Abstract
Transcription factors control the rate of transcription of genetic information from DNA to messenger RNA, by binding specific DNA sequences in promoter regions. Transcriptional gene control is a rate-limiting process that is tightly regulated and based on transient environmental signals which are translated into long-term changes in gene transcription. Post-translational modifications (PTMs) on transcription factors by phosphorylation or acetylation have profound effects not only on sub-cellular localization but also on substrate specificity through changes in DNA binding capacity. During times of cellular stress, specific transcription factors are in place to help protect the cell from damage by initiating the transcription of antioxidant response genes. Here we discuss PTMs caused by reactive oxygen species (ROS), such as H2O2, that can expeditiously regulate the activation of transcription factors involved in the oxidative stress response. Part of this rapid regulation are proteins involved in H2O2-related reduction and oxidation (redox) reactions such as redoxins, H2O2 scavengers described to interact with transcription factors. Redoxins have highly reactive cysteines of rate constants around 6–10−1 s−1 that engage in nucleophilic substitution of a thiol-disulfide with another thiol in inter-disulfide exchange reactions. We propose here that H2O2 signal transduction induced inter-disulfide exchange reactions between redoxin cysteines and cysteine thiols of transcription factors to allow for rapid and precise on and off switching of transcription factor activity. Thus, redoxins are essential modulators of stress response pathways beyond H2O2 scavenging capacity.
1. Introduction
Transcription factors (TFs) are proteins that play an essential role in controlling gene expression through binding of DNA thereby either promoting or suppressing transcription. By binding to specific segments of DNA in a gene's promoter region, a TF can up- or down-regulate gene expression by stabilizing or blocking binding of RNA polymerase, respectively [1], [2]. TFs may also recruit additional proteins into the transcription-DNA complex to aid in gene up-regulation (coactivators) or gene down-regulation (co-repressors) [3]. Additionally, when bound to a gene promoter, TFs can trigger histone proteins to be acetylated or deacetylated. Acetylation leads to a weaker association between DNA and the histone proteins, allows easier DNA access gene transcription. Deacetylation, on the other hand, leads to a tighter association between the DNA and histone proteins, making it more difficult to access the DNA, leading to down-regulated gene transcription [4].
TFs are transcribed in the nucleus and then translated in the cell’s cytoplasm, where most of the TFs, excluding nuclear TFs (hormone receptors), are usually then sequestered. Nuclear translocation is thus key for TF activity. This nuclear shuttling of TFs is accomplished in a number of ways, but often requires access to the nuclear localization signal (NLS) localized on the TFs [5]. Signaling transduction can induce rapid re-distribution of TFs from the cytosol to the nucleus through several different mechanisms. One is the interaction of the TF with the nuclear import and export machinery that is regulated by posttranslational modifications (PTMs). Here, a TF that resides in the cytosol and displays a low rate of nuclear import relative to its rate of nuclear export can rapidly move into the nucleus in response to a regulated increase in its association with an importin, decreased interaction with an exportin, or a combination of both. Alternatively, nuclear import can be reduced due to high-affinity binding of the TF to a cytosolic anchor protein or shielding of the NLS by a cytosolic protein partner. Both scenarios depend on the regulation of signal transduction-induced PTMs.
Oxidative stress is an imbalance between the systemic manifestation of ROS and a biological system's ability to quickly and efficiently detoxify the reactive intermediates or to repair the resulting damage. Disruption of the normal redox state of cells can cause toxic effects through the production of and free radicals and peroxides that damage all components of the cell, such as proteins, lipids, and DNA. Oxidative stress from oxidative metabolism causes DNA base damage, as well as DNA strand breaks. Base damage is mostly indirect and caused by ROS generated, e.g., ∙O2− (superoxide anion), ∙OH (hydroxyl radical) and H2O2 (hydrogen peroxide). TFs regulating oxidative stress responses are charged with gaging the rescue response in a way that promotes the expression of genes promoting cell survival or, in cases of overwhelming cell damage, cell death. In both scenarios, however, a speedy signaling response of TFs is required. Oxidative stress is a transmitter of signaling responses in part by inducing PTMs on proteins that are both, reversible and irreversible. Recently, reports have accumulated demonstrating that redoxins regulate TF activity through direct interaction. So far, more often than not, these interactions involve an electron transfer, oxidation of TF’s cysteine thiols through inter-disulfide exchange reactions, that way enabling a rapid and precise regulation of the stress response. Redoxins are proteins known to be involved in redox reactions and can reverse some oxidative stress induced PTMs on cysteine thiols such as thiol oxidations resulting in disulfide bridges. These redoxins include peroxiredoxins (PRDXs), thioredoxins (TRXs) and glutaredoxins (GRXs), which are all encompassed by the TRX-like superfamily. This family is a large, diverse group of proteins containing TRX folds that can alter the redox state of target proteins via the reversible oxidation of their active site dithiol. Recently, an increasing number of studies emerged describing TFs as target proteins of redoxins. Thus, in light of the fast kinetics involving redoxin thiol oxidation [6], this review will discuss cysteine oxidation PTMs as essential regulators of TFs in the oxidative stress response.
2. Cysteine modifications in redox signaling
2.1. Cellular reactive oxygen species (ROS)
ROS are chemically reactive species containing oxygen and include superoxide anions (∙O2-, containing a radical electron), singlet oxygen (1O2), hydrogen peroxide (H2O2), and hydroxyl radicals (∙OH) (Fig. 1). Some ROS are also called free radicals and are characterized by unpaired valence electrons such as the hydroxyl radicals (∙OH). Oxygen (O2) is thermodynamically highly reactive with having two unpaired electrons but kinetically considered stable because of spin restriction (Pauli exclusion principle). O2- is a product of the naturally occurring one-electron reduction of molecular oxygen (dioxygen, O2) (Fig. 1, I) is also very reactive, but relatively unstable with a half-life of seconds in aqueous solution [7]·H2O2 is a product of electron a reduction reaction where 2·O2- in the presence of 2H+ either spontaneously or enzymatically dismutate into one molecule of H2O2 (Fig. 1, II). The enzymatic reaction is catalyzed by superoxide dismutases (SODs) and has a rate constant of about 1.6 × 109 M−1 s−1, which is 3–4 magnitudes faster than the spontaneous non-enzymatic conversion [8].
Fig. 1.
Formation and elimination of reactive oxygen species. ROS are chemically reactive chemical species that contain oxygen. The reduction of oxygen (O2) produces superoxide, which in the presence of hydrogen can further dismutate to hydrogen peroxide (H2O2) (I)·H2O2, in turn, may be partially reduced to hydroxyl radical (∙OH) or fully reduced to water (H2O) (II)·H2O2 can react with ∙O2- (Haber Weiss reaction, Fig. 1, III) or ferrous iron (Fe2+) (Fenton reaction, Fig. 1, IV) and hydroxyl radicals (∙OH) and hydroxide ions (HO-) are generated.
In contrast to ∙O2-, the decay rate of H2O2 substantially longer depends on the circumstances. For example, in aqueous solution, H2O2 has been found to range from 1 to 8 h and in the atmosphere, it is believed to last up to 24 h [9]. In E.coli for example, H2O2 is formed at a rate of 10 μM s−1 and metabolized with a rate constant of kcat/Km = 4 × 107 M−1 s−1 by alkyl-hydroperoxide reductase C (AphC) with a titer of ~5 μM, resulting in a steady state concentration of 50 nM [10]. Several newer kinetic models have been developed using analytical equations to estimate cellular H2O2 concentrations in mammalian cells by simulating kinetic curves for clearance. These analyses demonstrate that intracellular H2O2 levels may be much lower in the pico-nanomolar ranges in contrast to earlier findings placing them in micromolar ranges [11]. Out of most ROS, H2O2 is stable for a longer time and thus plays an essential role as second messenger in thiol-based cell signaling, where it participates in both, thiol-disulfide interchange and thiol-oxidation reactions. These reactions are characterized by a wide range of rate constants depending on the local environment of the thiol group [12], [13]. The critical role for H2O2 in cell signaling stems from discoveries demonstrating phenotypic differences in outcomes depending on the localization of its production. When H2O2 reacts with ∙O2- (Haber Weiss reaction, Fig. 1, III) or ferrous iron (Fe2+) (Fenton reaction, Fig. 1, IV) hydroxy radicals (∙OH) and hydroxide ions (HO-) are generated. The HO- is a strong base and nucleophile, and the ∙OH, the neutral form of HO-, is a highly reactive radical and as such very short lived. Hydroxy radicals play an essential part in radical chemistry responsible for oxidative stress-induced DNA damage and lipid peroxidation [14], [15].
2.2. Cysteine oxidation
Cysteines contain a polarized sulfur atom of which the oxidation state can range from the fully reduced thiol/thiolate state to the fully oxidized sulfonic acid (Fig. 2). Many thiol reactions involve the nucleophilic attack of the deprotonated thiolate anion (S-) on an electrophilic center, that way making the overall reactivity of a thiol group strongly dependent on the pΚa of the cysteine side chain. Its position within a folded protein and the surrounding residues contribute to an individual cysteine's overall reactivity. Cysteine has a slightly alkaline dissociation constant (pΚa) of approximately 8.2 meaning that at physiological pH, it is more likely to be found in its protonated and therefore less reactive thiol state. In cases where a cysteine thiol is buried within the protein core, the pΚa has been found to be as high as ~9.5 [16]. Hydrogen bonding between the thiol backbone and the polypeptide backbone or nearby sidechains seems an essential step in lowering the pΚa, while positively charged amino acids, which have been previously thought responsible for lowering thiol pΚa, appear to play a less critical role in stabilizing the deprotonated thiolate [17]. Following this, a thiolate oxidizes to sulfenic acid (-SOH), which is the prerequisite for inter- or intramolecular disulfide bonding. Oxidation reactions of thiolate to sulfenic acid are reversible, and the disulfide bonds formed can be reduced upon reactions with thioredoxin reductase (TRXR) or glutathione reductase (GR) and NADPH as reducing equivalent. In the presence of H2O2, -SOH can be further oxidized to sulfinic acid (SO2H), and then from sulfinic acid to sulfonic acid (SO3H). These oxidations, however, are irreversible and result in permanent protein damage (Fig. 3) [18]. Importantly, for the 2-cys peroxiredoxins, reduction of sulfinic acid is reversible in ATP-dependent reaction catalyzed by the oxidoreductase sulfiredoxin (Srx) [19], [20]. Disulfide bond formation can significantly impact protein structure, stability, and function. Specifically, a disulfide is often required for proper protein folding, and in the absence of a bond, the resulting protein could be dysfunctional or inactive [21]. Oxidation of a thiol followed by disulfide bonding can have another vital function in redox signaling, and that is to regulate the activity of other proteins. For example, the below discussed thiol-disulfide exchange mechanism is one of the better-understood methods of protein regulation.
Fig. 2.
Stepwise oxidation of cysteine residues. Oxidation of the sulfur atom within a cysteine residue can result in the stepwise formation of a reactive cysteine thiolate, sulfenic acid, sulfinic acid, and finally sulfonic acid. With the aid of reducing systems including NADPH, as well as thioredoxin reductase (TRXR) or glutathione reductase (GR), the oxidation reactions leading to disulfide formation are reversible. On the other hand, oxidation to sulfinic (not for peroxiredoxins) or sulfonic acid is irreversible.
Fig. 3.
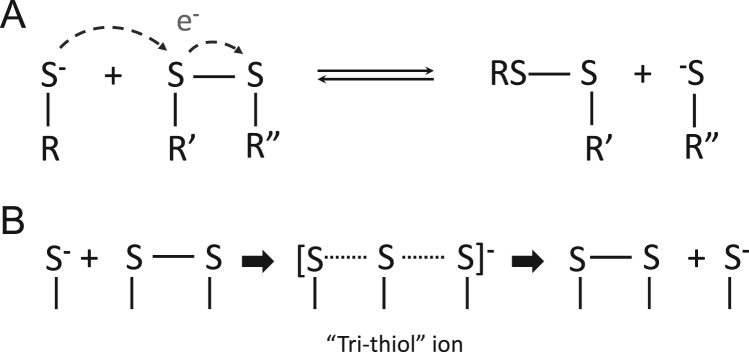
Thiol-disulfide exchange. A and B. A simplified model shows the thiolate group (nucleophile) on protein R to attack a sulfur atom of the protein disulfide bond of R′ and R′′. This creates a temporary "tri-thiol" ion (B) and ultimately displaces the other sulfur atom in the disulfide (protein R′′). This reaction leads in turn to the formation of a new disulfide bond between proteins R and R′, and a new thiolate ion (nucleophile) on protein R′′.
2.3. Thiol-exchange
Redox reactions involve the coordinated reduction (gain of electrons) of one species and the oxidation (loss of electrons) of another. Disulfide bonds formation between cysteines can be triggered in different ways. One by direct thiol oxidation through a low molecular weight two-electron oxidant, such as H2O2, or by thyil radical formation via a single-electron transfer, or by the nucleophilic substitution of a thiol-disulfide with another thiol, also called thiol-disulfide exchange reaction [22]. This reaction involves allosteric changes in protein structure and is thought to be responsible for intracellular redox homeostasis, and play an important role in antioxidant defense, in addition to its role in redox signaling [23]. Thiol-disulfide exchange is preceded by deprotonation (loss of H+) of a thiol resulting in a negatively charged thiolate anion (S-) that is highly reactive and will readily react with other nucleophiles to become more stable and neutralize their negative charge. During the thiol-disulfide exchange, a reactive thiolate anion ‘attacks’ one of the sulfur atoms of an existing disulfide bond. The original disulfide bond then begins to break as a new one forms, creating a trisulfide-like transition state, a “tri-thiol” ion [24]. Eventually, the new disulfide bond containing the ‘attacking' thiolate is formed, along with the release of a new thiolate anion (Fig. 3) [25]. This reaction is highly transient, with the newly formed thiolate free to go on and react, forming or reforming other bonds and triggering further downstream reactions [6]. There are several factors that affect the kinetics of thiol-disulfide exchange reactions such as the cysteine thiol surrounding pΚa, nucleophilicity in the context of pH, stability of the leaving group, the electrophilicity of the central disulfide sulfur, steric considerations such as H-bonding, coulombic and lastly, mechanical forces (reviewed comprehensively in [6]). Therefore, it is important to consider that kinetic differences for thiol exchange reactions exist which in turn provide specificity of redox signaling. For example, in an analysis using high-performance liquid chromatography bacterial disulfide interchange proteins (DsbD and DsbC) were examined for the kinetics of inter-disulfide exchange reactions. Notably, a difference of 3 magnitudes in the second order rate constant k( M−1 s−1) was found, DsbD 3.9 × 106 and DsdC 3.0 × 103, respectively [26].
3. Redoxins as regulators of redox-induced post-translational modifications
Accumulation of ROS can cause permanent damage and dysfunction on both the cellular and organismal levels. Fortunately, cells have a way to combat detrimental effects in the form of antioxidants that inhibit the oxidation of other molecules. When there is a buildup of ROS in a cell or the cellular environment, antioxidants restore redox homeostasis by neutralizing the ROS into non-reactive products and repairing any erroneously oxidized molecules before permanent cell damage can occur. Enzymatic antioxidants such as superoxide dismutase (SOD), catalase, thioredoxins, glutathione peroxidase, and peroxiredoxins, all function to detoxify any excess ROS, often working in combination and a stepwise fashion (Fig. 4) [27]. As an example, SOD can convert two superoxide anions into hydrogen peroxide (H2O2) and molecular oxygen (O2). To date, three mammalian SOD family members exist that differ in enzymatic cofactors and subcellular localization. For example, SODs that either use Cu/Zn localize in the cytosol (SOD1) or the extracellular space (SOD3) and the Mn utilizing SOD localizes to the mitochondria (SOD2)·H2O2 is converted into water and oxygen by peroxidases such as catalase, glutathione peroxidases (GPXs) and peroxiredoxins (PRDXs), which share similar rate constants reacting with H2O2: 107–108M−1 s−1.
Fig. 4.

Neutralization of ROS by different antioxidants. SOD: Superoxide dismutase; Cat: Catalase; GPX: Glutathione peroxidase; GSH: Glutathione; TRX: Thioredoxin; PRDX: Peroxiredoxin.
In contrast to GPXs and PRDXs which are distributed throughout the cell, catalase is confined to the peroxisomes where it removes H2O2. It is still unclear, however, what distinguishes these peroxidases in their specificity in redox signaling given that all share very similar rate constants towards H2O2. The answer may lie in the rate-limiting step of enzyme recovery. For example, in contrast to catalase, which does not rely on reducing systems for restoration, GPXs and PRDXs depend on the availability of NADPH, dithiols (2GSH and TRX), and reductases. Also, catalytic cysteines of eukaryotic PRDXs are highly susceptible to the sulfenic acid formation which can undergo further oxidation to sulfinic and sulfonic acid leading to enzyme inactivation, which in the case of the later is irreversible [28], [29]. Thus, in case of lack or inactivity of the antioxidant enzymes cysteine oxidation on proteins occurs then due to the increased local amounts of ROS. In thiol-exchange reactions, however, antioxidant enzymes, such as redoxins, directly participate in the electron transfer in cysteine thiol oxidation and disulfide formation (Fig. 3). Recent research has demonstrated that redoxins, such as peroxiredoxins are often partners in thiol-exchange reactions thus acting as a relay switch in redox cell signaling.
3.1. Glutaredoxin-controlled transcription factors
Glutaredoxins (GRXs) are a family of redox enzymes which use glutathione (GSH), an antioxidant found ubiquitously across species, as a cofactor [30]. Functioning similarly and with a similar structure to thioredoxins, glutaredoxins (GRXs) were first identified in Escherichia coli, and are found to exist in most living organisms, from prokaryotes up to humans [31], [32]. While there are many different isoforms, the classical GRXs are ten kDa proteins containing a C-P-Y-C motif within their active sites and comprise what is known as the glutaredoxin fold [33]. In short, the glutaredoxin fold is a structure consisting of four-stranded β-sheets, surrounded by three α-helices. Similar structures exist in the TRX binding site [34]. Reduction via the glutaredoxin system occurs when NADPH transfers electrons to glutathione reductase (GR), which then transfers electrons to GSH, GSH then functions as a cofactor for one of the GRX enzymes, allowing it to reduce target proteins via thiol exchanges [35]. For a more in-depth review of GRXs see Fernandes and Holmgren [36].
3.1.1. OXYR
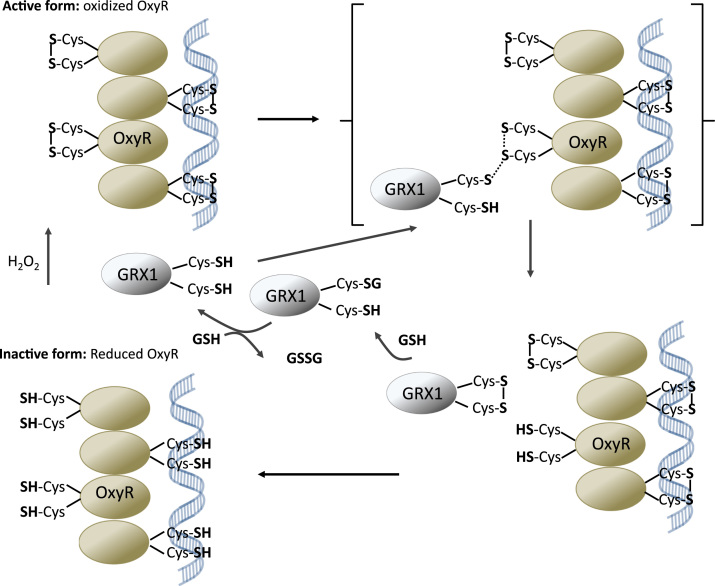
A member of the LysR family of bacterial transcription factors, OxyR is a transcription factor found in E. coli and is a critical protein in transcribing antioxidant genes [37]. OxyR’s activity is modified through a ROS induced disulfide bond between two of OxyR’s six cysteines (Cys199 and Cys208) and only activates transcription when in its structurally distinct oxidized form (Fig. 5). In its reduced form tetrameric OxyR binds to two pairs of adjacent DNA grooves in promoter regions of target genes thus preventing transcription. After oxidation of the Cys199 of OxyR by H2O2, sulfenic acid forms which the, in turn, attacks Cys208 to generate an intradisulfide bond. This structural change involves tetrameric OxyR that binds to four adjacent major DNA grooves that way allowing recruitment of RNA polymerase and activation of transcription [38]. The Cys199-Cys208 bond is then reduced by GRX1, which is regenerated by GSH. When activated, OxyR functions to enhance the transcription of genes involved in the combating the adverse effects of oxidative stress [39], [40].
Fig. 5.
H2O2 sensing by OxyR. In its reduced form OxyR binds to DNA promoter regions of target genes preventing transcription. After oxidation of the Cys199 of OxyR by H2O2, sulfenic acid forms that attacks Cys208 to form an intradisulfide. These structural changes in OxyR allow in turn recruitment of RNA polymerase and activation of transcription. The Cys199-Cys208 bond is then reduced by GRX1, which is regenerated by GSH.
The importance of reversible disulfide bridge formation between two proteins in cell signaling was demonstrated in 1998 by groundbreaking work from the Storz laboratory and has been ever since further defined by many publications and applied to other redoxins as discussed below [40], [41], [42], [43]. GRX1 triggers the recycling of a reversible disulfide bond, leading to the activation of OxyR. Mutational analysis confirmed that when OxyR cysteines 199 and 208 were mutated to serines, miRNA assays showed little to no expression of the OxyR target oxyS, further supporting the idea that Cys199 and Cys208 play a critical role in OxyR activation. Mass spectrometry revealed that the two peaks corresponding to alkylated Cys199 and Cys208 in the reduced protein disappeared and were replaced by a new peak corresponding to the sum of the two joined by a disulfide in the oxidized protein. This finding lent further support to the conclusion that a conformational change occurs to activate OxyR. Shown previously, the reaction of OxyR with H2O2 is fast and transient with a second-order rate constant of about 105 M−1 s−1 and activation and target expression peaking at about 10 min after Cys199-Cys208 disulfide bridges are reduced [37], [43]. Isogeneic E. coli strains deficient in the components the GRX (GRX reductase, GSH, GRX) and TRX (TRX reductase, NADPH, and TRX) pathways, the central disulfide reduction systems found in cells, were analyzed for OxyR activity. GRX1 mutants showed marked increased in oxyS RNA expression levels following H2O2 treatment as compared to the wild-type strain, while the thioredoxin mutants displayed an identical or decreased oxyS expression profile. Incubation of oxidized OxyR containing the Cys199-Cys208 disulfide with GRX1 and purified GSH, the required cofactor of GRX1′s catalytic activity, eliminated OxyR activity; treatment with H2O2 restored OxyR activity. Finally, an examination of the GRX1 promotor revealed an almost perfect match to the OxyR consensus sequence, indicating OxyR triggers the transcription of GRX1. These data strongly suggested that the glutaredoxin pathway regulates oxyr. H2O2 triggers the formation of a disulfide bond, activating OxyR and triggering transcription of OxyR targets including GRX1, GRX1 then goes on to reduce and deactivate OxyR through an intermolecular disulfide thiol exchange.
3.2. Peroxiredoxin-controlled transcription factors
Peroxiredoxins (PRDXs) are a family of six (PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6) hydrogen peroxide scavengers, antioxidant enzymes that reduce peroxides via the oxidation of a catalytic (or peroxidatic) cysteine to sulfenic acid [44]. They can function as signal transduction proteins that regulate stress-induced signaling cascades [45], [46]. The number of cysteines they contain classifies PRDXs, except for PRDX6 which is a 1-Cys PRDX, while the rest of the family are 2-Cys [47]. Classified as 2-Cys PRDXs, PRDX1-4, in addition to the catalytic cysteine, also contain a resolving cysteine that forms a disulfide bond with the sulfenic acid of the catalytic cysteine of a second PRDX molecule to form a head to tail dimer [48], which can then be reduced via thioredoxin (TRX) to reset catalytic function. PRDX1 homodimers can associate noncovalently forming doughnut-like assemblies that are usually decameric. The redox state of the peroxidatic cysteine (CP) in PRDX1, is the best-characterized factor regulating the oligomerization of PRDX1 subfamily members. When CP is reduced, a fully folded (FF) conformation is adapted through movement of the catalytic loop and C-terminal extension, thus stabilizing the decameric form. By reducing peroxides (ROOH) to ROH, CP is oxidized to SOH, and the active site becomes locally unfolded (LU) permitting disulfide bonding of the CP with the resolving cysteine (CR) from another PRDX1 subunit [49]. This conformational change favors the dissociation of decamers into dimers [50], [51]. In case of PRDX1's CP over-oxidation, PRDX1 decamers are also formed and thought to act as protein chaperones [45]. Recently, reports have accumulated describing PRDX1 and PRDX2 to bind proteins by either forming disulfide intermediates (thiol-exchange reactions) or non-covalent interactions often involving overoxidized CP of either PRDX1 or PRDX2. Examples of such binding partners include MKP5, JNK, p38α, MST1, and c-myc [46], [51], [52], [53], [54], [55], [56]. Covalent binding partners include STAT3, FOXO3, ASK1 and APE1 [54], [57], [58], [59]. Through these mechanisms, the PRDXs participate in regulating the activity of its binding partners by modulating redox-induced modification on important functional cysteines.
3.2.1. STAT3
Signal transducer and activator of transcription (STAT) proteins are transducers and transcription factors involved in the regulation of cellular inflammatory responses, cell survival, and proliferation. Seven STAT family members have been described in mammals: STAT1-4, STAT5A, STAT5B, and STAT6. STAT proteins are induced by activation of cytokine-Janus kinase (JAK) signaling [60], [61]. When triggered by various cytokines and growth factors, STAT3 is phosphorylated at multiple amino acids along the protein, forming homo- or heterodimers, which then translocate into the nucleus and become activated [62]. In addition to playing key roles in cellular processes such as cell growth and apoptosis, STAT3 has also been shown to play an essential role in the differentiation of helper T cells and is thereby involved in many autoimmune diseases [63].
Depend on cellular signaling pathways and cellular context, JAK-STAT pathways are affected by redox signaling, but at the same time, modulate ROS homeostasis [60]. While ROS trigger STAT3 phosphorylation as well a nuclear translocation, STAT3 can translocate into mitochondria and upregulate the activity of the electron transport chain which leads to an increase in ROS production, and STAT3-dependent antioxidant responses protect cells against oxidative stress [60], [64], [65], [66], [67].
PRDX2 is a fine-tuner of STAT3 DNA binding activity through a redox relay. PRDX2 acts as an H2O2 signal receptor and transmitter for STAT3 where oxidative equivalents flow from PRDX2 to STAT3 [59]. After the H2O2 cell stimulation, disulfide bond formation between PRDX2 and STAT3 was shortly followed by the forming of four distinct STAT3 oxidation products, disulfide-linked STAT3 dimers, and several tetramers. Out of the 14 STAT3 cysteines, loss of Cys418 within the DNA binding domain (DBD) had the most pronounced effect abrogating tetramers of higher molecular weights. Within the transactivation domain (TAD), loss of either Cys712 or Cys718 abolished STAT3 dimers. The Cys418 and Cys426 mutations diminished mixed disulfide intermediates >160 kDa, most likely reflecting PRDX2-conjugated STAT3 dimers on their way to forming tetramers, while the loss of Cys712 and Cys718 decreased mixed disulfide intermediates <160 kDa, most likely indicating PRDX2 conjugates of monomeric STAT3 on their way to form a dimer. These findings demonstrate that independent PRDX2-mediated oxidation pathways lead to STAT3 dimers and tetramers, respectively.
Furthermore, all STAT oxidation products derive from PRDX2-STAT3 intermediates. Most likely, the TAD is the initial or primary interaction site for PRDX2, as a C-terminally truncated STAT3 variant (lacking most of the TAD) did not show any H2O2-induced dimer or tetramer formation [68]. In aggregate, this suggests that it is the interaction with PRDX2 that triggers STAT3's subsequent oxidation thus allowing STAT3 homodimerization and DNA binding (Fig. 6A). Interestingly, chemical inhibition of thioredoxin reductase increased STAT3 oxidation and concomitantly decreased STAT3 activity, suggesting the possibility that TRX1 may be a direct mediator of STAT3 disulfide reduction [59]. However, while detection of TRX1-STAT3 disulfide exchange intermediates further supported this suggestion, TRX1 oligomers were not differentiated from PRDX2-STAT3 oligomers leaving open the possibility that TRX1 is a partner of the PRDX2-STAT3 complex to reduce PRDX2 dimers and not STAT3. Further research is needed to examine this hypothesis.
Fig. 6.
Redoxin induced transcription factor activation and inactivation. (A) The redoxin, PRDX2, oxidizes STAT3 via a thiol-disulfide exchange. (B) Based on (A) the following model for PRDX1 regulation of FOXO3 is proposed: PRDX1 dimer forms an oligomer with free FOXO3 via disulfide bonds.
3.2.2. FOXO3
The transcription factor Forkhead box O3 (FOXO3) is a downstream target of the PI3K/AKT signaling pathway [69], and a member of the Forkhead family of proteins, which are defined by the evolutionarily conserved DNA-binding domain, the Forkhead box (FOX). As a FOXO protein, FOXO3 is involved in mechanisms of the DNA damaged response, cell survival, and proliferation. FOXO3 has explicitly been shown to act as a tumor suppressor regulating the expression of genes involved in oxidative stress resistance, cell cycle arrest, and apoptosis [70], [71], [72], [73]. In response to oxidative stress, FOXO3 translocates to the nucleus, where it becomes activated, even in the presence of growth factors that generally inhibit its activity [74]. When inactive, FOXO3 remains sequestered in the cytoplasm through binding to phosphorylation by AKT at one or more of three AKT phosphorylation sites, T32, S253, and S315 because of association of FOXOs with 14–3-3 proteins [74], [75], [76], [77]. Phosphorylation, as well as dephosphorylation events, facilitate FOXO3 nuclear shuttling. The pro-apoptotic kinases JNK1, MST1, and p38α counteract FOXO3 AKT-induced cytoplasmic sequestration of FOXO3 by phosphorylating FOXO3 on Ser574, Ser209, and Ser7, respectively [78], [79], [80], [81]. On the other hand, PP2A has been shown to dephosphorylate FOXO3 on Akt phosphorylation sites T32 and S253 resulting in dissociation of 14-3-3 from FOXO3 and nuclear accumulation of FOXO3 [82]. Interestingly, JNK1, MST1, p38α as well as PP2A have been shown to be differentially regulated by PRDX1 under oxidative stress [52], [55], [56], [83], [84], [85].
In a study carried out by Hopkins et al. [58], PRDX1 has been shown to bind to, and subsequently regulate FOXO3 activity. In the absence of H2O2, PRDX1 binds to FOXO3 at low levels, but upon stimulation with H2O2, PRDX1 binding to FOXO3 increases. PRDX1-FOXO3 binding was time-dependent, with coupling occurring shortly after stimulation, only to decrease steadily after reaching its peak. Site-specific mutagenesis studies suggested that an oligomer is comprised of a PRDX1 dimer and a FOXO3 monomerfound. This heterotrimer involves FOXO3 cysteines 31 or 150, as previously indicated by a mass spectrometry analysis by Putker and al [86] as well as the dimer-building PRDX1 cysteines 52 and 173. As these PRDX1 cysteines, are essential in its peroxidase activity and have been implicated in other studies in redox relay reactions, these findings suggest a disulfide bond between PRDX1 and FOXO3, which could be part of a redox relay [54]. PRDX1 and FOXO3 oligomer analysis on non-reducing gels using FOXO3 cysteine mutants further supported this hypothesis. Compared to wild type FOXO3, the FOXO3 cysteine mutants (Cys31 and Cys150) showed elevated levels of phosphorylation at AKT phosphorylation sites, which were not significantly altered by H2O2 stimulation, suggesting cytoplasmic sequestration and inactivity of FOXO3. This result was confirmed by a) localization studies that showed nuclear wild type FOXO3 upon increasing amounts of H2O2 stimulation with cytoplasmic sequestration of FOXO3 cysteine mutants that abrogated binding to PRDX1 and b) loss of FOXO3 targets in cells expressing either expressing FOXO3 cysteine mutants. In aggregate, a functional mechanism for the redox specific regulation of FOXO3 by PRDX1 was possible where PRDX1 calibrates FOXO3 activity under oxidative stress (Fig. 5B). At low levels of H2O2, PRDX1-unbound FOXO3 is accessible for AKT-induced phosphorylation that sequesters FOXO3 in the cytoplasm and can lead to FOXO3 degradation. While H2O2 increases, a disulfide forms between PRDX1 and FOXO3, protecting FOXO3 from AKT-induced phosphorylation. Once H2O2 levels increase further, the PRDX1 peroxidatic cysteines become over-oxidized with the consequential release of FOXO3 to the nucleus due to phosphorylation by JNK, p38 or MST-1 (Fig. 6B).
3.3. Thioredoxin-controlled transcription factors
Thioredoxins (TRX), discovered in E. coli, are small dithiol redox proteins that play roles in a multitude of biological functions both inside and outside the cell, including the redox-driven regulation of gene transcription [87]. TRXs contain a highly conserved active site, consisting of two cysteine residues (C-G-P-C). TRX antioxidant activity is restored through the reversible oxidation of these conserved active site cysteines [88]. TRX also plays an essential role in regulating the functions of many target proteins, through the maintenance of the cell’s redox environment. When oxidized from its reduced form (TRX-SH-SH) to its oxidized form (TRX-S2), a disulfide bridge can form between the two active site cysteines, resulting in electrons being donated to the target protein, thereby reducing it, and regulating its activity through the previously described thiol-exchange [35]. Some of the targets of TRX include transcription factors, which can be both directly and indirectly controlled.
3.3.1. NF-kappa B
The transcription factor nuclear factor of kappa B (NF-κB) is not a single protein, but rather, is a transcription factor family. NF-κB is made up of two polypeptide subunits, p50 and p65, with each contributing to the binding specificity of the heterodimer [89]. As reviewed in Lenardo et al. [90], NF-κB is involved in the regulation of numerous cellular processes, including the immune and inflammatory responses, cell growth, and apoptosis. Oxidative stress can have activating and inhibiting effects on NF-κB which depends on the duration and the context of the exposure (reviewed comprehensively in [91]).
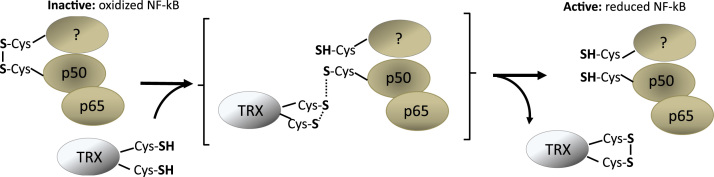
A redox-dependent regulation of NF-κB was discovered first in a study carried out by Matthews et al. in 1992 [92], and similarly by Hayashi et al. in 1993 [93], where both studies described the role of redox regulation in the activation and control of NF-κB by TRX. DNA binding assays reveanled that NF-κB's DNA binding affinity is enhanced in the presence of reducing reagents, while thiol-blocking compounds blocked NF-kB's DNA binding affinity. Systematic mutation analysis of the conserved cysteine residues of the NF-κB p50 subunit, identified Cys62 as the critical cysteine involved in NF-κB’s binding to DNA, as the Cys62Ser NF-κB protein showed reduced DNA binding affinity and insensitivity to thiol-modifying agents. In vitro, TRX promoted p50 dimers formation lead to DNA binding of NF-kB proteins, which was abolished in p50 Cys62 mutants. In resting cells, p50 Cys62 is highly oxidized in the cytoplasm but is rapidly reduced by TRX once NF-κB has migrated into the nucleus [94]. Besides TRX other enzymes have been reported to control the reduction of nuclear p50 Cys62. In resting cells, TRX mainly localizes to the cytoplasm, however, translocates into the nucleus upon cellular stimulation by TNFα. There, TRX it reduces p50 Cys62 to promote NF-κB DNA binding [92], [95]. Besides TRX, AP endonuclease 1/redox factor 1 (APE1/Ref-1), an enzyme with apurinic/apyrimidic (AP) endonuclease activity and essential for the base excision repair pathway [96] reduces oxidized cysteine residues within several transcription factors, including Cys62 of NF-κB p50 [94]. Interestingly, APE1/Ref-1 appears to act via two mechanisms. First, as a redox actor APE1/Ref-1 directly reduces cysteine thiols which require critical cysteine residues located within the N-terminal region of APE1/Ref-1 (Cys65) [97]. Second, APE1/Ref-1 regulates the DNA binding activity of NF-κB by promoting the reduction of p50 Cys62 by GSH as well as TRX [98]. It thus appears that APE1/Ref-1 both acts as a redox factor and a redox chaperone.
TRX reduces the oxidized Cys62 by donating protons in a structure-dependent fashion. The inter-molecular disulfide bridge between TRX and NF-kB was suggested to be transient because the binding of TRX to the NF-kB DNA-binding loop prevents NF-kB to recognize the target DNA. Based on biochemical reactions it was postulated that zinc ion (Zn2+) replaces the inter-molecular disulfide bridge and dissociates NF-kB from TRX leading then to NF-kB DNA binding and activity [99], [100]. Although recombinant p50 forms a homodimer involving Cys62 of another p50 subunit, the biological role of this homodimer remains elusive. Therefore, it entirely possible that Cys62 oxidation involves heterodimer disulfide formation with another NF-kB subunit or another protein (Fig. 7).
Fig. 7.
Redox-regulation of NF-kB by TRX. In resting cells, p50 Cys62 is highly oxidized in the cytoplasm but is rapidly reduced by TRX once NF-κB has migrated into the nucleus. After the reduction of Cys62, a zinc ion replaces the inter-molecular disulfide bridge and dissociates NF-kB from TRX leading to NF-kB DNA binding and activity. Recombinant p50 forms a homodimer involving Cys62 of another p50 protein. However, a biological role of this homodimer remains elusive. Therefore, it is entirely possible that Cys62 oxidation spurs disulfide-based heterodimers with p65 or even another protein.
3.3.2. SP1
Limited information is available for how TRX binding regulates specificity protein 1 (Sp1) activity. Sp1 is a member of the Sp family of transcription factors and contains a zinc finger motif which binds to GC boxes found in the promoter regions of its gene targets, triggering transcription [101]. Sp1 is susceptible to a number of post-translational modifications, such as phosphorylation and acetylation, which regulate its activity, allowing it to function as either an activator or repressor depending on the type and location of the PTM. [101]. Many of Sp1's targets are thought to be “housekeeping” genes, transcribing proteins involved in cellular metabolism [102]. As a zinc finger protein, the Zn2+ ions within each motif can coordinate with histone and cysteine residues present in the protein, thereby making Sp1 susceptible to redox regulation [103]. Moreover, Sp1, as well as Sp3, are induced by oxidative stress and activate the transcription of antioxidant proteins [104], [105], [106].
In a 2003 study by Bloomfield et al. [106], Sp1 was found to be a direct target of TRX regulation. In previous studies [107] it was found that the Trx promotor contains three conserved conscious sequences for Sp1. With this knowledge, and through the use of electrophoretic mobility shift assays (EMSAs), luciferase assays, and simple protein expression analyses, Bloomfield showed that TRX probably binds to Sp1 in a redox-dependent manner thus enhancing Sp1 DNA binding: TRX binding to Sp1 decreases as H2O2 concentration increases. Although the interaction between TRX and Sp1 have yet to be further specified, we speculate that in the presence of the TRX recovery system (thioredoxin reductase, NADPH) TRX binding to Sp1 involves electron transfer between the two proteins as an intricate part of regulation [106].
4. Conclusions and future directions
Binding to target DNA and triggering the expression of gene transcription involved in essential biological pathways, transcription factors are integral players in the proper cellular and organismal function and survival. Any dysregulation or dysfunction in a transcription factor can lead to a broad range of diseases including cancer, autoimmune disease, cardiovascular disease, neurological disease, and diabetes [108]. Maintenance of ROS levels is essential for organismal well-being and are controlled by enzymatic and non-enzymatic antioxidant defenses that scavenge oxidative aggression. ROS induce PTMs on protein thiols which in turn calibrate protein activities towards an adequate cellular response. Here we discussed the role of PTMs in regulating oxidative stress-responsive TFs. We described TFs that are participating in rapid redox reaction such as inter-disulfide exchange, that way titrating a response to changing redox level. As depicted in Fig. 5, PRDX1, as well as PRDX2, perform such relay function with FOXO3 and STAT3, respectively. Given the requirement of reducible disulfide bridges in the relay partners, it is conceivable to assume that with a rise in H2O2 levels, the relay is shut off due to non-reducible thiol modifications that prohibit further regulation of TFs by redoxins. The high-affinity redoxins have for ROS may be the key to this mechanism. For example, the rate constant for reactions between H2O2 and PRDXs ranges between 3.0 × 105 – 1.0 × 108 (M−1 s−1) [109], [110], in times of stress when H2O2 levels are high, a PRDX can quickly scavenge the H2O2, becoming oxidized, and in turn can promptly oxidize a target transcription factor. Depending on the manner in which the transcription factor is regulated that is activation or inactivation, this fast action can result in the fact up-regulation or down-regulation of target genes that may be key in to the stress response or reestablishing redox homeostasis. The redoxin-driven regulatory pathways may be critical sensors able to differentiate systems operating in a healthy state from systems affected by oxidative stress. By gaining a deeper understanding of the various mechanisms by which signaling during oxidative stress affects TF activity will provide a better understanding of disease pathologies related to disturbed redox homeostasis, such as cancer and neurodegenerative conditions.
There is still much to be explored concerning the relationship between redox, antioxidants, and transcription factor regulation, but with each discovery, our understanding of what drives essential cellular processes grow, opening up new avenues of investigation for when those processes fail.
Funding
This work was supported by BBA UPITT (C.A.N.), P30CA047904 (UPMC Hillman Cancer Center).
Contributor Information
Barbara L. Hopkins, Email: blh63@pitt.edu.
Carola A. Neumann, Email: neumannc@upmc.edu.
References
- 1.Latchman D.S. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 1997;29(12):1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 2.Gill G. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 2001;37:33–43. doi: 10.1042/bse0370033. [DOI] [PubMed] [Google Scholar]
- 3.Xu L., Glass C.K., Rosenfeld M.G. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 1999;9(2):140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar G.J., Fan H.Y., Kingston R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 5.Vandromme M., Gauthier-Rouviere C., Lamb N., Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 1996;21(2):59–64. [PubMed] [Google Scholar]
- 6.Nagy P. Kinetics and mechanisms of thiol-disulfide exchange covering direct substitution and thiol oxidation-mediated pathways. Antioxid. Redox Signal. 2013;18(13):1623–1641. doi: 10.1089/ars.2012.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov B.N. Cooperation of photosystem I with the plastoquinone pool in oxygen reduction in higher plant chloroplasts. Biochemistry. 2008;73(1):112–118. doi: 10.1134/s0006297908010173. [DOI] [PubMed] [Google Scholar]
- 8.Fukai T., Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011;15(6):1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.H.E.R.a.o.i.o.h.c. products, Hydrogen Peroxide, 2005. 〈https://www.heraproject.com/files/36-F-05-Shor_H2O2_version1.pdf〉.
- 10.Imlay J.A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 2013;11(7):443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim J.B., Huang B.K., Deen W.M., Sikes H.D. Analysis of the lifetime and spatial localization of hydrogen peroxide generated in the cytosol using a reduced kinetic model. Free Radic. Biol. Med. 2015;89:47–53. doi: 10.1016/j.freeradbiomed.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Santamarina S., Boronat S., Hidalgo E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry. 2014;53(16):2560–2580. doi: 10.1021/bi401700f. [DOI] [PubMed] [Google Scholar]
- 13.Forman H.J., Maiorino M., Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49(5):835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou Z., Chang H., Li H., Wang S. Induction of reactive oxygen species: an emerging approach for cancer therapy. Apoptosis. 2017;22(11):1321–1335. doi: 10.1007/s10495-017-1424-9. [DOI] [PubMed] [Google Scholar]
- 15.Shichiri M., Yoshida Y., Ishida N., Hagihara Y., Iwahashi H., Tamai H., Niki E. alpha-Tocopherol suppresses lipid peroxidation and behavioral and cognitive impairments in the Ts65Dn mouse model of Down syndrome. Free Radic. Biol. Med. 2011;50(12):1801–1811. doi: 10.1016/j.freeradbiomed.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404(5):902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremers C.M., Jakob U. Oxidant sensing by reversible disulfide bond formation. J. Biol. Chem. 2013;288(37):26489–26496. doi: 10.1074/jbc.R113.462929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113(7):4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biteau B., Labarre J., Toledano M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 20.Woo H.A., Jeong W., Chang T.S., Park K.J., Park S.J., Yang J.S., Rhee S.G. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J. Biol. Chem. 2005;280(5):3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 21.Betz S.F. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2(10):1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H.J., Ha S., Lee H.Y., Lee K.J. ROSics: chemistry and proteomics of cysteine modifications in redox biology. Mass Spectrom. Rev. 2015;34(2):184–208. doi: 10.1002/mas.21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45(5):549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Fernandes P.A., Ramos M.J. Theoretical insights into the mechanism for thiol/disulfide exchange. Chemistry. 2004;10(1):257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- 25.Bach R.D., Dmitrenko O., Thorpe C. Mechanism of thiolate-disulfide interchange reactions in biochemistry. J. Org. Chem. 2008;73(1):12–21. doi: 10.1021/jo702051f. [DOI] [PubMed] [Google Scholar]
- 26.Rozhkova A., Stirnimann C.U., Frei P., Grauschopf U., Brunisholz R., Grutter M.G., Capitani G., Glockshuber R. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23(8):1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weydert C.J., Cullen J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010;5(1):51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann C.A., Cao J.X., Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8(24):4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee S.G. Overview on peroxiredoxin. Mol. Cells. 2016;39(1):1–5. doi: 10.14348/molcells.2016.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A. Thioredoxin and glutaredoxin: small multi-functional redox proteins with active-site disulphide bonds. Biochem. Soc. Trans. 1988;16(2):95–96. doi: 10.1042/bst0160095. [DOI] [PubMed] [Google Scholar]
- 31.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA. 1976;73(7):2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padilla C.A., Martinez-Galisteo E., Barcena J.A., Spyrou G., Holmgren A. Purification from placenta, amino acid sequence, structure comparisons and cDNA cloning of human glutaredoxin. Eur. J. Biochem. 1995;227(1–2):27–34. doi: 10.1111/j.1432-1033.1995.tb20356.x. [DOI] [PubMed] [Google Scholar]
- 33.Bushweller J.H., Billeter M., Holmgren A., Wuthrich K. The nuclear magnetic resonance solution structure of the mixed disulfide between Escherichia coli glutaredoxin(C14S) and glutathione. J. Mol. Biol. 1994;235(5):1585–1597. doi: 10.1006/jmbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 34.Martin J.L. Thioredoxin--a fold for all reasons. Structure. 1995;3(3):245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 35.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2000;2(4):811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 36.Fernandes A.P., Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 2004;6(1):63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 37.Storz G., Tartaglia L.A., Ames B.N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248(4952):189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 38.Toledano M.B., Kullik I., Trinh F., Baird P.T., Schneider T.D., Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78(5):897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 39.Vazquez-Torres A. Redox active thiol sensors of oxidative and nitrosative stress. Antioxid. Redox Signal. 2012;17(9):1201–1214. doi: 10.1089/ars.2012.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S.E. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M., Aslund F., Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279(5357):1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 42.Lee C., Lee S.M., Mukhopadhyay P., Kim S.J., Lee S.C., Ahn W.S., Yu M.H., Storz G., Ryu S.E. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 2004;11(12):1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 43.Aslund F., Zheng M., Beckwith J., Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA. 1999;96(11):6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood Z.A., Schroder E., Robin Harris J., Poole L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28(1):32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 45.Neumann C.A., Cao J., Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8(24):4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvis R.M., Hughes S.M., Ledgerwood E.C. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53(7):1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Chae H.Z., Robison K., Poole L.B., Church G., Storz G., Rhee S.G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA. 1994;91(15):7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall A., Karplus P.A., Poole L.B. Typical 2-Cys peroxiredoxins--structures, mechanisms, and functions. Febs J. 2009;276(9):2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall A., Nelson K., Poole L.B., Karplus P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15(3):795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood Z.A., Poole L.B., Hantgan R.R., Karplus P.A. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41(17):5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 51.Pace P.E., Peskin A.V., Han M.H., Hampton M.B., Winterbourn C.C. Hyperoxidized peroxiredoxin 2 interacts with the protein disulfide- isomerase ERp46. Biochem. J. 2013;453(3):475–485. doi: 10.1042/BJ20130030. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y.J., Lee W.S., Ip C., Chae H.Z., Park E.M., Park Y.M. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66(14):7136–7142. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- 53.Mu Z.M., Yin X.Y., Prochownik E.V. Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J. Biol. Chem. 2002;277(45):43175–43184. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 54.Kim S.Y., Kim T.J., Lee K.Y. A novel function of peroxiredoxin 1 (Prx-1) in apoptosis signal-regulating kinase 1 (ASK1)-mediated signaling pathway. FEBS Lett. 2008;582(13):1913–1918. doi: 10.1016/j.febslet.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 55.Rawat S.J., Creasy C.L., Peterson J.R., Chernoff J. The tumor suppressor Mst1 promotes changes in the cellular redox state by phosphorylation and inactivation of peroxiredoxin-1 protein. J. Biol. Chem. 2013;288(12):8762–8771. doi: 10.1074/jbc.M112.414524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morinaka A., Funato Y., Uesugi K., Miki H. Oligomeric peroxiredoxin-I is an essential intermediate for p53 to activate MST1 kinase and apoptosis. Oncogene. 2011;30(40):4208–4218. doi: 10.1038/onc.2011.139. [DOI] [PubMed] [Google Scholar]
- 57.Nassour H., Wang Z., Saad A., Papaluca A., Brosseau N., Affar el B., Alaoui-Jamali M.A., Ramotar D. Peroxiredoxin 1 interacts with and blocks the redox factor APE1 from activating interleukin-8 expression. Sci. Rep. 2016;6:29389. doi: 10.1038/srep29389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hopkins B.L., Nadler M., Skoko J.J., Bertomeu T., Pelosi A., Shafaei P.M., Levine K., Schempf A., Pennarun B., Yang B., Datta D., Bucur O., Ndebele K., Oesterreich S., Yang D., Rizzo M. Giulia, Khosravi-Far R., Neumann C.A. A peroxidase peroxiredoxin 1-specific redox regulation of the novel FOXO3 microRNA target let-7. Antioxid. Redox Signal. 2018;28(1):62–77. doi: 10.1089/ars.2016.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobotta M.C., Liou W., Stocker S., Talwar D., Oehler M., Ruppert T., Scharf A.N., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11(1):64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 60.Bourgeais J., Gouilleux-Gruart V., Gouilleux F. Oxidative metabolism in cancer: a STAT affair? JAKSTAT. 2013;2(4):e25764. doi: 10.4161/jkst.25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim C.P., Cao X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006;2(11):536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 63.Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., Dong C. STAT3 regulates the cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282(13):9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 64.Carballo M., Conde M., El Bekay R., Martin-Nieto J., Camacho M.J., Monteseirin J., Conde J., Bedoya F.J., Sobrino F. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J. Biol. Chem. 1999;274(25):17580–17586. doi: 10.1074/jbc.274.25.17580. [DOI] [PubMed] [Google Scholar]
- 65.Li L., Wei W., Zhang Y., Tu G., Zhang Y., Yang J., Xing Y. SirT1, and STAT3 protect retinal pigmented epithelium cells against oxidative stress. Mol. Med. Rep. 2015;12(2):2231–2238. doi: 10.3892/mmr.2015.3570. [DOI] [PubMed] [Google Scholar]
- 66.Reed D.K., Arany I. Sex hormones differentially modulate STAT3-dependent antioxidant responses during oxidative stress in renal proximal tubule cells. In Vivo. 2014;28(6):1097–1100. [PubMed] [Google Scholar]
- 67.Yoon S., Woo S.U., Kang J.H., Kim K., Kwon M.H., Park S., Shin H.J., Gwak H.S., Chwae Y.J. STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy. 2010;6(8):1125–1138. doi: 10.4161/auto.6.8.13547. [DOI] [PubMed] [Google Scholar]
- 68.Darnell J.E., Jr. STATs and gene regulation. Science. 1997;277(5332):1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 69.Burgering B.M. A brief introduction to FOXOlogy. Oncogene. 2008;27(16):2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 70.Huang H., Tindall D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007;120(Pt 15):2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 71.Singh A., Plati J., Khosravi-Far R. Harnessing the tumor suppressor function of FOXO as an alternative therapeutic approach in cancer. Curr. Drug Targets. 2011;12(9):1311–1321. doi: 10.2174/138945011796150271. [DOI] [PubMed] [Google Scholar]
- 72.Yang J.Y., Hung M.C. Deciphering the role of forkhead transcription factors in cancer therapy. Curr. Drug Targets. 2011;12(9):1284–1290. doi: 10.2174/138945011796150299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plati J., Bucur O., Khosravi-Far R. Dysregulation of apoptotic signaling in cancer: molecular mechanisms and therapeutic opportunities. J. Cell. Biochem. 2008;104(4):1124–1149. doi: 10.1002/jcb.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lant B., Storey K.B. An overview of stress response and hypometabolic strategies in Caenorhabditis elegans: conserved and contrasting signals with the mammalian system. Int. J. Biol. Sci. 2010;6(1):9–50. doi: 10.7150/ijbs.6.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myatt S.S., Brosens J.J., Lam E.W. Sense and sensitivity: FOXO and ROS in cancer development and treatment. Antioxid. Redox Signal. 2011;14(4):675–687. doi: 10.1089/ars.2010.3383. [DOI] [PubMed] [Google Scholar]
- 76.Vurusaner B., Poli G., Basaga H. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic. Biol. Med. 2012;52(1):7–18. doi: 10.1016/j.freeradbiomed.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 77.Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 78.Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villen J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K., Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125(5):987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 79.Tikhanovich I., Kuravi S., Campbell R.V., Kharbanda K.K., Artigues A., Villar M.T., Weinman S.A. Regulation of FOXO3 by phosphorylation and methylation in hepatitis C virus infection and alcohol exposure. Hepatology. 2014;59(1):58–70. doi: 10.1002/hep.26618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ho K.K., McGuire V.A., Koo C.Y., Muir K.W., de Olano N., Maifoshie E., Kelly D.J., McGovern U.B., Monteiro L.J., Gomes A.R., Nebreda A.R., Campbell D.G., Arthur J.S., Lam E.W. Phosphorylation of FOXO3a on Ser-7 by p38 promotes its nuclear localization in response to doxorubicin. J. Biol. Chem. 2012;287(2):1545–1555. doi: 10.1074/jbc.M111.284224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sunayama J., Tsuruta F., Masuyama N., Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005;170(2):295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh A., Ye M., Bucur O., Zhu S., Tanya Santos M., Rabinovitz I., Wei W., Gao D., Hahn W.C., Khosravi-Far R. Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol. Biol. Cell. 2010;21(6):1140–1152. doi: 10.1091/mbc.E09-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao J., Schulte J., Knight A., Leslie N.R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C.A. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28(10):1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turner-Ivey B., Manevich Y., Schulte J., Kistner-Griffin E., Jezierska-Drutel A., Liu Y., Neumann C.A. Role for Prdx1 as a specific sensor in redox-regulated senescence in breast cancer. Oncogene. 2013;32(45):5302–5314. doi: 10.1038/onc.2012.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang S.W., Chang T.S., Lee T.H., Kim E.S., Yu D.Y., Rhee S.G. Cytosolic peroxiredoxin attenuates the activation of Jnk and p38 but potentiates that of Erk in Hela cells stimulated with tumor necrosis factor-alpha. J. Biol. Chem. 2004;279(4):2535–2543. doi: 10.1074/jbc.M307698200. [DOI] [PubMed] [Google Scholar]
- 86.Putker M., Vos H.R., van Dorenmalen K., de Ruiter H., Duran A.G., Snel B., Burgering B.M., Vermeulen M., Dansen T.B. Evolutionary acquisition of cysteines determines FOXO paralog-specific redox signaling. Antioxid. Redox Signal. 2015;22(1):15–28. doi: 10.1089/ars.2014.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arrigo A.P. Gene expression and the thiol redox state. Free Radic. Biol. Med. 1999;27(9–10):936–944. doi: 10.1016/s0891-5849(99)00175-6. [DOI] [PubMed] [Google Scholar]
- 88.Holmgren A. Thioredoxin. Annu. Rev. Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 89.Urban M.B., Schreck R., Baeuerle P.A. NF-kappa B contacts DNA by a heterodimer of the p50 and p65 subunit. Embo J. 1991;10(7):1817–1825. doi: 10.1002/j.1460-2075.1991.tb07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lenardo M.J., Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58(2):227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 91.Lingappan K. NF-kappaB in oxidative. Stress, Curr. Opin. Toxicol. 2018;7:81–86. doi: 10.1016/j.cotox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matthews J.R., Wakasugi N., Virelizier J.L., Yodoi J., Hay R.T. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20(15):3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hayashi T., Ueno Y., Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J. Biol. Chem. 1993;268(15):11380–11388. [PubMed] [Google Scholar]
- 94.Nishi T., Shimizu N., Hiramoto M., Sato I., Yamaguchi Y., Hasegawa M., Aizawa S., Tanaka H., Kataoka K., Watanabe H., Handa H. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J. Biol. Chem. 2002;277(46):44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 95.Hirota K., Murata M., Sachi Y., Nakamura H., Takeuchi J., Mori K., Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. J. Biol. Chem. 1999;274(39):27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 96.Demple B., Sung J.S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair. 2005;4(12):1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 97.Walker L.J., Robson C.N., Black E., Gillespie D., Hickson I.D. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell Biol. 1993;13(9):5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ando K., Hirao S., Kabe Y., Ogura Y., Sato I., Yamaguchi Y., Wada T., Handa H. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kappaB and AP-1, and activation of their DNA-binding activity. Nucleic Acids Res. 2008;36(13):4327–4336. doi: 10.1093/nar/gkn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okamoto T., Asamitsu K., Tetsuka T. Thioredoxin and mechanism of inflammatory response. Methods Enzymol. 2002;347:349–360. doi: 10.1016/s0076-6879(02)47035-8. [DOI] [PubMed] [Google Scholar]
- 100.Okamoto T., Sanda T., Asamitsu K. NF-kappa B signaling and carcinogenesis. Curr. Pharm. Des. 2007;13(5):447–462. doi: 10.2174/138161207780162944. [DOI] [PubMed] [Google Scholar]
- 101.Suske G. The Sp-family of transcription factors. Gene. 1999;238(2):291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 102.Wu X., Bishopric N.H., Discher D.J., Murphy B.J., Webster K.A. Physical and functional sensitivity of zinc finger transcription factors to redox change. Mol. Cell Biol. 1996;16(3):1035–1046. doi: 10.1128/mcb.16.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kadonaga J.T., Carner K.R., Masiarz F.R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 104.Ryu H., Lee J., Zaman K., Kubilis J., Ferrante R.J., Ross B.D., Neve R., Ratan R.R. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23(9):3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chhunchha B., Fatma N., Bhargavan B., Kubo E., Kumar A., Singh D.P. Specificity protein, Sp1-mediated increased expression of Prdx6 as a curcumin-induced antioxidant defense in lens epithelial cells against oxidative stress. Cell Death Dis. 2011;2:e234. doi: 10.1038/cddis.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bloomfield K.L., Osborne S.A., Kennedy D.D., Clarke F.M., Tonissen K.F. Thioredoxin-mediated redox control of the transcription factor Sp1 and regulation of the thioredoxin gene promoter. Gene. 2003;319:107–116. doi: 10.1016/s0378-1119(03)00799-6. [DOI] [PubMed] [Google Scholar]
- 107.Tonissen K.F., Wells J.R. Isolation and characterization of human thioredoxin-encoding genes. Gene. 1991;102(2):221–228. doi: 10.1016/0378-1119(91)90081-l. [DOI] [PubMed] [Google Scholar]
- 108.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Akerman S.E., Muller S. 2-Cys peroxiredoxin PfTrx-Px1 is involved in the antioxidant defence of Plasmodium falciparum. Mol. Biochem. Parasitol. 2003;130(2):75–81. doi: 10.1016/s0166-6851(03)00161-0. [DOI] [PubMed] [Google Scholar]
- 110.Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]