Abstract
Mitofusins (Mfn) are the important regulators of mitochondrial organization in mammalian cells; however, their roles during oocyte development remain unknown. In the present study, we generated mice with oocyte-specific knockout of Mfn1 or Mfn2 (Mfn1fl/fl;Zp3-Cre or Mfn2fl/fl;Zp3-Cre). We report that deletion of Mfn1, but not Mfn2, in oocytes leads to female mice sterility, associated with the defective folliculogenesis and impaired oocyte quality. In specific, follicles are arrested at secondary stage in Mfn1fl/fl;Zp3-Cre mice, accompanying with the reduced proliferation of granulosa cells. Moreover, alterations of mitochondrial structure and distribution pattern are readily observed in Mfn1-null oocytes. Consistent with this, mitochondrial activity and function are severely disrupted in oocytes from Mfn1fl/fl;Zp3-Cre mice. In addition, the differentially expressed genes in Mfn1-deleted oocytes are also identified by whole-transcriptome sequencing. In sum, these results demonstrate that Mfn1-modulated mitochondrial function is essential for oocyte development and folliculogenesis, providing a novel mechanism determining female fertility.
Keywords: Mitochondria, Folliculogenesis, Oocyte, Fertility, Reproduction
1. Introduction
Follicular development and oogenesis play a key role in the reproductive success, as the ovulation of healthy eggs is the necessary prerequisite to fertilization [1], [2]. To coordinate germ cell development and folliculogenesis, bidirectional communications between oocyte and somatic cells exist from the time of growth initiation until the final stages of maturation. Oocytes growth involves the accumulation of a large number of mRNAs, proteins, and organelles, while oocytes maturation consists of germinal vesicle breakdown, spindle formation, chromosome movement, and polar body extrusion. Both oocyte growth and maturation are energy-consuming processes [3], [4].
Mitochondria are essential for normal cellular functions such as energy generation, calcium homeostasis, signal transduction and apoptosis [5], [6], [7]. They synthesize the vital energy currency ATP through oxidative metabolism for many processes including folliculogenesis, oocyte maturation, fertilization, and early embryonic development [8], [9]. Emerging evidence indicates that mitochondrial function is a critical determinant of oocytes developmental potential [10], [11]. Mitochondrial dysfunction contributes to the meiotic defects in oocytes from obese mice [12], [13] and preimplantation embryo arrest in vitro [14]. Mitochondria are remarkably dynamic organelles, and their morphology is maintained by a balance between fusion and fission in mammal [15]. Drosophila fuzzy onions protein (Fzo), a large transmembrane protein with GTPase activity, is the first identified mediator in mitochondrial fusion [16]. Mitofusin1 and mitofusin2 (Mfn1 and Mfn2), the homologues of Fzo in yeast and Drosophila, are the critical regulators of mitochondrial fusionin mammalian cells [17]. Mice with deletions of Mfn1 or Mfn2 die in the midgestation [18]. Embryonic fibroblasts lacking Mfn1 and Mfn2 have defects in mitochondrial membrane potential and respiratory capacity [19]. Mutations in Mfn2 cause neurodegenerative diseases by disrupting mitochondrial organization [20]. Conditionally ablating cardiomyocyte Mfn1/2 induces mitochondrial fragmentation with eccentric remodeling and no cardiomyocyte dropout [21]. However, to date, the role of Mfn1/2 during oocyte development remains unknown.
To identify the in vivo function of Mfn1/2, oocytes specific-knockout (Mfn1fl/fl;Zp3-Cre or Mfn2fl/fl;Zp3-Cre) mouse strains were established in the present study. We discovered that specific deletion of Mfn1 in oocytes leads to female mice infertility. Furthermore, we found that Mfn1fl/fl;Zp3-Cre mice display the folliculogenesis defects and mitochondrial dysfunctions. Our findings demonstrate that Mfn1, but not Mfn2, is the crucial factor modulating mitochondrial function during oocyte development.
2. Results
2.1. Oocyte-specific deletion of Mfn1, not Mfn2, leads to female sterility in mice
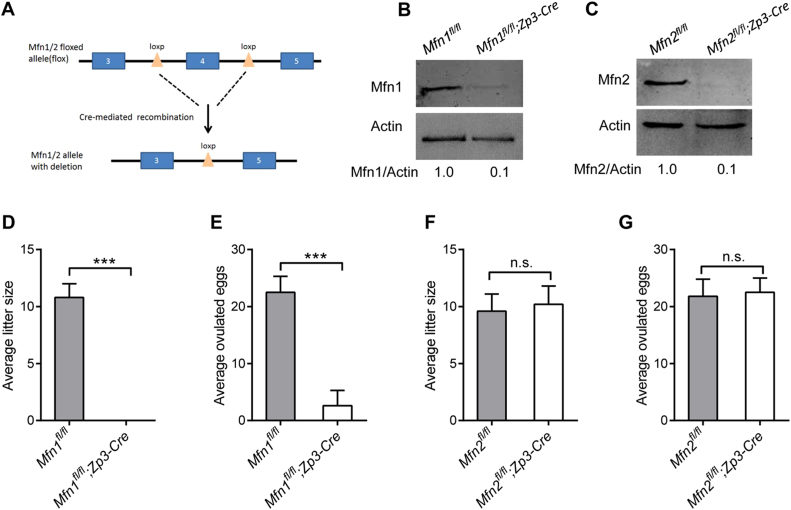
To investigate the role of Mfn1 and Mfn2 during oocyte development, we crossed Mfn1- or Mfn2-floxed mice (Mfn1fl/fl, Mfn2fl/fl) with Zp3-Cre transgenic mice to delete Mfn1/2 in growing oocytes (Fig. 1A). Immunoblotting analysis confirmed that Mfn1 and Mfn2 protein was nearly undetectable in oocytes from Mfn1fl/fl;Zp3-Cre and Mfn2fl/fl;Zp3-Cre mice, respectively (Fig. 1B-C). In fertility test, unlike Mfn1fl/fl mice, no pups were born by Mfn1fl/fl;Zp3-Cre females when crossed to wild-type (WT) males for at least 6 months, indicating Mfn1fl/fl;Zp3-Cre females are completely infertile (Fig. 1D). Similarly, no eggs were ovulated by Mfn1fl/fl;Zp3-Cre females compared to controls (Fig. 1E). In contrast, we found that the average litter size and the number of ovulate eggs in Mfn2fl/fl;Zp3-Cre mice were comparable to that in Mfn2fl/fl mice (Fig. 1F-G). Together, our results reveal that Mfn1, but not Mfn2, is crucial for female mice fertility.
Fig. 1.
Specific deletion of Mfn1 in oocytes leads to mouse infertility. (A) The schematic diagram of Mfn1/2 conditional knockout mouse construction. (B, C) The expression of Mfn1 and Mfn2 protein in oocytes from 3-week-old Mfn1/2fl/fl and Mfn1/2fl/fl;Zp3-Cre mice (100 oocyte per lane). Band intensity was calculated using ImageJ software, and the ratio of Mfn/Actin expression was normalized and values are indicated. The average litter size (D, F) and number of ovulated eggs (E, G) were evaluated in Mfn1/2fl/fl and Mfn1/2fl/fl;Zp3-Cre mice (n = 6 for each group). Data are expressed as mean ± SD. * **P < 0.001; n.s., none significant.
2.2. Mfn1fl/fl;Zp3-Cre mice display defects in follicle development
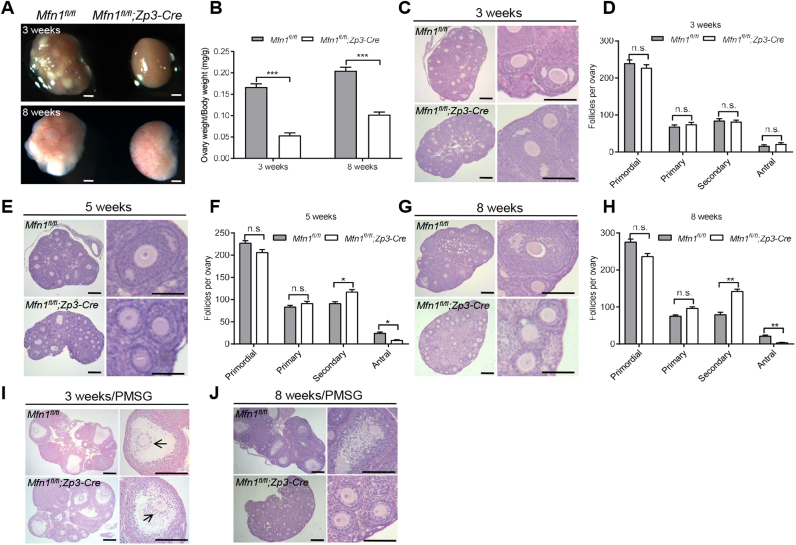
To find out how Mfn1 deletion causes female infertility, we assessed the follicle development by ovary sections. Compared with Mfn1fl/fl mice, Mfn1fl/fl;Zp3-Cre females had substantially smaller ovaries (Fig. 2A-B). There was progressive deterioration of follicular development in Mfn1fl/fl;Zp3-Cre ovaries with aging. The ovaries of 3-week-old Mfn1fl/fl;Zp3-Cre mice contained similar numbers of primordial, primary, bilayer secondary, and antral follicles relative to the control mice (Fig. 2C-D). By contrast, the ovaries of 5-week-old Mfn1fl/fl;Zp3-Cre mice were deficient in follicles beyond the secondary stage and contained fewer antral follicles than the control ovaries (Fig. 2E-F). Furthermore, at 8 weeks there were essentially no antral follicles in Mfn1fl/fl;Zp3-Cre females (Fig. 2G-H). This result might be caused by a defect of secondary-to-antral follicle transition. Next, we tested their responses to exogenous gonadotropins. Superovulation was performed on both Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre female mice. At 44 h after stimulating follicle growth with pregnant mare serum gonadotropin (PMSG), the ovaries of 3-week-old Mfn1fl/fl;Zp3-Cre mice showed the normal antral follicles with the cumulus-oocyte complex expansion (Fig. 2I, arrows); however, they still failed to ovulate following ovulation induction with human chorionic gonadotropin (hCG) (data not shown). In contrast, there were far fewer antral follicles in Mfn1fl/fl;Zp3-Cre ovaries even stimulated with PMSG (Fig. 2J). This suggested that the infertility of Mfn1fl/fl;Zp3-Cre females was due to defective follicular development and ovulation.
Fig. 2.
Mfn1fl/fl;Zp3-Cre mice display the defects in folliculogenesis. Representative images of ovaries (A) and ratio of ovary to body weight (B) from 3- and 8-week-old Mfn1fl/fland Mfn1fl/fl;Zp3-Cre mice. Histological sections of ovaries from 3-week-old (C), 5-week-old (E), and 8-week-old (G) Mfn1fl/fland Mfn1fl/fl;Zp3-Cre females. (D, F, H) Quantitative analysis of follicles at different stages in ovary sections from 3-, 5-, and 8-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice (n = 6 for each group). (I, J) Ovary histology of 3-week-old and 8-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cremice after PMSG injection (n = 4 for each group). Data are expressed as mean ± SD. *P< 0.05, **P< 0.01, ***P< 0.01, n.s., none significant. Scale bars, 100 µm.
2.3. Altered proliferation of granulosa cells in Mfn1fl/fl;Zp3-Cre mice
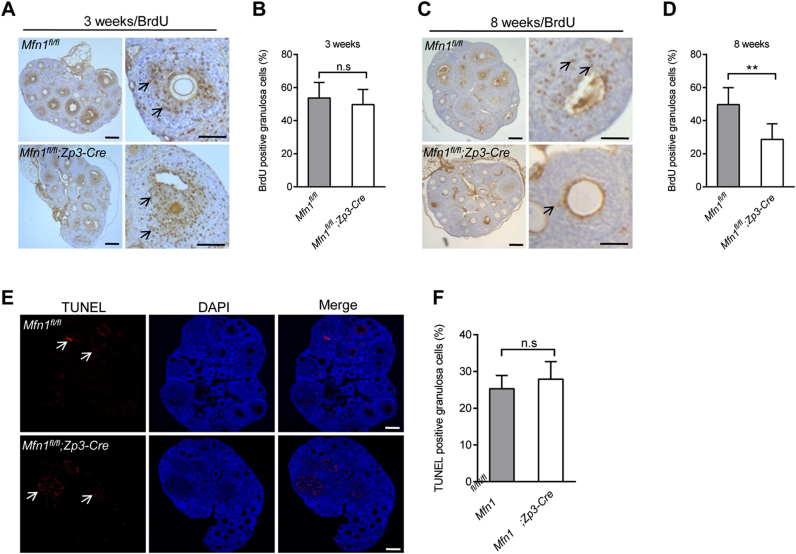
Granulosa cells provide necessary nutrients and steroids to the oocytes, playing vital roles in ovarian follicle development. To characterize the Mfn1fl/fl;Zp3-Cre ovaries at the cellular level, we examined the proliferative rate and apoptosis of granulosa cells in developing follicles based on BrdU incorporation and TUNEL assay. In antral follicles, there were no significant changes in proliferation and apoptosis of granulosa cells between Mfn1fl/fl and Mfn1fl/fl;Zp3- Cre mice (Fig. 3A-B, E-F; arrows). However, as shown in Fig. 3C-D, granulosa cells of 8-week- old Mfn1fl/fl;Zp3-Cre ovaries demonstrated the significantly decreased levels of proliferation. The results indicate that Mfn1 deletion in oocytes reduces the proliferation of surrounding somatic cells, which, in turn, might contribute to the defects of follicular development via the disruption of metabolite supply and/or the alterations in steroidogenesis [22].
Fig. 3.
The proliferation and apoptosis of granulosa cells in Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre ovary. (A, C) BrdU staining of ovaries from 3- and 8-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice. (B, D) Quantification of BrdU-positive granulosa cells in ovary sections. Arrows indicate the BrdU signals. (E) Apoptosis of granulosa cells in 8-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre ovaries was evaluated by TUNEL assay. (F) Quantitative analysis of TUNEL-positive cells in ovary section. The apoptotic cells were indicated by arrows. Data are expressed as mean ± SD. **P< 0.01, n.s., none significant. Scale bars, 100 µm.
2.4. Mfn1 deletion impairs the developmental competence of mouse oocytes
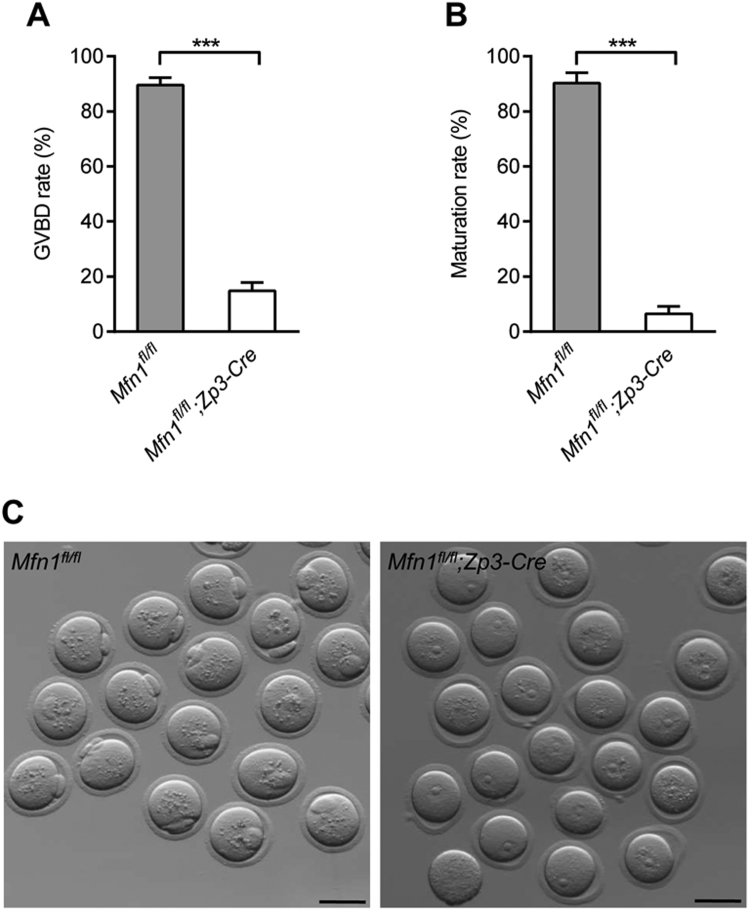
Following hormone stimulation, fully-grown oocytes reinitiate meiosis, as indicated by germinal vesicle breakdown (GVBD). With the chromatin condensation and microtubule organization, the oocytes proceed through the meiosis I division, extruding the first polar body (Pb1). Oocytes then become arrested at metaphase II (MII), waiting for fertilization. To evaluate the developmental potential of Mfn1-null oocytes, GV (germinal vesicle) oocytes isolated from 3-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice after PMSG injection were cultured in vitro. We found that GV oocytes derived from Mfn1fl/fl;Zp3-Cre mice were unable to resume meiosis normally, as evidenced by the dramatically reduced GVBD rate after 3 h culture (Fig. 4A). The ratio of Pb1 extrusion was still significantly lower in Mfn1fl/fl;Zp3- Cre oocytes than that in controls even after 14 h culture (Fig. 4B-C). Altogether, these observations imply that Mfn1 in mouse oocytes is closely associated with its developmental competence.
Fig. 4.
Oocytes from Mfn1fl/fl;Zp3-Cre mice are unable to complete meiotic maturation. Immature GV oocytes isolated from 3-week-old Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice were cultured in vitro to check the maturational progression. (A, B) Quantitative analysis of GVBD rate and Pb1 extrusion rate in Mfn1fl/fl (n = 238) and Mfn1fl/fl;Zp3-Cre (n = 214) oocytes. (C) Representative images of oocytes from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice after 14 h culture. The graph shows the mean ± SD of the results obtained in three independent experiments. ***P < 0.001. Scale bars, 80 µm.
2.5. Mitochondrial dysfunction in oocytes of Mfn1fl/fl;Zp3-Cre mice
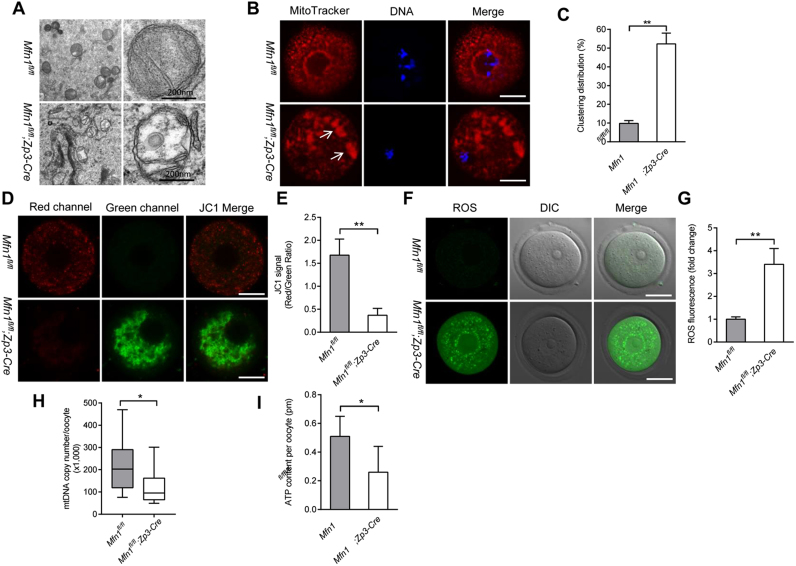
Considering the impaired quality of Mfn1fl/fl;Zp3-Cre oocytes, we decided to check whether the mitochondrial function was affected. First, transmission electron microscopy was performed on GV oocytes from 3-week-old mice. Most mitochondria in Mfn1fl/fl oocytes contain visible inner membrane, outer membrane, and well-aligned cristae. In striking contrast, ultrastructural aberrations were frequently observed in mitochondria of Mfn1fl/fl;Zp3-Cre oocytes, specifically the loss of cristae and formation of vacuoles (Fig. 5A). Mitochondria undergo stage-specific dynamic distribution during oocyte development [23], [24]. The apparent accumulation of mitochondria around nucleus was observed in the majority of normal GV oocytes, which is termed perinuclear distribution. With the completion of meiotic maturation, mitochondria display a polarized distribution pattern in oocytes. Interestingly, we noticed that the proportion of clustering mitochondrial distribution (Fig. 5B, arrows) in Mfn1fl/fl;Zp3-Cre oocytes was markedly elevated relative to controls (Fig. 5C), implying that mitochondrial organization during mouse oocyte development requires Mfn1.
Fig. 5.
Mitochondrial dysfunction in oocytes from Mfn1fl/fl;Zp3-Cre mice. (A) Representative electron micrographs of mitochondria from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes. (B) GV oocytes collected from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice were labeled with MitoTracker Red to visualize mitochondrial localization. (C) Quantification of the proportion of oocytes with clustering mitochondria distribution (n = 35 for each group). (D) Mitochondrial membrane potential in Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes was assessed by JC-1 staining. The green fluorescence shows the inactive mitochondria and the red fluorescence shows the active mitochondria in oocytes. (E) Histogram showing the JC-1 red/green fluorescence ratio (n = 20 for each group). (F) Representative images of CM-H2DCFAD (green) fluorescence in Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes. (G) Quantification of the relative ROS level (n = 20 for each group). (H) Quantitative analysis of the mtDNA copy number in single oocytes from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice (n = 28 for each group). For each box, the central bar represents the mean; upper and lower boundaries of boxes represent ± SD, and vertical lines extend to the maximal and minimal values. (I) The graph showing the ATP content per oocyte from Mfn1fl/flandMfn1fl/fl;Zp3-Cre mice (n = 30 for each group). Data are expressed as the mean ± SD from three independent experiments. *P< 0.05, **P< 0.01. Scale bars, 30 µm.
Mitochondrial membrane potential (Δψ) was assessed by JC1 staining. Fluorescent dye JC-1 shifts from green to red with increasing Δψ. As shown in Fig. 5D-E, the Δψ in oocyte was remarkably reduced when Mfn1 was deleted, indicative of the compromised mitochondrial activity. In support of this notion, we found that the reactive oxygen species (ROS) levels were increased in Mfn1fl/fl;Zp3-Cre oocytes (Fig. 5F-G), reflecting the oxidative stress. Furthermore, to evaluate the mitochondria number in oocytes, we quantified the mtDNA content. On average, Mfn1fl/fl oocytes possessed ~200,000 mtDNA molecules, consistent with a previous report [12]. However, the mtDNA copy number in Mfn1fl/fl;Zp3-Cre oocytes was reduced to ~100,000, indicating that Mfn1 deletion influences mitochondrial biogenesis or clearance during oogenesis. It has been reported that low mtDNA content is associated with the impaired quality of oocyte [25]. Mammalian oocyte displays a high turnover of ATP, which are mainly supplied by mitochondria. Consistent with this, the ATP content in Mfn1fl/fl;Zp3-Cre oocyte was lowered when compared with control cells (Fig. 5I). Taking together, these data suggest that Mfn1 is crucial for maintaining mitochondrial functions in oocytes, and thereby promoting the developmental potential.
2.6. Determination of differential expression genes in Mfn1fl/fl;Zp3-Cre oocyte
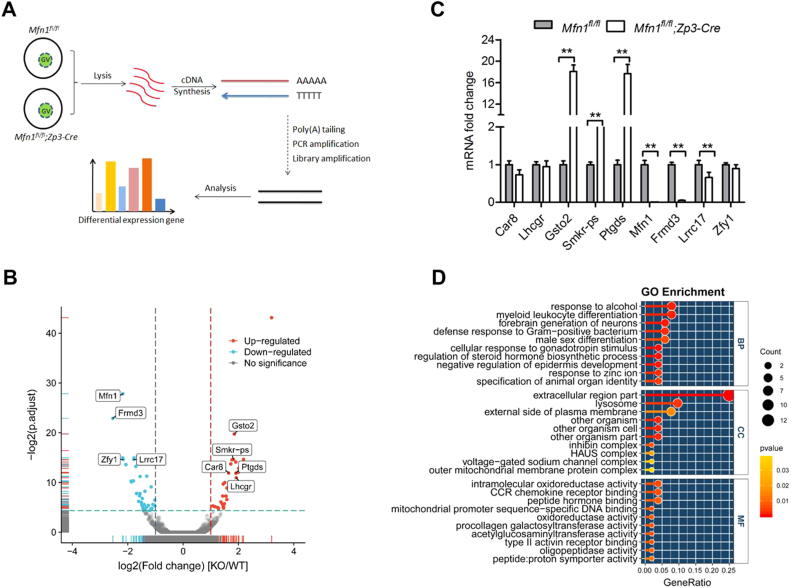
To explore the potential mechanism of Mfn1-dependent oogenesis and folliculogenesis, we performed whole-transcriptome sequencing (RNA-seq) on fully-grown GV oocytes from 3-week-old mice (Fig. 6A). Data analysis identified 70 significant differentially expressed genes, 38 of which were upregulated and 32 downregulated in Mfn1fl/fl;Zp3-Cre oocytes (Fig. 6B and Table S2). Accuracy of the sequencing data was validated by qRT-PCR of mRNA levels for randomly selected genes (Fig. 6C). Gene Ontology (GO) analysis showed that the multiple biological processes, specifically the metabolic activity, were affected (Fig. 6D). Of note, GSTO2 (glutathione S-transferase omega 2), a member of GST superfamily, exhibits glutathione-dependent thiol transferase and dehydroascorbate reductase activities characteristic of the glutaredoxins to modulate oxidative stress [26]. PTGDS (prostaglandin D synthase), coded by Ptgds gene, catalyzes the isomerization of various prostanoids to produce prostaglandin D2. It also has been reported that PTGDS potentially attenuates the activities of NADPH oxidases through interaction with NADPH to decrease the generation of excessive ROS [27]. Hence, the expression changes in these genes might mediate the effects of Mfn1 deletion on follicle/oocyte development. Our ongoing research is to clarify this issue.
Fig. 6.
Whole-genome transcription analysis of oocytes derived from Mfn1fl/fl;Zp3-Cre mice. (A) Illustration of the process of single-cell whole-transcriptome sequencing on oocyte from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice. (B) Volcano plot for the differentially expressed genes in Mfn1fl/fl;Zp3-Cre oocytes. Points labeled with the gene name represent the gene validated by qRT-PCR. (C) Validation of RNA-Seq data by quantitative RT-PCR. (D) GO analysis of the differentially expressed genes. Data are expressed as the mean ± SD from three independent experiments. **P< 0.01.
3. Discussion
The mitochondrial morphology and structure, which are remarkably changing under the control of fusion and fission to interact with each other in the different stage of cell, strongly correlate with functions such as ATP supply and signal transduction [28]. Multiple protein components identified in the remodeling of mitochondrial membranes, such as Mfn1/2 or Opa1 for fusion and Drp1 for fission, play vital roles in mitochondrial functions and development [29]. Outer membrane fusion is mediated by the mitofusins, large GTPases that traverse the outer mitochondrial membrane twice [30]. Mice with deletions of Mfn1 or Mfn2 die in the midgestation [18]. Mitochondrial fusion mediated by Mfn2 protects against neurodegenerative diseases such as peripheral neuropathy Charcot-Marie-Tooth subtype 2A [20]. Conditional deletion of Mfn2, not Mfn1, in cerebellum leads to defects in dendritic outgrowth, spine formation, and survival of Purkinje cells [31]. Moreover, by generation of Mfn1fl/fl;MVH-Cre mice, Zhang et al. found that Mfn1 deficiency leads to deficient spermatogenesis and male infertility [32]. Fusion of the inner mitochondrial membrane requires OPA1, a large GTPase tethered to the inner mitochondrial membrane facing the inter-membrane space. The function of OPA1 in fusion requires Mfn1, but interestingly not Mfn2 [33]. An emerging concept from the work to date suggests that Mfn1 has a ‘mito-centric’ role and that Mfn2 plays an important role in the interaction of mitochondria with surrounding organelles and intracellular structures [34]. In support of this notion, we found that the specific deletion of Mfn1 in oocytes results in the female sterility. However, in striking contrast, conditional knockout Mfn2 in oocytes had no effects on mouse fertility (Fig. 1). The findings suggest the differential roles of Mfn1/2 in the control of female reproduction. Our results are inconsistent with a previous report showing that microinjection of Mfn2 siRNA disturbs mouse oocyte maturation [35]. This contradiction is likely due to the different systems (in vivo knockout vs. in vitro knockdown) employed in respective studies.
By histology analysis, we noted that most follicles were arrested at the secondary stage in 5- to 8-week old Mfn1fl/fl;Zp3-Cre ovaries, resulting in the dramatic loss of antral follicles (Fig. 2). Moreover, these ovaries responded poorly to the gonadotropins stimulation (Fig. 2), indicating that Mfn1 deletion induced deficient folliculogenesis is likely independent of endocrine actions. Nonetheless, BrdU incorporation assay showed the decreased proliferation of granulosa cells in Mfn1fl/fl;Zp3-Cre ovaries (Fig. 3). The bidirectional communications existing between oocyte and somatic cells are not only required for the proliferation of granulosa cell [36], but also responsible for follicle development [37]. Therefore, it is conceivable that Mfn1 deletion may disrupt the metabolic coupling between oocytes and the surrounding cells, which in turn suppresses the proliferation of granulosa cells, consequently contributing to the developmental failure of mouse follicles.
During the transition from secondary to antral follicle in folliculogenesis, the dramatic changes of gene expression occurred in oocytes, which are crucial for oocyte development [38]. Herein, we revealed the arrest at secondary follicle stage in Mfn1fl/fl;Zp3-Cre ovary. Meanwhile, we observed the ultrastructural alterations and abnormal distribution pattern of mitochondria in Mfn1-null oocytes (Fig. 5A-B). These aberrations of mitochondrial structure in Mfn1fl/fl;Zp3-Cre oocytes are indicative of an increase of mitochondrial membrane permeability and impaired respiratory chain. Studies in mitochondria in somatic cells [39] and embryos [40], [41] have suggested that the magnitude of Δψm is associated with the level of mitochondrial respiration so the impaired respiratory function may account for the altered mitochondrial membrane potential in Mfn1fl/fl;Zp3-Cre oocytes. High frequency of mitochondrial aggregate has been shown to be associated with developmental retardation of oocytes [42]. Importantly, both ATP content and mtDNA copy number are markedly decreased in oocytes from Mfn1fl/fl;Zp3-Cre mice (Fig. 5H-I). This functional decline of mitochondria is also likely related to their structural abnormalities. Mitochondria are important source of ROS, as a by-product of oxidative phosphorylation in the cell [43]. Mitochondrial dysfunction inevitably enhances ROS production. In line with this conception, we detected the significantly elevated ROS levels in Mfn1fl/fl;Zp3-Cre oocytes (Fig. F-G). Such an oxidative stress environment may damage multiple components of the oocyte, including DNA, RNA, proteins and lipids, and thereby perturb diverse biological processes, finally impairing the developmental competence of Mfn1fl/fl;Zp3-Cre oocytes mentioned above. In addition, our whole-transcriptome sequencing data identified the differentially expressed genes in Mfn1fl/fl;Zp3-Cre oocytes. For example, glutathione S-transferase omega 2, coded by Gsto2, is involved in gamma-glutamyl cycle and has the activity of glutathione dehydrogenase to modulate redox homeostasis [44]. Hence, it is possible that upregulated expression of Gsto2 is a response to oxidative stress originated from metabolic disturbance in Mfn1-null oocytes. Our ongoing research is trying to dissect the molecular mechanisms mediating the effects of Mfn1 on follicle/oocyte development based on this RNA-Seq data.
In conclusion, we demonstrate that Mfn1, but not Mfn2, in oocytes is the critical factor affecting folliculogenesis and oocyte development. Mfn1 deletion results in the developmental arrest of follicles and mitochondrial dysfunction in oocytes. Our findings provide new insights into the mechanism determining female fertility.
4. Materials and methods
4.1. Mice
All experiments were performed in compliance with the guidelines of the Animal Care and Use Committee of Nanjing Medical University. Mfn1flox/flox and Mfn2flox/flox female micewere obtained from USA Mutant Mouse Resource & Research Centers (029901-UCD and 029902-UCD). All mice have a C57BL/6 genetic background. Primers used for genotyping are listed in Table S1.
4.2. Antibodies
Mouse monoclonal anti-Mfn1 (Cat#: ab57602) and anti-Mfn2 (Cat#: ab56889) were purchased from Abcam (Cambridge, MA, USA). Mouse monoclonal anti-β-actin antibodies (Cat#: A5441) were purchased from Sigma (St. Louis, MO, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Cat#: 31470) were purchased from Thermo Fisher Scientific. Except for those specifically stated, all chemicals and culture media in our research were purchased from Sigma.
4.3. Fertility assessment and ovarian histology analysis
For fertility tests, 8-week-old Mfn1(2)fl/fl;Zp3-Cre and Mfn1(2)fl/fl females were mated with normal 10-week-old WT males for 6 months (2 females: 1 male). The number of offspring from each pregnant female was recorded after birth. For histology analysis, ovaries were fixed in 10% formalin overnight at room temperature, embedded in paraffin, and then cut into 5–6 µm serial sections followed by staining with hematoxylin (31890610; Sigma, USA) and eosin (HHS16; Sigma, USA).
4.4. Oocytes collection and in vitro culture
To retrieve fully-grown GV oocytes, female mice were injected with 5 IU PMSG, and 48 h later, cumulus-oocytes-complex were obtained by manual rupturing of antral follicles. Denuded oocytes were obtained by repeatedly pipetting cumulus cells. For in vitro maturation, GV oocytes were cultured in M16 medium at 37 °C in a 5% CO2 incubator.
4.5. Western blotting
A total of 100 oocytes were heated at 100 °C for 5 min in protein lysis buffer (95% laemmli sample buffer and 5% β-mercaptoethanol) and stored at −20 °C until use. SDS-PAGE and immunoblots were performed following standard procedures using a Mini-PROTEAN Tetra Cell System (Bio-Rad). The protein bands were visualized using an ECL Plus Western Blotting Detection System.
4.6. BrdU incorporation and TUNEL assay
Mice were injected with BrdU (100 mg/Kg; B9285, Sigma) and sacrificed 2 h post injection. Ovary sections were immunostained with anti-BrdU antibody, and analyzed by microscope equipped with digital camera (Nikon). TUNEL assay was performed in ovary paraffin sections according to the instructions provided by the In Situ Cell Death Detection Kit, POD TUNEL (11684795910, Roche, Swiss). Fluorescence was detected using Leica DMIRB Inverted Fluorescence Microscope.
4.7. Immunofluorescence
Immunofluorescence staining was performed as we described previously [4]. Oocytes were fixedin 4% PFA for 30 min, permeabilized in 0.5% Triton-X 100 for 20 min, and then blocked in PBS with 1% BSA for 1 h. For mitochondria staining, oocytes were cultured in M2 medium containing 200 nM MitoTracker Red for 30 min at 37 °C. After counterstaining with DAPI, mitochondria distribution pattern in oocytes was observed under laser scanning confocal microscope, and the numbers of oocytes with clustering distribution pattern were recorded. Mitochondrial membrane potential was evaluated using MitoProbe JC-1 Assay Kit (M34152, Thermo, USA) following the protocol reported by Zeng et al. [45] with minor modifications. Briefly, oocytes were cultured in M2 medium with 2 μM JC-1 for 15 min at 37 °C, followed by washing with buffer for 10 min. Samples were immediately imaged in a glassbottom dish on a laser scanning confocal microscope (LSM 710; Carl Zeiss, Germany). JC-1 dye exhibits potential-dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green (~529 nm) to red (~590 nm). Therefore, mitochondrial depolarization is indicated by a decrease in the red/green fluorescence intensity ratio. Oocytes treated with 10 μM mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) for 2 h are set as positive control. The lowered red/green fluorescence ratio in CCCP-treated oocytes validates the JC-1 assay system works well in assessing membrane potential of mouse oocytes. ImageJ software was used to quantify the intensity of fluorescence.
4.8. Transmission electron microscopy
For analysis of mitochondrial structure, samples were processed as described previously [12]. Briefly, the paracentral tissue of mouse ovaries were cut into 1 mm [3] piece, and then fixed in 2.5% glutaraldehyde and 1% osmic acid, followed by dehydration and incubation in the embedding medium overnight. 70–90 nm sections were made by ultramicrotome. After stained by a lead citrate, the sections were analyzed under an electron transmission microscope at 10,000 magnifications.
4.9. ROS assessment
The ROS levels in oocytes were evaluated according to our previous report [46]. Oocytes were incubated in M2 medium containing 5 μM CM-H2DCFDA (C6827, Invitrogen) for 30 min at 37 °C in 5% CO2 incubator. Following three washes, 15 oocytes were transferred to a live cell-imaging dish, and immediately observed by using Laser Scanning Confocal (LSM700; Zeiss, Oberkochen, Germany)
4.10. Determination of mtDNA copy number and ATP content
mtDNA extraction and quantitative RT-PCR were conducted as we reported previously [12]. A single oocyte was loaded in a PCR tube with 10 µl lysis buffer and processed for qRT-PCR analysis. Five 10-fold serial dilutions of purified plasmid standard DNA were used to generate the standard curve. Mouse mtDNA-specific primers are listed in Table S1. ATP content in pools of 10–20 oocytes was measured using a Bioluminescent Somatic Cell Assay Kit (Sigma, USA) as we described previously [47]. A 5-point standard curve (0, 0. 1, 0.5, 1.0, 10, and 50 pmol of ATP) was generated in each assay and the ATP content was calculated by using the formula derived from the linear regression of the standard curve. All measurements were performed in triplicate.
4.11. Real-time RT-PCR
Total RNA was extracted from 50 oocytes using the RNeasy® Micro Kit (157030297, Invitrogen, USA) and cDNA synthesis was completed using QuanNova Reverse Transcription Kit (205311, Qiagen, Germany). qRT-PCR was conducted using a Power SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies) with ABI 7500 Real-Time PCR system (Applied Biosystems). Data were normalized against GAPDH and quantification of the fold change was determined by the comparative CT method, as we previously described [48]. The related primers are listed inTable S1.
4.12. RNA library construction and sequencing
Transcriptomic analysis of Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre GV-stage fully grown oocytes was carried out using a protocol for single cell RNA-Seqas described previously by Tang [49] with minor modifications. In brief, 3 sets of samples were collected for each genotype (3-week-old mice; 15 oocytes per sample) in lysis buffer. Reverse transcription was carried out on the whole cell lysate using SMARTer Ultra Low Input RNA for Illumina Sequencing-HV kits (Takara Bio Inc., Kusatsu, Japan) according to the manufacturer's instructions. Following amplification, cDNA was purified and subjected to library construction for the paired-end 125 bp (PE125) sequencing on Illumina HiSeq. 2500 platform.
4.13. Read alignment and differential expression analysis
Raw reads were trimmed to remove low-quality bases and adaptor sequences using Fastp (v0.18.0) with default parameters. Ribosomal RNAs were further removed by SortmeRNA (v2.1b) [50]. Filtered reads were then aligned against mouse reference (GRCm38) byapplication of Hisat2 (v2.05) software [51]. Mapped reads were processed through featureCount (v1.5.1) [52] at the gene level to account for the number of reads per gene in all samples. Differentially expressed genes between Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes were deemed significant using DESeq. 2 (1.14.1) [53] package of R at a |log2FoldChange|>1 and a false discovery rate (FDR)-adjusted p-value (p.adjust) < 0.05.
4.14. Statistical analysis
Data are presented as mean ± SD, unless otherwise indicated. Differences between two groups were analyzed by Student's t-test. Multiple comparisons were analyzed by one-way ANOVA test using GraphPad Prism 7.0. P < 0.05 was considered to be significant.
Acknowledgements
This work was supported by National Key R&D Program of China (NO. 2017YFC1001500 to QW and 2016YFA0503300 to XG), National Natural Science Foundation of China (NO. 31771657 and 31571543 to QW, and 81771641 to XG), the Science Foundation for Distinguished Young Scholars of Jiangsu Province (BK20180035 to QW).
Acknowledgments
Conflict of interest
The authors have nothing to disclose.
Author contributions
QW designed research; XH, SZ, CL, DQ, and JG performed research; XH, SZ, HZ, XG, and QW analyzed data; SZ and QW wrote paper.
Data availability
All sequencing data were deposited in the sequence read archive (SRA) database and the BioProject ID is PRJNA504853.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101110.
Contributor Information
Xuejiang Guo, Email: guo_xuejiang@njmu.edu.cn.
Qiang Wang, Email: qwang2012@njmu.edu.cn.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Baerwald A.R., Adams G.P., Pierson R.A. Ovarian antral folliculogenesis during the human menstrual cycle: a review. Hum. Reprod. Update. 2012;18:73–91. doi: 10.1093/humupd/dmr039. [DOI] [PubMed] [Google Scholar]
- 2.Keefe D., Kumar M., Kalmbach K. Oocyte competency is the key to embryo potential. Fertil. Steril. 2015;103:317–322. doi: 10.1016/j.fertnstert.2014.12.115. [DOI] [PubMed] [Google Scholar]
- 3.Gu L. Metabolic control of oocyte development: linking maternal nutrition and reproductive outcomes. Cell. Mol. life Sci. 2015;72:251–271. doi: 10.1007/s00018-014-1739-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng J. SIRT4 is essential for metabolic control and meiotic structure during mouse oocyte maturation. Aging Cell. 2018:e12789. doi: 10.1111/acel.12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L., Kepp O., Trojel-Hansen C., Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 2012;111:1198–1207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 6.Wallace D.C., Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Songsasen N. Dynamic changes in mitochondrial DNA, distribution and activity within cat oocytes during folliculogenesis. Reprod. Domest. Anim. = Zuchthyg. 2017;52(Suppl 2):71–76. doi: 10.1111/rda.12851. [DOI] [PubMed] [Google Scholar]
- 9.Ge H. The importance of mitochondrial metabolic activity and mitochondrial DNA replication during oocyte maturation in vitro on oocyte quality and subsequent embryo developmental competence. Mol. Reprod. Dev. 2012;79:392–401. doi: 10.1002/mrd.22042. [DOI] [PubMed] [Google Scholar]
- 10.Fissore R.A., Kurokawa M., Knott J., Zhang M., Smyth J. Mechanisms underlying oocyte activation and postovulatory ageing. Reproduction. 2002;124:745–754. doi: 10.1530/rep.0.1240745. [DOI] [PubMed] [Google Scholar]
- 11.Cecchino G.N., Seli E., Alves da Motta E.L., Garcia-Velasco J.A. The role of mitochondrial activity in female fertility and assisted reproductive technologies: overview and current insights. Reprod. Biomed. Online. 2018;36:686–697. doi: 10.1016/j.rbmo.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q. Maternal diabetes causes mitochondrial dysfunction and meiotic defects in murine oocytes. Mol. Endocrinol. 2009;23:1603–1612. doi: 10.1210/me.2009-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grindler N.M., Moley K.H. Maternal obesity, infertility and mitochondrial dysfunction: potential mechanisms emerging from mouse model systems. Mol. Hum. Reprod. 2013;19:486–494. doi: 10.1093/molehr/gat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thouas G.A., Trounson A.O., Wolvetang E.J., Jones G.M. Mitochondrial dysfunction in mouse oocytes results in preimplantation embryo arrest in vitro. Biol. Reprod. 2004;71:1936–1942. doi: 10.1095/biolreprod.104.033589. [DOI] [PubMed] [Google Scholar]
- 15.Chen H., Chan D.C. Mitochondrial dynamics in mammals. Curr. Top. Dev. Biol. 2004;59:119–144. doi: 10.1016/S0070-2153(04)59005-1. [DOI] [PubMed] [Google Scholar]
- 16.Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 17.Santel A., Fuller M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 18.Chen H. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., Chomyn A., Chan D.C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 20.Zuchner S. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 21.Song M., Mihara K., Chen Y., Scorrano L., Dorn G.W., 2nd Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–286. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanderhyden B.C., Macdonald E.A. Mouse oocytes regulate granulosa cell steroidogenesis throughout follicular development. Biol. Reprod. 1998;59:1296–1301. doi: 10.1095/biolreprod59.6.1296. [DOI] [PubMed] [Google Scholar]
- 23.Van Blerkom J., Davis P., Alexander S. Differential mitochondrial distribution in human pronuclear embryos leads to disproportionate inheritance between blastomeres: relationship to microtubular organization, ATP content and competence. Hum. Reprod. 2000;15:2621–2633. doi: 10.1093/humrep/15.12.2621. [DOI] [PubMed] [Google Scholar]
- 24.Nishi Y., Takeshita T., Sato K., Araki T. Change of the mitochondrial distribution in mouse ooplasm during in vitro maturation. J. Nippon Med. Sch. = Nippon Ika Daigaku zasshi. 2003;70:408–415. doi: 10.1272/jnms.70.408. [DOI] [PubMed] [Google Scholar]
- 25.May-Panloup P., Chretien M.F., Malthiery Y., Reynier P. Mitochondrial DNA in the oocyte and the developing embryo. Curr. Top. Dev. Biol. 2007;77:51–83. doi: 10.1016/S0070-2153(06)77003-X. [DOI] [PubMed] [Google Scholar]
- 26.Board P.G. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J. Biol. Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 27.Qin S. Thermodynamic and NMR analyses of NADPH binding to lipocalin-type prostaglandin D synthase. Biochem. Biophys. Res. Commun. 2015;468:234–239. doi: 10.1016/j.bbrc.2015.10.124. [DOI] [PubMed] [Google Scholar]
- 28.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 29.Chan D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 30.Rojo M., Legros F., Chateau D., Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- 31.Chen H., McCaffery J.M., Chan D.C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J. GASZ and mitofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep. 2016;17:220–234. doi: 10.15252/embr.201540846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandhok G., Lazarou M., Neumann B. Structure, function, and regulation of mitofusin-2 in health and disease. Biol. Rev. Camb. Philos. Soc. 2018;93:933–949. doi: 10.1111/brv.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J.H. Mitofusin-2 is required for mouse oocyte meiotic maturation. Sci. Rep. 2016;6:30970. doi: 10.1038/srep30970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilchrist R.B., Lane M., Thompson J.G. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Human. Reprod. Update. 2008;14:159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 37.Su Y.Q., Sugiura K., Eppig J.J. Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin. Reprod. Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan Z. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J. Genet. Genom. = Yi chuan xue bao. 2012;39:111–123. doi: 10.1016/j.jgg.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Duchen M.R. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Asp. Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Wilding M. Energy substrates, mitochondrial membrane potential and human preimplantation embryo division. Reprod. Biomed. Online. 2002;5:39–42. doi: 10.1016/s1472-6483(10)61595-7. [DOI] [PubMed] [Google Scholar]
- 41.Acton B.M., Jurisicova A., Jurisica I., Casper R.F. Alterations in mitochondrial membrane potential during preimplantation stages of mouse and human embryo development. Mol. Human. Reprod. 2004;10:23–32. doi: 10.1093/molehr/gah004. [DOI] [PubMed] [Google Scholar]
- 42.Nagai S. Correlation of abnormal mitochondrial distribution in mouse oocytes with reduced developmental competence. Tohoku J. Exp. Med. 2006;210:137–144. doi: 10.1620/tjem.210.137. [DOI] [PubMed] [Google Scholar]
- 43.Tiwari M. Apoptosis in mammalian oocytes: a review. Apoptosis: Int. J. Program. Cell Death. 2015;20:1019–1025. doi: 10.1007/s10495-015-1136-y. [DOI] [PubMed] [Google Scholar]
- 44.Masoudi M., Saadat I., Omidvari S., Saadat M. Genetic polymorphisms of GSTO2, GSTM1, and GSTT1 and risk of gastric cancer. Mol. Biol. Rep. 2009;36:781–784. doi: 10.1007/s11033-008-9245-0. [DOI] [PubMed] [Google Scholar]
- 45.Zeng H.T. Heparin and cAMP modulators interact during pre-in vitro maturation to affect mouse and human oocyte meiosis and developmental competence. Hum. Reprod. 2013;28:1536–1545. doi: 10.1093/humrep/det086. [DOI] [PubMed] [Google Scholar]
- 46.Han L. Melatonin protects against maternal obesity-associated oxidative stress and meiotic defects in oocytes via the SIRT3-SOD2-dependent pathway. J. Pineal Res. 2017;63 doi: 10.1111/jpi.12431. [DOI] [PubMed] [Google Scholar]
- 47.Hou X. Differing roles of pyruvate dehydrogenase kinases during mouse oocyte maturation. J. Cell Sci. 2015;128:2319–2329. doi: 10.1242/jcs.167049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han L. Embryonic defects induced by maternal obesity in mice derive from Stella insufficiency in oocytes. Nat. Genet. 2018;50:432–442. doi: 10.1038/s41588-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 49.Tang F. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kopylova E., Noe L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 51.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
All sequencing data were deposited in the sequence read archive (SRA) database and the BioProject ID is PRJNA504853.