Abstract
During embryonic retinal development, six types of retinal neurons are generated from multipotent progenitors in a strict spatiotemporal pattern. This pattern requires cell cycle exit (i.e. neurogenesis) and differentiation to be precisely regulated in a lineage‐specific manner. In zebrafish, the bHLH transcription factor NeuroD governs photoreceptor genesis through Notch signaling but also governs photoreceptor differentiation though distinct mechanisms that are currently unknown. Also unknown are the mechanisms that regulate NeuroD and the spatiotemporal pattern of photoreceptor development. Members of the miR‐17‐92 microRNA cluster regulate CNS neurogenesis, and a member of this cluster, miR‐18a, is predicted to target neuroD mRNA. The purpose of this study was to determine if, in the developing zebrafish retina, miR‐18a regulates NeuroD and if it plays a role in photoreceptor development. Quantitative RT‐PCR showed that, of the three miR‐18 family members (miR‐18a, b, and c), miR‐18a expression most closely parallels neuroD expression. Morpholino oligonucleotides and CRISPR/Cas9 gene editing were used for miR‐18a loss‐of‐function (LOF) and both resulted in larvae with more mature photoreceptors at 70 hpf without affecting cell proliferation. Western blot showed that miR‐18a LOF increases NeuroD protein levels and in vitro dual luciferase assay showed that miR‐18a directly interacts with the 3′ UTR of neuroD. Finally, tgif1 mutants have increased miR‐18a expression, less NeuroD protein and fewer mature photoreceptors, and the photoreceptor deficiency is rescued by miR‐18a knockdown. Together, these results show that, independent of neurogenesis, miR‐18a regulates the timing of photoreceptor differentiation and indicate that this occurs through post‐transcriptional regulation of NeuroD.
Keywords: miRNA, neurogenesis, bHLH, photoreceptors, retinal development, post‐transcriptional
Introduction
In the developing retina, six types of neurons are generated from a pool of multipotent, mitotic progenitors in a sequence that is highly conserved among vertebrates (Wallace, 2011; Bassett and Wallace, 2012; Centanin and Wittbrodt, 2014). For mature neurons to develop, progenitors must be specified to a particular fate, exit the cell cycle, and differentiate into mature neurons. These events are governed (in part) by transcription factors that regulate the expression of genes involved in the cell cycle and neuronal differentiation. The basic‐loop‐helix (bHLH) transcription factors play prominent roles in these events (Akagi et al., 2004; Ohsawa and Kageyama, 2008; Brzezinski et al., 2011; Mao et al., 2013; Pollak et al., 2013; Baker and Brown, 2018). Rod and cone photoreceptors are the neurons in the distal retinal layer that first collect visual information and, in zebrafish, the bHLH transcription factor NeuroD governs the cell cycle in photoreceptor progenitors through intercellular Notch signaling (Ochocinska and Hitchcock, 2007; Taylor et al., 2015). Following cell cycle exit, NeuroD also governs photoreceptor differentiation through separate mechanisms that are currently unknown.
In the embryonic zebrafish retina, neuroD mRNA is expressed from 30 h post‐fertilization (hpf) and, by 48 hpf, is expressed in all photoreceptor progenitors in the developing outer nuclear layer (ONL) (Ochocinska and Hitchcock, 2007). Most photoreceptor genesis and differentiation occurs between 48 and 72 hpf beginning in a small ventronasal region called the precocious ventral patch (Schmitt and Dowling, 1999), then spreading peripherally throughout the ONL with cones differentiating slightly before rods (Stenkamp, 2007). This tightly controlled spatiotemporal pattern of photoreceptor differentiation, despite the constitutive expression of neuroD throughout the ONL, suggests that post‐transcriptional mechanisms may regulate NeuroD and the timing of photoreceptor differentiation.
Post‐transcriptional regulation can occur through small ~22 nucleotide (nt) single‐stranded RNA molecules called microRNAs (miRNAS) that bind to the target mRNA through complementary base paring and regulate protein expression by blocking translation and/or causing mRNA degradation (Huntzinger and Izaurralde, 2011). Several miRNAs have been shown to regulate key aspects of brain and retinal development (La Torre et al., 2013; Andreeva and Cooper, 2014; Petri et al., 2014; Ohana et al., 2015; Sundermeier and Palczewski, 2016; Madelaine et al., 2017) and miRNAs are investigated here as potential regulators of NeuroD and photoreceptor genesis. MicroRNAs are initially expressed as primary transcripts called pri‐miRNAs, are then cleaved by the Drosha enzyme into shorter precursors (pre‐miRNAs) that fold into imperfect stem‐loop structures, and are ultimately cleaved in the cytoplasm by Dicer to become mature miRNAs (Zeng et al., 2005; Winter et al., 2009). Mature miRNAs typically function by binding via a specific “seed” sequence comprising ~6–8 nucleotides near the 5′ end of the miRNA to a complementary sequence in the 3′ untranslated region (UTR) of the target mRNA (Bartel, 2004; Broughton et al., 2016). Based on these complementary sequences, interactions between miRNAs and target mRNAs can be predicted (e.g. www.targetscan.org). A single miRNA can potentially regulate hundreds of different mRNAs and a single mRNA can be targeted by many different miRNAs (Peter, 2010), making it difficult to identify functional relationships between miRNAs and specific targets. Understanding the functions of miRNAs might, therefore, require combined approaches using morpholinos or siRNAs that block multiple functionally overlapping miRNAs (Flynt et al., 2017), as well as gene editing technologies (e.g. CRISPR/Cas9, TALENS) that disrupt individual miRNAs by generating insertion/deletion (indel) mutations in miRNA genes.
Many miRNAs are transcribed together as polycistronic clusters that are processed into functionally distinct miRNAs (Khuu et al., 2016). The miR‐17‐92 cluster generates 15 mature miRNAs including miR‐19b, which regulates NeuroD and insulin secretion in the pancreas (Zhang et al., 2011), and several miRNAs that regulate neurogenesis in the mouse neocortex (Bian et al., 2013). Another member of this cluster, miR‐18a, is also predicted to interact with neuroD (www.targetscan.org/fish_62/) but has not been studied in the developing brain or retina. Additionally, two other members of the miR‐18 subfamily, miR‐18b and miR‐18c, are at distinct genetic loci and not part of the miR‐17‐92 cluster but have identical seed sequences to miR‐18a and are also predicted to target neuroD.
Based on their predicted interactions with neuroD, miR‐18a, b, and c were examined as potential post‐transcriptional regulators of NeuroD during embryonic photoreceptor genesis. Quantitative PCR (qPCR) showed that, of the three miRNAs, the timing of pre‐miR‐18a and miR‐18a expression most closely parallels that of neuroD. Morpholino oligonucleotides targeted to miR‐18a, b, or c produced an identical phenotype with increased numbers of photoreceptors at 70 hpf. Focusing solely on miR‐18a, an in vitro dual luciferase assay showed that miR‐18a interacts directly with the 3′ UTR of neuroD mRNA. Mutation of miR‐18a using CRISPR/Cas9 gene editing reproduced the morphant phenotype, where more mature rod and cone photoreceptors are present at 70 hpf with no effect on cell proliferation. Western blot showed that when photoreceptor differentiation begins at 48 hpf, knockdown or mutation of miR‐18a results in higher levels of NeuroD protein. Finally, in tgif1‐mutant embryos that have higher levels of miR‐18a, there is less NeuroD protein and fewer mature photoreceptors, and the photoreceptor deficiency is rescued by miR‐18a knockdown. Taken together, these data show that during embryonic development, miR‐18a regulates the timing of differentiation in post‐mitotic photoreceptors and indicate that miR‐18a functions through post‐transcriptional regulation of NeuroD.
Methods
PCR Methods
AB wild‐type (WT) strain zebrafish, purchased from the Zebrafish International Research Center (ZIRC; University of Oregon, Portland, OR, USA), were used for the developmental experiments and to generate miR‐18a mutants. Embryos were collected within 15 min of spawning and incubated at 28.5°C on a 14/10‐h light/dark cycle. For standard qPCR used to amplify miR‐18a, b, and c precursor molecules, total RNA was collected from 40 whole embryo heads or 40 whole eyes (at 70 hpf) per biological replicate, using the Aurum Total RNA Mini Kit and following the manufacturer’s protocol (Bio‐Rad Laboratories, Inc., Hercules, CA, USA). Reverse transcription was performed using the Qiagen QuantiTect Reverse Transcription Kit by following the manufacturer’s protocol (Qiagen, Venlo, The Netherlands). Forward and reverse primers used to amplify miR‐18a, b, or c precursor sequences were as follows: pre‐miR‐18a F:GGCTTTGTGCTAAGGTGCATCTAG; R:CAGAAGGAGCACTTAGGGCAGTAG; pre‐miR‐18b F:CTGCTTATGCTAAGGTGCATTTAG; R:CTTATGCCAGAAGGGGCACTTAGG; pre‐miR‐18c F:GCCTTCCTGCTAAGGTGCATCTTG; R:CCTGCCAAAAGGAACATCTAGCGC. The primers used for qPCR analysis of neuroD mRNA expression were F:ATGCTGGAGTCTCAGAGCAGCTCG; R:AACTTTGCGCAGGCTCTCAAGCGC. Biological qPCR replicates were each performed in triplicate using 20 ng cDNA and IQ SYBR Green Supermix (Bio‐Rad Laboratories, Inc.) and run on a Bio‐Rad 384‐well real‐time PCR machine. Relative fold changes in expression levels were calculated using the comparative CT method and, when applicable, were compared for statistical significance using a Student’s t test with a significance level of P < 0.05.
For qPCR analysis of mature miR‐18a expression, a TaqMan custom qPCR assay was designed for mature miR‐18a and for the small nuclear RNA U6, to be used as the housekeeping gene for data normalization (ThermoFisher Scientific, Halethorp, MD, USA).
Total RNA, including small RNAs, was collected using a mirVana miRNA isolation kit (AM1560; ThermoFisher Scientific). For comparison of the precursor and mature miR expression, using the same samples, standard reverse transcription and qPCR were performed for pre‐miR‐18a amplification as described above and primer‐specific TaqMan reverse transcription and mature miRNA qPCR were performed using the manufacturer’s protocol (ThermoFisher Scientific). For mature miRNA qPCR, miR‐18a expression was normalized to U6 expression, relative to the 30 hpf sample, and was calculated using the comparative CT method.
miRNA Knockdown with Morpholino Oligonucleotides
Morpholino oligonucleotides (MO; Gene Tools, LLC, Philomath, OR. USA) targeted to the mature strand of miR‐18a, miR‐18b, or miR‐18c were used to induce miRNA knockdown. The miR‐18a morpholino [5′‐CTATCTGCACTAGATGCACCTTAG‐3′] was published previously and shown to effectively knock down miR‐18a in vivo (Friedman et al., 2009). The miR‐18b [5′‐CTATCTGCACTAAATGCACCTTAG‐3′] MO used here differs from the miR‐18a MO by only one nucleotide (underlined) and the miR‐18c MO [5′‐CTAACTACACAAGATGCACCTTAG‐3′] differs by only three nucleotides. Morpholino oligonucleotides were diluted in 1X Daneau buffer (Nasevicius and Ekker, 2000), and 3 ng MO were injected at the single cell stage as described previously (Ochocinska and Hitchcock, 2009).
Systemic Labeling with 5‐Bromo‐2′‐Deoxyuridine (BrdU), Immunohistochemistry, and Cell Counting
Cells in the S‐phase of the cell cycle were labeled by incubating embryos for 20 min, immediately prior to sacrifice, in ice‐cold 10 mM BrdU dissolved in an embryo‐rearing solution containing 15% dimethylsulfoxide (DMSO). Whole embryos were fixed and prepared for histology as previously described (Taylor et al., 2015), embedded in an optical cutting temperature (OCT) medium, and heads were sectioned at 10 μm and mounted on glass slides (Superfrost plus; Fisher Scientific, Pittsburgh, PA). Immunolabeling was performed using previously published protocols (Luo et al., 2012) on cross sections through the central retina in the vicinity of the optic nerve. For BrdU immunolabeling, DNA was denatured by incubating sections in 100°C sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 30 min, cooled at room temperature for 20 min, and processed with standard immunolabeling techniques. The primary and secondary antibodies and dilution factors used here were: mouse anti‐BrdU, 1:100 (347580; BD Biosciences, Franklin Lakes, NJ, USA); Zpr‐1, 1:200 (anti‐Arrestin 3, red‐green double cones, ZIRC); goat anti‐mouse Alexa Fluor 488; and goat anti‐mouse Alexa Fluor 555, 1:500 (Life Technologies, Carlsbad, CA, USA). Nuclei were counterstained with 20 mM Hoechst 33342 (ThermoFisher Scientific) prior to adding coverslips. BrdU‐labeled cells and cones were counted in the one central‐most cross section for each larval fish. Cell counts were compared using a Student’s t test, with a P < 0.05 indicating statistically significant differences.
Luciferase Assay
To test the interaction between miR‐18a and the 3′ UTR of neuroD mRNA, an in vitro dual luciferase assay was performed following published protocols (Jin et al., 2012). Briefly, a custom oligonucleotide corresponding to a 66 bp portion of the neuroD 3′ UTR containing the predicted target sequence for miR‐18a (underlined) [“neuroDWT” 5′‐GGAGAAAAGAGAATTGGTTGATTCTCGTTCACCTTATGTATTGTATTCTATAGCGCTTCTACGTTG‐3′] was generated (ThermoFisher Scientific) and inserted into the pGL3 vector immediately 3′ of the firefly luciferase gene (E1741; Promega, Madison, WI, USA). A negative control pGL3 vector was also created containing the same neuroD 3′ UTR sequence, but with the predicted miR‐18a target site mutated to TTTTTTT [“neuroDMut”]. Following bacterial transformation, culture, and plasmid purification, HEK 293 cells, grown to 20–40% confluence, were transfected with either the neuroDWT or neuroDMut plasmid along with the pRL‐TK vector that constitutively expresses Renilla luciferase to serve as an internal transformation control. Each transfection group was co‐transfected with either hsa‐miR‐18a‐5p mimic (identical to zebrafish mature miR‐18a) or hsa‐let7a‐5p mimic, for which neither vector had a predicted target site and served as a negative control (Exiqon/Qiagen, Venlo, The Netherlands). Following transfection, 48‐h incubation, and cell lysis, luciferase expression levels were assayed on a luminometer using a Dual‐Luciferase Reporter Assay System (E1910; Promega). Using this approach, direct interaction between the miRNA mimic and the cloned 3′ UTR neuroD sequence is expected to reduce the level of firefly luciferase levels. For each experimental group, firefly luciferase was normalized to constitutive Renilla luciferase levels and the results were compared using a Student’s t test.
Generating miR‐18a Mutants
CRISPR/Cas9 genome editing was used to generate mutations in the miR‐18a gene (Taylor et al., 2015). Briefly, the sgRNA target sequence was identified within the miR‐18a precursor sequence using ZiFiT software (available in the public domain at www.zifit.partners/org). Cas9 mRNA and sgRNA were generated (Hwang et al., 2013), and single cell‐stage embryos were injected with 1 nL solution containing 100 pg/nL sg RNA and 150 pg/nl Cas9 mRNA. F0 injected fish were raised to maturity. Genomic DNA was purified from caudal fins, and screening primers (F: CCAGGAAAGATGGGAGTAGTTG; R: CTCACACTGCAGTAGATGACAG) were used to amplify a 626 bp region around the sgRNA target site using standard PCR and 100 ng template DNA. CRISPR‐induced insertions and deletions were detected using the T7 endonuclease assay according to established protocols (available in the public domain at www.crisprflydesign.org). Briefly, 200 ng of purified PCR product was used for the analysis and, following denaturation and reannealing, subjected to a 15‐min digest at 37°C with 10 U T7 endonuclease I (New England Biolabs, Ipswich, MA, USA). Digested DNA was run on a 2% agarose gel and indels were identified by the presence of a double band around 200–300 bp. F0 adult fish that were positive for indels were outcrossed with the AB WT fish and, using the same methods as above, the T7 assay was used to identify indels in F1 generation adults. PCR products from T7‐positive F1 adults were subcloned into the pGEM‐T Easy vector (Promega) and six clones were sequenced for each fish. Mutations were detected using a pairwise blast (NCBI, Bethesda, DM, USA) against the WT DNA.
In one F1 adult fish, a 25 bp insertion was detected in the miR‐18a precursor sequence, and this introduced an AleI restriction enzyme cut site that was used for subsequent genotyping. The same screening primers (above) were used for this method and AleI digest using standard protocols (New England Biolabs) cuts the mutant PCR product into 244 and 407 bp segments that are easily visualized using gel electrophoresis. F1 generation heterozygous adult fish were outcrossed and then F2 generation heterozygotes incrossed to produce a homozygous line of miR‐18a mutants, and these fish were incrossed to produce homozygous mutant embryos used here.
Western Blot Analysis
Protein samples were obtained by pooling whole heads of embryos or larvae at 48 or 70 hpf, respectively, in RIPA lysis buffer (89900; ThermoFisher Scientific) containing 1x protease and phosphatase inhibitor cocktail (5872; Cell Signaling Technology, Danvers, MA, USA). Proteins were separated in a 12% SDS‐PAGE pre‐cast gel (4561043; Bio‐Rad Laboratories, Inc.) and transferred to a PVDF membrane (Sigma‐Aldrich Corp., St. Louis, MO, USA). The membrane was incubated in 5% bovine serum albumin (BSA) with 0.05% Tween‐20 for 2 h to block non‐specific binding of the antibodies and then incubated overnight at 4C with rabbit anti‐NeuroD antibodies (Ochocinska and Hitchcock, 2009) diluted 1:1,000 in 2.5% blocking solution. Blots were rinsed with TBS with 0.05% Tween‐20 and incubated with horseradish peroxidase‐conjugated secondary IgG (1:2,000) for 1 h at room temperature. Bands were visualized using the enhanced chemiluminescence assay detection system (34075; ThermoFisher Scientific). For loading controls, blots were stripped in the stripping buffer (21059; ThermoFisher Scientific) for 5 min, processed as described above, and labeled with mouse anti‐βactin antibodies (1:5,000) (NB10074340T, Novus Biologicals, LLC, Littleton, CO, USA). Images were captured using the FluorChem E Imaging System (Bio‐Techne, Minneapolis, MN, USA) and band intensity was quantified relative to βactin.
In situ Hybridization
In situ hybridization with rhodopsin probes was used to identify rod photoreceptors. A DIG‐labeled antisense riboprobe for zebrafish rhodopsin was generated from a 976 bp PCR product containing a T3 polymerase promoter sequence (lowercase, underlined) on the reverse primer (aattaaccctcactaaagggCTTCGAAGGGGTTCTTGCCGC) following published methods (David and Wedlich, 2001); for similar primer lengths, a T7 polymerase promoter sequence (lowercase, underlined) was added to the forward primer (taatacgactcactatagggGAGGGACCGGCATTCTACGTG). The antisense DIG‐labeled probe was generated using T3 polymerase and in situ hybridization performed as previously described (Barthel and Raymond, 1993; Ochocinska and Hitchcock, 2007; Taylor et al., 2015). Control and morphant or mutant sections were mounted on the same slides and color reactions were developed for identical periods of time. Cells were counted in cross sections and compared as described above.
For in situ hybridization labeling of miR‐18a in tissue sections, a miRCURY LNA detection probe (Exiqon/Qiagen), labled with DIG at the 5′ and 3′ ends, was designed to hybridize with the mature miR‐18a sequence. Standard in situ hybridization methods were used, as described above, using a 0.25 μM probe working concentration at a hybridization temperature of 58°C.
Results
miR‐18a Expression Closely Parallels NeuroD Expression
In the developing brain and retina, neuroD mRNA expression increases markedly between 30 and 70 hpf (Fig. 1A), and most photoreceptors are generated between 48 and 72 hpf (Stenkamp, 2007). As a first step to determine if miR‐18 might regulate photoreceptor development, qPCR was used to analyze the expression at time points consistent with neuroD and photoreceptor genesis. The miRNAs miR‐18a, mir‐18b, and mir‐18c differ in sequence by only 1–3 nucleotides, are expressed from distinct genetic loci, and have a conserved seed sequence that is predicted to interact with neuroD mRNA. Quantitative PCR analysis of mature miRNAs requires specialized kits (e.g. TaqMan, Applied Biosystems) and the nearly identical sequences among closely related miRNAs (e.g. miR‐18a, b and c) can result in cross‐amplification. The longer precursor molecules (pre‐miRNAs) for similar miRNAs, however, have unique sequences and can be analyzed with standard qPCR and, if expression levels are proportional to mature miRNAs, can be used as a fast and accurate proxy for mature miRNA expression. To determine if pre‐miR‐18a expression can be used as a proxy for mature miR‐18a, total RNA (including short RNAs) was purified from whole head tissue of zebrafish embryos at 30, 48, and 70 hpf with a miRVana miRNA isolation kit and then, on the same samples, standard qPCR was performed for pre‐miR‐18a and TaqMan qPCR for mature miR‐18a. The results showed that the levels of both mature pre‐miR‐18a and miR‐18a increase steadily and proportionally between 30 and 70 hpf (Fig. 1B), indicating that pre‐miR‐18a can be used as a proxy for mature miR‐18a expression. Standard qPCR was then used to compare the expression of the 83–87 nt pre‐miRNAs for miR‐18a, b, and c that each have unique sequences, despite the nearly identical mature miRNA sequences. In brain and retina tissues, pre‐miR‐18a expression increases steadily between 30 and 70 hpf. This closely matches the increase in neuroD expression during the same time period. In comparison, pre‐miR‐18b expression remains substantially lower at all time points and miR‐18c expression decreases after 30 hpf (Fig. 1C). By 70 hpf, eyes are large enough for easy dissection and qPCR analysis of eye tissue only, and this showed that pre‐miR‐18b expression is substantially lower in the eye compared with pre‐miR‐18a or c (Fig. 1D). These results show that among the three pre‐miRs, pre‐miR‐18a expression most closely parallels that of neuroD.
Figure 1.
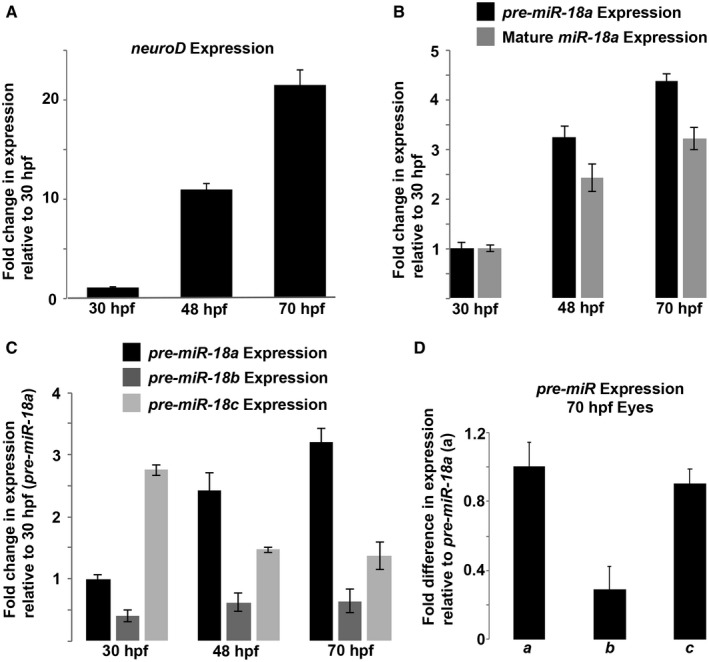
During development, pre‐miR‐18a expression increases proportionally along with neuroD mRNA and mature miR‐18a. (A) neuroD mRNA expression in the developing brain and retina between 30 and 70 hpf. (B) Fold changes in the expression of pre‐miR‐18a and mature miR‐18a in the developing brain and retina between 30 and 70 hpf. (C) Fold changes in the expression of pre‐miR‐18a, pre‐miR‐18b, and pre‐miR‐18c in the developing brain and retina between 30 and 70 hpf. (D) Fold difference in the expression between pre‐miR‐18a (a), pre‐miR‐18b (b), and pre‐miR‐18c (c) in the eyes only at 70 hpf. Error bars represent standard deviation; single biological replicates were used per time point, n = 40 whole heads (A–C) or 40 whole eyes (D) from the AB WT embryos per sample.
Morpholino‐Induced Knockdown of miR‐18a, miR‐18b, or miR‐18c Increases the Number of Mature Photoreceptors at 70 hpf
NeuroD is required for photoreceptor progenitors to exit the cell cycle and differentiate, and if NeuroD levels are regulated by miR‐18 miRNAs, knockdown of these molecules is expected to affect the rate of photoreceptor development. To determine if miR‐18 miRNAs regulate photoreceptor development, morpholino oligonucleotides targeted to the mature sequences of miR‐18a, mir‐18b, or miR‐18c were injected into embryos at the single cell stage. Compared with embryos injected with standard control morpholinos, morpholinos targeting each of the three miR‐18 types resulted in a greater number of cone photoreceptors at 70 hpf (Fig. 2A,C); miR‐18a knockdown was verified by in situ hybridization (Fig. 2B). None of the three morpholinos altered the number of BrdU+ cells (Fig. 2D), indicating that the miR‐18 miRNAs regulate photoreceptor differentiation, but do not regulate the cell cycle. The high degree of sequence similarity between the three miR‐18 types and the identical effect of morpholinos targeted to each suggest that each morpholino might comprehensively knock down miR‐18a, b, and c. This potential cross‐reactivity makes it difficult to determine which miR‐18 is the most important regulator of photoreceptor development, but the results indicate that among post‐mitotic cells of the photoreceptor lineage, miR‐18 miRNAs regulate differentiation.
Figure 2.
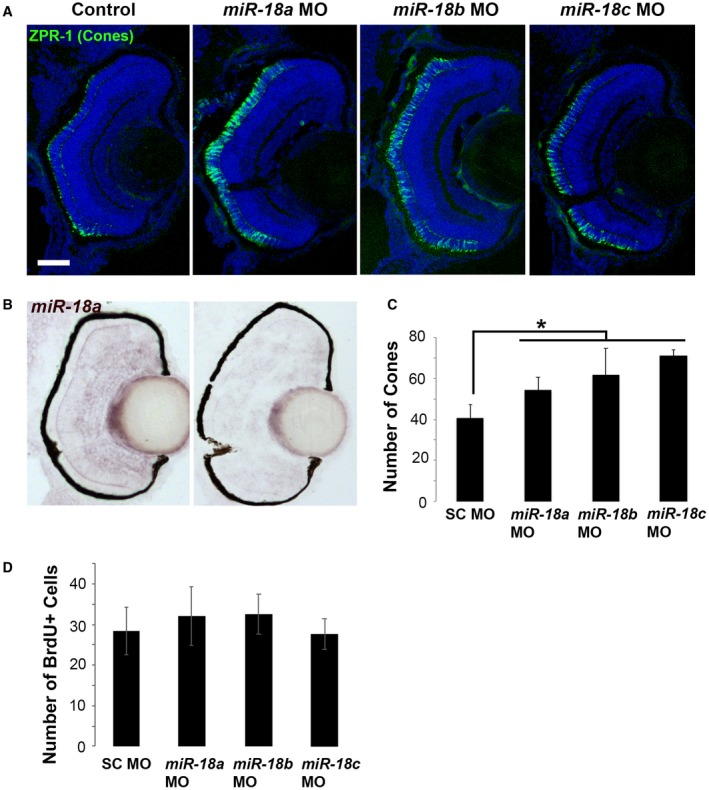
Knockdown with morpholinos targeted to miR‐18a, miR‐18b, or miR‐18c results in more differentiated cone photoreceptors. (A) ZPR‐1 (cone) immunolabeling in 70 hpf larvae that were injected at the single‐cell stage with standard control, miR‐18a, miR‐18b, or miR‐18c morpholinos; note that, due to sequence similarities, each morpholino might comprehensively knock down miR‐18a, b, and c. (B) In situ hybridization for miR‐18a, comparing expression in larvae injected with standard control morpholino (left) with miR‐18a knockdown (right). (C) Cone photoreceptor counts presented as the mean of one eye per fish (n = 3) counted in the centermost cross section in the vicinity of the optic nerve. (D) Total number of BrdU‐labeled cells presented as the mean of one eye per fish (n = 3) counted in the centermost cross section in the vicinity of the optic nerve. Error bars show standard deviation and counts were statistically compared using a Student’s t test. [Colour figure can be viewed at wileyonlinelibrary.com]
miR‐18a Directly Interacts with NeuroD mRNA
The similarity in the timing of expression between pre‐miR‐18a, mature miR‐18a, and neuroD suggests that, in the developing retina, miR‐18a might regulate NeuroD. To determine if miR‐18a directly interacts with neuroD mRNA, a dual luciferase assay was performed on HEK 293 cells transfected with pGL3‐control firefly luciferase vector into which a 66 nt portion of the neuroD 3′ UTR was cloned immediately 3′ of the luciferase gene. The wild‐type vector had the normal predicted target site for miR‐18a on the neuroD 3′ UTR (CACCTTA) and a negative control vector was created with this predicted target site mutated to TTTTTTT (Fig. 3A). As an internal transfection control, cells were co‐transfected with the pRL‐TK vector with a constitutive expression of Renilla luciferase. Following cell culture and transfection, cell lysates were treated with either miR‐18a mimic or a negative control miRNA mimic (let‐7a) not predicted to interact with the neuroD 3′ UTR (Fig. 3B). Binding of the miRNA to the cloned neuroD sequence was predicted to suppress the level of firefly luciferase expression relative to Renilla luciferase. In the wild‐type neuroD vector, relative to negative controls, miR‐18a resulted in a significant decrease in firefly luciferase expression (Student’s t test, P = 0.001; Fig. 3C). In the mutated neuroD vector, relative to negative controls, miR‐18a did not significantly affect firefly luciferase expression (Student’s t test, P = 0.20). These results indicate that miR‐18a binds to the 3′ UTR of neuroD at the predicted target site. In vectors with the wild‐type compared with the mutant‐predicted target site, however, the effect of miR‐18a on luciferase expression did not differ significantly (Student’s t test, P = 0.08). This suggests that, in addition to the predicted target site, miR‐18a might also interact with other regions of the 3′ UTR neuroD sequence. Together, these data suggest that miR‐18a functions to negatively regulate NeuroD translation.
Figure 3.

Dual luciferase assay showing direct interaction between miR‐18a and its predicted target site in the 3′ UTR of neuroD mRNA. (A) The 66 bp portion of the neuroD 3′ UTR sequence inserted into the pGL3 vector with the predicted intact (top) or mutated (bottom) miR‐18a target site underlined. (B) Sequences for the miR‐18a and let‐7a (negative control) mimics that were co‐transfected into the cells. The seed sequence of the miR‐18a mimic is underlined with a solid line, which is complementary to the underlined target sequence in (A); the seed sequence of the let‐7a mimic, used as a negative control, is underlined with a dashed line, and is not complementary to any portion of the neuroD 3′ UTR sequence in (A). (C) Firefly luciferase from the pGL3 vector shown relative to constitutive firefly luciferase from the pRL‐TK vector, compared between treatments with the let‐7a mimic (negative control) and miR‐18a mimic and shown for both the intact predicted miR‐18a target site and the mutated target site. Error bars show standard deviation; values for let‐7a and miR‐18a mimic were compared with a Student’s t test on n = 3 samples per group; asterisks indicate P < 0.05.
miR‐18a Mutants Generated by CRISPR/CAS9 Gene Editing
To determine the role of miR‐18a, independent of miR‐18b or c, in regulating NeuroD and photoreceptor differentiation, CRISPR/Cas9 gene editing was used to generate a miR‐18a ‐/‐ mutant line. An appropriate CRISPR target site was not available within the 22 bp sequence coding for the mature miRNA molecule, so a target site was chosen within the sequence for the larger precursor molecule (pre‐miR‐18a), and mutations here were predicted to interfere with processing by Dicer into the mature miRNA. This method produced animals with a 25 nt insertion within the sequence (Fig. 4A) that normally produces the stem‐loop precursor molecule (Fig. 4B). This insertion introduced a restriction site for AleI that is not present in the WT DNA (Fig. 4A), and restriction analysis was subsequently used for genotyping (Fig. 4C). To ensure that off‐target mutations, including possible mutations in miR‐18b or miR‐18c precursor sequences, were not present in experimental fish, F0 and F1 generation fish were selected for mutations specifically at the miR‐18a locus and outcrossed with the wild‐type fish. The loci for miR‐18b and miR‐18c are on separate chromosomes from miR‐18a and are thus inherited independently, and the analogous sequences for these differ from miR‐18a. Fish selected for miR‐18a mutations and outcrossed over two generations are thus highly unlikely to also have mutations in miR‐18b or miR‐18c. F2 generation heterozogous fish were then incrossed to produce homozygous miR‐18a mutants, and F3 generation homozygous mutants were incrossed to produce homozygous mutant embryos for this study. TaqMan qPCR, specific for the mature 22 nt miR‐18a, was then used to compare the expression of mature miR‐18a in mutant and wild‐type fish. In these mutants compared with wild‐type fish at 70 hpf, mature miR‐18a abundance was reduced by more than 14‐fold (Fig. 4D), demonstrating that miR‐18a mutants lack mature miR‐18a.
Figure 4.
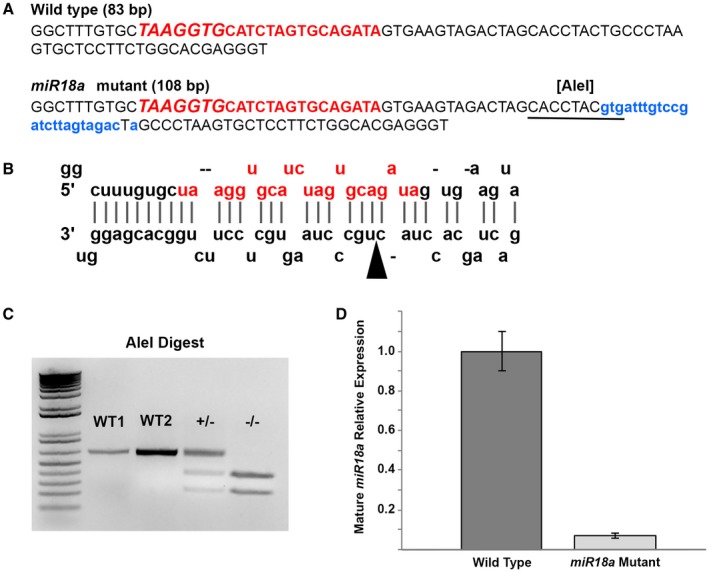
Characterization of the miR‐18a ‐/‐ mutant line. (A) Comparison between the WT and mutant genomic sequences corresponding to pre‐miR‐18a and in the mutant, the 25 bp insertion is shown in lowercase, blue lettering, and the introduced AleI restriction cut site is underlined. The uppercase red lettering shows the genomic sequence corresponding to mature miR‐18a, with the seed sequence in larger italics. (B) The predicted stem loop arrangement for the WT pre‐miR‐18a RNA molecule with the mature miR‐18a sequence shown in red (adapted from www.mirbase.org); in the mutant sequence, the 25‐base insertion location is indicated by the arrowhead. (C) Genotyping of miR‐18a mutants using AleI restriction digest. In the WT fish, the 626 bp PCR product remains uncut, with clear intensity distinctions between 200 ng (W1) and 400 ng (WT2) PCR products. In heterozygous mutants (+/−), 50% (~200 ng) of the PCR product is cut into smaller fragments and in homozygous mutants (−/−) 100% (400 ng) of the PCR product is cut into smaller fragments. (D) TaqMan qPCR showing, compared with the WT at 70 hpf, the relative absence of mature miR‐18a in mutant fish. Error bars represent standard deviation on a single biological replicate of n = 40 embryo heads per sample. [Colour figure can be viewed at wileyonlinelibrary.com]
miR‐18a Regulates NeuroD and the Timing of Photoreceptor Differentiation
To determine if miR‐18a regulates NeuroD, Western blot was used to compare NeuroD protein levels in 48 hpf embryos (heads) between fish injected with standard control or miR‐18a morpholinos, and between the WT and miR‐18a‐mutant fish. Knockdown of miR‐18a resulted in a 32% increase in NeuroD protein and miR‐18a mutation resulted in a 20% increase in NeuroD protein indicating that in the developing brain and retina, miR‐18a suppresses the level of NeuroD (Fig. 5A,B). These data also indicate that broader knockdown of miR‐18(a, b and c) with morpholinos may have a greater effect on NeuroD protein levels than miR‐18a mutation, where miR‐18b and c are still functional. Then, to determine if miR‐18a, independent of miR‐18b or c, regulates photoreceptor differentiation, the numbers of mature photoreceptors were compared between the WT and miR‐18a‐mutant fish. Immunohistochemistry for the red/green cone marker Arrestin‐3a was used to label a subset of cone photoreceptors, while in situ hybridization for the mature rod marker rhodopsin was used to label rods. Larvae were placed in 10 mM BrdU solution for 20 min prior to sacrifice at 70 hpf. The miR‐18a mutation resulted in a significantly greater number of both mature red/green cones and rods, whereas the numbers of BrdU‐labeled cells remained invariant (Fig. 5C–F). This indicates that within the photoreceptor lineage, miR‐18a regulates photoreceptor differentiation but does not regulate the cell cycle. By 6 dpf, the numbers of mature photoreceptors do not differ between mutant and wild‐type fish (Fig. 5F), indicating that miR‐18a does not regulate cell fate or the total numbers of photoreceptors generated. Taken together, these results indicate that, among post‐mitotic cells already determined to become photoreceptors, miR‐18a functions to regulate the timing of photoreceptor differentiation.
Figure 5.
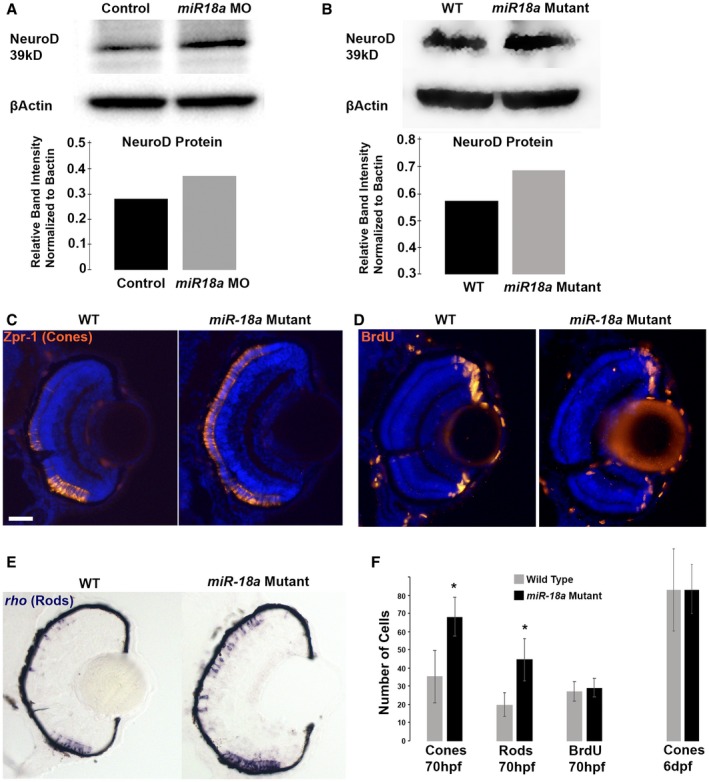
Loss of miR‐18a increases NeuroD protein levels and the number of differentiated photoreceptors. Western blot on 48 hpf embryo heads (n = 40) comparing NeuroD protein levels between standard control MO‐injected and miR‐18a MO‐injected embryos (A) and between the WT and miR‐18a‐/‐‐mutant embryos (B) with corresponding quantification graphs. In the WT compared with the miR‐18a‐mutant larvae at 70 hpf, immunolabeling for mature cone photoreceptors (C: Zpr‐1) and cells in the S‐phase of the cell cycle (D: BrdU); and in situ hybridization for rod photoreceptors (E: rhodopsin); scale bar = 0.50 μm. (F) Quantification of cones (n ≥ 14 larvae), rods (n ≥ 8 larvae), and BrdU+ cells (n ≥ 7 larvae) in 70 dpf retinas. Error bars represent standard deviation; cell counts compared with a Student’s t test and asterisks indicate P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
Increased miR‐18a Expression Suppresses NeuroD Protein Levels and Photoreceptor Differentiation
TGIF1 is a transcriptional repressor in the TGFβ signaling pathway (Lenkowski et al., 2013) and, compared with the WT larvae at 70 hpl, tgif1‐mutant larvae were observed to have fewer differentiated cone photoreceptors (Fig. 6A,B). miR‐18a was investigated as the possible mediator of this phenotype and, in 70 hpf tgif1 mutants compared with the WT, in situ hybridization indicated increased retinal expression of miR‐18a (Fig. 6A,B) while qPCR showed higher levels of pre‐miR‐18a expression (Fig. 6C). To determine if increased miR‐18a expression mediates the loss‐of‐cone phenotype in tgif1 mutants, morpholinos were used to knock down miR‐18a in tgif1‐mutant larvae. Compared with standard control morpholino‐injected larvae (SC MO) at 70 hpf, mir‐18a knockdown in tgif1 mutants fully rescued the deficiency in cone differentiation (Fig. 6A,B), indicating that the lack of cone phenotype is mediated through miR‐18a. To determine if, in the tgif1‐mutant retina, miR‐18a post‐transcriptionally regulates NeuroD, qPCR and Western blot were used to compare mRNA expression and NeuroD protein, respectively, between the WT and tgif1‐mutant larvae. Compared with the WT at 70 hpf, tgif1 mutants have identical levels of neuroD expression (Fig. 6D) but reduced NeuroD protein (Fig. 6E,F). These results show that, in tgif1 mutants, NeuroD protein levels are suppressed at the post‐transcriptional level and suggest that this regulation is mediated through increased levels of miR‐18a.
Figure 6.
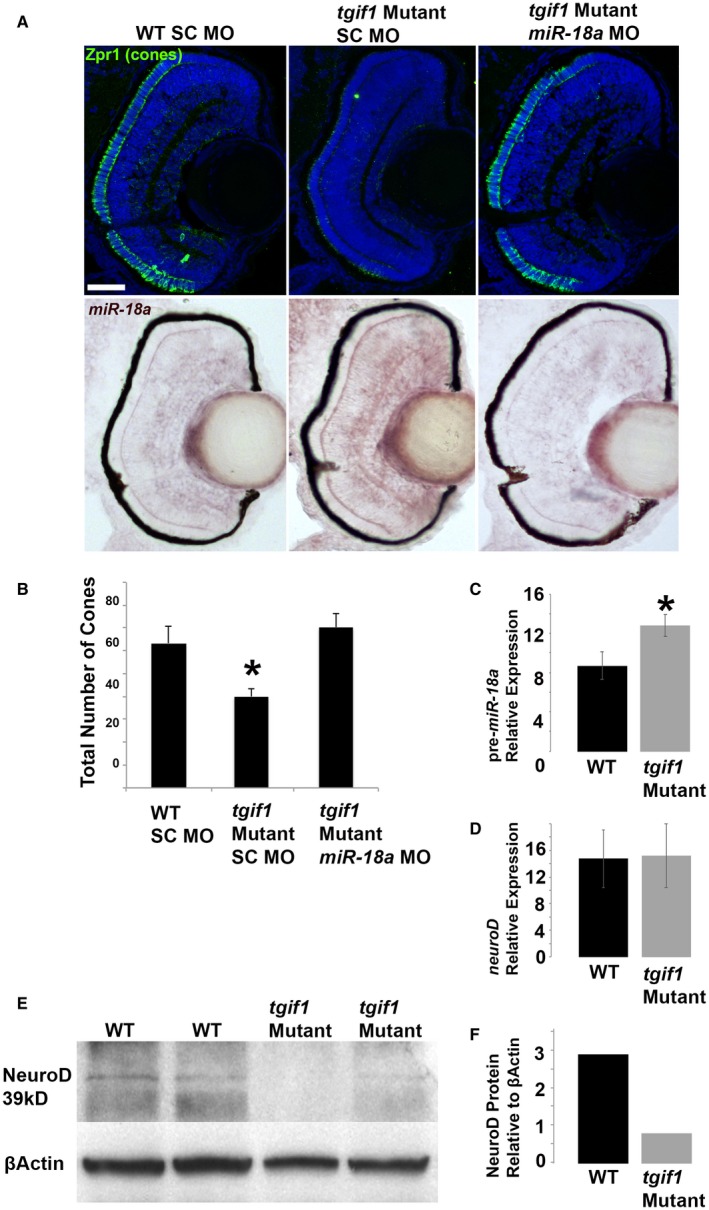
tgif1 mutant larvae have fewer photoreceptors, increased miR‐18a expression, and reduced NeuroD protein levels. (A) Immunolabeling for cone photoreceptors (Zpr‐1) at 70 hpf in the WT larvae injected with standard control morpholinos (SC MO), tgif1‐mutant larvae injected with SC MO, tgif1‐mutant larvae injected with miR‐18a morpholinos, with corresponding images showing in situ hybridization for miR‐18a in the same retinas; scale bar = 50 μm. (B) Cone photoreceptor counts in retinal cross sections (n = 3 larvae each) in fish corresponding to the images in (A). (C) Standard qPCR showing pre‐miR‐18a expression in 70 hpf WT larvae compared with tgif1 mutants (n = 40 heads); normalized to βactin and shown relative to let‐7b expression. (D) Standard qPCR comparing neuroD mRNA expression in 70 hpf larvae between WT and tgif1 mutants (n = 40 heads); normalized to βactin and shown relative to ccnb1 expression. (E) Western blot showing NeuroD protein levels in 70 hpf WT compared with tgif1‐mutant fish (n = 40 heads). (F) Quantification of the average band intensities in E. All error bars represent standard deviation and comparisons were made with Student’s t tests (asterisks indicate P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
Generating the correct type and number of neurons in the developing retina requires precise temporal and spatial regulation of mechanisms that specify progenitor cell fate, determine the timing of cell cycle exit, and regulate differentiation (recent reviews: Cepko, 2014; Mattar and Cayouette, 2015; Stenkamp, 2015; Wang and Cepko, 2016). This control is partially accomplished through transcription factors that regulate mRNA expression levels (reviewed in Gregory‐Evans et al., 2013; Brzezinski and Reh, 2015). NeuroD is a bHLH transcription factor that, within the photoreceptor lineage, governs the cell cycle through Delta‐Notch signaling and, in newly generated photoreceptors, governs differentiation through a separate mechanism (Taylor et al., 2015). Little is currently known about how NeuroD is regulated, but there is evidence that in the retina of Medaka fish the transcription factor Six6 influences neuroD expression (Conte et al., 2010), and Insm1a functions upstream of NeuroD in zebrafish (Forbes‐Osborne et al., 2013). Here we identify a mechanism through which miR‐18a post‐transcriptionally regulates NeuroD protein levels and the timing of photoreceptor differentiation. We propose that the fine‐tuning of NeuroD protein levels by miR‐18a provides a mechanism through which NeuroD differentially governs both cell cycle exit in photoreceptor progenitors and the timing of photoreceptor differentiation.
Within the photoreceptor lineage, NeuroD governs two sequential events—cell cycle exit and differentiation—through distinct mechanisms (Taylor et al., 2015). Retinal expression of neuroD mRNA increases steadily between 30 and 70 hpf, with expression occurring throughout the retinal neuroepithelium by 38 hpf, and then in all developing photoreceptors in the ONL by 48 hpf (Ochocinska and Hitchcock, 2007). Despite this relative uniform expression of neuroD among photoreceptor progenitors, the spatiotemporal pattern of events governed by NeuroD is complex. Among photoreceptor progenitors, NeuroD governs the cell cycle through intercellular Delta‐Notch signaling, with the first photoreceptor progenitors beginning to exit the cell cycle around 48 hpf (Stenkamp, 2007). At this time, neuroD mRNA is expressed in all ONL cells (Ochocinska and Hitchcock, 2007), but photoreceptor genesis begins in only a small ventronasal patch around this time (Schmitt and Dowling, 1999). Photoreceptor genesis then spreads peripherally until most ONL cells have exited the cell cycle by about 60 hpf (Stenkamp, 2007). Photoreceptor differentiation, coincident with the expression of mature photoreceptor markers Rhodopsin (rods) and Arrestin3a (red/green cones), is also governed by NeuroD but lags slightly behind cell cycle exit and is completed by 72 hpf (Stenkamp, 2007). This clear and tightly controlled spatiotemporal pattern of photoreceptor genesis and differentiation, despite the uniform expression of neuroD among photoreceptor progenitors, suggests that post‐transcriptional mechanisms could regulate NeuroD function.
MicroRNAs post‐transcriptionally regulate protein levels by binding to the 3′ UTR of target mRNA and blocking translation (Zeng et al., 2003; Valencia‐Sanchez et al., 2006; Djuranovic et al., 2012; Iwakawa and Tomari, 2015) and in some cases causing mRNA degradation (Huntzinger and Izaurralde, 2011). In the developing CNS, miRNAs regulate stem and progenitor cell proliferation, cell fate specification and neural differentiation (Shi et al., 2010; Pham and Gallicano, 2012), as well as the timing of retinal neurogenesis (La Torre et al., 2013). The microRNAs miR‐18a, miR‐18b, and miR‐18c share a seven‐base seed region that is predicted to interact with the 3′ UTR of neuroD and, if expressed in the retina, could post‐transcriptionally regulate NeuroD and photoreceptor genesis. The genes for these three miR‐18 molecules are on different chromosomes, and their expression is, therefore, presumably controlled by different regulatory mechanisms.
MicroRNAs are initially expressed as long primary transcripts and then cleaved into smaller pre‐miRNAs in the nucleus by the Drosha enzyme complex (Zeng et al., 2005). Then, in the cytoplasm, pre‐miRNAs are cleaved again and processed into single‐stranded mature miRNAs by the RNAse DICER and the RISC loading complex (Winter et al., 2009). MicroRNA biogenesis is a complex process but, for many miRNAs, precursor and mature miRNA expression levels are closely correlated (Nepal et al., 2016; Powrózek et al., 2018). Accordingly, we show that in the developing brain and retina, pre‐miR‐18a expression increases proportionally with, and can serve as an accurate proxy for, mature miR‐18a expression. This is an advantage because, compared with mature miRNAs that must be amplified with special qPCR kits (e.g. TaqMan) that might not fully discriminate between nearly identical miRNAs (e.g. miR‐18a, b, c), the longer pre‐miRNAs have unique sequences and can be easily discriminated using standard qPCR. Taking advantage of this, we show that each of the precursor molecules for miR‐18a, miR‐18b, and miR‐18c is expressed in the developing brain and retina, but the timing of their expression differs, which might be key to their functions. Most photoreceptor genesis occurs between 48 and 72 hpf and, like neuroD, pre‐miR‐18a and mature miR‐18a expression increase steadily between 30 and 70 hpf. In comparison, pre‐miR‐18c expression peaks at 30 hpf and then is rapidly downregulated, while pre‐miR‐18b expression remains substantially lower (than pre‐miR‐18a or c) throughout embryonic development. These data indicate that, in the brain and retina, miR‐18a, b, and c function during distinct developmental time frames, but miR‐18a expression most closely correlates with the timing of neuroD expression and photoreceptor genesis.
Even though the expression data suggest that miR‐18a, b, and c function during different developmental events, morpholinos targeted to each of these miRNAs result in an identical phenotype in which 70 hpf larvae have significantly more mature photoreceptors. This suggests that, due to their nearly identical sequences, each morpholino knocks down all three miRNAs and these miRNAs may have overlapping functions. Redundancy among miRNAs occurs commonly and may be important for cooperative translational repression (Fischer et al., 2015). Knockdown of multiple, redundant miRNAs by a single morpholino has been documented for other miRNA groups (Flynt et al., 2009), indicating that morpholino oligonucleotides can be an effective tool for understanding cooperative function of multiple miRNAs (Flynt et al., 2017). In contrast, removing individual miRNAs using gene editing (e.g. CRISPR/Cas9) or knockout techniques sometimes does not produce a phenotype (Olive et al., 2015), because redundant miRNAs might partially or fully compensate for the functional loss of a single miRNA (Ventura et al., 2008; Bao et al., 2012; Gurtan and Sharp, 2013). This redundancy, however, does not indicate that familial miRNAs are merely functional replicates of one another. Expression regulation and feedback mechanisms under different circumstances can confer distinct roles for what are considered redundant miRNAs (Olive et al., 2015). Accordingly, differential expression of pre‐miR‐18a, b, and c in the brain and retina suggests that these miRNAs may have functional specializations during distinct developmental events.
Based on the overlap in expression between miR‐18a and neuroD, miR‐18a was investigated, independent of miR‐18b or c, as a potential regulator of NeuroD and photoreceptor genesis. First, an in vitro double luciferase assay showed that miR‐18a suppresses translation through direct interaction with the 3′ UTR of neuroD, indicating that miR‐18a post‐transcriptionally regulates NeuroD. This is consistent with the widely demonstrated roles of miRNAs to suppress translation through direct interaction with the 3′ UTR region of target mRNAs (Zeng et al., 2003; Humphreys et al., 2005; Valencia‐Sanchez et al., 2006; van den Berg et al., 2008). Then, to determine if miR‐18a regulates NeuroD and photoreceptor differentiation, CRISPR‐Cas9 gene editing was used to generate a stable mutant line that lacks mature miR‐18a. In miR‐18a morphants and mutants, at 48 hpf when photoreceptor differentiation begins, Western blot showed that NeuroD protein levels are increased by 32% and 20%, respectively. This indicates that, in the wild‐type retina during the time of photoreceptor differentiation, miR‐18a suppresses NeuroD protein levels. This also suggests that concurrent knockdown of miR‐18a, b, and c by the miR‐18a morpholino has a greater effect on NeuroD protein than mutation of miR‐18a alone. Finally, identical to miR‐18(a, b and c) morphants, 70 hpf miR‐18a‐mutant larvae have significantly more mature photoreceptors, whereas the numbers of cells in the cell cycle are equivalent. By 6 days post‐fertilization, after embryonic retinal development is complete, the total number of mature rods and cones does not differ between the miR‐18a‐mutant and the WT fish. Taken together, these data indicate that, among post‐mitotic cells within the photoreceptor lineage, miR‐18a regulates the timing of photoreceptor differentiation. This is consistent with miR‐18a functioning through NeuroD, which also does not regulate photoreceptor fate but, within the photoreceptor lineage, governs differentiation (Ochocinska and Hitchcock, 2009; Taylor et al., 2015). These results also demonstrate that while, based on their sequences, miR‐18a, b, and c may be considered functionally redundant, miR‐18b and c do not fully compensate for the loss of miR‐18a.
Tgif1‐mutant larvae were observed to have increased expression of miR‐18a throughout the retina, providing an opportunity to determine the effects of miR‐18a gain‐of‐function. In tgif1 mutants compared with the WT, despite equivalent neuroD mRNA expression, NeuroD protein levels are lower and there are fewer differentiated photoreceptors. Knockdown of miR‐18a in tgif1 mutants fully rescues the photoreceptor deficiency, indicating that the elevated miR‐18a expression in these mutants can account for the absence of differentiated photoreceptors. The Tgif1 protein is a transcriptional co‐repressor in the TGFβ pathway that, in the adult zebrafish retina, is critical for injury‐induced photoreceptor regeneration (Lenkowski et al., 2013). The role of the TGFβ pathway has not been investigated during embryonic retinal development, but our results suggest a negative regulation of miR‐18a downstream of TGFβ/Tgif1 to regulate NeuroD and photoreceptor differentiation (Fig. 7). These data show that miR‐18a gain‐of‐function in the tgif1 mutants produces a phenotype opposite to that of the miR‐18a morphants or mutants and, taken together, show that miR‐18a post‐transcriptionally regulates NeuroD and, thereby, governs photoreceptor differentiation.
Figure 7.
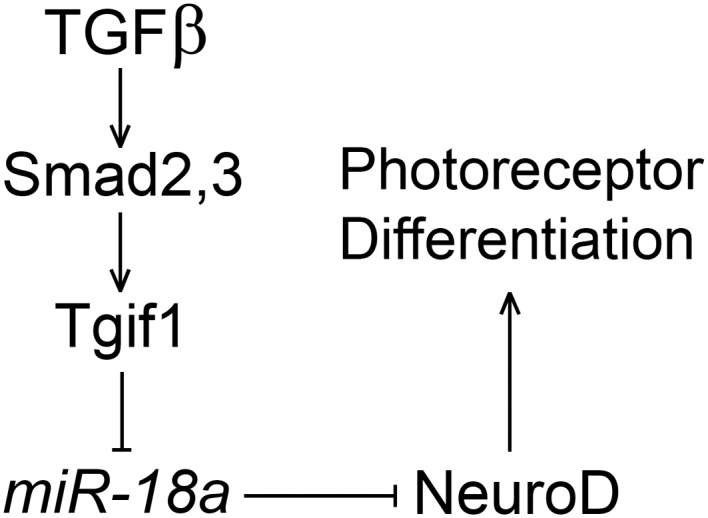
Hypothesized regulatory relationships between TGFβ, Tgif1, miR‐18a, and NeuroD during photoreceptor differentiation. Tgif1 functions downstream of Smad2 and Smad3 in the TGFβ signaling pathway (Lenkowski and Raymond, 2014). As a transcriptional corepressor, Tgif1 negatively regulates the expression of miR‐18a, and miR‐18a suppresses NeuroD protein levels to regulate the timing of photoreceptor differentiation.
In mutant or morphant fish lacking miR‐18a, photoreceptors differentiate at a faster rate without any obvious defects in retinal morphology, suggesting that miR‐18a inhibition could have therapeutic potential. Recent studies demonstrate regenerative potential in the mouse retina, in which EGF stimulates Müller glia to proliferate (Ueki and Reh, 2013) and Ascl1a confers reprogramming in Müller glia that generate neuronal progenitor cells (Pollak et al., 2013). Few of these Müller glia‐derived progenitors differentiate into photoreceptors, but neural regeneration can be augmented by also treating with a histone deacetylase inhibitor that promotes chromatin accessibility at important gene loci in the Müller glia (Jorstad et al., 2017). Creating a permissive environment is therefore critical for neurogenesis and photoreceptor regeneration might be therapeutically augmented by creating an environment that favors photoreceptor differentiation (e.g. through miR‐18a inhibition). This could be accomplished using RNA silencing or CRISPR/Cas9 gene editing, both of which have been successfully employed in vivo to knock down miRNAs (Chang et al., 2016; Shah et al., 2016). Therapeutic approaches using these methods are becoming more feasible as improvements are made in molecule delivery to target tissues using viral vectors (Zhu et al., 2017) and in creating CRISPR tools that can be activated in vivo (Dow et al., 2015; Hirosawa et al., 2017). Using viral vectors, CRISPR‐based cellular reprogramming was recently shown to prevent photoreceptor degeneration in a mouse model for the retinal disease retinitis pigmentosa (Zhu et al., 2017).
In conclusion, the data presented here demonstrate that during normal retinal development, miR‐18a regulates the timing of photoreceptor differentiation, and indicate that miR‐18a functions through post‐transcriptional regulation of NeuroD protein levels. This is consistent with the known role of NeuroD in governing differentiation in post‐mitotic photoreceptors (Taylor et al., 2015) and the functions of some miRNAs to effectively uncouple transcription and translation in order to ensure the correct spatiotemporal expression of proteins (Parchem et al., 2015; Bao et al., 2016; McLaughlin et al., 2018). We propose that within the photoreceptor lineage, following cell cycle exit, fine‐tuning of NeuroD protein levels by miR‐18a regulates the spatiotemporal pattern of photoreceptor differentiation. The importance of the spatiotemporal pattern of photoreceptor genesis is not yet understood, but could affect the development of the correct distribution, positioning, and, in zebrafish, the rigid spatial mosaic of cone photoreceptors (Raymond and Barthel, 2004; Allison et al., 2010; Raymond et al., 2014) that are essential for normal visual function.
Author Contributions
SMT performed this research at the University of Michigan and the University of West Florida. EG and PM performed this research at the University of West Florida. Writing and editing were performed by SMT, EG, and PFH.
The authors thank Laura Kakuk‐Atkins and Dilip Pawar for technical assistance. Fish lines and reagents provided by ZIRC were supported by NIH‐NCRR Grant P40 RR01.
Conflict of Interest
The authors declare no competing financial interests.
Literature Cited
- Akagi, T. , Inoue, T. , Miyoshi, G. , Bessho, Y. , Takahashi, M. , Lee, J.E. et al. (2004) Requirement of multiple basic helix‐loop‐helix genes for retinal neuronal subtype specification. The Journal of Biological Chemistry, 279(27), 28492–28498. Available at: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Allison, W.T. , Barthel, L.K. , Skebo, K.M. , Takechi, M. , Kawamura, S. and Raymond, P.A. (2010) Ontogeny of cone photoreceptor mosaics in zebrafish. The Journal of Comparative Neurology, 518(20), 4182–4195. Available at: 10.1002/cne.22447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva, K. and Cooper, N.G.F. (2014) MicroRNAs in the neural retina. International Journal of Genomics, 2014(10), 165897–165814. Available at: 10.1155/2014/165897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N.E. and Brown, N.L. (2018) All in the family: proneural bHLH genes and neuronal diversity. Development (Cambridge, England), 145(9), dev159426–159429. Available at: 10.1242/dev.159426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, J. , Li, D. , Wang, L. , Wu, J. , Hu, Y. , Wang, Z. et al. (2012) MicroRNA‐449 and microRNA‐34b/c function redundantly in murine testes by targeting E2F transcription factor‐retinoblastoma protein (E2F‐pRb) pathway. The Journal of Biological Chemistry, 287(26), 21686–21698. Available at: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, J. , Vitting‐Seerup, K. , Waage, J. , Tang, C. , Ge, Y. , Porse, B.T. et al. (2016) UPF2‐dependent nonsense‐mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3'UTR transcripts. PLoS Genetics, 12(5), e1005863 Available at: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. Available at: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Barthel, L.K. and Raymond, P.A. (1993) Subcellular localization of α‐tubulin and opsin mRNA in the goldfish retina using digoxigenin‐labeled cRNA probes detected by alkaline phosphatase and HRP histochemistry. Journal of Neuroscience Methods, 50(2), 145–152. Available at: 10.1016/0165-0270(93)90002-9. [DOI] [PubMed] [Google Scholar]
- Bassett, E.A. and Wallace, V.A. (2012) Cell fate determination in the vertebrate retina. Trends in Neurosciences, 35(9), 565–573. Available at: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- van den Berg, A. , Mols, J. and Han, J. (2008) RISC‐target interaction: cleavage and translational suppression. Biochimica Et Biophysica Acta, 1779(11), 668–677. Available at: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, S. , Hong, J. , Li, Q. , Schebelle, L. , Pollock, A. , Knauss, J.L. et al. (2013) MicroRNA cluster miR‐17‐92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Reports, 3(5), 1398–1406. Available at: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton, J.P. , Lovci, M.T. , Huang, J.L. , Yeo, G.W. and Pasquinelli, A.E. (2016) Pairing beyond the seed supports microRNA targeting specificity. Molecular Cell, 64(2), 320–333. Available at: 10.1016/j.molcel.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski, J.A. and Reh, T.A. (2015) Photoreceptor cell fate specification in vertebrates. Development (Cambridge, England), 142(19), 3263–3273. Available at: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski, J.A. , Kim, E.J. , Johnson, J.E. and Reh, T.A. (2011) Ascl1 expression defines a subpopulation of lineage‐restricted progenitors in the mammalian retina. Development (Cambridge, England), 138(16), 3519–3531. Available at: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin, L. and Wittbrodt, J. (2014) Retinal neurogenesis. Development (Cambridge, England), 141(2), 241–244. Available at: 10.1242/dev.083642. [DOI] [PubMed] [Google Scholar]
- Cepko, C. (2014) Intrinsically different retinal progenitor cells produce specific types of progeny. Nature Chemical Biology, 15(9), 615–627. Available at: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chang, H. , Yi, B. , Ma, R. , Zhang, X. , Zhao, H. and Xi, Y. (2016) CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Scientific Reports, 6(1), 22312 Available at: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte, I. , Marco‐Ferreres, R. , Beccari, L. , Cisneros, E. , Ruiz, J.M. , Tabanera, N. et al. (2010) Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development (Cambridge, England), 137(14), 2307–2317. Available at: 10.1242/dev.045294. [DOI] [PubMed] [Google Scholar]
- David, R. and Wedlich, D. (2001) PCR‐based RNA probes: a quick and sensitive method to improve whole mount embryo in situ hybridizations. BioTechniques, 30(4), 769–775. Available at: 10.2144/01304st02. [DOI] [PubMed] [Google Scholar]
- Djuranovic, S. , Nahvi, A. and Green, R. (2012) miRNA‐mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science (New York, NY), 336(6078), 237–240. Available at: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, L.E. , Fisher, J. , O'Rourke, K.P. , Muley, A. , Kastenhuber, E.R. , Livshits, G. et al. (2015) Inducible in vivo genome editing with CRISPR‐Cas9. Nature Biotechnology, 33(4), 390–394. Available at: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, S. , Handrick, R. , Aschrafi, A. and Otte, K. (2015) Unveiling the principle of microRNA‐mediated redundancy in cellular pathway regulation. RNA Biology, 12(3), 238–247. Available at: 10.1080/15476286.2015.1017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt, A.S. , Thatcher, E.J. , Burkewitz, K. , Li, N. , Liu, Y. and Patton, J.G. (2009) miR‐8microRNAs regulate the response to osmotic stress in zebrafish embryos. The Journal of Cell Biology, 185(1), 115–127. Available at: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt, A.S. , Rao, M. and Patton, J.G. (2017). Blocking zebrafish microRNAs with morpholinos. Methods in Molecular Biology (Clifton, N.J.), 1565, 59–78. Available at: 10.1007/978-1-4939-6817-6_6. [DOI] [PubMed] [Google Scholar]
- Forbes‐Osborne, M.A. , Wilson, S.G. and Morris, A.C. (2013) Insulinoma‐associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Developmental Biology, 380(2), 157–171. Available at: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, L.M. , Dror, A.A. , Mor, E. , Tenne, T. , Toren, G. , Satoh, T. et al. (2009) MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proceedings of the National Academy of Sciences of the United States of America, 106(19), 7915–7920. Available at: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory‐Evans, C.Y. , Wallace, V.A. and Gregory‐Evans, K. (2013) Gene networks: dissecting pathways in retinal development and disease. Progress in Retinal and Eye Research, 33, 40–66. Available at: 10.1016/j.preteyeres.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Gurtan, A.M. and Sharp, P.A. (2013) The role of miRNAs in regulating gene expression networks. Journal of Molecular Biology, 425(19), 3582–3600. Available at: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa, M. , Fujita, Y. , Parr, C.J.C. , Hayashi, K. , Kashida, S. , Hotta, A. et al. (2017) Cell‐type‐specific genome editing with a microRNA‐responsive CRISPR–Cas9 switch. Nucleic Acids Research, 45(13), e118–e118. Available at: 10.1093/nar/gkx309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, D.T. , Westman, B.J. , Martin, D.I.K. and Preiss, T. (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proceedings of the National Academy of Sciences of the United States of America, 102(47), 16961–16966. Available at: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger, E. and Izaurralde, E. (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews Genetics, 12(2), 99–110. Available at: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Hwang, W.Y. , Fu, Y. , Reyon, D. , Maeder, M.L. , Tsai, S.Q. , Sander, J.D. et al. (2013) Efficient genome editing in zebrafish using a CRISPR‐Cas system. Nature Biotechnology, 31(3), 227–229. Available at: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H.‐O. and Tomari, Y. (2015) The Functions of microRNAs: mRNA decay and translational repression. Trends in Cell Biology, 25(11), 651–665. Available at: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Chen, Z. , Liu, X. and Zhou, X. (2012). Evaluating the microRNA targeting sites by luciferase reporter gene assay In: Ying S. Y., (Ed.) Methods in Molecular Biology (Clifton, N.J.). Totowa, NJ: Humana Press, Vol. 936, pp. 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad, N.L. , Wilken, M.S. , Grimes, W.N. , Wohl, S.G. , VandenBosch, L.S. , Yoshimatsu, T. et al. (2017) Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature, 548, 103 Available at: 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuu, C. , Utheim, T.P. and Sehic, A. (2016) The three paralogous microRNA clusters in development and disease, miR‐17‐92, miR‐106a‐363, and miR‐106b‐25. Scientifica, 2016, 1379643 Available at: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Torre, A. , Georgi, S. and Reh, T.A. (2013) Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America, 110(26), E2362–2370. Available at: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkowski, J.R. and Raymond, P.A. (2014) Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish. Progress in Retinal and Eye Research, 40, 94–123. Available at: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkowski, J.R. , Qin, Z. , Sifuentes, C.J. , Thummel, R. , Soto, C.M. , Moens, C.B. et al. (2013) Retinal regeneration in adult zebrafish requires regulation of TGFβ signaling. Glia, 61(10), 1687–1697. Available at: 10.1002/glia.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Uribe, R.A. , Hayton, S. , Calinescu, A.‐A. , Gross, J.M. and Hitchcock, P.F. (2012) Midkine‐A functions upstream of Id2a to regulate cell cycle kinetics in the developing vertebrate retina. Neural Development, 7(1), 33 Available at: 10.1038/nprot.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madelaine, R. , Sloan, S.A. , Huber, N. , Notwell, J.H. , Leung, L.C. , Skariah, G. et al. (2017) MicroRNA‐9 couples brain neurogenesis and angiogenesis. Cell Reports, 20(7), 1533–1542. Available at: 10.1016/j.celrep.2017.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C.‐A. , Cho, J.‐H. , Wang, J. , Gao, Z. , Pan, P. , Tsai, W.‐W. et al. (2013) Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development (Cambridge, England), 140(3), 541–551. Available at: 10.1242/dev.085886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar, P. and Cayouette, M. (2015) Mechanisms of temporal identity regulation in mouse retinal progenitor cells. Neurogenesis, 2(1), e1125409 Available at: 10.1080/23262133.2015.1125409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, J.M. , Smith, D.F.Q. , Catrina, I.E. and Bratu, D.P. (2018) P‐bodies and the miRNA pathway regulate translational repression of bicoid mRNA during Drosophila melanogaster oogenesis. bioRxiv, 283630 Available at: 10.1101/283630. [DOI] [Google Scholar]
- Nasevicius, A. and Ekker, S.C. (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nature Genetics, 26(2), 216–220. Available at: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nepal, C. , Coolen, M. , Hadzhiev, Y. , Cussigh, D. , Mydel, P. , Steen, V.M. et al. (2016) Transcriptional, post‐transcriptional and chromatin‐associated regulation of pri‐miRNAs, pre‐miRNAs and moRNAs. Nucleic Acids Research, 44(7), 3070–3081. Available at: 10.1093/nar/gkv1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochocinska, M.J. and Hitchcock, P.F. (2007) Dynamic expression of the basic helix‐loop‐helix transcription factor neuroD in the rod and cone photoreceptor lineages in the retina of the embryonic and larval zebrafish. The Journal of Comparative Neurology, 501(1), 1–12. Available at: 10.1002/cne.21150. [DOI] [PubMed] [Google Scholar]
- Ochocinska, M.J. and Hitchcock, P.F. (2009) NeuroD regulates proliferation of photoreceptor progenitors in the retina of the zebrafish. Mechanisms of Development, 126(3–4), 128–141. Available at: 10.1016/j.mod.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohana, R. , Weiman‐Kelman, B. , Raviv, S. , Tamm, E.R. , Pasmanik‐Chor, M. , Rinon, A. et al. (2015) MicroRNAs are essential for differentiation of the retinal pigmented epithelium and maturation of adjacent photoreceptors. Development (Cambridge, England), 142(14), 2487–2498. Available at: 10.1242/dev.121533. [DOI] [PubMed] [Google Scholar]
- Ohsawa, R. and Kageyama, R. (2008) Regulation of retinal cell fate specification by multiple transcription factors. Brain Research, 1192, 90–98. Available at: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Olive, V. , Minella, A.C. and He, L. (2015) Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Science Signaling, 8(368), re2–re2. Available at: 10.1126/scisignal.2005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem, R.J. , Moore, N. , Fish, J.L. , Parchem, J.G. , Braga, T.T. , Shenoy, A. et al. (2015) miR‐302 Is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Reports, 12(5), 760–773. Available at: 10.1016/j.celrep.2015.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, M.E. (2010) Targeting of mRNAs by multiple miRNAs: the next step. Oncogene, 29(15), 2161–2164. Available at: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- Petri, R. , Malmevik, J. , Fasching, L. , Åkerblom, M. and Jakobsson, J. (2014) miRNAs in brain development. Experimental Cell Research, 321(1), 84–89. Available at: 10.1016/j.yexcr.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Pham, J.T. and Gallicano, G.I. (2012) Specification of neural cell fate and regulation of neural stem cell proliferation by microRNAs. American Journal of Stem Cells, 1(3), 182–195. papers3://publication/uuid/2D0AFD65-6542-4CF3-A25A-AE617D04EA3C. [PMC free article] [PubMed] [Google Scholar]
- Pollak, J. , Wilken, M.S. , Ueki, Y. , Cox, K.E. , Sullivan, J.M. , Taylor, R.J. et al. (2013) ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development (Cambridge, England), 140(12), 2619–2631. Available at: 10.1242/dev.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrózek, T. , Mlak, R. , Dziedzic, M. , Małecka‐Massalska, T. and Sagan, D. (2018) Investigation of relationship between precursor of miRNA‐944 and its mature form in lung squamous‐cell carcinoma—the diagnostic value. Pathology ‐ Research and Practice, 214(3), 368–373. Available at: 10.1016/j.prp.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Raymond, P.A. and Barthel, L.K. (2004) A moving wave patterns the cone photoreceptor mosaic array in the zebrafish retina. The International Journal of Developmental Biology, 48(8–9), 935–945. Available at: 10.1387/ijdb.041873pr. [DOI] [PubMed] [Google Scholar]
- Raymond, P.A. , Colvin, S.M. , Jabeen, Z. , Nagashima, M. , Barthel, L.K. , Hadidjojo, J. et al. (2014) Patterning the cone mosaic array in zebrafish retina requires specification of ultraviolet‐sensitive cones. PLoS ONE, 9(1), e85325 Available at: 10.1371/journal.pone.0085325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, E.A. and Dowling, J.E. (1999) Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. The Journal of Comparative Neurology, 404(4), 515–536. [PubMed] [Google Scholar]
- Shah, M.Y. , Ferrajoli, A. , Sood, A.K. , Lopez‐Berestein, G. and Calin, G.A. (2016) microRNA therapeutics in cancer—an emerging concept. EBioMedicine, 12, 34–42. Available at: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Zhao, X. , Hsieh, J. , Wichterle, H. , Impey, S. , Banerjee, S. et al. (2010) MicroRNA regulation of neural stem cells and neurogenesis. Journal of Neuroscience, 30(45), 14931–14936. Available at: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp, D.L. (2007) Neurogenesis in the fish retina. International Review of Cytology, 259, 173–224. Available at: 10.1016/S0074-7696(06)59005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp, D.L. (2015) Development of the vertebrate eye and retina. Progress in Molecular Biology and Translational Science, 134, 397–414. Available at: 10.1016/bs.pmbts.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier, T.R. and Palczewski, K. (2016) The impact of microRNA gene regulation on the survival and function of mature cell types in the eye. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 30(1), 23–33. Available at: 10.1096/fj.15-279745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.M. , Alvarez‐Delfin, K. , Saade, C. , Thomas, J.L. , Thummel, R. , Fadool, J.M. et al. (2015) The bHLH transcription factor NeuroD governs photoreceptor genesis and regeneration through Delta‐Notch signaling. Investigative Ophthalmology & Visual Science, 56(12), 7496–7515. Available at: 10.1167/iovs.15-17616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, Y. and Reh, T.A. (2013) EGF stimulates müller glial proliferation via a BMP‐dependent mechanism. Glia, 61(5), 778–789. Available at: 10.1002/glia.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia‐Sanchez, M.A. , Liu, J. , Hannon, G.J. and Parker, R. (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Development, 20(5), 515–524. Available at: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Ventura, A. , Young, A.G. , Winslow, M.M. , Lintault, L. , Meissner, A. , Erkeland, S.J. et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR‐17 through 92 family of miRNA clusters. Cell, 132(5), 875–886. Available at: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, V.A. (2011) Concise review: making a retina—from the building blocks to clinical applications. Stem cells (Dayton, Ohio), 29(3), 412–417. Available at: 10.1002/stem.602. [DOI] [PubMed] [Google Scholar]
- Wang, S. and Cepko, C.L. (2016) Photoreceptor fate determination in the vertebrate retina. Investigative Ophthalmology & Visual Science, 57(5), ORSFe1 Available at: 10.1167/iovs.15-17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, J. , Jung, S. , Keller, S. , Gregory, R.I. and Diederichs, S. (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biology, 11(3), 228–234. Available at: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Zeng, Y. , Yi, R. and Cullen, B.R. (2003) MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proceedings of the National Academy of Sciences of the United States of America, 100(17), 9779–9784. Available at: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. , Yi, R. and Cullen, B.R. (2005) Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. The EMBO Journal, 24(1), 138–148. Available at: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.‐W. , Zhang, L.‐Q. , Ding, L. , Wang, F. , Sun, Y.‐J. , An, Y. et al. (2011) MicroRNA‐19b downregulates insulin 1 through targeting transcription factor NeuroD1. FEBS Letters, 585(16), 2592–2598. Available at: 10.1016/j.febslet.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Ming, C. , Fu, X. , Duan, Y. , Hoang, D.A. , Rutgard, J. et al. (2017) Gene and mutation independent therapy via CRISPR‐Cas9 mediated cellular reprogramming in rod photoreceptors. Cell Research, 27(6), 830–833. Available at: 10.1038/cr.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]


