Abstract
In this mini-review, the major types of photolytic labeling reagents are presented together with their reaction mechanisms. The applications of photolytic labeling in protein drug discovery and development are then discussed; these have expanded from studies of protein-protein interactions in vivo to protein-matrix interactions in lyophilized solids. The mini-review concludes with recommendations for further development of the approach, which include the need for new and more chemically diverse photo-reactive reagents and better understanding of the mechanisms of photolytic labeling reactions in various media.
Keywords: photolytic labeling, protein drug, phenyl azide, carbene, diazirine, benzophenone, protein-protein interaction, lyophilized solid
Introduction
Photolytic labeling, also known as photoaffinity labeling and photo-crosslinking, is a series of chemical reactions that are activated upon exposure to light at a certain wavelength. With covalent bond formation after UV activation, transient protein-protein or protein-ligand interactions can be captured.1,2 The use of photo-reactive reagents can be dated back to 1969 when the 4-azido-2-nitrophenyl group was first used to label bovine γ-globulin and human serum albumin to study antibodies in vivo3 Today, the application of photolytic labeling has been expanded to several areas including the identification of membrane protein targets, the elucidation of protein structure in solution and the characterization of proteins in pharmaceutical solids.4–6 Photo-reactive reagents typically are stable before UV activation and become reactive under specific activation conditions. Moreover, photolytic labeling generates covalently modified products that are stable enough to be analyzed with common analytical methods. These properties have allowed photolytic labeling to be increasingly used to study protein interactions both in vitro and in vivo.
Selecting appropriate photo-reactive reagents has been important in the success of photolytic labeling studies. In this mini-review, the reaction mechanisms of common photo-reactive reagents will be introduced and the reported applications of these reactions in the discovery and development of protein drugs will be reviewed. The applications include studies of protein-protein interactions, protein-ligand binding, structure of the native protein and its oligomers, and protein-matrix interactions in lyophilized solids.
Photo-Reactive Reagents
1. Phenyl Azide
Phenyl azide labels have been incorporated in the protein structure to study protein-protein interactions, often using modified amino acids such as p-azido-L-phenylalanine.7 The absorption maximum for the phenyl azide group is ~260 nm when the phenyl ring is unsubstituted.8 In studies involving proteins, the short wavelength required to activate phenyl azides may cause damage to protein structures.9 The reaction mechanism has been studied in a rigid glassy system consisting of diethyl ether, isopentane and ethanol at 77K.10 After UV activation, singlet and triplet phenylnitrene were proposed as transient intermediates. Phenylnitrene can insert into C-H, N-H or O-H bonds and form covalently labeled products. Phenylnitrene can also undergo ring expansion to form addition products with nucleophiles such as primary or secondary amines. Trace amounts of azobenzene were detected as side products of the reaction.10 The oxidation of phenylnitrene with formation of nitrobenzene has also been reported.11 The reaction products can be affected by system temperature and by substituent groups on the aromatic ring.12 With many possible reaction pathways (Fig. 1), the labeling yields are often low after irradiation.13 Nitrenes have shown different reactivities towards naturally occurring amino acids, with a preference for cysteine and aromatic amino acids, suggesting that labeling may favor interactions involving these amino acids.14
Fig. 1.
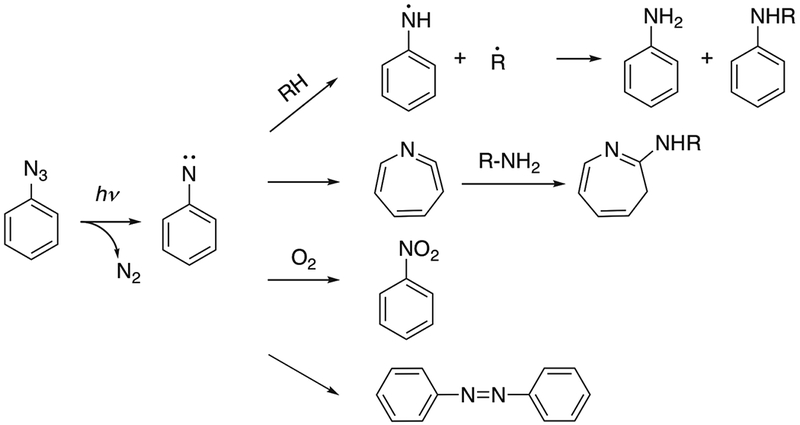
Reaction pathway of phenyl azide reagents. Adapted from [10] and [13].
2. Carbene
Carbenes can form covalent bonds with molecules in their vicinity by inserting into C-C bonds, X-H (X=C, O, N, S) bonds or adding onto C=C bonds.15–17 Photolytic labeling with carbenes is a fast, relatively non-specific radical reaction. Carbenes can be generated from diazirine or diazo groups after UV activation at a certain wavelength.18 Stable diazo compounds such as diazo ketones have been reported to undergo the Wolf rearrangement (see Fig. 2B), which leads to the formation of ketene products.19 Ketenes can be further hydrolyzed by water to form carboxylic acids,20 or can react with other nucleophiles. The inherent instability and low labeling yield of diazo compounds has limited their application in photolytic labeling studies.8
Fig. 2.

Reaction pathway of diazirine with trifluoromethylphenyldiazirine as an example (A) and diazo ketone showing Wolff rearrangement (B). Adapted from [16] and [19].
Unlike diazo compounds, diazirines tend to be relatively stable, allowing their broad application in protein interaction studies.21 In the absence of activating light, diazirines are stable at room temperature. Activation requires absorption of light at relatively long wavelengths (330-370 nm), causing less damage to protein structure than with phenyl azides. It has been shown that the diazirine radical reaction also occurs in amorphous solid powders.22 In addition to the expected photolytic labeling products, diazirine adducts with water and phosphate from buffer salt were detected in solution and solid-state reactions in these studies.23 “Dead-end” products, resulting from deactivation of the carbene without intermolecular reaction, accounted for a large fraction of the photolytic labeling products in these samples.23 The structures of these dead-end products remain unknown. Ketones can form at the carbene carbon after oxidation by O2 or hydrolysis of N-H insertion products.16 Photolysis of the diazirine can induce conversion to diazo compounds, which have low reactivity and can lead to undesired side products.24 A trifluoromethyl group can be introduced into the diazirine carbon to stabilize the carbene and prevent rearrangement.25 The reaction pathways of diazirines are shown in Fig. 2, using trifluoromethylphenyldiazirine (TFMD) as an example.
The selectivity of the carbene towards different amino acid side chains has been studied in fiberglass coupled with 3-(trifluoromethyl)-3-(m-isothiocyanophenyl) diazirine.26 In this study, the carbene preferentially interacted with cysteine and aromatic amino acids while relatively low affinity was observed for amino acids with aliphatic side chains. Although carbene reactions are thought to be relatively non-specific in solution, it remains unclear whether the preferential reactivity observed in the fiberglass system also occurs in amorphous powders containing proteins.
3. Benzophenone
Benzophenones have also been used as photo-reactive reagents in protein interaction studies. For example, p-benzoyl-L-phenylalanine (pBpA) has been incorporated into proteins in vitro and in vivo as a photo-reactive analog of phenylalanine to study protein-ligand binding and protein-protein interactions.27–29 Benzophenone can be activated with UV light at 350–360 nm to form a reactive ketyl radical that reacts preferentially with C-H bonds. A new C-C bond forms between the ketone carbon from benzophenone and the carbon from the C-H bond (see Fig. 3).30 C-H bonds adjacent to N or S atoms, especially those in methionine, are favored reaction sites for benzophenone compounds.31–33 Proteins are less damaged at the wavelengths required for the activation of benzophenone than with the shorter wavelengths needed for phenyl azides, although benzophenone occasionally requires a relatively long irradiation period.34 The structure of benzophenone makes it stable in many solvents. However, one of the disadvantages of using benzophenone is that its structure does not mimic those of natural amino acids without aromatic rings. As a result, protein structure may be disrupted with the incorporation of pBpA or other benzophenones.
Fig. 3.
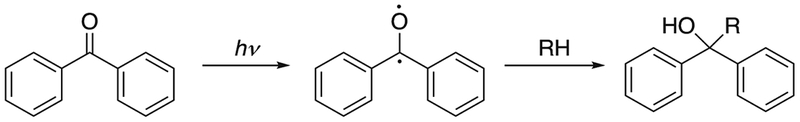
Reaction pathway of benzophenone. Adapted from [30].
4. Other Reagents
Click-chemistry is an addition reaction with high specificity. A typical example is an azide-alkyne cycloaddition to form triazole compounds with copper as a catalyst.35 While the high specificity may be desirable in some cases, the requirement of metal ions may not be suitable for protein interaction studies. A metal-free click chemistry reaction has been achieved in human Jurkat cells.36 N-azidoacetyl-sialic acid was metabolically introduced into cell surface glycoproteins. Biotinylated cyclopropenone was chemically synthesized with the reactive triple bond masked, and the click-chemistry addition between the azide and dibenzocyclooctyne was activated by UV exposure at 350 nm (Fig. 4). The labeling products were evaluated with fluorescence microscopy.
Fig. 4.
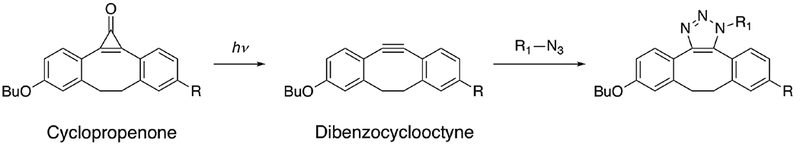
An example of photo-triggered click chemistry reaction in living cells. Adapted from [36].
2-aryl-5-carboxytetrazole (ACT) has been used as a photo-reactive label for drug target identification.37 ACT was covalently attached to drug candidates such that the modification did not significantly change their binding affinity and specificity. A reaction mechanism was proposed (Fig. 5) in which, after ligand binding, a carboxy-nitrile imine intermediate is generated following UV exposure at 302 nm, which then reacts with a nucleophile (e.g., the carboxylate group on Glu side chain) to form a covalent bond with the target protein near the active site. Binding site analysis was achieved with mass spectrometry (MS). The efficiency in capturing desired targets in vivo using the ACT label was comparable with the that of diazirine-labeled ligands in this study.
Fig. 5.
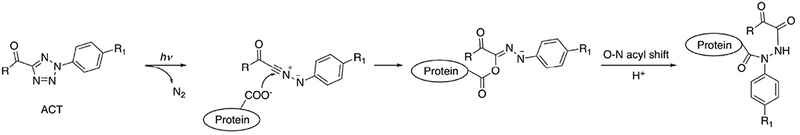
Proposed mechanism of ACT reaction. Adapted from [37].
S-adenosyl-L-methionine (AdoMet) is a natural substrate for methyltransferases. The methyl group on the sulfur atom can be transferred by methyltransferase to biological targets in the cell.38 With UV exposure, AdoMet itself was shown to specifically label several methyltransferases.39,40 This method has been used to study the binding site of AdoMet.41,42 An analog of AdoMet has also been used to specifically label DNA.43
Applications
1. Protein-protein interactions
Photo reactive reagents have been used to study protein-protein interactions in vivo. These reagents tend to have structures similar to those of natural amino acids and can be site-specifically incorporated into the protein. UV irradiation in situ induces the formation of covalent protein-protein adducts, allowing transient protein-protein interactions to be captured.
L-photo-leucine and L-photo-methionine are two diazirine compounds that can be chemically synthesized (see Fig. 7A) and are available commercially. Their structures are similar to the corresponding natural amino acids. Photo-leucine and photo-methionine have been successfully incorporated into membrane proteins through unmodified mammalian translation machinery of COS7 (monkey kidney) cells.1 These two unnatural amino acids were nontoxic to these cells. Different protein-protein complexes linked with covalent bonds were detected by western blotting. Photolytic labeling enabled a new protein-protein interaction between membrane protein PGRMC1 and Insig-1 to be identified, which is involved in the regulation of cellular lipid homeostasis. In a study of Alzheimer’s disease, photo-leucine and photo-methionine were used to confirm the preferential binding of soluble oligomeric assemblies of amyloid β peptides (ADDLs) with a subunit of AMPA receptors in neurons.44 A similar method utilizing these two photo-reactive reagents was used to detect a direct interaction between subunits of RNA polymerase II and a mediator protein complex in eukaryotic cells.45
Fig. 7.
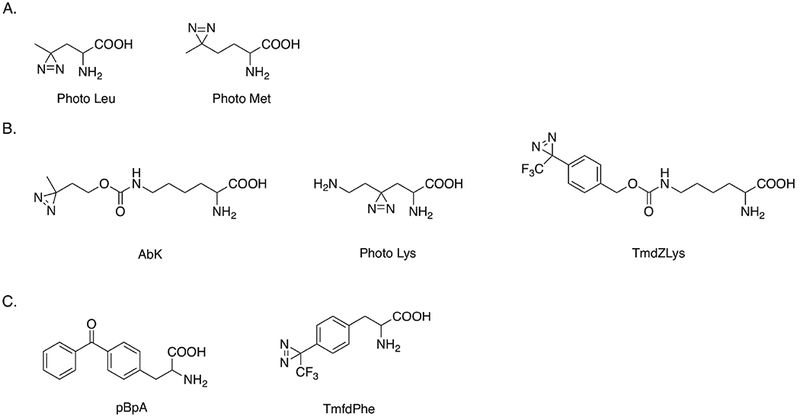
Structures of photo-reactive amino acids site specifically incorporated into proteins in vivo. Adapted from [1,27,46–48,50].
Several diazirine compounds with structures similar to lysine have been used to study protein-protein interactions. 3’-azibutyl-N-carbamoyl-lysine (AbK) has the diazirine functional group attached at the end of the lysine side chain. An approach using a combination of AbK photolytic labeling and stable isotope labeling of amino acids in cell culture (SILAC) allowed protein interactions with histone subunits to be identified in HEK293T cells.46 A second photolytically labeled lysine that retains the primary amine on the side chain was site-specifically incorporated into histone and used to identify histone and chromatin binding proteins during post-translational modifications in HeLa cells.47 In order to expand the spatial range in which interactions could be detected, Nε-[((4-(3-(trifluoromethyl)-3H-diazirin-3-yl)-benzyl)oxy)carbonyl]-L-lysine (TmdZLys) was prepared to detect possible protein interactions, with a spacer arm of 15 Å.48 The structures of AbK, photo lysine and TmdZlys are shown in Fig. 7B.
The benzophenone compound p-benzoyl-L-phenylalanine (pBpA) has been used for protein-protein interaction studies in vitro and in vivo following procedures similar to those described above.27 pBpA has also been used to study protein-DNA binding.49 Another diazirine derivative of phenylalanine, 4’-[3-(trifluoromethyl)-3H-diazin-3-yl]-L-phenylalanine (TmfdPhe), has been genetically encoded in Escherichia coli and could be used in photolytic labeling in vivo50, though results have not been reported. Structures of pBpA and TmfdPhe are shown in Fig. 7C.
2. Protein-ligand binding
Photolytic labeling can be used to identify the interactions involved in ligand binding. For ligands that interact with proteins non-covalently, it is often difficult to detect their target in situ. Ligands with photo-reactive functional groups can be activated after UV exposure to form covalent bonds with the target protein. The ligand-protein complex can then be analyzed with methods such as SDS-PAGE and liquid chromatography mass spectrometry (LC-MS). Known ligands with photo-reactive functional groups can also be used to discover off-target interactions, and to study selectivity in ligand-protein binding.
In early development, many drugs are designed to be ligands that are selective for specific targets. Off-target interactions are generally undesirable, as they can lead to unexpected side effects.51 However, the study of off-target interactions may expand the understanding and effective usage of existing drugs, and may enable the discovery of new drugs or facilitate screening of drug candidates for a range of targets early in development.52 Dasatinib is a chemotherapeutic agent that targets the Src/Abl family of tyrosine kinases, and is used for the treatment of imatinib-resistant chronic myelogenous leukemia (CML).53 DA-2, a dasatinib analog with its core structure maintained, was used to establish a proteome profiling method to identify off-target interactions of dasatinib in vivo.54 DA-2 differs from dasatinib in that a diazirine and an alkyne were introduced through chemical synthesis. DA-2 was internalized by both K562 and HepG2 cancer cells and bound to various intracellular targets. Following UV exposure, covalent bonds formed between DA-2 and the target proteins through the diazirine moiety. Two click reporters, rhodamine and biotin, each modified to contain an azide group, were then reacted with the DA-2 alkyne through click chemistry, allowing the ligand-complexes to be purified and analyzed. Several serine/threonine kinases were identified with this method, and their activities were validated by pull-down/immunoblotting experiments and kinase inhibition assays. In this study, the photolytic labeling method was able to identify more putative targets than an immobilized dasatinib affinity matrix. Similar diazirine and alkyne modifications have been made to the inhibitor MLN8237, which targets the ATP-binding site of kinases. The off-target interactions of MLN8237 were identified.55
Angiotensin II (AngII) is an octapeptide hormone that activates AT1 and AT2 receptors and functions in the regulation of the cardiovascular system.56 Some truncated analogs of AngII have been found to be biological active, but their receptors are difficult to distinguish from one another. Two photo-reactive analogs of the hexapeptide AngIV, [N3-Phe6]AngIV and [Bpa6]AngIV, were developed with the photo-reactive functional group at the phenylalanine residue.57 [N3-Phe6]AngIV underwent the phenyl azide photo-reaction (Fig. 1) and [Bpa6]AngIV underwent the benzophenone photo-reaction (Fig. 3). [N3-Phe6]AngIV and [Bpa6]AngIV showed high affinity for the AT4 receptor from bovine endothelial membrane. Subsequent analysis of AT4 showed that this receptor is a 186 kDa integral membrane glycoprotein.
3. Structures of proteins and their oligomers
Photolytic labeling can be used to form covalent bonds between nearby residues within proteins themselves. Following UV exposure, analysis of the digested peptides provides information on protein structure. Three hetero-bifunctional probes that are both isotopically-labeled and photo-reactive have been developed to study protein structure.5 All have an NHS ester in their structure that reacts preferentially with primary amines, allowing derivatization of lysine groups in the protein. The photo-reactive functional groups on the probes were phenyl azide (ABAS-12C6/13C6), diazirine (SDA-12C5/13C5) and benzophenone (CBS-12C6/13C6). Proteins were first incubated with the probe in the dark to allow reaction with the NHS ester, followed by UV exposure to induce covalent bond formation within ~ 5 or 7 A of the lysine nitrogen atom. ABAS and CBS were able to detect crosslinks in a well-established system of RNase S. The probe ABAS was also used to study the structure of a disordered protein, α-synuclein, which is implicated in neurodegenerative disease. Multiple interactions within the protein were identified.
Transient protein-interactions, such as oligomerization dependent on post-translational modifications, can be difficult to capture. To address this problem, photo methionine was site-specifically incorporated into the MH2 domain of the Smad2 signaling protein, which forms homo-trimers in response to the phosphorylation of serine residues.58 This method was compatible with both solid-phase peptide synthesis and expressed protein ligation. The transient MH2-MH2 interaction was captured with photolytic labeling. In a study of Aβ16-22, its photo-reactive analogs were generated and incubated to form aggregates.59 Photolytic labeling of the diazirine analog within the aggregates was successfully used to characterize the inter-peptide interactions. The insertion sites were identified with LC-MS analysis.
4. Protein-matrix interactions in lyophilized solids
Lyophilization, also known as freeze-drying, is widely used to create dried powders of therapeutic proteins with the goal of preserving the protein and extending the shelf-life of the product.60 Excipients such as disaccharides are usually included in the formulation, and are thought to contribute to stabilization by interacting with the protein in the solid matrix.61 Understanding the interactions between the lyophilized matrix and the protein could contribute to rational formulation design and reduce the time-to-market for new protein drugs. At present, however, there is a lack of methods able to detect non-covalent protein-matrix interactions in the solid state. Our group has shown that photolytic labeling reactions occur in solid powders and can provide information on the interactions between the protein and the lyophilized matrix.
In one approach to these studies, the photo-reactive reagent can be incorporated in the lyophilized matrix as an excipient. For example, photo leucine has been included in lyophilized apomyoglobin formulations at molar ratios of apomyoglobin to photo leucine from 1:20 to 1:100.6 Sucrose was also included in the matrix. After lyophilization, the solid samples were exposed to UV at 365 nm to induce photo-reaction. The photolytic labeling products formed from apomyoglobin and photo leucine were analyzed at both intact and digested protein levels, and the sites of labeling were localized to specific peptide fragments. The distribution of the photo leucine label on apomyoglobin depended on the ratio of photo leucine to apomyglobin in the sample, and gave an indication of the local interactions between apomyoglobin and photo leucine in the lyophilized matrix.
In an alternative approach, the photo-reactive reagent can be site-specifically incorporated into the protein to study its interaction with surrounding environment in the lyophilized solids. Succinimidyl 4,4’-azipentanoate (SDA) was used to react with lysine residues in order to introduce a diazirine functional group into myoglobin.22 The addition of the diazirine label did not disturb the overall structure of myoglobin. Diazirine-labeled myoglobin was then lyophilized with either raffinose or Gdn HCl to detect protein-excipient interactions in the solid state. After proteolytic digestion of the labeled products, peptide-peptide, peptide-water and peptide-excipient adducts were identified by LC-MS.
Future Directions and Challenges
It has been nearly 50 years since photolytic labeling was first introduced to study protein-protein interactions in vivo3 Since that time, photo-reactive reagents have been mostly used in solution environments to study protein-related interactions. Successful application of the method to study protein-matrix interactions in solid samples has been reported only recently.6 The photolytic labeling reagents that have been discussed in this mini-review are summarized in Table 1, together with their advantages and disadvantages, as well as applications that have been reported. This section describes potential future developments of photolytic labeling technologies in protein drug development, as well as challenges that may be associated with these developments.
Table 1.
Photo-reactive reagents and their applications
| Reagent1 | Applications2 | Advantages | Disadvantages | Refs |
|---|---|---|---|---|
| Phenyl Azides | ||||
| p-azido-L-phenylalanine | PPI | - Structural similarities to phenylalanine | - Short wavelength light required - Low labeling yield - Unexpected side products |
7 |
| ABAS-12C6/13C6 | PS | 5 | ||
| TmfdPhe | NR | 50 | ||
| Carbenes | ||||
| Photo Leu | PPI, PMI | - Small size - High labeling efficiency - Activation wavelength safe for proteins |
- Unexpected side products | 1, 6, 44, 45 |
| Photo Met | PPI | 1, 44, 45 | ||
| AbK | PPI | 46 | ||
| Photo Lys | PPI | 47 | ||
| TmdZLys | PPI | 48 | ||
| TFMD | PA | 59 | ||
| NHS-SDA | PS, PMI | 5, 22 | ||
| DA-2 | PLB | 54 | ||
| Benzophenones | ||||
| pBpA | PPI, PLB, PDB | - Genetically encoded | - Limited structural flexibility | 27–29, 49 |
| CBS-12C6/13C6 | PS | 5 | ||
| Other Reagents | ||||
| Cyclopropenone | Click | - Largely unknown | - Largely unknown | 36 |
| ACT | Target ID | 37 | ||
| AdoMet | Labeling | 39–42 | ||
See text for abbreviations and full chemical names.
Abbreviations: PPI = protein-protein interactions, PS = protein structure determination, NR = none reported, PMI = protein-matrix interactions in solid samples, PA = protein aggregates, PLB = protein-ligand binding, PDB = protein-DNA binding, Click = photo-activated click chemistry, Target ID = drug target identification, Labeling = labeling of methyltransferases and DNA.
As noted above, photo-reactive reagents can be introduced into protein structure site-specifically through cell translation machinery. Protein-protein complexes can then form in situ through photo-activated covalent bonds, which allow them to remain stable through common detection methods. An advantage of this approach is that the location of the photo-reactive functional group can be controlled. However, this approach is limited by the availability of photo-reactive amino acid analogs, and by the ability of the derivatized protein to be taken up by cells. Photo leucine and photo methionine are commonly used reagents and are preferred for their small size and minor change to the side chain structure of the corresponding amino acids. Other reagents such as pBpA have also been used, but their structures mimic those of natural amino acids less closely. Some photo-reactive reagents simply cannot be recognized and utilized by cells if their structures differ too greatly from natural amino acids. Thus, there is a need for a broader range of photolytically labeled amino acid analogs that can be efficiently expressed in proteins of interest and recognized by cells.
The photo-reactive moiety can also be introduced into protein through a post-translational chemical reaction. As noted above, the bifunctional probe succinimidyl 4,4’-azipentanoate (SDA) has been used in a number of protein-protein interaction studies, and contains a diazirine group and an NHS ester. The process of introducing the diazirine functional group into the protein via the reaction of NHS with lysine is relatively simple, but changes the size and charge of the lysine side chain. If lysine plays a critical role in the interaction to be studied, the results can be misleading. Moreover, the diazirine label cannot be introduced site-specifically with this approach. In addition to its expected reaction with lysine, the NHS ester may also react with serine, tyrosine and threonine.63 Due to the heterogeneity of labeling, a large number of photolytically labeled species may be produced that differ in the extent and sites of labeling. This can make it difficult to analyze the derivatized protein and the products of its photolytic reaction at either the whole protein or peptide level. Similarly, quantitation of photolytic labeling products is complicated by this heterogeneity. Thus, there is a need for site-specific chemical labeling approaches that can be applied post-translationally to produce uniformly labeled protein.
The selection of appropriate photo-reactive reagents for a given study is also a concern, since the use of different photo-reactive reagents has been shown to give different results in some cases. Benzophenone, diazirine and phenyl azide labeling were compared in a study to identify the target proteins of PPAR-e and Tubulin-i.64 The investigators found that the target proteins that were identified depended on the type of photo-reactive reagent that was used. In a study of Aβ16-22 amyloid structure, three analogs containing phenyl azide (PA), phenyl trifluoromethyldiazirine (TFMD) and benzophenone (BP) were generated. The TFMD analog generated only intermolecular peptide-peptide products and provided straightforward information on aggregate structure. The information from PA and BP was less clear and more difficult to interpret.59 A better understanding of the mechanisms of photolytic labeling reactions in various media would help in interpreting the results. Most commercial photolytic labeling reagents undergo radical reaction. However, the mechanisms are not fully understood in specific reaction environments, including the solid state, and the effects of parameters such as temperature, pH, viscosity and crowding are unknown. Some photo-reactive reagents show preferences towards particular amino acids, as mentioned previously, which can bias the results if these preferences are not recognized. Knowing these preferences and the factors controlling photolytic reactions would enable better use of this approach and further expansion of its application. Similarly, very little is known about the toxicities of photolytic labeling reagents or their more subtle effects on cellular processes. While photolytic reagents that are expressed or introduced into cells are often assumed to be non-toxic if the cells remain viable, it is possible that the pathways of interest are affected by the non-native functional groups, even before photolytic activation. In vitro and in vitro results using photolytic labeling could be interpreted with greater confidence if these effects were better understood.
Photo-reactive functional groups can also be incorporated into other molecules that are relevant for the discovery and development of protein drugs. For example, in formulation development, excipients with photo-reactive functional groups could be incorporated into the formulation to study protein-excipient interactions. Following the photolytic reaction, the resulting protein-excipient complex could be analyzed with methods such as HPLC and LC-MS with high resolution. The results would shed light on the protein-excipient interactions associated with protein stability and could lead to a better understanding of protein-excipient interaction mechanisms, all of which would benefit formulation design in the future. To facilitate these studies, there is a need for photolytically labeled compounds that mimic the excipients commonly used in protein formulation, including sugars and stabilizing amino acids (e.g., histidine).
Fig. 6.
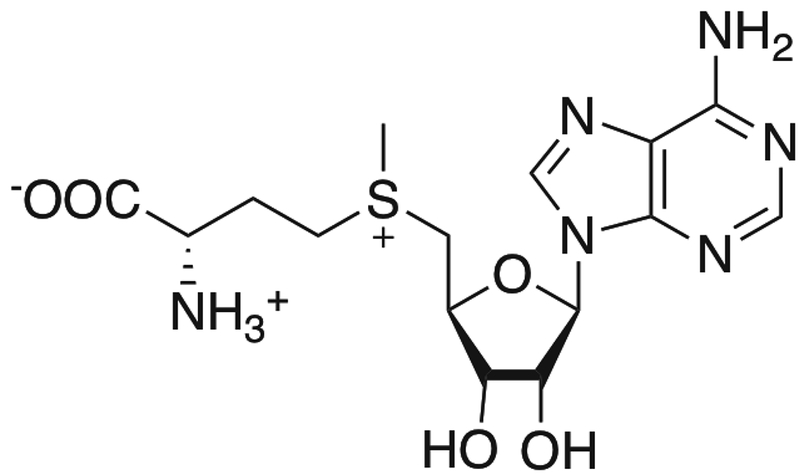
AdoMet structure. Adapted from [38].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005;2(4):261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 2.Rao S, Horwitz SB, Ringel I. Direct Photoaffinity Labeling of Tubulin With Taxol. JNCI JNatl Cancer Inst. 1992;84(10):785–788. 10.1093/jnci/84.10.785. [DOI] [PubMed] [Google Scholar]
- 3.FLEET GWJ PORTER KNOWLES RR JR. Affinity Labelling of Antibodies with Aryl Nitrene as Reactive Group. Nature. 1969;224:511 10.1038/224511a0. [DOI] [Google Scholar]
- 4.Johnson GL, MacAndrew VI, Pilch PF. Identification of the glucagon receptor in rat liver membranes by photoaffinity crosslinking. Proc Natl Acad Sci. 1981;78(2):875–878. http://www.pnas.org/content/78/2/875.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodie NI, Makepeace KAT, Petrotchenko E V, Borchers CH. Isotopically-coded short-range hetero-bifunctional photo-reactive crosslinkers for studying protein structure. J Proteomics. 2015;118(Supplement C):12–20. doi: 10.1016/j.jprot.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Iyer LK, Moorthy BS, Topp EM. Photolytic Labeling to Probe Molecular Interactions in Lyophilized Powders. MolPharm. 2013;10(12):4629–4639. doi: 10.1021/mp4004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002; 124(31): 9026–9027. [DOI] [PubMed] [Google Scholar]
- 8.Neberle A, de Graan PNEBT-M in E. [11] General principles for photoaffinity labeling of peptide hormone receptors In: Hormone Action Part I: Peptide Hormones. Vol 109 Academic Press; 1985:129–156. doi: 10.1016/0076-6879(85)09081-4. [DOI] [Google Scholar]
- 9.Davies MJ. Singlet oxygen-mediated damage to proteins and its consequences. Biochem Biophys Res Commun. 2003;305(3):761–770. doi: 10.1016/S0006-291X(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 10.Leyva E, Platz MS, Persy G, Wirz J. Photochemistry of phenyl azide: the role of singlet and triplet phenylnitrene as transient intermediates. J Am Chem Soc. 1986;108(13):3783–3790. doi: 10.1021/ja00273a037. [DOI] [Google Scholar]
- 11.Bergeron RJ, Dionis JB, Ingeno MJ. Synthesis of a parabactin photoaffinity label. J Org Chem. 1987;52(1): 144–149. doi: 10.1021/jo00377a026. [DOI] [Google Scholar]
- 12.L’abbe G Decomposition and addition reactions of organic azides. Chem Rev. 1969;69(3):345–363. doi: 10.1021/cr60259a004. [DOI] [Google Scholar]
- 13.Geurink PP, Prely LM, van der Marel GA, Bischoff R, Overkleeft HS. Photoaffinity Labeling in Activity-Based Protein Profiling BT - Activity-Based Protein Profiling. In: Sieber SA, ed. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012:85–113. doi: 10.1007/128_2011_286. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz MA. Studying the Cytoskeleton by Label Transfer Crosslinking: Uses and Limitations BT - Photochemical Probes in Biochemistry. In: Nielsen PE, ed. Dordrecht: Springer Netherlands; 1989:157–168. doi: 10.1007/978-94-009-0925-0_11. [DOI] [Google Scholar]
- 15.Buehler CA. Carbenes in insertion and addition reactions. J Chem Educ. 1972;49(4):239. doi: 10.1021/ed049p239. [DOI] [Google Scholar]
- 16.Smith E, Collins I. Photoaffinity labeling in target- and binding-site identification. Future Med Chem. 2015;7(2):159–183. doi: 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner J, Senn H, Richards FM. 3-Trifluoromethyl-3-phenyldiazirine. A new carbene generating group for photolabeling reagents. J Biol Chem . 1980;255(8):3313–3318. http://www.jbc.org/content/255/8/3313.abstract. [PubMed] [Google Scholar]
- 18.Cowell GW, Ledwith A. Developments in the chemistry of diazo-alkanes. Q Rev Chem Soc. 1970;24(1):119–167. doi: 10.1039/QR9702400119. [DOI] [Google Scholar]
- 19.Kirmse W 100 Years of the Wolff Rearrangement. European J Org Chem. 2002;2002(14):2193–2256. doi:. [DOI] [Google Scholar]
- 20.Louie MK, Francisco JS, Verdicchio M, Klippenstein SJ, Sinha A. Hydrolysis of Ketene Catalyzed by Formic Acid: Modification of Reaction Mechanism, Energetics, and Kinetics with Organic Acid Catalysis. JPhys Chem A. 2015;119(19):4347–4357. doi: 10.1021/jp5076725. [DOI] [PubMed] [Google Scholar]
- 21.Dubinsky L, Krom BP, Meijler MM. Diazirine based photoaffinity labeling. Bioorg Med Chem. 2012;20(2):554–570. doi: 10.1016/j.bmc.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 22.Iyer LK, Moorthy BS, Topp EM. Photolytic Crosslinking to Probe Protein-Protein and Protein-Matrix Interactions In Lyophilized Powders. MolPharm. 2015;12(9):3237–3249. doi: 10.1021/acs.molpharmaceut.5b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Topp EM. Quantitative Analysis of Peptide-Matrix Interactions in Lyophilized Solids Using Photolytic Labeling. Mol Pharm. May 2018. doi: 10.1021/acs.molpharmaceut.8b00283. [DOI] [PubMed] [Google Scholar]
- 24.Liu MTH. The thermolysis and photolysis of diazirines. Chem Soc Rev. 1982;11(2):127–140. doi: 10.1039/CS9821100127. [DOI] [Google Scholar]
- 25.Das J Aliphatic Diazirines as Photoaffinity Probes for Proteins: Recent Developments. Chem Rev. 2011;111(8):4405–4417. doi: 10.1021/cr1002722. [DOI] [PubMed] [Google Scholar]
- 26.Sigrist H, Mühlemann M, Dolder M. Philicity of amino acid side-chains for photogenerated carbenes. JPhotochem PhotobiolB Biol. 1990;7(2):277–287. doi: 10.1016/1011-1344(90)85162-P. [DOI] [Google Scholar]
- 27.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2(5):377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 28.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002;99(17): 11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettelkau J, Ihling CH, Frohberg P, van Werven L, Jahn O, Sinz A. Reliable identification of cross-linked products in protein interaction studies by 13C-labeled p-benzoylphenylalanine. J Am Soc Mass Spectrom. 2014;25(9):1628–1641. doi: 10.1007/s13361-014-0944-6. [DOI] [PubMed] [Google Scholar]
- 30.Dorman G, Prestwich GD. Benzophenone Photophores in Biochemistry. Biochemistry. 1994;33(19):5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 31.Jahn O, Eckart K, Brauns O, Tezval H, Spiess J. The binding protein of corticotropin-releasing factor: Ligand-binding site and subunit structure. Proc Natl Acad Sci. 2002;99(19): 12055–12060. http://www.pnas.org/content/99/19/12055.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kage R, Leeman SE, Krause JE, Costello CE, Boyd ND. Identification of methionine as the site of covalent attachment of a p-benzoyl-phenylalanine-containing analogue of substance P on the substance P (NK-1) receptor. JBiol Chem. 1996;271(42):25797–25800. [DOI] [PubMed] [Google Scholar]
- 33.Angela W E TB F MD, Michael R. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBSLett. 2006;580(7):1872–1876. doi: 10.1016/j.febslet.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 34.PG D, György D, EJ T, MD M, Anu C. Benzophenone Photoprobes for Phosphoinositides, Peptides and Drugs. Photochem Photobiol. 2008;65(2):222–234. doi: 10.1111/j.1751-1097.1997.tb08548.x. [DOI] [PubMed] [Google Scholar]
- 35.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chem Soc Rev. 2007;36(8): 1249–1262. doi: 10.1039/B613014N. [DOI] [PubMed] [Google Scholar]
- 36.Poloukhtine AA, Mbua NE, Wolfert MA, Boons G- J, Popik V V. Selective Labeling of Living Cells by a Photo-Triggered Click Reaction. J Am Chem Soc. 2009;131(43):15769–15776. doi: 10.1021/ja9054096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herner A, Marjanovic J, Lewandowski TM, et al. 2-Aryl-5-carboxytetrazole as a New Photoaffinity Label for Drug Target Identification. J Am Chem Soc. 2016;138(44): 14609–14615. doi: 10.1021/jacs.6b06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struck A- W, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem. 2012;13(18):2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 39.Som S, Friedman S. Direct photolabeling of the EcoRII methyltransferase with S-adenosyl-L-methionine. J Biol Chem . 1990;265(8):4278–4283. http://www.jbc.org/content/265/8/4278.abstract. [PubMed] [Google Scholar]
- 40.Finta C, Sulima U, Venetianer P, Kiss A. Purification of the KpnI DNA methyltransferase and photolabeling of the enzyme with S-adenosyl-l-methionine. Gene. 1995;164(1):65–69. doi: 10.1016/0378-1119(95)00439-D. [DOI] [PubMed] [Google Scholar]
- 41.Subbaramaiah K, Simms SA. Photolabeling of CheR methyltransferase with S-adenosyl-L-methionine (AdoMet). Studies on the AdoMet binding site. J Biol Chem . 1992;267(12):8636–8642. http://www.jbc.org/content/267/12/8636.abstract. [PubMed] [Google Scholar]
- 42.Takata Y, Fujioka M. Identification of a tyrosine residue in rat guanidinoacetate methyltransferase that is photolabeled with S-adenosyl-L-methionine. Biochemistry. 1992;31(17):4369–4374. doi: 10.1021/bi00132a030. [DOI] [PubMed] [Google Scholar]
- 43.Klimašauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25(3):99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhao W- Q, Santini F, Breese R, et al. Inhibition of Calcineurin-mediated Endocytosis and α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors Prevents Amyloid β Oligomer-induced Synaptic Disruption. J Biol Chem . 2010;285(10):7619–7632. doi: 10.1074/jbc.M109.057182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct Interaction of RNA Polymerase II and Mediator Required for Transcription in Vivo. Science (80-). 2011;331(6023):1451–1454. http://science.sciencemag.org/content/331/6023/1451.abstract. [DOI] [PubMed] [Google Scholar]
- 46.Kleiner RE, Hang LE, Molloy KR, Chait BT, Kapoor TM. A Chemical Proteomics Approach to Reveal Direct Protein-Protein Interactions in Living Cells. Cell Chem Biol. 2018;25(1):110–120. e3. doi: 10.1016/j.chembiol.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang T, Li X- M, Bao X, Fung YME, Li XD. Photo-lysine captures proteins that bind lysine post-translational modifications. Nat Chem Biol. 2016;12(2):70–72. doi: 10.1038/nchembio.1990. [DOI] [PubMed] [Google Scholar]
- 48.Yanagisawa T, Hino N, Iraha F, Mukai T, Sakamoto K, Yokoyama S. Wide-range protein photo-crosslinking achieved by a genetically encoded N?-(benzyloxycarbonyl)lysine derivative with a diazirinyl moiety. MolBiosyst 2012;8(4): 1131–1135. doi: 10.1039/C2MB05321G. [DOI] [PubMed] [Google Scholar]
- 49.Lee HS, Dimla RD, Schultz PG. Protein-DNA photo-crosslinking with a genetically encoded benzophenone-containing amino acid. BioorgMed Chem Lett. 2009;19(17):5222–5224. doi: 10.1016/j.bmcl.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.TE M, Wenshe L, Daniel S, V MA, G SP . A Genetically Encoded Diazirine Photocrosslinker in Escherichia coli. ChemBioChem. 2007;8(18):2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- 51.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(ll):682–690. doi: 10.1038/nchembio.ll8. [DOI] [PubMed] [Google Scholar]
- 52.Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus Imatinib in Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia. NEngl JMed. 2010;362(24):2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 54.Shi H, Zhang C- J, Chen GYJ, Yao SQ. Cell-Based Proteome Profiling of Potential Dasatinib Targets by Use of Affinity-Based Probes. J Am Chem Soc. 2012;134(6):3001–3014. doi: 10.1021/ja208518u. [DOI] [PubMed] [Google Scholar]
- 55.Su Y, Pan S, Li Z, et al. Multiplex Imaging and Cellular Target Identification of Kinase Inhibitors via an Affinity-Based Proteome Profiling Approach. Sci Rep. 2015;5:7724 10.1038/srep07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International Union of Pharmacology. XXTTT. The Angiotensin II Receptors. Pharmacol Rev. 2000;52(3):415–472. http://pharmrev.aspetjournals.org/content/52/3/415.abstract. [PubMed] [Google Scholar]
- 57.Bernier SG, Bellemare JML, Escher E, Guillemette G. Characterization of AT4 Receptor from Bovine Aortic Endothelium with Photosensitive Analogues of Angiotensin IV. Biochemistry. 1998;37(12):4280–4287. doi: 10.1021/bi972863j. [DOI] [PubMed] [Google Scholar]
- 58.Vila-Perelló M, Pratt MR, Tulin F, Muir TW. Covalent Capture of Phospho-Dependent Protein Oligomerization by Site-Specific Incorporation of a Diazirine Photo-Cross-Linker. J Am Chem Soc. 2007;129(26):8068–8069. doi: 10.1021/ja072013j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Preston GW, Radford SE, Ashcroft AE, Wilson AJ. Analysis of Amyloid Nanostructures Using Photo-cross-linking: In Situ Comparison of Three Widely Used Photo-cross-linkers. ACS Chem Biol. 2014;9(3):761–768. doi: 10.1021/cb400731s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyophilization Wang W. and development of solid protein pharmaceuticals. Int J Pharm. 2000;203(1):1–60. doi: 10.1016/S0378-5173(00)00423-3. [DOI] [PubMed] [Google Scholar]
- 61.Carpenter JF, Pikal MJ, Chang BS, Randolph TW. Rational Design of Stable Lyophilized Protein Formulations: Some Practical Advice. Pharm Res. 1997;14(8):969–975. doi: 10.1023/A:1012180707283. [DOI] [PubMed] [Google Scholar]
- 62.Hermanson GT. Bioconjugate Techniques. Academic press; 2013. [Google Scholar]
- 63.Kalkhof S, Sinz A. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. AnalBioanal Chem. 2008;392(1):305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 64.Park J, Koh M, Koo JY, Lee S, Park SB. Investigation of Specific Binding Proteins to Photoaffinity Linkers for Efficient Deconvolution of Target Protein. ACS Chem Biol. 2016;11(1):44–52. doi: 10.1021/acschembio.5b00671. [DOI] [PubMed] [Google Scholar]


