SUMMARY
Nutrient restriction (NR) decreases the incidence and growth of many types of tumors, yet the underlying mechanisms are not fully understood. In this study, we identified Headcase (Hdc) and Unkempt (Unk) as two NR-specific tumor suppressor proteins that form a complex to restrict cell cycle progression and tissue growth in response to NR in Drosophila. Loss of Hdc or Unk does not confer apparent growth advantage under normal nutrient conditions but leads to accelerated cell cycle progression and tissue overgrowth under NR. Hdc and Unk bind to the TORC1 component Raptor and preferentially regulate S6 phosphorylation in a TORC1-dependent manner. We further show that HECA and UNK, the human counterparts of Drosophila Hdc and Unk, respectively, have a conserved function in regulating S6 phosphorylation and tissue growth. The identification of Hdc and Unk as two NR-specific tumor suppressors provides insight into molecular mechanisms underlying the anti-tumorigenic effects of NR.
In Brief
The molecular mechanisms underlying nutrient restriction resistance remain unclear. Li et al. find that Hdc and Unk function in the mTOR signaling pathway to restrict tissue growth and cell cycle progression in response to nutrient restriction.
Graphical Abstract

INTRODUCTION
Nutrient restriction (NR) without malnutrition is the most robust environmental intervention that is known to slow aging and aging-associated diseases (including cancer) in a variety of species (Lee and Longo, 2016; Mair and Dillin, 2008; Piper et al., 2011). Indeed, NR has been shown to delay the incidence and suppress the growth of various types of tumors (Buono and Longo, 2018; Meynet and Ricci, 2014).The protective effects of NR on tumor incidence and growth were first reported in the early 1900s and have since been observed in numerous epidemiological studies and laboratory rodent and non-human models (Buono and Longo, 2018; Meynet and Ricci, 2014). To date, the anti-tumorigenic effects of NR have been well established, and its potential implications in both cancer prevention and treatment have been suggested (Buono and Longo, 2018; Meynet and Ricci, 2014). Despite these advances, our understanding of the molecular mechanisms underlying the anti-tumorigenic effects of NR remains fragmented.
It has been shown that both systemic changes in the host and tumor intrinsic signaling events contribute to NR-induced tumor suppression (Buono and Longo, 2018; Hursting et al., 2013; Kalaany and Sabatini, 2009; Marsh et al., 2008; Meynet and Ricci, 2014; Mukherjee et al., 2004; Mulrooney et al., 2011). Indeed, a consistent response of animals to NR is a reduction in the levels of circulating growth factors and hormones, especially insulin and insulin-like growth factors (IGFs) (Breese et al., 1991; Ruggeri et al., 1989; Sonntag et al., 1999). Insulin and IGFs activate the downstream phosphatidylinositol-3-kinase (PI3K) signaling pathway to regulate metabolism and cell proliferation (Fruman et al., 2017). Consistently, cells with PI3K activation, either by activating mutations of PI3K or loss of the tumor suppressor PTEN, are resistant to NR in both mammalian (Kalaany and Sabatini, 2009) and Drosophila (Nowak et al., 2013) tumor models. PI3K signaling activation results in activation of the protein kinase AKT, which directly phosphorylates and inactivates the tumor suppressor tuberous sclerosis complex (TSC), leading to activation of the target of Rapamycin complex 1 (TORC1) (Fruman et al., 2017). Consistent with this notion, TORC1 activation has been shown to play a key role in NR resistance of PTEN or TSC null tissues in Drosophila (Nowak et al., 2013, 2018). In spite of these important discoveries, it still remains unclear how TORC1 activation leads to NR resistance. Moreover, it is also unclear whether there are NR-specific tumor suppressors that, in contrast to PTEN and TSC1/2, restrict tumor growth only in response to NR.
In this study, we identify Hdc and Unk as two NR-specific tumor suppressors that restrict cell cycle progression and tissue growth in response to NR. We found that hdc and unk mutant cells do not show apparent growth advantage in comparison to wild-type cells under normal nutrient conditions. However, the mutant cells proliferate much faster than wild-type cells under NR. We further found that the Hdc-Unk complex binds to TORC1 component Raptor and regulates S6 phosphorylation in a TORC1-dependent manner. Interestingly, the physical interaction between Unk and Raptor is highly sensitive to insulin and TORC1 activities, suggesting potential mechanisms underlying the NR-specific function of Hdc and Unk. We also demonstrated that HECA and UNK, the human homolog of Hdc and Unk, respectively, also form a complex that binds to mTORC1 component RPTOR and regulates S6 phosphorylation. Our identification of Hdc and Unk as two NR-specific tumor suppressors sheds light on molecular mechanisms underlying the anti-tumor-igenic effects of NR.
RESULTS
Identification of hdc as a Negative Growth Regulator
In a genetic screen for negative growth regulators using the eyeless-flippase (FLP) recessive cell lethal technique that eliminates most of the homozygous wild-type tissue in the mosaic eyes (Newsome et al., 2000), we identified a lethal mutation (82-A43) that caused enlarged eyes (Figures S1A–S1C). We mapped 82-A43 to molecular coordinates 26103647; 26215013 based on its noncomplementation with Df(3R) ED6332. Tests with available mutations in this interval showed that 82-A43 failed to complement 4 independent alleles of headcase (hdc): hdcFus−6, hdcBG00237, hdcEY02460, and hdcKG09851. Indeed, sequencing analysis revealed a nonsense mutation in 82-A43 that was predicted to change Gln523 of Hdc to a stop codon (Figure S1D). We therefore renamed 82-A43 as hdcA43.
Hdc encodes a cytoplasmic protein with no recognizable functional domains and was first identified as a gene that is specifically expressed in all imaginal cells (Weaver and White, 1995). Genetic analysis has implicated Hdc in several developmental processes, such as metamorphosis (Weaver and White, 1995), tracheal branching (Steneberg et al., 1998), and dendrite pruning (Loncle and Williams, 2012). Hdc was also reported to regulate neuronal cell differentiation downstream of the mTOR pathway but, surprisingly, not tissue growth nor any known mTOR targets (Avet-Rochex et al., 2014). An unusual feature of this gene is that the Hdc mRNA generates two overlapping proteins (70 kDa and 120 kDa) as a result of translational readthrough of an internal UAA stop codon at residue 650 (Figures S1D and S1E) (Steneberg et al., 1998; Steneberg and Samakovlis, 2001). Analysis of homozygous hdcA43 mutant larval extracts revealed reduced levels of 70-kDa and 120-kDa species (Figure S1E). Consistent with this finding, immunostaining with anti-Hdc antibody revealed decreased but detectable levels of Hdc in hdcA43clones (Figures S1F–S1G’). Thus, hdcA43 is likely a hypomorphic allele.
Using the flippase recognition target (FRT)-FLP-mediated genomic deletion strategy (Parks et al., 2004), we generated another allele of hdc that deletes the first two coding exons of hdc (Figure S1D). Unlike the hypomorphic allele hdcA43, no Hdc expression could be detected by western blotting of hdcdel mutant larval extracts (Figure S1E) or immunostaining of hdcdel mutant clones (Figures S1H–S1I’), suggesting hdcdel is a null allele. We therefore used hdcdel for all subsequent studies. Both hdcA43 and hdcdel alleles exhibited pupal lethality, suggesting the essential role of Hdc during Drosophila development.
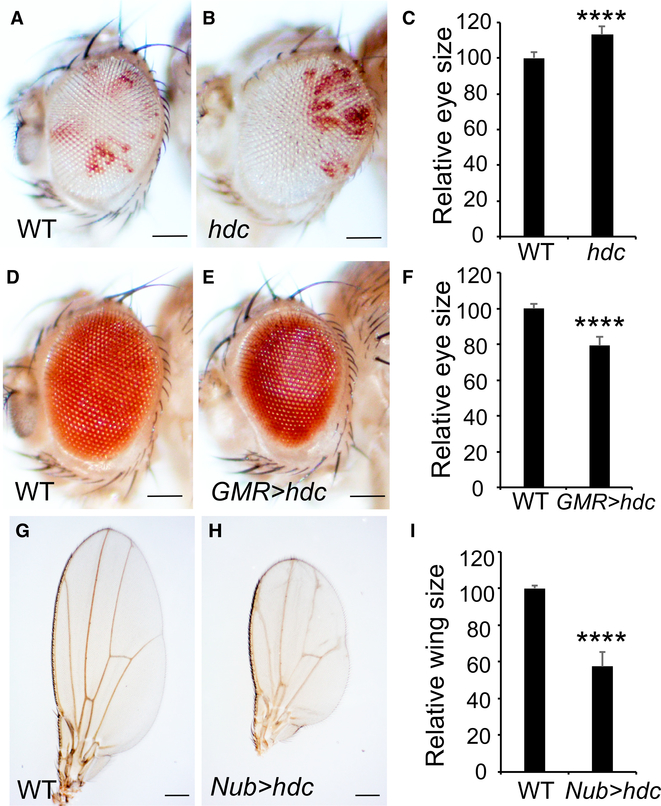
Like hdcA43, mutant eyes composed predominantly of hdcdel mutant cells were markedly larger compared to control eyes (Figures 1A–1C). Conversely, overexpression of Hdc using the eye-specific GMR-Gal4 driver or the wing-specific Nub-Gal4 driver led to a decrease in eye size (Figures 1D–1F) or wing size (Figures 1G–1I), respectively. Taken together, the loss- and gain-of-function phenotypes of Hdc implicate it as a tumor suppressor in Drosophila.
Figure 1. Hdc Is a Negative Regulator of Tissue Growth.
(A–C) Adult eyes comprised predominantly of control cells (A) or hdcdel cells (B). Quantification of relative eye sizes (C). For each analysis (A and B), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
(D–F) Images of compound eyes from a control eye (D) and an eye with Hdc overexpression (E). Quantification of relative eye sizes (F). For each analysis (D and E), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
(G–I) Adult wings from the indicated genotypes. Quantification of relative wing sizes (I). For each analysis (G and H), a total of 10 fly wings (n = 10) were used. The scale bars represent 0.2 mm.
Data in (C), (F), and (I) are represented as means ± SD; ****p < 0.0001.
Given that the wild-type Hdc mRNA generates two protein products, we investigated whether the two protein species may function differently in growth control. We therefore generated an expression construct that could only encode the 70-kDa species (Hdc70kDa) by deleting the Hdc cDNA sequence after the 650UAA codon. In comparison, we generated an expression construct that could only encode the 120-kDa species (Hdc120kDa) by replacing the UAA codon at 650 with a GCA (alanine) codon. While overexpression of Hdc70kDa by the GMR-Gal4 driver had little effect on eye size (Figure S5D), overexpression of Hdc120kDa resulted in a small eye phenotype that is comparable to that induced by wild-type Hdc (Figure S5D). This result is consistent with a previous report that Hdc120kDa (corresponding to the stop codon readthrough product) is a stronger inhibitor of trachea terminal branching compared to Hdc70kDa (Steneberg and Samakovlis, 2001).
Hdc Regulates Tissue Growth in Response to NR
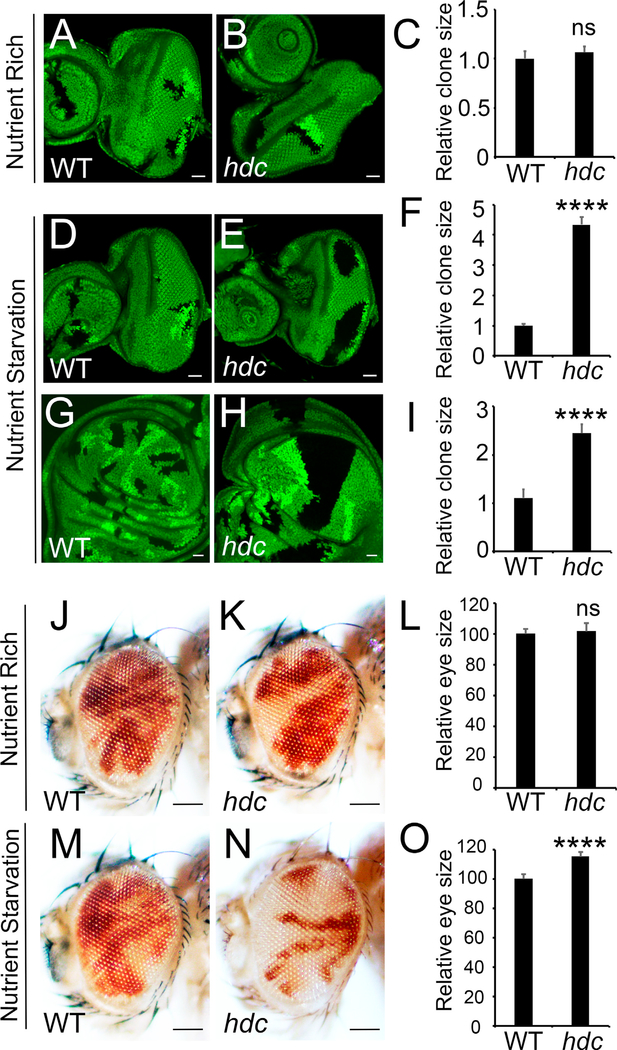
To further characterize the function of Hdc in growth control, we generated hdc mutant clones in eye discs using the hsFlp/FRT technique without the recessive cell lethal. This allows us to directly compare the growth of mutant tissue versus wild-type twin spots control. Surprisingly, we found that hdc clones showed comparable size to their twin spots (Figure 2B). This result is in contrast to the enlarged eyes generated by the eyeless-FLP recessive cell lethal technique, which are predominantly composed of mutant cells. We reason that Hdc may represent a context-dependent tumor suppressor whose function can be revealed only by the very sensitive eyeless-FLP recessive cell lethal technique under normal circumstances. We then tested various stress conditions and found that Hdc restricts tissue growth in response to NR. Indeed, when animals were raised in media with reduced yeast supply (main source of protein in fly media), hdc mutant clones were much larger than their neighboring wild-type twin spots in both eye disc (Figures 2E and 2F) and wing disc (Figures 2H and 2I), an indication of strong overgrowth. Next, we examined eye size of adult fly containing random hdc mutant clones induced by the eyFlp/ FRT system. In this system, hdc mutant cells were marked in white, while wild-type control cells were marked in red. Consistently, we found that the eye sizes of hdc mutant flies were slightly but significantly larger than that of wild-type control flies when animals were raised under NR conditions (Figures 2M–2O). In addition, under NR conditions, adult eyes were composed predominantly of hdc mutant tissue (Figure 2N). In contrast, overrepresentation of hdc mutant tissue in adult eyes was less observed under nutrient-rich conditions (Figure 2K). Taken together, these results suggest that Hdc regulates tissue growth in response to NR.
Figure 2. Hdc Regulates Tissue Growth in Response to Nutrient Restriction.
(A–I) Third instar eye discs containing control or hdc mutant clones (marked by the absence of GFP) from larvae reared under nutrient-rich (20 g/l yeast) conditions (A and B) or nutrient-starvation (5 g/l yeast) conditions (D and E). Third instar wing discs containing control (G) or hdc mutant (H) clones (marked by the absence of GFP) from larvae reared under nutrient-starvation (5 g/l yeast) conditions. Quantification of corresponding relative clone size compared with wild-type neighboring twin spots (C, F, and I). For each analysis (A, B, D, E, G, and H), a total of 9 clones (n = 9) were measured. The scale bars represent 20 μm.
(J–L) Adult eyes containing control clones (J) or hdc mutant clones (K, marked by the absence of red color) raised under normal conditions. Quantification of relative eye sizes (L). For each analysis (J and K), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
(M–O) Adult eyes containing control clones (M) or hdc mutant clones (N, marked by the absence of red color) raised under nutrient starvation conditions. Quantification of relative eye sizes (O). For each analysis (M and N), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
Data in (C), (F), (I), (L), and (O) are represented as means ± SD; ****p < 0.0001.
Hdc Regulates Cell Cycle Progression
The size of an organ is determined by coordinated regulation between cell size and cell number. We therefore asked whether Hdc controls organ size through regulating cell size or cell number, or both. To address this question, we first generated hdc mutant bristles in wing margin. We found that hdc mutant bristles are indistinguishable in cell size from wild-type cells under both nutrient-rich and nutrient-starvation conditions (Figures S2B and S2C). We next generated hdc mutant clones in eye discs and compared cell size between hdc mutant and their wildtype control. Consistently, loss of hdc does not cause any appreciable effect on cell size in eye discs (Figures S2D–S2E’). Based on the above observations, we asked whether Hdc regulates cell number. For this purpose, we counted trichome numbers in wild-type control wings (Figure S2F) versus wings overexpressing hdc (Figure S2G). Quantification analysis revealed a significant decrease of trichome numbers in adult fly wings overexpressing hdc (Figure S2H). We further examined the effect of Hdc overexpression on trichome density, a more sensitive measurement for cell size changes. Interestingly, we found a mild increase of trichome density in Hdc-overexpressing wings (Figures S2I–S2K), indicating decreased cell sizes. Taken together, we concluded that the primary function of Hdc in tissue growth is to regulate cell number, with a mild effect on cell size when overexpressed.
The reduced cell number in hdc-overexpressing wings could be a result of defects in either cell survival or cell proliferation, or both. To distinguish between these possibilities, we overexpressed hdc in the posterior compartment of wing disc using the en-Gal4 driver and examined cell death by TUNEL staining (Figure S3B). As a control, we overexpressed expanded (ex), a known tumor suppressor that can induce strong cell death when overexpressed (Blaumueller and Mlodzik, 2000) (Figure S3A). TUNEL staining revealed strong ectopic cell death in ex-overexpressing wing discs as expected (Figures S3A and S3A’) but not in hdc-overexpressing discs (Figures S3B and S3B’). Consistently, expression of baculovirus P35, a caspase inhibitor that prevents cell death in Drosophila (Hay et al., 1994), has no effect on hdc-overexpression-induced eye size reduction (Figures S3C–S3F). Therefore, we concluded that Hdc does not regulate cell death.
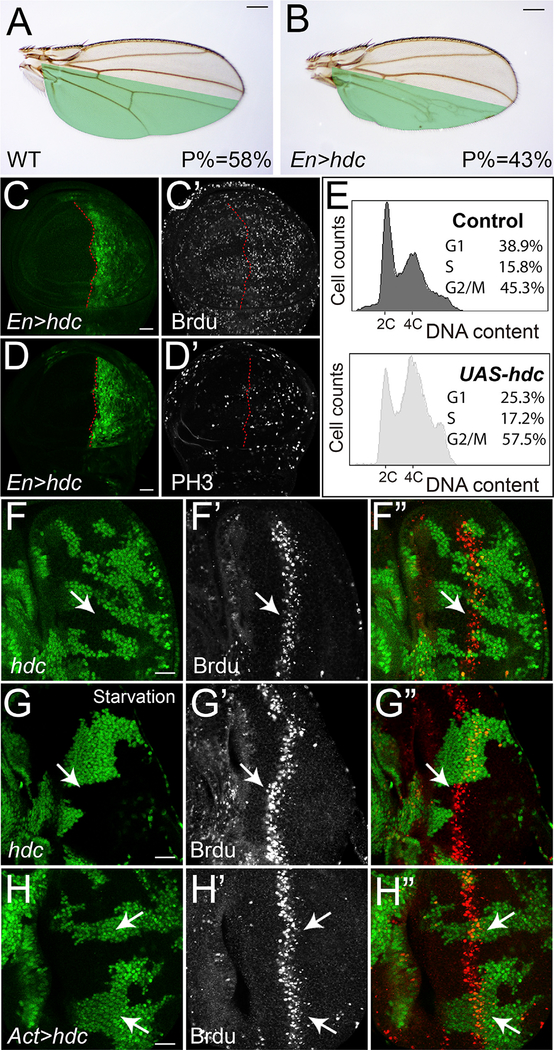
Next, we examined the effect of Hdc on cell proliferation using Brdu labeling (S phase marker) and PH3 antibody staining (M phase marker). If Hdc restricts tissue growth through inhibiting cell proliferation, we would expect reduced Brdu or PH3 labeling in hdc-overexpressing cells. Surprisingly, we found more cells with Brdu labeling (Figures 3C and 3C’) and PH3 staining (Figures 3D and 3D’) in the posterior compartment of wing disc where hdc is overexpressed, despite a significant decrease in posterior compartment size (Figures 3A and 3B). Consistently, DNA content analysis by flow cytometry revealed increased population of S and G2/M fractions in hdc-overexpressing cells (Figure 3E). While increased Brdu and PH3 labeling generally indicates cells undergoing ectopic proliferation, the concurrent reduction of wing size does not support the occurrence of over-proliferation for hdc-overexpressing cells. Alternatively, the conflicting phenomenon observed here can be explained as a result of accelerated progression through G1, causing more cells retained in S and G2/M phases. To test this possibility, we further examined the effect of Hdc on cell cycle progression in Drosophila eye discs. During Drosophila eye disc development, a group of precursor cells re-enter S phase for another round of division called second mitotic wave (SMW) (Baker, 2001). As a result, the synchronized S-phase cells in the SMW form a stripe that can be visualized by Brdu labeling. If Hdc is required for proper cell cycle progression, we would expect abnormal Brdu labeling in hdc mutant cells in the SMW. Indeed, under starvation conditions, hdc mutant cells labeled with Brdu are more anterior than wild-type control cells in the SMW, indicating accelerated cell cycle progression in hdc mutant cells (Figures 3G–3G’’ and S4D–S4D’’). Of note, the effect of hdc loss of function on Brdu labeling in SMW cells is less obvious under nutrient-rich conditions (Figures 3F–3F’’), suggesting the regulation of Hdc on cell cycle progression is nutrient sensitive. Conversely, hdc-overexpressing cells with Brdu labeling are more posterior than wild-type control cells in the SMW, indicating delayed cell cycle progression (Figures 3H–3H’’). To complement the Hdc overexpression results in wing discs, we also RNAi knocked down Hdc expression in the posterior compartment of wing disc and examined resulted cell cycle changes. Although we could not detect visible changes of PH3 staining (Figures S4C–S4C’’), we did observe a mild decrease of Brdu labeling (Figures S4B–S4B’’), which is consistent with Hdc-overexpression effects. Interestingly, despite our observed effect of hdc loss of function on clone sizes (Figure 2H) and Brdu labeling (Figure S4B), the adult wing sizes appeared normal (data not shown), suggesting hdc loss of function has a less impact on adult wing sizes. Taken together, our results suggest that Hdc plays a critical role in cell cycle progression during mitotic cell proliferation.
Figure 3. Hdc Regulates Cell Cycle Progression.
(A and B) A control wing (A) and a wing with Hdc overexpression (B) in the posterior compartment (marked in green) driven by Engrailed-Gal4 (En-Gal4). The average ratio of the size of posterior compartment to the whole wing (P%) was measured using ImageJ (n = 10). The scale bars represent 0.2 mm.
(C–D’) Third instar wing discs containing hdc-overexpressing clones (GFP-positive) were stained for Brdu (C’) and PH3 (D’). The scale bars represent 20 μm.
(E) Flow cytometric analysis of dissociated wing imaginal discs containing Hdc overexpression cells. Histograms showing DNA content of control cells (top) and hdc-overexpressing cells (bottom). The percentage of cells in each cell cycle phase were analyzed.
(F–G’’) A third instar eye disc containing hdc mutant clones (GFP-negative) raised under nutrient-rich conditions (F–G’’) or nutrient-starvation conditions (G–G’’) was stained for Brdu. The scale bar represents 20 μm.
(H–H’’) A third instar eye disc containing hdc-overexpressing clones (GFP-positive) was stained for Brdu (H’). The scale bar represents 20 μm.
Hdc Functions with Unk to Inhibit Tissue Growth
To understand the molecular mechanism by which Hdc regulates tissue growth and cell cycle progression, we performed a yeast two-hybrid screen for Hdc binding proteins. Since the 70-kDa species of Hdc (Hdc70kDa) is conserved in mammals, we used the corresponding N-terminal region of Hdc as bait. From this screen, we isolated 16 independent cDNA clones of Unkempt (Unk). The overlapping sequences of these Unk clones define Unk94−154 as an Hdc binding region (Figure S5A). Conversely, in another yeast two-hybrid screen using the full-length Unk as bait, we isolated 9 independent cDNA clones of Hdc. The overlapping sequences of these clones define Hdc421−639 as an Unk binding region (Figure S5A). Consistent with the yeast two-hybrid results and a previous report (Avet-Rochex et al., 2014), epitope-tagged Hdc (both Hdc70kDa and Hdc120kDa species) and Unk co-immunoprecipitated with each other in Drosophila S2R+ cells (Figures S5B and S5C). These results suggest that Hdc physically associates with Unk.
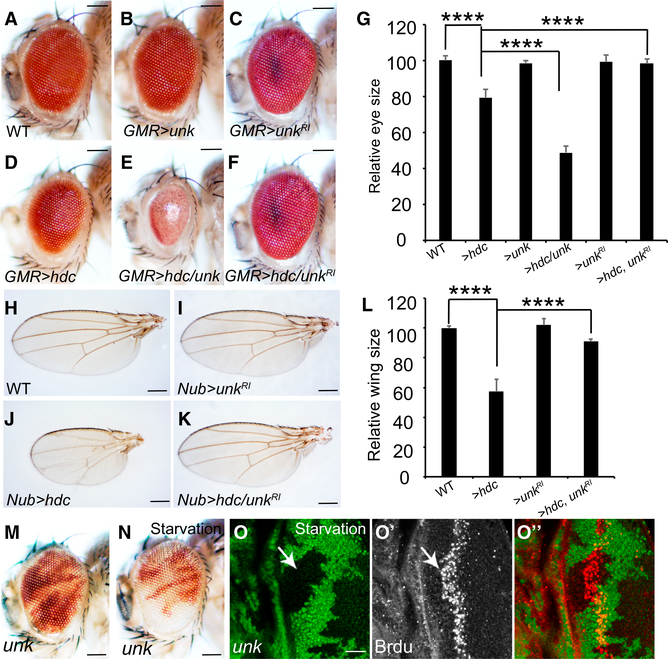
unk is an essential gene in Drosophila (Mohler et al., 1992), but its involvement in growth control has not been documented. Like hdc, Unk was reported to function downstream of the mTOR pathway to regulate neuronal cell differentiation but not tissue growth nor any known mTOR targets (Avet-Rochex et al., 2014). To explore the possibility that Unk may function together with Hdc in growth control, we first examined the consequence of Unk overexpression. While overexpression of Unk alone by the GMR-Gal4 driver had little effect on eye size (Figure 4B), it greatly exacerbated the small eye phenotype induced by Hdc overexpression (Figure 4E), suggesting Unk functions together with Hdc to restrict tissue growth. This synergistic genetic interaction was not only observed between Unk and wildtype Hdc (producing both 70-kDa and 120-kDa species) but also between Unk and Hdc70kDa or between Unk and Hdc120kDa (Figure S5D). This is consistent with our yeast two-hybrid results defining Hdc421−639, which is included in both Hdc70kDa and Hdc120kDa, as an Unk binding region. Next, we asked whether Unk is required for the inhibition function of Hdc on tissue growth. Strikingly, while RNAi knockdown of Unk alone has little effect on eye size (Figure 4C) or wing size (Figure 4I), it greatly rescued hdc-overexpression-induced small eye (Figures 4A–4G) or wing phenotypes (Figures 4H–4L) to almost normal, suggesting the inhibition function of Hdc on tissue growth is Unk dependent. Taken together, we concluded that Hdc and Unk function together as a protein complex to inhibit tissue growth.
Figure 4. Hdc Functions Together with Unk to Regulate Tissue Growth.
(A–G) Images of compound eyes from the indicated genotypes. Quantification of relative eye sizes (G). For each analysis (A, B, C, D, E, and F), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
(H–L) Images of compound wings from the indicated genotypes. Quantification of relative wing sizes (L). For each analysis (H, I, J, and K), a total of 10 fly wings (n = 10) were used. The scale bars represent 0.2 mm.
(M and N) Adult eyes containing unk mutant clones under nutrient-rich (M) or nutrient-starvation conditions (N). The scale bars represent 0.1 mm.
(O–O’’) A third instar eye disc containing unk mutant clones under nutrient starvation was stained for Brdu. The scale bar represents 20 μm. Data in (G) and (L) are represented as means ± SD; ****p < 0.0001.
Unk Regulates Tissue Growth and Cell Cycle Progression
If Hdc and Unk function together to regulate tissue growth, one would expect these genes to exhibit similar loss-of-function phenotypes. A previous study showed that unk mutant cells proliferate normally as wild-type control cells under normal nutrient conditions (Avet-Rochex et al., 2014). Given our observed strong genetic interactions between Hdc and Unk in both fly eye and wing growth, we asked whether unk mutant cells also have proliferation advantage under NR conditions like hdc mutant cells. To test this possibility, we examined unk mutant clone size in adult eyes raised under normal and nutrient-starvation conditions, respectively. Similarly, unk mutant cells overproliferated under nutrient-starvation conditions but not under nutrient-rich conditions (Figures 4M and 4N). Further, unk mutant eyes generated by the eyeless-FLP recessive cell lethal technique are larger than wild-type controls (Figure S5E), confirming its function as a negative growth regulator. In addition, we examined the role of Unk on cell cycle progression. We found that under nutrient-starvation conditions, unk mutant cells exhibited accelerated cell cycle progression, as unk mutant cells showed more anterior Brdu labeling than that of wild-type control cells in the SMW (Figures 4O–4O’’). Taken together, we concluded that like Hdc, Unk regulates cell cycle progression and cell proliferation in response to NR.
Hdc and Unk Form a Protein Complex with Raptor
Notably, in a systematic protein interaction study focused on the TOR pathway using quantitative affinity purification and mass spectrometry, Unk was reported to co-purify with multiple TORC1 pathway components in Drosophila Kc167 cells (Glatter et al., 2011), including TOR, Raptor, and 4E-BP. Furthermore, Unk has been identified as a transcriptional target of TORC1 in several independent studies (Avet-Rochex et al., 2014; Guertin et al., 2006; Tiebe et al., 2015). These findings suggest that Hdc and Unk may function through the TORC1 pathway to regulate cell cycle progression. To test this model, we first examined reported physical interactions between Unk and TORC1 pathway components. Although we could not detect appreciable co-immunoprecipitation between Unk and TOR or 4E-BP (data not shown), we observed robust interaction between Unk and Raptor in Drosophila S2R+ cells (Figure 5A). We also tested interactions between Raptor and Hdc and found that Hdc70kDa isoform, but not Hdc120kDa isoform, co-immuno-precipitated with Raptor (Figure 5B; data not shown). Interestingly, when Unk expression was knocked down by RNAi, less Hdc70kDa was co-immunoprecipitated by Raptor (Figure 5C). Conversely, in the presence of Unk co-expression, significantly more Hdc70kDa was co-immunoprecipitated by Raptor (Figure 5C). These results suggest that Hdc70kDa, Unk, and Raptor form a protein complex and Unk potentiates Hdc70kDa-Raptor interaction.
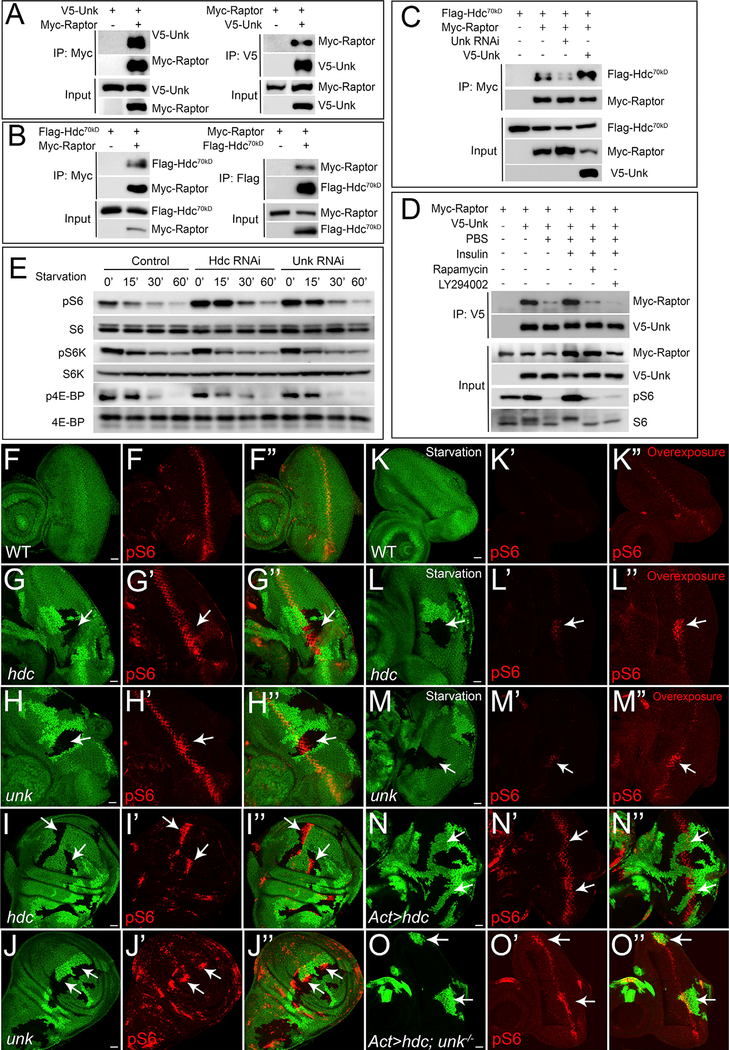
Figure 5. Hdc and Unk Form a Protein Complex with Raptor and Regulate S6 Phosphorylation.
Physical interaction between Unk and Raptor. Immunoprecipitates of S2R+ cell lysate expressing the indicated combination of V5-Unk and Myc-Raptor constructs were probed with the indicated antibodies.
Physical interaction between Hdc70kDa and Raptor. Immunoprecipitates of S2R+ cell lysate expressing the indicated combination of Flag-Hdc70kDa and Myc-Raptor constructs were probed with the indicated antibodies.
Unk potentiates Hdc70kDa-Raptor interaction. S2R+ cells expressing the indicated constructs were analyzed by co-immunoprecipitation (co-IP).
The interaction between Unk and Raptor is nutrient sensitive. S2R+ cells expressing the indicated constructs were analyzed by co-IP.
RNAi of Hdc or Unk induces S6 phosphorylation. S2R+ cells were incubated with double-stranded RNA (dsRNA) of GFP, hdc, or unk for 3 days and treated with PBS starvation for indicated time before western blot. Total cell lysates were probed with anti-phospho-S6, anti-phospho-S6K (T389), and anti-phospho-4E-BP1 (T37/46) antibodies.
(F–F’’) A wild-type control eye disc was stained with pS6 antibody (red).
(G–J’’) Eye discs or wing discs containing mutant clones of the indicated genotypes were stained with pS6 antibody (red). In all panels, mutant clones were marked by loss of GFP. Note increased pS6 staining in hdc or unk mutant clones (arrows).
(K–M’’) Same as (F)–(H’’), except all eye discs were treated with 15 min PBS starvation prior to fixation.
(N–N’’) An eye disc containing hdc-overexpressing clones (GFP-positive) was stained with pS6 antibody (red).
(O–O’’) An eye disc containing unk mutant clones overexpressing hdc (GFP-positive) was stained with pS6 antibody (red).
Images (F), (G’), and (H’) and images (K’), (L’), and (M’) were acquired using the same parameter settings. The scale bars represent 20 μm.
A previous study reported that Unk associates with TORC1 components in an insulin-sensitive manner (Glatter et al., 2011). We therefore asked whether the physical interactions between Hdc/Unk and Raptor is nutrient sensitive. Indeed, we found that while Unk strongly interacts with Raptor in S2R+ cells supplied with a nutrient-rich medium, a short PBS starvation largely abolished their binding (Figure 5D). Strikingly, the decreased binding between Unk and Raptor upon PBS starvation was completely restored upon insulin treatment, and further treatment with either Rapamycin or LY294002 inhibited insulin-stimulated Unk-Raptor binding (Figure 5D). These results suggest that the physical interaction between Unk and Raptor is nutrient sensitive and regulated by both insulin/PI3K and TORC1 activities. Unlike the interaction between Unk and Raptor, we did not detect visible changes for the bindings between Unk and Hdc nor between Raptor and Hdc under various conditions (data not shown), suggesting these interactions may not be regulated by nutritional status.
Hdc and Unk Regulate S6 Phosphorylation
The physical interactions between Hdc, Unk, and Raptor suggest these proteins function together in a protein complex. This prompted us to further examine whether Hdc and Unk, like Raptor, regulates TORC1 activity. To this end, we took advantage of a recently developed anti-phospho-dRpS6 (pS6) antibody that can detect TORC1 activity in vivo (Kim et al., 2017; Romero-Pozuelo et al., 2017). Interestingly, we found increased pS6 staining in hdc mutant clones in both eye (Figures 5G–5G’’) and wing discs (Figures 5I–5I’’), as well as in unk mutant eye discs (Figures 5H–5H’’) and wing discs (Figures 5J–5J’’). Conversely, overexpression of Hdc reduced the pS6 level in the SMW (Figures 5N–5N’’). Simultaneously, knockout of unk in hdc-overexpressing clones reversed the pS6 level (Figures 5O–5O’’), further suggesting the regulation of Hdc on pS6 is Unk dependent. Taken together, these results suggest that the Hdc-Unk protein complex, like the Tsc1-Tsc2 protein complex, negatively regulates TORC1 activity in vivo. Despite these similarities, we noted an important difference between the two protein complexes. In contrast to a uniform increase of the pS6 level in cells lacking Tsc1 or Tsc2 (Kim et al., 2017; Romero-Pozuelo et al., 2017), the increase of the pS6 level in hdc or unk mutant cells is restricted to the SMW in eye discs (Figures 5G–5H’’) and is patchy in wing discs (Figures 5I–5J’’). The observed difference between Hdc-Unk and Tsc1-Tsc2 in pS6 expression suggests that Hdc and Unk may preferentially mediate part of the TORC1 function in regulating cell cycle progression.
Given that Hdc and Unk regulate cell proliferation in response to nutrient starvation, we therefore asked whether the regulation of pS6 by Hdc and Unk is nutrient sensitive. To address this point, we starved eye discs in PBS and examined the resulting pS6 levels. We found that a short PBS starvation led to a rapid decrease of the pS6 level, suggesting pS6 is sensitive to starvation in vivo (Figures 5K–5K’’). Then, we used this ex vivo assay to examine the effect of Hdc and Unk on the pS6 level in eye discs under starvation conditions. Interestingly, we found that while the pS6 levels in wild-type control cells were greatly diminished upon starvation, a high level of pS6 was observed in both hdc (Figures 5L–5L’’) and unk (Figures 5M–5M’’) mutant clones, suggesting the regulation of Hdc and Unk on S6 phosphorylation is nutrient sensitive. Consistent with our in vivo results, RNAi knockdown of Hdc or Unk in Drosophila S2R+ cells resulted in increased S6 phosphorylation (Figure 5E). We also examined the effect of Hdc and Unk on the two well-known TORC1 substrates, S6K and 4E-BP, respectively. Interestingly, we did not find a robust effect of Hdc or Unk on S6K and 4E-BP phosphorylation under the same experimental conditions (Figure 5E). Prompted by this observation, we asked whether the regulation of Hdc or Unk on S6 phosphorylation is S6K dependent. We found that while S6K knockdown significantly decreased S6 phosphorylation, synergistically knocking down Hdc or Unk partially rescued S6 phosphorylation (Figure S6), indicating Hdc and Unk may regulate S6 phosphorylation partially bypass S6K. Taken together, our results suggest that Hdc and Unk preferentially regulate TORC1 downstream target S6 activity.
The Regulation of S6 Activity by Hdc and Unk Is Tor and Raptor Dependent
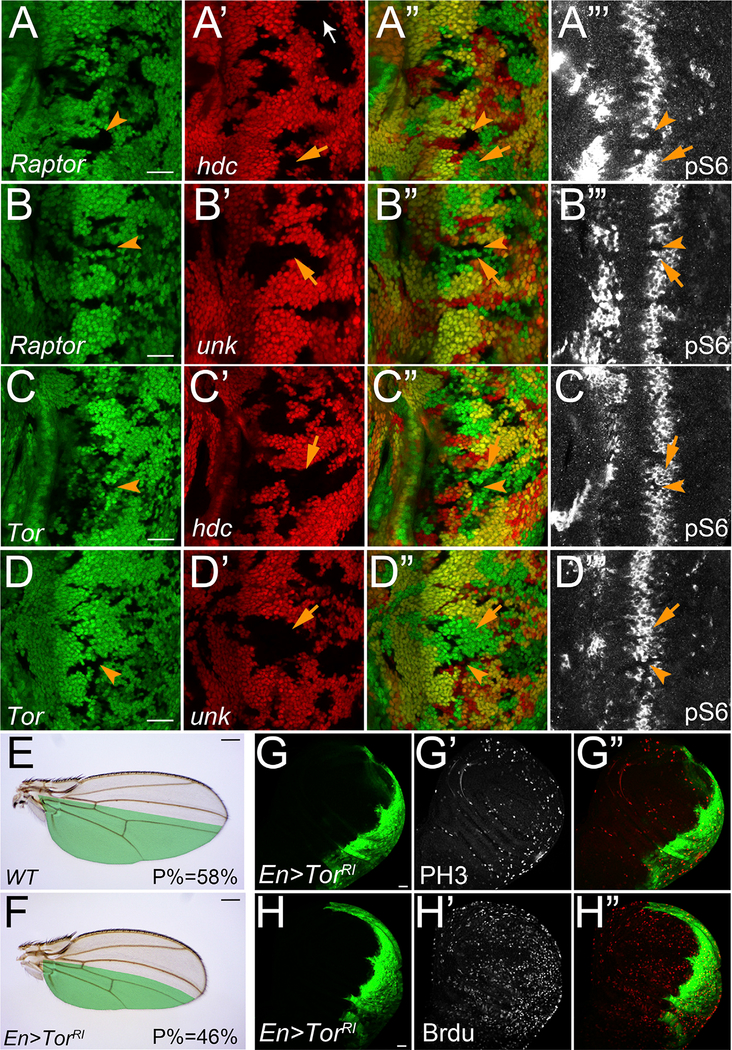
Having established a regulation between Hdc-Unk and S6 phosphorylation, as well as the physical interactions among Hdc, Unk, and Raptor, we next examined the genetic interactions between Hdc-Unk and Raptor. For this purpose, we used a null mutant of raptor (Zhang, 2009) and examined its role in regulating the pS6 level. Consistent with the critical role of Raptor in mediating TORC1 activity, loss of function of raptor completely inhibited S6 activity, as evidenced by abolished S6 phosphorylation in raptor mutant cells (Figure S7A–S7A’’). In addition, loss of function of raptor completely suppressed the ectopic increase of pS6 in Tsc1 mutant cells, further demonstrating its essential role for TORC1 to control S6 phosphorylation (Figures S7B–S7B’’’). Next, we examined the pS6 level in raptor; hdc or raptor; unk double mutant clones. Clearly, raptor loss of function completely abolished pS6 in hdc (Figures 6A–6A’’’) or unk mutant cells (Figures 6B–6B’’’). Similarly, loss of function of Tor also suppressed pS6 in hdc (Figures 6C–6C’’’) or unk mutant cells (Figures 6D–6D’’’). These results therefore suggest that Hdc and Unk regulate S6 activity in a Tor and Raptor-dependent manner.
Figure 6. Hdc- and Unk-Mediated S6 Phosphorylation Is Tor and Raptor Dependent.
(A–D’’’) Eye discs containing mutant clones of the indicated genotypes were stained with pS6 antibody (white). In all panels, mutant clones of raptor or Tor were marked by loss of GFP, and mutant clones of hdc or unk were marked by loss of RFP. The scale bars represent 20 μm.
(E and F) A control wing (E) and a wing with TorRNAi overexpression (F) in the posterior compartment (marked in green) driven by Engrailed-Gal4 (En-Gal4). The average ratio of the size of posterior compartment to the whole wing (P%) was measured using ImageJ (n = 10). The scale bars represent 0.2 mm.
(G–H’) Third instar wing discs containing TorRNAi-overexpressing clones (GFP-positive) were stained for PH3 (G’) and Brdu (H’). The scale bars represent 20 μm.
The results so far indicate that Hdc and Unk may function through Tor and Raptor to regulate cell cycle progression. If this is the case, we would expect that inhibition of Tor or Raptor activities may lead to similar cell cycle progression delay as shown in hdc-overexpressing clones (Figures 3C–3D’). Consistent with its prominent role in growth control, RNAi knockdown of Tor caused a severe size reduction (Figure 6F). Concurrent with this size reduction, we found that Brdu and PH3 labeling were strongly increased in Tor RNAi clones (Figures 6G–6H’’), suggesting delayed cell cycle progression. Knocking down Raptor led to similar accumulation of cells with ectopic Brdu and PH3 labeling (Figures S7C–S7D’’). Taken together, these results suggest that Hdc and Unk regulate cell cycle progression through Tor and Raptor.
Conserved Function of Hdc-Unk Human Counterparts: HECA and UNK
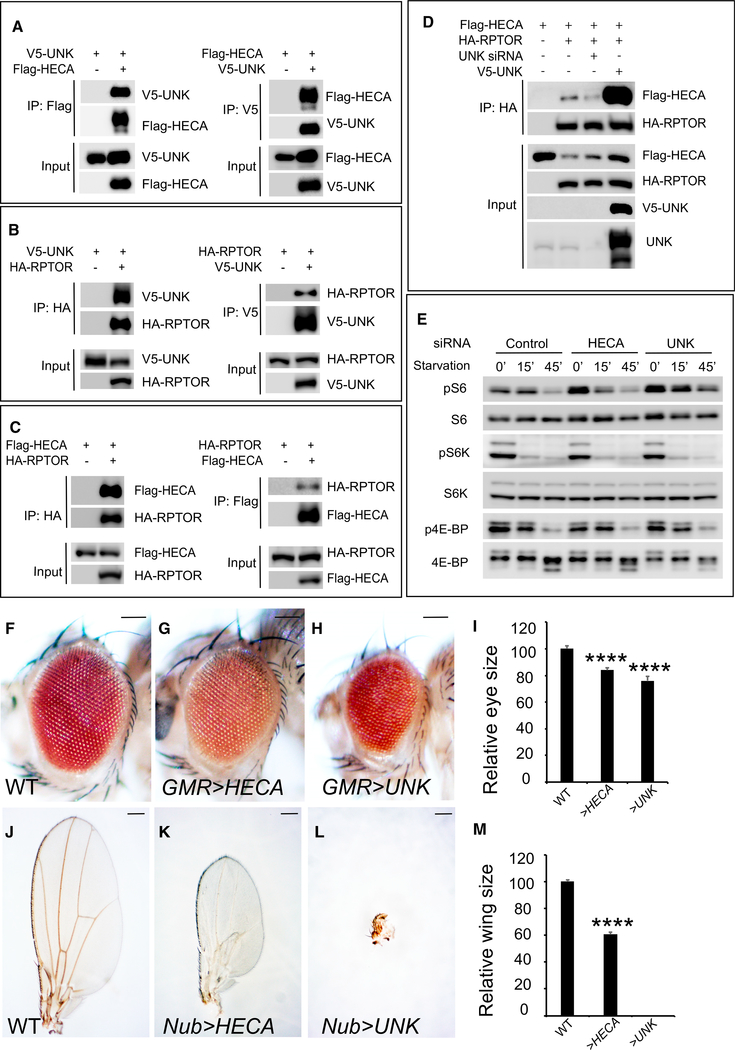
The TOR signaling network is evolutionarily conserved from Drosophila to mammals. We therefore asked whether HECA and UNK, the human counterparts of Hdc and Unk, respectively, have a conserved function in regulating TORC1 activity in mammals. To answer this question, we first examined the physical interactions between HECA and UNK. We found that epitope-tagged HECA and UNK co-immunoprecipitated with each other in HEK293 cells (Figure 7A), suggesting HECA physically associates with UNK. Next, we examined the interactions between UNK and RPTOR, the human homolog of Drosophila Raptor (Kim et al., 2002). Like their Drosophila counterparts, human UNK and RPTOR strongly bind with each other in HEK293 cells (Figure 7B). We also tested interactions between HECA and RPTOR and found that HECA and RPTOR co-immunoprecipitated with each other (Figure 7C). Consistent with a role of Drosophila Unk in potentiating Hdc-Raptor interaction in S2R+ cells, we found that human UNK potentiates HECA and RPTOR interactions in HEK293 cells, as evidenced by decreased HECARPTOR binding when UNK is knocked down and increased HECA-RPTOR binding in the presence of UNK co-expression (Figure 7D). Taken together, HECA, UNK, and RPTOR, like their Drosophila counterparts, form a protein complex in HEK293 cells.
Figure 7. Human HECA and UNK Have Conserved Functions.
(A) Physical interaction between HECA and UNK. Immunoprecipitates of HEK293 cell lysate expressing the indicated combination of Flag-HECA and V5-UNK constructs were probed with the indicated antibodies.
(B) Physical interaction between UNK and RPTOR. Immunoprecipitates of HEK293 cell lysate expressing the indicated combination of V5-UNK and HA-RPTOR constructs were probed with the indicated antibodies.
(C) Physical interaction between HECA and RPTOR. Immunoprecipitates of HEK293 cell lysate expressing the indicated combination of Flag-HECA and HA-RPTOR constructs were probed with the indicated antibodies.
(D) UNK potentiates HECA-RPTOR interaction. HEK293 cells expressing the indicated constructs were analyzed by co-IP.
(E) siRNA of HECA or UNK induces S6 phosphorylation. HEK293 cells were transfected with siRNA of control, HECA, or UNK and treated with PBS starvation. Total cell lysates were probed with anti-phospho-S6, anti-phospho-S6K (T389), and anti-phospho-4E-BP1 (T37/46) antibodies.
(F–I) Images of compound eyes from the indicated genotypes. Quantification of relative eye sizes (I). For each analysis (F, G, and H), a total of 10 fly eyes (n = 10) were used. The scale bars represent 0.1 mm.
(J–M) Images of compound wings from the indicated genotypes. Quantification of relative wing sizes (M). For each analysis (J, K, and L), a total of 10 fly wings (n = 10) were used. The scale bars represent 0.2 mm.
Data in (I) and (M) are represented as means ± SD; ****p < 0.0001
The observed physical interactions among HECA, UNK, and RPTOR suggest HECA and UNK may have a conserved function in regulating TORC1 activity in mammals. To test this, we knocked down HECA and UNK by small interfering RNA (siRNA) in HEK293 cells and examined their effects on TORC1 activity. We found that knockdown of HECA or UNK by siRNA led to strong S6 phosphorylation elevation but not S6K or 4E-BP phosphorylation (Figure 7E). These results suggest that HECA and UNK play critical roles in regulating TORC1 substrate S6 in mammals.
Several lines of evidence suggested that HECA may function as a tumor suppressor in human cancers. The human HECA gene locus is mapped to chromosome 6q23–24, a region of recurrent loss of heterozygosity in lung and head and neck cancers (Bailey-Wilson et al., 2004; Bockmühl et al., 1998). Decreased expression of HECA was reported in pancreatic and renal cell carcinoma cell lines (Makino et al., 2001), as well as primary tumors from oral squamous cell carcinoma (OSCC) patients (Dowejko et al., 2009, 2012). We therefore asked whether HECA regulates tissue growth in vivo. To test this, we generated transgenic flies carrying full-length human HECA cDNA. We found that overexpression of HECA in fly eyes or wings led to a decrease in eye size (Figure 7G) or wing size (Figure 7K), respectively. Therefore, our results showed in vivo evidence that HECA negatively regulates tissue growth. Like HECA, overexpression of UNK in fly eyes also led to decreased eye size (Figure 7H). Strikingly, when UNK was overexpressed in wing, the wing growth was completely suppressed (Figure 7L). Taken together, our results demonstrated that HECA and UNK, like their Drosophila counterparts, regulate tissue growth.
DISCUSSION
Here, we identified Hdc and Unk as two nutrient-sensitive tumor suppressors that restrict cell cycle progression and hence tissue growth in response to NR. An interesting feature of Hdc and Unk is that their tumor-suppressing function is manifested only under nutrient-poor conditions. This feature distinguishes Hdc and Unk from the other nutrient-sensitive tumor suppressors such as PTEN and TSC1/2, which restrict tissue grow under both nutrient-rich and nutrient-poor conditions. This feature also provides a plausible explanation for the results of a previous study in which the role of Hdc and Unk in neuronal cell differentiation, but not in tissue growth, was revealed (Avet-Rochex et al., 2014). Indeed, we also did not observe a growth advantage of hdc or unk mutant clones under nutrient-rich conditions. However, when nutrients are limited, both hdc and unk mutant cells proliferate much faster than wild-type twin-spot control cells, resulting in overgrowth of mutant tissue. Thus, our results suggest that in addition to NR non-specific tumor suppressors like PTEN and TSC1/2, NR also involves NR-specific tumor suppressors to antagonize tumor growth. The identification of NR-specific tumor suppressors raises the possibility that mutation of these tumor suppressor genes may also contribute to the NR resistance of some types of tumors.
The unk gene has been identified as a transcriptional target of TORC1 in multiple studies (Avet-Rochex et al., 2014; Guertin et al., 2006; Tiebe et al., 2015). It has been shown that upon TORC1 inhibition, the transcription factors REPTOR and REPTOR-BP enter the nucleus to upregulate unk expression (Tiebe et al., 2015). Consistent with the role of Unk in NR response, REPTOR and REPTOR-BP knockout flies are also very sensitive to starvation (Tiebe et al., 2015). Interestingly, Unk was co-purified with multiple TORC1 pathway components in a systematic protein interaction study (Glatter et al., 2011), but its potential role in regulating TORC1 signaling has not been well investigated. Here we uncover the function of Hdc and Unk in regulating TORC1 signaling with evidence including (1) Hdc, Unk, and Raptor form a protein complex, (2) Hdc and Unk regulate S6 phosphorylation both in vivo and in cell culture, and (3) Raptor is required for Hdc and Unk to regulate S6 phosphorylation. In contrast to Tsc1/2, loss of Hdc or Unk affects the phosphorylation level of S6 but not S6K and 4E-BP. Thus, Hdc and Unk preferentially regulate S6 phosphorylation in a TORC1-dependent manner.
An interesting finding from this study concerns the profound effects of Hdc and Unk on cell cycle progression. Consistent with the role of Hdc in cell cycle progression, Weaver and White (1995) found that the expression pattern of hdc correlates with the timing of imaginal cells entering into mitotic cell cycle. Previous studies in both mammals and Drosophila have uncovered an important role for mTOR in cell cycle progression (Dowling et al., 2010; Fingar et al., 2002; Kim et al., 2017; Romero-Pozuelo et al., 2017). Interestingly, cell cycle marker analysis revealed that S6 phosphorylation, and therefore TORC1 activity, is selectively increased in S-phase cells in Drosophila imaginal discs (Kim et al., 2017; Romero-Pozuelo et al., 2017). Thus, during Drosophila disc development, S6 phosphorylation is spatially regulated by TORC1 and correlates to proper cell cycle progression. However, both S6K and 4E-BP are not important for TORC1 to regulate cell cycle progression during Drosophila eye development (Kim et al., 2017), suggesting that there may be other substrates mediating TORC1 signaling in cell cycle regulation. Interestingly, we found that Hdc and Unk regulate both cell cycle progression and S6 phosphorylation, but not S6K or 4E-BP phosphorylation, in a TORC1-dependent manner. Taken together, our results raise the possibility that the Hdc-Unk complex, through its physical interactions with Raptor, preferentially mediate the effects of TORC1 on cell cycle progression but not cell size. Future studies will be necessary to provide more evidence and understand the underlying molecular mechanisms.
A question for further investigation concerns how Hdc and Unk regulate tissue growth specifically in response to NR. Based on the observed nutrient-sensitive interactions between Unk and Raptor, an attractive mechanism could be that the growth inhibition function of Unk is regulated by its physical interaction with Raptor. Under nutrient-rich conditions, Raptor interacts with Unk and suppresses its activity, which in turn suppresses the Unk-dependent growth inhibition function of Hdc. In contrast, upon nutrient starvation, Unk is released from Raptor and its growth inhibition function is activated. Activated Unk therefore functions with Hdc to suppress cell cycle progression and eventually tissue growth. Future studies are warranted to test this model.
Given the conserved role of human HECA and UNK in regulating S6 phosphorylation, our results suggest that HECA and UNK may function as tumor suppressors in mammals. Consistent with this notion, silencing HECA expression in OSCC cells causes accelerated cell division, and overexpression of HECA slows down the proliferation of OSCC cells (Dowejko et al., 2009, 2012). Although the human ortholog of Unk has not been studied in the context of cell proliferation, we showed that both HECA and UNK are able to inhibit tissue growth in vivo in the Drosophila model. Thus, it is worthwhile in the future to investigate the growth control function of HECA and UNK, especially their contribution to tumorigenesis with aberrant mTOR signaling.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianzhong Yu (jianzhongyu@ksu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Genetics
hdcA43 was generated by the EMS mutagenesis. hdcdel was generated by the FRT-FLP-mediated genomic deletion strategy using two FRT-containing Exelexis lines (d05275 and e03709) according to standard procedures. A full-length hdc cDNA (LD44381) was used to make UAS-hdc transgenic flies. A full-length unk cDNA (RE58038) was used to make UAS-unk transgenic flies. raptordel was generated by homologous recombination strategy (Gong and Golic, 2003; Zhang, 2009). unkex24 (Avet-Rochex et al., 2014), tsc129 (Gao and Pan, 2001), and Tor2L1 (Oldham et al., 2000) mutant flies were previously reported. UAS-p35 was obtained from Bloomington Stock Center (stock number: 5073). All RNAi flies were obtained from Bloomington Stock Center (UAS-unkRNAi: 57026, UAS-hdcRNAi: 30489, UAS-TorRNAi: 35578, UAS-raptorRNAi: 31529).
Double mutant clones in the eye imaginal discs were generated using flies containing double FRT chromosomes with GFP and RFP markers together with an eye-specific FLP source as described before (Yu and Pan, 2018).
Drosophila cell culture
Drosophila S2R+ cells were propagated in Drosophila Schneider’s Medium (GIBCO) supplemented with 10% FBS and antibiotics. FLAG-Hdc was constructed in the pAc5.1/V5-HisB vector by adding a FLAG epitope (DYKDDDDK) to the N terminus. V5-Unk was constructed in the pAc5.1/V5-HisB vector by adding a V5 epitope (GKPIPNPLLGLDST) to the N terminus. Myc-Raptor was constructed in the pAc5.1/V5-HisB vector by adding a Myc epitope (EQKLISEEDL) to the N terminus.
For RNAi experiments, S2R+ cells were propagated in 12-well plates overnight. The medium from each well was removed next day and replaced with 0.5 mL of serum-free medium containing dsRNA (5 μg) against specific Drosophila genes or GFP (as control). After 1 hour incubation, 1 mL of complete medium was added. The S2R+ cells were then left to grow for 3 days. For starvation experiments, the S2R+ cells were incubated in PBS for indicated time prior to western blot analysis.
Mammalian cell culture
HEK293 cells (ATCC® CRL-10852) were maintained in DMEM, 10% FBS, and antibiotics (Invitrogen). Flag-HECA was constructed in the pcDNA3.1+ vector by addition of an N-terminal Flag epitope (DYKDDDDK) to the full-length human HECA cDNA clone (NM_016217.2, Genescript). V5-UNK was constructed in the pcDNA3.1+ vector by addition of an N-terminal V5 epitope (GKPIPNPLLGLDST) to the full-length human UNK cDNA clone (NM_001080419.2, Genescript). HA-RPTOR was a gift from David Sabatini (Addgene plasmid # 8513). All clones were verified by sequencing.
For siRNA experiments, siRNA against HECA and UNK (Dharmacon) were transfected into HEK293 cells according to the manufacturer’s recommendation. For starvation experiments, the HEK293 cells were incubated in PBS for indicated time prior to western blot analysis.
METHOD DETAILS
Fly food and nutrient starvation
We use the Cornmeal-Molasses-Yeast medium recipe from the Bloomington Drosophila Stock Center with minor adjustment. For nutrient starvation experiments, fly eggs were distributed to food with reduced yeast supply (30% yeast of standard fly food) and allowed to hatch after induction of eyFlp/FRT or hsFlp/FRT clones.
Immunohistochemistry
Third instar larval imaginal discs were dissected for immunohistochemistry according to standard antibody protocol. For pS6 staining, all imaginal discs were dissected in Schneider’s medium and transferred immediately to 4% PFA for fixation except PBS starvation experiments whereas eye discs were incubated in PBS for 15 minutes prior to fixation.
Yeast two-hybrid screens
Yeast two-hybrid analysis was performed as a service by Hybrigenics S.A. (Paris, France). 79.1 million interactions were tested using Drosophila full length Unk (amino acids 1–599) as a bait, and 89.5 million interactions were tested using N-terminal port of Drosophila Hdc (amino acids 1–650) as a bait.
Immunoprecipitation
Immunoprecipitation was carried out as previously described (Yu et al., 2010) using NP-40 lysis buffer (150 mM NaCl, 1% Triton X-100, 10 mM Tris [pH 7.4], 1 mM EGTA, 0.5% NP-40) supplemented with 1 mM PMSF and protease inhibitors (Roche).
Flow Cytometry
Clones of cells expressing hdc were induced at 30 hours after egg deposition and were marked by GFP. In both cases, clones, and larvae were dissected 40 h later. Larvae will be cultured for an additional 72 hours after clone induction and imaginal wing disc cells were dissociated for FACS analysis as previously described (Neufeld et al., 1998). Cell cycle analysis of FACS was performed on a BD LSRFortessa™ X-20 machine and the data was analyzed by BD FACSDiva™ software.
Image analysis
All confocal images were acquired on a Carl Zeiss 700 microscope and analyzed using ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was determined by Student’s t test. Significance is indicated as **p < 0.01, ****p < 0.0001. Error bars represent the standard deviation.
N numbers are as follows:
Figure 1C: For each analysis (A and B), a total of 10 fly eyes (n = 10) were used.
Figure 1F: For each analysis (D and E), a total of 10 fly eyes (n = 10) were used.
Figure 1I: For each analysis (G and H), a total of 10 fly wings (n = 10) were used.
Figures 2C, 2F, and 2I: For each analysis (A, B, D, E, G, and H), a total of 9 clones (n = 9) were measured.
Figure 2L: For each analysis (J and K), a total of 10 fly eyes (n = 10) were used.
Figure 2O: For each analysis (M and N), a total of 10 fly eyes (n = 10) were used.
Figure 4G: For each analysis (A, B, C, D, E, and F), a total of 10 fly eyes (n = 10) were used.
Figure 4L: For each analysis (H, I, J, and K), a total of 10 fly wings (n = 10) were used.
Figure 7I: For each analysis (F, G, and H), a total of 10 fly eyes (n = 10) were used.
Figure 7M: For each analysis (J, K, and L), a total of 10 fly wings (n = 10) were used.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Hdc | Developmental Studies Hybridoma Bank | U33 |
| mouse anti-Dlg | Developmental Studies Hybridoma Bank | 4F3 |
| mouse anti-Brdu | Developmental Studies Hybridoma Bank | G3G4 |
| rabbit anti-PH3 | Cell Signaling Technology | cat# 9701 |
| rabbit anti-phospho-S6K (T389) | Cell Signaling Technology | cat# 9209 |
| rabbit anti-human S6K | Cell Signaling Technology | cat# 9202 |
| rabbit anti-phospho-4E-BP1 (T37/46) | Cell Signaling Technology | cat# 2855 |
| rabbit anti-human 4E-BP1 | Cell Signaling Technology | cat# 9452 |
| rabbit anti-phospho-human S6 (S235/236) | Cell Signaling Technology | cat# 2211 |
| mouse anti-S6 Ribosomal Protein (54D2) | Cell Signaling Technology | cat# 2317 |
| Drosophila phospho-S6 | Romero-Pozuelo et al., 2017 | N/A |
| Drosophila S6K | Stewart et al., 1996 | N/A |
| Drosophila 4E-BP | Miron et al., 2001 | N/A |
| human UNK | Abcam | cat# ab205021 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5-Bromo-2′-deoxyuridine | Sigma | B5002 |
| Schneider’s Drosophila Media | GIBCO | 21720024 |
| DMEM | GIBCO | 11995–065 |
| Fetal bovine serum | GIBCO | 16000044 |
| Effectene Transfection Reagent | QIAGEN | 301425 |
| LipoD293 In Vitro DNA Transfection Reagent | SignaGen Laboratories | SL100668 |
| Experimental Models: Cell Lines | ||
| Drosophila S2R+ cells | Laboratory of Dr. Jianzhong Yu | N/A |
| HEK293 cells | ATCC® | CRL-10852 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: hdcA43 | This paper | N/A |
| D. melanogaster: hdcdel | This paper | N/A |
| D. melanogaster: UAS-hdc | This paper | N/A |
| D. melanogaster: unkex24 | Avet-Rochex et al., 2014 | N/A |
| D. melanogaster: UAS-unkRNAi | Bloomington Stock Center | 57026 |
| D. melanogaster: UAS-hdcRNAi | Bloomington Stock Center | 30489 |
| D. melanogaster: UAS-TorRNAi | Bloomington Stock Center | 35578 |
| D. melanogaster: UAS-raptorRNAi | Bloomington Stock Center | 31529 |
| D. melanogaster: UAS-unk | This paper | N/A |
| D. melanogaster: raptordel | Zhang, 2009 | N/A |
| D. melanogaster: Tor2L1 | Oldham et al., 2000 | |
| D. melanogaster: tsc129 | Gao and Pan, 2001 | N/A |
| D. melanogaster: UAS-p35 | Bloomington Stock Center | 5073 |
| Recombinant DNA | ||
| pAc5.1-FLAG-Hdc | This paper | N/A |
| pAc5.1-V5-Unk | This paper | N/A |
| pAc5.1- Myc-Raptor | This paper | N/A |
| pcDNA3.1-FLAG-HECA | This paper | N/A |
| pcDNA3.1- V5-UNK | This paper | N/A |
| prk5-HA-RPTOR | Addgene | plasmid # 8513 |
| Software and Algorithms | ||
| Fijiwings | Dobens and Dobens, 2013 | N/A |
Highlights.
Hdc and Unk are two nutrient-restriction-specific growth regulators
Hdc and Unk form a protein complex with Raptor to restrict cell cycle progression
The interaction between Unk and Raptor is nutrient sensitive
Hdc and Unk preferentially regulate TORC1 downstream target S6 phosphorylation
ACKNOWLEDGMENTS
We thank Duojia Pan and Aurelio Teleman for kindly providing us with reagents and fly stocks. We thank Duojia Pan and Yonggang Zheng for helpful discussions and critical reading of this manuscript. We thank Bloomington Stock Center for fly stocks. We thank the Confocal Microscopy Core and the Molecular Biology Core funded by KSU-CVM. This work was supported in part by a grant from the Kansas INBRE (P20 GM103418) and a start-up fund and SUCCESS-FYI Intramural Grant from Kansas State University College of Veterinary Medicine. J.Y. and Y.X. were also supported by the Johnson Cancer Research Center at Kansas State University.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.086.
REFERENCES
- Avet-Rochex A, Carvajal N, Christoforou CP, Yeung K, Maierbrugger KT, Hobbs C, Lalli G, Cagin U, Plachot C, McNeill H, and Bateman JM (2014). Unkempt is negatively regulated by mTOR and uncouples neuronal differentiation from growth control. PLoS Genet. 10, e1004624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, de Andrade M, Wiest JS, Fain P, Schwartz AG, You M, Franklin W, et al. (2004). A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am. J. Hum. Genet 75, 460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE (2001). Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol 12, 499–507. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, and Mlodzik M (2000). The Drosophila tumor suppressor expanded regulates growth, apoptosis, and patterning during development. Mech. Dev 92, 251–262. [DOI] [PubMed] [Google Scholar]
- Bockmühl U, Wolf G, Schmidt S, Schwendel A, Jahnke V, Dietel M, and Petersen I (1998). Genomic alterations associated with malignancy in head and neck cancer. Head Neck 20, 145–151. [DOI] [PubMed] [Google Scholar]
- Breese CR, Ingram RL, and Sonntag WE (1991). Influence of age and long-term dietary restriction on plasma insulin-like growth factor-1 (IGF-1), IGF-1 gene expression, and IGF-1 binding proteins. J. Gerontol 46, B180–B187. [DOI] [PubMed] [Google Scholar]
- Buono R, and Longo VD (2018). Starvation, Stress Resistance, and Cancer. Trends Endocrinol. Metab 29, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobens AC, and Dobens LL (2013). FijiWings: an open source toolkit for semiautomated morphometric analysis of insect wings. G3 (Bethesda) 3, 1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowejko A, Bauer RJ, Müller-Richter UD, and Reichert TE (2009). The human homolog of the Drosophila headcase protein slows down cell division of head and neck cancer cells. Carcinogenesis 30, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Dowejko A, Bauer R, Bauer K, Müller-Richter UD, and Reichert TE (2012). The human HECA interacts with cyclins and CDKs to antagonize Wnt-mediated proliferation and chemoresistance of head and neck cancer cells. Exp. Cell Res 318, 489–499. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. (2010). mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, and Blenis J (2002). Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, and Abraham RT (2017). The PI3K Pathway in Human Disease. Cell 170, 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, and Pan D (2001). TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15, 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter T, Schittenhelm RB, Rinner O, Roguska K, Wepf A, Jünger MA, Köhler K, Jevtov I, Choi H, Schmidt A, et al. (2011). Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol. Syst. Biol 7, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, and Golic KG (2003). Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Guntur KV, Bell GW, Thoreen CC, and Sabatini DM (2006). Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol 16, 958–970. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, and Rubin GM (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Dunlap SM, Ford NA, Hursting MJ, and Lashinger LM (2013). Calorie restriction and cancer prevention: a mechanistic perspective. Cancer Metab. 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, and Sabatini DM (2009). Tumours with PI3K activation are resistant to dietary restriction. Nature 458, 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, and Sabatini DM (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- Kim W, Jang YG, Yang J, and Chung J (2017). Spatial Activation of TORC1 Is Regulated by Hedgehog and E2F1 Signaling in the Drosophila Eye. Dev. Cell 42, 363–375.e4. [DOI] [PubMed] [Google Scholar]
- Lee C, and Longo V (2016). Dietary restriction with and without caloric restriction for healthy aging. F1000Res. 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, and Williams DW (2012). An interaction screen identifies head-case as a regulator of large-scale pruning. J. Neurosci 32, 17086–17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, and Dillin A (2008). Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem 77, 727–754. [DOI] [PubMed] [Google Scholar]
- Makino N, Yamato T, Inoue H, Furukawa T, Abe T, Yokoyama T, Yatsuoka T, Fukushige S, Orikasa S, Takahashi T, and Horii A (2001). Isolation and characterization of the human gene homologous to the Drosophila headcase (hdc) gene in chromosome bands 6q23-q24, a region of common deletion in human pancreatic cancer. DNA Seq. 11, 547–553. [DOI] [PubMed] [Google Scholar]
- Marsh J, Mukherjee P, and Seyfried TN (2008). Akt-dependent proapoptotic effects of dietary restriction on late-stage management of a phosphatase and tensin homologue/tuberous sclerosis complex 2-deficient mouse astrocytoma. Clin. Cancer Res 14, 7751–7762. [DOI] [PubMed] [Google Scholar]
- Meynet O, and Ricci JE (2014). Caloric restriction and cancer: molecular mechanisms and clinical implications. Trends Mol. Med 20, 419–427. [DOI] [PubMed] [Google Scholar]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, and Sonenberg N (2001). The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signaling and cell growth in Drosophila. Nat. Cell Biol 3, 596–601. [DOI] [PubMed] [Google Scholar]
- Mohler J, Weiss N, Murli S, Mohammadi S, Vani K, Vasilakis G, Song CH, Epstein A, Kuang T, English J, et al. (1992). The embryonically active gene, unkempt, of Drosophila encodes a Cys3His finger protein. Genetics 131, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Abate LE, and Seyfried TN (2004). Antiangiogenic and proapoptotic effects of dietary restriction on experimental mouse and human brain tumors. Clin. Cancer Res 10, 5622–5629. [DOI] [PubMed] [Google Scholar]
- Mulrooney TJ, Marsh J, Urits I, Seyfried TN, and Mukherjee P (2011). Influence of caloric restriction on constitutive expression of NF-κB in an experimental mouse astrocytoma. PLoS ONE 6, e18085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, and Edgar BA (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, and Dickson BJ (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851–860. [DOI] [PubMed] [Google Scholar]
- Nowak K, Seisenbacher G, Hafen E, and Stocker H (2013). Nutrient restriction enhances the proliferative potential of cells lacking the tumor suppressor PTEN in mitotic tissues. eLife 2, e00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak K, Gupta A, and Stocker H (2018). FoxO restricts growth and differentiation of cells with elevated TORC1 activity under nutrient restriction. PLoS Genet. 14, e1007347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, and Hafen E (2000). Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14, 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. (2004). Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet 36, 288–292. [DOI] [PubMed] [Google Scholar]
- Piper MD, Partridge L, Raubenheimer D, and Simpson SJ (2011). Dietary restriction and aging: a unifying perspective. Cell Metab. 14, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Pozuelo J, Demetriades C, Schroeder P, and Teleman AA (2017). CycD/Cdk4 and Discontinuities in Dpp Signaling Activate TORC1 in the Drosophila Wing Disc. Dev. Cell 42, 376–387.e5. [DOI] [PubMed] [Google Scholar]
- Ruggeri BA, Klurfeld DM, Kritchevsky D, and Furlanetto RW (1989). Caloric restriction and 7,12-dimethylbenz(a)anthracene-induced mammary tumor growth in rats: alterations in circulating insulin, insulin-like growth factors I and II, and epidermal growth factor. Cancer Res. 49, 4130–4134. [PubMed] [Google Scholar]
- Sonntag WE, Lynch CD, Cefalu WT, Ingram RL, Bennett SA, Thornton PL, and Khan AS (1999). Pleiotropic effects of growth hormone and insulin-like growth factor (IGF)-1 on biological aging: inferences from moderate caloric-restricted animals. J. Gerontol. A Biol. Sci. Med. Sci 54, B521–B538. [DOI] [PubMed] [Google Scholar]
- Steneberg P, and Samakovlis C (2001). A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneberg P, Englund C, Kronhamn J, Weaver TA, and Samakovlis C (1998). Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the Drosophila trachea. Genes Dev. 12, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Berry CO, Zilberman F, Thomas G, and Kozma SC (1996). The Drosophila p70s6k homolog exhibits conserved regulatory elements and rapamycin sensitivity. Proc. Natl. Acad. Sci. USA 93, 10791–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiebe M, Lutz M, De La Garza A, Buechling T, Boutros M, and Teleman AA (2015). REPTOR and REPTOR-BP Regulate Organismal Metabolism and Transcription Downstream of TORC1. Dev. Cell 33, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver TA, and White RA (1995). headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development 121, 4149–4160. [DOI] [PubMed] [Google Scholar]
- Yu J, and Pan D (2018). Validating upstream regulators of Yorkie activity in Hippo signaling through scalloped-based genetic epistasis. Development 145, dev157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, and Pan D (2010). Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2009). Genetic and Biochemical Characterization of the TSC-Rheb-TOR Signaling Pathway in Drosophila. Doctoral thesis (Johns Hopkins University). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.