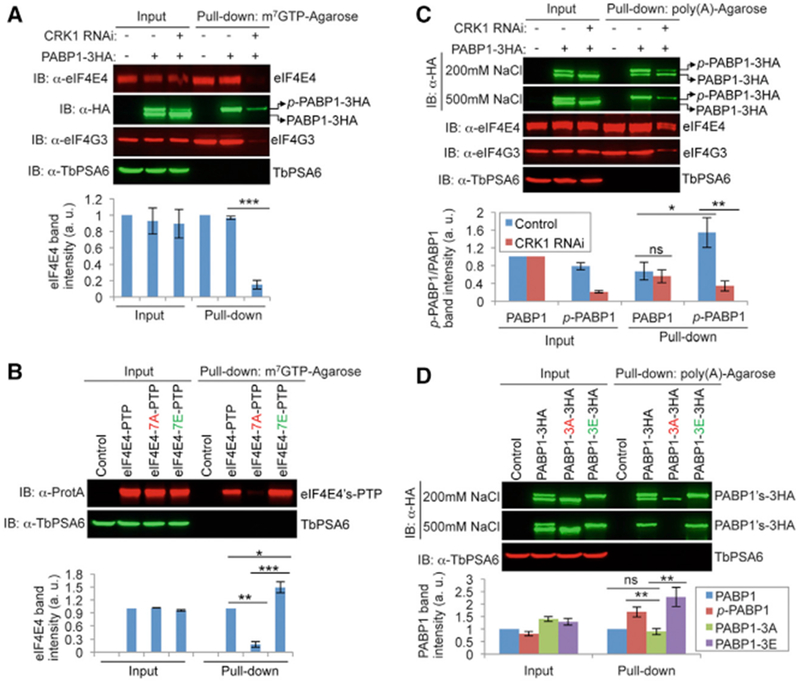
(A) Phosphorylation of eIF4E4 is required for association with the m7G cap. Shown is the m7GTP-agarose pull-down of eIF4E4 in control nd CRK1 RNAi cells. The effect on co-precipitation of PABP1 and eIF4G3 was also assessed. TbPSA6 served as a negative control.
(B) Phosphorylation of eIF4E4 is required for association with the m7GTP cap. Pull-down assays were performed as above.
(C) Phosphorylation of PABP1 by CRK1 is required for association with the poly(A) sequence. Shown is the poly(A) agarose pull-down of PABP1 in control and CRK1 RNAi cells. The effect on co-precipitation of eIF4E4 and eIF4G3 was also assessed.
(D) Phosphorylation of PABP1 is required for association with poly(A). Poly(A) pull-down assays were carried out using cells expressing wild-type and mutant PABP1 proteins.
In all of the panels, the graph below the western blots shows the quantification of band intensity. Error bars indicate SDs from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no statistical significance.
The effect of PABP1 phosphorylation on its association with the poly(A) sequence was investigated using poly(A) pull-down assays. Under medium stringency (200 mM NaCl) conditions, a lower amount of p-PABP1 was pulled down from CRK1 RNAi cells than from the control cells, whereas there was no significant difference in the amount of non-phospho-PABP1 pulled down from the control and RNAi cells (Figure 6C). Moreover, a significantly higher amount of p-PABP1 than PABP1 was pulled down (Figure 6C). Under high stringency (500 mM NaCl) conditions, only p-PABP1 was pulled down (Figure 6C). These results suggest that p-PABP1 has stronger affinity to the poly(A) sequence than PABP1. The association of PABP1 with the poly(A) sequence is not cell-cycle dependent (Figures S3C and S3D). In addition, a significantly lower amount of eIF4E4 and eIF4G3 was co-precipitated with p-PABP1 by poly(A) beads from CRK1 RNAi cells than from the control cells (Figure 6C), suggesting that CRK1 depletion impaired the association of the eIF4E4-PABP1-eIF4G3 complex with the poly(A) sequence. Furthermore, a poly(A) pull-down assay was carried out for the PABP1-3A and PABP1-3E mutants. Under medium stringency conditions, a significantly higher amount of p-PABP1 and PABP1-3E than PABP1-3A was pulled down (Figure 6D), and under high stringency conditions, PABP1-3A was not pulled down (Figure 6D), indicating that the phosphorylation of PABP1 enhances its association with the poly(A) sequence.
CRK1 Phosphorylation of eIF4E4 and PABP1 Is Required for Protein Translation
Because CRK1 phosphorylates eIF4E4 and PABP1 (Figure 1), we investigated whether CRK1 is required for protein translation. To monitor protein synthesis, we carried out L-azidohomoalanine (AHA) incorporation assays (Dieterich et al., 2006), which detect the incorporated AHA during protein synthesis by fluorescence microscopy, and quantified the fluorescence intensity of AHA-incorporated proteins in individual cells. CRK1 RNAi cells possessed a significantly lower cellular fluorescence intensity than the non-induced control cells (Figures 7A and S4A), similar to the eIF4E4 cKO cells and PABP1 RNAi cells, which served as positive controls (Figures 7A and S4A), demonstrating that CRK1 is required for protein translation. Using the same approach, we showed that cells expressing the phospho-deficient eIF4E4 mutant (eIF4E4-7A) and PABP1 mutant (PABP1-3A) had a significantly lower level of incorporated AHA than the control cells (Figures 7B, 7C, S4B, and S4C), demonstrating that phosphorylation of both proteins is important for protein translation. Cells expressing eIF4E4-7E had a significantly higher level of incorporated AHA than the control cells (Figure 7B), suggesting that constitutive phosphorylation of eIF4E4 further promotes protein translation.
Figure 7. Phosphorylation of eIF4E4 and PABP1 by CRK1 Is Required for Protein Translation.
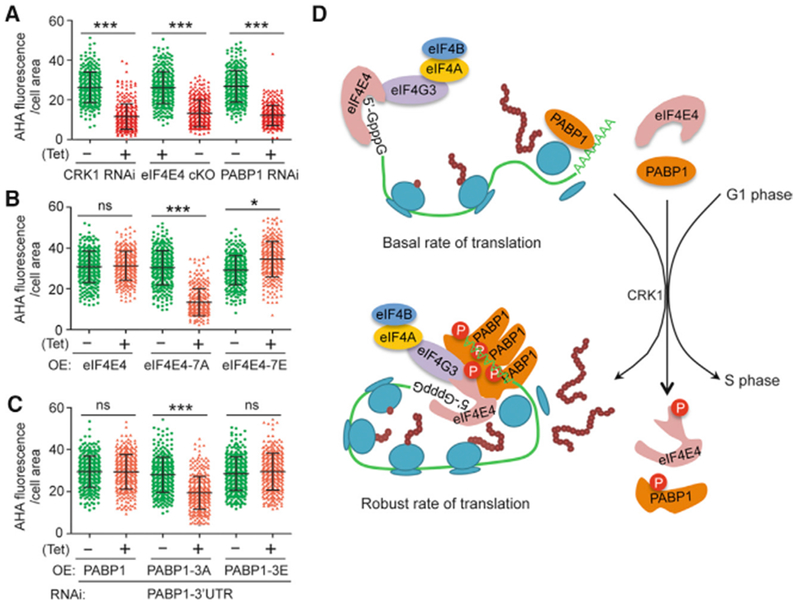
(A) CRK1 is required for protein translation. Shown is the quantification of AHA fluorescence intensity per cell area of the control and RNAi cells. To measure AHA fluorescence intensity, 100 cells for each cell line were used. ***p < 0.001.
(B) Phosphorylation of eIF4E4 is required for protein translation. Quantification of AHA fluorescence intensity was carried out as described above. ***p < 0.001; ns, no statistical significance.
(C) Phosphorylation of PABP1 is required for protein translation. Quantification of AHA fluorescence intensity was carried out as described above. ***p < 0.001; ns, no statistical significance.
(D) Model of CRK1-mediated phosphorylation of eIF4E4 and PABP1 on protein translation and G1/S transition. Phosphorylation of eIF4E4 and PABP1 promotes the association of eIF4E4 with the m7G cap and of PABP1 with poly(A). It also promotes the interactions among translation initiation factors and self-interaction of PABP1. This may facilitate or strengthen the assembly of the translation initiation factor complex, the cooperative binding of PABP1 to the poly(A) tail, the association of eIF4E4 with the m7G cap, and the circulation of mRNA, thereby promoting protein translation and the G1/S cell-cycle transition.
DISCUSSION
The earliest evidence for the requirement of protein translation for the cell-cycle transition from G1 to S phase was obtained with the budding yeast PRT1/eIF3b mutant (Hanic-Joyce et al., 1987) and the Cdc33/eIF4E mutant (Brenner et al., 1988). Additional evidence came from genetic ablation of other translation initiation factors, such as eIF2a, eIF4A, eIF2b, eIF3i, and eIF1, which arrests yeast cells at G1 phase (Yu et al., 2006). Moreover, pharmacological inhibition of the TOR kinase causes a G1 arrest in yeast (Heitman et al., 1991) due to a blockade in translation initiation (Barbet et al., 1996), and a G1 arrest in humans (Wicker et al., 1990) due to the inhibition of eIF4E-BP phosphorylation and thus defective protein translation (Burnett et al., 1998). In T. brucei, depletion of the TOR complex 1 (TORC1) also inhibits protein translation and causes a G1 arrest (Barquilla et al., 2008). The cell-cycle defects caused by cKO of eIF4E4 and RNAi of PABP1 (Figure 2) further confirm that protein translation is required for the G1/S cell-cycle transition in T. brucei. Such a control mechanism for the G1/S transition is physiologically relevant, as numerous proteins need to be synthesized during S and G2 phases to prepare the cells for duplicating and segregating the genome and organelles. It is thus not surprising that this control scheme is conserved from T. brucei, one of the earliest divergent eukaryotes, to humans.
The activation of cap-dependent translation in G1 phase through mTOR-mediated phosphorylation of eIF4E-BP and MnK-mediated phosphorylation of eIF4E has been well characterized in mammals, but the mechanism for TOR-mediated translation initiation and G1/S cell-cycle progression in yeast remains poorly understood (Pyronnet and Sonenberg, 2001). Using the early branching T. brucei as a model system, we uncovered a regulatory mechanism for the activation of cap-dependent translation in G1 phase, which involves the phosphorylation of eIF4E4 and PABP1 by the G1 cyclin-dependent kinase CRK1 (Figure 7D). Phosphorylation of eIF4E4 and PABP1 promotes protein-protein interactions among translation initiation factors (Figure 5) and enhances their respective binding to the m7G cap structure and the poly(A) sequence (Figure 6), thereby activating protein translation (Figures 7A–7C) and promoting the G1/S cell-cycle transition (Figures 2, 3, and 4). This CRK1-mediated regulation of cap-dependent translation during the G1/S cell-cycle transition is distinct from the mitotic CDK1-mediated activation of cap-dependent translation during mitosis in humans, in which CDK1 substitutes for mTOR to phosphorylate eIF4E-BP (Shuda et al., 2015). In T. brucei, TORC1 is required for activating protein translation during G1 phase (Barquilla et al., 2008), but a structural homolog of eIF4E-BP is missing in the genome (Zinoviev and Shapira, 2012), and whether a functional homolog of eIF4E-BP exists remains unknown. Therefore, it is unclear whether T. brucei still adopts a canonical TOR-mediated translation control mechanism as in mammals or has evolved a distinct TOR-mediated pathway. Moreover, a close homolog of MnK has not been found in T. brucei (Naula et al., 2005; Parsons et al., 2005), and eIF4E4 in T. brucei lacks the analogous residue of Ser209 of human eIF4E (Freire et al., 2011), raising the question of whether eIF4E4 undergoes similar regulation as in humans. Conversely, in yeast and humans, whether the G1 CDK regulates translation initiation by phosphorylating eIF4E and PABP is also unknown. The eIF4E and PABP homologs from yeast and humans contain putative CDK phosphosites (Figure 1A), but their candidacy as CDK substrates and the effect of phosphorylation of these sites on translation remain to be investigated.
The effect of Ser209 phosphorylation on the binding of eIF4E to the m7G cap has been controversial (Scheper and Proud, 2002). Earlier work suggests that phosphorylation of Ser209 increases the binding of eIF4E to capped mRNA (Minich et al., 1994), consistent with the fact that hyperphosphorylation of eIF4E usually correlates with higher rates of protein synthesis (Lamphear and Panniers, 1990). However, another study suggests that phosphorylation of eIF4E reduces its binding to capped mRNA, primarily due to an increased dissociation rate (Scheper et al., 2002). Our work showed that the association of eIF4E4 with the m7G cap is strengthened by CRK1 phosphorylation of seven Ser/Thr residues at the N terminus of eIF4E4 (Figures 6A and 6B). However, the m7GTP-agarose pull-down assay used in this study does not test the direct binding of eIF4E4 to the m7G cap structure, as the eIF4E4 protein used for the assay was expressed in T. brucei and still forms a complex with other translation initiation factors, which could have significantly influenced the binding of eIF4E4 to the m7G cap. Previous work demonstrated that the interaction of eIF4G with eIF4E stabilizes the binding of eIF4E to capped mRNA (Haghighat and Sonenberg, 1997; von Der Haar et al., 2000). The interaction of eIF4G3 with eIF4E4 was impaired by the dephosphorylation of eIF4E4 (Figures 5B and 5F). Therefore, it is likely that the dissociation of eIF4G3 from eIF4E4 significantly reduced eIF4E4 binding to the m7G cap. Moreover, the CRK1 phosphosites on eIF4E4 are mostly located in the N-terminal extension, which is not present in the eIF4E homologs from yeast and humans (Figure 1A). Thus, these phosphosites are unlikely to contribute directly to cap binding, but the possibility that phosphorylation may influence the overall structure of eIF4E4 and thus affects its capability of binding to the m7G cap cannot be ruled out.
PABP1 is a phosphoprotein in various organisms, including yeast, plants, and animals (Drawbridge et al., 1990; Gallie et al., 1997; Zhou et al., 2013). The phosphorylated form of PABP purified from plants binds to the poly(A) RNA cooperatively (Le et al., 2000), and human PABP1 also binds to poly(A) cooperatively (Melo et al., 2003). In the Xenopus oocyte, four residues within the proline-rich region of the embryotic PAB (ePAB) are phosphorylated, and hyperphosphorylated ePAB associates with polysomes or cap complexes (Friend et al., 2012). T. brucei PABP1 is phosphorylated at three sites within the proline-rich region (Figure 1). The phosphorylated form of PABP1 appears to associate with the poly(A) sequence with higher affinity than the non-phosphorylated form of PABP1 (Figures 6C and 6D) and with the cap-binding translation initiation factor complex (Figures 5A, 5E, and 6A), and it is important for translation (Figure 7C). Although the poly(A) agarose pull-down assay is unable to test the direct binding of PABP1 to the poly(A) sequence, our results demonstrated that the phosphorylation state of PABP1 affects its association with poly(A) (Figures 6C and 6D). Furthermore, because phosphorylation of PABP1 in the proline-rich linker region is necessary for self-interaction (Figures 5G and 5H), it suggests that the phosphorylation of PABP1 may promote cooperative binding to the poly(A) sequence, as in the case of human PABP1, which self-interacts through the proline-rich linker, thereby binding cooperatively to the poly(A) sequence (Melo et al., 2003).
The phosphorylation state of eIF4E4 and PABP1 not only influences their respective association with the m7G cap and poly(A) but it also affects protein-protein interactions between themselves and with other translation initiation factors. Interaction between PABP1 and eIF4E4 depends on the phosphorylation of either of them by CRK1 (Figures 5A, 5C, and 5E), but the interaction between PABP1 and eIF4G3 does not require CRK1 phosphorylation of PABP1 (Figure 5D). Interaction between PABP1 and eIF4E4 (Zinoviev et al., 2011) appears to be found only in the kinetoplastid parasites (Zinoviev and Shapira, 2012); however, the physiological significance of this interaction remains unknown. We speculate that the effect of the phosphorylation state of eIF4E4 and PABP1 on their pairwise interaction and on the eIF4E4-eIF4G3 interaction may affect translation initiation in the following ways. Phosphorylation may provide the specificity for the interaction among the translation initiation factors, thereby controlling the proper assembly of the translation initiation machinery. Phosphorylation may regulate the strength of the interaction among the translation initiation factors in response to the internal cues that promote the G1/S cell-cycle transition and/or global protein synthesis. Through regulating the PABP1-eIF4E4 interaction, phosphorylation by CRK1 may facilitate the circulation of mRNA; and through regulating the interaction between eIF4E4 and eIF4G3, phosphorylation by CRK1 may enhance the binding of eIF4E4 to the m7G cap, both of which promote translation.
The discovery of the G1 CDK-mediated regulation of translation initiation during the G1/S cell-cycle transition in the early divergent T. brucei raises the question of whether it represents an ancient control mechanism and is conserved across the eukaryotic organisms from yeast to humans. It also raises a question of whether this control mechanism operates in addition to the TORC1-mediated control of translation initiation to provide another layer of regulation or substitutes for TORC1 to promote translation initiation. Despite these open questions, the present work uncovers a mechanism for cell-cycle-dependent translational control and reveals a non-canonical role for the G1 CDK in regulating protein translation to promote the G1/S cell-cycle transition.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ziyin Li (Ziyin.Li@uth.tmc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The procyclic form of T. brucei strain Lister427 was grown at 27°C in SDM-79 medium containing 10% heat-inactivated fetal bovine serum. The procyclic form of T. brucei strain 29-13 (Wirtz et al., 1999) was cultured at 27°C in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Inc), 15 μg/ml G418, and 50 μg/ml hygromycin. Cells were sub-cultured by 1/10 dilution with fresh medium whenever the cell density reached 5×106/ml.
METHOD DETAILS
Purification of recombinant proteins and in vitro kinase assay
The full-length coding sequence of CRK1 was cloned into the pCDF-Duet1 vector (Novagen), and the full-length coding sequence of CYC2, the cyclin partner of CRK1, was cloned into the pGEX-4T-3 vector (Clontech). The two proteins were co-expressed in E. coli BL21 Rosetta strain (Novagen) and purified by passing through the glutathione-Sepharose 4B beads as described previously (Hu et al., 2016). The full-length coding sequence of eIF4E4 and PABP1 was cloned into pGEX-4T-3 vector. Site-directed mutagenesis was carried out using the QuickChange Site-directed mutagenesis kit (Agilent Technologies) to mutate the seven phosphosites in eIF4E4 to alanine (eIF4E4-7A) and glutamate (eIF4E4-7E) and the three phosphosites in PABP1 to alanine (PABP1-3A) and glutamate (PABP1-3E). These plasmids were each transformed into the E. coli BL21 Rosetta strain, and recombinant proteins were purified through binding to the glutathione-Sepharose 4B beads according to the manufacture’s instruction manual.
In vitro kinase assay was carried out as described previously (Hu et al., 2016). Purified recombinant proteins were incubated in the kinase buffer (50 mM HEPES, pH 7.5, 0.1 M NaCl, 10 mM MgCl2) containing 1 mM ATP for 1 h at room temperature. Kinase reaction was stopped by adding 2 mM EDTA. Samples were separated by SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with anti-phospho-Threonine-Proline mAb (Cell Signaling Technologies, Cat#: 9391), which detects the phosphorylated serine and threonine followed by a proline. In vitro phosphorylated eIF4E4 and PABP1 were also excised from the SDS-PAGE gel, and analyzed by LC-MS/MS to identify the phosphosites at the Clinical and Translational Proteomics Service Center of the University of Texas Health Science Center at Houston.
Mass spectrometry and data analysis
The SDS-PAGE gel samples containing the desired protein band were processed with USCF In-gel digestion protocol including reduction and alkylation of cysteine. After trypsin digestion overnight, the peptides were extracted from the gel with 50% acetonitrile and 5% formic acid. After extraction, samples were dried using speedvac, and then reconstituted in 2% acetonitrile with 0.1% formic acid and injected on to Thermo LTQ Orbitrap XL. Samples in 2% (v/v) acetonitrile 0.1% (v/v) formic acid were analyzed on a LTQ Orbitrap XL (Thermo-Fisher Scientific, Bremen, Germany) interfaced with an Eksigent nano-LC 2D plus ChipLC system (Eksigent Technologies, Dublin, CA). The sample was loaded onto a ChromXP C18-CL trap column (200μm i.d. × 0.5 mm length, 3 μm particle size) at flow rate 3 μl/min. Reversed-phase C18 chromatographic separation of peptides was carried on a on a ChromXP C18-CL column (75mm i.d × 10 cm length, 3μm) at 300 nl/min, column temperature was control at 60°C. Gradient conditions were: 3%–8% B for 5 min; 8%–30% B for 80min; 30%–90% B for 10 min; 90% B held for 10 min; 90% –3% for 5 min (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile), the total run time was 125 min. The LTQ Orbitrap was operated in the data dependent mode to simultaneously measure full scan MS spectra in the Orbitrap and the five most intense ions in the LTQ by CID, respectively. In each cycle, MS1 was acquired at target value 1E6 with resolution R = 100,000 (m/z 400) followed by top 5 MS2 scan at target value 3E4. The mass spectrometric setting as follows: spray voltage was 1.6 KV, charge state screening and rejection of singly charged ion were enabled. Ion selection thresholds were 8000 for MS2; 35% normalized collision energy; activation Q was 0.25, and dynamic exclusion was employed for 30 s. Raw data files were processed and search using Thermo Proteome Discoverer software or the Mascot search engine. Protein search was against Tb927-Trypanasoma database. The search conditions used peptide tolerance of 10 ppm and MS/MS tolerance of 0.8 Da with the enzyme trypsin and 2 missed cleavages.
RNA interference and conditional gene knock-out
To generate the PABP1 RNAi cell line, a 500-bp DNA fragment (nucleotides 501-1000) corresponding to the middle portion of the coding sequence of PABP1 was cloned into the Stem-Loop RNAi vector pSL (Hu et al., 2016). The pSL-PABP1 plasmid was linearized with Notl and transfected into the 29-13 strain by electroporation. Transfectants were selected with 2.5 μg/ml phleomycin in SDM-79 medium containing 15 μg/ml G418 and 50 μg/ml hygromycin, and cloned by limiting dilution in a 96-well plate. The CRK1 RNAi cell line was reported previously (Hu et al., 2016; Liu et al., 2013). RNAi was induced by incubating the cells with 1.0 μg/ml tetracycline. Growth of cells was monitored daily by counting the cells with a hemacytometer.
To generate eIF4E4 cKO cell line, the full-length eIF4E4 coding sequence was cloned into pLew100-PTP vector, and the resulting plasmid was linearized with NotI and transfected into the 29-13 strain by integrating the plasmid into the rDNA spacer locus. Successful transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution. Subsequently, one allele of eIF4E4 was replaced with the puromycin-resistance gene, and transfectants were further selected under 1.0 μg/ml puromycin and cloned by limiting dilution. The single-allele eIF4E4 knockout cell line thus obtained was cultured in the presence of 0.5 μg/ml tetracycline to induce the expression of the ectopic PTP-tagged eIF4E4, and the second allele of eIF4E4 was replaced by the blasticidin-resistance gene. This conditional double knockout cell line was maintained by culturing the cells in the presence of 0.5 μg/ml tetracycline. Knockout of both alleles of eIF4E4 was confirmed by PCR and western blotting with anti-eIF4E4 antibody. To examine the growth defect of eIF4E4 cKO cell line, tetracycline was washed off and cells were cultured in tetracycline-free medium.
PABP1 RNAi by targeting the 3′UTR and complementation
For RNAi complementation experiments, a new PABP1 RNAi cell line was generated by targeting against the 3′UTR of PABP1. A 500-bp fragment from the 3′UTR of PABP1 gene, which does not overlap with the downstream gene, was cloned into the stem-loop RNAi vector pSL-PAC, which was modified from the pSL RNAi vector by replacing the phleomycin resistance gene with the puromycin resistance gene. The resulting construct pSL-PABP1-3′UTR-PAC was linearized with NotI, and transfected into the 29-13 cell line. Transfectants were selected under 1 μg/ml puromycin in addition to 15 μg/ml G418 and 50 μg/ml hygromycin B, and further cloned by limiting dilution in a 96-well plate.
Full-length PABP1 was cloned into pLew100-3HA vector. To generate the phospho-deficient mutant (PABP1-3A) and the phospho-mimic mutant (PABP1-3E), site directed mutagenesis was carried out to mutate the three phosphosites to alanine or glutamic acid using pLew100-PABP1-3HA as the template. These plasmids were each linearized with NotI and transfected into the cell line containing the pSL-PABP1-3′UTR-PAC construct. Transfectants were selected under 2.5 μg/ml phleomycin in addition to 1 μg/ml puromycin, 15 μg/ml G418, and 50 μg/ml hygromycin B, and cloned by limiting dilution in a 96-well plate. Subsequently, the clonal cell line was used for endogenous PTP tagging of PABP1 at the N terminus, which was used to monitor the efficiency of PABP1 RNAi (see below).
In situ epitope tagging of proteins
Epitope tagging of PABP1 and eIF4G3 from their respective endogenous locus was carried out using the PCR-based method (Shen et al., 2001). PABP1 was tagged with a C-terminal PTP epitope (puromycin resistance) in PABP1 RNAi cell line, with an N-terminal PTP epitope (blasticidin resistance) in PABP1-3′UTR RNAi cell line, and with a C-terminal triple HA epitope (puromycin resistance) in 427 cell line, CRK1 RNAi cell line, and cell lines overexpressing wild-type and mutant eIF4E4. eIF4G3 was tagged with a C-terminal triple HA epitope (puromycin resistance) in CRK1 RNAi cell line and cell lines overexpressing wild-type and mutant eIF4E4. For assessing BABP1 self-interaction, one allele of PABP1 was tagged with a C-terminal triple HA epitope (puromycin resistance), and the other allele of PABP1 was tagged with a C-terminal PTP epitope (blasticidine resistance) in CRK1 RNAi cell line. Transfectants were selected with appropriate antibiotics and cloned by limiting dilution in a 96-well plate.
EdU incorporation assay
Analysis of DNA replication by detecting the incorporation of EdU into DNA was carried out using the Click-iT™ EdU Alexa Fluor™ 488 imaging kit (Thermo Fisher, Cat# C10337). T. brucei cells were incubated with SDM-79 medium containing 30 μM EdU for 6 h. Cells were collected by centrifugation, washed once with PBS, and fixed with 4% paraformaldehyde in PBS. Cells were then adhered to coverslips for EdU detection according to the manufacturer’s instructions. Cells were visualized under a fluorescence microscope, and EdU-positive cells were counted.
Cell cycle synchronization and flow cytometry
Cell cycle synchronization of T. brucei by starvation was performed as described previously (Archer et al., 2011). Briefly, cells were seeded at a density of 1 × 106 cells/ml in 50 mL SDM-79 medium, and were cultured in the same medium at 27°C for 72 h. Cells were then harvested by centrifugation at 2000 rpm (2500 × g) for 10 min, re-suspended in fresh SDM-79 medium, and cultured for another 3 h to allow cells to recover from starvation (Archer et al., 2011). Cells were then collected and analyzed by flow cytometry or used for co-immunoprecipitation, m7G cap-binding assay, and poly(A)-binding assay.
Flow cytometry analysis of synchronized and asynchronous cells was carried out as previously described (Hu et al., 2016). Briefly, cells were washed three times with PBS, fixed in ethanol at 4°C for 1 h. Cells were washed twice with PBS and then suspended in PBS. DNase-free RNase (10 μg/ml) and propidium iodide (20 μg/ml) were added to the cell suspension, and the DNA content of the cells was analyzed using a FACScan analytical flow cytometer (BD Biosciences). The percentage of cells in each phase of the cell cycle (G1, S, and G2/M) was determined by Kaluza Analysis software (Beckman Coulter).
Co-immunoprecipitation and quantitative western blotting
Co-immunoprecipitation was carried out according to our previous procedures (Hu et al., 2016). Briefly, cells (5 × 107) were lysed by incubating with 1.0 mL immunoprecipitation buffer (25 mM Tris-HCl, pH7.6, 100 mM NaCl, 1 mM DTT, 1% NP-40, and Complete Mini EDTA-free protease inhibitor cocktail) for 30 min on ice. Cell lysate was centrifugated at 14000 rpm (18,407 × g) in a microcentrifuge for 15 min at 4°C. Cleared cell lysate was incubated with 50 μl settled IgG Sepharose 6 Fast Flow affinity resin (GE Healthcare, Cat#: 17-0969-01) or 50 μl settled EZview™ anti-HA affinity gel (Sigma-Aldrich, Cat# E6779) for 2 h at 4°C, and immunoprecipitates were washed three times with the immunoprecipitation buffer. Proteins bound to the IgG beads or the anti-HA beads were eluted with 10% SDS, separated by SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with appropriate antibodies. For RNase A treatment, the cleared cell lysate was incubated with 40 μg/ml RNase A (Roche) at 37°C for 30 min, and then used for co-immunoprecipitation as described above.
For quantitative western blotting, an equal number (5 × 105) of cells were lysed, and cell lysate was fractionated on SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with the primary antibodies for 1 hour at room temperature. The following primary antibodies were used: anti-HA mAb (1:1,000 dilution, Sigma-Aldrich, Clone HA-7), anti-Protein A pAb (1:1,000 dilution, Sigma-Aldrich), anti-phospho-Threonine-Proline mAb (1:1,000 dilution, Cell Signaling), anti-eIF4E4 pAb (1:1,000 dilution) (Pereira et al., 2013), anti-eIF4G3 pAb (1:500 dilution) (Pereira et al., 2013). After washing three times with TBST, the membrane was incubated with FITC-conjugated anti-mouse IgG (1:400 dilution, Sigma-Aldrich) or IRDye® 680LT anti-rabbit IgG (1:2,500 dilution, Li-Cor Cooperate), and western blot signals were captured using the Bio-Rad ChemiDoc™ MP imaging system. Quantitation of protein band intensity was carried out with the ImageJ software (NIH).
Phos-tag™ and lambda protein phosphatase treatments
For separation of phosphorylated proteins from non-phosphorylated proteins on SDS-PAGE, 50 μM Phos-tag™ (Wako Pure Chemical Industrials, Inc) was added to the solutions when preparing the SDS acrylamide gel. Western blotting using the Phostag™-containing SDS-PAGE gel was carried out according to standard procedures. For lambda protein phosphatase (λPPase) treatment, cells were first lysed with the immunoprecipitation buffer (see above) on ice for 30 min. Subsequently, 100 units of lambda protein phosphatase (New England Biolabs, Cat#: P0753), 5μl 10 × NEBuffer for Protein MetalloPhosphatases, and 1 mM MnCl2 were added into the cell lysate (40 μl) and incubated at 30°C for 30 min.
m7GTP agarose pull-down and poly(A) agarose pull-down assays
For m7GTP agarose pull-down of eIF4E4 from T. brucei cell lysate, cells (2 × 107) were lysed in 0.5 mL immunoprecipitation buffer (see above) on ice for 30 min. Cleared cell lysate was incubated with 50 μl γ-aminophenyl-m7GTP (C10-spacer)-agarose (Jena Bioscience, Cat#: AC-155) with gentle rotation at 4°C for 2 h. Beads were washed three times with the immunoprecipitation buffer. Bound proteins were eluted by incubating with SDS-PAGE sampling buffer. Western blotting was carried out with anti-eIF4E4 polyclonal antibody to detect cap-bound eIF4E4 and with anti-HA antibody to detect 3HA-tagged PABP1 that was co-precipitated with eIF4E4.
For poly(A) agarose pull-down of PABP1 from T. brucei cell lysate, cells (2 × 107) expressing endogenously 3HA-tagged PABP1 were lysed in 0.5 mL immunoprecipitation buffer (25 mM Tris-HCl, pH7.6, 200 mM NaCl, 1 mM DTT, 1% NP-40, and protease inhibitor cocktail). Cleared cell lysate was incubated with 50 μl poly(A)-agarose (Jena Bioscience, Cat#: AC-159S) with gentle rotation at 4°C for 2 h. Bound proteins were similarly eluted as described above. Western blotting was performed with anti-HA antibody to detect poly(A)-bound PABP1-3HA and with anti-eIF4E4 antibody to detect eIF4E4 that was co-precipitated with PABP1-3HA.
L-Azidohomoalanine incorporation assay
L-Azidohomoalanine (AHA) incorporation assay was carried out using the Click-iT® AHA Alexa Fluor® 488 Protein Synthesis HCS Assay kit (Thermo Fisher, Cat#: C10289). T. brucei cells were cultured in methionine-free SDM-79 medium for 2 h and then incubated with 50 μM AHA in methionine-free SDM-79 medium for an additional 6 h. Cells were harvested by centrifugation, washed with PBS, and adhered onto glass coverslips. Cell fixation, permeabilization, and AHA detection were performed according to the manufacturer’s instructions. Cells were visualized under a fluorescence microscope, and images were taken under the same exposure time for control cells and RNAi cells or overexpression cells (eIF4E4 and mutants) or RNAi complementation cells (PABP1 and mutants). Fluorescence intensity of individual cells was measured with the ImageJ software, and the data were further processed by subtracting the background signal and then dividing with the area of the cell to generate the AHA fluorescence per cell area.
Immunofluorescence microscopy
Cells were fixed with cold methanol (−20°C) for 30 min, rehydrated with PBS, and then blocked with 3% BSA in PBS for 1 h at room temperature. Cells on the coverslip were incubated with YL 1/2 monoclonal antibody (1: 1,000 dilution) (Sherwin et al., 1987) for 1 h at room temperature. Cells were then washed three times with PBS, and incubated with FITC-conjugated anti-rat IgG (1:400 dilution, Sigma-Aldrich) for 1 h at room temperature. Cells were washed three times with PBS, mounted with DAPI-containing VectaShield mounting medium (Vector Labs), and imaged under an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60x1.42-NA lens. Images were acquired using the Slidebook 5 software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using the Student’s t test in the Microsoft Excel software. Detailed n values for each panel in the figures were stated in the corresponding legends. For immunofluorescence microscopy, images were randomly taken and all cells in each image were counted.
DATA AND SOFTWARE AVAILABILITY
Mass spectrometry data: the list of all identified phosphopeptides in eIF4E4and PABP1 has been deposited in the PeptideAtlas database with the accession number PASS01291.
The original dataset has been deposited in Mendeley database with the following https://doi.org/10.17632/xwk46s3vg7.1
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-HA | Sigma-Aldrich | Cat# H3663; RRID:AB_262051 |
| Rabbit polyclonal anti-Protein A | Sigma-Aldrich | Cat# P3775; RRID:AB_261038 |
| Mouse monoclonal anti-phospho-Threonine-Proline | Cell Signaling | Cat# 9391; RRID:AB_331801 |
| Mouse monoclonal anti-GST | Sigma-Aldrich | Cat# G7781; RRID:AB_259965 |
| Rabbit polyclonal anti-TbPSA6 | Li et al., 2002 | N/A |
| Rabbit polyclonal anti-eIF4E4 | Gift by O.P. de Melo Neto | N/A |
| Rabbit polyclonal anti-eIF4G3 | Gift by O.P. de Melo Neto | N/A |
| Rat monoclonal YL1/2 | EMD Millipore | Cat# MAB1864; RRID:AB_2210391 |
| IRDye 680LT anti-rabbit IgG | Li-Cor | 925-68022 |
| FITC-conjugated anti-rat IgG | Sigma-Aldrich | Cat# F6258; RRID:AB_259695 |
| FITC-conjugated anti-mouse IgG | Sigma-Aldrich | Cat# F0257; RRID:AB_259378 |
| Critical Commercial Assays | ||
| Phos-tag | Wako Pure Chemical, Inc | 300-93523 |
| Click-iT EdU Alexa Fluor 488 imaging kit | ThermoFisher | C10337 |
| Poly(A)-agarose | Jena Bioscience | AC-159S |
| γ-Aminophenyl-m7GTP (C10-spacer)-Agarose | Jena Bioscience | AC-155 |
| lambda protein phosphatase | New England Biolabs | P0753 |
| RNase A | Sigma-Aldrich | R6513 |
| IgG Sepharose 6 Fast Flow affinity resin | GE Healthcare | 17-0969-01 |
| EZview anti-HA affinity gel | Sigma-Aldrich | Cat# E6779; RRID:AB_10109562 |
| Complete Mini EDTA-free protease inhibitor cocktail | Roche | 11836170001 |
| QuickChange Site-directed mutagenesis kit | Agilent Technologies | 200524 |
| Glutathione-Sepharose 4B beads | GE Healthcare | 17075601 |
| Experimental Models: Cell lines | ||
| Trypanosoma brucei 29-13 cell line | ATCC | PRA-381 |
| Trypanosoma brucei Lister427 | ATCC | NR-42010 |
| Oligonucleotides | ||
| Extensive primer sequences provided in Table S1 | This study | N/A |
| Deposited data | ||
| Raw mass spectrometry data for eIF4E4 and PABP1 phosphosite identification: The PeptideAtlas repository, accession number: PASS01291 | This study | https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/PASS_View?identifier=PASS01291 |
| Original dataset: Mendeley database | This study | https://data.mendeley.com/datasets/xwk46s3vg7/1 |
| Recombinant DNA | ||
| pC-eIF4E4-PTP-NEO | This study | N/A |
| pC-PABP1-PTP-NEO | This study | N/A |
| pC-PABP1-3HA-PAC | This study | N/A |
| pSL-PABP1-BLE | This study | N/A |
| pSL-PABP1-3′UTR-PAC | This study | N/A |
| pLew100-eIF4E4-PTP-BLE | This study | N/A |
| pLew100-eIF4E4-S102A-PTP-BLE | This study | N/A |
| pLew100-eIF4E4-S102E-PTP-BLE | This study | N/A |
| pLew100-eIF4E4-7A-PTP-BLE | This study | N/A |
| pLew100-eIF4E4-7E-PTP-BLE | This study | N/A |
| pLew100-eIF4E4-3HA-BLE | This study | N/A |
| pLew100-eIF4E4-7A-3HA-BLE | This study | N/A |
| pLew100-eIF4E4-7E-3HA-BLE | This study | N/A |
| pLew100-PABP1-3HA-BLE | This study | N/A |
| pLew100-PABP1-3A-3HA-BLE | This study | N/A |
| pLew100-PABP1-3E-3HA-BLE | This study | N/A |
| pGEX-4T-3-CYC2 | Hu et al., 2016 | N/A |
| pCDF-CRK1 | Hu et al., 2016 | N/A |
| pGEX-4T-3-eIF4E4 | This study | N/A |
| pGEX-4T-3-eIF4E4-7A | This study | N/A |
| pGEX-4T-3-PABP1 | This study | N/A |
| pGEX-4T-3-PABP1-3A | This study | N/A |
Highlights.
CRK1 phosphorylation of eIF4E4 and PABP1 promotes the G1/S cell-cycle transition
Binding of eIF4E4 to the m7G cap is enhanced by CRK1-mediated phosphorylation
Phosphorylation of PABP1 by CRK1 promotes its association with the poly(A) sequence
Phosphorylation of eIF4E4 and PABP1 couples translation with the cell-cycle transition
ACKNOWLEDGMENTS
We are grateful to Dr. Osvaldo P. de Melo Neto of the Centro de Pesquisas Aggeu Magalhães of Brazil for the anti-eIF4E4 and anti-eIF4G3 antibodies. This work was supported by the National Institutes of Health, United States grants R01AI118736 and R01AI101437 to Z.L., NIH R01AI21786 to C.C.W., and in part, by the Clinical and Translational Proteomics Service Center of the University of Texas Health Science Center at Houston. This paper is dedicated to the memory of Dr. Ching C. Wang.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.063.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Altmann M, and Trachsel H (1989). Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 17, 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R, and Clayton C (2011). The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6, e18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, and Hall MN (1996). TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A, Crespo JL, and Navarro M (2008). Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc. Natl. Acad. Sci. USA 105, 14579–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau AM, and Sonenberg N (1987). Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem 262, 11134–11139. [PubMed] [Google Scholar]
- Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, and Matsumoto K (1988). CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol 8, 3556–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X, Haas DW, and Hagedorn CH (1993). Novel phosphorylation sites of eukaryotic initiation factor-4F and evidence that phosphorylation stabilizes interactions of the p25 and p220 subunits. J. Biol. Chem 268, 4975–4978. [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, and Sabatini DM (1998). RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, and Beyaert R (2000). Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5, 597–605. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, and Schuman EM (2006). Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawbridge J, Grainger JL, and Winkler MM (1990). Identification and characterization of the poly(A)-binding proteins from the sea urchin: a quantitative analysis. Mol. Cell. Biol 10, 3994–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, and Penman S (1970). Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol 50, 655–670. [DOI] [PubMed] [Google Scholar]
- Flynn A, and Proud CG (1995). Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J. Biol. Chem 270, 21684–21688. [DOI] [PubMed] [Google Scholar]
- Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, and de Melo Neto OP (2011). The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol 176, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, Sturm NR, Campbell DA, and de Melo Neto OP (2017). The Role of Cytoplasmic mRNA Cap-Binding Protein Complexes in Trypanosoma brucei and Other Trypanosomatids. Pathogens 6, E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, and Seli E (2012). Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem. J 445, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Le H, Caldwell C, Tanguay RL, Hoang NX, and Browning KS (1997). The phosphorylation state of translation initiation factors is regu-lated developmental and following heat shock in wheat. J. Biol. Chem 272,1046–1053. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, and Sonenberg N (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, and Sonenberg N (2001). Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, and Sonenberg N (1997). eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem 272, 21677–21680. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, and Sonenberg N (1995). Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14, 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanic-Joyce PJ, Johnston GC, and Singer RA (1987). Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp. Cell Res. 172, 134–145. [DOI] [PubMed] [Google Scholar]
- Heesom KJ, Gampel A, Mellor H, and Denton RM. (2001). Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr. Biol 11, 1374–1379. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, and Hall MN (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 258, 905–909. [DOI] [PubMed] [Google Scholar]
- Herbert TP, Tee AR, and Proud CG (2002). The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J. Biol. Chem 277, 11591–11596. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, and Morgan DO (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 825, 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Gourguechon S, Wang CC, and Li Z (2016). The G1 Cyclin-dependent Kinase CRK1 in Trypanosoma brucei Regulates Anterograde Protein Transport by Phosphorylating the COPII Subunit Sec31. J. Biol. Chem 291, 15527–15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, Stepinski J, Stolarski R, Darzynkiewicz E, and Rhoads RE (1995). Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J. Biol. Chem 270, 14597–14603. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, and Koike T (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757. [DOI] [PubMed] [Google Scholar]
- Klein C, Terrao M, Inchaustegui Gil D, and Clayton C (2015). Polysomes of Trypanosoma brucei: Association with Initiation Factorsand RNA-Binding Proteins. PLoS One 10, e0135973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad CG (1963). Protein Synthesis and Rna Synthesis during Mitosis in Animal Cells. J. Cell Biol. 19, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2006). Rethinking some mechanisms invoked to explain translational regulation in eukaryotes. Gene 882, 1–11. [DOI] [PubMed] [Google Scholar]
- Kramer S, Bannerman-Chukualim B, Ellis L, Boulden EA, Kelly S, Field MC, and Carrington M (2013). Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS One 8, e54004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, and Panniers R (1990). Cap binding protein complex that restores protein synthesis in heat-shocked Ehrlich cell lysates contains highly phosphorylated eIF-4E. J. Biol. Chem 265, 5333–5336. [PubMed] [Google Scholar]
- Le H, Browning KS, and Gallie DR (2000). The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (elF) 4F, eIFiso4F, and eIF4B. J. Biol. Chem 275, 17452–17462. [DOI] [PubMed] [Google Scholar]
- Li Z, Zou CB, Yao Y, Hoyt MA, McDonough S, Mackey ZB, Coffino P, and Wang CC (2002). An easily dissociated 26S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J. Biol. Chem 277, 15486–15498. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hu H, and Li Z (2013). The cooperative roles of PHO80-like cyclins in regulating the G1/S transition and posterior cytoskeletal morphogenesis in Trypanosoma brucei. Mol. Microbiol 90, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, and Sonenberg N (1995). The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol 15, 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo EO, Dhalia R, Martins de Sa C, Standart N, and de Melo Neto OP (2003). Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly (A) and efficient cap distal translational repression. J. Biol. Chem 278, 46357–6368. [DOI] [PubMed] [Google Scholar]
- Minich WB, Balasta ML, Goss DJ, and Rhoads RE (1994). Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl. Acad. Sci. USA 91, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura DM, Reis CR,Xavier CC, daCosta Lima TD, Lima RP, Carrington M, and de Melo Neto OP (2015). Two related trypanosomatid eIF4G homologues have functional differences compatible with distinct roles during translation initiation. RNA Biol. 12, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naula C, Parsons M, and Mottram JC (2005). Protein kinases as drug targets in trypanosomes and Leishmania. Biochim. Biophys. Acta 1754,151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M, Worthey EA, Ward PN, and Mottram JC (2005). Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MM, Malvezzi AM, Nascimento LM, Lima TD, Alves VS, Palma ML, Freire ER, Moura DM, Reis CR, and de Melo Neto OP (2013). The eIF4E subunits of two distinct trypanosomatid eIF4F complexes are subjected to differential post-translational modifications associated to distinct growth phases in culture. Mol. Biochem. Parasitol 190, 82–86. [DOI] [PubMed] [Google Scholar]
- Prescott DM, and Bender MA (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260–268. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, and Sonenberg N (2001). Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev 11, 13–18. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, and Sonenberg N (2000). A cell cycle-dependent internal ribosome entry site. Mol. Cell 5, 607–616. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, and Sonenberg N (2001). Suppression of cap-dependent translation in mitosis. Genes Dev. 15, 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, and Blagoev B (2011). System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal 4, rs3. [DOI] [PubMed] [Google Scholar]
- Sachs AB, and Davis RW (1989). The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58, 857–867. [DOI] [PubMed] [Google Scholar]
- Scheper GC, and Proud CG (2002). Does phosphorylation of thecap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem 269, 5350–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, and Proud CG (2002). Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem 277, 3303–3309. [DOI] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, and Mann M (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyrand Ser/Thr-based signaling. Cell Rep. 8,1583–1594. [DOI] [PubMed] [Google Scholar]
- Shatkin AJ (1976). Capping of eucaryotic mRNAs. Cell 9, 645–653. [DOI] [PubMed] [Google Scholar]
- Shen S, Arhin GK, Ullu E, and Tschudi C (2001). In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113, 171–173. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, and Gull K (1987). Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Velásquez C, Cheng E, Cordek DG, Kwun HJ, Chang Y, and Moore PS (2015). CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci. USA 112, 5875–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, and Villén J (2013). Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Stern-Ginossar N, Weissman JS, and Vale RD (2015). Regulation of mRNA translation during mitosis. eLife 4, e07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, and Wang CC (2004). The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem 279, 20519–20528. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DM, and Ferguson MA (2013). Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 12, 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Der Haar T, Ball PD, and McCarthy JE (2000). Stabilization of eukaryotic initiation factor 4E binding to the mRNA 50-Cap by domains of eIF4G. J. Biol. Chem 275, 30551–30555. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, and Sachs AB (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Wicker LS, Boltz RC Jr., Matt V, Nichols EA, Peterson LB, and Sigal NH (1990). Suppression of B cell activation by cyclosporin A, FK506 and rapamycin. Eur. J. Immunol 20, 2277–2283. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, and Cross GA (1999). Atightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol 99, 89–101. [DOI] [PubMed] [Google Scholar]
- Yu L, Peña Castillo L, Mnaimneh S, Hughes TR, and Brown GW (2006). A survey of essential gene function in the yeast cell division cycle. Mol. Biol. Cell 17, 4736–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, and Mohammed S (2013). Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271. [DOI] [PubMed] [Google Scholar]
- Zinoviev A, and Shapira M (2012). Evolutionary conservation and diversification of the translation initiation apparatus in trypanosomatids. Comp. Funct. Genomics 2012, 813718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Léger M, Wagner G, and Shapira M (2011). A novel 4E-inter-acting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 39, 8404–8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data: the list of all identified phosphopeptides in eIF4E4and PABP1 has been deposited in the PeptideAtlas database with the accession number PASS01291.
The original dataset has been deposited in Mendeley database with the following https://doi.org/10.17632/xwk46s3vg7.1


