Summary
Clusters of Neisseria meningitidis (Nm) urethritis among primarily heterosexual males in multiple United States cities have been attributed to a unique non-encapsulated meningococcal clade (the U.S. Nm urethritis clade, US_NmUC) within the hypervirulent clonal complex 11. Resistance to antimicrobial peptides (AMPs) is a key feature of urogenital pathogenesis of the closely related species, N. gonorrhoeae. The US_NmUC isolates were found to be highly resistant to the model AMP, polymyxin B (PmB, MICs 64–256 μg/ml). The isolates also demonstrated stable subpopulations of heteroresistant colonies that showed near total resistant to PmB (MICs 384–1024 μg/ml) and colistin (MIC 256 μg/ml) as well as enhanced LL-37 resistance. This is the first observation of heteroresistance in N. meningitidis. Consistent with previous findings, overall PmB resistance in US_NmUC isolates was due to active Mtr efflux and LptA-mediated lipid A modification. However, whole genome sequencing, variant analyses and directed mutagenesis revealed that the heteroresistance phenotypes and very high level AMP resistance were the result of point mutations and IS1655 element movement in the pilMNOPQ operon, encoding the type IV pilin biogenesis apparatus. Cross-resistance to other classes of antibiotics was also observed in the heteroresistant colonies. High-level resistance to AMPs may contribute to the pathogenesis of US_NmUC.
Keywords: Neisseria meningitidis, meningococcal urethritis, heteroresistance, polymyxin B, antimicrobial peptide, pilQ
Graphical Abstract
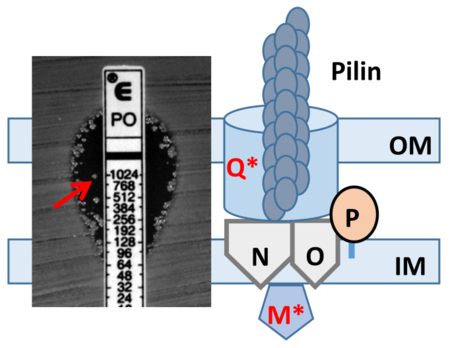
A unique meningococcal clade (US_NmUC) is causing urethritis clusters in multiple U.S. cities. US_NmUC isolates were resistant to the model antimicrobial peptide polymyxin B (PmB) and further expressed heteroresistant colonies highly resistant to PmB and colistin. Stable heteroresistance was caused by mutations and IS1655 insertions in the pilMNOPQ operon, which mediates the type IV pilin biogenesis, and conferred cross-resistance to other classes of antibiotics. This is the first observation of heteroresistance in N. meningitidis.
Introduction
Neisseria meningitidis (Nm), an obligate human pathogen, is carried asymptomatically in the nasopharynx of 5–10% of adults and is transmitted by close contact with respiratory droplets of oral or nasal secretions. Nm is also a leading cause of meningitis and rapidly fatal sepsis in otherwise healthy individuals that can cause large epidemic outbreaks (Rouphael & Stephens, 2012, Stephens et al., 2007). While capsular polysaccharide conjugate and protein-based meningococcal vaccines provide protection, invasive meningococcal disease is a continued worldwide problem.
Nm and Neisseria gonorrhoeae (Ng) are the only two Neisseria species that are human pathogens and these organisms most commonly colonize respiratory and urogenital tracts, respectively. Historically, Nm has not been a significant cause of urogenital disease and was infrequently recovered from the urogenital tract (cervix, vagina, and urethra) and rectum. However, there have been sporadic case reports in which Nm was isolated from patients with urethritis, cervicitis, vaginitis, proctitis, pelvic inflammatory disease, and postpartum endometritis dating back to the 1940s (Givan et al., 1977, Conde-Glez & Calderon, 1991, Maini et al., 1992). Recently, sustained sexually transmitted meningococcal urethritis outbreaks have been reported (Bazan et al., 2016, Bazan et al., 2017, Toh et al., 2017, Tzeng et al., 2017, Retchless et al., 2018). In one study, seventy-five Nm urethritis cases detected between January and November of 2015 in Columbus OH, represented 20% of all men who presented during that time with urethral GNID and growth of oxidase-positive Gram-negative diplococci (Bazan et al., 2017). Similar Nm-associated urethritis clusters have now been observed in multiple U.S. cities (Bazan et al., 2016, Bazan et al., 2017, Toh et al., 2017, Retchless et al., 2018). These urethritis-associated Nm isolates are members of a novel nongroupable US Nm urethritis clade (US_NmUC) in the cc11/ET-15 hyperinvasive lineage (Tzeng et al., 2017). The US_NmUC isolates do not make capsules (Toh et al., 2017, Tzeng et al., 2017) due to an IS1301 insertion that caused a multi-gene deletion at the capsule biosynthesis locus (Tzeng et al., 2017). Unlike many Nm isolates, the US_NmUC isolates are capable of efficient nitrite dependent anaerobic growth. This is due to a gene conversion event that introduced gonococcal aniA-norB genes, which encode enzymes that catalyze conversion of nitrite to nitric oxide and then nitrous oxide (Tzeng et al., 2017). Thus, the emergence of US_NmUC as a urethritis pathogen is likely the result of multiple evolutionary genetic events that allow better assimilation into the same niche first adopted by gonococci (Tzeng et al., 2017, Retchless et al., 2018).
Resistance to host-derived antimicrobial peptides (AMPs) is a key feature of neisserial pathogenesis at mucosal surfaces (Johnson & Criss, 2011, Tzeng & Stephens, 2015). A hallmark of gonococcal (and meningococcal) urethritis is the influx of PMNs, which employ both oxidative (production of reactive oxygen species) and non-oxidative (release of AMPs) killing mechanisms (Johnson & Criss, 2011, Criss & Seifert, 2012). In both Nm and Ng, resistance to PMN-derived and epithelial derived AMPs is mainly due to the activity of the LptA-mediated lipid A modification by phosphoethanolamine and the Mtr efflux pump (Tzeng et al., 2005). We have previously performed mariner random mutagenesis screening and identified transposon mutants in pilM and pilP that increased AMP resistance (Tzeng et al., 2005). In this report we show that the US_NmUC isolates are highly resistant to the AMP polymyxin B (PmB), a well-recognized surrogate for endogenous AMPs, and also exhibit “heteroresistance” (subpopulations of higher resistant colonies in the zone of inhibition) to PmB and colistin (polymyxin E). Increased AMP resistance of these isolates is linked to mutations in pilM and pilQ. PilQ is a member of the secretin family of proteins, and a major component of the outer membrane (Berry et al., 2012). PilM is a cytoplasmic ATP-binding protein that together with PilN/O/P proteins forms the inner membrane platform of the type IV pilus biogenesis complex (Ayers et al., 2009). Nm heteroresistance selected by PmB exposure, in addition to enhance resistance to colistin and LL-37, also conferred cross-resistance to several antibiotics, suggesting entry of these antibiotics is PilQ dependent. Heteroresistance to PmB has been described in several other Gram-negative bacterial pathogens, (Li et al., 2006, Lo-Ten-Foe et al., 2007, Hermes et al., 2013, Hjort et al., 2016, Jayol et al., 2015, El-Halfawy & Valvano, 2013) but this is the first demonstration of this phenomenon in Neisseriae.
Results
US_NmUC isolates demonstrate heteroresistance to polymyxin B and colistin
AMP resistance is an important pathogenic trait for both Ng and Nm. PmB E-test strips were used to determine the PmB MICs and revealed that the non-encapsulated US_NmUC isolates were highly resistant (Supplemental Table S1; Figure 1A). Of 52 CNM isolates, 41 (79%) had PmB MIC of 128–256 μg/ml and MICs of 10/11 remaining isolates were 64–96 μg/ml. One isolate, CNM34, had a significantly lower PmB MIC (16 μg/ml). Whole genome analysis of this isolate revealed a 2-bp deletion in mtrC, which encodes a key component of the Mtr efflux pump (Shafer et al., 1998). Two US_NmUC isolates from Atlanta had MICs of 96 and 128 μg/ml, respectively (Table S1). For comparison, the PmB MIC of a well-characterized unencapsulated meningococcal M7 strain (Swartley & Stephens, 1994, Tzeng et al., 2005) is 64 μg/ml. The PmB MICs of a clinical gonococcal isolate recovered during the Columbus urethritis outbreak (CNG20) and two gonococcal reference strains (FA19 and FA1090) were 48, 48 and 96 μg/ml, respectively; while the PmB MIC of a multi-drug resistant (MDR) gonococcal isolate from Japan, H041, (Ohnishi et al., 2011) is 192 μg/ml. These data suggest that the non-encapsulated US_NmUC isolates display equal or greater AMP resistance than gonococci.
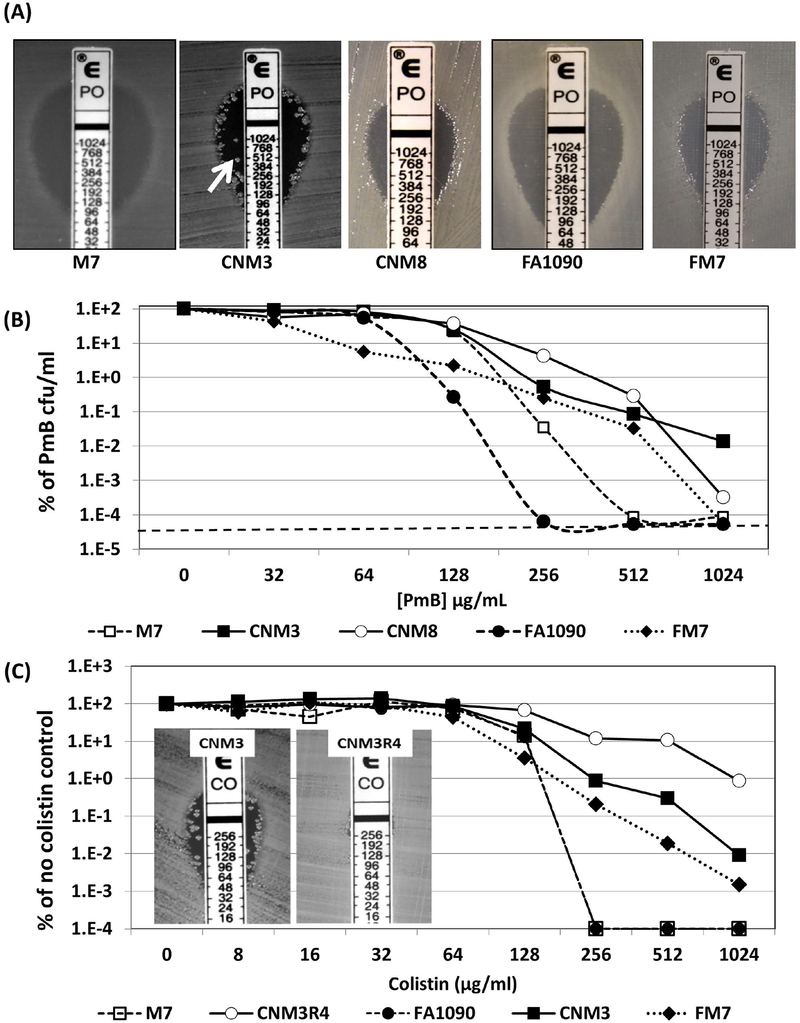
Figure 1.
(A) E-test pictures of two US_NmUC isolates (CNM3 and CNM8), two non-encapsulated Nm reference strains (M7 and FM7) and Ng reference strains GA1090. The arrow points to a heteroresistant colony in the zone of growth inhibition. (B) Population analysis profiling (PAP) assays of PmB with strains M7 (□), CNM3 (■), CNM8 (○), FM7 (◆) and FA1090 (●). The CFU counts of plates without PmB were set as 100% for normalization. The dotted line indicates the limit of detection. Each PmB concentration was assayed in triplicate and the experiments were repeated at least three times. (C) PAP assays of colistin performed similarly to PmB PAP assays with strains CNM3 (■), PmB heteroresistant mutant 3R4 (○), FM7 (◆), M7 (□) and FA1090 (●). Each concentration was tested in duplicates and the assays repeated twice.
Many of the US_NmUC isolates displayed PmB heteroresistance (Li et al., 2006, Lo-Ten-Foe et al., 2007); i.e., they yielded subpopulations of highly PmB resistant colonies in the zone of inhibition in disc diffusion and E-test assays (e.g. CNM3 shown in Figure 1A). These isolates also yielded higher MIC values in microbroth dilution assays. For example, the PmB MIC of CNM3 was 256 μg/ml by E-test, but was 1,024 μg/ml using the microbroth dilution assay. In contrast, neither the gonococcal strains tested nor M7 exhibited heteroresistance. However, during E testing, we did observe heteroresistant colonies of FAM18, a serogroup C cc11 reference strain, and a serogroup W cc22 invasive isolate GA18736 from Georgia (E-test pictures shown in supplemental figure 1), indicating that heteroresistance in Nm is not only found in US_NmUC isolates.
Heteroresistance of the US_NmUC isolates was confirmed using population analysis profiling (PAP) assays (El-Halfawy & Valvano, 2015). CNM3, CNM8 and the non-encapsulated FAM18 derivative FM7 was not eliminated by > 16-fold increases in PmB concentration (Figure 1B). In contrast, growth inhibition of M7 and FA1090 occurred across a narrow PmB concentration range. MC58 and FA19 also did not exhibit heteroresistance (data not shown). Interestingly, although CNM8 and FM7 failed to form colonies in the zone of inhibition in E tests (Figure 1A), these strains were heteroresistant in the PAP assay (Figure 1B).
We also examined whether the isolates exhibiting PmB heteroresistance would display analogous phenotypes toward another clinically used AMP, colistin (polymyxin E). As shown in Figure 1C, CNM3 and FM7 displayed the heteroresistance profile toward colistin; while the growth of M7 and FA1090 was inhibited across a narrow range. These data were consistent with the PmB resistance profiles of these strains. Further, the colistin E-test of CNM3 also showed colonies within the zone of inhibition (insert in Fig. 1C).
Heteroresistance to PmB is stable in US_NmUC isolates
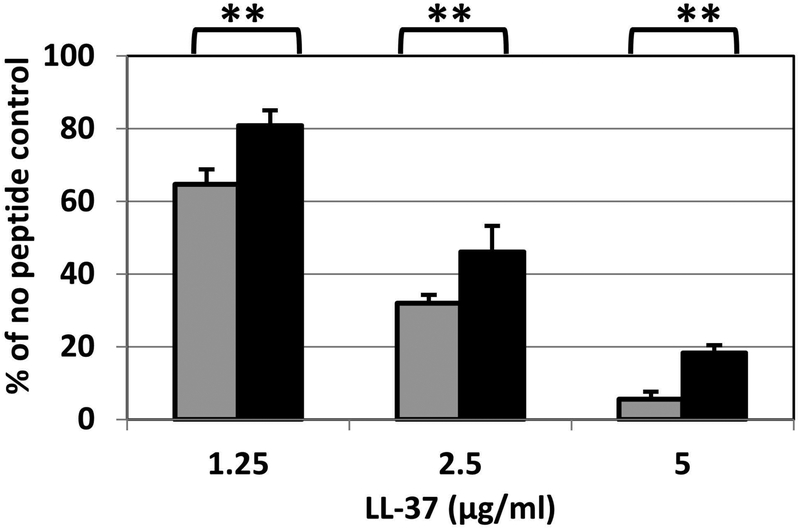
Heteroresistance to PmB, colistin and other antimicrobial agents in some bacteria is transient and reverts in the absence of continuous antimicrobial pressure (Napier et al., 2014, El-Halfawy & Valvano, 2015). To test if heteroresistance in the US_NmUC isolates was reversible, colonies picked from the zones of inhibition of CNM isolates (Table 1) were repeatedly passed on GCB agar plates in the absence of PmB. The elevated PmB MICs of these colonies were retained, suggesting that their enhanced PmB resistance was stable and likely due to genetic change(s). One of the recovered heteroresistant mutants, 3R4, was examined for resistance to colistin using PAP and E-test. The mutant was more resistant to colistin with a MIC greater than 256 μg/ml, whereas the MIC of the parental isolate CNM3 to colistin was ~ 48–64 μg/ml (Figure 1C). The resistance to LL-37 was also compared. The strains were treated with varying concentrations of LL-37 for 30 min followed by plating for viable CFU counts. The 3R4 mutant was more resistant to LL-37 in all concentrations tested than the parental strain (Figure 2), thus supporting the hypothesis that PmB heteroresistance confers cross-resistance to host endogenous antimicrobial peptides.
Table 1.
Polymyxin resistant mutants derived from the US_NmUC isolates
| Mutant | PmB MIC (parental) | Gene | Position | Changesa Parent/mutant | Outcomeb | Transformation frequencyc (parental) |
|---|---|---|---|---|---|---|
| 3R3, 3R4, 3R5, 3R6, 3R7 | 1,024 (128) |
pilQ | T167 | ctTcg/ctcg | fs F56S* | <1.3×10−8
(3.4×10−4) |
| 14R2 | 1,024 (192) |
pilQ | A2266 | acc/Ccc | T756P | <4.9×10−9
(1.9×10−5) |
| 17R1 | 1,024 (128) |
pilQ | A1465 | acc/Ccc | T489P |
3.2×10−7
(2.1×10−5) |
| 32R1, 32R2 | 1,024 (128) |
pilQ | G2229 | ggg/gggG | fs G743 → 815 aa | <1.4×10−8
(4.1×10−5) |
| 33R1 | 1,024 (96) |
pilQ | A1947 | Aaaaaa/aaaaa | fs AVLG/PSWG 655* |
1.3×10−7
(2.2×10−5) |
| 37R1 | 384 (96) |
pilM | A16 | IS1655 insertion | Disruption | <3.9×10−8
(3.6×10−5) |
| 45R2 | 384 (128) |
pilM | C126 | caa/Taa | fs Q43* | <7.2×10−9
(7.9×10−5) |
For point mutation, the wildtype sequence is shown on the left and the mutant on the right. The changed nucleotide was in capital letter.
Amino acid changes from the wild type to the mutant and the residue number are indicated. An asterisk indicates a stop codon immediately following the residue. The frameshift in 32R1 and 32R2 removed the original stop codon and yielded a larger PilQ protein. The lengths of PilQ and PilM proteins are 769 and 371, respectively.
Transformation was performed using 1 μg chromosomal DNA carrying a tonB::Ω(Sp) mutation. Frequencies (n=3) were calculated as the ratio of SpR cfu/ml to total cfu/ml per 1 μg DNA. The mutants with numbers in bold were transformable at low frequencies; while no transformants were recovered from the others.
Figure 2.
Sensitivity to LL-37 of the CNM3 clade isolate (gray) and its heteroresistant mutant 3R4 (black). Bacterial cells were incubated with LL-37 in RPMI at the indicated concentrations for 30 min and the number of viable CFU were determined by plating onto GC agar plates. Each conditions were assayed in duplicate at least twice. The averages and standard deviations of two independent assays are presented. Student’s t test was used to determine the statistical significance of survival of the mutant with respect to that of the wild type strain (**, P < 0.01).
Identification of heteroresistance associated mutations using genome sequencing
Genomes of eight heteroresistant colonies (3R3, 14R2, 17R1, 32R1, 32R2, 33R1, 37R1 and 45R2) derived from 7 CNM isolates (first number in strain designation is the parental CNM number, Table 1) were sequenced. Variants were identified by aligning the raw sequence reads against a CNM10 reference genome using PATRIC (www.patricbrc.org). Separately, assembled contigs were also compared to the CNM10 genome using the genome comparator (www.pubmlst.org).
Variants (Supplemental Data set S1) were examined for genes with previously reported roles in antimicrobial resistance. Alterations in several pil genes were identified in multiple heteroresistant mutants, pilU (33R1), pilM (45R2), pilQ (14R2, 17R1) and the pilS cassettes (silent incomplete pilin coding fragments responsible for antigenic variation of PilE) (17R1, 32R1, 32R2, 33R1 and 37R1) (Supplemental Data set S1). The class II pilE gene encoded near katA was intact in all mutants. Multiple repeated sequence motifs and slipped strand mispairing (SSM) events were detected, but none of these were in loci with known roles in AMP resistance. Overall, point mutations in pilM or pilQ were identified in seven of the eight heteroresistant mutants; while pilM is disrupted by an IS element in the remaining mutant (Table 1).
Frameshift mutations were identified in pilQ in the 3R4, 32R1, 32R2 and 33R1 mutants. 3R4 had a deletion (T) at position 167 that resulted in a premature stop at residue 57. Five additional colonies recovered from CNM3 (3R4–3R7) in independent experiments had the same deletion. 32R1 and 32R2 were isolated from CNM32 and both had a 1-bp insertion (G) at position 2229 that extended the PilQ coding region (815 aa vs. 769 aa). 33R1 had a 1-bp deletion (A) at position 1947 that truncated PilQ from 769 to 655 residues. The insertion in 32R1 changed a G3 to a G4 track and the deletion in 33R1 resulted in an A6-to-A5 transition. Whether these two short homopolymeric tracks have an increased sequence instability associated with the slipped strand mispairing phase variation events is not clear (Saunders et al., 2000, Snyder et al., 2001). The 14R2 and 17R1 mutants had C-A conversions at positions 2266 and 1465 that resulted in T to P residue changes at residue 756 and 489, respectively. Since the pilQ frameshift and these missense mutations yielded high levels of PmB resistance (MIC 1,024 μg/ml), these observations suggest that the missense mutations in 14R2 and 17R1 likely disrupt PilQ’s function as a multimeric protein complex. Two mutants had mutations in pilM, the first gene in the pilMNOPQ operon (Carbonnelle et al., 2005). 45R2 had a C-T conversion that resulted in a stop codon at residue 44; while pilM in 37R1 was disrupted by an IS1655 insertion at position 16. The pilM mutants had lower PmB MICs (256–512 μg/ml) than the pilQ mutants.
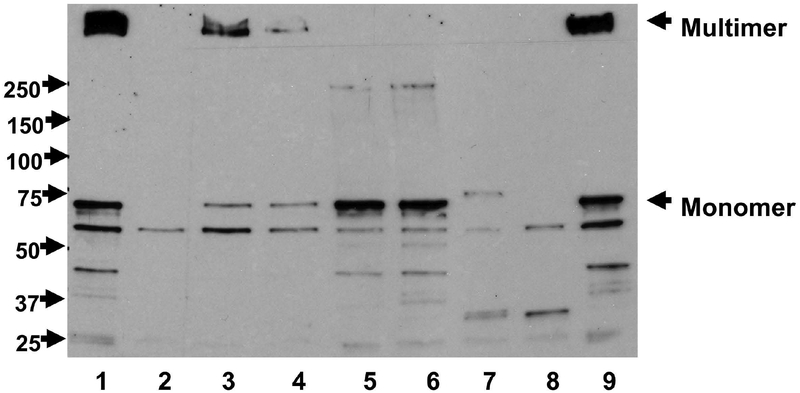
To further evaluate the effects of various pilQ and pilM mutations, we performed PilQ Western blots on total cellular extracts of the heteroresistant mutants and the wild type strain. As shown in Figure 3, a high molecular weight multimer band, a monomer band and several smaller bands, presumably degraded products, were detected by the PilQ monoclonal antibodies, like the previously reported pattern (Nandi et al., 2015). The pilQ frameshift mutants (3R4 and 33R1) resulting in premature truncation eliminated both of multimer and monomer bands (lanes 2 and 8). A weak and larger monomer band was detected for 32R1 (lane 7), in which the frameshift mutation resulting in a predicted larger protein; however, no multimer band was detected in this mutant. Two mutants with pilQ T to P residue changes (14R2 and 17R1) yielded no multimer bands, while maintaining monomer bands at comparable intensities as the wild type strain (lanes 5 and 6). Finally, the pilM::IS1655 mutant (37R1) and the pilM frameshift mutant (45R2) showed reduced levels of PilQ in both multimer and monomer forms (lanes 3 and 4), indicating a probable polar effect on PilQ protein expression, a phenomenon also observed in gonococci (Nandi et al., 2015).
Figure 3.
PilQ expression determined by Western blots. Equal amounts of whole cell lysates were resolved on 8% SDS-PAGE gels and transferred to PVDF membranes. The membrane was probed with PilQ antisera (Tonjum et al., 1998). Lanes: 1, WT; 2, 3R4 (pilQ-frameshift); 3, 37R1 (pilM::IS1655); 4, 45R2 (pilM-frameshift); 5, 14R2 (pilQ/T756P); 6, 17R1 (pilQ/T489P); 7, 32R1 (pilQ-frameshift); 8, 33R1 (pilQ-frameshift); 9, WT. The locations of PilQ multimer and monomer were marked on the right. Protein MW ladders were labeled on the left.
Independent pilM and pilQ mutations conferred enhanced PmB resistance
We previously showed that separate mariner transposon mutants in pilM and pilP caused increased PmB resistance (Tzeng et al., 2005). Transferring the pilM::aphA3 mutation into CNM3 was sufficient to increase the PmB MIC and the introduction of pilQ::aphA3 into CNM3 also conferred the higher PmB MICs observed in pilQ frame shift mutants. To independently confirm the effects of pilQ T→P point mutations, constructs with the aphA3(KnR) cassette inserted immediately downstream of the pilQ stop codon and carrying 14R2 or 17R1 point mutations in pilQ were generated. Five transformants were sequenced. All five CNM3–14R2 transformants carried the expected mutation via homologous recombination, whereas 2 of 5 CNM3–17R1 transformants contained the desired mutation. E-tests confirmed that the transformants carrying the T→P mutation have enhanced PmB resistance (Supplemental Figure 1), while PmB MICs of the transformants with a wild type pilQ sequence were identical to that of CNM3. Introducing these mutations in the respective parental isolate (CNM14 and CNM17) also produced the same PmB resistance results as the original mutants.
The pilM and pilQ mutations reduced transformation efficiency
Disruption of type IV pilin biogenesis apparatus is known to cause competence deficiency (Georgiadou et al., 2012). Thus, we expected the PmB heteroresistant mutants to have defects in transformation efficiency. Transformation efficiencies of the mutants were examined using chromosomal DNA carrying a tonB::Ω(Sp) mutation. As show in Table 1, all parental isolates showed transformation efficiency in the range of 10−4 – 10−5 per μg DNA. Significant reductions (>3 logs) were observed in all PmB resistant mutants. Most of the mutants were not transformable. The 17R1 (PilQ/T489P) mutant and the 33R1 mutant with a truncated PilQ remained transformable but with a 2-order of magnitude reduction in efficiency. Despite the differences in transformation efficiency, all pilQ mutations yielded similar higher levels of PmB resistance than the pilM mutants, both mutants were incompetent in transformation. There was no correlation between PmB resistance and transformation phenotypes.
Heteroresistance requires the Mtr efflux pump and the LptA transferase
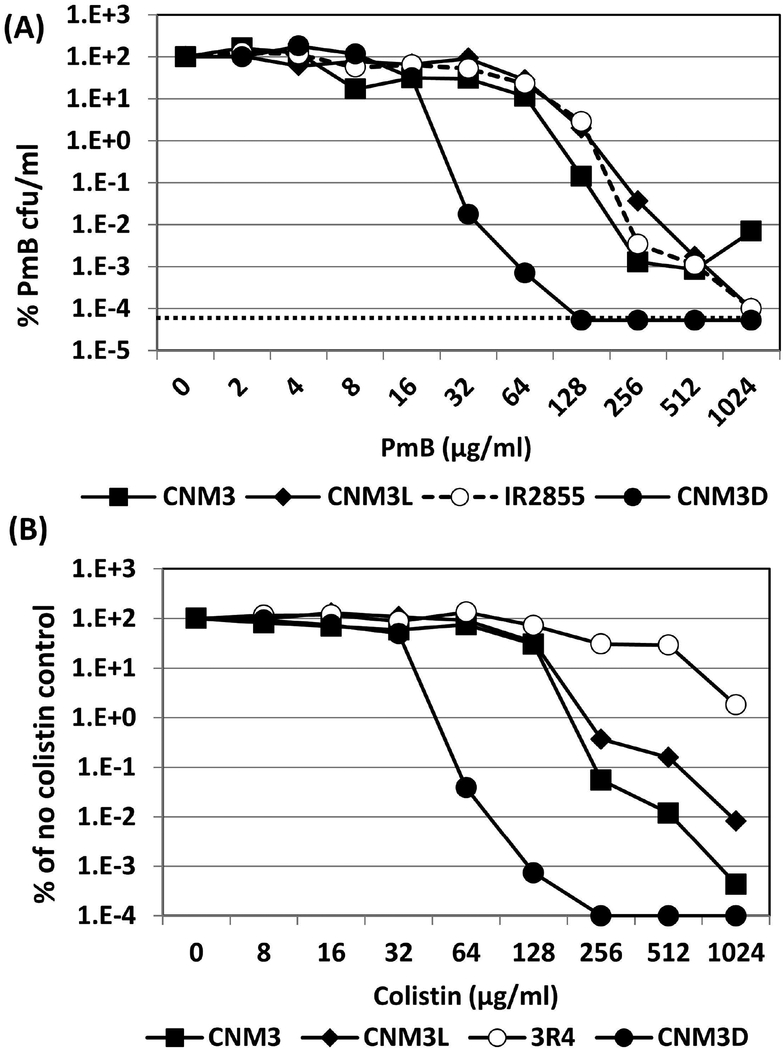
N. meningitidis intrinsic PmB resistance is mediated by LptA-mediated lipid A modification with phosphoethanolamine, the Mtr efflux pump and the capsule (Tzeng et al., 2005, Spinosa et al., 2007, Jones et al., 2009). To test if heteroresistance required these mechanisms, we introduced mtrD or lptA mutations in CNM3. Inactivation of mtrD and lptA reduced the PmB MIC levels to 24–32 μg/ml and 0.1 μg/ml, respectively (E-test data shown in supplemental figure 1). Neither mutants formed heteroresistant colonies in E-test and disc diffusion assays. In addition, PAP assays showed no heteroresistance toward PmB or colistin in the mtrD mutant (CNM3D) (Figure 4). Thus, development of heteroresistance in these strains is dependent upon intrinsic determinants of PmB resistance (Tzeng et al., 2005).
Figure 4.
(A) Population analysis profiling (PAP) assays of PmB resistance. Strains CNM3 (■), its mutL (CNM3L, ◆) and mtrD (CNM3D, ●) mutants and the mutator strain IR2855 (○) were compared. The CFU counts of plates without PmB were set as 100% for normalization. Each PmB concentration was assayed in triplicate and the experiments were repeated at least three times. The dotted line indicates the limit of detection. (B) PAP assays of colistin resistance. Strains CNM3 (■), its mutL (CNM3L, ◆), mtrD (CNM3D, ●) and heteroresistant mutant 3R4 (○) were examined analogously to the PmB PAP assays.
The US_NmUC isolates did not have higher spontaneous mutation or slipped strand mispairing frequencies
The number of point mutations and changes in monomeric tracks we observed suggested that the heteroresistant isolates might have higher mutation rates. We examined the spontaneous mutation rate in CNM3 using rifampin plating assays. The spontaneous rifampin resistance rate of CNM3 was low (median 2.2×10−10) and comparable to a low switcher strain IR2781 (median 1.7×10−9) (Richardson & Stojiljkovic, 2001). For comparison, IR2855, a mutL mutator strain that has a ~1000-fold higher mutation rate (median 3.7×10−7) than CNM3, exhibited a PAP profile similar to CNM3 (Figure 4A). The DNA mismatch repair (MMR) pathway is an important determinant of overall mutability and phase variation frequency in Nm (Richardson & Stojiljkovic, 2001). We generated a mutL mutation in CNM3 (CNM3L) to test if heteroresistance was influenced by the MMR pathway. The spontaneous rifampin resistance rate in CNM3L was more than 100-fold higher compared to CNM3 but these strains had similar PAP profile (Figure 4A). Thus, the MMR system did not appear to have a major role in the PmB heteroresistance phenotype.
Interestingly, it has been reported that cationic antimicrobial peptides, LL-37 and colistin, increased iron-induced mutagenesis in P. aeruginosa (Limoli et al., 2014, Rodriguez-Rojas et al., 2015). As we have performed experiments using iron-rich GC media, it was plausible that the heteroresistant mutations were enhanced in the presence of PmB and possibly other AMPs in N. meningitidis. We examined whether the antimicrobial peptide affected mutagenic phenotype in the clade isolate by comparing spontaneous rifampin mutation rates of meningococci grown on standard GC plates (Fe+3 is supplemented at 12 μg/ml) with or without colistin at 128 or 256 μg/ml (MIC50 as determined by the PAP assay is ~128 μg/ml). When compared to meningococci grown in the absence of colistin, no significant increases in spontaneous rifampin mutation rates upon exposure to colistin under the iron-replete condition were observed (data not shown).
We also measured the frequency of slipped-strand mispairing using the universal rate of switching cassette (UROS) assay (Alexander et al., 2004b). The UROS cassette contains a poly (G)8 tract within the Ω(Sp) cassette of aadA that is in the off phase. Thus, SpR colonies form when aadA is switched into the on phase by slipped strand mispairing. The UROS cassette did not show a higher slipped strand mispairing rate in CNM3, when compared to that of strain IR5426, (UROS cassette in the high switcher/mutator IR2855) (Alexander et al., 2004b).
PmB heteroresistant mutants have reduced susceptibility to multiple antibiotics
We tested if the resistance to other antimicrobial agents was altered in the PmB heteroresistant mutant 3R4, using E-tests (Table 2) and disc diffusion (supplemental table 2) on GC agar plates. CNM3 and 3R4 had similar susceptibility to levofloxacin, meropenem and azithromycin, which is one of the two current standards of care antibiotic treatment for gonorrhea. However, 3R4 has reduced susceptibility to penicillin G, ceftriaxone, cefotaxime, streptomycin, kanamycin, chloramphenicol and tetracycline (Table 2). The resistances of 3R4 to ceftriaxone and cefotaxime were two-fold higher than CNM3 by E-test, although its MICs remained in the sensitive range (Table 2). Interestingly, CNM3 has reduced susceptibility to several antibiotics such as azithromycin, meropenem, penicillin, and cefuroxime when compared to the Ng reference strain FA19 and to a clinical isolate CNG20. The resistance of CNM3 to penicillin G, cefuroxime and azithromycin was like that of the gonococcal isolate MS11, which has elevated resistance due to the presence of the penA, mtrR, and penB mutations (Ropp et al., 2002, Ohneck et al., 2011). Thus, the pilQ defect in the PmB heteroresistant mutant indeed influenced, albeit modestly, the susceptibility to many unrelated antibiotics.
Table 2.
Comparison of antibiotic resistance levels of the PmB heteroresistant mutant#
| Antibiotics | CNM3 | CNM3R4 | MS11 | CNG20 | FA19 |
|---|---|---|---|---|---|
| E test (μg/ml) | |||||
| Colistin | 53.3 ± 9.2 | 256** | 256 | 48 | 32 |
| Penicillin G | 0.22 ± 0.08 | 0.46 ± 0.10** | 0.25 | 0.064 | 0.012 |
| ceftriaxone | 0.002 ± 0.000 | 0.004 ± 0.001** | 0.002 | 0.002 | 0.002 |
| Cefotaxime | 0.01 ± 0.003 | 0.021 ± 0.004** | 0.012 | 0.004 | 0.002 |
| Azithromycin | 0.70 ± 011 | 0.85 ± 0.14 | 0.19 | 0.032 | 0.064 |
| Streptomycin | 7.8 ± 0.7 | 10.9 ± 2.0** | 1024 | 4 | 6 |
| Kanamycin | 10.9 ± 2.0 | 14.3 ± 2.1** | 8 | 8 | 4 |
| Chloramphenicol | 0.63 ± 0.13 | 0.81 ± 0.18* | 2 | 0.125 | 0.25 |
| Meropenem | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.064 | 0.094 | 0.064 |
| Tetracycline | 0.19 ± 0.0 | 0.26 ± 0.1* | 0.5 | 0.25 | 0.094 |
| Levofloxacin | 0.008 ± 0.0 | 0.007 ± 0.0 | 0.004 | 0.002 | 0.003 |
Data are presented as the mean ± standard deviation (N=3–8). The MIC values of antibiotics in bold are statistically different between CNM3 and 3R4 by student’s t test (**, p < 0.01; *, p < 0.05). A single data point is shown for each gonococcal strain as comparison.
Altered resistances to penicillin G, chloramphenicol and tetracycline between CNM3 and 3R4 were also compared in the mtrD and the lptA mutant backgrounds and no differences were observed in these backgrounds (Table 3). These data suggest that the higher intracellular antibiotic levels caused by either efflux pump inactivation (mtrD) or compromised membrane integrity (lptA) cannot be effectively reduced by blocking antibiotic entry through a pilQ mutation.
Table 3.
Comparison of antibiotic resistance levels of the PmB heteroresistant derivative in the mtrD or lptA mutant background
| Antibiotics | CNM3D | CNM3DQ | CNM3A | CNM3AQ |
|---|---|---|---|---|
| E test (μg/ml) | ||||
| Penicillin G | 0.053 ± 0.01 | 0.058 ± 0.01 | 0.079 ± 0.021 | 0.115 ± 0.018 |
| Chloramphenicol | 0.38 ± 0.0 | 0.38 ± 0.0 | 0.38 ± 0.0 | 0.38 ± 0.0 |
| Tetracycline | 0.182 ± 0.151 | 0.115 ± 0.018 | 0.094 ± 0.0 | 0.084 ± 0.017 |
Data are presented as the mean ± standard deviation (n=3).
Discussion
The US_NmUC has emerged as urogenital pathogen. Historically, Nm is not recognized as a significant cause of urogenital infection and the occasional meningococci recovered from such isolated cases have been from diverse serogroups and lineages (Harrison et al., 2017, Ma et al., 2017). The recent urethritis outbreaks and clusters caused by US_NmUC suggest that with novel genetic and phenotypic changes, this meningococcal clade is being transmitted efficiently between sexual partners and can successfully resist local innate immune responses (Bazan et al., 2016, Tzeng et al., 2017, Bazan et al., 2017, Toh et al., 2017, Retchless et al., 2018).
Like N. gonorrhoeae, the US_NmUC isolates trigger a potent local inflammatory response characterized by urethral discharge and presence of many polymorphonuclear leukocytes (PMNs) in the inflammatory exudates (Johnson & Criss, 2011). These US_NmUC isolates exhibited bacterial resistance to host killing. In this report, we investigate the basis of high-level resistance to AMPs of these US_NmUC isolates. Resistance to host-derived AMPs is an important pathogenic trait for both Ng and Nm (Johnson & Criss, 2011). Recent studies have indicated that PMNs primarily direct non-oxidative antimicrobial activities against Ng (Johnson & Criss, 2011). Non-encapsulated Nm and Ng are generally more sensitive to the action of AMPs than encapsulated meningococci (Tzeng et al., 2005). The ability of US NmUC isolates to resist killing by human AMPs, either produced locally by the mucosal epithelia where they serve as a primary defense mechanism or to resist AMP-mediated non-oxidative killing by PMNs at this site, may have important advantages. Survival in PMN’s may also serve as vehicles for dissemination in urethral exudates. Many of the non-encapsulated US_NmUC isolates displayed greater PmB resistance (MICs 96–256 μg/ml) than Ng and considerable higher resistance (MICs 384–1,024 μg/ml) was observed for the heteroresistant subpopulations.
Heteroresistance to PmB or colistin has been described in Acinetobacter baumannii (Li et al., 2006), Enterobacter cloacae (Lo-Ten-Foe et al., 2007), P. aeruginosa (Hermes et al., 2013), Salmonella typhimurium (Hjort et al., 2016), Klebsiella pneumonia (Jayol et al., 2015, Band et al., 2018) and Burkholderia cenocepacia (El-Halfawy & Valvano, 2013) and can lead to treatment failure in clinical settings and in experimental models (Band et al., 2016, Band et al., 2018). Interestingly, the PmB heteroresistant subpopulation of B. cenocepacia can protect the more sensitive B. cenocepacia population as well as sensitive P. aeruginosa and Escherichia coli from killing by PmB and various bactericidal antibiotics (El-Halfawy & Valvano, 2013). However, heteroresistance to AMPs has not been previously reported for Neisseria. Further, we found that meningococcal heteroresistance was not transiently induced upon exposure to AMPs (Napier et al., 2014), but was instead stable in the absence of PmB, and we have identified the genetic alteration(s) aiding PmB resistance.
Mutations in the pilin biogenesis apparatus appear to restrict the entry of PmB and other antimicrobial agents, and responsible for heteroresistance in Nm. PilQ secretin, in addition to promoting pilin biogenesis and DNA transformation, facilitates the entry of small molecules into the bacterial cell. High-level heteroresistance to PmB can produce cross-resistance to several other important antimicrobial agents [Table 2 and (Napier et al., 2014)]. The increase in MICs of PmB and to a variety of antibiotics in the heteroresistant mutants compared to the wild type strain are consistent with those observed in gonococcal pilQ mutants (Chen et al., 2004, Zhao et al., 2005, Johnson et al., 2014), highlighting the importance of a functional PilQ in the entry of antibiotics.
PilQ facilitates the entry of a variety of antibiotics in the gonococci, including penicillin, ceftriaxone, vancomycin, tetracycline, rifampin, and ciprofloxacin (Ropp et al., 2002, Zhao et al., 2005, Nandi et al., 2015). An in vitro spontaneous mutation screen of two gonococcal isolates containing mosaic penA sequence with MICs to ceftriaxone ranging from 0.03 to 0.06 μg/ml identified mutants with increased MICs to ceftriaxone almost 10-fold (0.5 μg/ml). Genetic analysis showed an identical 2-bp insertion in pilQ in each of the mutants (Johnson et al., 2014). Further, spontaneous penicillin resistant gonococcal clones were selected at a frequency of ~10−6 and all had a non-piliated morphology (Nandi et al., 2015). The mutations were found to be clustered within the C-terminal domain (residue 400 to 731) of PilQ and all pilQ mutants increased the MIC of penicillin by 2.5- to 3-fold (Nandi et al., 2015).
Of the five gene mutations known to contribute to high-level penicillin resistance in Ng (Ropp et al., 2002), penB, mtrR and pilQ2, have been shown to also play a role in PmB susceptibility (Tzeng et al., 2005). The pilQ2 allele is an E666K point mutation in pilQ (Zhao et al., 2005). Interestingly, the increased resistance due to acquisition of the pilQ2 mutation is observed only in strains containing the mtrR and penB resistance determinants (Zhao et al., 2005). The diffusion of antibiotics through PilQ become significant only when influx through porins is limited due to disruptions in the porin gene (penB) or up-regulation of Mtr pump efflux (mtrR), (Zhao et al., 2005). Similarly, diffusion of antibiotics through PilQ is likely only a small fraction of the antibiotic influx in N. meningitidis, as reflected by the modest changes in MIC values in the heteroresistant mutants. We observed that the pilQ mutations caused enhanced PmB resistance only in the wild type background. When resistance levels were reduced ~4–8 fold by mutations in the Mtr efflux pump, we did not observe any clear difference in PmB resistance with the combined pilQ/mtr mutation.
Defects in the mismatch repair (MMR) pathway responsible for removing insertion/deletion loops (Lahue et al., 1989) have generally been associated with meningococcal mutator phenotype (Richardson & Stojiljkovic, 2001, Richardson et al., 2002). Since PmB resistant mutants due to frameshift and point mutations in pilQ and pilM were recovered readily from the heteroresistant clade isolates, a possible mutator phenotype and a defect in MMR pathway was explored. Nevertheless, the clade isolates did not show enhanced mutation rate using the standard spontaneous rifampin resistance plating assay and have relatively low slipped strand mispairing rates. We examined the contribution of the MMR pathway by introducing a mutL mutation, and confirmed that no effect on heteroresistance was detected in the mutL mutant. However, the standard rifampin plating assay that measures the spontaneous mutation rate in the essential rpoB gene, encoding a subunit of RNA polymerase, is likely constrained to only detect the rate yielding viable mutants. Thus, it is possible that the mutation rates obtained with rifampin resistance do not reflect the rates of those mutations needed for antimicrobial peptide resistance. Antimicrobial peptides such as colistin and LL-37 have been reported to enhance mutation rates in P. aeruginosa (Limoli et al., 2014, Rodriguez-Rojas et al., 2015). However, we did not detect significant changes in rifampin mutation rates when meningococci were grown on iron-rich GC agar plates in the presence of colistin, a condition shown to influence mutagenic phenotype in P. aeruginosa (Rodriguez-Rojas et al., 2015).
Other repair pathways correcting DNA lesions include the base excision repair (BER) (MutY, Fpg/MutM, Nth), the nucleotide excision repair (NER) (UvrA/B/C), the recombinational repair (RecA/B/C/D), and translesion synthesis (DinB) (Davidsen et al., 2007b). Several of these DNA repair proteins and others have been characterized to influence mutation rates (Alexander et al., 2004a, Martin et al., 2004, Davidsen et al., 2007b, Davidsen et al., 2007a), thus additional studies are needed to have a detailed understanding about the contribution of other DNA repair pathways in the development of heteroresistance.
The 37R1 heteroresistant mutant had a pilM disruption by a newly inserted IS1655. IS1655 is a 1080-bp long element that would generate a 3-bp target duplication upon insertion (Kiss et al., 2007), which was indeed observed in 37R1. A whole genome comparison study of disease and carriage strains has suggested that Nm can be separated from Neisseria lactamica and Ng based on the respective IS repertoires and that IS1655 is restricted to Nm (Schoen et al., 2008). The authors noted that none of the six Nm strains analyzed have IS1655 at the same chromosomal location, suggesting a high mobility of IS1655. Inspecting the complete genome of the US_NmUC isolate CNM10, we found 11 intact copies and one truncated copy of IS1655. As a comparison, the serogroup B strain MC58 has 14 copies; while the cc11 reference FAM18 has 9 intact and 1 truncated copies. Since the genome of 37R1 had multiple contig breaks, it is uncertain whether 37R1 has the same copy number of IS1655 as CNM10.
Because the pilin biogenesis mutations disrupt normal piliation, the increases in AMP resistance because of such mutations might not be biologically significant, considering the importance of pili in colonization and infections. However, mutations in pilQ have been shown to result in complex phenotypes (Helm et al., 2007), it is possible that certain pilQ mutations allow for increased AMP resistance and retain pathogenic potential. Interestingly, experimental infections of male volunteers using a nonpiliated gonococcal pilE mutant showed that the pilus was not required for infection, although the symptoms were less severe in infections with the nonpiliated variant (Hobbs et al., 2011).
Ng has been proposed to have originated from Nm, a pharyngeal colonizer that switched to primarily colonizing the urogenital tracts, resulting in lower frequency of gene flow between Nm and Ng due to ecological separation within the human host (Vazquez et al., 1993). The ability of US_NmUC isolates to withstand killing by AMPs produced either by epithelial cells or by PMNs is likely an important selective advantage and requirement for dissemination in urethral exudates. Further, the ability of the clade isolates to effectively colonize the urogenital tract raises the concern of horizontal gene transfer of antimicrobial resistance determinants, considering the wide-spread antimicrobial resistance in Ng (Unemo & Shafer, 2014). US_NmUC isolates are intermediate in sensitivity to penicillin, sensitive to azithromycin and ceftriaxone and have uniformly responded to gonococcal treatment regimens. However, the heteroresistance phenotype further demonstrates the propensity for enhanced antibiotic resistance by spontaneous mutations of the pilin biogenesis genes and/or IS movement in this clade. Based on emergence of antibiotic resistance in the gonococcus, we will continue to need to monitor the antimicrobial resistance of this novel meningococcal urethritis clade.
Experimental Procedures
Bacterial isolates and growth conditions
Bacterial strains used in this study are listed in Table 4. These stains included 52 N. meningitidis urethritis clade isolates collected from men between January 2015 and September 2015 at Columbus Public Health (CPH), Columbus, Ohio (Tzeng et al., 2017) with the CNM3 isolate being the major representative of this collection. The initial demographic features of these cases and the collection protocol have been previously reported (Bazan et al., 2016, Bazan et al., 2017). Two additional clade isolates from Atlanta, Georgia, ATL#1 and ATL#2, a serogroup A isolate, IR2855, a serogroup W clinical isolate, GA18736, as well as genetically defined derivatives of well-characterized N. meningitidis strains IR2781 (NMB) (Stephens et al., 1991) and FAM18 were also used. One gonococcal isolate (CNG20) recovered from the same period and gonococcal reference strains, FA19, FA1090, MS11 and H041 were also used for comparisons. Neisseria were cultured with 5% CO2 at 37°C on GC base (GCB; Difco) agar containing supplements of 0.4% glucose and 0.68 mM Fe(NO3)3, or GC broth with the same supplements and 0.043% NaHCO3. Brain heart infusion (BHI) medium with 1.25% fetal bovine serum was used when kanamycin selection was required. Escherichia coli strains were routinely grown in Luria Bertani broth for cloning and propagation of plasmids. N. meningitidis was transformed by the procedure of Janik et al. (Janik et al., 1976). E. coli strains were transformed by chemical competence or by electroporation with a GenePulser (Bio-Rad) according to the manufacturer’s protocol. When necessary, Neisseria (E. coli) were grown in the presence of antibiotic concentrations (μg/ml): kanamycin (Kn) 80 (50), chloramphenicol, 5 (34), tetracycline, 5 and spectinomycin (Sp), 60 (100).
Table 4.
Bacterial strains used in this study.
| Strains | Description | Source |
|---|---|---|
| IR2781 | N. meningitidis serogroup B strain NMB | (Richardson & Stojiljkovic, 2001) |
| FM7 | Non-encapsulated N. meningitidis serogroup C strain FAM18 | This study |
| M7 | Non-encapsulated serogroup B strain IR2781 | (Swartley & Stephens, 1994) |
| GA18736 | N. meningitidis serogroup W 2002 clinical isolate | Laboratory collection |
| IR2855 | N. meningitidis serogroup A clinical isolate | (Richardson & Stojiljkovic, 2001) |
| IR5426 | hpuB::UROS derivative of IR2855 | (Alexander et al., 2004b) |
| CNM3 | N. meningitidis US_NmUC isolate | (Tzeng et al., 2017) |
| CNM8 | N. meningitidis US_NmUC isolate | (Tzeng et al., 2017) |
| CNM3uros | hpuB::UROS derivative of CNM3 | This study |
| 3R4 | N. meningitidis PmB heteroresistant derivative of CNM3 | This study |
| 14R2 | N. meningitidis PmB heteroresistant derivative of CNM14 | This study |
| 17R1 | N. meningitidis PmB heteroresistant derivative of CNM17 | This study |
| 32R1 | N. meningitidis PmB heteroresistant derivative of CNM32 | This study |
| 32R2 | N. meningitidis PmB heteroresistant derivative of CNM32 | This study |
| 33R1 | N. meningitidis PmB heteroresistant derivative of CNM33 | This study |
| 37R1 | N. meningitidis PmB heteroresistant derivative of CNM37 | This study |
| 45R2 | N. meningitidis PmB heteroresistant derivative of CNM45 | This study |
| CNM3–14R2 | N. meningitidis pilQ mutation of 14R2 incorporated into CNM3 | This study |
| CNM3–17R1 | N. meningitidis pilQ mutation of 17R1 incorporated into CNM3 | This study |
| CNM3L | N. meningitidis CNM3/mutL::aphA3 | This study |
| CNM3E | N. meningitidis CNM3 with mtrE:: Ω(Kn) mutation | This study |
| CNM3D | N. meningitidis CNM3 with mtrD::Ω(Sp) mutation | This study |
| CNM3A | N. meningitidis CNM3 with lptA:: Ω(Sp) mutation | This study |
| CNM3EM | N. meningitidis CNM3E with pilM:: aphA3 mutation | This study |
| CNM3DQ | N. meningitidis with the pilQ mutation of 14R2 incorporated into CNM3D | This study |
| CNM3AQ | N. meningitidis with the pilQ mutation of 14R2 incorporated into CNM3A | This study |
| FA1090 | N. gonorrhoeae reference strain | Laboratory collection |
| FA19 | N. gonorrhoeae reference strain | Laboratory collection |
| CNG20 | N. gonorrhoeae urethritis isolate | (Tzeng et al., 2017) |
| H041 | MDR N. gonorrhoeae clinical isolate | (Ohnishi et al., 2011) |
Susceptibility assays
The minimum inhibitory concentrations of PmB and antibiotics were determined by E test. Cell suspensions from overnight GC plates adjusted to OD550 of 0.3 were swabbed onto GC agar plates. Discs soaked with 10 μl of PmB solutions (25.6 mg/ml) or discs with defined levels of antibiotics (BBL) were overlaid and the plates were incubated overnight at 37°C in 5% CO2. E test strips (bioMerieux) were performed in a similar fashion. MIC values of PmB were reported as μg/ml. Microbroth dilution assays using 96-well microtiter plates were performed using GC broth with standard supplements. Two-fold serial dilutions of PmB concentrations starting at 1,024 μg/mL were tested. The sensitivity to LL-37 was determined using 96-well microtiter plate.
Population analysis profiling (PAP) assays
Overnight plate cultures of meningococcal strains were resuspended in GC broth and adjusted to optical density of 0.3 at 550 nm. Ten-fold serial diluted bacterial suspensions were prepared and duplicate aliquots of 40 μl of suspensions were spotted onto GC agar plates with 2-fold incremented concentrations of polymyxin B. Bacterial growth at each of these concentrations is quantified by CFU count after overnight incubation. An isolate would be considered heteroresistant when the lowest antibiotic concentration giving maximum growth inhibition is >8-fold higher than the highest non-inhibitory concentration (El-Halfawy & Valvano, 2015).
Western blot
Expression of PilQ in whole-cell extracts was examined by Western blot. Briefly, strains grown on GC plates overnight at 37°C were collected by centrifugation. Whole cell lysates of equal cell densities were prepared in SDS loading buffer, resolved by 8% SDS-PAGE, and transferred to nitrocellulose membranes by semi-dry transfer. The antisera against PilQ (Tonjum et al., 1998) were used at 1:5000 dilutions and anti-rabbit IgG-HRP conjugate secondary antibody (Bio-Rad) was used at 1:10,000 dilution. The blot was developed using a 1:5 dilution of pico chemiluminescent substrate (Pierce).
LL-37 killing assay
Overnight cultures were harvested into RPMI and adjusted to OD550 of 0.3. The standardized suspensions were diluted 100-fold and then 50-fold to have approximately 105 CFU/ml. Each assay was started by the addition of 90 μl of cells into a well containing 10 μl of LL-37 to reach the desired final concentrations, 1.25 to 10 μg/ml. The microtiter plate was incubated at 37°C and 5% CO2. Two 20-μl aliquots of the sample were removed after 30 min and the number of viable CFU were determined by plating onto GC agar plates. Experiments were performed in duplicate wells on several occasions. Student’s t test was used to determine the statistical significance of survival of the mutant with respect to that of the wild type strain, with P values of < 0.05 considered significant.
Whole genome sequencing (WGS) and variant analysis
WGS of all 52 CNM isolates has been performed by Illumina at CDC (Tzeng et al., 2017). The single contig genome of isolate CNM10 sequenced by Pacific Biosciences (PacBio) technology (Tzeng et al., 2017) was used as the reference genome in variant analysis. Polymyxin B resistant derivatives within the zone of inhibitions were recovered from seven CNM urethritis isolates (Table 1) were sequenced by MiSeq, yielding ~200X coverage of paired end 250-bp reads, assembled using SPAdes (Bankevich et al., 2012) and annotated by RAST(Aziz et al., 2008). The variant analysis service provided at www.patricbrc.org was utilized with the BWA-men (Li & Durbin, 2009) and SAMtools (Li et al., 2009) programs as the aligner and the SNP caller, respectively. The Illumina raw reads of the mutants were analyzed against a complete CNM10 genome as the reference. In addition, the assembled contigs were compared to the reference CNM10 genome using the genome comparator tool available at the PUBMLST site (www.pubmlst.org) to identify allele differences. Variants were further confirmed by targeted PCR amplification and sequencing.
Construction of lptA, mtrD/E, pilM, pilQ, and mutL mutants
The CNM3 isolate was transformed with pKA314 (Tzeng et al., 2004) to generate the lptA::Ω(Sp) mutant, CNM3A. The mtrE:: Ω(Kn) mutation was PCR amplified from the M7mtrE mutant with primers mrtDF2-ER and mtrE3R1-ER and the mtrD:: Ω(Sp) mutation from strain XZ134 (Tzeng et al., 2005) using primers mtrCF1 and mtrE3R1-ER. The purified PCR products were then used to transform CNM3. The lptA/mtrE double mutant, CNM3EA, was subsequently generated by transforming the mtrE mutant with pKA314. A PCR product with the mutL::Ω(Sp) mutation was obtained with primers mutL1a and mutL1b (Richardson & Stojiljkovic, 2001) and used to transform the CNM3 isolate.
The pilM::aphA3 mutation was created by the overlapping PCR method. Primer pairs of YT113 and pilM-5Ra and primer pairs of pilM-3Fa and pilMR2 were used to generate 5,627-bp and 3,460-bp fragments, respectively. The aphA3(Kn) cassette was amplified using primers aphA3-SmF and aphA3-SmR. First overlapping PCR was performed with the aphA3 cassette and the 3’ fragment and then the resulting product was used for the second overlapping with the 5’ fragment. The resulting construct deleted 756-bp pilM sequence.
To introduce the pilQ point mutations found in the PmB heteroresistant mutants 14R2 and 17R1, a construct with aphA3 cassette inserted 25 bp downstream of the pilQ stop codon was created by overlapping PCR. Primers pilQ-F1 and pilQ-5RA3 were used to amplify an 1144-bp 5’ fragment from chromosomal DNAs of isolate 14R2 and 17R1 that carries the respective point mutations. The first overlapping PCR combined the 5’ fragment with the aphA3 cassette and the resulting product was subsequently used for overlapping PCR with a 3’ 849-bp fragment of primers pilQ-3FA3 and pilQ-3R2. The final ~2.8 kb product was used to transform either the CNM14 or CNM17 parental isolates as well as the CNM3 isolate and kanamycin resistant colonies were saved. PCR products were generated from the colonies and sequenced to determine whether the point mutations were recombined into the transformants. Transformants with the desired point mutations or with a wild type sequence were saved for comparison. The CNM3DQ and CNM3AQ mutants were created by transformation of strains CNM3D and CNM3A with the overlapping PCR products carrying the 14R2 pilQ mutation.
Determination of spontaneous mutation and slipped strand mispairing frequencies
Overnight GC plate cultures were resuspended in GC broth and standardized by the OD550 readings. For spontaneous mutation rates, 40 μl of serial dilutions of cell suspensions were spotted onto GC plates for total CFU counts. Approximately 1010 cells were plated onto GC plates containing 3 μg rifampin/ml (Richardson & Stojiljkovic, 2001). Spontaneous rifampin mutation rates were obtained as the ratio of rifampin-resistant cells to the total number of cells. Serial dilutions of UROS-containing strains were plated on non-selective GC plates for total counts and on selective (60 μg/ml spectinomycin) plates for switch-on CFU counts. Frequencies of phase variation were determined as described previously (Alexander et al., 2004b) and are represented as medians of three independent measurements.
Transformation efficiency
Plate-grown meningococcal strains were suspended in GC broth supplemented with 5 mM MgCl2. One μg of chromosomal DNA (tonB::ΩSp) from Nm strain NMB was added to aliquots (100 μl) of cell suspension at an OD550 of 1 and then incubated for 1 hr at 37oC. Pre-warmed GC broth with complete supplements (500 μl) and DNase I (2 units) was added and the incubation continued for another 30 minutes. Serial dilutions were made and aliquots of 50 μl were spotted onto non-selective GC plates and the colony forming units (cfu) determined after overnight growth. 500-μl of the transformation mixtures and 100-μl aliquots of 10−1 and 10−2 dilutions of the mutants and the parental strains, respectively, were plated onto selective (Sp) plates. The efficiencies were calculated as the ratio of cfu/ml from the selection plate to the cfu/ml of non-selective plates.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grant R01AI107116 (YT), R01AI127863 (ANT, JAB, DSS and YT), R21AI128313 (DSS and YT) and R01AI116706 (DEN). We thank Dr. William Shafer for providing gonococcal MS11 and H041 isolates and helpful discussion. This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/). We are grateful to Dr. Ashild Vik for the PilQ antisera. All authors have no conflict of interest to declare.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- Alexander HL, Rasmussen AW & Stojiljkovic I, (2004a) Identification of Neisseria meningitidis genetic loci involved in the modulation of phase variation frequencies. Infect. Immun 72: 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander HL, Richardson AR & Stojiljkovic I, (2004b) Natural transformation and phase variation modulation in Neisseria meningitidis. Mol. Microbiol 52: 771–783. [DOI] [PubMed] [Google Scholar]
- Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL & Burrows LL, (2009) PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol 394: 128–142. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A & Zagnitko O, (2008) The RAST Server: rapid annotations using subsystems technology. BMC genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, Burd EM & Weiss DS, (2016) Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1: 16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band VI, Satola SW, Burd EM, Farley MM, Jacob JT & Weiss DS, (2018) Carbapenem-Resistant Klebsiella pneumoniae Exhibiting Clinically Undetected Colistin Heteroresistance Leads to Treatment Failure in a Murine Model of Infection. mBio 9: 02448–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA & Pevzner PA, (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JA, Peterson AS, Kirkcaldy RD, Briere EC, Maierhofer C, Turner AN, Licon DB, Parker N, Dennison A, Ervin M, Johnson L, Weberman B, Hackert P, Wang X, Kretz CB, Abrams AJ, Trees DL, Del Rio C, Stephens DS, Tzeng YL, DiOrio M & Roberts MW, (2016) Notes from the Field: Increase in Neisseria meningitidis-Associated Urethritis Among Men at Two Sentinel Clinics - Columbus, Ohio, and Oakland County, Michigan, 2015. MMWR. Morbidity and mortality weekly report 65: 550–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JA, Turner AN, Kirkcaldy RD, Retchless AC, Kretz CB, Briere E, Tzeng YL, Stephens DS, Maierhofer C, Del Rio C, Abrams AJ, Trees DL, Ervin M, Licon DB, Fields KS, Roberts MW, Dennison A & Wang X, (2017) Large Cluster of Neisseria meningitidis Urethritis in Columbus, Ohio, 2015. Clin Infect Dis 65: 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JL, Phelan MM, Collins RF, Adomavicius T, Tonjum T, Frye SA, Bird L, Owens R, Ford RC, Lian LY & Derrick JP, (2012) Structure and assembly of a trans-periplasmic channel for type IV pili in Neisseria meningitidis. PLoS Pathog 8: e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Prouvensier L, Nassif X & Pelicic V, (2005) Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol 55: 54–64. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Tobiason DM, Thomas CE, Shafer WM, Seifert HS & Sparling PF, (2004) A mutant form of the Neisseria gonorrhoeae pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J. Bacteriol 186: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Glez CJ & Calderon E, (1991) Urogenital infection due to meningococcus in men and women. Sexually transmitted diseases 18: 72–75. [DOI] [PubMed] [Google Scholar]
- Criss AK & Seifert HS, (2012) A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat Rev Microbiol 10: 178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, Amundsen EK, Rodland EA & Tonjum T, (2007a) DNA repair profiles of disease-associated isolates of Neisseria meningitidis. FEMS Immunol. Med. Microbiol 49: 243–251. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tuven HK, Bjoras M, Rodland EA & Tonjum T, (2007b) Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol 189: 5728–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Halfawy OM & Valvano MA, (2013) Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One 8: e68874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Halfawy OM & Valvano MA, (2015) Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28: 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou M, Castagnini M, Karimova G, Ladant D & Pelicic V, (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol 84: 857–873. [DOI] [PubMed] [Google Scholar]
- Givan KF, Thomas BW & Johnston AG, (1977) Isolation of Neisseria meningitidis from the urethra, cervix, and anal canal: further observations. The British journal of venereal diseases 53: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Cole K, Peters J, Cresswell F, Dean G, Eyre DW, Paul J & Maiden MC, (2017) Genomic analysis of urogenital and rectal Neisseria meningitidis isolates reveals encapsulated hyperinvasive meningococci and coincident multidrug-resistant gonococci. Sex Transm Infect 93: 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm RA, Barnhart MM & Seifert HS, (2007) pilQ Missense mutations have diverse effects on PilQ multimer formation, piliation, and pilus function in Neisseria gonorrhoeae. J Bacteriol 189: 3198–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes DM, Pormann Pitt C, Lutz L, Teixeira AB, Ribeiro VB, Netto B, Martins AF, Zavascki AP & Barth AL, (2013) Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and -r esistant Pseudomonas aeruginosa. Journal of medical microbiology 62: 1184–1189. [DOI] [PubMed] [Google Scholar]
- Hjort K, Nicoloff H & Andersson DI, (2016) Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol 102: 274–289. [DOI] [PubMed] [Google Scholar]
- Hobbs MM, Sparling PF, Cohen MS, Shafer WM, Deal CD & Jerse AE, (2011) Experimental Gonococcal Infection in Male Volunteers: Cumulative Experience with Neisseria gonorrhoeae Strains FA1090 and MS11mkC. Frontiers in microbiology 2: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik A, Juni E & Heym GA, (1976) Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol 4: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayol A, Nordmann P, Brink A & Poirel L, (2015) Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59: 2780–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB & Criss AK, (2011) Resistance of Neisseria gonorrhoeae to neutrophils. Frontiers in microbiology 2: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SR, Grad Y, Ganakammal SR, Burroughs M, Frace M, Lipsitch M, Weil R & Trees D, (2014) In Vitro selection of Neisseria gonorrhoeae mutants with elevated MIC values and increased resistance to cephalosporins. Antimicrob Agents Chemother 58: 6986–6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A, Georg M, Maudsdotter L & Jonsson AB, (2009) Endotoxin, capsule, and bacterial attachment contribute to Neisseria meningitidis resistance to the human antimicrobial peptide LL-37. J. Bacteriol 191: 3861–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J, Nagy Z, Toth G, Kiss GB, Jakab J, Chandler M & Olasz F, (2007) Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol Microbiol 63: 1731–1747. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Au KG & Modrich P, (1989) DNA mismatch correction in a defined system. Science 245: 160–164. [DOI] [PubMed] [Google Scholar]
- Li H & Durbin R, (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R & S. Genome Project Data Processing, (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE & Liolios L, (2006) Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50: 2946–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T & Wozniak DJ, (2014) Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog 10: e1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA & van Keulen PH, (2007) Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother 51: 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KC, Unemo M, Jeverica S, Kirkcaldy RD, Takahashi H, Ohnishi M & Grad YH, (2017) Genomic Characterization of Urethritis-Associated Neisseria meningitidis Shows that a Wide Range of N. meningitidis Strains Can Cause Urethritis. J Clin Microbiol 55: 3374–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maini M, French P, Prince M & Bingham JS, (1992) Urethritis due to Neisseria meningitidis in a London genitourinary medicine clinic population. International journal of STD & AIDS 3: 423–425. [DOI] [PubMed] [Google Scholar]
- Martin P, Sun L, Hood DW & Moxon ER, (2004) Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 150: 3001–3012. [DOI] [PubMed] [Google Scholar]
- Nandi S, Swanson S, Tomberg J & Nicholas RA, (2015) Diffusion of antibiotics through the PilQ secretin in Neisseria gonorrhoeae occurs through the immature, sodium dodecyl sulfate-labile form. J Bacteriol 197: 1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier BA, Band V, Burd EM & Weiss DS, (2014) Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58: 5594–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, Jerse AE & Shafer WM, (2011) A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2: e00187–00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama S, Kitawaki J & Unemo M, (2011) Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother 55: 3538–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless AC, Kretz CB, Chang HY, Bazan JA, Abrams AJ, Norris Turner A, Jenkins LT, Trees DL, Tzeng YL, Stephens DS, MacNeil JR & Wang X, (2018) Expansion of a urethritis-associated Neisseria meningitidis clade in the United States with concurrent acquisition of N. gonorrhoeae alleles. BMC genomics 19: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR & Stojiljkovic I, (2001) Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol 40: 645–655. [DOI] [PubMed] [Google Scholar]
- Richardson AR, Yu Z, Popovic T & Stojiljkovic I, (2002) Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc. Natl. Acad. Sci. U S A 99: 6103–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rojas A, Makarova O, Muller U & Rolff J, (2015) Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria. PLoS Genet 11: e1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp PA, Hu M, Olesky M & Nicholas RA, (2002) Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouphael NG & Stephens DS, (2012) Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Biol 799: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R & Moxon ER, (2000) Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol 37: 207–215. [DOI] [PubMed] [Google Scholar]
- Schoen C, Blom J, Claus H, Schramm-Gluck A, Brandt P, Muller T, Goesmann A, Joseph B, Konietzny S, Kurzai O, Schmitt C, Friedrich T, Linke B, Vogel U & Frosch M, (2008) Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A 105: 3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer WM, Qu X, Waring AJ & Lehrer RI, (1998) Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U S A 95: 1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LA, Butcher SA & Saunders NJ, (2001) Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147: 2321–2332. [DOI] [PubMed] [Google Scholar]
- Spinosa MR, Progida C, Tala A, Cogli L, Alifano P & Bucci C, (2007) The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun 75: 3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, Greenwood B & Brandtzaeg P, (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369: 2196–2210. [DOI] [PubMed] [Google Scholar]
- Stephens DS, Swartley JS, Kathariou S & Morse SA, (1991) Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun 59: 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartley JS & Stephens DS, (1994) Identification of a genetic locus involved in the biosynthesis of N-acetyl-D-mannosamine, a precursor of the (α 2-->8)-linked polysialic acid capsule of serogroup B Neisseria meningitidis. J. Bacteriol 176: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh E, Gangaiah D, Batteiger BE, Williams JA, Arno JN, Tai A, Batteiger TA & Nelson DE, (2017) Neisseria meningitidis ST11 Complex Isolates Associated with Nongonococcal Urethritis, Indiana, USA, 2015–2016. Emerg Infect Dis 23: 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Caugant DA, Dunham SA & Koomey M, (1998) Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol. Microbiol 29: 111–124. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM & Stephens DS, (2005) Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol 187: 5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Bazan JA, Turner AN, Wang X, Retchless AC, Read TD, Toh E, Nelson DE, Del Rio C & Stephens DS, (2017) Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc Natl Acad Sci U S A 114: 4237–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Datta A, Ambrose KD, Davies JK, Carlson RW, Stephens DS & Kahler CM, (2004) The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J. Biol. Chem 279: 35053–35062. [DOI] [PubMed] [Google Scholar]
- Tzeng YL & Stephens DS, (2015) Antimicrobial peptide resistance in Neisseria meningitidis. Biochim Biophys Acta 1848: 3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M & Shafer WM, (2014) Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27: 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JA, de la Fuente L, Berron S, O’Rourke M, Smith NH, Zhou J & Spratt BG, (1993) Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol 3: 567–572. [DOI] [PubMed] [Google Scholar]
- Zhao S, Tobiason DM, Hu M, Seifert HS & Nicholas RA, (2005) The penC mutation conferring antibiotic resistance in Neisseria gonorrhoeae arises from a mutation in the PilQ secretin that interferes with multimer stability. Mol Microbiol 57: 1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.