Abstract
Background
We previously isolated a Klebsiella pneumoniae strain, NTUH-K2044, from a community-acquired pyogenic liver abscess (PLA) patient. Analysis of the NTUH-K2044 genome revealed that this strain harbors 2 putative type VI secretion system (T6SS)-encoding gene clusters.
Methods
The distribution of T6SS genes in the PLA and intestinal-colonizing K pneumoniae clinical isolates was examined. icmF1-, icmF2-, icmF1/icmF2-, and hcp-deficient K pneumoniae strains were constructed using an unmarked deletion method. The roles of T6SSs in antibacterial activity, type-1 fimbriae expression, cell adhesion, and invasion and intestinal colonization were determined.
Results
The prevalence of T6SSs is higher in the PLA strains than in the intestinal-colonizing strains (37 of 42 vs 54 of 130). Deletion of icmF1/icmF2 and hcp genes significantly reduced interbacterial and intrabacterial killing. Strain deleted for icmF1 and icmF2 exhibited decreased transcriptional expression of type-1 fimbriae and reduced adherence to and invasion of human colorectal epithelial cells and was attenuated for in vivo competition to enable colonization of the host gut. Finally, Hcp expression in K pneumoniae was silenced by the histone-like nucleoid structuring protein via direct binding.
Conclusions
These results provide new insights into T6SS-mediated bacterial competition and attachment in K pneumoniae and could facilitate the prevention of K pneumoniae infection.
Keywords: H-NS protein, intestinal colonization, Klebsiella pneumoniae, type-1 fimbriae, type VI secretion system (T6SS)
Community-acquired pyogenic liver abscess (PLA) caused by Klebsiella pneumoniae has recently emerged [1, 2]. For patients with K pneumoniae PLA, the mortality rate is 10%, and for patients who also have metastatic meningitis, the mortality rate is 30%–40% [3, 4]. Therefore, early diagnosis and appropriate therapy are critical.
Many Gram-negative bacteria encode a molecular machine called the type VI secretion system (T6SS), which appears to function in bacterial pathogenesis as a nano-syringe that translocates effector proteins into eukaryotic and prokaryotic target cells [5–7]. For some enteric pathogens, such as Salmonella Typhimurium and Vibrio cholerae, T6SSs are required for overcoming microbiota-mediated colonization resistance, leading to successful host infection [8, 9].
Intracellular multiplication F (IcmF) family proteins are conserved integral inner membrane proteins of T6SS that are involved in effector proteins delivery into target cells [10]. Although the exact functions of most T6SS proteins are unknown, many are not secreted but are required for the secretion of T6SS substrates, such as hemolysin-coregulated protein (Hcp) and valine-glycine repeat protein (VgrG) [11, 12]. Some evidence suggests the existence of an organized T6SS mechanism that likely accounts for the host-pathogen interaction of K pneumoniae. Lawlor et al [13] demonstrated that K pneumoniae strains with mutations in the core proteins of the T6SS showed decreased ability to infect mouse spleen. Klebsiella pneumoniae Kp52.145, which harbors a deletion in a gene encoding a phospholipase D family protein (PLD1) located within a T6SS locus, was avirulent in a mouse pneumonia model [14].
Type-1 fimbriae are produced by many strains of K pneumoniae and are involved in adhesion to eukaryotic cells [15]. We have shown that a PLA K pneumoniae NTUH-K2044 strain expressed type-1 fimbriae-encoding genes when cultivated in Luria-Bertani (LB) broth with shaking. In addition, inversion of a putative invertible deoxyribonucleic acid (DNA) element containing the fimA promoter regulates the expression of type-1 fimbriae-encoding genes in K pneumoniae [16, 17].
The histone-like nucleoid-structuring protein (H-NS) is a DNA-binding protein and functions as a silencer by directly binding to target AT-rich DNA [18]. Previous studies have noted that T6SS gene clusters are tightly regulated in several Gram-negative pathogens [19], and H-NS has been reported to be a negative regulator of T6SS gene transcription [20–22].
In this study, we (1) examined the distribution of T6SS genes in PLA and intestinal-colonizing K pneumoniae clinical isolates, (2) investigated the role of T6SSs in interbacterial and intrabacterial competition, (3) demonstrated the roles of T6SSs in type-1 fimbriae expression, cell adhesion, and invasion, (4) explored the function of T6SSs in intestinal colonization of mice, and (5) confirmed that H-NS functions as a silencer of T6SS expression in K pneumoniae.
METHODS
Ethics Statement
All animal procedures were approved (application number 20150487) by the Institutional Animal Care and Use Committee of National Taiwan University College of Medicine. All animal procedures were conducted in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and Taiwan’s Animal Protection Act.
Bacterial Strains and Culture Conditions
A total of 42 clinical PLA K pneumoniae isolates and 130 intestinal-colonizing strains were collected as described previously [23, 24]. The bacterial strains, plasmids, and primers used in this study are listed in Supplementary Table S1.
Type VI Secretion System Genetic Loci Determination
To identify the T6SS genetic loci in K pneumoniae, polymerase chain reaction (PCR) was performed using primer pairs designed specifically for hcp, icmF1, and icmF2. Polymerase chain reaction was performed as described previously [25].
Gene Deletion and Complementation
Klebsiella pneumoniae strains with hcp, icmF1, and icmF2 single gene deletions, the icmF1/icmF2 double gene deletion, and the hns/icmF1/icmF2 triple gene deletion were constructed using a previously described unmarked deletion method [23]. For complementation, hcp, icmF2, and icmF1 genes were amplified by PCR and cloned into the pGEM-T or pACYC184 plasmid, respectively. These resulting plasmids were transformed into their corresponding deletion mutants by electroporation. All of the deletion mutants and complementation strains were confirmed by PCR and sequencing.
Interbacterial and Intrabacterial Competitive Growth Assays
Cultures of predator and prey strains were mixed together at a multiplicity of infection of 10:1, and 25 mL of the mixed bacterial culture were spotted onto LB agar for 5 hours. Bacterial spots were harvested, and the colony-forming units (CFU) per milliliter of surviving prey and predator strains were measured by plating serial dilutions on selective agar. The output/input ratio of the prey to predator strains was interpreted as survival.
Adhesion and Invasion Assays
Human colorectal epithelial Caco-2 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum and 1% nonessential amino acids (Gibco/BRL). Adhesion and invasion assays were performed as described previously [17, 26].
Fimbrial Switch Orientation Assays
A modification of a previously described method was used to determine the orientation of the K pneumoniae fim switch [17, 27].
Mouse Inoculation Experiments
For in vivo competition, the mice experiments were followed according to previously described methods [28]. The output/input ratio of the test/virulent strains, the competitive index (CI) was interpreted as colonization ability. To investigate the contribution of T6SSs in the intestinal colonization and systemic dissemination, mice were infected by intragastric inoculation (3 × 106 CFU each) of the wild-type or the icmF1/icmF2 mutant strain. Surviving animals were sacrificed on the second day or the sixth day after challenge; organ homogenates (including liver, spleen, and intestine) were cultured for quantification of CFU.
Reverse-Transcription Polymearase Chain Reaction Analysis
To identify the transcriptional unit of T6SS locus I, total ribonucleic acid (RNA) was extracted from NTUH-K2044 [23], complementary DNA was prepared, and PCR was performed as described previously [25].
Quantitative Real-Time Reverse-Transcription Polymearase Chain Reaction Analysis
To measure the expression of the T6SS locus, total RNA from cultures of K pneumoniae strains was extracted, and reverse-transcription PCR was performed as described previously [28].
Gel Retardation Assays
The DNA fragments of the T6SS locus I were generated by PCR from NTUH-K2044 chromosomal DNA. The PCR fragments were purified, and 200 ng of DNA was incubated with 1 or 1.5 μg of recombinant H-NS in binding buffer, and these assays were performed as described previously [29]. Supershift assays were conducted by adding anti-His antibodies (Sigma-Aldrich) for 30 minutes at room temperate.
Preparation and Analysis of Proteins in Whole-Cell Extracts and Culture Supernatant
Equal amounts of cell lysates were generated by sonicating the cell pellets (~1 × 109 CFU). Supernatants and secreted proteins were collected as described previously [30] and verified by Western blotting.
Statistical Analyses
Statistical significance was assessed by 1-way analysis of variance or 2-tailed Student’s t tests using Prism 5 (GraphPad) software.
RESULTS
Frequencies of Type VI Secretion System Loci in Klebsiella pneumoniae Clinical Isolates
From the previously sequenced genome of K pneumoniae NTUH-K2044, it was shown that this strain possesses 2 contiguous putative T6SS loci: locus I and locus III [31]. Locus I appears to be complete, because it contains both the putative hcp and vgrG secreted protein-encoding genes as well as icmF, a structural protein-encoding gene, which is designated as icmF1. In locus III, the hcp, impB, and impC genes are missing; however, it does contain putative vgrG and icmF genes, the latter of which is designated as icmF2.
To assess the prevalence of these T6SS loci, we focused on 3 genes: icmF1, icmF2, and hcp. Pyogenic liver abscess and intestinal-colonizing K pneumoniae isolates were examined for the presence of these genes by PCR. The frequency of the icmF1, icmF2, and hcp genes was significantly higher in the PLA isolates (37 of 42, 88.1%) than in the intestinal isolates (54 of 130, 41.5%). icmF1 and icmF2 genes-deficient strains were more prevalent among the intestinal isolates (29 of 130, 22.3%) than those among the PLA isolates (0 of 42, 0%) (Table 1).
Table 1.
Prevalence of T6SS Genetic Loci in PLA-Associated and Intestinal-Colonizing Klebsiella pneumoniae Strains
| T6SS Genetic Loci | ||||||
|---|---|---|---|---|---|---|
| Group | hcp a | icmF1 b | icmF2 b | hcp, icmF1, icmF2 (+)b | hcp, icmF1, icmF2 (−)c | icmF1, icmF2 (−)a |
| PLA | 42 of 42 | 39 of 42 | 40 of 42 | 37 of 42 (88.10%) | 0 of 42 | 0 of 42 |
| Intestinal colonizing | 104 of 130 | 70 of 130 | 86 of 130 | 54 of 130 (41.54%) | 14 of 130 | 29 of 130 |
Abbreviations: PLA, pyogenic liver abscess; T6SS, type VI secretion system.
a P < .01, bP < .001, and cP < .05 by χ2 test.
IcMF1, IcmF2, and Hcp Are Required for Interbacterial and Intrabacterial Killing
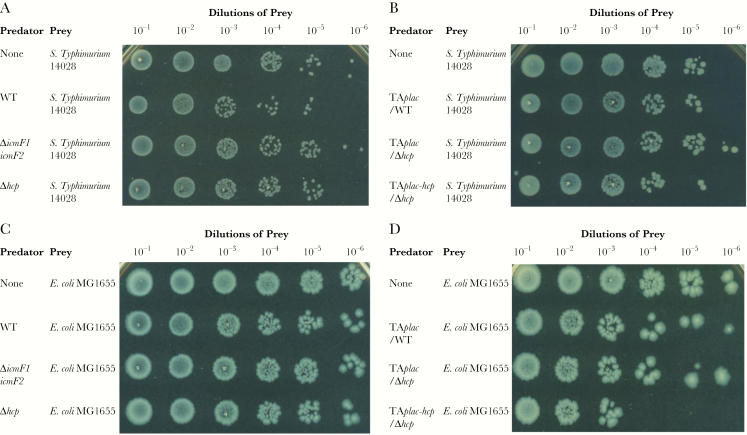
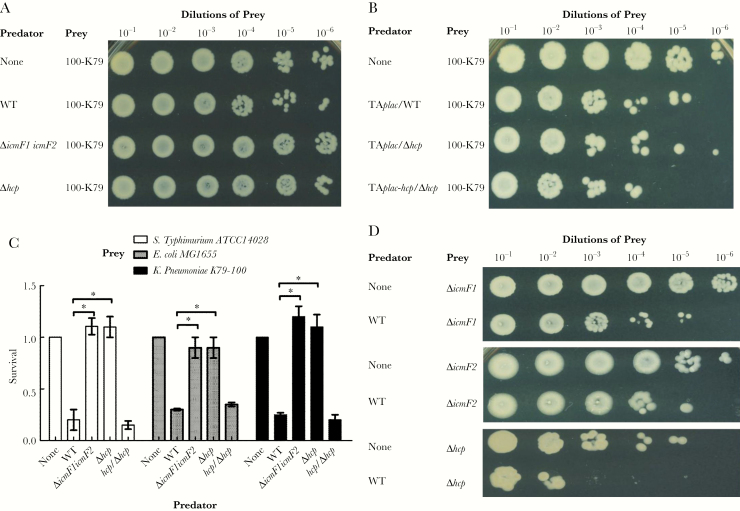
To examine the contributions of the 2 distinct IcmF proteins and the Hcp effector protein toward antibacterial activity, we isolated spontaneous streptomycin-resistant prey and performed in vitro competition assays. Growth of Salmonella Typhimurium 14028 or Escherichia coli MG1655 was significantly reduced (3-fold) in the presence of wild-type NTUH-K2044 when compared with both the icmF1/icmF2 mutant and hcp mutant strains (Figures 1A and C and 2C). The ability of the hcp mutant strain to kill either Salmonella Typhimurium 14028 or E coli MG1655 was restored by complementation with a plasmid containing the hcp gene under a lac promoter (Figure 1B and D). To test whether the T6SSs are involved in intrabacterial competition, 4 spontaneous streptomycin-resistant K pneumoniae strains (a T6SS-null colonization strain [100-K79] and the NTUH-K2044 icmF1, icmF2 and hcp mutants) were used as prey in a competition assay with wild-type NTUH-K2044. We obtained similar results to the previous assay; the wild-type strain was better able to eliminate the T6SS-null intestinal K pneumoniae isolate 100-K79 when compared with the T6SS-related mutant strains. The ability of the hcp mutant strain to kill K pneumoniae 100-K79 was restored by complementation (Figure 2A–C). The growth of the icmF1, icmF2 and hcp mutant strains was significantly reduced in the presence of the wild-type strain by more than 1 log (Figure 2D). In these competition assays, survival of predator strains was similar for each strain (Supplementary Figure S1). Taken together, these results show that K pneumoniae competes against Salmonella Typhimurium, E coli, and T6SS-null K pneumoniae in a T6SS-dependent manner, suggesting that the Hcp-mediated antibacterial activity of T6SSs confers a competitive advantage to K pneumoniae.
Figure 1.
Interspecies killing by Klebsiella pneumoniae NTUH-K2044 in a T6SS-dependent manner. (A–D) Interbacterial competitive growth assays showing that a functional T6SS is essential for interbacterial killing by K pneumoniae NTUH-K2044. Surviving Salmonella Typhimurium 14028 (A) and Escherichia coli MG1655 (C) after 5 hours of coincubation with K pneumoniae NTUH-K2044 wild-type (WT), T6SS-related gene mutants, the hcp mutant carrying a functional copy of hcp on a plasmid (TAplac-hcp), and the wild-type or hcp mutant containing the empty vector (TAplac) (B and D). Recovered mixtures were plated onto Luria-Bertani medium supplemented with 100 μg/mL streptomycin.
Figure 2.
The role of the NTUH-K2044 T6SS in intraspecies and interspecies competitive growth. (A and B) Intrabacterial competitive growth assays showing that a functional T6SS is essential for intrabacterial killing. Surviving Klebsiella pneumoniae 100-K79 (A) after 5 hours of coincubation with various K pneumoniae NTUH-K2044 strains, including wild-type (WT), T6SS-related gene mutant, hcp mutant carrying a functional copy of hcp on a plasmid (TAplac-hcp), and wild-type or hcp mutant containing the empty vector (TAplac) strains (B). (C) Survival rates of the prey strains Salmonella Typhimurium 14028 (white bars), Escherichia coli MG1655 (gray bars), or K pneumoniae 100-K79 (black bars) after 5 hours of coincubation with the predator strains: K pneumoniae NTUH-K2044 wild-type, T6SS-related gene mutants, and hcp complemented strains. The data represent the means of 3 independent trials, and the error bars represent the standard deviations. *,P < .05 by 1-way analysis of variance (compared with the wild-type strain). (D) Survival of the T6SS-related gene mutant strains after 5 hours of coincubation with NTUH-K2044 wild-type. The recovered mixtures were plated on Luria-Bertani medium supplemented with 100 μg/mL streptomycin.
Type VI Secretion Systems in Klebsiella pneumoniae Is Involved in Adhesion to and Invasion of Human Caco-2 Colonic Epithelial Cells
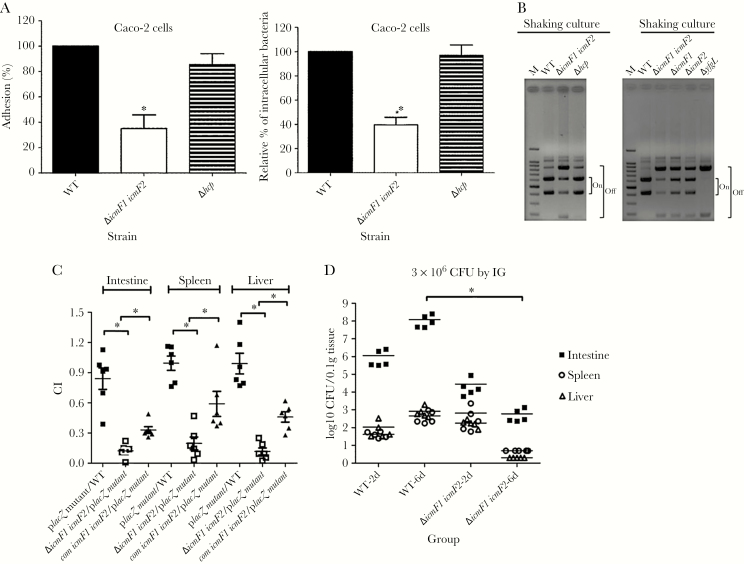
Several studies in E coli and Campylobacter jejuni have demonstrated that T6SSs promote host cell adhesion and invasion, 2 crucial processes required for host colonization [32–34]. To explore the roles of T6SSs in K pneumoniae in these processes, we evaluated the cell adherence and internalization potential of the wild-type and T6SS-related gene deletion strains in Caco-2 cells. The icmF1/icmF2 mutant exhibited a highly significant, 3-fold decrease in adherence to Caco-2 cells. Moreover, the capacity of the icmF1/icmF2 mutant to invade Caco-2 cells was significantly reduced, by approximately 60%, compared with that of the wild-type strain. In contrast, the adherence and invasion capacity of the hcp mutant strain were similar to those of the wild-type strain (Figure 3A). These results suggest that the T6SSs machinery contributes to K pneumoniae adherence to and invasion of Caco-2 cells.
Figure 3.
The function of the T6SS of Klebsiella pneumoniae in adhesion to and invasion of Caco-2 cells, type-1 fimbriae production, and in vivo colonization. (A) Adhesion and invasion of Caco-2 cells by the K pneumoniae NTUH-K2044 wild-type (WT) and derived T6SS-related gene mutant strains. The adhesion rate is expressed as the proportion of the NTUH-K2044 wild-type strain that adhered. The invasion rate is expressed as the proportion of the NTUH-K2044 wild-type strain that invaded. The data represent the means of 3 independent trials, and the error bars represent the standard deviations (SDs). *, P < .05 by 1-way analysis of variance (compared with the wild-type strain). (B) Orientation of the fim phase-switch element in the NTUH-K2044 and T6SS-related gene mutant strains after overnight culture in Luria-Bertani broth with shaking. Depending on the orientation of the fim invertible element, paired fragments of different sizes are amplified (613 and 404 base pairs [bp] when in the “on” orientation, and 840 and 177 bp when in the “off” orientation). Lane M contains the deoxyribonucleic acid molecular size marker. The yfgL-deletion mutant was included as a negative control and showed downregulated type-1 fimbriae expression. (C) Survival of the K pneumoniae icmF1/icmF2 mutant and its complementation strains in the in vivo bacterial competition assay against a fully virulent placZ mutant. The icmF1/icmF2 mutant or its complementation strain and the fully virulent placZ-deletion mutant were mixed at a 1:1 ratio (1 × 106 colony-forming units [CFU] for each mice of the wild-type group and icmF1/icmF2 mutant group; 1 × 107 CFU for each mice of the icmF1/icmF2 complementation group) and inoculated intragastrically into 5-week-old BALB/c mice (6 mice for each group). The ratio of lacZ+/lacZ−K pneumoniae in the spleen, liver, and intestine was determined from a single mouse when it either died or was sacrificed on the seventh day after infection. Each symbol represents the competitive index (CI) for each inoculum; and the medians and SDs of the values are shown (closed squares for the placZ mutant group vs the wild-type group, the CI of intestine, spleen, and liver were 0.841 ± 0.261, 0.995 ± 0.175, and 0.990 ± 0.252, respectively; open squares for the icmF1/icmF2 mutant vs placZ mutant, the CI of the intestine, spleen, and liver were 0.123 ± 0.069, 0.197 ± 0.155, and 0.118 ± 0.085, respectively; P = .031, compared with the wild-type group, Wilcoxon’s signed-rank test; closed triangles for the icmF1/icmF2 complementation strain vs placZ mutant, the CI of the intestine, spleen, and liver were 0.330 ± 0.082, 0.590 ± 0.305, and 0.460 ± 0.125, respectively; P = .031, compared with the icmF1/icmF2 mutant group, Wilcoxon’s signed-rank test). (D) Five-week-old BALB/c mice (5 mice for each group) were inoculated by intragastric inoculation with equivalent doses (3 × 106 CFU) of the NTUH-K2044 wild type or its isogenic icmF1/icmF2 mutant strain. Surviving animals were sacrificed on the second day or sixth day after challenge. Bacterial loads were measured in the liver, spleen, and intestine. Log10 CFU was standardized per 0.1 gram wet organ weight. The medians and SDs of the values are shown; *, P < .05 by Wilcoxon’s signed-rank test.
Transcriptional Regulation of the Klebsiella pneumoniae fim Gene Cluster by the Type VI Secretion System
A previous study reported that 2 T6SS genes, hcp and clpV, influence the expression of type-1 fimbriae and contribute to the pathogenesis of avian-pathogenic E coli [35]; therefore, we compared the fim phase switching of our T6SS-related gene deletion strains with that of the wild-type strain in a fimbrial switch orientation assay. When either icmF1 or icmF2 was deleted, it appeared that there was as much in the “off” orientation as in the “on” orientation, indicating that type-1 fimbriae expression was downregulated in both the icmF1 and icmF2 mutants (Figure 3B). Compared with the wild-type strain, more DNA fragments corresponding to the off orientation were detected in the icmF1/icmF2 mutant, indicating a significant reduction in type-1 fimbriae expression in this mutant. In contrast, more DNA fragments corresponding to the on orientation were detected in the wild-type and hcp mutant strains. We obtained similar results in the mannose-sensitive yeast agglutination (MSYA) assay. Consistent with the fim transcription results, the MSYA assays demonstrated that the icmF1 and icmF2 mutants synthesized 8-fold lower titers of type-1 fimbriae than the wild-type strain (Supplementary Table S2). Likewise, the icmF1/icmF2 mutant synthesized 16-fold lower titers of type-1 fimbriae than the wild-type parent strain. In contrast, the level of type-1 fimbriae expressed in the hcp mutant was similar to that of the wild-type strain. Therefore, the attenuated adherence of the icmF1/icmF2 mutant might partly be due to a reduction in the production of type-1 fimbriae.
Klebsiella pneumoniae Type VI Secretion System Is Important for Bacterial Colonization and Dissemination In Vivo During Infection
In a previous study, we generated an isogenic lacZ mutant with a promoter deletion, which was used as the wild-type strain in a competition assay [28]. Using the same assay, the icmF1/icmF2 mutant strain showed a lower competitive index than the wild-type or placZ mutant. Complementation of the icmF1/icmF2 mutant with 2 plasmids containing the icmF1 and icmF2 genes partially restored the ability for in vivo competition (P < .05) (Figure 3C). To determine the kinetics of the gastrointestinal colonization and systemic dissemination, we examined the bacterial load in mice challenged with equivalent doses (3 × 106 CFU) of the wild-type and the icmF1/icmF2 mutant strains. The second day after bacterial inoculation, bacterial loads in mice infected with icmF1/icmF2 mutant compared with those in mice infected with the wild-type strain showed no significant difference. When organs were examined in surviving animals on the sixth day after bacterial inoculation, the mice infected with the icmF1/icmF2 yielded significantly fewer colony counts in the intestine, spleen, and liver compared with those of the wild-type (P < .05) (Figure 3D). These data demonstrate that mutants impaired for T6SSs have decreased virulence, with reduced the intestinal colonization and decreased bacterial dissemination.
Type VI Secretion System Locus I Transcriptional Unit in Klebsiella pneumoniae NTUH-K2044
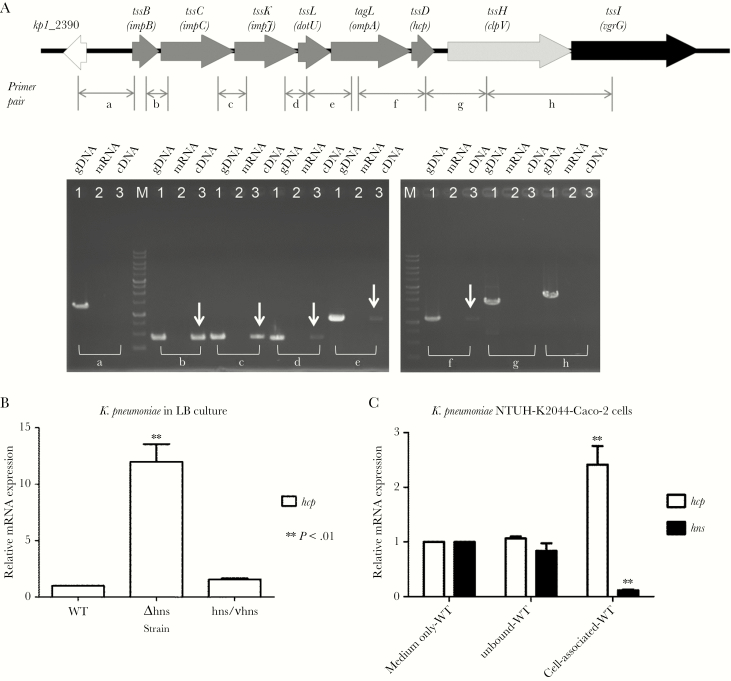
Reverse-transcription PCR to determine the transcriptional unit of the T6SS locus I was performed with total RNA from the NTUH-K2044 strain as template and several primer pairs (a–h) that hybridize within 2 consecutive open reading frames (ORFs). In the analysis, positive results were obtained for pairs b, c, d, e, and f (Figure 4A), suggesting that 6 ORFs (impB, impC, impJ, dotU, ompA, and hcp) formed a single transcriptional unit, whereas clpV and vgrG1 were transcribed independently.
Figure 4.
Histone-like nucleoid-structuring protein (H-NS) represses the transcription of the type VI secretion gene clusters. (A) Schematic representation of T6SS locus I in Klebsiella pneumoniae NTUH-K2044 (top). The arrow heads indicate the direction of transcription. The primer pairs (a–h) used to amplify the junctions between the open reading frames by reverse-transcription polymerase chain reaction (RT-PCR) are shown. The bottom part of the panel shows the RT-PCR products obtained in an ethidium bromide-stained agarose gel, with genomic deoxyribonucleic acid (DNA) as a positive control. The assay described above was repeated 3 times, and representative gels are shown. (B and C) Downregulation of T6SS gene transcription levels by H-NS. (B) Transcription levels of hcp were measured in the NTUH-K2044 wild-type (WT) and isogenic hns deletion mutant strains after culture in Luria-Bertani broth with shaking by quantitative RT-PCR (qRT-PCR). (C) Transcription levels of hcp and hns were measured in the NTUH-K2044 wild-type strain during coculturing with Caco-2 cells by qRT-PCR. The data represent the means of 3 independent trials, and the error bars represent the standard deviations. **, P < .01 by 1-way analysis of variance (compared with the wild-type strain cultured in medium only or the unbound wild-type strain).
Histone-Like Nucleoid-Structuring Silences the Transcriptional Expression of Type VI Secretion System Locus I in NTUH-K2044
We sought to determine whether an hns deletion impacts the transcriptional expression of the T6SS loci in the NTUH-K2044 strain. Deletion of hns resulted in increased hcp gene transcription when compared with that of the parental strain when the strain was cultivated in LB broth (Figure 4B). To determine whether bacteria-host interaction affects the transcriptional expression of hns and hcp in K pneumoniae, we measured expression with or without Caco-2 cells. hns transcript levels in the wild-type strain were lower (<10%) during coculture with Caco-2 cells than during culture in medium only without Caco-2 cells (Figure 4C). In contrast, hcp transcript levels were higher (approximately 2-fold) when cultured with Caco-2 cells, than when this strain was cultured in medium only. These results demonstrate that H-NS silences the transcriptional expression of hcp in K pneumoniae.
Binding of Histone-Like Nucleoid Structuring to the Type VI Secretion System Locus I in NTUH-K2044
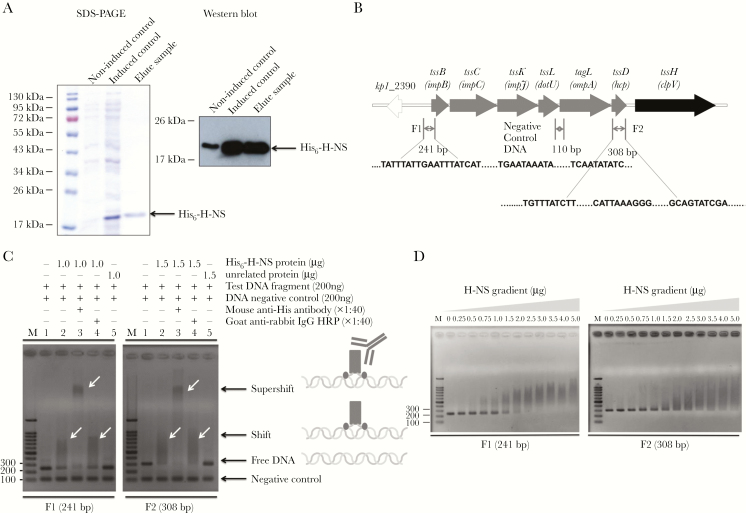
In silico analyses using Virtual Footprint software [36] predicted a large number of putative H-NS binding sites within the NTUH-K2044 T6SS loci distributed throughout the promoter region and within the coding sequences. To test whether H-NS binds to T6SS locus I in vitro, a His-tagged version of the Hcp protein of NTUH-K2044 was expressed and purified (Figure 5A). We then selected 2 DNA fragments that bear putative H-NS binding sites, including a fragment encompassing the partial promoter region of T6SS locus I (F1) and a fragment within the hcp gene (F2), as well as a control fragment within the junction of the dotU and ompA genes, for which no H-NS binding site is predicted (Figure 5B). These 3 fragments were PCR amplified and used in gel retardation assays with purified NTUH-K2044 H-NS. Figure 5C showed that the F1 and F2 fragments were retarded in the presence of H-NS, whereas the negative control fragment was not retarded. Image analyses of the shifts estimated that H-NS binds to fragments F1 and F2 in a dose-dependent manner (Figure 5D). Therefore, we concluded that H-NS specifically binds to different regions in K pneumoniae NTUH-K2044 T6SS locus I.
Figure 5.
Histone-like nucleoid-structuring protein (H-NS) binds to several regions in the T6SS locus I of NTUH-K2044. (A) Purity of the recombinant NTUH-K2044 H-NS protein. The purified H-NS protein was separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining, and the molecular weight is indicated beside the protein markers. Western blot showing the purified recombinant Hcp protein separated by SDS-PAGE, transferred to a nitrocellulose membrane, and detected using a mouse anti-His antibody (1:5000) and rabbit antimouse immunoglobulin G (IgG)-horseradish peroxidase ([HRP] 1:10000). (B) Schematic representation of T6SS locus I of NTUH-K2044. Gel retardation assays using 2 deoxyribonucleic acid (DNA) fragments corresponding to regions of the T6SS gene cluster (F1 and F2). The 110 base-pair (bp) fragment corresponds to a negative control (no H-NS binding site predicted). (C) The different substrates used in the gel retardation assays are represented. Binding reactions (indicated at the top of each lane) were resolved in a 1.5% agarose gels at 100 V. Deoxyribonucleic acid was detected after staining with ethidium bromide. Free DNA and DNA-protein complexes are indicated. (D) The DNA fragments were mixed with increasing concentrations of purified the NTUH-K2044 H-NS (0–5 μg). Binding reactions using increasing concentrations of H-NS (indicated at the top of each lane) were resolved in a 1.5% agarose gel. The assays described above were repeated 3 times, and representative gels are shown.
Histone-Like Nucleoid-Structuring Represses Hcp Expression and Secretion in NTUH-K2044
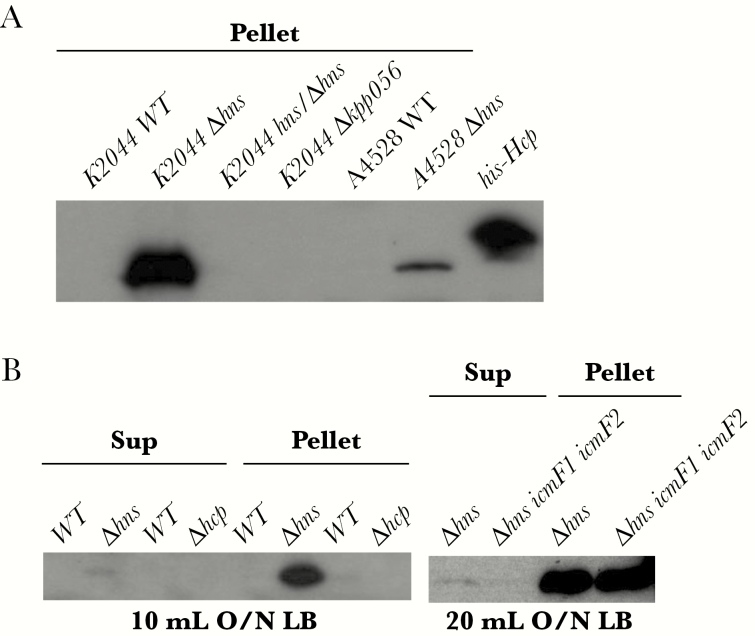
Finally, we tested the effects of an hns deletion on the expression and secretion of Hcp. The Hcp protein was absent from lysates prepared from wild-type NTUH-K2044 cells. Deletion of hns resulted in increased expression of Hcp compared with that in the parental strain, whereas deletion of another hns homolog, kpp056, did not increase expression. Consistently, we obtained similar results with another PLA-associated K pneumoniae K2 strain, NTUH-A4528 [37] (Figure 6A). A slight increase in Hcp protein levels was observed in the culture supernatant of the hns mutant compared with the levels in the wild-type NTUH-K2044, indicating that it is unable to secrete the T6SS substrate Hcp, even in the absence of the repressor H-NS (Figure 6B). These results suggest that Hcp expression is repressed by H-NS in K pneumoniae.
Figure 6.
Effects of Klebsiella pneumoniae histone-like nucleoid-structuring protein (H-NS) protein on Hcp expression. (A) Western blot analysis to compare the expression of Hcp protein in whole-cell lysates of different K pneumoniae strains (as indicated). Hcp protein was expressed in the hns-deletion mutants. Anti-Hcp antisera were obtained from rabbits immunized with the recombinant Hcp protein (1:100000; LTK BioLaboratories) and used as the primary antibodies, and rabbit horseradish peroxidase-conjugated antigoat immunoglobulin G antibody (1:10000; Jackson ImmunoResearch) was used as the secondary antibody. Blots were visualized and developed with an enhanced chemiluminescence system (Amersham Biosciences). (B) Western blot analysis to compare the expression of Hcp protein in supernatants (Sup) and whole-cell lysates of different K pneumoniae strains (as indicated). Hcp expression was repressed by H-NS in K pneumoniae. LB, Luria-Bertani; WT, wild type.
DISCUSSION
Type VI secretion system has been found in more than 25% of all sequenced Gram-negative bacteria [38]. In a previous study, we demonstrated that PLA-related strains have a pathogenesis-specific genotype and transcriptional profile [23, 39]. Strains of the K1 and K2 capsular types have been identified as the predominant virulent types and are prevalent among K pneumoniae PLA isolates [2, 40]. Transcellular translocation is exploited by K pneumoniae to migrate from the gut flora into other tissues, resulting in systemic infections [41]. Recent studies have also demonstrated that several pathogenic bacteria use a T6SS to interact with and compete against bacterial competitors [8, 42–44]. It has been speculated that these pathogens have evolved mechanisms to kill bacterial competitors and establish infection. In this study, we demonstrated the higher prevalence of T6SSs in the PLA strains when compared with that in the intestinal-colonizing isolates (88.1% vs 41.5%), and we showed that deletion of icmF1 and icmF2 in a PLA K pneumoniae strain reduced interspecies and intraspecies antibacterial competitiveness in vitro. Therefore, our findings suggest that K pneumoniae outcompetes Salmonella Typhimurium and E coli in a T6SS-dependent manner and that T6SSs may aid bacterial competition in PLA strains.
IcmF is necessary for the efficient functioning of T6SS in Legionella pneumophila, and a protein homolog is required for Hcp secretion from Agrobacterium tumefaciens [45, 46]. Comparison of the amino acid sequences of the 2 IcmF proteins from NTUH-K2044 by using Basic Local Alignment Search Tool (BLAST) revealed 41.1% sequence identity. Analysis using the Simple Modular Architecture Research tool (SMART) showed that these 2 predicted IcmF proteins belong to the IcmF-related protein family and contain 2 conserved Pfam domains. Wild-type NTUH-K2044 showed intrabacterial killing ability and could compete against the icmF1, icmF2, and hcp mutant strains. In addition, compared with the icmF1 or icmF2 mutant, a significant reduction in type-1 fimbriae expression was observed in the icmF1/icmF2 mutant. Thus, we concluded that these 2 IcmF proteins are important for a fully functioning T6SS in NTUH-K2044. Hcp has been proposed as a core component and hallmark secreted protein of T6SS [11]. The Hcp protein from NTUH-K2044 was predicted to be a T6SS effector Hcp1 family protein, and analysis of the amino acid sequence of this protein revealed high sequence identity (99%–100%) to Hcp proteins in other Klebsiella spp and Enterobacteriaceae. When the suppressor H-NS was deleted from NTUH-K2044, Hcp was expressed. Therefore, we suggest that Hcp is a hallmark secreted protein of T6SSs in K pneumoniae.
Bacterial adherence to epithelial cell surfaces is believed to be an important first step in the initiation of infection. Our recent study demonstrated that type-1 fimbriae are involved in the adherence of PLA K pneumoniae [17]. We demonstrated here that deletion of icmF1 and icmF2 in a PLA K pneumoniae also led to decreased expression of fim transcripts. Hence, T6SSs mediate in the transcriptional expression of type-1 fimbriae and are essential for adhesion to and invasion of host cells in PLA K pneumoniae.
Upon in vivo infection, the competition index of the icmF1/icmF2 mutant was reduced when compared with the parental strain. Moreover, the ability for the intestinal colonization and systemic dissemination of the icmF1/icmF2 mutant was attenuated during the first 6 days of infection. These findings indicate that deletion of the K pneumoniae genes encoding the T6SS biosynthetic machinery results in decreased bacterial colonization and restricted bacterial dissemination into distant organs. We conclude that T6SSs confer a competitive advantage to PLA K pneumoniae strains against the gut microbiota and facilitates the establishment of infection in the host tissues, thus enhancing pathogenicity.
The H-NS functions as an architectural component of the nucleoid and a pleiotropic regulator of gene expression [47, 48]. In a recent study, Ares et al [49] demonstrated that the H-NS nucleoid protein is an activator of type 3 fimbriae and a repressor of capsular polysaccharide expression in K pneumoniae. In this study, we showed that transcript levels of hcp were downregulated by H-NS and that this global regulator could interact directly with the promoter region and the coding sequences of T6SS locus I in K pneumoniae. These findings indicate that H-NS functions as a repressor of the T6SS secretory protein Hcp in K pneumoniae.
CONCLUSIONS
In conclusion, we report the following in this study: the prevalence of T6SSs is higher in the PLA K pneumoniae strains than in the intestinal-colonizing strains; T6SS antibacterial activity is essential for K pneumoniae survival and competition against host microbiota; T6SS-mediated type-1 fimbriae expression, cell adherence, invasion, and subsequent in vivo colonization are critical for establishing an infection; and H-NS functions as a silencer of T6SS expression in K pneumoniae. This report demonstrates that the antibacterial activity and cell invasiveness conferred by T6SS is important for K pneumoniae to establish itself within the host gut, thereby opening new and exciting research perspectives.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Author contributions. P.-F. H. and J.-T. W. conceived and designed the experiments. P.- F. H., Y.-R. L., T.-L. L., and L.-Y. L. performed the experiments. P.- F. H., Y.-R. L., and L.-Y. L. analyzed the data. P.-F. H. and J.-T. W. wrote the paper.
Financial support. This work was funded by the Ministry of Science and Technology, National Taiwan University, Academia Sinica, National Taiwan University Hospital, and the Liver Disease Prevention and Treatment Research Foundation of Taiwan.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chiu CT, Lin DY, Liaw YF. Metastatic septic endophthalmitis in pyogenic liver abscess. J Clin Gastroenterol 1988; 10:524–7. [DOI] [PubMed] [Google Scholar]
- 2. Tsai FC, Huang YT, Chang LY, Wang JT. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 2008; 14:1592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med 1991; 151:1557–9. [PubMed] [Google Scholar]
- 4. Fung CP, Chang FY, Lee SC, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis?Gut 2002; 50:420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol 2008; 11:3–8. [DOI] [PubMed] [Google Scholar]
- 6. Cascales E. The type VI secretion toolkit. EMBO Rep 2008; 9:735–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 2008; 154:1570–83. [DOI] [PubMed] [Google Scholar]
- 8. Sana TG, Flaugnatti N, Lugo KA, et al. Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 2016; 113:E5044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng AT, Ottemann KM, Yildiz FH. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 2015; 11:e1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sexton JA, Miller JL, Yoneda A, Kehl-Fie TE, Vogel JP. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun 2004; 72:5983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 2013; 8:e61086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol 2010; 192:155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005; 58:1054–73. [DOI] [PubMed] [Google Scholar]
- 14. Lery LM, Frangeul L, Tomas A, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 2014; 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tarkkanen AM, Allen BL, Williams PH, et al. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect Immun 1992; 60:1187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76:4055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsieh PF, Hsu CR, Chen CT, Lin TL, Wang JT. The Klebsiella pneumoniae YfgL (BamB) lipoprotein contributes to outer membrane protein biogenesis, type-1 fimbriae expression, anti-phagocytosis, and in vivo virulence. Virulence 2016; 7:587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ali SS, Xia B, Liu J, Navarre WW. Silencing of foreign DNA in bacteria. Curr Opin Microbiol 2012; 15:175–81. [DOI] [PubMed] [Google Scholar]
- 19. Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 2012; 66:453–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salomon D, Klimko JA, Orth K. H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology 2014; 160:1867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J, Xiao J, Zhang Y, et al. A new target for the old regulator: H-NS suppress T6SS secretory protein EvpP, the major virulence factor in the fish pathogen Edwardsiella tarda. Lett Appl Microbiol 2014; 59:557–64. [DOI] [PubMed] [Google Scholar]
- 22. Brunet YR, Khodr A, Logger L, et al. H-NS silencing of the Salmonella pathogenicity island 6-encoded type VI secretion system limits Salmonella enterica serovar Typhimurium interbacterial killing. Infect Immun 2015; 83:2738–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2008; 197:1717–27. [DOI] [PubMed] [Google Scholar]
- 24. Lin TL, Pan YJ, Hsieh PF, Hsu CR, Wu MC, Wang JT. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep 2016; 6:31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2006; 193:645–54. [DOI] [PubMed] [Google Scholar]
- 26. Sahly H, Podschun R, Oelschlaeger TA, et al. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun 2000; 68:6744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Struve C, Forestier C, Krogfelt KA. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 2003; 149:167–76. [DOI] [PubMed] [Google Scholar]
- 28. Hsieh PF, Lin HH, Lin TL, Wang JT. CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J Infect Dis 2010; 202:52–64. [DOI] [PubMed] [Google Scholar]
- 29. Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc 2007; 2:1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A 2013; 110:2623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarris PF, Zoumadakis C, Panopoulos NJ, Scoulica EV. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect Genet Evol 2011; 11:157–66. [DOI] [PubMed] [Google Scholar]
- 32. de Pace F, Boldrin de Paiva J, Nakazato G, et al. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology 2011; 157:2954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Y, Tao J, Yu H, et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun 2012; 80:1243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lertpiriyapong K, Gamazon ER, Feng Y, et al. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 2012; 7:e42842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, da Silveira WD. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun 2010; 78:4990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Münch R, Hiller K, Grote A, et al. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 2005; 21:4187–9. [DOI] [PubMed] [Google Scholar]
- 37. Hsieh PF, Lin TL, Yang FL, et al. Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 2012; 7:e33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources?BMC Genomics 2009; 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chung DR, Lee SS, Lee HR, et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect 2007; 54:578–83. [DOI] [PubMed] [Google Scholar]
- 41. Hsu CR, Pan YJ, Liu JY, Chen CT, Lin TL, Wang JT. Klebsiella pneumoniae translocates across the intestinal epithelium via Rho GTPase- and phosphatidylinositol 3-kinase/Akt-dependent cell invasion. Infect Immun 2015; 83:769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 2013; 152:884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Repizo GD, Gagné S, Foucault-Grunenwald ML, et al. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 2015; 10:e0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwarz S, West TE, Boyer F, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 2010; 6:e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. VanRheenen SM, Duménil G, Isberg RR. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun 2004; 72:5972–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma LS, Lin JS, Lai EM. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 2009; 191:4316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 2003; 11:511–8. [DOI] [PubMed] [Google Scholar]
- 48. Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol 2004; 2:391–400. [DOI] [PubMed] [Google Scholar]
- 49. Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, et al. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol 2016; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.