Abstract
We describe a method for visualizing mRNAs in living mouse. Nascent transcripts and cytoplasmic mRNAs were labeled via lentiviral expression of MS2 coat protein (MCP) tagged with fluorescent protein (MCP-XFP) in knock-in mice whose β-actin mRNAs contained MCP binding stem loops (MBS). Then the mRNA molecules were imaged in the live cerebral cortex through an optical cranial window by intravital two-photon microscopy. By means of the controlled expression of MCP-XFP, single mRNA particles could be detected differentially in the nucleus and cytoplasm of a specific cell type. Consequently, this method is useful for investigating the cell-type-dependent dynamics of mRNAs underlying the structure and function of the brain.
Keywords: Transcriptional dynamics, mRNA transport, viral transfer, intravital microscopy
1. Introduction
Synthesis of mRNAs in and their subsequent transport out of the nucleus vary considerably across cell types as well as upon different environmental cues. The heterogeneity is thought to play significant roles in the development and function of the brain, which consists of diverse cell types within a highly ordered architecture [1–3]. However, the regulation of gene expression underlying the organization and plasticity in the central nervous system (CNS) is largely unknown. For elucidating transcriptional dynamics and intracellular localization of mRNAs in the course of physiology and pathology, nascent transcripts and mRNA-protein complexes (mRNPs) must be observed in live animals, which has not been demonstrated to date.
A technique of labeling mRNAs in vivo, namely MBS-MCP, has been widely employed for livecell imaging [4]. By virtue of 24 repeats of bacteriophage MS2 binding stem loops (MBS) inserted into the 3’ untranslated region (UTR) of a gene of interest, which can bind up to 48 MS2 coat protein (MCP) tagged with fluorescent protein (MCP-XFP), sufficient fluorescence is provided for visualizing single endogenous mRNA molecules in real time. In particular, since the MBS-MCP method labels nascent transcripts while preserving the intrinsic cis-regulatory elements, it enables measuring the native transcriptional dynamics of the gene of interest. The MBS-MCP strategy has been implemented in various model organisms, such as Drosophila embryo [5,6] and zebrafish [7]. Also, murine models have been created for studying mammalian mRNA dynamics, e.g., a knock-in (Kl) mouse containing MBS in the β-actin mRNA (‘Actb-MBS’) [8] and a transgenic mouse expressing MCP-GFP under the control of ubiquitin C promoter (UBC) [9]. In the hybrid offspring (‘MBS×MCP’), every β-actin mRNAs are fluorescently labeled providing a useful resource for monitoring mRNAs in the native tissue environment. Using acute hippocampal slices from the animal, in vivo dynamics of mRNAs has been investigated. However, labeling mRNAs in all cells or tissues is undesirable for discovering cell-type- and tissue-dependent dynamics of a multi-functional (e.g., pleiotropic) gene. As a binary system, MBS-MCP affords combinatorial labeling, which can be exploited for targeting specific cells and tissue types so as to elucidate the heterogeneity of mRNA dynamics in the brain.
Here we demonstrate an advanced intravital MBS-MCP method by which mRNAs can be visualized in specific cell types of the living mouse brain. A construct encoding MCP-XFP reporter was delivered via lentiviral (LV) vectors into the brain of the Actb-MBS mouse, and then the labeled β-actin mRNAs in the cortex were imaged through an optical cranial window by intravital two-photon microscopy (TPM).
2. Material and methods
Cloning of LV transgene:
The insert (e.g., Syn-NLS-MCP-mKate2 and Syn-MCP-mKate2) was cloned and amplified by PCR using Pfx DNA polymerase (Thermo Fisher Scientific) and purified by gel electrophoresis. The LV vector (2 μg, Addgene #14883) was digested and gel purified. The insert and the vector were ligated by incubating a mixture containing 100-ng DNA (the vector to insert ratio of ~1:3) and T4 DNA ligase (NEB) at 16°C overnight. Bacterial cells were transformed and incubated overnight. 10 colonies were selected and grown in LB media overnight. The result of ligation was verified by digestion and gel electrophoresis, and also by sequencing.
Production of LV:
High titer LVs were produced in a BSL-2 class facility, as described previously [10]. Approximately ~300 μg of transfer vector was necessary for high-titer LVs, which was obtained by growing bacteria in 500-mL LB media overnight and maxi-prep. 293T cells were seeded in twelve 15-cm dish (~5×106 cells per dish) approximately 8 hours prior to transfection. Medium (IMDM + 10% FBS + 0.25% PS + 1% glutamine, 20 mL final volume per dish) was changed 2 hours prior to transfection. The plasmid mixture was prepared for transfection containing (per dish): 25 pg of transfer vector, 12.5 μg of pMDLg/pRRE and 6.25 μg of pREV, and 8 pg of pVSV-G. Then 50 μL of 2.5M CaCI2, 1.2 mL of double distilled H20, and 1.25 mL of 2xHBSS were added, mixed by inversion, and incubated at room temperature for 10 minutes to allow DNA-CaP04 precipitate to form. The precipitate was added dropwise to the 293T cells. After incubation for 12–16 hours, the precipitate was removed and media was changed (15 mL final volume per dish). After incubation overnight, the supernatant was collected for the first harvest (15 mL×12 dishes=180 mL). Fresh 15-mL media was added and incubated overnight. The supernatant was collected for the second harvest. The supernatants were pooled (a total of 360 mL) and cleaned up with 0.45-pm filter. At this point the titer was >106 viral particles/mL.
Concentration of LV:
The range and efficiency of transduction can be poor when the titer of LV is too low; for the reference, injecting 1- μL LV solution containing ~106 viral particles would yield the maximum transduction volume of ~1 mm3 (containing ~106 cells), assuming the multiplicity of infection (MOI, the number of viral particles per cell) of 1. In order to concentrate LVs, twelve ultracentrifuge tubes (6 tubes/ rounds × 2 rounds) were loaded with 30-mL solution each and ultracentrifuged at 20,000 rpm for 2 hours at 18 °C. Supernatants was discarded into 10% bleach and tubes were dried by inverting on paper towel. The pellet was re-suspended in 200 μL of 1 × HBSS. The titer of LV was measured by qRT-PCR with Lenti-X™ titration kit (Takara Bio Co.). The titer increased ~150-fold after concentration, yielding >109 viral particles/mL. LV can be aliquoted and stored at −80 °C for a long term (~1 year). The biological titer cannot be measured for a cell-type-specific promoter inactive in the standard cell lines (e.g., Syn), so the actual transducing units may be smaller than the number of viral particles.
Stereotaxic injection:
Animals were handled in accordance with the procedures approved by the Institute Animal Care and Use Committee (IACUC) at Hunter College. LV was injected using a stereotaxic frame, as described previously [11]. An Actb-MBS mouse at a postnatal age of P21-P48 was anesthetized with 1.5% isoflurane at 0.5 liter/min oxygen delivery. The mouse was placed in a stereotaxic frame and the head was firmly secured with ear bars. A small flap of skin was removed with a scalpel from the dorsal skull of the visual cortex. A hole of 300 μm in diameter was drilled above the target area (2.5 mm lateral and 0.6 mm anterior to the Lambdoid suture for the primary visual cortex). The tip of 32-gauge needle was lowered to a depth of 300 pm below the pia mater. Then a 1- μL solution of LV particles was injected slowly over 10 min. After 2 minutes, the needle was withdrawn slowly and the wound was closed by clipping the skin. The animal was recovered and MCP-mKate2 expression matured in 2 weeks after LV injection.
Installing cranial optical window:
An optical cranial window was placed, as described in the literature [12]. A mouse was anaesthetized with 1.5% isoflurane, the head immobilized, and placed on a heating blanket to maintain the body temperature at 37°C. An eye ointment was applied to prevent drying. The hair was shaved from the back of the neck up to the eyes using a rodent trimmer. The mouse was placed in a stereotaxic frame and the head was firmly secured with ear bars. The area of operation was sterilized by wiping twice with betadine and then 70% ethanol. The skin over the head was cut using scissors. The exposed area was scraped with a scalpel for better adhesion. A 2-mm guide circle was drawn with a pencil and drilled around the circle until a thin layer of skull is left. A drop of PBS was put on the area and the thinned skull was removed using the tip of a thin forceps. In case of bleeding, cotton soaked in PBS was applied until it stopped. After the dura was dry, a sterile 3-mm glass coverslip was placed on top of the dura mater and glue was applied around the coverslip. Dental acrylic mix was applied around the edges of the cover slip to cover the entire skull surface. A titanium head bar or razor blade was placed on the acrylic resin and allowed 10 mins to harden.
Intravital imaging by TPM:
Before imaging, a mouse was anaesthetized with 1.5% isoflurane and the head was immobilized. For two-photon excitation of mKate2, a beam of ~100-fs pulses at 1140-nm wavelength from an optical parametric amplifier (Coherent, Chameleon Ultra) was employed. For two-color imaging of GFP and mKate2, the 1140-nm beam was combined with another at 850-nm wavelength from an independent mode-locked Ti:Sapphire laser (Spectra-Physics, Tsunami). The beam was focused with an objective lens (Nikon, CFI75 16 × 0.8NA) and the average power was approximately 10 to 50 mW at the sample. The signal was detected with photomultiplier tubes (Hamamatsu, H7422–40). For image acquisition, the pixel dwell time was set to approximately 1 μs/ μm2. In order to distinguish mKate2 signal from autofluorescence, a separate channel was monitored where the mKate2 emission was blocked. Time-lapse images were acquired to measure the elongation of mRNAs for >200 seconds, which was approximately three times longer for RNA polymerase II to transcribe the entire MBS sequence (~1.3 kbp) at the typical rate of ~1 kbp/min [13].
Single-particle tracking of mRNAs:
Single-particle tracking was performed. For precise quantification of transcriptional dynamics, the magnitude of motion artifacts (i.e., non-biological fluctuations of MCP-XFP intensity) was determined, which arise from axial drift of the brain relative to the laser focus due to breathing and heartbeats. The correlated variations of the characteristics of single particles, e.g., the coordinates of centroid, the brightness, the radius of gyration, and the eccentricity [14,15], were evaluated to determine the motion-induced defocus (Fig. 3c) as well as spherical aberration and astigmatism.
Figure 3.
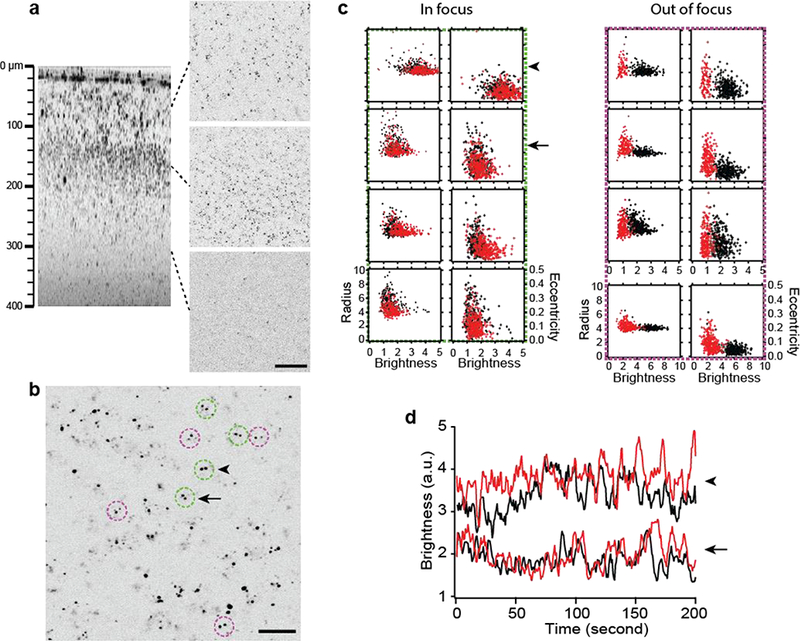
Nascent transcripts in the live brain visualized with LV-Syn-NLS-MCP-mKate2. (a) The cerebral cortex injected with LV-Syn-NLS-MCP-mKate2 at a depth of 300 μm; axial (left) and lateral cross sections at three different depths (right). Scale bar, 50 μm. (b) Lateral section showing cells with biallelic transcription (doublets in circles). Scale bar, 20 μm. (c) The radius of gyration and the eccentricity versus the brightness of the transcription sites, recorded for a period of 200 seconds, for 8 cells in (b). Each scatter plot displays two alleles (red and black) within the same cell. Green and magenta dashed boxes denote 4 cells each with in- and out-offocus transcription sites, respectively. The brightness of a transcription site varied with the axial position relative to the focal plane, but the effect of motion artifacts could be discriminated by correlated increases in the radius and eccentricity. (d) The time-resolved brightness of transcription sites in two neighboring cells (arrow and arrowhead in (b) and (c)) with negligible motion artifacts and differential defocus.
3. Results
Validating LV transduction of MCP-XFP.
The relatively large packaging capacity (~9 Kb) of LV vector allows adding sequences to manipulate the expression of transgene. We tested the human synapsin 1 (Syn) promoter for neuron-specific expression [18]. A cell-type-specific promoter can drive the expression of the MCP-XFP reporter such that β-actin mRNAs are visualized only in a certain population of cells. The MBS-containing mRNAs of the housekeeping gene are otherwise present in all cells of Actb-MBS mouse. It also limits the labeling of mRNAs to a specific cortical area where the promoter is activated [16,17]. In addition, a small sequence can be included encoding an amino acid signal to instruct the translocation of the reporter protein, which is useful for unraveling mRNA dynamics that depends on the intracellular location. We examined the effect of the SV40 nuclear localization signal (NLS) [19], which has been employed to remove free MCP-XFPs and thus lower non-specific background in the cytoplasm [20]. A karyophilic signal can also increase the nuclear background while compromising the efficiency of labeling cytoplasmic mRNAs. Finally, an appropriate reporter to tag MCP can be selected from a palette of fluorescent proteins developed specifically for enhanced tissue imaging, e.g., far-red (RFP) and near-infrared fluorescent proteins (NIRFP). We employed a monomeric RFP, mKate2 [21], for intravital TPM imaging. The designed MCP-XFP transgene was verified by comparing to the expression of Syn-H2B-GFP. Fig. 2 depicts a representative region of the visual cortex, where LV-Syn-H2B-GFP and LV-Syn-NLS-MCP-mKate2 were co-injected as a cocktail at 1:1 stoichiometry at a depth of approximately 300 μm. H2B-GFP and MCP-mKate2 fluorescence were observed across ~1 mm3 and co-localized in the same cortical depth (the layer 2/3) and cell type (Syn+ cells), confirming the titer of LV and the desired expression of MCP-mKate2. Given the MOI of approximately 1, the process of LV transduction is intrinsically stochastic, causing variations in the intermediate steps including the entry to the host cells and the formation of pre-integration complex. However, the variability of MCP expression, which appears not only across the cells within the same animal but also in different animals, does not translate proportionally to the measurement of mRNAs due to the high affinity binding between MBS and MCP (Kd~1 nM).
Figure 2.
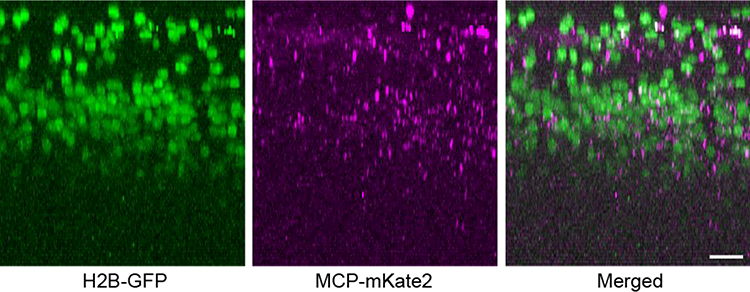
The biological titer of LV and the range of promoter confirmed by co-injection of LV-Syn-H2B-GFP and LV-Syn-NLS-MCP-mKate2. Scale bars, 30 μm.
Observing nuclear mRNAs.
LV delivery of Syn-NLS-MCP-mKate2 preferentially labeled mRNAs in the nucleus, allowing transcriptional dynamics to be probed in situ (Fig. 3). Labeled mRNAs appeared as bright, punctate particles with a size close to the optical resolution. The emission in the far-red wavelength range allowed the background to be negligible. The 3D spatial distribution of mRNAs was measured, illustrating the state of transcriptional activation of the gene under the physiological condition of the brain (Fig. 3a). For β-actin, they were uniform across the layer 2/3 enriched with Syn+ neuronal cells. In addition to isolated single particles, which were presumably single transcription sites or cytoplasmic mRNA granules, many were present as doublets suggesting sites of biallelic transcription (Fig. 3b, dashed circles). The strength of transcriptional activities at the individual sites was reported by the brightness. For precise quantification of transcriptional activity, the degree of defocus at the transcription sites was determined from the variations of the single-particle properties (Fig. 3c). Examining alleles that were in focus with negligible motion artifacts (arrow and arrowhead in Fig. 3b), we found that the dynamics even in the same cell were not necessarily synchronized (Fig. 3d), indicating allele-specific transcription.
Observing cytoplasmic mRNAs.
LV delivery of Syn-MCP-mKate2 (without NLS) preferentially labeled cytoplasmic mRNAs, ideal for measuring the motion of mRNAs dependent on the cellular context. The pattern of labeled mRNAs was remarkably different from that of NLS-containing transgene, with more mRNAs visualized in the cytoplasm and neurites. There was also low, diffuse fluorescence background in these regions. A variety of motion types could be observed, including directed (Fig. 4 and Movie 1) and Brownian-like movements (Movie 2). An example of directed motion is depicted in Fig 4. Mobile and stationary mRNA particles were distinguished by their coefficients of variation (Fig. 4c). Typically only a small fraction (<5%) of mRNA particles were mobile in live brains, fewer than in cultured cells [22]. Directed motions included temporary pause and/or reversal of directions (Fig. 4d). The velocity of mRNAs was approximately 0.02 – 0.11 μm/sec, which was apparently slower than the values previously measured in cultured cells [23,24].
Figure 4.
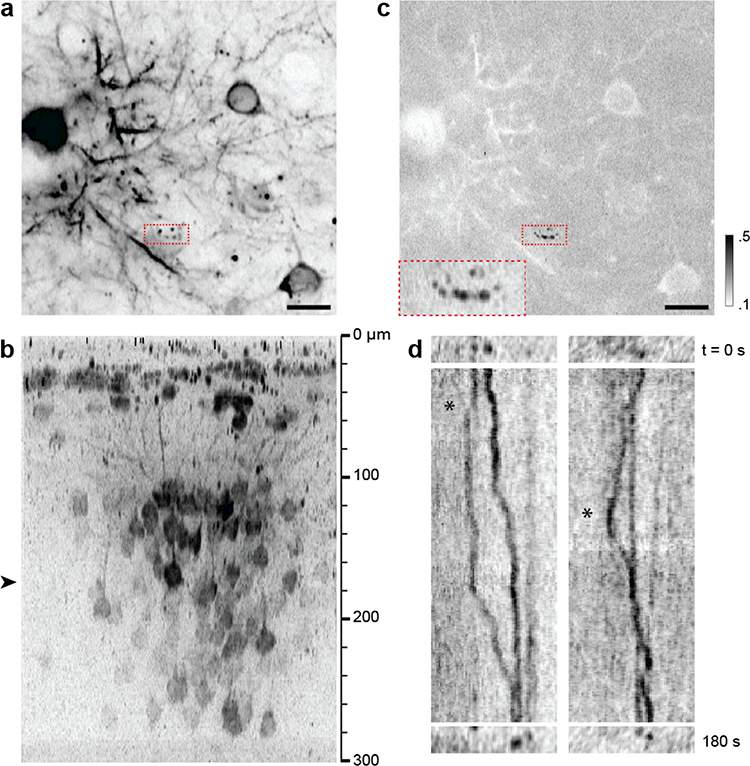
Cytoplasmic mRNAs in the live brain visualized with LV-Syn-MCP-mKate2 (without NLS). (a) Lateral section showing mobile mRNA particles (red dashed box). Scale bar, 15 μm. (b) Axial cross section where the approximate depth of (a) is denoted with an arrowhead. (c) The coefficient of variation of the region distinguishes mobile and stationary mRNA species (black and white, respectively). Inset: A magnified view of the red dashed box revealing hotspots where mRNAs pause. (d) Kymographs of mRNA particles showing reversal of directions (asterisks, see Movie 1).
4. Discussion
We have presented LV-mediated intravital MBS-MCP for imaging neuronal ß-actin mRNAs in the living mouse brain. The generation of additional transgenic animals expressing the MCP is no longer necessary. Furthermore, due to significantly less cost and time LV transduction requires compared to creating transgenic animals, a number of candidate MCP reporters can be tested in rapid turnaround times. The versatility of the new intravital method has been demonstrated for preferential labeling of the molecules either in the nucleus for real-time transcriptional dynamics or in the cytoplasm for distinct intracellular trafficking, which is ideal for discovering any cell-type-specific dynamics of mRNA throughout its lifetime, e.g., release of nascent transcripts, assembly and nuclear export of mRNPs, and intracellular localization. Also, remarkable improvements over the previous MBS-MCP such as the MBS×MCP models [9] include the negligible non-specific background fluorescence in the nucleus. Arising from free MCP-XFPs, it has been considered as a major drawback of MBS-MCP although in principle the high affinity binding between MBS and MCP (Kd ~1 nM) should provide a low background sufficient for detecting single nascent transcripts. It is plausible that LV-mediated intravital MBS-MCP prevents the overexpression of MCP reporter unlike infecting cultured cells or transgenesis of embryos.
The ability to resolve transcriptional dynamics at individual alleles of single cells is key for characterizing the variability on distinct levels, e.g., transient bursting, allelic and cell-to-cell variations [25–30], which may play important roles in brain function. The relevance of heterogeneous mRNA dynamics for health and disease can be better elucidated by interrogating mRNAs in live animals, where cellular identities are undisturbed, than using fixed or live cultured cells. The intravital MBS-MCP demonstrated here can be a powerful tool for investigating this important aspect of gene expression.
Supplementary Material
Figure 1.
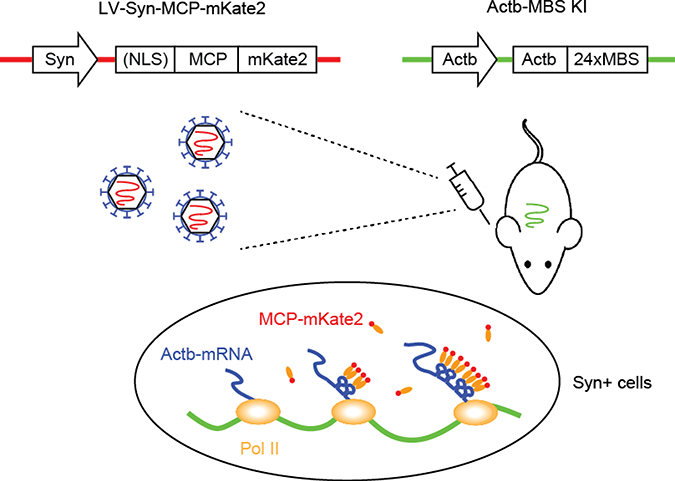
Schematic of LV-mediated intravital MBS-MCP.
Highlights:
The brain is characterized by the diversity of cell types
Variability (i.e., noise) in the synthesis and intracellular movement of mRNAs is thought to play a crucial role in regulating the structure and function of the brain
The dynamics of mRNA is observed in the brain of living mouse, where cellular identities are undisturbed, by intravital two-photon microscopy
Context-dependent mRNA dynamics can be resolved by engineering viral construct for MBS-MCP
Acknowledgments
We thank Xiuhua Meng for help with cloning and Dr. Vladislav Verkhusha for advice on far-red fluorescent proteins. C.N. received support from the National Institute of Health MBRS program at Hunter College (GM060665). This work was supported by funds from the National Institute of Health (EB013571 to H.L. and R.H.S., NS083085 to R.H.S. and GM121198 to H.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA, A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function, Journal of Neuroscience 28(1) (2008) 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang Y, Chen KN, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng SY, Liddelow SA, Zhang CL, Daneman R, Maniatis T, Barres BA, Wu JQ, An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex, Journal of Neuroscience 34(36) (2014) 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres BA, Quake SR, A survey of human brain transcriptome diversity at the single cell level, Proceedings of the National Academy of Sciences of the United States of America 112(23) 2015. 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM, Localization of ASH1 mRNA particles in living yeast, Mol. Cell 2(4) (1998) 437–445. [DOI] [PubMed] [Google Scholar]
- [5].Forrest KM, Gavis ER, Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila, Current Biology 13(14) (2003) 1159–1168. [DOI] [PubMed] [Google Scholar]
- [6].Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M, Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos, Proceedings of the National Academy of Sciences of the United States of America 111 (29) (2014) 10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Campbell PD, Chao JA, Singer RH, Marlow FL, Dynamic visualization of transcription and RNA subcellular localization in zebrafish, Development 142(7) (2015) 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH, A transgenic mouse for in vivo detection of endogenous labeled mRNA, Nature Methods 8(2) (2011) 165–U96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng XH, Singer RH, Visualization of dynamics of single endogenous mRNA labeled in live mouse, Science 343(6169) (2014) 422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tiscornia G, Singer O, Verma IM, Production and purification of lentiviral vectors, Nature Protocols 1(1) (2006) 241–245. [DOI] [PubMed] [Google Scholar]
- [11].Cetin A, Komai S, Eliava M, Seeburg PH, Osten P, Stereotaxic gene delivery in the rodent brain, Nature Protocols 1(6) (2006) 3166–3173. [DOI] [PubMed] [Google Scholar]
- [12].Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Huebener M, Keck T, Knott G, Lee W-CA, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L, Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window, Nature Protocols 4(8) (2009) 1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH, In vivo dynamics of RNA polymerase II transcription, Nat Struct Mol Biol 14(9) (2007) 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Crocker JC, Grier DG, Methods of digital video microscopy for colloidal studies, Journal of Colloid and Interface Science 179(1) (1996) 298–310. [Google Scholar]
- [15].Chenouard N, Smal I, de Chaumont F, Maska M, Sbalzarini IF, Gong YH, Cardinale J, Carthel C, Coraluppi S, Winter M, Cohen AR, Godinez WJ, Rohr K, Kalaidzidis Y, Liang L, Duncan J, Shen HY, Xu YK, Magnusson KEG, Jalden J, Blau HM, Paul-Gilloteaux P, Roudot P, Kervrann C, Waharte F, Tinevez JY, Shorte SL, Willemse J, Celler K, van Wezel GP, Dan HW, Tsai YS, de Solorzano CO, Olivo-Marin JC, Meijering E, Objective comparison of particle tracking methods, Nature Methods 11(3) (2014) 281–U247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gong SC, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N, A gene expression atlas of the central nervous system based on bacterial artificial chromosomes, Nature 425(6961) (2003) 917–925. [DOI] [PubMed] [Google Scholar]
- [17].Gerfen CR, Paletzki R, Heintz N, GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits, Neuron 80(6) (2013) 1368–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kugler S, Kilic E, Bahr M, Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area, Gene Therapy 10(4) (2003) 337–347. [DOI] [PubMed] [Google Scholar]
- [19].Kalderon D, Roberts BL, Richardson WD, Smith AE, A Short amino-acide sequence about to specify nuclear location, Cell 39(3) (1984) 499–509. [DOI] [PubMed] [Google Scholar]
- [20].Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E, Single mRNA molecules demonstrate probabilistic movement in living mammalian cells, Current Biology 13(2) (2003) 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM, Far-red fluorescent tags for protein imaging in living tissues, Biochemical Journal 418 (2009) 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, Singer RH, Glutamate-induced RNA localization and translation in neurons, Proceedings of the National Academy of Sciences of the United States of America 113(44) (2016) E6877–E6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH, Dynamics of single mRNPs in nuclei of living cells, Science 304(5678) (2004) 1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Monnier N, Barry Z, Park HY, Su KC, Katz Z, English BP, Dey A, Pan K, Cheeseman IM, Robert H, Bathe M, Inferring transient particle transport dynamics in live cells, Nature Methods 12(9) (2015) 838-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Swain PS, Elowitz MB, Siggia ED, Intrinsic and extrinsic contributions to stochasticity in gene expression, Proceedings of the National Academy of Sciences of the United States of America 99(20) (2002) 12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Raser JM, O'Shea EK, Control of stochasticity in eukaryotic gene expression, Science 304(5678) (2004) 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gregg C, Zhang JW, Weissbourd B, Luo SJ, Schroth GP, Haig D, Dulac C, High resolution analysis of parent-of-origin allelic expression in the mouse brain, Science 329(5992) (2010) 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smith RM, Webb A, Papp AC, Newman LC, Handelman SK, Suhy A, Mascarenhas R, Oberdick J, Sadee W, Whole transcriptome RNA-Seq allelic expression in human brain, Bmc Genomics 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH, Single-molecule analysis of gene expression using two-color RNA labeling in live yeast, Nature Methods 10(2) (2013) 119–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR, Kinetic competition during the transcription cycle results in stochastic RNA processing, Elife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


