ABSTRACT
Glucose produced from photosynthesis is a key nutrient signal regulating root meristem activity in plants; however, the underlying mechanisms remain poorly understood. Here, we show that, by modulating reactive oxygen species (ROS) levels, the conserved macroautophagy/autophagy degradation pathway contributes to glucose-regulated root meristem maintenance. In Arabidopsis thaliana roots, a short exposure to elevated glucose temporarily suppresses constitutive autophagosome formation. The autophagy-defective autophagy-related gene (atg) mutants have enhanced tolerance to glucose, established downstream of the glucose sensors, and accumulate less glucose-induced ROS in the root tips. Moreover, the enhanced root meristem activities in the atg mutants are associated with improved auxin gradients and auxin responses. By acting with AT4G39850/ABCD1 (ATP-binding cassette D1; Formerly PXA1/peroxisomal ABC transporter 1), autophagy plays an indispensable role in the glucose-promoted degradation of root peroxisomes, and the atg mutant phenotype is partially rescued by the overexpression of ABCD1. Together, our findings suggest that autophagy is an essential mechanism for glucose-mediated maintenance of the root meristem.
Abbreviation: ABA: abscisic acid; ABCD1: ATP-binding cassette D1; ABO: ABA overly sensitive; AsA: ascorbic acid; ATG: autophagy related; CFP: cyan fluorescent protein; Co-IP: co-immunoprecipitation; DAB: 3’,3’-diaininobenzidine; DCFH-DA: 2’,7’-dichlorodihydrofluorescin diacetate; DR5: a synthetic auxin response element consists of tandem direct repeats of 11 bp that included the auxin-responsive TGTCTC element; DZ: differentiation zone; EZ, elongation zone; GFP, green fluorescent protein; GSH, glutathione; GUS: β-glucuronidase; HXK1: hexokinase 1; H2O2: hydrogen peroxide; IAA: indole-3-acetic acid; IBA: indole-3-butyric acid; KIN10/11: SNF1 kinase homolog 10/11; MDC: monodansylcadaverine; MS: Murashige and Skoog; MZ: meristem zone; NBT: nitroblue tetrazolium; NPA: 1-N-naphtylphthalamic acid; OxIAA: 2-oxindole-3-acetic acid; PIN: PIN-FORMED; PLT: PLETHORA; QC: quiescent center; RGS1: Regulator of G-protein signaling 1; ROS: reactive oxygen species; SCR: SCARECROW; SHR, SHORT-ROOT; SKL: Ser-Lys-Leu; SnRK1: SNF1-related kinase 1; TOR: target of rapamycin; UPB1: UPBEAT1; WOX5: WUSCHEL related homeobox 5; Y2H: yeast two-hybrid; YFP: yellow fluorescent protein
KEYWORDS: Autophagy, glucose, peroxisome, reactive oxygen species, root meristem
Introduction
Glucose produced during photosynthesis functions not only as the fundamental fuel for cellular metabolism in plants, but also as a hormone-like signaling molecule that regulates diverse developmental and physiological activities [1–4]. In plants, 3 distinct pathways have been found to mediate glucose sensing and signal transduction: (1) the hexokinase (HXK)-dependent direct sensing pathway; (2) the glycolysis-dependent indirect sensing pathway; and (3) extracellular sugar sensing by the heterotrimeric G-proteins [1,5–7]. AT4G29130/HXK1 (hexokinase 1) in Arabidopsis thaliana has been the first plant sugar sensor found to play a dual role in glucose metabolism and sensing [8]. Besides HXK1, several other HXKs and HXK-like proteins in Arabidopsis appear to share either overlapping or differing functions in glucose metabolism and signaling [4,8,9]. Intracellular sugar levels can also be perceived by AT3G01090/KIN10 (SNF1 kinase homolog 10, formerly also termed SnRK1.1) and AT3G29160/KIN11 (SNF1 kinase homolog 11), in Arabidopsis, and AT1G50030/TOR (target of rapamycin) [4,10,11]. The TOR kinase system is especially important for sensing nutrient starvation, energy and oxygen deprivation, or other energy-related environmental cues, and the subsequent transduction of this signal through glycolysis and the electron transport chain in the mitochondria [11]. Furthermore, AT3G26090/RGS1 (REGULATOR OF G-PROTEIN SIGNALING 1), a protein with 7 transmembrane domains, acts as a glucose receptor and plays a critical role in the perception of external glucose levels [6,12]. Extensive studies have suggested that either HXK1-dependent or -independent glucose signaling interacts with several phytohormones, such as ethylene, cytokinin, abscisic acid (ABA), auxin, and the brassinosteroids, to form a dynamic and integrated signaling network and facilitate adaptive growth, development, and stress responses in plants [8,13–17].
Roots are vital systems that enable plants to cope with the continuously changing environment, in addition to taking up water and nutrients from the soil [18]. Optimal root growth and architecture are predominately determined by the root meristem, whose stem cell activity is maintained by several factors, including hormonal levels (e.g., auxin), nutrient availability (e.g., glucose), and reactive oxygen species (ROS) homeostasis (e.g., H2O2 and superoxide) [11,19,20]. Auxin is one of the most important hormonal regulators for root growth and root meristem maintenance [21–23]. Through the action of polar transporters such as the PIN-FORMED proteins (PINs), auxin accumulates in the primary root tips and contributes to the patterning of the root and the regulation of root cell division [22,24]. The inhibition of polar auxin transport in the pin mutants strongly affects root meristem activity and subsequent root development. The auxin gradient regulates root growth by coordinating the action of the PLETHORA (PLT) transcription factors [25] and the quiescent center (QC)-specifying proteins AT3G54220/SCR (SCARECROW) [26], AT4G37650/SHR (SHORT-ROOT) [27], and AT3G11260/WOX5 (WUSCHEL related homeobox 5) [28].
Besides auxin and glucose, ROS serve as additional key signaling molecules regulating meristem activity in plants. In particular, ROS are actively produced in the root tips, and their levels drive the direction and extent of root growth [29]. As byproducts of cellular metabolism, low levels of ROS act as key secondary messengers, controlling many important developmental processes including the maintenance of the root meristem [30,31]; however, the excessive accumulation of ROS may cause irreversible oxidative damage to cellular components. Several mutants with an altered ROS balance and redox distribution have significant differences in their primary root growth [30,32–34]. In Arabidopsis, the higher accumulation of ROS in the root tips of the mutants of AT5G04895/ABO6 (ABA overly sensitive 6) and AT4G11690/ABO8 (ABA overly sensitive 8), abo6 and abo8 respectively, which lack a DExH box RNA helicase and a pentatricopeptide repeat protein, respectively, leads to increased ABA sensitivity in the roots, altering their growth and reducing their meristem activity [35,36]. Moreover, the addition of the reducing agent glutathione (GSH) can partially rescue the root meristem phenotypes in these mutants, confirming that mitochondrial ROS in the root tips is an important retrograde signal for the maintenance of their meristematic activity [35,36]. Consistent with this, increasing evidence suggests that root growth in Arabidopsis is promoted by the application of GSH, but retarded by the depletion of antioxidants both in vitro or in vivo [19,37]. Although previous studies have revealed that glucose promotes ROS production [38], the mechanism by which ROS regulate glucose-mediated root growth and meristem activity is still largely unknown. The oxidation of active indole-3-acetic acid (IAA) into the inactive catabolic form 2-oxindole-3-acetic acid (OxIAA) by ROS occurs in the Arabidopsis root tips and is proposed to be a crucial step for auxin catabolism and root growth [39,40]; however, it is still unclear how ROS homeostasis is properly regulated to help maintain meristem activity in the roots.
Autophagy is a highly conserved biological process in which cellular components are degraded to remove damaged materials and maintain ROS homeostasis in all eukaryotic cells [41,42]. During autophagy, the substrates are sequestered and enclosed within cytoplasmic double-membrane organelles called autophagosomes, then delivered to the vacuole for degradation [43,44]. In Arabidopsis, autophagy is activated by several environmental cues [45,46] and recent research has revealed the crucial roles of autophagy in various aspects of the plant life cycle, including seedling establishment, development, and reproduction [47]. Here, we show that the autophagy-defective mutants in Arabidopsis display improved root growth and enhanced root meristem activity under high-glucose conditions. Furthermore, our results demonstrate that autophagy plays a key role in the glucose-mediated regulation of root meristem activity by maintaining the cellular homeostasis of ROS and auxin in Arabidopsis.
Results
Short-term glucose treatment temporarily suppresses constitutive autophagy in Arabidopsis roots
To investigate the potential role of autophagy in the plant root response to high glucose, we used green fluorescent protein (GFP)-tagged AT2G45170/ATG8E [48,49] as a marker to examine autophagosome formation in Arabidopsis root cells. Five-day-old seedlings expressing GFP-ATG8E were transferred from a 1/2-strength Murashige and Skoog (MS) medium to 1/2MS supplemented with 0, 1, or 3% glucose for 24 and 48 h, and the GFP fluorescence in their root cells was observed by confocal microscopy. Consistent with previous findings [50], we observed that autophagy occurs constitutively in the regions of the meristem zone (MZ), the elongation zone (EZ), and the differentiation zone (DZ) of Arabidopsis root tips (Figure 1(a)). In seedlings moved to 0% glucose medium, the formation of GFP-labeled punctuate autophagosomes was markedly induced in the primary root tips; however, the punctate structures observed in the MZ, EZ, and DZ were significantly reduced in the seedlings moved to 3% glucose medium (Figure 1(a,b)).
Figure 1.

Exogenous glucose affects constitutive autophagy in Arabidopsis roots. (a) Confocal microscopy showing the autophagosomes in the meristem zone (MZ), elongation zone (EZ), and differentiation zone (DZ) of GFP-ATG8E [48,49] roots under different glucose conditions. Five-day-old GFP-ATG8E seedlings grown on 1/2 MS agar medium containing 1% sucrose were transferred to 1/2 MS medium containing 0, 1, or 3% (w:v) glucose for 24 or 48 h. Bars: 50 μm. (b) Number of puncta per section in the MZ, EZ, and DZ cells in (a). The experiments with the same experimental design were repeated 3 times (biological repeats) with similar results. Values are means ± SD (n = 8 roots) calculated from one experiment. Asterisks indicate significant differences from the GFP-ATG8E seedlings grown on 1% glucose (*P < 0.05 and **P < 0.01, Student t test). ‘a’ and ‘b’ indicate values that are significantly higher or lower than the control, respectively. (c) Immunoblot analysis showing the processing of the GFP-ATG8E protein in GFP-ATG8E roots upon treatment with various concentrations (0, 1, 3, or 5%) of glucose. One-week-old GFP-ATG8E seedlings grown on normal 1/2 MS medium were transferred to 1/2 MS medium containing glucose for 24 h or 48 h. Anti-GFP antibodies were used for the protein blotting analysis. The GFP-ATG8E (GFP8E) fusion and free GFP levels are indicated on the right. The numbers on the left indicate the molecular mass (kD) of the size markers. The Coomassie Brilliant Blue-stained total proteins (CBB) are shown as the loading control.
We next assessed the release of free GFP to detect the degradation of GFP-ATG8E, a widely used approach for determining the rate of autophagy [44]. Upon induction of autophagy, the ATG8 proteins are lipidated with phosphatidylethanolamine (PE) to initiate autophagosome formation. The outer membrane of the autophagosome subsequently fuses with the vacuolar membrane to transport the contents of the autophagic bodies into the vacuole, where GFP-ATG8E is degraded to release a free, relatively stable GFP; therefore, the levels of free GFP reflect the rate of autophagy [44,51]. Consistent with the confocal microscopy observations, western blotting using anti-GFP antibodies revealed that the accumulation of free GFP was substantially reduced in the 24-h high-glucose conditions (3% and 5%) compared with the levels in 0% and 1% glucose conditions; however, when the treatment was extended to 48 h, the ratio of free GFP to GFP-ATG8E largely recovered (Figure 1(c)). These findings were further confirmed by analyzing the protein abundances of AT3G61960/ATG1A and ATG8s, as detected by the anti-ATG1A and anti-AT4G21980/ATG8A antibodies (Figure S1), which accumulate in the autophagy defective mutants because they failed to be degraded by the autophagy pathway [52]. These results suggested that the high-glucose applications temporarily suppressed constitutive autophagy in the Arabidopsis root tips.
Root inhibition in response to high glucose is reduced in autophagy-defective mutants
To evaluate the involvement of autophagy in the glucose response, we tested the sensitivity of the autophagy-defective mutants to glucose. To this end, wild-type, atg2-1, atg5-1, and atg7-3 [44] seeds were germinated on 1/2 MS medium containing 0, 1, or 3% glucose, and their phenotypes were recorded after 7 days of treatment. The 7-day-old atg seedlings showed increased sensitivities to sugar deprivation (0% glucose), producing shorter primary roots than the wild type (Figure 2(a)). In comparison, when grown on medium with 1% glucose, the atg mutants formed slightly longer roots than the wild type, which was further exacerbated when the seedlings were grown on 3% glucose (Figure 2(a)). Consistent with the phenotypic observations, the relative root lengths of the atg mutants were significantly shorter than those of the wild type on the glucose-deficient medium (0%), but they were significantly longer than the wild type in the glucose-treated conditions (Figures 2(b,c) and S2). These results demonstrated that the roots of the atg mutants showed enhanced tolerance to the inhibitory effects of elevated glucose, in a dose-dependent and time-dependent manner.
Figure 2.

Autophagy-defective mutants show enhanced root meristem activity in response to high glucose treatment. (a) One-week-old wild-type (WT), atg2-1, atg5-1, and atg7-3 seedlings grown on 1/2 MS medium with 0, 1, or 3% (w:v) glucose. Bars: 5 mm. (b) Relative root lengths of WT, atg2-1, atg5-1, and atg7-3 seedlings grown on 1/2 MS medium with 0, 1, 2, 3, 4, 5, or 6% (w:v) glucose for 7 d. Root lengths are expressed relative those of the WT on 1% glucose. (c) Time course (3, 5, 7, and 9 d) of WT, atg2-1, atg5-1, and atg7-3 seedlings grown on 1/2 MS medium with 3% (w:v) glucose. Root lengths are expressed relative to the corresponding WT seedlings on 1/2 MS medium with 1% glucose. (d) Root meristem zones in the WT and the atg mutants following a 7-d 0, 1, or 3% glucose treatment. Arrow pairs indicate the meristematic zone. Bars: 50 μm. (e) The cell lengths and cell numbers of the root meristem zones in (d). The seeds of WT, atg2-1, atg5-1, and atg7-3 were sown on 1/2 MS medium with 0, 1, or 3% (w:v) glucose. The root lengths and cell numbers were recorded at 7 days after germination. Three independent experiments were carried out with similar results, and representative data from one experiment are shown. For each experiment, 15 roots from 3 different plates were measured. Values are means ± SD (n = 15). Asterisks indicate significant differences from the WT (*P < 0.05 and **P < 0.01, Student t test). ‘a’ and ‘b’ indicate values that are significantly higher or lower, respectively, in the mutants than the WT.
We further measured the contents of different sugars including sucrose, glucose, fructose, and galactose in wild-type, atg5-1, and atg7-3 seedlings in response to various glucose treatments. As shown in Figure S3, at 0% glucose, the atg5-1 and atg7-3 mutants showed few differences in the contents of most of the sugars, such as glucose, fructose, and galactose, compared to that of wild-type seedlings. By contrast, in all conditions, including 0% glucose, the atg5-1 and atg7-3 mutants had clearly higher sucrose contents than that of wild-type seedlings (Figure S3), suggesting that the improved tolerance of atg mutants to high glucose is not due to a deficient supply of sugars in planta.
Primary root growth is regulated by the MZ, which contains the stem cell niche, and comprises the region between the QC and the first elongated cell in the cortex [53,54]. To further elucidate the glucose-resistant phenotype of the atg mutants, we analyzed the MZ of their roots in response to glucose treatment. The atg mutants had shorter MZs containing fewer cells than the wild-type roots after germinating and growing on 0% glucose for 7 days (Figure 2(d,e)). In contrast, on 1% glucose, the MZs of the atg mutants were slightly longer and contained more cells than the wild type. In particular, the wild-type roots grown on 3% glucose showed strongly reduced MZ length and fewer cells, but the atg mutants produced significantly longer MZs containing more cells (Figure 2(d,e)). Together, these findings indicate that the glucose-resistant phenotypes of the atg mutants are due to the higher activity of root meristems.
The autophagy-mediated glucose response acts downstream of the energy sensors
Given that the glucose sensor HXK1 plays an important role in glucose signaling [8], we attempted to investigate the effect of HXK1 on the autophagy-mediated glucose response by crossing the transgenic HXK1 overexpressor (HXK1-OE) line to the atg5-1 and atg7-3 mutants. As shown in Figure 3(a), the HXK1-OE line was hypersensitive to 3% glucose, confirming previous findings [5]. Similar to the atg5-1 and atg7-3 mutants, the atg5 HXK1-OE and atg7 HXK1-OE lines showed insensitivity to 3% glucose stress, as indicated by their relative root lengths (Figure 3(a,b)). Moreover, on 3% glucose, the atg5 HXK1-OE and atg7 HXK1-OE lines had similar root meristem activities to their respective atg parents (Figure 3(c)), with significantly longer MZs containing more cells than those of the wild-type plants (Figure 3(d)). These results suggest that the autophagy-mediated glucose response is likely regulated downstream of HXK1 in the HXK1-dependent glucose signaling pathway.
Figure 3.

The autophagy-mediated glucose response acts downstream of HXK1. (a) The autophagy defects in the atg mutants rescued the hypersensitivity of the HXK1-OE line to elevated glucose. Bars: 5 mm. (b) Relative root lengths of WT (Col-0 and Be), atg5-1, atg5 HXK1-OE, atg7-3, atg7 HXK1-OE, and HXK1-OE seedlings in (a). Root lengths are expressed relative to each genotype on 1% glucose. (c) Lengths of the root meristems (indicated by pairs of arrow) of WT (Col-0 and Be), atg5-1, atg5 HXK1-OE, atg7-3, atg7 HXK1-OE, and HXK1-OE seedlings grown for 7 d on 1/2 MS medium with 1% or 3% (w:v) glucose. Bars: 50 μm. (d) Relative number of cells in the root meristem zones in (c). Seeds of the WT (Col-0 and Be), atg5-1, atg5 HXK1-OE, atg7-3, atg7 HXK1-OE, and HXK1-OE plants were germinated on 1/2 MS agar medium with 1% or 3% (w:v) glucose. The seedlings were photographed 7 d after germination. The experiments with the same experimental design were repeated 3 times (biological repeats) with similar results. Values are means ± SD (n = 15) calculated from one experiment. Asterisks with an ‘a’ indicate significant differences from that of WT (Col-0 and Be), and those with a ‘b’ indicate significant differences from that of HXK1-OE (**P < 0.01, Student t test).
Because the energy sensor AT3G01090/KIN10 (SnRK1.1) plays a central role in regulating energy and stress signaling and is inactivated by glucose [55], we performed further genetic analyses of KIN10 and autophagy in response to glucose treatment using the atg5 KIN10-OE and atg7 KIN10-OE lines [56]. In contrast to the wild type, atg5-1, and atg7-3 plants, the root growth of KIN10-OE was highly sensitive to 3% glucose (Figure S4). Similar to the parental atg lines, the atg5 KIN10-OE and atg7 KIN10-OE plants showed an enhanced tolerance to the 3% glucose treatment (Figure S4). We have recently reported that KIN10 is an upstream positive regulator of autophagy in Arabidopsis [56]. Here, we further confirmed that KIN10 function in autophagosome formation requires downstream components of the autophagy pathway, by monodansylcadaverine (MDC) staining of the root cells of wild-type, atg5-1, atg7-3, KIN10 overexpressor (KIN10-OE), atg5 KIN10-OE, and atg7 KIN10-OE plants under various glucose conditions (Figure S5). Together, these findings indicate that the glucose-sensitive phenotype of KIN10-OE is largely dependent on the autophagy pathway.
Glucose-induced ROS accumulation is attenuated in the atg mutants
Given that ROS are important signaling molecules that regulate root growth by controlling cell division and differentiation [30,32,33], we examined the ROS levels in the roots of wild-type and atg seedlings upon glucose treatment. After being moved to medium with 1% glucose for 6 h, the root tips of the wild-type, atg5-1, and atg7-3 seedlings produced comparable levels of H2O2 and superoxide, as revealed by in situ staining with 3ʹ,3ʹ-diaininobenzidine (DAB; Figure 4(a)), nitroblue tetrazolium (NBT; Figure 4(b)), and 2ʹ,7ʹ-dichlorodihydrofluorescin diacetate (DCFH-DA; Figure 4(c)). The 3% glucose treatment promoted the accumulation of H2O2 and superoxide in the wild-type root tips; however, the root tips of the atg5-1 and atg7-3 mutants accumulated much lower levels of ROS than the wild type under this treatment (Figure 4(a–f)).
Figure 4.

Inhibition of root elongation by high glucose levels involves ROS. (a–c) DAB staining for H2O2 (a), NBT staining for superoxide (b), and DCFH-DA staining for H2O2 (c) in the primary root tips of the wild type (WT) and the atg5-1 and atg7-3 mutants upon treatment with 1% or 3% glucose for 6 h. Bars: 100 μm. (d–f) Relative fluorescence intensities calculated from (a–c). (g) Recovery of high-glucose-induced root inhibition in the WT by the addition of GSH. Seeds of WT, atg2-1, atg5-1, and atg7-3 were germinated on 1/2 MS medium with 1% or 3% (w:v) glucose, with or without 500 μM GSH, for 7 d. Bars: 5 mm. (h) Relative root lengths of WT, atg2-1, atg5-1, and atg7-3 in (g). Root lengths are expressed relative to that of the WT on the 1/2 MS medium containing 1% glucose. For the staining assays, 5-day-old WT, atg5-1 and atg7-3 seedlings grown on 1/2 MS agar medium containing 1% sucrose were transferred to 1/2 MS medium with 1% or 3% glucose for 6 h. For the root elongation assays, seeds of WT, atg5-1 and atg7-3 were germinated on 1/2 MS medium containing 1% or 3% glucose with or without 500 μM GSH. The root lengths were measured at 7 days after germination. The experiments were repeated 3 times with similar results. Values are means ± SD (n = 15 seedlings) from one experiment. The asterisks with “a” indicate significant higher in the WT seedlings treated with 3% glucose than those in WT under 1% glucose. The asterisks with “b” indicate significantly lower values in the atg mutants than that of WT under 3% glucose treatment (**P < 0.01, Student t test). Asterisks in (h) indicate significant differences between the WT and the atg mutants, with and without GSH (**P < 0.01, Student t test).
To further confirm the involvement of ROS in regulating glucose-inhibited root elongation, we investigated whether the addition of reducing agents, GSH, or AsA would affect the phenotypic difference between the wild type and the atg mutants under the elevated glucose conditions. The addition of 500 μM GSH or 50 μM AsA significantly increased the root growth of the wild type under both 1% and 3% glucose; therefore, the relative root lengths of the atg mutants were more similar to the wild type under these conditions (Figures 4(g,h) and S6). We also observed that 500 μM GSH increased the MZ lengths and number of cells to a greater extent in the wild type than the atg mutants under the 3% glucose treatment (Figure S7). These findings suggest that ROS are important for the regulation of the autophagy-associated glucose response.
Glucose-mediated suppression of root elongation is correlated with auxin levels
The growth hormone auxin plays an essential role in controlling meristematic activity and growth in the roots [57]. We therefore hypothesized that the improved activity of the root meristem and the reduced ROS accumulation in the atg mutants in response to high glucose likely arise from the elevation of auxin levels. To test this, transgenic lines expressing DR5pro:GFP and DR5pro:GUS were crossed to the atg5-1 and atg7-3 mutants, and the resulting transgenic plants were further analyzed using confocal microscopy and histochemical staining. As shown in Figures 5(a) and S8a, the expression levels of DR5pro:GFP and DR5pro:GUS were reduced in the root tips of the wild type following treatment with high levels of glucose, but this difference was less pronounced in the atg5-1 and atg7-3 mutants. A statistical analysis confirmed that the atg mutants had significantly higher levels of DR5pro:GFP signal than those of the wild type on 1, 3, and 5% glucose (Figure 5(c)). Consistent with this, the expression levels of DR5pro:GFP and DR5pro:GUS were significantly lower in the atg mutant roots under glucose-deprived conditions (0% glucose), compared to that of wild-type roots (Figures 5(a,c) and S8a)
Figure 5.
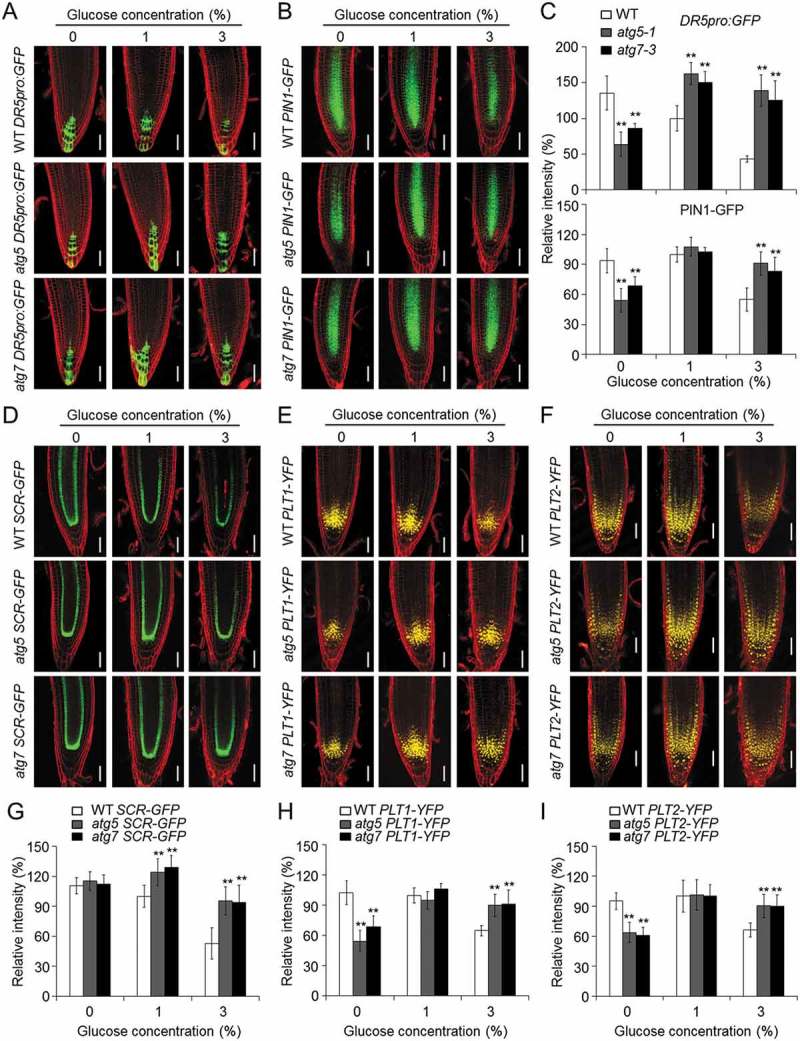
Disruption of autophagy attenuates the glucose-suppressed expression of DR5, PIN1, SCR, PLT1, and PLT2. (a,b) The expression levels of DR5pro:GFP (a) and PIN1-GFP (b) in the wild-type (WT), atg5-1, and atg7-3 backgrounds (WT DR5pro:GFP, atg5 DR5pro:GFP, atg7 DR5pro:GFP, WT PIN1-GFP, atg5 PIN1-GFP, and atg7 PIN1-GFP) at 7 d after germination on 1/2 MS medium with 0, 1, or 3% (w:v) glucose. Bars: 50 μm. (C) The relative fluorescence intensities of DR5pro:GFP and PIN1-GFP in (a) and (b), respectively. Three independent experiments were performed with similar results. Values are means ± SD (n = 15) from one representative experiment. Asterisks indicate significant differences from WT (*P < 0.05 and **P < 0.01, Student t test). (d–f) The expression levels of SCR-GFP (d), PLT1-YFP (e), and PLT2-YFP (f) in the wild-type (WT), atg5-1, and atg7-3 backgrounds (WT SCR-GFP, atg5 SCR-GFP, atg7 SCR-GFP, WT PLT1-YFP, atg5 PLT1-YFP, atg7 PLT1-YFP, WT PLT2-YFP, atg5 PLT2-YFP, and atg7 PLT2-YFP) 7 d after germination on 1/2 MS medium with 0, 1, or 3% (w:v) glucose. Bars: 50 μm. (g–i) The relative fluorescence intensities of SCR-GFP (g), PLT1-YFP (h), and PLT2-YFP (i) in the WT, atg5-1, and atg7-3 backgrounds (WT pSCR-GFP, atg5 pSCR-GFP, atg7 pSCR-GFP, WT PLT1-YFP, atg5 PLT1-YFP, atg7 PLT1-YFP, WT PLT2-YFP, atg5 PLT2-YFP, and atg7 PLT2-YFP) following a 7-d treatment of 0, 1, or 3% glucose. The fluorescence intensities are expressed relative to that of the 1% glucose-treated WT SCR-GFP, WT PLT1-YFP, and WT PLT2-YFP seedlings, respectively. The seeds of all genotypes were germinated on 1/2 MS medium containing 0, 1 or 3% glucose and the root fluorescence was measured at 7 days after germination. Three independent experiments were conducted with similar results. Values are means ± SD (n = 10) calculated from one experiment. Asterisks indicate significant differences from each WT control (*P < 0.05 and **P < 0.01, Student t test).
Auxin transporters are important for modulating root growth. To further elucidate the association of auxin with the autophagy-mediated glucose response, we examined the expression of the auxin transporter AT1G73590/PIN1 in wild-type and atg root tips treated with 0, 1 or 3% glucose. Similar to DR5, the expression level and relative fluorescence intensity of PIN1-GFP were both significantly lower in the atg5-1 and atg7-3 root tips on 0% glucose and higher in the root tips on 3% glucose, compared with wild type (Figure 5(b,c)). 1-N-naphtylphthalamic acid (NPA) is a polar auxin transport inhibitor that has been widely used to study auxin-dependent biological activities [53]. When 3 μM NPA was added to the growth media containing 1% or 3% glucose, the root growth of the wild type was differentially inhibited (Figure S8(b,c)). The NPA treatment compromised the glucose-tolerant phenotypes of the atg mutants, which instead exhibited comparable root lengths to that of the wild type following treatments of 1% or 3% glucose (Figure S8(b,c)). These results suggest that both auxin levels and the response to auxin are improved in the atg mutants following glucose treatment.
Disruption of autophagy attenuates the glucose-suppressed expression of SCR, PLT1, and PLT2
Root meristem activity and root growth are regulated by specific transcription factors, including SCR and the PLTs. SCR is a member of the GRAS family of transcription factors and plays a central role in asymmetric stem cell divisions as well as regulating the specification and maintenance of the QC stem cells [26,54]. The AP2 transcription factors AT3G20840/PLT1 and AT1G51190/PLT2 are predominately located in the QC and are necessary for the maintenance of the root apical meristem [54]. As shown in Figure 5(d–i), the relative fluorescence intensities of SCR-GFP, but not PLT1-YFP, and PLT2-YFP, were significantly higher in the atg5-1 and atg7-3 mutant backgrounds than in the wild-type background with the 1% glucose treatment; however, after the 3% glucose treatment, the expression levels of all of the fusion proteins were greatly decreased in the wild-type root tips, which was not observed in the atg mutants (Figure 5(d–i)). Consistent with their hypersensitivity in the sugar-deprived medium, both atg5-1 and atg7-3 mutants showed significantly decreased levels of PLT1-YFP and PLT2-YFP compared to that of the wild type (Figure 5(e,f,h,i)). Together, these results indicate that the loss of autophagy attenuates the glucose-mediated inhibition of root meristem activity, possibly by modulating the expression of SCR, PLT1, and PLT2.
Autophagy-defective mutants accumulate more peroxisomes in root cells following high-glucose treatment
Given that both ROS and auxin are likely involved in the autophagy-mediated glucose response, we therefore proposed that the peroxisome, an organelle essential for the production of both ROS and IAA [58], may integrate the effects of ROS and IAA on the glucose response in plants. To verify this, we introduced the peroxisomal marker eCFP-SKL [59] into the atg5-1 and atg7-3 mutants to evaluate the stability of peroxisomes in response to different glucose applications. In response to the 1% glucose treatment, there were significantly more peroxisomes in the shoot and root cells of atg5-1 and atg7-3 than in the wild type (Figure 6(a–c)). In response to the 3% glucose treatment, there were significantly fewer peroxisomes in the eCFP-SKL roots in the wild-type background (Figure 6(b,c)), but not the shoots (Figure 6(a,c)). By contrast, more eCFP-SKL puncta accumulated in the root cells of the atg5-1 eCFP-SKL and atg7-3 eCFP-SKL seedlings in response to the 3% glucose treatment (Figure 6(b,c)). A western blotting analysis was used to further confirm that more eCFP-SKL proteins accumulated in the shoots of the atg5-1 and atg7-3 mutants than in the wild type, and that their levels remained consistent in the mutants regardless of the glucose treatment (Figure 6(d)). In particular, the levels of the eCFP-SKL protein were lower in the wild-type roots in response to the high-glucose treatments (3% or 5%; Figure 6(e)). Consistent with the confocal data, the levels of the eCFP-SKL protein were higher in the atg mutants treated with the various concentrations of glucose (1, 3, or 5%; Figure 6(e)). Taken together, these findings imply that the glucose-promoted peroxisome degradation in roots requires a functional autophagy pathway.
Figure 6.

Autophagy is required for glucose-promoted degradation of peroxisomes in Arabidopsis roots. (a,b) Confocal microscopy images showing eCFP-SKL-labelled peroxisomes in the shoot (a) and root (b) cells of wild-type (WT, WT eCFP-SKL), atg5-1 (atg5 eCFP-SKL), and atg7-3 (atg7 eCFP-SKL) plants. Bars: 50 μm. (c) Numbers of peroxisomes quantified from confocal images of WT eCFP-SKL, atg5 eCFP-SKL, and atg7 eCFP-SKL shoot (upper panel) and root (lower panel) cells. Five-day-old seedlings grown on 1/2 MS agar medium containing 1% sucrose were treated with 1% or 3% (w:v) glucose for 24 h. Areas of the same size were randomly selected from confocal images and CFP-SKL-labelled puncta were quantified using ImageJ. Three independent experiments were performed with similar results. Values are representative means ± SD (n = 15 roots) from one experiment. Asterisks with an ‘a’ indicate significantly fewer puncta than those of the WT in the 1% glucose condition, while those with a ‘b’ indicate significantly more puncta in the atg mutants than in the WT (**P < 0.01, Student t test). (d,e) Immunoblot analysis showing the level of eCFP-SKL in the shoots (d) and roots (e) of WT eCFP-SKL, atg5 eCFP-SKL, and atg7 eCFP-SKL seedlings following a 24-h treatment with 0, 1, 3, or 5% glucose. One-week-old seedlings expressing the eCFP-SKL transgene germinated on 1/2 MS medium were transferred to 1/2 MS medium containing the various glucose concentrations for 24 h. Anti-GFP antibodies were used for the protein blotting analysis. The numbers on the left indicate the molecular mass (kD) of the size markers. A Coomassie Brilliant Blue-stained gel (CBB) is shown as the loading control.
ATG8E physically interacts with ABCD1 and overexpression of ABCD1 partially rescues the glucose-associated phenotypes of the atg mutants
During peroxisomal β-oxidation, the transmembrane protein ABCD1 plays multiple roles in plant lipid metabolism and signaling, including the transport of indole-3-butyric acid (IBA) for subsequent conversion via β-oxidation into the active form indole-3-acetic acid (IAA). The C-Terminal Walker B motif of the second nucleotide binding domain of ABCD1 is a functional region essential for ABCD1 activity in planta [58,60]. To establish the molecular link between the peroxisomes and the autophagy-mediated glucose response, we first examined the possible interaction between ATG8E and the Walker B motif of ABCD1 (ABCD1-B) by yeast two-hybrid (Y2H) and coimmunoprecipitation (CoIP) assays. As shown in Figure 7(a,b), the ATG8E protein could physically interact with ABCD1-B in vitro and in vivo.
We next examined the sensitivity of the wild type and the atg mutants (atg2-1, atg5-1, and atg7-3) to IBA, the precursor of IAA during peroxisomal β-oxidation [61]. Previous studies have suggested that mutants defective in β-oxidation were less sensitive to exogenous IBA [61]. In comparison with the wild-type seedlings, the atg mutants showed increased sensitivity to IBA, as indicated by their significantly shorter relative root lengths following the IBA treatment (Figure S9). Moreover, the addition of IBA into the 3% glucose MS medium significantly rescued the glucose-tolerant root elongation phenotype of the atg mutants, resulting in similar sensitivities to the wild type (Figure S9). To further explore the function of peroxisomes in the plant response to elevated glucose, ABCD1-overexpression lines (ABCD1-OEs) were generated by cloning the ABCD1 full-length cDNA into the BamHI sites of the pBI-eGFP vector, driven by the cauliflower mosaic virus 35S promoter. Phenotypic assays showed that the ABCD1-OE lines were sensitive to both the elevated glucose and IBA treatments (Figures 7(c,d) and S10). ABCD1-OE-1 was then crossed to atg5-1 and atg7-3 to obtain the atg5 ABCD1-OE and atg7 ABCD1-OE lines for phenotypic analysis. As shown in Figure 7(c,d), the sensitivities of the atg5 ABCD1-OE and atg7 ABCD1-OE lines under both the 3% glucose and 3% glucose plus IBA conditions resembled the wild type, rather than the enhanced-tolerance phenotypes of the atg5-1 and atg7-3 single mutants.
Figure 7.
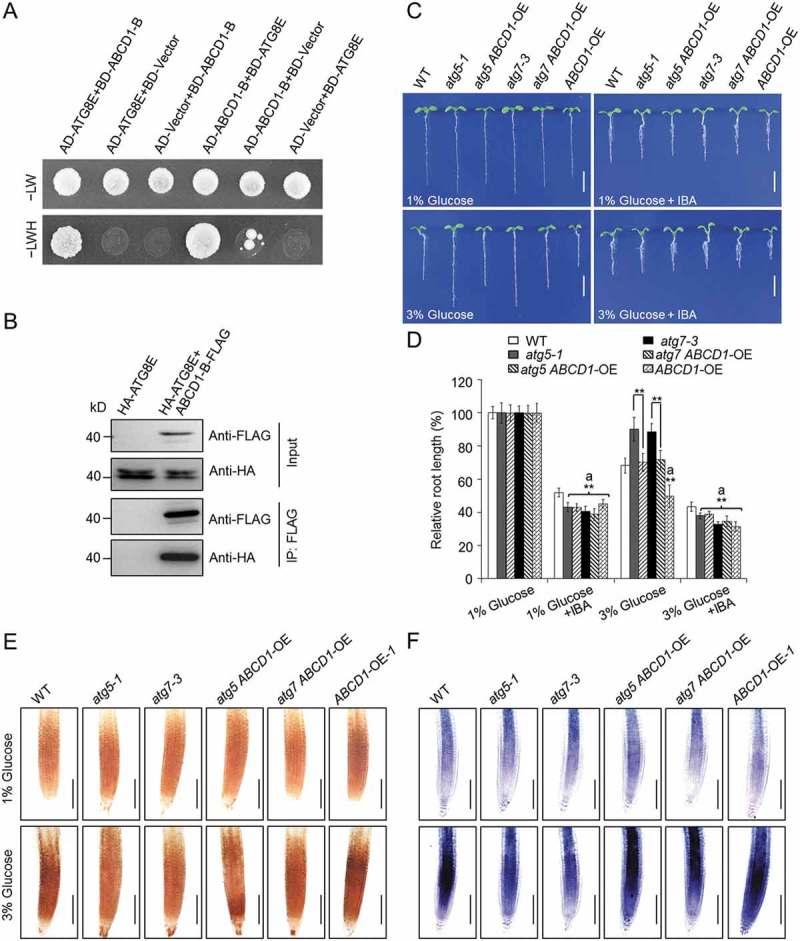
ABCD1 is involved in autophagy-mediated glucose response. (a) Yeast two-hybrid assay showing the interaction between ATG8E and the Walker B motif of the second nucleotide-binding domain (NBD) of the ABCD1 protein. The yeast strains were selected on SD/-Trp-Leu-His-Ade medium (–LWH) that are dependent on two-hybrid protein interactions. (b) In vivo CoIP analysis showing the physical interaction between ATG8E and the Walker B motif of the second NBD of the ABCD1 protein. HA-tagged ATG8E (HA-ATG8E) was coexpressed with ABCD1-B-FLAG in Arabidopsis protoplasts and immunoprecipitated by FLAG affinity magnetic beads. (c,d) Phenotypes (c) and relative root lengths (d) of WT, atg5-1, atg5 ABCD1-OE, atg7-3, atg7 ABCD1-OE, and ABCD1-OE lines germinated for 7 d on 1/2 MS medium containing 1% or 3% (w:v) glucose, with or without 10 μM IBA. Bars: 5 mm. Root lengths are expressed relative to those of the genotypes on the 1/2 MS medium containing 1% glucose. The seeds of all genotypes were germinated on 1/2 MS medium containing 1% or 3% glucose with or without 10 μM IBA and the root lengths were measured at 7 days after germination. The experiments were repeated 3 times with similar results. Values are means ± SD (n = 15 seedlings) from one experiment. Asterisks with an ‘a’ indicate values significantly lower than that of WT (**P < 0.01, Student t test). (e,f) DAB staining for H2O2 (e) and NBT staining for superoxide (f) in the primary root tips of the wild-type (WT), atg5-1, atg7-3, atg5 ABCD1-OE, atg7 ABCD1-OE, and ABCD1-OE seedlings following a 6-h treatment with 1% or 3% glucose. Bars: 100 μm. The 5-d-old seedlings of all genotypes grown on 1/2 MS agar medium containing 1% sucrose were transferred to 1/2 MS medium with 1% or 3% glucose for 6 h.
To evaluate the association of the high-glucose-mediated phenotypes with the ROS levels in the atg mutants and atg ABCD1-OE lines, we measured the levels of H2O2 and superoxide in the wild-type, atg5-1, atg7-3, atg5 ABCD1-OE, atg7 ABCD1-OE, and ABCD1-OE-1 lines following treatment with 1% or 3% glucose. The DAB and NBT staining revealed little difference among the genotypes with 1% glucose; however, after the 3% glucose treatment, the H2O2 and superoxide levels were both greatly elevated in the ABCD1-OE lines, compared with those of the wild type and the atg5-1 and atg7-3 mutants (Figure 7(e,f)). Consistent with the phenotypic observations, we further found that the overexpression of ABCD1 rescued the reduced ROS levels in the atg mutants; the atg5 ABCD1-OE and atg7 ABCD1-OE lines accumulated notably more ROS than the atg single mutants under the 3% glucose condition (Figure 7(e,f)).
Discussion
Besides its fundamental role as an energy store, glucose can also serve as an important signaling molecule that modulates diverse physiological processes in plant cells [1,3,4]. Recent findings revealed that glucose is involved in the regulation of root meristem activity [11,53], but the molecular mechanism underlying the inhibition of root elongation by high levels of glucose is still unclear. In present study, we provide several lines of evidence to support the hypothesis that autophagy is a central mechanism in the regulation of glucose-mediated root meristem activity, acting by modulating the cellular levels of ROS and auxin in Arabidopsis.
The root apical meristem is important for the growth and patterning of roots, and its stem cell activity is maintained by several factors including glucose [11,19]. On the other hand, high concentrations of glucose inhibit root growth by reducing the root meristem length and cell number [53]. A recent study reveals that glucose-inhibited root meristem activity is regulated by TOR signaling, which acts through glycolysis and mitochondrial bioenergetics to provide the essential energy for its cell proliferation and growth [11]. Moreover, the TOR kinase directly phosphorylates the AT2G36010/E2F3 (E2F transcription factor 3), which transcriptionally activates the S-phase genes, bypassing the conventional CYC-CDK-RBR-E2F cascade that regulates the cell cycle [62] to directly control meristem activity in the root tips [11].
Through phenotypic, genetic, and biochemical analyses, we revealed that autophagy is involved in the regulation of root meristem activity and acts downstream of the HXK1 and KIN10 sensors, providing further evidence to link autophagy to the energy signaling pathways in the control of root growth. Previous findings have suggested that TOR is a negative regulator of autophagy in eukaryotic cells [63,64]. More recently, we and another group have demonstrated that the energy sensor KIN10 plays a key role in the positive activation of autophagy in Arabidopsis, possibly by inhibiting TOR activity [56,65]. It is therefore conceivable that, in response to high glucose, the TOR signaling pathway activates at least 2 distinct mechanisms: 1) the unconventional suppression of E2F3 to suspend cell proliferation, and 2) the inhibition of autophagy to modulate cellular dynamics. This suggests that 2 parallel pathways may regulate root meristems for plant growth and patterning. In agreement with this proposal, genome-wide expression profiling revealed that the expression of several core ATG genes are significantly upregulated and downregulated by the KIN10 and TOR kinases, respectively [11,55].
In plants, autophagy is generally considered to be induced in response to various developmental or environmental cues [45,66]. Under favorable conditions, the defective autophagy of the atg mutants does not affect root elongation [67]; however, under nutrient-depleted conditions (e.g., in the sugar-free growth medium), the root system of the atg mutants is significantly stunted in comparison with those of the wild-type seedlings [50]. We observed that, in the MZ, EZ, and DZ regions of Arabidopsis roots expressing eGFP-ATG8E, autophagosome formation occurs constitutively, and is further strengthened upon exposure to low sugar conditions (Figure 1(a)). This observation is consistent with the previous work of Inoue et al. [50] and confirms the hypothesis that constitutive autophagy is essential for root elongation and development under sugar-deprived conditions. In contrast, our data further revealed that the short-term (24 h) application of high levels of glucose temporarily suppressed the constitutive autophagy in the root tips, but that this process was largely restored by 48 h after the treatment (Figure 1). The temporary inhibition of autophagy during the 24-h glucose treatment, indicated by the decreased numbers of autophagosome puncta and the degradation of the eGFP-ATG8E fusion protein, may be explained by the activation of the glucose response via the TOR-dependent signaling pathway. More interestingly, a recent study reveals that, in response to most biotic and abiotic stresses, the induction of autophagy requires the downregulation of TOR activity; however, this does not appear to be the case under oxidative and endoplasmic reticulum stresses [68]. This distinction supports the presence of a TOR-independent pathway for autophagy activation in plants. Similarly, our results showed that the application of high levels of exogenous glucose had few long-term effects on the constitutive formation of autophagosomes in the Arabidopsis roots. It is therefore possible that, in the long term, constitutive autophagy is maintained despite the inhibitory effects of glucose by a TOR-independent mechanism such as oxidative stress. Consistent with this hypothesis, we found that ROS accumulation was remarkably induced in the wild-type roots by the application of high levels of glucose (Figure 4).
We suggest that ROS act not as only modulators of autophagy activation in response to glucose, but also function downstream of autophagy-mediated glucose signaling. Compared with the high accumulation of ROS in the wild-type roots, the atg mutants accumulated much less ROS in response to the glucose treatments (Figure 4), a phenotype that could be greatly rescued by the overexpression of ABCD1 (Figure 7). ABCD1 is an ABC transporter essential for β-oxidation and the production of IAA in the peroxisomes [69]. Consistent with this, we analyzed DR5pro:GFP, DR5pro:GUS and PIN1-GFP transgenic lines to reveal that the inhibition of the auxin gradient and auxin transport following the glucose treatments was attenuated in the atg mutants in comparison with the wild type (Figures 5(a–c) and S8). In the Arabidopsis root tips, IAA has been previously reported to be oxidized to its inactive form, OxIAA, for degradation when ROS accumulate [39,40]. These results support the hypothesis that the elevated abundance and transport of auxin in the atg mutants is likely caused by the reduced accumulation of ROS in these roots. The appropriate levels of ROS and auxin likely form a coordinated signal complex that contributes to the proper maintenance of root growth under high-glucose conditions.
Increasing evidence suggests that ROS play a key role in controlling cell division and differentiation during root growth and development [30,32]. In Arabidopsis and rice (Oryza sativa), the levels of ROS in the root tips determine their growth patterns [29,70]. In particular, the growth of the primary root changes when an imbalance of ROS occurs in the root tips [30,32–34]. The transcription factor AT2G47270/UPB1 (UPBEAT1) regulates cell proliferation and differentiation in Arabidopsis root tips, and the reduced and elevated H2O2 contents of the upb1 mutant and the UPB1 overexpression lines, respectively, lead to longer and shorter primary roots [32]. Thus, an appropriate ROS distribution is important for root growth and patterning [33]. We also suggest that ROS are required for the autophagy-mediated glucose response, which is to be expected because autophagy modulates cellular ROS levels during plant responses to various environmental cues, including oxidative stress [44,71].
Based on the findings that both ROS and auxin are involved in the autophagy-mediated glucose response, we proposed that multifunctional organelles called peroxisomes likely regulate the glucose response by integrating the ROS and IAA signaling pathways. This hypothesis was confirmed by analyses of the glucose-promoted degradation of these organelles in the peroxisomal marker line eCFP-SKL (Figure 6), the direct interaction between ATG8E and ABCD1 (Figure 7(a,b)), and the phenotypic characterization of the ABCD1-overexpressing lines, as well as their combinations with the atg mutants (Figures 7(c,d) and S10). In particular, we observed that, in terms of root elongation, the atg mutants were more sensitive than the wild type to IBA, the precursor of IAA in the peroxisomal β-oxidation process [61], under both 1% and 3% glucose conditions (Figure S9). This suggests that similar to the mechanism in the shoots and hypocotyls [72,73], autophagy is likely involved in the quality control of peroxisomes in Arabidopsis roots. These results highlight the importance of functional peroxisomes in the maintenance of cellular ROS and IAA levels, and in the subsequent autophagy-mediated regulation of root meristems under elevated glucose conditions.
In conclusion, our findings demonstrate that the Arabidopsis sensors HXK1 and KIN10 perceive the high-glucose stress and signal to the constitutive autophagy system in roots, possibly through the direct or indirect action of TOR (Figure 8). In the wild-type roots, constitutive autophagy modulates the function of the peroxisomes in response to high glucose and contributes to the production of ROS and IAA, which determine the root meristem activity. In the downstream events of glucose signaling, autophagy-mediated ROS levels serve as a central regulator, playing dual roles in the regulation of the root meristem. On the one hand, the imbalance of different ROS species or the accumulation of ROS induced by the high levels of glucose oxidize the active IAA for degradation, impair root meristem activity, and subsequently inhibit root growth. On the other hand, high levels of ROS also enhance the constitutive autophagy machinery in a TOR-independent manner to attenuate the oxidative damage caused by excess ROS and maintain root meristem function under these stress conditions (Figure 8). By contrast, the autophagy deficiency of the atg mutants disrupts the transmission of the high-glucose signal to the peroxisomes, which may alleviate both the ROS oxidization of IAA and the enhanced constitutive autophagy, thereby increasing root growth under high-glucose conditions (Figure 8). Taken together, these results demonstrate that autophagy plays a key role in regulating glucose-mediated root meristem activity.
Figure 8.

Proposed model for the role of autophagy in regulating the glucose-mediated suppression of root meristem activity. Glucose is sensed by HXK1, which suppresses the autophagy machinery through an unknown mechanism. In response to glucose signaling, autophagy modulates the homeostasis of cellular ROS and promotes the degradation of the oxidatively damaged peroxisome. In roots, the peroxisome mediates the biosynthesis of auxin and determines the meristem activity in the root tips; therefore, its degradation leads to reduced root growth.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana accession Columbia-0 (Col-0), Bensheim (Be), and Landsberg erecta (Ler) were used as the wild-type lines in this study. The autophagy-deficient mutants (atg2-1, atg5-1, atg7-3, and atg10-1; ecotype Col-0), KIN10-OE, atg5 KIN10-OE, and atg7 KIN10-OE (ecotype Ler) used in this study have been described previously [44]. The HXK1 overexpression (HXK1-OE) transgenic seeds (CS70282; ecotype Be) were obtained from The Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org). For the genetic analysis, atg5-1 and atg7-3 were crossed with HXK1-OE to generate the atg5 HXK1-OE and atg7 HXK1-OE lines. The gene information is listed in Table S2.
For the glucose sensitivity assays, the Arabidopsis seeds were surface-sterilized with 20% bleach containing 0.1% Tween 20 (Sigma, P2287) for 15 min, washed with distilled water 5 times, then plated on 1/2 MS (Sigma, M5519) agar with various concentrations of glucose, with or without IBA (Sigma, I5386), NPA (Sigma-Aldrich, 33,371), GSH (Sigma-Aldrich, G4251), or AsA (Sigma, A4544). The plates were incubated at 4°C for 3 days, then transferred to a growth room under a 16-h light (125 μmol m−2 s−1)/8-h dark photoperiod at 23°C for 7 d. Primary root length was measured from digital images using ImageJ software (http://rsb.info.nih.gov/ij/).
For DAB, NBT, and DCFH-DA staining assays, 5-d-old seedlings of the wild type and atg mutants grown on 1/2 MS agar medium containing 1% sucrose (Sigma, S9378) were transferred to 1/2 MS agar medium containing 1% or 3% (w:v) glucose for 6 h. For confocal microscopy analysis of peroxisomes in root cells and western blotting assays, 5 or 7 d seedlings grown on 1/2 MS agar medium containing 1% sucrose were transferred to 1/2 MS agar medium containing various concentrations of glucose for 24 h. For analyzing the levels of DR5, PIN1, SCR, PLT1, and PLT2 markers, seeds of all genotypes were germinated on 1/2 MS agar medium containing 0, 1, or 3% (w:v) glucose for 7 days, and then observed by confocal microscopy.
Confocal microscopy
Transgenic plants expressing a GFP-ATG8E fusion were used to monitor autophagosome formation [44,48,49]. Five-day-old GFP-ATG8E seedlings were transferred to 1/2 MS medium supplemented with 0, 1, or 3% (w:v) glucose for 24 h. The primary root cells were observed using a Zeiss7 DUO NLO confocal laser scanning microscope (Carl Zeiss, Germany).
The generation of transgenic Arabidopsis plants harboring DR5pro:GFP [74], PLT1-YFP [75], PLT2-YFP [75], SCR-GFP [76], PIN1-GFP [74], or eCFP-SKL [59] has been previously described. To combine these reporter genes with the atg mutants, the transgenic lines were crossed to atg5-1 and atg7-3. The F2 seedlings were genotyped by PCR for the atg mutation using the gene-specific or T-DNA-specific primer pairs listed in Table S1. Confocal microscopy was used to screen the reporter lines in the F3 seedlings with atg5 or atg7 homozygous backgrounds. Propidium iodide fluorescence was used to visualize the cells in the root tips, as previously described [36]. The roots of the seedlings were stained with 10 μM propidium iodide (Sigma, P4170) then visualized with a Zeiss7 DUO NLO confocal laser scanning microscope. For imaging GFP, YFP, CFP, and propidium iodide, excitation wavelengths of 488, 514, 458, and 543 nm were used, respectively, while their emissions were detected at 500 to 530, 500 to 530, 480 to 520, and 585 to 615 nm, respectively. The fluorescence intensities were qualified with the histogram produced on each confocal section using the same microscope settings.
Western blotting
Protein extraction and western blotting were carried out as described previously [44]. Samples were harvested, ground in liquid nitrogen, and homogenized in ice-cold extraction buffer (50 mM sodium phosphate, pH 7.0, 200 mM NaCl, 10 mM MgCl2, 0.2% β-mercaptoethanol, 10% glycerol) supplemented with a protease inhibitor cocktail (Roche, 04693132001). Samples were incubated on ice for 30 min, then centrifuged at 12,000 g for 30 min at 4°C. The supernatant was transferred to a new microcentrifuge tube for electrophoresis. Protein samples were separated using SDS-PAGE and transferred onto a Hybond-C membrane (Amersham, 10600016). The immunoblots were incubated in a blocking solution (1xTBST [50 mM Tris, 150 mM NaCl, pH 7.5, 0.1% Tween 20] containing 5% [w:v] nonfat dried milk). Primary antibodies were diluted in blocking solution as follows: 1:2000 for anti-GFP (Cell Signaling Technology, 2955), 1:8000 for anti-ATG1A [49], 1:1500 for anti-AT4G21980/ATG8A (Abcam, ab77003), 1:1000 for anti-ACTIN (AT3G18780/ACT2)(ABclonal, AC009), 1:5000 for anti-HA (Sigma, H6533), and 1:5000 for anti-FLAG (Sigma, A8592). The protein information is listed in Table S2.
Measurement of root meristem zone lengths and cell numbers
The root meristem zone length and cell number were determined as described previously [77]. Briefly, 1-week-old wild-type, atg2-1, atg5-1, and atg7-3 seedlings grown on 1/2 MS agar medium supplemented with 1% or 3% glucose were harvested. The root tips were stained with 10 μM propidium iodide and imaged using a Zeiss7 DUO NLO confocal laser scanning microscope. Root meristem lengths were measured from the QC to the first elongated cell in the cortex using ImageJ. The total number of cells in the meristematic zone was recorded.
Measurement of sugar contents by GC-MS
Seven-day-old seedlings grown on 1/2 MS medium containing 1% sucrose and 1% agar (m/v) were transferred to 1/2 MS agar medium containing various concentrations of glucose for 24 h and saccharides were quantified by GC-MS as described previously [78]. Briefly, ribitol (Sigma, 02240) was added as the interior label quantitative standard (1 μg/sample). The supernatant was transferred to a new microcentrifuge tube for derivatization by silylation. Glucose, sucrose, fructose, and galactose were used as standards (0.02 mg/ml) to confirm the retention time. All the samples and standards after silylation were analyzed via GC-MS system using the default settings. The temperature program of GC started at 85°C for 3 min, followed a 5°C per min increase to 285°C and a 20°C per min increase to 310°C, which was then held for 7 min. The contents of each sample were calculated according to linear relationship between peak areas of the interior label and the sample.
Histochemical staining
Histochemical staining for GUS activity was carried out as described previously [79]. To generate the atg5 DR5pro:GUS and atg7 DR5pro:GUS lines, a transgenic plant carrying DR5pro:GUS [73] was crossed with the atg5-1 and atg7-3 mutants. The transgenic F2 seedlings were selected on 1/2 MS containing 50 mg/L kanamycin and genotyped for the atg mutations using the gene-specific or T-DNA-specific primer pairs listed in Table S1. Staining was performed for 2 h using root samples from at least 15 seedlings, followed by destaining in 70% ethanol. The root tips were observed in clearing solution (chloral hydrate: water: glycerol, 8: 2: 1) with a DM5000B microscope (Leica Microsystems, Germany).
ROS staining was performed as described previously [30,36], with minor modifications. For DAB staining, the seedlings were steeped in 0.5 mg/mL DAB (Sigma-Aldrich, D8001) in 50 mM Tris-HCl, pH 5.0, for 2 h at room temperature. For the NBT staining, seedlings were incubated in 1 mM NBT (Sigma-Aldrich, N6876) dissolved in 20 mM potassium phosphate and 0.1 M NaCl at pH 6.2 for 15 min. The seedlings were then washed 3 times with dH2O, and cleared in 70% boiling ethanol for 10 min. The root tips were observed with a Leica DM5000B microscope. For the DCFH-DA staining, the seedlings were incubated in darkness in a buffer containing 50 μM DCFH-DA (Sigma, D6883) and 20 mM potassium phosphate at pH 6.0 for 15 min. The fluorescence was then detected using a Zeiss7 DUO NLO confocal microscope with excitation at 488 nm, and the emissions were measured at a wavelength of 525 nm.
MDC staining was carried out as described previously [44]. Briefly, 7-d-old seedlings germinated on 1/2 MS medium were transferred to 1/2 MS medium containing 0, 1, or 3% glucose for 24 h and subsequently stained with 0.05 mM MDC (Sigma, D4008) in PBS for 10 min. After being washed 2 times with PBS, the root cells were then observed with a Leica DM5000B microscope.
Plasmid construction
To generate the pGADT7-ATG8E, pGBKT7-ATG8E, pGADT7-ABCD1 B walker motif and pGBKT7-ABCD1 B Walker motif constructs, the complete ATG8E coding sequence and ABCD1 B Walker motif coding sequence were recombined into the vectors pGADT7 and pGBKT7 (Clontech, 630442 and 630443). To construct the HA-GFP-ATG8E and FLAG-GFP-ABCD1-B Walker motif expression vectors, the full-length cDNA fragments of ATG8E and the coding sequence of ABCD1-B (B Walker motif) were inserted into pUC120-HA-GFP and pUC119-FLAG-GFP to fuse with 2X HA and 2X FLAG, respectively.
Y2H and CoIP assays
For Y2H assays, preparation of the yeast competent cells and yeast transformation were conducted as described previously [80]. Plasmids for transient expression analysis were extracted using a Maxi Kit (Omega, D6922-02). The yeast strain AH109 was transformed with pGADT7 and pGBKT7 as a negative control. After mating of the yeast strains, Y2H assays were performed by plating a dilution series of yeast cells on selective media. The transformants were selected on synthetic defined SD/-Trp-Leu or SD/-Trp-Leu-His-Ade dropout medium. The transformants growing on SD/-Trp-Leu-His-Ade dropout medium indicate interaction between the corresponding proteins.
For the CoIP assays, 4-week-old Col-0 seedlings were harvested and protoplasts were extracted from samples as described previously [81]. Protoplasts were transfected with the indicated plasmids and cultured for 16 h for protein expression. The cells were then collected and lysed in immunoprecipitation (IP) buffer (10 mM HEPES, pH 7.4, 2 mM EDTA, 150 mM NaCl, 10% glycerol) with 0.5% Triton X-100 (Sigma, T8787). Approximately 10% of the total lysis was kept for input, and the remainder was incubated with FLAG affinity beads (Sigma-Aldrich, M8823) for 4 h at 4°C. The beads were then immobilized and washed 5 times with IP buffer containing 0.1% Triton X-100, followed by sample buffer elution at 95°C for 5 min. Eluted proteins were separated on a 10% SDS-PAGE gel.
Generation of ABCD1-OE transgenic plants
The full-length coding sequence of ABCD1 was amplified with RT-PCR on a VeritiTM 96-well Fast Thermal Cycler (Applied Biosystems, 4375305, USA) using the gene-specific primer pairs listed in Table S1. The PCR products were cloned into the BamHI sites of the binary vector pBI-eGFP [48] to generate the 35S:ABCD1 construct. The plasmid was mobilized from Escherichia coli to Agrobacterium tumefaciens strain LBA4404 by triparental mating, and the resultant Agrobacterium was used to transform the wild type (Col-0) and the atg5 and atg7 mutants. Putative transgenic plants expressing 35S:ABCD1 were selected on 1/2 MS medium containing 1% sucrose and kanamycin (50 g/mL).
Statistical analysis
The significance of the difference between 2 groups was determined using a Student t test. The level of statistical significance is indicated by asterisks (*P < 0.05 and **P < 0.01). The numbers of samples are indicated in the figure legends.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31725004, 31670276, 31370298, and 31461143001 to S. X.].
Acknowledgments
We thank the ABRC for the provision of all atg mutant seeds; C.W. Yang for providing the DR5pro:GFP, DR5pro:GUS, PLT1-YFP, PLT2-YFP, and SCR-GFP lines; N. Yao for the PIN1-GFP line; L.W. Jiang for the eCFP-SKL line, and F.Q. Li for anti-ATG1A antibodies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- [1].Sheen J, Zhou L, Jang J.. Sugars as signaling molecules. Curr Opin Plant Biol. 1999;2(5):410–418. [DOI] [PubMed] [Google Scholar]
- [2].Leon P. Sugar and hormone connections. Trends Plant Sci. 2003;8(3):110–116. [DOI] [PubMed] [Google Scholar]
- [3].Gibson SI, Sugar and phytohormone response pathways: navigating a signalling network. J Exp Bot. 2004;55(395):253–264. [DOI] [PubMed] [Google Scholar]
- [4].Sheen J. Master regulators in plant glucose signaling networks. J Plant Biol. 2014;57(2):67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jang JC, León P, Zhou L, et al. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9(1):5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen J, Jones AM. AtRGS1 Function in Arabidopsis thaliana. Method Enzymol. 2004;389:338–350. [DOI] [PubMed] [Google Scholar]
- [7].Shahri W, Ahmad SS, Tahir I. Sugar signaling in plant growth and development. In: hakeem KR, Rehman RU, Tahir I, editors. Plant signaling: understanding the molecular crosstalk. New Delhi (ND): Springer India; 2014. p. 93–116. [Google Scholar]
- [8].Moore B. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300(5617):332–336. [DOI] [PubMed] [Google Scholar]
- [9].Karve A, Xia X, Moore BD. Arabidopsis Hexokinase-Like1 and Hexokinase1 form a critical node in mediating plant glucose and ethylene responses. Plant Physiol. 2012;158(4):1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Halford NG, Hey S, Jhurreea D, et al. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot. 2003;54(382):467–475. [DOI] [PubMed] [Google Scholar]
- [11].Xiong Y, McCormack M, Li L, et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496(7444):181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Urano D, Phan N, Jones JC, et al. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012;14(10):1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhou L, Jang JC, Jones TL, et al. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA. 1998;95(17):10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mishra BS, Singh M, Aggrawal P, et al. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One. 2009;4(2):e4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carvalho RF, Carvalho SD, Duque P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010;154(2):772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kushwah S, Jones AM, Laxmi A. Cytokinin interplay with ethylene, auxin, and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiol. 2011;156(4):1851–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gupta A, Singh M, Laxmi A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015;168(1):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rellán-Álvarez R, Lobet G, Dinneny JR. Environmental control of root system biology. Annu Rev Plant Biol. 2016;67(1):619–642. [DOI] [PubMed] [Google Scholar]
- [19].Sánchez-Fernández R, Fricker M, Corben LB, et al. Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc Natl Acad Sci USA. 1997;94(6):2745–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pacifici E, Polverari L, Sabatini S. Plant hormone cross-talk: the pivot of root growth. J Exp Bot. 2015;66(4):1113–1121. [DOI] [PubMed] [Google Scholar]
- [21].Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010;2(6):a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annu Rev Plant Biol. 2012;63(1):563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee Y, Lee WS, Kim SH. Hormonal regulation of stem cell maintenance in roots. J Exp Bot. 2013;64(5):1153–1165. [DOI] [PubMed] [Google Scholar]
- [24].Sabatini S, Beis D, Wolkenfelt H, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99(5):463–472. [DOI] [PubMed] [Google Scholar]
- [25].Galinha C, Hofhuis H, Luijten M, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449(7165):1053–1057. [DOI] [PubMed] [Google Scholar]
- [26].Sabatini S, Heidstra R, Wildwater M, et al. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Gene Dev. 2003;17(3):354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Benfey PN, Linstead PJ, Roberts K, et al. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119(1):57–70. [DOI] [PubMed] [Google Scholar]
- [28].Haecker A, Gross-Hardt R, Geiges B, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131(3):657–668. [DOI] [PubMed] [Google Scholar]
- [29].Liszkay A, van der Zalm E, Schopfer P. Production of reactive oxygen intermediates (O2˙−, H2O2, and ˙OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004;136(2):3114–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dunand C, Crevecoeur M, Penel C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol. 2007;174(2):332–341. [DOI] [PubMed] [Google Scholar]
- [31].Del Pozo JC. Reactive oxygen species: from harmful molecules to fine-tuning regulators of stem cell niche maintenance. PLoS Genet. 2016;12(9):e1006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143(4):606–616. [DOI] [PubMed] [Google Scholar]
- [33].Yu Q, Tian H, Yue K, et al. A P-loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PLoS Genet. 2016;12(9):e1006175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zeng J, Dong Z, Wu H, et al. Redox regulation of plant stem cell fate. Embo J. 2017;36(19):2844–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].He J, Duan Y, Hua D, et al. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012;24(5):1815–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang L, Zhang J, He J, et al. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 2014;10(12):e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Vernoux T, Wilson RC, Seeley KA, et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during during postembryonic root development. Plant Cell. 2000;12(1):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Couée I, Sulmon C, Gouesbet G, et al. Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot. 2006;57(3):449–459. [DOI] [PubMed] [Google Scholar]
- [39].Peer WA, Cheng Y, Murphy AS. Evidence of oxidative attenuation of auxin signalling. J Exp Bot. 2013;64(9):2629–2639. [DOI] [PubMed] [Google Scholar]
- [40].Pěnčík A, Simonovik B, Petersson SV, et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell. 2013;25(10):3858–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Sign. 2014;20(3):460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol. 2007;10(6):587–593. [DOI] [PubMed] [Google Scholar]
- [44].Chen L, Liao B, Qi H, et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy. 2015;11(12):2233–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Han S, Yu B, Wang Y, et al. Role of plant autophagy in stress response. Protein Cell. 2011;2(10):784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. [DOI] [PubMed] [Google Scholar]
- [47].Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–what’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. [DOI] [PubMed] [Google Scholar]
- [48].Xiao S, Gao W, Chen QF, et al. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22(5):1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Qi H, Xia F, Xie L, et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell. 2017;29(4):890–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Inoue Y, Suzuki T, Hattori M, et al. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47(12):1641–1652. [DOI] [PubMed] [Google Scholar]
- [51].Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62(3):483–493. [DOI] [PubMed] [Google Scholar]
- [52].Suttangkakul A, Li F, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23(10):3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yuan T, Xu H, Zhang K, et al. Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ. 2014;37(6):1338–1350. [DOI] [PubMed] [Google Scholar]
- [54].Drisch RC, Stahl Y. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front Plant Sci. 2015;6:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Baena-Gonzalez E, Rolland F, Thevelein JM, et al. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448(7156):938–942. [DOI] [PubMed] [Google Scholar]
- [56].Chen L, Su ZZ, Huang L, et al. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci. 2017;8:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kerk NM, Jiang K, Feldman LJ. Auxin metabolism in the root apical meristem. Plant Physiol. 2000;122(3):925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Park S, Gidda SK, James CN, et al. The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell. 2013;25(5):1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51(6):1126–1136. [DOI] [PubMed] [Google Scholar]
- [60].Dietrich D, Schmuths H, De Marcos Lousa C, et al. Mutations in the Arabidopsis peroxisomal ABC transporter COMATOSE allow differentiation between multiple functions in planta: insights from an allelic series. Mol Biol Cell. 2009;20(1):530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156(3):1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].de Jager SM, Scofield S, Huntley RP, et al. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol. 2009;71(4):345–365. [DOI] [PubMed] [Google Scholar]
- [63].Pattingre S, Espert L, Biard-Piechaczyk M, et al. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90(2):313–323. [DOI] [PubMed] [Google Scholar]
- [64].Liu Y, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One. 2010;5(7):e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Soto-Burgos J, Bassham DC. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One. 2017;12(8):e182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Üstün S, Hafrén A, Hofius D. Autophagy as a mediator of life and death in plants. Curr Opin Plant Biol. 2017;40:122–130. [DOI] [PubMed] [Google Scholar]
- [67].Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584(7):1350–1358. [DOI] [PubMed] [Google Scholar]
- [68].Pu Y, Luo X, Bassham DC. TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci. 2017;8:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kunz HH, Scharnewski M, Feussner K, et al. The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell. 2009;21(9):2733–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xu L, Zhao H, Ruan W, et al. ABNORMAL INFLORESCENCE MERISTEM1 Functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in Rice. Plant Cell. 2017;29(3):560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pérez-Pérez ME, Lemaire SD, Crespo JL. Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 2012;160(1):156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kim J, Lee H, Lee HN, et al. Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell. 2013;25(12):4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shibata M, Oikawa K, Yoshimoto K, et al. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013;25(12):4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Peng Y, Ma W, Chen L, et al. Control of root meristem size by DA1-RELATED PROTEIN2 in Arabidopsis. Plant Physiol. 2013;161(3):1542–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xu P, Yuan D, Liu M, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 2013;161(4):1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wysocka-Diller JW, Helariutta Y, Fukaki H, et al. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127(3):595–603. [DOI] [PubMed] [Google Scholar]
- [77].Casamitjana-Martínez E, Hofhuis HF, Xu J, et al. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol. 2003;13(16):1435–1441. [DOI] [PubMed] [Google Scholar]
- [78].Lisec J, Schauer N, Kopka J, et al. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. [DOI] [PubMed] [Google Scholar]
- [79].Zheng SX, Xiao S, Chye ML. The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen inducible and subject to circadian regulation. J Exp Bot. 2012;63(8):2985–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21(1):83–84. [DOI] [PubMed] [Google Scholar]
- [81].Yoo S, Cho Y, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


