Abstract
When training a machine learning algorithm for a supervised-learning task in some clinical applications, uncertainty in the correct labels of some patients may adversely affect the performance of the algorithm. For example, even clinical experts may have less confidence when assigning a medical diagnosis to some patients because of ambiguity in the patient’s case or imperfect reliability of the diagnostic criteria. As a result, some cases used in algorithm training may be mis-labeled, adversely affecting the algorithm’s performance. However, experts may also be able to quantify their diagnostic uncertainty in these cases. We present a robust method implemented with Support Vector Machines to account for such clinical diagnostic uncertainty when training an algorithm to detect patients who develop the acute respiratory distress syndrome (ARDS). ARDS is a syndrome of the critically ill that is diagnosed using clinical criteria known to be imperfect. We represent uncertainty in the diagnosis of ARDS as a graded weight of confidence associated with each training label. We also performed a novel time-series sampling method to address the problem of inter-correlation among the longitudinal clinical data from each patient used in model training to limit overfitting. Preliminary results show that we can achieve meaningful improvement in the performance of algorithm to detect patients with ARDS on a hold-out sample, when we compare our method that accounts for the uncertainty of training labels with a conventional SVM algorithm.
Keywords: Machine learning, support vector machine, label uncertainty, acute respiratory distress syndrome, sampling from longitudinal electronic health records (EHR)
I. Introduction
The Acute Respiratory Distress Syndrome (ARDS) is a critical illness syndrome affecting 200,000 patients in United States each year [1]. While the mortality rate of patients with ARDS is 30%, multiple evidence-based management strategies can be provided to patients with ARDS to improve their outcomes [2]. However, recent evidence suggests that patients with ARDS are not recognized when they develop this syndrome, and consequently, do not receive the evidence-based therapies proven to reduce mortality [3]. The inability of healthcare providers to process the massive streams of clinical data generated while caring for these patients has been specifically cited as a potential reason for poor ARDS recognition [4]. Algorithms that analyze electronic health record (EHR) data and alert providers when patients develop signs of ARDS have been proposed as a potential way to improve early ARDS detection [5, 6].
At present, simple rule-based electronic algorithms have been described that analyze EHR data to screen patients for ARDS [7, 8]. Current systems search the text of radiology reports for language consistent with ARDS to identify patients. For these systems to be successful, however, chest imaging must be obtained at the time when ARDS develops and a radiologist must accurately interpret the radiology image in a timely manner using language that could be interpreted as consistent with ARDS. These dependencies are problematic for successful implementation in clinical practice. Systems that rely solely on routinely collected clinical data to identify at risk patients could alert clinicians to those patients who warrant further evaluation, specifically triggering chest imaging for timely ARDS diagnosis.
An additional challenge in the development of an ARDS detection algorithm is the creation of reference patient cohorts to train the algorithm. ARDS is a clinical diagnosis that requires a nuanced interpretation of each patient’s clinical data by clinical experts. Some patients are difficult to classify with available clinical data even for highly trained experts [9]. Previous research has shown how errors in the labeling of ARDS and non-ARDS patients can substantially degrade clinical study results [10]. One potential solution is to allow clinical experts to classify patients as equivocal when a diagnosis of ARDS is uncertain. Using this approach, researchers have previously shown that known ARDS risk factors have stronger associations with ARDS development when equivocal patients were excluded [11].
When training an algorithm to detect ARDS, rather than excluding patients with diagnostic uncertainty, an alternative approach is to use this additional infonnation about diagnostic certainty during training, which could lead to more efficiently learning and better generalize to new patient cases. Learning with uncertainty is a recent machine learning paradigm that may be well suited for the task of training an ARDS detection algorithm [12]. The standard machine-learning classification task is to learn a function f(x) : X → Y, which maps input training data x ∈ X to class y ∈ Y, where X represents a feature space of each patient’s covariates and Y is the classification label. The model is trained on well-defined input data of labeled training examples. However, in certain clinical applications, there may be uncertainty in the training labels themselves that could adversely affect model training. In the example of ARDS, there may be challenging cases where the physician has difficulty determining a patient’s diagnosis due to clinical ambiguity. As a result, this uncertainty and subsequent mislabeling of training data could adversely affect model training.
Varying methods have been proposed to address the issue of training with label uncertainty. Frenay and Verleysen considered uncertainty as a stochastic process of noise in the label and proposed a statistical taxonomy of definitions for various label noise typically presented in classification with machine learning [13]. Natarajan et al addressed the challenges of learning with noisy labels and developed an algorithm for risk minimization under certain conditions using an unbiased estimator and logistic regression to account for labels independently corrupted by random noise [14]. Duan and Wu proposed the concept of flipping probability used to model inaccurate labels in real-world applications [15] and suggested several methods to optimize for noise tolerance. Vembu and Zilles developed an iterative learning scheme to address label uncertainty, which they recognize as disagreement among annotators in generation of classification labels [16].
Although these methods propose novel solutions to address label uncertainty, many of them are theoretical and were not tested on real-world data (primarily benchmarked on artificially generated data and referenced datasets) or simply consider uncertainty in the label as random noise. Such an approach may not be well suited for biomedical or clinical applications where a clinical expert might also be able to provide a level of confidence in a patient’s label.In the current study, when clinical experts reviewed each patients’ clinical data to determine whether they developed ARDS, they also provide their level of confidence in the diagnosis. This uncertainty rating was represented as the confidence of the label’s annotation. Using a support vector machines learning model, the confidence weighting of the label is used as additional information in the training process. This approach is a form of instance-weighted SVM, although instead of learning weights based on characteristics of the data [17], or weights based on the class label [18], we use a clinical expert’s confidence in the diagnosis weights during SVM training. This approach incorporates a more realistic representation of uncertainty in real-world applications, avoids discarding uncertain data, and balances the influence of such uncertain inputs in the learning algorithm.
The current study also addresses the problem of using highly correlated longitudinal clinical data in machine-learning model training, which is often ignored in applications of machine learning in biomedical domains. With the increased use of electronic health records, clinical data are often available in a longitudinal format, where specific metrics of health (e.g. vital signs, or laboratory values) are measured intermittently over time. Analysis of such data requires additional consideration of the stochastic dependency and time-series nature of these data [19], and they should not be considered independent and identically distributed (i.i.d.) [20], as the data is obviously not. By ignoring the inter-dependency of the time-series data and the i.i.d assumption, training may result in a biased model that overfits to the available data and yield unrealistically large values of specificity, sensitivity, and AUROC [21, 22]. Methods exist to deal with correlated data in traditional machine learning, such as using a Markov switching process model [23], or partially linear regression model [24] for longitudinal time-series data analysis or a correlation-based fast filter method [25] for choosing among highly correlated features in the model selection process. Beyond the scope of generalized machine learning problems, additional methods to analyze time series properties exist in many domain-specific applications, such as stock market prediction with support vector machine and case-based reasoning [26], or time-delay neural networks [27] and dynamic time warping [28] for speech recognition.
Several techniques, such as dynamic sampling within Markov chain Monte Carlo methods [29] and Bayesian Changepoint Detection [30], are established for analyzing the dependency structure of multivariate time series data. However, methods addressing stochastic dependency are largely underdeveloped for applications on longitudinal clinical data. We address the problem by viewing patients’ time-series data as a mixing process and consider the data structure as a stationary process with exponentially weakening dependency, and sample instances in a strategic manner to minimize intercorrelation. This approach provides a way to measure the decay in correlation [31] among data on an individual patient over time, and informs a novel sampling strategy to minimize the correlation among data sampled from the same patient for model training.
II. Methods
A. Data Generation
The patient cohort included consecutive adult patients hospitalized in January of 2016 with moderate hypoxia, defined as requiring more than 3 L of supplemental oxygen by nasal cannula for at least 2 hours. The cohort was enriched with additional patients who developed acute hypoxic respiratory failure (PaO2/FiO2 ratio of < 300 mm Hg while receiving invasive mechanical ventilation) in February and March of 2016 who are higher risk for developing ARDS. In total, 401 patients were used to develop the ARDS detection algorithm.
A group of expert clinicians reviewed all patients for the development of ARDS based on the Berlin definition [32]. As ARDS is a clinical diagnosis without a simple gold standard, we were unable to benchmark expert performance. However, because the inter-rater reliability of ARDS diagnosis is known to be only moderate in patients with acute hypoxic respiratory failure [33], these patients were reviewed independently by 3 experts, and their ratings were averaged. In addition to determining whether the diagnosis was present (yes or no) and record the time of ARDS onset among positive cases, the experts were also asked to provide their confidence level in the diagnosis label (high, moderate, low, equivocal). This 4-point confidence scale was carefully tested on the experts prior to use in this study, and felt to reasonably capture the range of uncertain that they might have when reviewing patient cases. Their diagnosis label and confidence level could then be converted to a 1-8 scale, as illustrated in Figure 1, where 1 = no ARDS with high confidence, 8 = ARDS with high confidence.
Fig. 1:
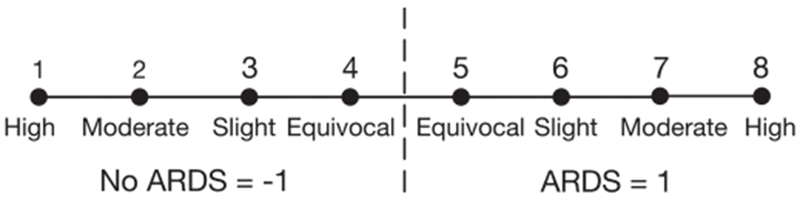
Accounting for uncertainty in a classification label using a clinical expert’s confidence in the diagnosis of ARDS. Critical care trained clinicians were asked to independently review patients’ EHR data and determine if any individuals in the cohort had ARDS, while also rating their confidence of the diagnosis using the following scale: equivocal, slight, moderate, or high.
In patients who developed ARDS, data collected before the time of onset were labeled as no ARDS, while data collected after the time of onset were labeled as ARDS. In total, 48 of the patients in the cohort were diagnosed with ARDS with a confidence of 5 or higher after expert review.
Time-stamped vital signs and laboratory values were extracted from each patient’s Electronic Health Record (EHR) from the first six days of hospitalization and included as clinical features (covariates) to train the ARDS algorithm. Only routinely acquired vital signs and laboratory values with potential for association with ARDS were included, based on guidance from clinical experts. Further details of the clinical variables in the model could be made available upon request. This approach minimized the total number of features in the model to 24 variables commonly used in clinical practice and statistical feature selection techniques were not utilized prior to model training. Patients were observed every 2 hours with previous data carried forward until a new value was recorded. If clinical data was missing on a patient because the vital sign or laboratory tests was not performed, it was imputed as a normal value. This is standard approach when developing clinical predictions models and assumes data is not collected because the treating clinician had a low suspicion that it would be abnormal [34, 35].
B. Sampling from Longitudinal Data and Inter-Correlation
Longitudinal patient data with repeated measurements over time have strong inter-dependency between each instance for a given patient. Ignoring these dependencies during training may lead to a biased estimator and a flawed learning model.
Inter-dependency among longitudinal data has been previously conceptualized as a system under mixing conditions [23]. For a given stochastic process, mixing indicates asymptotically independency implying that for a stationary process X, the dependency between X(t1) and X(t2) becomes negligible as |t1 − t2| increases towards infinity [36]. This mixing structure, while assuming that the dependency weakens in time, often exponentially, allows local dependency among the data points, and as such matches the reality of the majority of time-series processed in medicine as well as many other applications [37].
In order to address the interdependency of the data, we assumed that each patient’s time-series data used to develop the ARDS detection algorithm was a mixing stochastic process and we sampled data according to the quantitative assessment of the correlation decay among the data points. This approach limits the degree of inter-correlation on the data points sampled within the same patient and allows a more realistic assessment of model accuracy and reliability.
To implement this sampling strategy, we first calculated pairwise correlation distance matrices to represent dependency over the span of each patient’s time-series data. Given an m-by-n matrix for each patient’s data, where m is the number of times the patient was observed, and each observation is treated as 1-by-n row vectors, the correlation distance between vectors Xa and Xb for a single pair of observations is defined as:
where:
Using this correlation distance formula, an m-by-m correlation distance matrix can be derived for all observations on the patient, taken pairwise.
The sampling procedure begins by examining the correlation distances between Xt and 〈Xt〉 was generated, where Xt corresponds to an instance at the start of a patient’s time-series data and 〈Xt〉 is the span of all subsequent time-points. Then a sampling threshold η is set, which represents the point in which the inter-dependency between data becomes more limited. We chose the threshold value of η to be , based on literature that suggests values of approximately as an estimate of the width of a correlation-type function [38]. We also explored other values of η to understand their effect on the model building process. Figure 2 shows the effect of different sampling thresholds on model performance, including the difference in model accuracy in the training to testing set and AUROC of the testing set. This empirical analysis confirmed that optimal results are achieved when the sampling threshold is approximately 0.7 and supports the literature suggested value of .
Fig. 2:
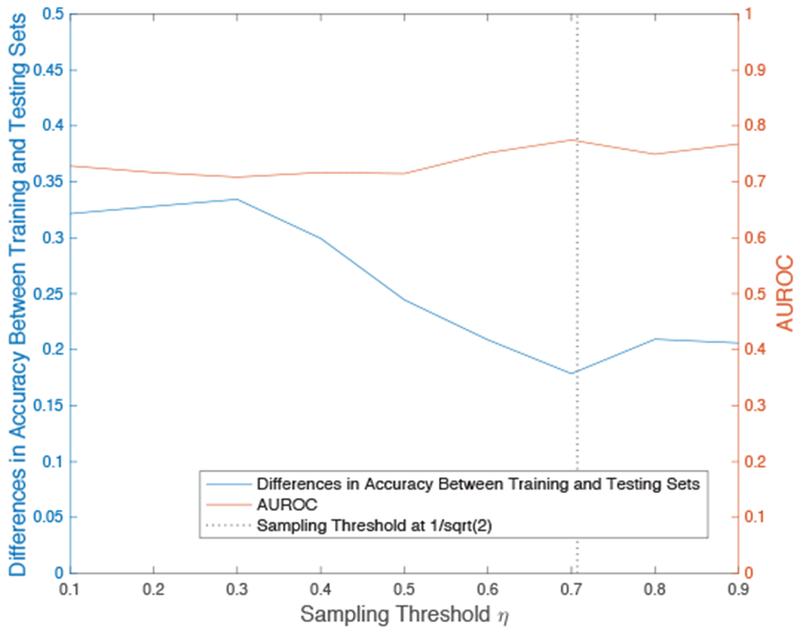
Effects of different sampling thresholds on prediction generalizability with SVM. With our sampling strategy, SVM performs very well on the training data at any threshold. We indicate the loss in training accuracy when the same model makes a prediction on a hold-out testing set to properly assess the effects of changing the sampling threshold and empirically determine the value for optimal results.
During the data sampling process for each patient, Xt is selected as the start of a patient’s time-series data. A pairwise correlation distance matrix is then calculated between Xt and 〈Xt〉, and a data point Xt1 is sampled as the first instance with a correlation distance of below η from 〈Xt〉. This selected point Xt1 and subsequent time points beyond Xt1, 〈Xt1〉, are used to re-calculate a new pairwise correlation distance matrix. A data point Xt2 is then selected in a similar manner as Xt1 from data points in 〈Xt1〉 with a correlation distance below the threshold of η. The sampling method is repeated until no further instances of 〈Xtn〉 are below the threshold from Xtn.
For this specific dataset, we did not utilize the sampling strategy described above for patient instances with the classification label of ARDS = 1. After inspection of the data, we observed the correlation decay to behave differently according to the label, with the data remained highly correlated over time when ARDS = 1 while correlation decay occurring when ARDS = −1. Therefore, this sampling approach was only performed on the data when ARDS = −1 while all instances were sampled when ARDS = 1. This approach effectively samples all positive examples while undersampling negative examples, which was also necessary given the significant class imbalance of the two labels [39]. The sampling strategy is shown in pseudocode as Algorithm 1 and the average decay of correlation from all patients is shown in Figure 5 with error bars representing standard error of the mean.
Fig. 5:

Average decay of correlation from all patients. Error bars represent standard error of the mean and each point represents correlation in relation to time (hours) from the initial observation sampled on each patient.
C. Formulation of SVM with Label Uncertainty
We implement the following formulation of Support Vector Machine [40] to account for label uncertainty in the classification model in the following manner:
subject to:
| (1) |
Algorithm 1:
Pseudocode for our algorithm to sample time-series data and reduce inter-dependency.
| Input : All available time-series data 〈Xt〉 from each patient. | |
| 1 | for each patient do |
| 2 | partition data into separate bins according to the classification label; |
| 3 | if size of either bins is ≤ 4 then sample all available data; |
| 4 | |
| 5 | else |
| 6 | 1) select Xt at the start of the time-series data and sample this instance; |
| 7 | 2) calculate the pairwise correlation distance from Xt to 〈Xt〉; |
| 8 | 3) sample the first row in 〈Xt〉 with a correlation distance < η and set as the new Xt; |
| 9 | repeat |
| 10 | 1) set 〈Xt〉 as all points subsequent to Xt; |
| 11 | 2) calculate the pairwise correlation distance matrix from Xt to 〈Xt〉; |
| 12 | 3) sample the first row where the correlation distance is < η and set as the new Xt; |
| 13 | until pairwise distance of Xt to 〈Xt〉 > η; |
| 14 | end |
| 15 | end |
| Output: Partial data {Xt,Xt1,Xt2, … ,Xtn} with reduced inter-correlation from each patient. |
where:
This formulation incorporates the slack variable ξi to permit some misclassification and also includes the penalty parameter C to establish soft-margin decision boundaries because ARDS and non-ARDS examples are not linearly separable. In this implementation, support vectors that are based on patients’ data with high label confidence are given more weight and influence in the SVM decision boundary. Uncertainty in the label (li), as shown in Figure 1, is incorporated within (zi) to directly influence the box constraint (C). The formula for zi combines two linear transformations for uncertainty in the label annotation (li) and generate a scalable weight to that specific observation. In this application, we set α = 4.5, β = 3.0, γ = 20, and δ = 90, which scales li, with a range of 1-8, into the weighting zi, with a range between 40-100 in increments of 20. As a result, labels with high confidence (eg. li = 1 or 8) receive the weight zi = 100, while equivocal labels (eg. li = 4 or 5) receive the weight zi = 40. zi is then normalized to 1. This formula for zi adjusts sample weighting based on li and rescales the C parameter as Ci for each observation in a patient’s data structure so that the classifier puts more emphasis on points with high confidence.
To ensure that our proposed sampling strategy and threshold still maintains for SVM with label uncertainty, we repeat the previous analysis to show the effect of different sampling thresholds on prediction generalizability. Figure 3 confirms that optimal results are achieved when the sampling threshold is approximately 0.7, which supports the previous analysis and the literature suggested value of .
Fig. 3:
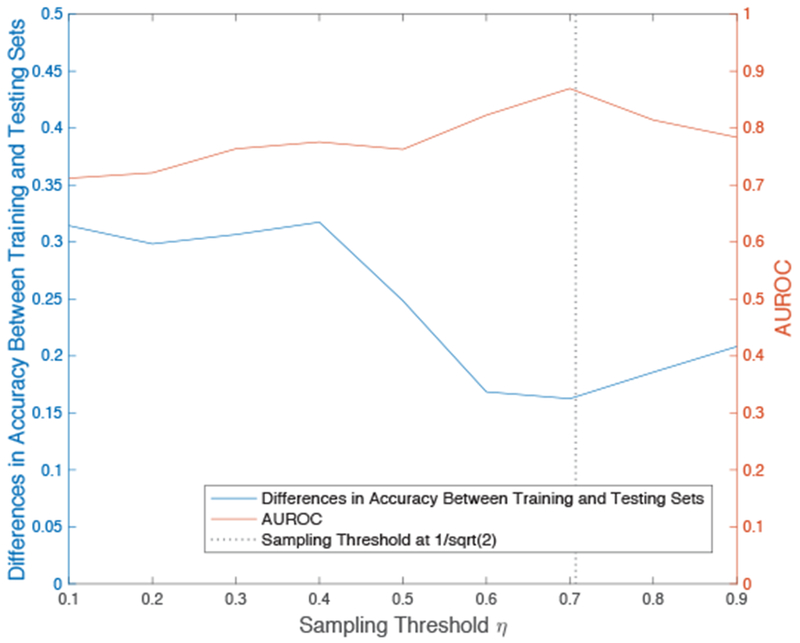
Effects of different sampling thresholds on prediction generalizability with SVM and label uncertainty. We confirm that the sampling strategy and threshold effects observed in Figure 2 is maintained when the SVM model is formulated to account for label uncertainty.
D. Model Building, Cross-Validation and Model Testing
In this study, the primary learning algorithms we compare are linear SVM with and without label uncertainty. Prior to building these models, the data was first normalized to prevent features with large dynamic ranges from dominating the separating hyperplane. Then the training data was sampled using the proposed sampling method described previously to minimize correlation between data points on the same patient. Prior to sampling, the training set contained 13,722 total instances, 736 of which were positive. After sampling, there were 1,893 total instances, 736 of which were positive.
5-fold cross validation was performed on the training data to find the optimal value of the hyper-parameter C using grid search [41] over C ∈ {0.001,0.01,0.1,1,10,100,1000}. We then re-trained the model on the entire training set using this optimal C parameter. This updated model was then used to classify patients in the hold-out dataset using all their data (i.e. no sampling was performed on the holdout data). The model predictions for each patient in the holdout sample, i.e. ARDS = 1 or −1, are then compared against the label assigned by the majority of experts reviewing the patient. We also compare the performance of our proposed SVM method with Logistic Regression and Random Forest (using the same subsampled training/testing bins and 5-fold cross validation partitions) to determine if the achieved results are equivalent or superior to other state-of-the-art methods.
A simplified protocol of this analysis, including data pre-processing, sampling from the training data to limit inter-correlation, hyper-parameter optimization with 5-fold cross-validation, and hold-out testing is shown in the flowchart of Figure 4.
Fig. 4:
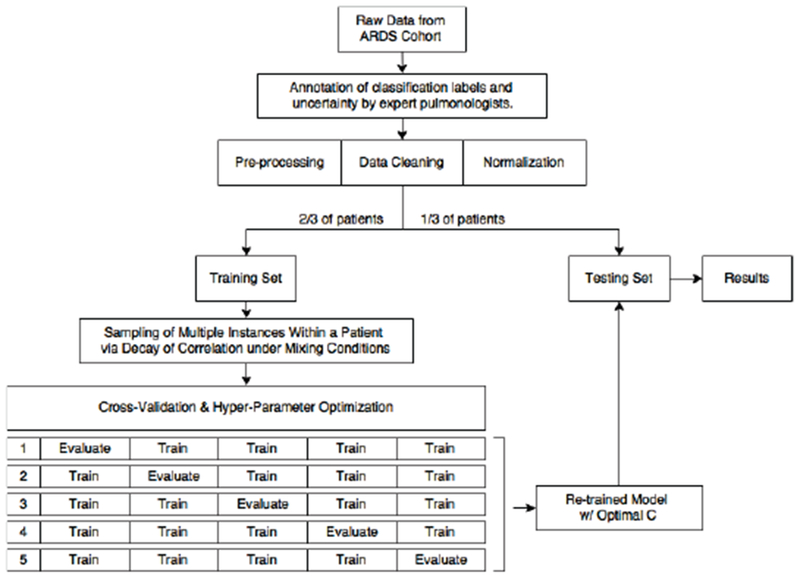
Flowchart of this study’s protocol with 5-fold cross-validation and hyper-parameter optimization using grid search. All samples from the same patient are kept exclusively in either the training or testing set. Hyper-parameter optimization was implemented for separately each model (with and without label uncertainty weight) to give an accurate assessment of performance.
III. Results
A total of 401 patient cases were available from the study cohort. Within this dataset, 48 were positive for ARDS and the remaining 353 were negative. Two-thirds of the patients were used in the model training process while the remaining one-third were kept as a hold-out set for testing. All samples from the same patients are kept exclusively in either the training or testing set (not both) to avoid bias in the data.
The average correlation decay for each patient’s data is shown in Figure 5. On average, the correlation between Xt and 〈Xt〉 drops below η at a distance in time of around 22 hours. Figure 6 shows the decay of correlation to be different when the data was analyzed separately according to the classification label: decay of correlation is observed when ARDS = −1 but not observed when ARDS = 1. Therefore, the sampling under η approach was performed on the data when ARDS = −1, which reduce the number of negative examples for model training. Due to the lower number examples, and lack of correlation decay when ARDS = 1, sampling was not performed as it would have further exacerbated the class imbalance.
Fig. 6:
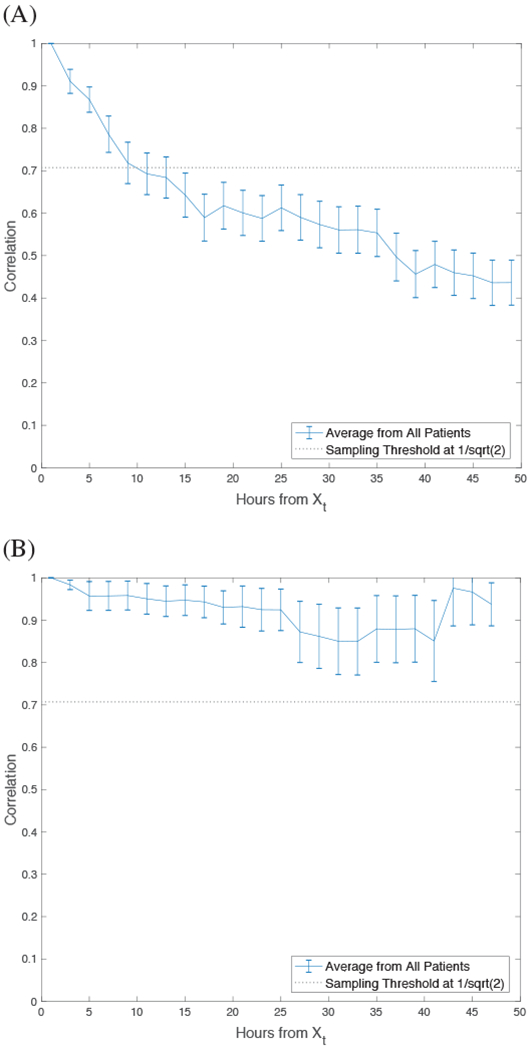
Average decay of correlation from all patients during (A) negative diagnosis of ARDS and (B) positive diagnosis of ARDS. Error bars represent standard error of the mean and each point represents correlation in relation to time (hours) from the initial observation sampled on each patient.
When the SVM was trained to account for uncertainty in the label, we observed over 10% improvement of AUROC (0.8548 versus 0.7542) compared to the conventional SVM learning algorithm (Figure 7) when judged in the holdout sample. When the algorithms were benchmarked at a sensitivity of 95% and 90% (to ensure few ARDS cases are missed), the SVM model that accounted for label uncertainty also had improved specificity and outperforms the standard model (Table 1). These sensitivity levels were set to high levels because it is important clinically for a model to have a high sensitivity and not miss cases of ARDS.
Fig. 7:
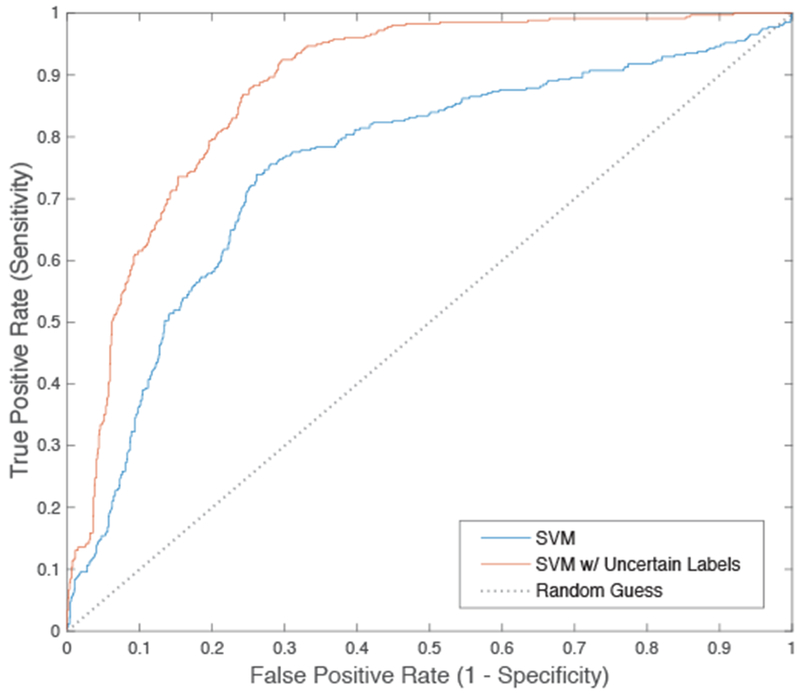
ROC curve comparing SVM with and without label uncertainty. Performance metrics are reported in Table 1.
TABLE I:
Performance of Logistic Regression, Random Forrest, SVM, SVM with a class-weighted cost function, and SVM with label uncertainty.
| Sampling Based on the Proposed Correlation Decay Method |
Random Sampling for Balanced (2:1) Training Data |
No Sampling |
||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | AUROC | Specificity at 95% Sensitivity | Specificity at 90% Sensitivity | Accuracy | AUROC | Accuracy | AUROC | |
| Logistic Regression | 0.7263 | 0.7265 | 0.3007 | 0.4267 | 0.6982 | 0.6979 | 0.6621 | 0.6454 |
| Random Forest | 0.7434 | 0.7488 | 0.3392 | 0.4751 | 0.7111 | 0.7254 | 0.6873 | 0.6903 |
| SVM | 0.7492 | 0.7542 | 0.3797 | 0.5114 | 0.7253 | 0.7361 | 0.6920 | 0.7152 |
| SVM w/ Class-Weighted Cost Function | 0.7804 | 0.8113 | 0.4571 | 0.5918 | 0.7478 | 0.7703 | 0.7094 | 0.7122 |
| SVM w/ Uncertain Labels | 0.8157 | 0.8548 | 0.5285 | 0.6450 | 0.7698 | 0.7989 | 0.7188 | 0.7431 |
We benchmarked our proposed SVM method utilizing uncertainty in the label to SVM with a misclassification cost function proportional to the weight of imbalance in the datasets and other standard classification models, such as Random Forest and Logistic Regression, in Table 1. We also compared our sampling strategy to an alternative method that utilizes random sampling on negative examples to yield a 2:1 negative to positive ratio from each patient to provide a more balanced dataset. In addition, we also examined performance without sampling (using all available data).
IV. Discussion
We present a robust machine learning algorithm to detect Acute Respiratory Distress Syndrome among hospitalized patients using routinely collected electronic health record data. We report an increase of 10% in AUROC in a hold-out data set when label uncertainty is incorporated in the learning process as a weight on classification penalty, when compared to a conventional SVM learning model.
Our proposed SVM model was trained by incorporating a clinical expert’s uncertainty in each patient’s classification label as a constraining weight of confidence on the SVM’s box constraint. Rather than treating label uncertainty as simple stochastic noise, this approach leverages information about the degree of uncertainty of each label, as provided by clinical experts, to improve the efficiency of model training. Our implementation of label weighting (zi) directly influences the C parameter and rescales the cost of misclassification according to uncertainty associated with each label (li). Support vectors that are based on the data from patients with high label confidence are given more influence in the SVM decision boundary while instances with more uncertainty are assigned less weight when determining the SVM hyperplane. In future works, alternative mappings between the label uncertainty (li) provided by clinical experts and label weighting (zi) used to find the SVM decision boundary should also be explored.
In addition, we performed a novel time-series sampling method, guided by the theory of mixing in stochastic processes, to limit the amount of correlation among data points on the same patient over time. Due to the time-series structure of a patient’s longitudinal health data, each instance is not independent from another. We explored whether the data could be represented under mixing conditions and implemented a novel sampling strategy for minimizing inter-correlation among data points in the training data. For the data to be represented under mixing conditions, the correlation between data on the same patient should decay over time such that CF,G(n) → 0 as n → ∞. A plot of the correlation function of the data in Figure 6 supported this assumption overall, but not for the data with a classification label of ARDS = 1.
It may not be appropriate to assume all data types can be represented under mixing conditions, therefore, plotting the correlation function of the data is essential prior to utilizing the sampling algorithm. When patients were diagnosed with ARDS, we found their data to have very high inter-correlation with little observable decay indicating a strong mixing process. Therefore, the proposed sampling method would have been unsuccessful in reducing inter-correlation and would yield very little data instances available for training. This finding made sense when interpreted from a clinical point of view. When a patient is admitted to the emergency room for pulmonary injury (eg. sepsis) and has not yet reached the critical stage of ARDS, their condition rapidly changes as a result of clinical intervention or decline of health, resulting in less stability and inter-correlation in the recorded data. If the patient develops ARDS, less rapid change in the data would be observed since ARDS is recognized as the final pathway of pulmonary damage [42].
Since there were significantly more negative than positive examples, we decided against using the sampling strategy when ARDS = 1, which ensured a more balanced number of positive and negative examples in the training data. As minimal correlation decay was observed among the data when ARDS = 1, implementing the sampling strategy for those data instances would have led to further imbalance among positive and negative examples, and limited the model’s ability to learn a good decision boundary. Our sampling approach utilized a pairwise correlation distance matrix to quantify dependency within the data structure. There are many ways to quantify the measurement of dependency between Xt to 〈Xt〉. Bradley et al provides a comprehensive list of mathematical definitions for dependency coefficients to define these mixing conditions [43] and measure decay of correlations [31]. In the future work, we will perform a more comprehensive examination of the data structure using formalized definitions of mixing, such as quantifying dependency with the α-mixing coefficient.
Our sampling method outperforms using all available data (no sampling) from the EHR by producing a much balanced dataset for training and minimizing dependencies in each patient’s time series data, making it closer to the state of being i.i.d. We also compared our sampling algorithm to randomly sampling on negative examples to yield a 2:1 negative to positive ratio from each patient. This random sampling method also provides a balanced dataset for training, and as a result, we observed an increase in accuracy and AUROC from all algorithms when compared to training without sampling. However, compared to our proposed sampling strategy, random sampling doesn’t achieve as high performance metrics because it does not account for correlation and may be sampling repeated measurements with strong dependencies, and therefore is not as robust as our method.
This study used a linear SVM for the ARDS model. In preliminary work not shown, we found that an SVM with a non-linear kernel (RBF) had less consistent results. Although the SVM with RBF kernel generally outperformed linear SVM on training dataset, it had inferior performance (accuracy and AUROC) on the hold-out set. Even with 5-fold cross-validation and grid-search hyper-parameter optimization (of C and gamma), we found the performance of the SVM with RBF kernel to be lower on the test set, and standard deviation of the results (after multiple random train-test splits) to be 2-3 fold larger than the linear SVM. We speculate that overfitting possibly occurred because of lower sample size and the number of variables used as features for machine learning. Because linear SVM was more robust, we chose to focus on using label uncertainty in the modeling process using only linear SVM.
With more clinical data, it would be worthwhile to investigate whether incorporating both label uncertainty and a non-linear SVM model would lead to improved model performance. The electronic health record may contain additional data that could be added to our model. Evaluating the performance of the training approach that considers label uncertainty in a higher dimensional space would be of value; however, to limit the possibility of overfitting with our current small dataset size, we have focused on using features that are routinely used for clinical evaluation of ARDS in the current study.
In additional future work, we plan to re-formulate the SVM model to account for both label uncertainty and privileged information to improve algorithm training [44]. Learning with privileged information (LUPI) also utilizes information available only in the training stage to help establish decision boundaries. Privileged information, which is information available during training but not when the model is deployed in real-time, may also be frequently available when developing machine-learning algorithms for healthcare applications and could also be relevant for ARDS detection.
We believe our paper makes a significant contribution towards solving traditional classification problems in the context of biomedical and clinical applications. In medicine, there is almost always a degree of uncertainty when assigning a patient to a medical diagnosis. Yet, that diagnosis label may then be used as the classification label or predictive outcome during a machine learning task. Typically, the diagnostic uncertainty associated with the label is not considered during model building. We show how an expert clinicians’ confidence in a diagnosis label can be used as vital information in the model training process. Exploiting the known diagnostic uncertainty within a medical domain is a generalizable approach that could be used in many medical applications. For example, sepsis is a clinical condition where early recognition is import for optimal patient care. However, diagnostic uncertainty is common [45], limiting ability to develop robust algorithms for sepsis detection. Incorporating label uncertainty when training an algorithm for sepsis detection may improve algorithm performance in a manner similar to ARDS.
It would also likely be of value to further develop approaches to incorporate label uncertainty into other machine learning frameworks besides SVM, such as random forest and neural networks. Since uncertainty in medical diagnosis occurs so commonly in clinical practice, accounting for label uncertainty with these learning algorithms may be highly applicable in other healthcare applications.
V. Conclusion
This paper introduces and tests a method of implementing uncertainty in the classification label in machine learning for detection of ARDS. It also describes a novel sampling strategy to reduce inter-correlation among longitudinal clinical data to prevent the creation of a biased model. using these novel approaches, we successfully trained an ARDS classification algorithm with significantly increased performance compared to a standard approach.
Acknowledgement
This work was partially supported by the National Science Foundation under Grant No. 1722801 and by the National Institute of Health under Grant NHLBI K01HL136687.
Contributor Information
Narathip Reamaroon, Department of Computational Medicine & Bioinformatics, University of Michigan, Ann Arbor, MI 48109 USA nreamaro@umich.edu.
Michael W. Sjoding, Department of Internal Medicine, the Institute of Healthcare Policy & Innovation, and the Michigan Center for Integrative Research in Critical Care, University of Michigan, Ann Arbor, MI 48109 USA msjoding@umich.edu
Kaiwen Lin, Department of Computational Medicine & Bioinformatics, University of Michigan, Ann Arbor, MI 48109 USA (linkw@umich.edu).
Theodore J. Iwashyna, Department of Internal Medicine, the VA Center for Clinical Management Research, and the Institute for Social Research, University of Michigan, Ann Arbor, MI, 48109 USA (tiwashyn@umich.edu)
Kayvan Najarian, Department of Computational Medicine and Bioinformatics, the Department of Emergency Medicine, and the Michigan Center for Integrative Research in Critical Care, University of Michigan, Ann Arbor, MI 48109 USA (kayvan@umich.edu).
References
- [1].Rubenfeld GD et al. , Incidence and outcomes of acute lung injury, N Engl J Med, vol. 353(16), pp. 1685–1693. October 2005. [DOI] [PubMed] [Google Scholar]
- [2].Sweeney RM, McAuley DF. Acute respiratory distress syndrome, Lancet, vol. 388(10058), pp. 2416–2430. November 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bellani G et al. , Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries, JAMA, vol. 315(8), pp. 788–800. February 2016. [DOI] [PubMed] [Google Scholar]
- [4].Clark BJ, Moss M, The Acute Respiratory Distress Syndrome: Dialing in the Evidence? JAMA, vol. 315(8), pp. 759–761. February 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sjoding MW, Hyzy RC, Recognition and Appropriate Treatment of the Acute Respiratory Distress Syndrome Remains Unacceptably Low, Crit Care Med, vol. 44(8), pp. 1611–1612. August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sjoding MW, Translating evidence into practice in acute respiratory distress syndrome: teamwork, clinical decision support, and behavioral economic interventions, Curr Opin Crit Care. July 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Herasevich V et al. , Validation of an electronic surveillance system for acute lung injury, Intensive Care Med, vol 35(6), pp. 1018–1023. June 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koenig HC et al. , Performance of an automated electronic acute lung injury screening system in intensive care unit patients, Crit Care Med, vol. 39(1), pp. 98–104. January 2001. [DOI] [PubMed] [Google Scholar]
- [9].Rubenfeld GD et al. , Interobserver variability in applying a radiographic definition for ARDS, Chest, vol. 116(5), pp. 1347–53. November 1999. [DOI] [PubMed] [Google Scholar]
- [10].Sjoding MW et al. , Acute Respiratory Distress Syndrome Measurement Error: Potential Effect on Clinical Study Results, Ann Am Thorac Soc, vol. 13(7), pp. 1123–8. July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shah et at CV, An alternative method of acute lung injury classification for use in observational studies, Chest; vol. 138(5), pp. 1054–1061. November 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nettleton DF, Orriols-Puig A, Fornells A, A study of the effect of different types of noise on the precision of supervised learning techniques, Artificial Intelligence Reviews, Vol. 33(40), 2010. [Google Scholar]
- [13].Frenay B and Verleysen M, ”Classification in the Presence of Label Noise: A Survey,” IEEE Transactions on Neural Networks and Learning Systems, vol. 25(5), pp. 845–869. May 2014. [DOI] [PubMed] [Google Scholar]
- [14].Natarajan N et al. , Learning with noisy labels,” in Neural Information Processing Systems, pp. 1196–1204. December 2013 [Google Scholar]
- [15].Duan Y and Wu O, Learning With Auxiliary Less-Noisy Labels, IEEE Transactions on Neural Networks and Learning Systems, vol. 28(7), pp. 1716–1721. May 2017. [DOI] [PubMed] [Google Scholar]
- [16].Vembu S and Zilles S, ”Interactive Learning from Multiple Noisy Labels,” in Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Springer International Publishing; 2016, pp. 493–508. [Google Scholar]
- [17].Yang X, Song Q, Want Y, A weighted support vector machine for data classification, International Journal of Pattern Recognition and Artificial Intelligence, vol 21, November 5 (2007). [Google Scholar]
- [18].Osuna E, Freund R, Girosi F. An improved training algorithm for support vector machines, in Neural Networks for Signal Processing [1997] VII. Proceedings of the 1997 IEEE Workshop (pp. 276–285). [Google Scholar]
- [19].Shawe-Taylor J et al. , ”Structural risk minimization over data-dependent hierarchies,” IEEE Transactions on Information Theory, vol. 44(5), pp. 1926–1940, September 1998. [Google Scholar]
- [20].Bellazzi R and Riva A, Learning conditional probabilities with longitudinal data, in Working Notes of the IJCAI Workshop Building Probabilistic Networks: Where Do the Numbers Come From, 1995. (pp. 7–15). [Google Scholar]
- [21].Najarian K et al. , PAC learning in nonlinear FIR models, International Journal of Adaptive Control and Signal Processing, vol. 15(1), pp. 37–52. February 2001. [Google Scholar]
- [22].Vapnik V, ”An overview of statistical learning theory,” IEEE Transactions on Neural Networks, vol. 10(5), pp. 988–999. September 1999. [DOI] [PubMed] [Google Scholar]
- [23].Wulsin D, Fox E, Litt B, Parsing epileptic events using a Markov switching process model for correlated time series, in Proceedings of the 30th International Conference on Machine Learning, PMLR 28(1):356–364, 2013. [Google Scholar]
- [24].Fan G and Liang H, Empirical likelihood for longitudinal partially linear model with -mixing errors, J Syst Sci Complex, vol. 26(2), pp. 232248 April 2013. [Google Scholar]
- [25].Yu L and Liu H, Feature Selection for High-Dimensional Data: A Fast Correlation-Based Filter Solution, in Proceedings of the Twentieth International Conference on Machine Learning, vol. 2, pp. 856–863. 2003. [Google Scholar]
- [26].Kim KJ, Financial time series forecasting using support vector machines, Neurocomputing, Vol. 55(2), pp. 307–319, September 2003. [Google Scholar]
- [27].Waibel A, Modular Construction of Time-Delay Neural Networks for Speech Recognition, Neural Computation, vol. 1(1) pp. 39–46, 1989. [Google Scholar]
- [28].Berndt DJ, Clifford J, Using Dynamic Time Warping to Find patterns in Time Series, Knowledge Discovery and Data Mining Workshop. Vol. 10, No. 16, 1994. [Google Scholar]
- [29].Berzuini C, Best NG, Gilks WR, Larizza C, Dynamic conditional independence models and Markov Chain Monte Carlo Methods, Journal of the American Statistical Association, 92(440), pp.1403–1412, 1997. [Google Scholar]
- [30].Xuan X, Murphy K, Modeling Changing Dependency Structure in Multivariate Time Series, in Proceedings of the 24th international conference on Machine learning (pp. 1055–1062), ACM, 2007. [Google Scholar]
- [31].Kastoryano MJ and Eisert J, ”Rapid mixing implies exponential decay of correlations,” Journal of Mathematical Physics, vol. 54(10), pp. 102201 October 2013. [Google Scholar]
- [32].Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS, Acute respiratory distress syndrome: the Berlin Definition, JAMA: the journal of the American Medical Association, Vol. 307(23), pp. 252633, 2012. [DOI] [PubMed] [Google Scholar]
- [33].Sjoding MW, Hofer TP, Co I, Courey A, Cooke CR, Iwashyna TJ, Inter-observer reliability of the Berlin ARDS definition and strategies to improve the reliability of ARDS diagnosis, Chest, 2017, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Knaus WA et al. , The APACHE III prognostic system: risk prediction of hospital mortality for critically III hospitalized adults, Chest. vol. 100(6), pp. 1619–36. December 1991. [DOI] [PubMed] [Google Scholar]
- [35].Churpek MM et al. , Multicenter development and validation of a risk stratification tool for ward patients, Am J Respir Crit Care Med, vol. 190(6), pp. 649–55. September 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Vidyasagar M, Learning and Generalization with Applications to Neural Networks, Springer-Verlag, March 2013. [Google Scholar]
- [37].Verbeke G, Linear Mixed Models for Longitudinal Data, in Linear Mixed Models in Practice, Springer; New York: 1997, pp. 63–153. [Google Scholar]
- [38].Binh LN, ”Problems on Tx for Advanced Modulation Formats for Long-Haul Transmission Systems” in Advanced Digital Optical Communications, 2nd ed Boca Raton: CRC Press, ch. 3, pp. 129, 2015. [Google Scholar]
- [39].He H, Garcia EA, Learning from Imbalanced Data, IEEE Transactions on Knowledge and Data Engineering,. 21(9), 2009. [Google Scholar]
- [40].Cortes C and Vapnik V, Support-vector networks, Machine Learning, vol. 20(3), pp. 273–97. [Google Scholar]
- [41].Pedregosa F et al. , Scikit-learn: Machine Learning in Python, Journal of Machine Learning Research, Vol. 12, pp. 2825–2830, October 2011. [Google Scholar]
- [42].Ware LB, Matthay MA, The acute respiratory distress syndrome, N Engl J Med, vol. 342(18), pp. 1334–49. May 2000. [DOI] [PubMed] [Google Scholar]
- [43].Bradley RC, Basic properties of strong mixing conditions. A survey and some open questions, Probability Surveys, vol. 21;2(2), pp. 107–44. April 2005. [Google Scholar]
- [44].Vapnik V and Vashist A, A new learning paradigm: learning using privileged information, Neural Networks. vol. 22(5), pp. 544–557. August 2009. [DOI] [PubMed] [Google Scholar]
- [45].Walkey A, Unreliable Syndromes, Unreliable Studies, Annals of the American Thoracic Society, vol. 13(7), pp. 1010–1011. July 2016. [DOI] [PubMed] [Google Scholar]


