Abstract
Inflammatory cell activation drives diverse cellular programming during hepatic diseases. Hypoxia-inducible factors (HIFs) have recently been identified as important regulators of immunity and inflammation. In non-alcoholic steatohepatitis (NASH), HIF-1α is upregulated in hepatocytes where it induces steatosis; however, the role of HIF-1α in macrophages under metabolic stress has not been explored. In this study, we found increased HIF-1α levels in hepatic macrophages in methionine-choline-deficient (MCD) diet-fed mice and in macrophages of NASH patients compared to controls. The HIF-1α increase was concomitant with elevated levels of autophagy markers BNIP3, Beclin-1, LC3-II, and p62 both in mouse and human macrophages. LysMCre-HIFdPAfl/fl mice, which have HIF-1α levels stabilized in macrophages, showed higher steatosis and liver inflammation compared to HIFdPAfl/fl mice on MCD diet. In vitro and ex vivo experiments reveal that saturated fatty acid, palmitic acid (PA), induces both HIF-1α and impairs autophagic flux in macrophages. Using siRNA-mediated knock-down and over-expression of HIF-1α in macrophages, we demonstrated that PA impairs autophagy via HIF-1α. We found that HIF-1α mediates NF-κB activation and MCP-1 production and that HIF-1α mediated impairment of macrophage autophagy increases IL-1β production contributing to MCD diet-induced NASH.
Conclusions
Palmitic acid impairs autophagy via HIF-1α activation in macrophages. HIF-1α and impaired autophagy are present in NASH in vivo in mouse macrophages and in human blood monocytes. We identified that HIF-1α activation and decreased autophagic flux stimulate inflammation in macrophages through upregulation of NF-κB activation. These results suggest that macrophage activation in NASH involves a complex interplay between HIF-1α and autophagy as these pathways promote proinflammatory overactivation in MCD diet-induced NASH.
Keywords: Hypoxia-Inducible Factor-1 alpha, Non-alcoholic steatohepatitis, macrophage, autophagy
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases worldwide with increasing incidence in many developing countries (1). In patients with metabolic syndrome, NAFLD evolves from steatosis to non-alcoholic steatohepatitis (NASH) and ultimately to cirrhosis (2). The clinical co-morbidities of NAFLD/NASH include obesity, insulin resistance, and obstructive sleep apnea leading to periodic hypoxia (3). Recent studies demonstrate that hypoxia promotes liver triglyceride accumulation, necroinflammation, and fibrosis during NAFLD (4–6). These effects are mediated by the activation of hypoxia-inducible factors (HIFs) and their target genes that modify hepatocyte lipid metabolism (6, 7).
During hypoxia, HIFs regulate the transcription of hundreds of genes involved in a variety of cellular processes, including energy metabolism, cell survival, tumor invasion, angiogenesis and inflammation (5, 8). The best described HIF, HIF-1, is a heterodimeric complex composed of an α- and β-subunit. The α-subunit expression depends on oxygen levels, while the β-subunit is constitutively expressed (9). In addition to hypoxia, HIF-1α is activated by non-canonical pathways such as oxidative stress, growth factors, lipopolysaccharide (LPS), insulin, and TNF-α (10, 11). Liver HIF expression is increased in alcoholic fatty liver disease (ALD) and NAFLD (6, 7, 12). Although studies reported HIF-1α upregulation in ALD, the role of hepatocyte-specific HIF-1α in the progression of ALD remains controversial (6, 13). In a mouse model of NASH, induced by a methionine-choline-deficient (MCD) diet, the activation of HIF-1α in the liver and hepatocytes plays a role in the pathogenesis (14).
In NASH, free fatty acids (FFAs) are important mediators of lipotoxicity leading to cell injury and inducing hepatocyte damage via different pathways including endoplasmic reticulum stress. This process is counterbalanced by the upregulation of autophagic pathways that maintain cellular survival and homeostasis (15, 16). Impaired autophagy is a key element of hepatocyte damage in NASH (17). Cellular stressors, such as hypoxia and HIF activation can also interact with autophagy in hepatocytes (5). In vitro data demonstrates that HIF-1α protects against fatty acid-induced toxicity in hepatocytes (16). However, the role of these pathways is undefined in macrophages in NASH.
Accumulation of macrophages and overproduction of pro-inflammatory cytokines contribute to NASH pathogenesis (18). In addition, hypoxia potentiates the pro-inflammatory activation of primary human macrophages induced by palmitate (19). Here, we explored the role of HIF-1α in macrophages in a mouse model of NASH and in circulating monocytes from patients with NASH. Our results show that HIF-1α is activated in macrophages in NASH and correlates with impaired autophagic flux. We demonstrate that saturated fatty acid treatment of macrophages impairs autophagy via HIF-1α activation. We also describe an effect of macrophage-specific HIF-1α in NFκB activation that contributes to NASH development.
Materials and Methods
Animal Studies
The University of Massachusetts Medical School (UMMS) Institutional Animal Care and Use Committee approved the animal studies. Wild-type (C57BL/6J) and LysMCre mice, were purchased from Jackson Laboratories (Bar Harbor, Maine). Stabilization of HIF-1α in vivo was achieved through a Cre-lox approach as described (6, 20) with HIF1dPA mice, a gift from Dr. William Kim (University of North Carolina, Chapel Hill). Male and female mice (8–10 weeks old) were fed an MCD diet for five or eight weeks (Dyets Inc., PA, USA), depending on the experiment. Control mice received the MCS diet (MCD diet supplemented with DL-methionine and choline bitartrate) (Dyets, Inc., PA, USA). Male 8–12 weeks old wild-type mice were fed a High-Fat High-Cholesterol High-Sugar (HF-HC-HS) diet (Research Diets, New Brunswick, NJ, USA) with 42 g/L of carbohydrates (55% fructose and 45% sucrose) in drinking water for 24 weeks while control animals received chow diet.
Patient Samples
Paraffin-embedded blocks of human livers were obtained from the National Institutes of Health Liver Tissue Cell Distribution System (Minneapolis, MN; Pittsburgh, PA; Richmond, VA). Liver samples were from ten patients with a clinical diagnosis of NASH and four healthy controls. Human peripheral blood was collected, with informed consent, from ten healthy donors and twelve NASH patients with approval from the UMMS Institutional Review Board for Protection of Human Subjects in Research. The criteria used to define the patients with NASH were based on alanine aminotransferase (ALT) levels (>49 U/L) and, in some cases, biopsy.
Histopathological Analysis
Liver sections were stained with hematoxylin/eosin. Frozen sections were prepared from liver tissue frozen in OCT media and stained with Oil-Red-O. Immunohistochemistry staining for F4/80 (Thermo Fisher Scientific, MA1-91124) was performed on formalin-fixed, paraffin-embedded livers. The total number of F4/80-positive cells from five high-powered fields was counted per liver section. At least three liver sections were included in each group.
Cell Treatments
Macrophage isolation (bone marrow-derived, human, hepatic and THP-1) is described in the Supplementary Materials and Methods. Cells were stimulated in vitro with palmitic acid (PA, 250μM, Sigma-Aldrich) coupled with bovine serum albumin (BSA, Sigma-Aldrich) for different time periods. BSA was used as a vehicle control for PA. In selected studies, cells were treated with either the autophagy inducer, rapamycin (100 nM, Sigma-Aldrich), or the autophagy inhibitor, 3-MA (5mM, Sigma-Aldrich), for 30 minutes prior to PA treatment, or treated with the autophagy inhibitor bafilomycin (100 nM, Sigma-Aldrich) for the last 2 hours of the PA treatment.
Fluorescence and Confocal Microscopy
THP-1 cells were seeded in 6-well plates with microscope coverslips. After the indicated treatments, cells were fixed with 10% paraformaldehyde and permeabilized in PBS containing 0.5% Triton X-100 (Sigma-Aldrich). The cells were incubated at 4°C overnight with rabbit anti-LC3 (Cell Signaling Technology, #12741). The secondary antibody was Alexa Fluor 594 goat anti-rabbit IgG antibody (Thermo Fisher Scientific, A-11012). Images were acquired using fluorescence microscopy. The percentage of autophagic cells with LC3 puncta (>10) was determined by counting ≥100 cells from three independent experiments. The size and number of LC3 puncta/cell were calculated using the ImageJ program.
RNA Analysis
Total RNA was extracted using the RNeasy Mini kit (Qiagen, MD, USA) and reverse transcribed into cDNA using iScript Reverse Transcription Supermix (Bio-Rad, CA, USA). Real-time quantitative polymerase chain reaction (qPCR) was performed with the iCycler (Bio-Rad, CA, USA) using SYBR Green (Bio-Rad, CA, USA). The human and mouse primer sequences are described in the Supplementary Materials and Methods.
Immunoblot Analysis
Whole cell or nuclear lysates were isolated from mouse liver tissue or human THP-1 macrophages. Protein samples were separated by SDS-PAGE electrophoresis, transferred onto a nitrocellulose membrane, and probed with indicated primary antibodies followed by the appropriate secondary HRP-conjugated IgG antibody (Santa Cruz, CA, USA). The following antibodies were used: HIF-1α (Abcam, #ab179483), Beclin-1 (Cell Signaling Technology, #3738), BNIP3 (Abcam, #ab109362), LC3 (Cell Signaling Technology, #12741), p62 (Cell Signaling Technology, #8025), p-NF-κB p65 (Cell Signaling Technology, #3031), and p65 (Cell Signaling Technology, #8242). Protein bands were visualized using the Fujifilm LAS-4000 luminescent image analyzer.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay was performed on equal amounts (5–10 μg) of nuclear protein extracts from THP-1 macrophages using 50,000 cpm of γ-32P-labeled HIF-1α consensus oligonucleotide as we previously described (Santa Cruz, CA, USA) (14). All reactions were run on a 4% polyacrylamide gel. The dried gel was exposed to an X-ray film, the signal was intensified at −80°C, and densitometry was performed using ImageJ software.
Cell transfection for gene silencing and overexpression
For siRNA experiments, THP-1 macrophages were transfected with HIF-1α siRNA (Santa Cruz, CA, USA), ATG5 siRNA (Thermo Fisher Scientific, MA, USA), or scrambled siRNA as a control for 24 h. Silencing was achieved using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s protocol. For overexpression studies of HIF-1α, THP-1 macrophages were transfected with 6 μg of either the HIF-1α P402/P564A-pcDNA3 (21) or control vector for 24 hours. HA-HIF-1α P402/P564A-pcDNA3 was a gift from Dr. William Kaelin (Addgene plasmid #18955). Overexpression was achieved using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, MA, USA). Transfection efficiency of HIF-1α was evaluated by qPCR on mRNA.
ELISA
TNF-α, IL-1β, and IL-6 concentrations in culture supernatants were measured by ELISA according to the manufacturer’s instructions (BD PharMingen, CA, USA).
Statistical Analysis
Statistical significance was determined using a two-tailed student’s t-test or one-way ANOVA test. Data are presented as the mean±SEM and P<0.05 was considered statistically significant. All statistical analysis was performed using GraphPad Prism 5 software.
Results
Hepatic macrophages have increased HIF-1α expression and impaired autophagic flux in NASH
To evaluate the role of HIF-1α in macrophages in NASH pathogenesis, we used an MCD diet-induced mouse model of NASH. In this model, NASH is induced in MCD diet-fed mice as exhibited by steatosis indicated by Oil-Red-O staining (Fig. 1A) and prominent necroinflammation. Different mechanisms regulate steatosis in hepatocytes, including HIF-1α (6). Here, we observed significant increases in HIF-1α mRNA levels both in total liver (5-fold) and in isolated liver mononuclear cells (LMNCs, 6-fold) in MCD diet-induced NASH compared to control animals fed the MCS diet (Fig.1B). Mice fed a HF-HC-HS diet demonstrated a trend towards increasing HIF-1α in the liver and LMNCs similar to that seen in the MCD diet model (Supplementary Fig. 1).
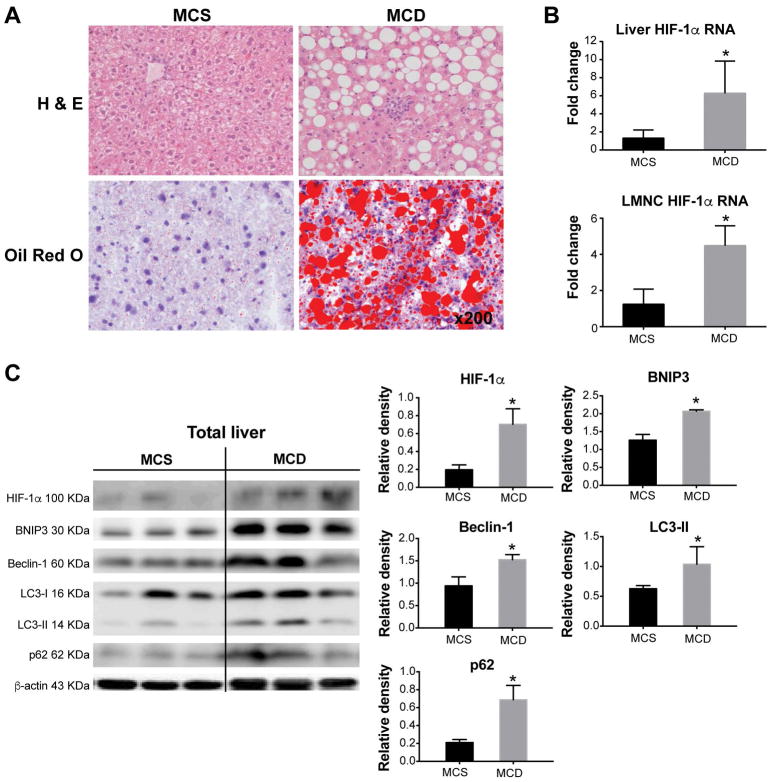
Figure 1. HIF-1α increases in total liver in MCD diet-fed mice and is associated with decreased autophagic flux.
Wild-type C57BL/6 mice were fed an MCD or MCS diet for 8 weeks. (A) Liver sections were stained with H&E or Oil-Red-O to assess liver injury and steatosis. (B) RNA isolated from either total liver or liver mononuclear cells (LMNC) was used to measure HIF-1α expression by qPCR. (C) The protein levels of HIF-1α, BNIP3, Beclin-1, LC3-II and p62 in total liver were assessed by western blot, quantified using ImageJ analysis and normalized to β-actin. Mice experiments are representative of 3–10 mice per experimental group. *P<0.05.
Studies support a role for autophagy in the pathogenesis of NASH, and autophagic flux is decreased in both hepatocytes and macrophages in obesity (22–24). Because HIF-1α is reported to induce autophagy under various metabolic stresses (25), we hypothesized that metabolic stress in NASH could be related to dysregulation of HIF-1α and the autophagy axis. In total livers, we detected a significant increase in HIF-1α protein levels and found concomitant increases in the expression of autophagy markers, BNIP3, Beclin-1, LC3-II and p62 in MCD diet-fed mice compared to MCS controls (Fig. 1C). BNIP3, a transcriptional target of HIF-1α, and Beclin-1 are inducers of autophagy (26), while LC3-II and p62 levels are established markers of autophagy induction or impaired autophagic flux, respectively (27).
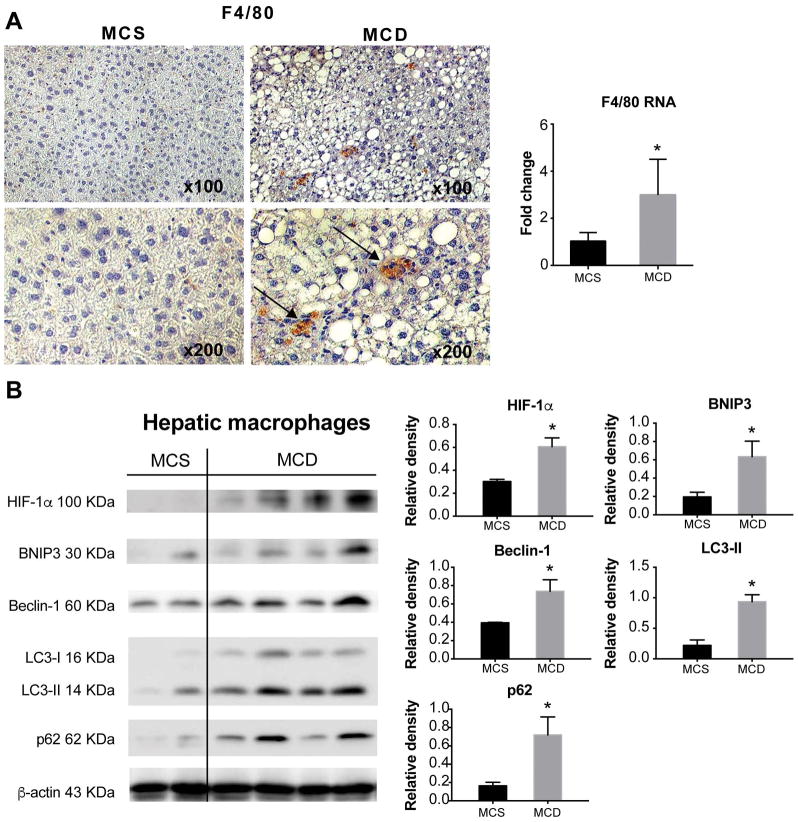
Consistent with the presence of inflammation in NASH, we found increased macrophage infiltration confirmed by F4/80 staining in livers of MCD diet-fed mice compared to MCS controls (Fig. 2A). While increased HIF-1α expression was found in hepatocytes in both NASH and ALD (6, 14), the role of HIF-1α in macrophages in steatohepatitis has not been elucidated. Thus, we analyzed the protein levels of HIF-1α and autophagy markers in isolated hepatic macrophages. The levels of HIF-1α and the autophagy markers, Beclin-1, LC3-II, BNIP3 and p62 were all significantly higher in hepatic macrophages with NASH compared to controls (Fig. 2B). These results indicated that macrophage regulation in NASH involves increased HIF-1α expression and decreased autophagic flux.
Figure 2. Hepatic macrophages of MCD diet-fed mice have increased HIF-1α and decreased autophagic flux.
Liver sections from MCD or MCS diet-fed mice were subjected to F4/80 immunohistochemistry (black arrows indicate the F4/80 positive cells), the F4/80 RNA levels were measured by qPCR in corresponding liver samples. (B) The protein levels of HIF-1α, BNIP3, Beclin-1, LC3-II and p62 were assessed in isolated hepatic macrophages by western blot. Image density was quantified using ImageJ analysis and normalized to β-actin. Mice experiments are representative of 3–10 mice per experimental group. *P<0.05.
Hepatic macrophages and circulating monocytes in NASH patients show increased HIF-1α expression and impaired autophagic flux
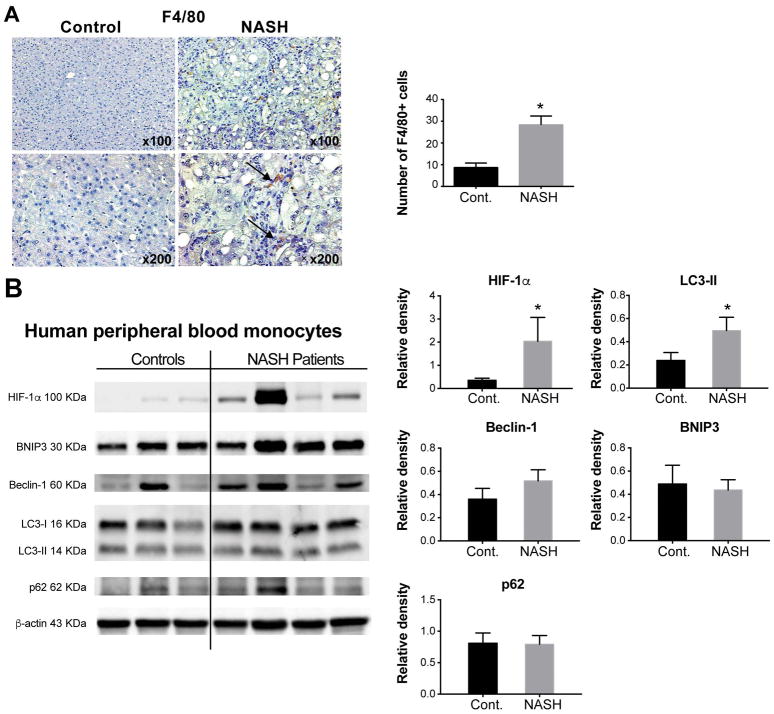
To validate our findings in the NASH mouse model, we tested human samples. In the livers of NASH patients, we detected significantly increased numbers of F4/80+ macrophages (Fig. 3A) compared with healthy controls, consistent with our findings in the livers of MCD diet-fed mice. Next, we compared HIF-1α expression and autophagy markers in circulating monocytes from NASH patients and healthy controls and found elevated protein levels of HIF-1α and LC3-II in NASH patients, but no difference in BNIP3, Beclin-1 and p62 levels (Fig. 3B). Thus, our results indicate autophagic flux is impaired in peripheral monocytes of NASH patients.
Figure 3. HIF-1α increases in both hepatic macrophages and circulating monocytes in NASH patients and correlates with autophagy.
Liver tissues and blood samples were collected from either NASH patients or healthy controls. (A) Liver sections were subjected to F4/80 immunohistochemistry (black arrows indicate the F4/80 positive cells). The total number of F4/80-positive cells from five high-powered fields was counted per liver section by microscopy. At least three liver sections were included in each group. (B) Circulating monocytes were isolated from blood samples, the protein levels of HIF-1α, BNIP3, Beclin-1, LC3 I, LC3 II and P62 in monocytes were assessed by western blot, quantified using ImageJ analysis and normalized to beta-actin. Representative blots are shown from 10 healthy donors and 12 patients with diagnosed NASH. *P<0.05. Cont, control.
Stabilizing HIF-1α in myeloid cells potentiates inflammation and steatosis induced by MCD diet
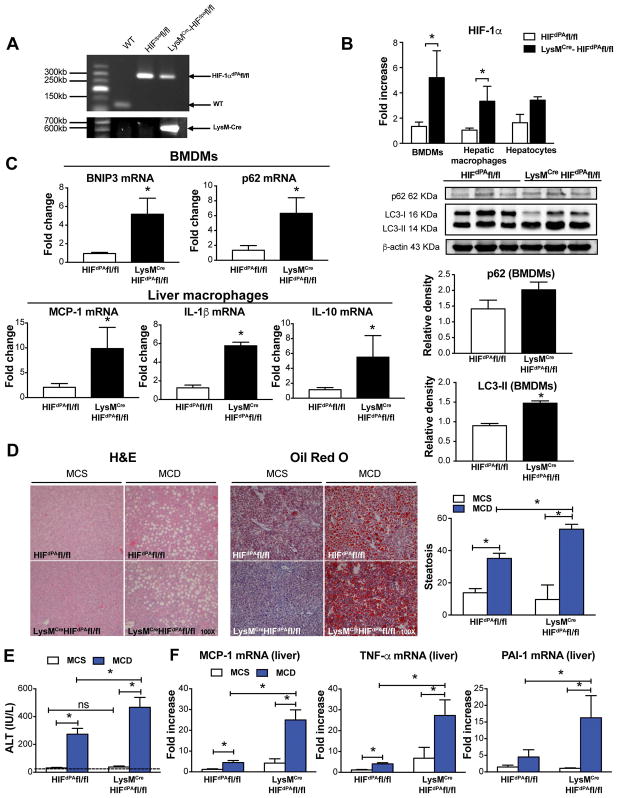
To explore the role of HIF-1α in macrophages and its correlation with autophagy in NASH pathogenesis in vivo, we used LysMCreHIFdPAfl/fl mice, in which levels of HIF-1α are stabilized in myeloid cells leading to higher levels of HIF-1α compared to wild-type mice. Using PCR, we confirmed the genotype of LysMCreHIFdPAfl/fl mice and the respective control mice, HIFdPAfl/fl (Fig. 4A). The increased HIF-1α mRNA levels in BMDMs and hepatic macrophages (Fig. 4B) but not in hepatocytes confirmed the myeloid cell-specificity of the HIF-1α increase. Previous evidence suggests that HIF-1α can either induce or impair autophagy depending on the disease (28, 29). To explore whether HIF-1α induces autophagy in macrophages, we examined the levels of the autophagy markers BNIP3 (both a HIF-1α target gene and an autophagy inducer), LC3-II, and p62. In BMDMs from LysMCreHIFdPAfl/fl mice, BNIP3 and p62 mRNA levels were increased as well as LC3-II protein levels; however, p62 protein levels were not significantly increased (Fig. 4C). We also found increased mRNA levels of MCP-1, IL-1β, and IL-10 in liver macrophages from LysMCreHIFdPAfl/fl compared to HIFdPAfl/fl mice (Fig. 4C). After 5 weeks on the MCD diet, LysMCreHIFdPAfl/fl mice exhibited a greater extent of liver steatosis (Fig. 4D) and ALT levels (Fig. 4E) compared to HIFdPAfl/fl mice. The increase in steatosis and liver injury was observed together with liver inflammation as indicated by increased MCP-1, TNF-α and PAI-1 mRNA levels (Fig. 4F). Altogether, these results suggest that increased macrophage HIF-1α levels contribute to MCD diet-induced NASH through impairment of autophagic flux and increased proinflammatory cytokines.
Figure 4. Macrophage-specific HIF-1α contributes to steatosis and liver inflammation induced by the MCD diet.
LysMCreHIFdPAfl/fl mice were generated by cross-breeding LysMCre mice, which express Cre recombinase in myeloid cells under the control of the lysozyme M promoter, with HIFdPAfl/fl mice, which lacks proline hydroxylation sites for HIF degradation. (A) Genotyping analysis of the HIFdPAfl/fl and LysMCreHIFdPAfl/fl mice. (B) The mRNA levels of HIF-1α in BMDMs, hepatic macrophages, and hepatocytes from HIFdPAfl/fl and LysMCreHIFdPAfl/fl mice were measured by qRT-PCR. (C) BNIP3 and p62 mRNA levels in BMDMs and MCP-1, IL-1β, and IL-10 mRNA expression in liver macrophages were measured by qRT-PCR. LC3-II and p62 protein levels were measured by western blot, quantified using ImageJ analysis, and normalized to β-actin. (D) Liver sections were stained with H&E or Oil-Red-O to assess liver injury and steatosis. (E) ALT levels were assessed from serum samples. (F) mRNA levels of MCP-1, TNF-α and PAI-1 were measured in the total liver by qRT-PCR. Mice experiments are representative of 3–10 mice per experimental group. *P<0.05.
Palmitic acid activates HIF-1α and impairs autophagic flux in macrophages
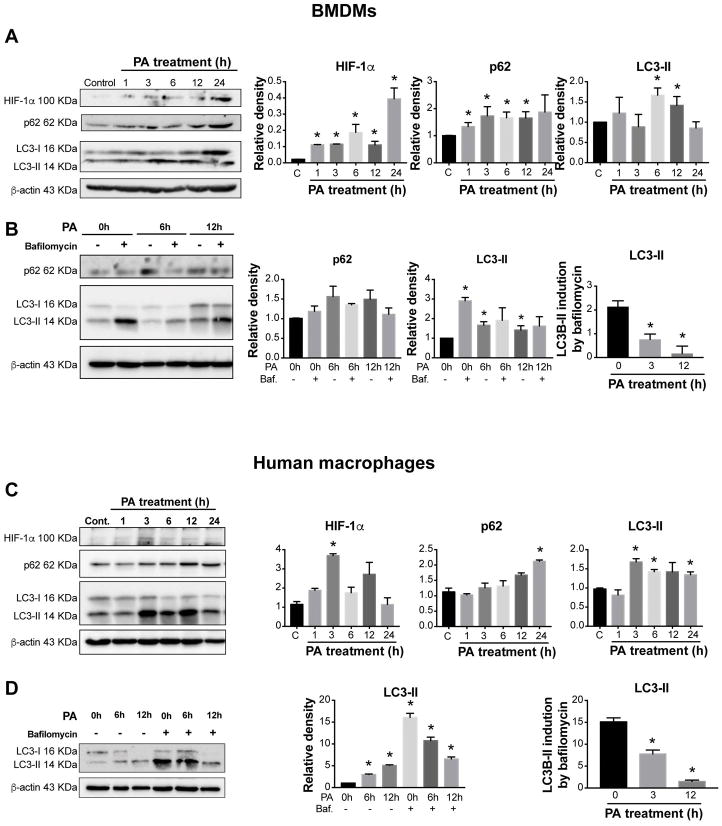
NASH progression is associated with high serum FFA levels. In particular, saturated FFAs such as palmitic acid (PA) have greater lipotoxicity compared to unsaturated FFAs (30) and induce HIF-1α activation in hepatocytes (31). Thus, we investigated whether PA activates HIF-1α in macrophages. PA treatment of primary hepatic and bone marrow-derived macrophages (BMDMs) increased HIF-1α mRNA levels followed by upregulation of HIF-1α target genes, BNIP3 and PAI-1 (Supplementary Fig. 2A and 2B). HIF-1α protein levels were also increased in BMDMs (Fig. 5A) and human macrophages (Fig. 5C) upon PA treatment and correlated with increases in p62 and LC3-II levels.
Figure 5. HIF-1α increases in BMDMs and human macrophages and correlates with decreased autophagic flux.
(A) Bone marrow from WT mice was isolated, and BMDMs generated after 7 days in the presence of L929 cell supernatant. Cells were stimulated with 250 μM PA for different time points and HIF-1α, p62, and LC3-II were assessed by Western blot. (B) BMDMs were cultivated in control medium or under PA stimulation for 6 or 12 hours with or without bafilomycin (Baf, 100 nM) (added at 0h or for the last 2 hours for 6 and 12 h incubations). (C) Human monocytes from healthy volunteers were isolated and differentiated to macrophages and HIF-1α, p62, and LC3-II were assessed in PA-stimulated cells by Western blot, quantified using ImageJ analysis and normalized to β-actin. (D) LC3-II levels of human macrophages were assessed in the presence or absence of bafilomycin (100nM) (added as in B) in PA-stimulated cells. Representative blots are shown from at least three independent experiments. *P<0.05. C, control.
To determine whether the PA-induced accumulation of LC3-II resulted from autophagosome formation or impairment of autophagosome degradation, we used bafilomycin to inhibit late-stage autophagy by preventing the fusion of autophagosomes and lysosomes (32). BMDMs and human macrophages were cultured in control medium or under PA stimulation in the presence or absence of bafilomycin. Consistent with the inhibition of autophagosome degradation, bafilomycin treatment alone resulted in LC3-II accumulation in both cell types and the combination of bafilomycin plus PA did not further increase LC3-II levels over that induced by PA alone (Fig. 5B, D). These results suggested that the LC3-II accumulation induced by PA is due to impaired autophagosome degradation and not increased autophagosome formation. The decrease in autophagic flux in PA-treated cells was confirmed by densitometric analysis of LC3-II expression in the presence or absence of bafilomycin to determine the LC3-II induction by bafilomycin. Briefly, we normalized the band intensities of LC3-II to β-actin and calculated the ratio of normalized LC3-II bands in bafilomycin-treated to untreated human macrophages (Fig. 5B, D).
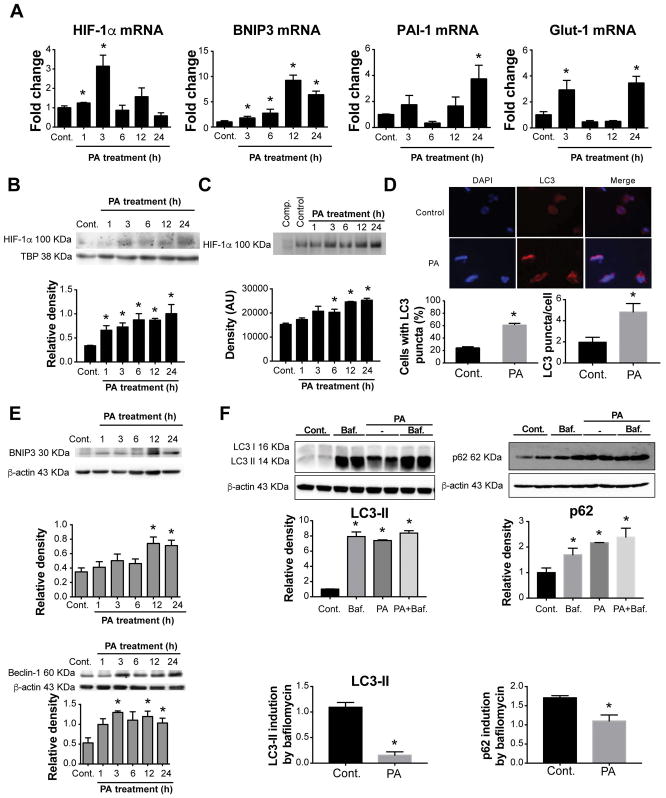
We next examined the effects of PA treatment on human THP-1 macrophages. Similar to primary murine macrophages, PA treatment in human THP-1 macrophages induced a rapid increase in HIF-1α mRNA levels peaking at 3 hours followed by upregulation of HIF-1α target genes, BNIP3, PAI-1, and Glut-1, 12–24 hours after stimulation (Fig. 6A). HIF-1α protein levels (Fig. 6B) and DNA-binding activity (Fig. 6C) were also increased in human THP-1 macrophages upon PA treatment. We found that PA treatment resulted in significant increases in BNIP3, Beclin-1, LC3-II and p62 levels in macrophages (Fig. 6E and 6F). Using fluorescence microscopy, we demonstrated a dramatic increase in the percentage of cells with LC3-II puncta, as a readout of autophagy, after PA stimulation (Fig. 6D).
Figure 6. Palmitic acid activates HIF-1α and decreases autophagic flux in THP-1 macrophages.
THP-1 cells were differentiated by 100nM PMA for 48h and treated with either 250 μM PA for the indicated time points, or BSA as a control. (A) The mRNA expression of HIF-1α and its target genes BNIP3, PAI-1 and Glut-1 were measured by qPCR. (B) Nuclear HIF-1α protein expression was measured via western blot and quantified using ImageJ analysis. (C) HIF-1α DNA-binding activity was tested and quantified using the electrophoretic mobility shift assay. (D) The detection of LC3 puncta in THP-1 cells was performed with a rabbit anti-LC3 antibody and immunofluorescence staining. THP-1 cells with more than 10 punctate dots were considered as positive cells (right), and the number and size of LC3 puncta/cell were quantified (left). Quantification of LC3-positive THP-1 cells and representative blots were performed in three independent experiments. (E) Protein levels of BNIP3 and Beclin-1 were assessed by western blot, quantified using ImageJ analysis, and normalized to β-actin. (F) Cells were cultivated in control medium or under PA stimulation for 12 hours with or without bafilomycin (Baf, 100 nM) for the last 2 hours. LC3 II and p62 protein levels were measured by western blot and quantified using ImageJ analysis. *P<0.05. Cont., control.
Bafilomycin alone resulted in the accumulation of LC3-II and p62 in THP-1 macrophages. PA increased LC3-II levels similar to bafilomycin alone, and the combination of bafilomycin plus PA did not further increase LC3-II levels (Fig. 6F), demonstrating a decrease in autophagic flux represented by decreased LC3-II or p62 induction by bafilomycin in PA-treated cells. These results are consistent with previous findings using primary macrophages and suggest that PA stimulation impairs autophagosome degradation in THP-1 macrophages.
To test if macrophage HIF-1α activation and impaired autophagy in NASH was PA specific, we treated BMDMs with low and high doses of lipopolysaccharide (LPS), a well-known pathogen-associated molecular pattern involved in NASH proinflammatory responses (11, 33). Both doses of LPS increased HIF-1α mRNA levels, its target gene, PAI-1, as well as autophagy components BNIP3 and p62 (Supplementary Fig. 2C). These results demonstrate that, similar to PA, LPS also increases HIF-1α expression and regulates the expression of autophagy components.
HIF-1α regulates autophagy in PA-stimulated macrophages
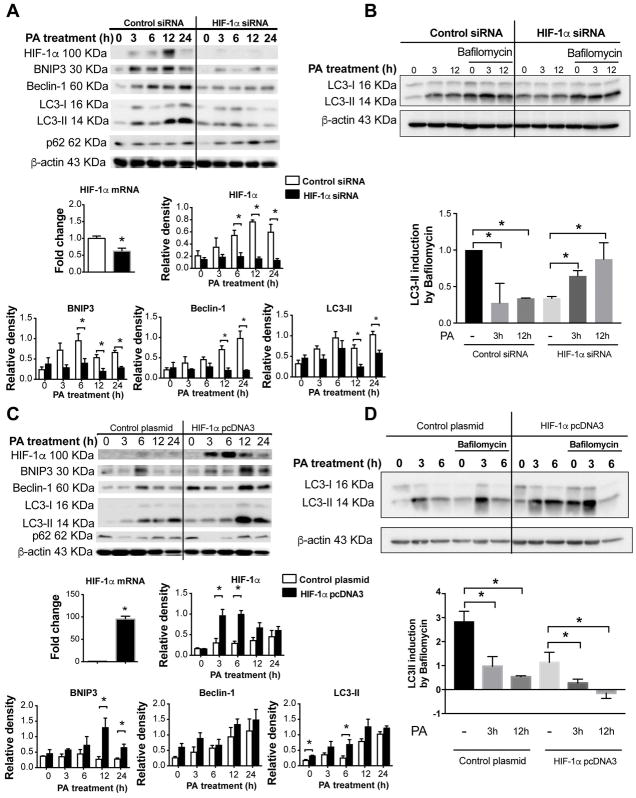
To test whether HIF-1α regulates autophagy in macrophages under PA-induced metabolic stress, we examined macrophages in which HIF-1α was knocked down using siRNA (Fig. 7A, B) or enhanced by plasmid transfection (Fig. 7C, D). Transfection of the control scrambled siRNA or empty plasmid did not alter the protein levels of HIF-1α, BNIP3, Beclin-1, and LC3-II, while PA treatment increased the expression of all of these proteins. Silencing HIF-1α prevented the PA-induced HIF-1α increase and dramatically downregulated BNIP3, Beclin-1 and LC3-II expression (Fig. 7A). HIF-1α overexpression significantly increased PA-induced levels of both HIF-1α and autophagy components BNIP3, Beclin-1 and LC3-II (Fig. 7C). Following PA stimulation, HIF-1α induction peaked at 6–12 hours and BNIP3, Beclin-1 and LC3-II around 6–24 hours (Fig. 7A, C).
Figure 7. HIF-1α is involved in impaired autophagy in palmitic acid-stimulated THP-1 macrophages.
HIF-1α mRNA and protein levels were downregulated using HIF-1α siRNA (A) or overexpressed using the HIF-1α-bearing pcDNA3 expression vector (B) in THP-1 macrophages. HIF-1α mRNA levels were assessed by qPCR. Protein levels of HIF-1α, BNIP3, Beclin-1, and LC3-II were assessed at indicated time points by western blot after transfection, quantified using ImageJ analysis and normalized to β-actin. HIF-1α siRNA (C) or overexpressed (D) THP-1 macrophages were cultivated in control medium or under PA stimulation with or without bafilomycin (100 nM) for the last 2 hours. LC3-II and P62 protein levels were measured by western blot, and the LC3-II ratio was calculated.
Next, we tested the role of HIF-1α in autophagic flux. In bafilomycin-treated THP-1 macrophages, PA increased LC3-II induction in the HIF-1α knockdown cells, but not in the control cells (Fig. 7B). HIF-1α overexpression produced the opposite effect (Fig. 7D); PA decreased LC3-II induction in bafilomycin-treated macrophages that overexpressed HIF-1α compared to control cells. Together, these results indicate that HIF-1α is involved in decreasing autophagic flux following PA stimulation.
HIF-1α-mediated macrophage autophagy inhibits palmitic acid-induced proinflammatory responses and downregulates NF-κB activation
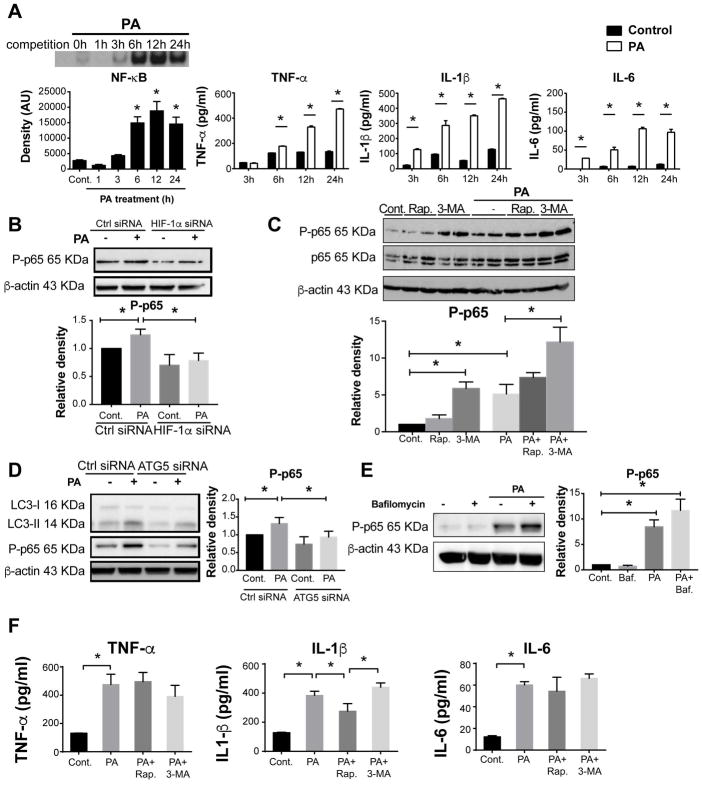
To investigate the role of HIF-1α in regulating inflammatory responses in macrophages, we evaluated the nuclear factor (NF)-κB pathway, a prototypical proinflammatory signaling pathway (34). Upon PA treatment, the DNA-binding activity of NF-κB and the levels of proinflammatory cytokines TNF-α, IL-1β, and IL-6 were increased in a time-dependent manner in THP-1 macrophages (Fig. 8A). These findings demonstrated that PA potently induces proinflammatory responses in macrophages through activation of the NF-κB pathway.
Figure 8. HIF-1α inhibits palmitic acid-induced NF-κB activation.
(A) NF-κB DNA-binding activity was measured after PA stimulation at indicated time points using EMSA. TNF-α, IL-1β and IL-6 levels in the supernatants of THP-1 cells upon PA treatment were measured using an ELISA assay. (B) Phosphorylated p65 (P-p65) levels in THP-1 macrophages treated with HIF-1α siRNA were measured by western blot and quantified using ImageJ analysis. (C) Cells were pretreated for 30 min with an autophagy inducer, rapamycin (Rap, 100 nM) or inhibitor, 3-methyladenine (3-MA, 5 mM) prior to 6h-PA stimulation. Protein levels of P-p65 and P65 were measured by western blot and quantified using ImageJ analysis. (D) Cells were transfected using control or ATG5 siRNA, and P-p65 levels were measured by western blot and quantified using ImageJ analysis. (E) Cells were cultivated in control medium or under PA stimulation for 6 hours with or without bafilomycin (Baf, 100 nM) (added at 0h or for the last 2 hours for 6 and 12 h incubations), and P-p65 levels were measured by western blot. (F) TNF-α, IL-1β and IL-6 levels in the supernatants were measured by ELISA. Representative blots and ELISAs are shown from at least three independent experiments *P<0.05.
To determine whether HIF-1α regulates NF-κB activation in PA-stimulated macrophages, we evaluated changes in p65 phosphorylation, a readout of NF-κB activation, after HIF-1α silencing. In HIF-1α knockdown cells, PA-induced p65 phosphorylation was completely abolished (Fig. 8B), suggesting that HIF-1α regulates not only PA-induced autophagy but also inflammation in macrophages. Similar effects were observed in liver macrophages from LysMCreHIFdPAfl/fl mice that showed enhanced MCP-1 and IL-1β (Fig. 4C).
To clarify whether HIF-1α modulation in the macrophage proinflammatory response to PA was associated with autophagy, we applied either an autophagy inducer (rapamycin) or an inhibitor (3-MA) in THP-1 cells with or without PA treatment. Inhibiting autophagy in macrophages with 3-MA alone increased NF-κB activation, and this activation was further increased with 3-MA plus PA treatment (Fig. 8C). In contrast, NF-κB activation was not modulated by rapamycin, suggesting that inhibition of autophagy can augment NF-κB in normal macrophages. However, when we silenced ATG5, a protein responsible for autophagosome formation, we found a decrease in LC3-II expression and p65 phosphorylation both in untreated cells and cells treated with PA (Fig. 8D). Because ATG5 silencing and 3-MA treatment both inhibit early stages of autophagy but had opposite effects in our experiments, we tested the effect of inhibiting the late stages of autophagy on NF-κB activation using bafilomycin. Bafilomycin treatment failed to modulate p65 phosphorylation in media or PA-stimulated macrophages (Fig. 8E) suggesting that impaired autophagy does not affect NF-κB activation in macrophages.
We also measured the levels of TNF-α, IL-1β and IL-6 in supernatants from cells treated with PA, rapamycin, or 3-MA. We found that the PA-induced increases in TNF-α and IL-6 levels were not modulated significantly by induction or inhibition of autophagy. However, PA-induced IL-1β protein levels were inhibited by rapamycin, but inhibition of autophagy with 3-MA had no significant additional effect compared with PA alone (Fig. 8F). These data imply that the induction of macrophage autophagy may suppress IL-1β induction by PA without any modulation on NF-κB activation. Our results suggest that HIF-1α is involved in NF-κB activation, macrophage pro-inflammatory responses, and impairment of PA-induced autophagy.
DISCUSSION
NASH pathogenesis is linked to hepatocyte steatosis and necroinflammation, and liver macrophages play an important role in both of these processes (18). Previous studies demonstrated that steatosis is linked to metabolic signals such as FFAs that induce cellular stress responses in hepatocytes including HIF-1α and autophagy (16, 31). However, the role of HIF-1α and autophagy in macrophages is not well understood, despite their crucial involvement in the disease progression of NASH. In this study, we found HIF-1α activation and decreased autophagic flux in liver macrophages in mice and circulating monocytes in humans with NASH. We show that HIF-1α expression in macrophages correlates with decreased autophagic flux and increased proinflammatory cytokine production which contributes to NASH pathogenesis in the MCD diet-induced mouse model. Our mechanistic studies reveal that PA-induced HIF-1α expression is a potent mediator of autophagy impairment in macrophages in NASH. Finally, we discovered that PA-induced NF-κB activation is mediated by HIF-1α but not by autophagic flux impairment.
We previously reported that HIF-1α is a key regulator of cellular responses to hypoxia during alcohol-induced steatosis (5). However, HIF-1α also responds to stimuli under normoxia, including thrombin, growth factors, LPS, insulin and cytokines such as interleukins and TNF-α (10, 11). Previous studies found that hepatocyte-specific HIF-1α contributes to lipid accumulation and liver injury in ALD and in a mouse model of NASH (5). Here, we found increased HIF-1α expression both in total livers and isolated LMNCs from MCD diet-fed mice, suggesting that HIF-1α may play a role in liver inflammation as well as steatosis. We found increased HIF-1α levels in the F4/80+ macrophages from livers of MCD diet-fed mice compared with MCS controls. These observations prompted us to explore the mechanistic role of HIF-1α in macrophages.
Recent studies investigated the interaction between HIF-1α and autophagy in various diseases (25, 28, 29). Autophagy is a highly-conserved cellular response that mediates degradation and recycling of damaged proteins and cellular organelles. Autophagy may also play a role in obese mice with hepatic steatosis. Studies show that activation of autophagy protects hepatocytes in liver steatosis both in vivo and in vitro (31, 35). Loss of autophagy in macrophages correlates with macrophage M1 polarization (22, 27). However, how macrophage autophagy is regulated in NASH and its correlation with pathogenesis is not fully understood.
In our study, the increase of HIF-1α in macrophages in NASH was followed by upregulation of its target gene, BNIP3. BNIP3 is a member of the BH3-only subfamily of Bcl-2 family proteins that antagonizes the activity of the pro-survival proteins, Bcl-2 and Bcl-XL. BNIP3 increases under stressors such as hypoxia through HIF-1-dependent or -independent mechanisms (26). Recent studies have shown that BNIP3 can compete with Beclin-1 for binding to Bcl-2, and when Beclin-1 is not bound to Bcl-2, it induces autophagy (26). Beclin-1, a central regulator of autophagy, regulates the lipid kinase Vps-34 protein and promotes the formation of Beclin-1-Vps34-Vps15 core complexes in the initial stages of autophagosome formation (36). Recently, Song and colleagues found that under chronic hypoxia and/or high-glucose conditions, impaired autophagy drives Neuro-2a cell damage (37). We show that increased BNIP3 was accompanied by the upregulation of Beclin-1 and increased LC3-II, an established marker of autophagosome formation (27). Notably, LC3-II accumulation may indicate induction of autophagosome formation or inhibition of autophagosome degradation, also known as autophagic flux. Using bafilomycin, an inhibitor of late-stage autophagy (32), we verified that the PA-induced LC3-II accumulation results from decreased autophagic flux rather than activation of autophagosome formation. This finding is consistent with another report demonstrating that HFD and PA impair autophagic flux through lysosomal dysfunction in hepatocytes (38).
We previously reported that autophagic flux is inhibited in NASH macrophages, by analyzing the level of p62/SQSTM1, a selective substrate of autophagy (27). Our results demonstrate an increase in mouse hepatic macrophages after PA treatment in vitro and ex vivo. However, p62 levels were not modulated in human peripheral blood monocytes of NASH patients, probably due to phenotypic differences observed in this cell, such as the increase in non-classical phenotypes that have been related with NAFLD progression (39, 40). Interpreting changes in p62 levels is complex because changes can be cell type- and context-specific. Increased p62 levels may result from transcriptional upregulation observed under certain conditions or indicate the possible inhibition of autophagosome degradation (27). Furthermore, we found that LPS altered the mRNA levels of key genes in autophagy, such as p62 and HIF-1α, suggesting that the effect on autophagic flux is not unique to PA. This data is consistent with a study by Baker et al. that showed LPS blocks lysosome fusion with endosomes or autophagosomes in macrophages, suggesting a possible inhibition of autophagic flux (41).
In whole liver tissue, increased HIF-1α and decreased autophagic flux mediate NASH pathogenesis (16, 31). In addition, an increase of liver macrophage infiltrate, and its importance for NASH progression has been reported (18). Here, we used THP-1 macrophages in which HIF-1α was silenced or overexpressed prior to PA treatment and a mouse model where HIF-1α was stabilized in macrophages to show for the first time that HIF-1α in macrophages regulates NASH pathogenesis by decreasing autophagic flux.
Macrophages have a pivotal role in inflammatory responses in NASH. Our results show that PA treatment resulted in NF-κB activation and increased production of proinflammatory cytokines in macrophages and that HIF-1α mediates PA-induced NF-κB activation. Lin et al., reported that autophagy alleviates steatosis and injury in the HFD-induced non-alcoholic fatty liver condition in mice (42) and others showed that macrophage autophagy protects against liver diseases (43, 44). Our results suggest that impaired autophagy mediated by HIF-1α causes inflammation during NASH. Recent studies have investigated the mutual regulation of autophagy and NF-κB, which is a central regulator of proinflammatory cytokines in NASH. Autophagy can degrade nuclear shuttle protein-interacting kinase and IκB kinase, and thus prevent NF-κB activation when the chaperone HSP90 is inhibited (45). In addition, autophagy is required for the activation of NF-κB (46). In our study, inducing autophagy with rapamycin did not modulate NF-κB activation. Rapamycin decreased PA-induced IL-1β but not TNFα and IL-6 production, suggesting a role for autophagy in inflammasome activation. This finding agrees with reports that activation of autophagy targets ubiquitinated inflammasomes for destruction, limiting IL-1β production (47); knocking down Atg16L1, an autophagy component, in macrophages enhances LPS-induced IL-1β (48); and that macrophage autophagy downregulates IL-1β limiting acute toxic liver injury (44). Besides suppressing autophagy, 3-MA also increased NF-κB activation with PA stimulation. Upon silencing ATG5, an early stage autophagy inducer, we observed a different effect from 3-MA. This effect could be related to the dual effect of 3-MA in modulating autophagy via Inhibition Class I and III phosphoinositide 3-kinase (49). Inhibiting late stages of autophagy with bafilomycin was unable to modulate basal and PA-induced NF-κB activation. These results suggest that macrophage autophagy does not modulate PA-induced NF-κB activation but interferes with IL-1β production probably through targeting inflammasomes.
Our study demonstrates a role for macrophage-specific HIF-1α in a mouse model of NASH. Although MCD diet-fed mice developed lipid accumulation, hepatic inflammation, and fibrosis, they did not exhibit weight gain or insulin resistance as observed in human NASH patients indicating a limitation of the human relevance of our study. However, we found increased HIF-1α expression both in human blood monocytes from NASH patients and in the livers and LMNCs from HF-HC-HS diet-fed mice that may imply that the effects observed following increased HIF-1α expression could also be present in a human-like model.
In conclusion, our findings summarized in Supplementary Fig. 4 indicate that in NASH the activation of HIF-1α in macrophages by PA has a central role in disease progression. HIF-1α activation in macrophages decreases autophagic flux and increases IL-1β production. HIF-1α also activates NF-κB, which is important for MCP-1 expression. Both IL-1β and MCP-1 induce liver steatosis and inflammation (6, 50). Our work points to a central role of macrophage-specific HIF-1α in MCD diet-induced NASH. Given the prevailing presence of inflammation in advanced NASH, future work should evaluate whether macrophage-specific targeting of HIF-1α activation has potential therapeutic applications.
Supplementary Material
Acknowledgments
This work was supported by: NIH R01AA011576 and R01AA020744 to GS, NSFC 81500452 to XW, F30 AA024680 to PL, F31 AA025545 to AIV, F30 AA022283 to AS, NIH Liver Tissue Cell Distribution System (Minneapolis, MN; Pittsburgh, PA; Richmond, VA) NIH N01-DK-7-004/HHSN267007004C
The authors thank Dr. Jeffrey Nickerson, Dr. Jean Underwood, and the Cell & Development Biology Confocal Core at UMass Medical School for support in the use of the Leica TCS SP5 II Laser Scanning Confocal Microscope. Liver tissue was obtained from the National Institutes of Health Liver Tissue Cell Distribution System (Minneapolis, MN; Pittsburg, PA; Richmond, VA). We thank Donna Giansiracusa from the Gastroenterology research office for assistance with patient samples. The authors greatly appreciate the participation of patients and volunteers for this study. The authors thank Melanie Trombly and Candice Dufour for critical reading of the paper and Istvan Furi and Vivek Chowdhary for their help with hepatic macrophage isolation.
Abbreviations
- HIFs
Hypoxia-inducible factors
- NASH
non-alcoholic steatohepatitis
- PA
palmitic acid
- NAFLD
Non-alcoholic fatty liver disease
- ALD
alcoholic fatty liver disease
- MCD
methionine-choline-deficient
- FBS
fetal bovine serum
- PMA
phorbolmyristate acetate
- LAS
Leica Application Suite
- LMNCs
liver mononuclear cells
- LPS
lipopolysaccharide
- FFAs
free fatty acids
- EMSA
electrophoretic mobility shift assay
- ELISA
enzyme-linked immunosorbent assay
- ALT
alanine aminotransferase
- HF-HC-HS
high-fat high-cholesterol high-sugar
- HFD
high-fat diet
References
- 1.Chitturi S, Wong VW, Farrell G. Nonalcoholic fatty liver in Asia: Firmly entrenched and rapidly gaining ground. J Gastroenterol Hepatol. 2011;26(Suppl 1):163–172. doi: 10.1111/j.1440-1746.2010.06548.x. [DOI] [PubMed] [Google Scholar]
- 2.Bugianesi E, Moscatiello S, Ciaravella MF, Marchesini G. Insulin resistance in nonalcoholic fatty liver disease. Curr Pharm Des. 2010;16:1941–1951. doi: 10.2174/138161210791208875. [DOI] [PubMed] [Google Scholar]
- 3.Paschetta E, Belci P, Alisi A, Liccardo D, Cutrera R, Musso G, Nobili V. OSAS-related inflammatory mechanisms of liver injury in nonalcoholic fatty liver disease. Mediators Inflamm. 2015;2015:815721. doi: 10.1155/2015/815721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musso G, Cassader M, Olivetti C, Rosina F, Carbone G, Gambino R. Association of obstructive sleep apnoea with the presence and severity of non-alcoholic fatty liver disease. A systematic review and meta-analysis. Obes Rev. 2013;14:417–431. doi: 10.1111/obr.12020. [DOI] [PubMed] [Google Scholar]
- 5.Nath B, Szabo G. Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology. 2012;55:622–633. doi: 10.1002/hep.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nath B, Levin I, Csak T, Petrasek J, Mueller C, Kodys K, Catalano D, et al. Hepatocyte-specific hypoxia-inducible factor-1alpha is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology. 2011;53:1526–1537. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu A, Taylor M, Xue X, Matsubara T, Metzger D, Chambon P, Gonzalez FJ, et al. Hypoxia-inducible transcription factor 2alpha promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 2011;54:472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenneth NS, Rocha S. Regulation of gene expression by hypoxia. Biochem J. 2008;414:19–29. doi: 10.1042/BJ20081055. [DOI] [PubMed] [Google Scholar]
- 10.Blouin CC, Page EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 11.Haddad JJ, Land SC. A non-hypoxic, ROS-sensitive pathway mediates TNF-alpha-dependent regulation of HIF-1alpha. FEBS Lett. 2001;505:269–274. doi: 10.1016/s0014-5793(01)02833-2. [DOI] [PubMed] [Google Scholar]
- 12.Carabelli J, Burgueno AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, Sookoian S. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–1338. doi: 10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiyama Y, Goda N, Kanai M, Niwa D, Osanai K, Yamamoto Y, Senoo-Matsuda N, et al. HIF-1alpha induction suppresses excessive lipid accumulation in alcoholic fatty liver in mice. J Hepatol. 2012;56:441–447. doi: 10.1016/j.jhep.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35:532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo W, Noh KH, Ahn JH, Yu JH, Seo JA, Kim SG, Choi KM, et al. HIF-1alpha expression as a protective strategy of HepG2 cells against fatty acid-induced toxicity. J Cell Biochem. 2014;115:1147–1158. doi: 10.1002/jcb.24757. [DOI] [PubMed] [Google Scholar]
- 17.Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tosello-Trampont AC, Landes SG, Nguyen V, Novobrantseva TI, Hahn YS. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet-induced mouse model through tumor necrosis factor-alpha production. J Biol Chem. 2012;287:40161–40172. doi: 10.1074/jbc.M112.417014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snodgrass RG, Boss M, Zezina E, Weigert A, Dehne N, Fleming I, Brune B, et al. Hypoxia Potentiates Palmitate-induced Pro-inflammatory Activation of Primary Human Macrophages. J Biol Chem. 2016;291:413–424. doi: 10.1074/jbc.M115.686709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambade A, Satishchandran A, Saha B, Gyongyosi B, Lowe P, Kodys K, Catalano D, et al. Hepatocellular carcinoma is accelerated by NASH involving M2 macrophage polarization mediated by hif-1alphainduced IL-10. Oncoimmunology. 2016;5:e1221557. doi: 10.1080/2162402X.2016.1221557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Q, Bartz S, Mao M, Li L, Kaelin WG., Jr The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Rodriguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillon J, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Masia D, Cosin-Roger J, Calatayud S, Hernandez C, Alos R, Hinojosa J, Apostolova N, et al. Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD. Mucosal Immunol. 2014;7:929–938. doi: 10.1038/mi.2013.108. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Hunter JE, Zhang J, Kris-Etherton PM. Cardiovascular disease risk of dietary stearic acid compared with trans, other saturated, and unsaturated fatty acids: a systematic review. Am J Clin Nutr. 2010;91:46–63. doi: 10.3945/ajcn.2009.27661. [DOI] [PubMed] [Google Scholar]
- 31.Cai N, Zhao X, Jing Y, Sun K, Jiao S, Chen X, Yang H, et al. Autophagy protects against palmitate-induced apoptosis in hepatocytes. Cell Biosci. 2014;4:28. doi: 10.1186/2045-3701-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 2012;586:1584–1591. doi: 10.1016/j.febslet.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 37.Song Y, Du Y, Zou W, Luo Y, Zhang X, Fu J. Involvement of impaired autophagy and mitophagy in Neuro-2a cell damage under hypoxic and/or high-glucose conditions. Sci Rep. 2018;8:3301. doi: 10.1038/s41598-018-20162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyagawa K, Oe S, Honma Y, Izumi H, Baba R, Harada M. Lipid-Induced Endoplasmic Reticulum Stress Impairs Selective Autophagy at the Step of Autophagosome-Lysosome Fusion in Hepatocytes. Am J Pathol. 2016;186:1861–1873. doi: 10.1016/j.ajpath.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Gadd VL, Patel PJ, Jose S, Horsfall L, Powell EE, Irvine KM. Altered Peripheral Blood Monocyte Phenotype and Function in Chronic Liver Disease: Implications for Hepatic Recruitment and Systemic Inflammation. PLoS One. 2016;11:e0157771. doi: 10.1371/journal.pone.0157771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Oeztuerk S, Kratzer W, Boehm BO. A Nonclassical Monocyte Phenotype in Peripheral Blood is Associated with Nonalcoholic Fatty Liver Disease: A Report from an EMIL Subcohort. Horm Metab Res. 2016;48:54–61. doi: 10.1055/s-0035-1547233. [DOI] [PubMed] [Google Scholar]
- 41.Baker B, Geng S, Chen K, Diao N, Yuan R, Xu X, Dougherty S, et al. Alteration of lysosome fusion and low-grade inflammation mediated by super-low-dose endotoxin. J Biol Chem. 2015;290:6670–6678. doi: 10.1074/jbc.M114.611442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, Dong XC, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lodder J, Denaes T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, Lotersztajn S, et al. Macrophage autophagy protects against liver fibrosis in mice. Autophagy. 2015;11:1280–1292. doi: 10.1080/15548627.2015.1058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilyas G, Zhao E, Liu K, Lin Y, Tesfa L, Tanaka KE, Czaja MJ. Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1beta. J Hepatol. 2016;64:118–127. doi: 10.1016/j.jhep.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao G. Autophagy and NF-kappaB: fight for fate. Cytokine Growth Factor Rev. 2007;18:233–243. doi: 10.1016/j.cytogfr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Criollo A, Chereau F, Malik SA, Niso-Santano M, Marino G, Galluzzi L, Maiuri MC, et al. Autophagy is required for the activation of NFkappaB. Cell Cycle. 2012;11:194–199. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 47.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 49.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stienstra R, Saudale F, Duval C, Keshtkar S, Groener JE, van Rooijen N, Staels B, et al. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology. 2010;51:511–522. doi: 10.1002/hep.23337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.