Abstract
Long non-coding RNAs (lncRNAs) are transcripts which are usually more than 200 nt in length, and which do not have the protein-coding capacity. LncRNAs can be categorized based on their generation from distinct DNA elements, or derived from specific RNA processing pathways. During the past several decades, dramatic progress has been made in understanding the regulatory functions of lncRNAs in diverse biological processes, including RNA processing and editing, cell fate determination, dosage compensation, genomic imprinting and development etc. Dysregulation of lncRNAs is involved in multiple human diseases, especially neurological disorders. In this review, we summarize the recent progress made with regards to the function of lncRNAs and associated molecular mechanisms, focusing on neuronal development and neurological disorders.
Keywords: long non-coding RNA, gene expression, neuronal development, neurological disorders, mechanism
Introduction
Over the past several decades, advances in genomic sequencing technology and findings from large-scale consortia have facilitated our understanding of the complexity and flexibility of mammalian genomes. The majority of mammalian genomes are transcribed, whereas only a few transcripts encode proteins, the majority of transcripts are non-coding RNAs (ncRNAs) (Roberts et al., 2014). Based on the length of transcripts, ncRNAs are usually classified into two categories: small non-coding RNAs and long non-coding RNAs (lncRNAs). Small ncRNAs are usually <200 nucleotides, including microRNAs, Piwi-interacting RNAs and small nuclear RNAs (snoRNAs). lncRNAs are >200 nucleotides and frequently transcribed by polymerase II, and share some features, e.g., 5′-capping, 3′-polyadenylation, alternative splicing and sequence conservation with mRNA (Ponting et al., 2009; Nagano and Fraser, 2011).
Although lncRNAs generally lack protein coding capacity, spatiotemporal-specific expression patterns have highlighted the diverse functions and complicated mechanisms of lncRNAs (Cao et al., 2018). Currently, it is widely accepted that lncRNAs play an important function in a variety of biological processes, including regulating gene expression, both at the transcriptional and the post-transcriptional level, shaping the chromatin conformation and imprinting the genomic loci (Lee and Bartolomei, 2013; Chen, 2016; Cao et al., 2018), and multiple diseases such as neurological disorders, cancer, and immunological diseases (Bian and Sun, 2011; Huarte, 2015; Wan et al., 2017). In this review, we summarize the recent progress made regarding the functions of lncRNAs, especially the functions and associated mechanisms related to neurological disorders.
Characterization of LncRNA
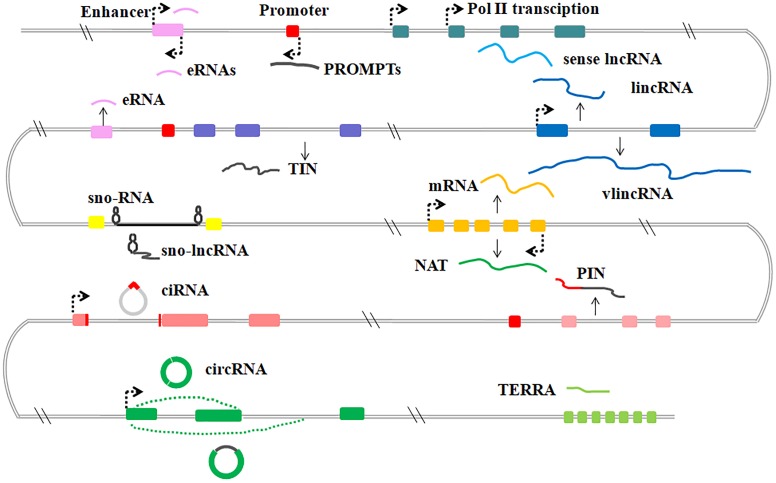
LncRNAs are generally transcribed from various genomic contexts and tend to have fewer exons than protein-coding transcripts (Iyer et al., 2015). Although there are still many challenges in annotation and interpretation of lncRNAs, because of the lack of an unambiguous classification framework, the existing lncRNAs can be subdivided into several categories based on their positional relation to protein coding genes, DNA elements or diverse mechanisms of processing (St Laurent et al., 2015; Kopp and Mendell, 2018) (Figure 1).
FIGURE 1.
Graphic illustration of the classification of lncRNAs in mammalian. General classification of lncRNAs. eRNAs, enhancer RNAs; PROMPTs, promoter upstream transcripts; lincRNAs, large intergenic ncRNAs; vlincRNA, very long intergenic ncRNAs; TIN, totally intronic long non-coding RNAs; sno-lncRNAs, small nucleolar RNA (snoRNA)-ended lncRNAs; NATs, natural antisense transcripts; PIN, partially intronic lncRNAs; ciRNAs, circular intronic RNAs; circRNAs, circular RNAs; TERRA, telomeric repeat-containing RNA.
Sense lncRNAs are transcribed from the sense DNA strand, and have overlapping regions with protein-coding genes, including un-spliced sense partially intronic RNAs (PINs) and spliced transcripts resembling mRNAs (St Laurent et al., 2015). Further, natural antisense transcripts (NATs) of protein-coding genes have also been identified and many NATs share some opposite strand DNA sequences with the sense transcripts (Katayama et al., 2005). Some studies also indicate that NATs have either positive or negative effects on the corresponding sense transcripts or nearby protein-coding transcripts (Faghihi et al., 2010; Modarresi et al., 2012). For example, human brain-derived neurotrophic factor antisense RNA (BDNF-AS) was originally identified as natural antisense transcripts of neuronal transcriptional factor BDNF, shares 225 complementary nucleotides with BDNF mRNA and regulates the expression of BDNF both in vivo and in vitro (Modarresi et al., 2012;Fatemi et al., 2015).
Other studies indicate that intronic regions of coding genes produce a lot of lncRNAs. These intronic lncRNAs form the largest class of lncRNAs and are expressed independently from the pre-mRNA of protein coding genes. Many intronic lncRNAs fail to be debranched after splicing and form a covalent circle without 3′ linear appendages, these circular intronic ncRNAs (ciRNAs) were found to play a regulatory role on their host genes (Zhang et al., 2013). In addition, circRNAs derived from the internal exons of pre-mRNAs through back-splicing, have also been found in various cell lines and tissues (Wu H. et al., 2017). These circular ncRNAs usually present tissue- and developmental stage-specific expression, such as the intensively studied cerebellar degeneration-related protein 1(CDR1as) (Memczak et al., 2013).
A relatively well-characterized subclass of lncRNAs is large/long intergenic or intervening non-coding RNAs (lincRNAs), and transcribed from the intergenic regions. LincRNAs have no overlapping sequences with transcripts of either protein-coding genes or other types of genes (Clark and Blackshaw, 2014). At the molecular level, most annotated lincRNAs have mRNA-like features including 5′-cap structures, 3′-poly(A) tails, exon–exon splice junctions and association with ribosomes (Cabili et al., 2011). Compared with mRNA counterparts, lincRNAs exhibit a more tissue-specific expression, a greater nuclear localization and less evolutionary conservation (Djebali et al., 2012).
Promoter upstream transcripts (PROMPTs) localize in a fairly narrow region between ∼500 and ∼2500 nucleotides upstream of transcription start sites of nearby active protein-coding genes (Preker et al., 2011; Lloret-Llinares et al., 2016). It was reported that the expression levels of certain PROMPTs are altered in stress conditions, such as DNA damage responses and osmotic responses (Lloret-Llinares et al., 2016; Song et al., 2018). Enhancer-related lncRNAs (eRNAs) are bidirectional transcripts of enhancers and have enhancer-like functions. Increased binding of DNA hydroxylase Tet1 and histone methyltransferases Mll3/Mll4 and DNA hypomethylation and H3K27ac modifications at enhancers, may activate eRNAs transcription. Both PROMPTs and eRNAs are targets of the RNA exosome and display similarities during processing (Andersson et al., 2014; Wu H. et al., 2017).
Emerging evidence indicates that telomeric repeat-containing RNA (TERRA) is a heterogeneous lncRNA consisting of a combination of subtelomeric and telomeric sequences. These sequences are mostly transcribed from intrachromosomal telomeric repeats by pol II and polyadenylated at 3′ region (Luke and Lingner, 2009). The length and expression level of human TERRA is influenced by the telomere length. The vast majority of mouse TERRA-binding sites were found in distal intergenic and intronic regions, where TERRA may regulate expression of target genes (Chu et al., 2017; Diman and Decottignies, 2018).
SnoRNA-ended lncRNAs (sno-lncRNAs) are transcripts of one intron flanked by two snoRNA genes that can be further processed to form snoRNA. sno-lncRNAs can be stabilized by snoRNPs formed by snoRNAs and specific protein components. SLERT is a representative Box H/ACA snoRNA-ended lncRNA and has been reported to be translocated to the nucleus by snoRNAs to function in pre-rRNA biogenesis (Wu H. et al., 2017).
Physiological Functions of LncRNA
Loss- and gain-of-function studies revealed that many lncRNAs are involved in various biological processes during development. Many lncRNAs have been found to regulate transcription via chromatin modulation, by working as molecular scaffolds for protein–protein interactions or interacting with chromatin modifying complexes and recruiting chromatin modifying complexes to specific loci, to activate or repress target gene expression. Some lncRNAs could affect transcription by modulating the binding of the general transcription machinery and regulatory factors (Wang and Chang, 2011; Fang and Fullwood, 2016; Wan et al., 2017; Lekka and Hall, 2018). Aside from modulating chromatin states, nuclear lncRNAs are involved in the RNA processing (Tripathi et al., 2010), turnover, silencing, translation and decay of mRNAs (Gong and Maquat, 2011; Carrieri et al., 2012; Geisler and Coller, 2013), or act as miRNA decoys to neutralize miRNA-mediated mRNAs silencing and interact with signaling molecules, to modulate signaling pathways (Faghihi et al., 2010; Liu et al., 2015). In addition, some lncRNAs are determined to be precursors of certain miRNAs at particular stages of development (Dykes and Emanueli, 2017) (Table 1).
Table 1.
Diverse mechanisms of lncRNAs playing function.
| Mechanism | lncRNA | Function | Relationship with target (Cis/Trans) | Reference |
|---|---|---|---|---|
| Chromatin remodeling | ANRIL | Repression at the CDKN2A/B locus | Cis/trans | Congrains et al., 2013 |
| BCAR4 | SNIP1 and PNUTS recruitment | Trans | Xing Z. et al., 2014 | |
| Braveheart | Activation of MesP1 | Trans | Klattenhoff et al., 2013 | |
| cga eRNA | Formation of looping between enhancer and promoter | Cis | Pnueli et al., 2015 | |
| COOLAIR | Repression at the FLC locus | Cis | Marquardt et al., 2014 | |
| FENDRR | PRC2 and TrxG/MLL complexes recruitment | Trans | Grote et al., 2013 | |
| H19 | Genomic imprinting | Cis/trans | Cai and Cullen, 2007 | |
| HAUNT | Repression at the HOXA locus | Cis | Yin et al., 2015 | |
| HOTAIR | Repression at the HOXD locus | Trans | Cai et al., 2014 | |
| HOTTIP | Activation at the HOXA locus | Cis | Lian et al., 2016 | |
| HOXD-AS1 | Recruitment of WDR5 to target genes | Trans | Gu et al., 2017 | |
| Kcnq1ot1 | Imprinting at the KCNQ1 cluster | Cis | Chiesa et al., 2012 | |
| lncTCF7 | Recruitment of SWI/SNF complex to TCF7 | Cis | Wu B. et al., 2018 | |
| MEG3 | Accumulation of p53 protein | Cis/trans | Zhou et al., 2007 | |
| Oct4P4 | Repression at the Oct4 locus | Trans | Scarola et al., 2015 | |
| PAPAS | rRNA synthesis | Cis | Zhao Z.L. et al., 2018 | |
| PARTICLE | Repression of methionine adenosyltransferase 2A | Cis/trans | O’leary et al., 2017 | |
| TERRA | Felomeric heterochromatin formation | Cis/trans | Luke and Lingner, 2009 | |
| TSIX | X inactivation | Cis | Sado et al., 2005 | |
| XIST | X inactivation | Cis | Cerase et al., 2015 | |
| Six3OS | Recruit histone modification enzymes to Six3 target genes | Trans | Rapicavoli et al., 2011 | |
| DNA methylation | APTR | Recruitment of PRC2 to CDKN1A/p21 | Trans | Negishi et al., 2014 |
| DALI | DNA methylation on promoter regions of target genes | Trans | Chalei et al., 2014 | |
| DUM | DNA methylation on Dppa2 | Cis | Wang et al., 2015a | |
| Evf2/Dlx6as | Transcriptional repression of Evf2 and Dlx5 | Cis/trans | Berghoff et al., 2013 | |
| FIRRE | H3K27me3 methylation maintenance | Trans | Yang et al., 2015 | |
| miRNA binding | CDR1as | miR-7 decoy | Trans | Yu et al., 2016 |
| lincRNA-ROR | miR-145 binding | Trans | Wang et al., 2013 | |
| lncRNA-ATB | miR-200s binding | Trans | Li et al., 2017 | |
| UCA1 | miR-184 sponge | NA | Li and Hu, 2015 | |
| TUG1 | miR-144/145 binding | Trans | Li et al., 2016 | |
| Transcriptional regulation | Uph | Repression at the Hand2 locus | Cis | Kopp and Mendell, 2018 |
| Paupar | Negative regulation Pax6 expression | Cis/trans | Vance et al., 2014 | |
| PANDA | Repression of NF-YA-mediated transcription | NA | Puvvula et al., 2014 | |
| Airn | Imprinting at the IGF2R cluster | Cis | Kopp and Mendell, 2018 | |
| RMST | Transcriptional coregulator of SOX2 | Trans | Ng et al., 2013a. | |
| Post transcriptional regulation | MALAT1 | Ser/Arg splicing factor regulation | Trans | Wu Y.T. et al., 2015 |
| PCA3 | PRUNE2 editing and stability | Cis | Salameh et al., 2015 | |
| CCAT2 | Alternative splicing of Glutaminase (GLS) | Trans | Xin Y. et al., 2017 | |
| MIAT | Alternative splicing of DISC1, ERBB4 | Trans | Sun et al., 2018 | |
| Sirt1 AS | Promotion of Sirt1 mRNA stability | Cis | Wang et al., 2014 | |
| TINCR | Stability of multiple mRNAs | Trans | Kretz, 2013 | |
| 1/2-sbsRNA | Activation of STAU1-mediated decay | Trans | Gong and Maquat, 2011 | |
| BACE1-AS | Positive regulation of BACE1 | Cis | Modarresi et al., 2011 | |
| aHIF | Nuclear membrane trafficking | Cis | Cayre et al., 2003 | |
| AS UCHL1 | UCHL1 mRNA translation | Cis | Carrieri et al., 2015 | |
| lincRNA-p21 | Translational suppression | Cis | Dimitrova et al., 2014 | |
| ZEB2 NAT | Activation of ZEB2 translation | Cis | Bernardes De Jesus et al., 2018 | |
| NORAD | Inhibition of PUM protein activity | NA | Lee et al., 2016 | |
| SNHG4/5/6 | Localization of MTA2 protein in the nuclear | NA | Chang et al., 2016; Zhao et al., 2016 | |
| LINK-A | Recruitment of BRK to GPNMB | NA | Wu D. et al., 2017 | |
| NEAT1 | Formation of nuclear paraspeckles | Trans | Clemson et al., 2009 | |
| AOC4P | Degradation of vimentin | Trans | Wang et al., 2015b | |
| ASBEL | Localization of ANA/BTG3 mRNA in the nuclear | Cis | Zhao J. et al., 2018 | |
| GAS5 | Repression of glucocorticoid receptor-mediated transcription | NA | Kino et al., 2010 | |
LncRNAs and Stem Cell Pluripotency and Differentiation
Accumulating evidence suggests that lncRNAs exert critical functions in pluripotency maintenance, reprogramming and lineage differentiation of stem cells (Wang and Chang, 2011; Ghosal et al., 2013). The long intergenic non-protein coding RNA regulator of reprogramming (lincRNA-ROR), increases the reprogramming efficiency of human induced pluripotent stem cells (iPSCs) and promotes the maintenance of embryonic stem cells (ESCs) pluripotency (Loewer et al., 2010). Similar to a miRNA sponge, lincRNA-ROR forms a regulatory feedback loop with miR-145 and OCT4, SOX2, and NANOG, and regulates ESC pluripotency (Wang et al., 2013). MIAT (myocardial infarction associated transcript) is a co-activator of Oct4 and participates in OCT4 and NANOG regulatory networks in mouse ESCs. Loss of MIAT reduces the expression of Oct4, Sox2, and Klf4, and inhibits ESCs proliferation (Sheik Mohamed et al., 2010).
LncRNAs and Development
Genomic imprinting is an important epigenetic mechanism and is crucial for normal development in mammals. It restricts gene expression on one of the two parental chromosomes in diploid cells and affects both male and female descendants (Barlow and Bartolomei, 2014). H19, a maternally expressed 2.3 kb lncRNA, is generated from the highly conserved and imprinted vertebrate gene cluster insulin-like growth factor 2 (Igf2)/H19. H19 transcripts are the precursors of miR-675-3p and miR-675-5p (Cai and Cullen, 2007). Before parturition, H19 slows the growth of the placenta down partially, by down-regulating the RNA binding protein HuR. The decreased HuR cannot block the processing of miR-675, which further decreases the growth regulator Igf1r with Igf2 as its main ligand (Keniry et al., 2012). Another two well-characterized lncRNAs that have been found to regulate genomic imprinting are Kcnq1ot1 (KCNQ1 opposite strand transcript 1) and Airn (antisense of IGF2R non-protein coding RNA), both are paternally expressed and regulate transcriptional silencing through a multilayered silencing pathway (Perry and Ulitsky, 2016).
Besides genomic imprinting, dosage compensation plays a vital role in equalizing the dosage of X-linked genes between males and females in heterogametic species. Xist is a 17 ∼ 20 kb lncRNA and transcribed from the X inactivation center. During female development, Xist initiates X-chromosome inactivation (XCI), by progressively coating the future inactive X chromosome (Xi) and then utilizing its conserved A-repeat domain to bind PRC2, to form a transcriptionally silent nuclear compartment. The compartment is enriched by H3K27me3 and responsible for the chromosome-wide gene repression in the Xi (Chery and Larschan, 2014). TSIX, transcribed from the active chromosome (Xa), represses Xist at the early steps of X inactivation (Gendrel and Heard, 2014). Another lncRNA JPX/ENOX, which is transcribed from Jpx/Enox gene, that resides 10 kb upstream of Xist, also involves in XCI through repressing the TSIX expression from the Xi and evicting nuclear protein CCCTC-binding factor (CTCF) away from promoter of XIST to activate the XIST expression from Xi (Sun et al., 2013).
LncRNAs in Neurodevelopment
A great number of lncRNAs are expressed during neural development and in the brain. Using high-throughput technologies, in situ hybridization, microarray analysis and RNA sequencing (RNA-seq), researchers have found that most of the lncRNAs examined (849 out of 1328) are expressed in specific cell types, subcellular compartments and different regions of the brain (Ng et al., 2013b; Shi et al., 2017). LncRNAs display differential expressions across the cortical layers, and region-specific expressions in the subventricular zone, dentate gyrus and olfactory bulb of mice (Belgard et al., 2011; Ramos et al., 2013). Based on a unique custom microarray platform, 8 lncRNAs were identified to be expressed in an age-dependent manner, from 36 surgically resected human neocortical samples, ranging from infancy to adulthood (Lipovich et al., 2014). However, the function of those lncRNAs need to be validated, through loss-of-function assays, RNA-protein association assays or assessments of RNA-chromatin association.
Some lncRNAs are also found to participate in neural cell fate determination, neuronal-glia fate switching and oligodendrocyte elaboration. An antisense transcript of the distal-less homeobox 1 (Dlx1), Dlx1AS, was discovered to be up-regulated during GABAergic differentiation and down-regulated during oligodendrocyte differentiation (Mercer et al., 2010). A subsequent study found that Dlx1AS participated in neurogenesis, implying its function in neuronal differentiation by regulating expression of its homeobox gene neighbors (Ramos et al., 2013). Evf2, a cloud-forming Dlx5/6 ultra-conserved eRNA, influences the formation of GABAergic interneurons in both a mouse and human forebrain. In the developing ventral forebrain, Evf2 regulates Dlx5, Dlx6 and glutamate decarboxylase 1 (Gad1) expression by recruiting DLX1 and/or DLX2 and methyl CpG binding protein 2 (MeCP2) to specific DNA regulatory elements. GAD1 is an enzyme responsible for catalyzing glutamate to form GABA. Evf2 mouse mutants reduced GABAergic interneurons in the early postnatal dentate gyrus and hippocampus (Bond et al., 2009; Berghoff et al., 2013; Cajigas et al., 2018).
The vertebrate retina is comprised of three well-organized cell type-specific neuron layers, interconnected by synapses (Ng et al., 2013b). Six3OS is the long non-coding opposite strand transcript (lncOST) of the homeodomain factor Six3. During mammalian eye development, Six3 regulates both early eye formation and postnatal retinal cell specification. Knockdown of Six3OS leads to a decrease of bipolar cells and an increase of Müller glia, similar to the results in the knockdown of Six3. In contrast, overexpression of Six3OS decreased syntaxin positive cells. Gene perturbation studies revealed that Six3OS participates in retinal cell specification as a molecular scaffold, to regulate Six3 activity rather than expression (Rapicavoli et al., 2011). TUG1 (taurine upregulated gene 1), a spliced and polyadenylated lncRNA, is highly conserved in humans and mice. TUG1 may participate in rod-photoreceptor genesis and inhibits cone-photoreceptor gene expression globally, by altering the chromatin configurations of photoreceptor-specific transcription factors Crx and Nrl (Young et al., 2005).
LncRNAs in Neurologic Disorders
Emerging evidence has shown that the dysregulation of lncRNAs is related to multiple neurological disorders, such as schizophrenia (Scholz et al., 2010), autism spectrum disorder (ASD) (Wang et al., 2015c), Parkinson’s (Ni et al., 2017), Huntington’s (Sunwoo et al., 2017) and Alzheimer’s diseases (Faghihi et al., 2008).
Schizophrenia (SCZ) is a debilitating mental disorder with a broad spectrum of neurocognitive impairments. Abundant data suggests that both genetic and environmental factors contribute to the pathophysiology of SCZ (Seidman and Mirsky, 2017). Several lncRNAs have been used as biomarkers and therapeutic targets for SCZ (Chen et al., 2016). lncRNA MIAT, also known as Gomafu or RNCR2, is down-regulated in SCZ upon neuronal activation (Sun et al., 2018). Previous studies found MIAT either acts as a competitive endogenous RNA (ceRNA) for miR-150-5p, miR-24, miR-22-3p or miR-150, to influence cell proliferation, apoptosis and migration, or participates in various signaling pathways by enhancing Nrf2 (nuclear factor erythroid 2-related factor 2) and Oct4 expression. Subsequent studies revealed that MIAT can directly bind to the splicing regulator quaking homolog (QKI) and splicing factor 1 (SF1), to modulate several gene expressions in the neuron. In SCZ patient brains, DISC1 (disrupted in schizophrenia 1), ERBB4 (v-erb-a erythroblastic leukemia viral oncogene homolog 4) and their alternatively spliced variants were all down-regulated due to MIAT up-regulation. MIAT could act as a scaffold to affect alternative splicing of those SCZ-associated genes (Roberts et al., 2014; Liu et al., 2018; Sun et al., 2018).
Autism spectrum disorder (ASD) is a heterogeneous group of neurodevelopmental disorders characterized by impaired reciprocal social interactions, communication, and repetitive stereotyped behaviors (Tang et al., 2017). 222 differentially expressed lncRNAs have been identified from autistic brain tissues. 90% of these lncRNAs are oriented in or around known genes related to neurodevelopmental and psychiatric diseases, such as UBE3A (ubiquitin protein ligase E3A), which is associated with Angelman syndrome, that shares common features with ASD. At the same time, it has been found that the number of lncRNAs differentially expressed within a control sample, was much greater than that within an autistic sample (1375 lncRNAs vs. 236 lncRNAs, respectively) (Ziats and Rennert, 2013). A genome-wide association study (GWAS) of ASD identified a 3.9 kb lncRNA designated MSNP1AS, which is encoded by the opposite strand of the moesin pseudogene 1 (MSNP1). The sense transcript MSN encodes the moesin protein that regulates neuronal architecture and immune responses. MSNP1AS was found to be significantly upregulated in a postmortem ASD temporal cortex, and overexpression of MSNP1AS led to significant decreases in MSN, moesin, neurite number and length in cultured neurons. Thus, MSNP1AS contributes to ASD risk, by possibly influencing the sense transcript MSN expression negatively (Wilkinson and Campbell, 2013).
BACE1-AS is a conserved non-coding antisense transcript of β-secretase 1 (BACE1) and has been shown to be closely associated with Alzheimer’s disease (AD). BACE1 is responsible for the generation of β-amyloid and the amyloid plagues in the brain, which are the primary pathophysiology of AD. BACE1-AS is markedly up-regulated in AD brains and promotes the stability of BACE1 through stabilizing BACE1 mRNA, thereby increasing the BACE1 protein and Aβ1–42 levels (Faghihi et al., 2008). Knock down of BACE1-AS in vivo resulted in the down-regulation of both BACE1 and BACE1-AS, along with reduced β-amyloid in the brain. In addition, the brain cytoplasmic RNA BC200 (BCYRN1), GDNF gene antisense transcript (GDNF-AS) and Sox2 overlapping transcript (Sox2OT), all participate in progress and development in AD brains (Wan et al., 2017).
Huntington disease (HD) is a hereditary neurodegenerative disease with symptoms including dementia, chorea, and psychiatric disturbances. HD is caused by a CAG trinucleotide abnormal expansion in the first exon of the huntingtin gene and its probability of occurrence is 1/10000. Microarray data found that the expression of four lncRNAs significantly changed in HD brains: NEAT1 (nuclear paraspeckle assembly transcript 1) and TUG1 are upregulated, and DGCR5 (DiGeorge syndrome critical region gene 5) and MEG3 (maternally expressed 3) are downregulated. The up-regulation of NEAT1 in HD might contribute to the pathogenic alteration of the transcriptional status, by sequestrating various paraspeckle proteins (Sunwoo et al., 2017), whereas TUG1 is possibly activated by p53 and then interacts with PRC2, to affect downstream HD-associated genes. DGCR5 and MEG3, are both direct targets of REST. Their down-regulation may result from the aberrant accumulation of REST in the nuclei of striatal neurons in HD (Johnson, 2012; Hwang and Zukin, 2018). Using the whole genome chromatin immunoprecipitation sequencing (ChIP-Seq) method, HAR1, a deeply conserved genomic region that is directly bound by REST was confirmed. This region encodes a pair of structured lncRNAs as well, HAR1F and HAR1R. Both HAR1F and HAR1R were downregulated in the striatum of HD patients (Johnson et al., 2010).
Parkinson’s disease (PD) is one of most prevalent neurodegenerative disorders, characterized by progressive impairments of motor abilities caused by the loss of dopamine-producing cells in the brain. Antisense ubiquitin carboxy-terminal hydrolase L1 (AS-Uchl1) was discovered to up-regulate the translation of UchL1 protein at a post-transcriptional level depending on a 5′ overlapping sequence and an embedded inverted SINEB2 sequence (Carrieri et al., 2012). AS-Uch1 is strongly down-regulated in neurochemical models of PD as a component of the Nurr1-dependent gene network and the subsequent reduced translation of UCHL1 protein, lead to the perturbation of the ubiquitin-proteasome system (Carrieri et al., 2015). H19 upstream conserved 1 and 2 (Huc1 and Huc2), lincRNA-p21, MALAT1, SNHG1, and TncRNA were differentially expressed in PD patients (Kraus et al., 2017). As these lncRNAs are associated with synaptogenesis, proliferation and apoptosis, the expression of these lncRNAs precede the course of PD, suggesting they may be biomarkers of PD (Kraus et al., 2017).
Mechanisms of LncRNAs in Biological Processes
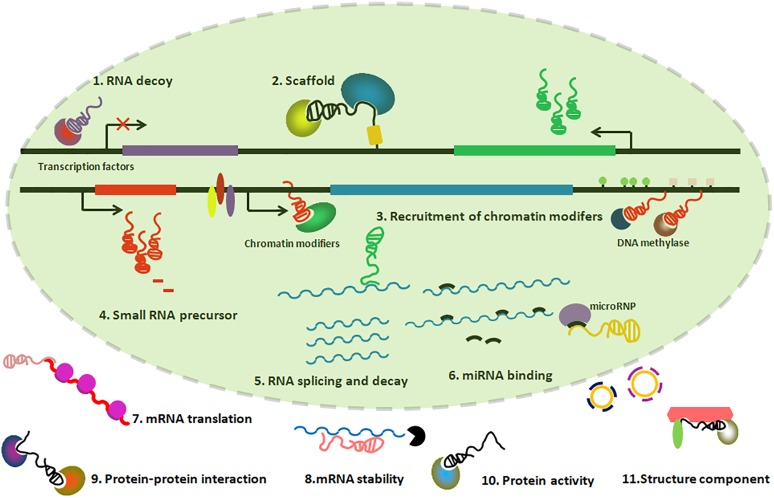
lncRNAs could provide functions through differential mechanisms, including serving as molecular scaffolds, molecular signals, guiding chromatin modifiers, and miRNA sponges, etc. (Figure 2).
FIGURE 2.
Graphic illustration of Mechanisms of lncRNAs playing functions. (1) lncRNAs can titrate transcription factors away from chromatin; (2) lncRNAs can serve as a scaffold to form ribonucleoprotein complexes; (3) lncRNAs can recruit chromatin-modifying enzymes to target genes; (4) lncRNAs can be precursors of small regulatory RNAs; (5–8) lncRNAs can regulate processes such as RNA splicing, translation and decay, in addition to miRNA binding; (9–11) lncRNAs can participate in protein–protein interactions, regulate protein activity and present as a structural component in the cytoplasm.
Molecular Scaffolds
Xist, a 17 ∼ 20 kb lncRNA transcribed from the X inactivation center, initiates X-chromosome inactivation (XCI) by progressively coating the future inactive X chromosome (Xi). Xist can bind to chromatin-modifying complexes PRC2 through the conserved A-repeat domain and form a transcriptionally silent nuclear compartment. This compartment is responsible for the chromosome-wide gene repression in the Xi (Chery and Larschan, 2014). Another example of the lncRNA acting as a molecular scaffold is the HOTAIR, which are transcripts of the antisense strand of HOXC gene cluster, which can modulate nearby gene expression by interacting with PRC2 and lysine specific demethylase 1 (LSD1) (Tsai et al., 2010).
lncRNA tsRMST, an isoform of RMST (rhabdomyosarcoma 2 associated transcript) was highly expressed in human iPSCs and ESCs. Further studies revealed that tsRMST down-regulation leads to NANOG and the PRC2 complex component SUZ12 fail to bind to the promoters of several inactive genes. These genes are thereby activated and promote ectoderm and endoderm differentiation (Ng et al., 2012; Yu and Kuo, 2016).
Molecular Signals
Genomic imprinting restricts gene expression on one of the two parental chromosomes and the parental-specific gene expression in diploid cells, and affects both male and female descendants (Barlow and Bartolomei, 2014). H19 transcripts are the precursors of miR-675-3p and miR-675-5p (Cai and Cullen, 2007). Before parturition, H19 slows the growth of the placenta down partially, through down-regulating the RNA binding protein HuR. The decreased HuR fails to block the processing of miR-675, which further decreases the growth regulator Igf1r (Keniry et al., 2012).
MALAT1 (metastasis-associated lung adenocarcinoma transcript (1) is initially found as an abundant lncRNA in nuclear speckles to regulate processes of mRNA alternative splicing by modulating the levels of serine/arginine splicing factors (Tripathi et al., 2010). Recent studies confirmed that MALAT1 is a sensitive prognostic marker for lung cancer metastasis and linked to several other human cancers (Gutschner et al., 2013).
Guiding Chromatin Modifiers
Genome regulation via DNA methylation and post-translational histone modifications by the activity of chromatin modifiers, is a well-documented function of lncRNA in eukaryotes (Bohmdorfer and Wierzbicki, 2015; Nanda et al., 2016). Altered DNA methylation patterns at CpG islands and mutations in chromatin modifiers, may result in oncogenesis (Haladyna et al., 2015; Nanda et al., 2016).
The first lncRNA identified to interact with both maintenance and de novo methylases, Dum, is tightly associated with myogenesis and transcriptionally induced by MyoD upon myoblast differentiation. Dum can recruit DNA methyltransferase1/3a/3b complex to the Dppa2 promoter, through intra-chromosomal looping, mediated by RAD21 and NIPBL, resulting in two CpG loci hypermethylation and Dppa2 silencing (Wang et al., 2015a). Recent studies of HOTAIR suggest that under heypoxia, HOTAIR expression is up-regulated in several cancer cells induced by the hypoxia-inducible factors (HIFs), recruiting hypoxia-response elements (HRE) to bind on the HOTAIR promoter. Along with HIFs, histone acetyltransferase CREB-binding protein (CBP/p300) and histone H3K4 specific methyltransferases, mixed lineage leukemia (MLL) family, are enriched in the HRE region of the HOTAIR promoter (Bhan et al., 2017).
Additionally, N-Myc can directly bind to the JMJD1A promoter to upregulate JMJD1A expression in neuroblastoma cells. The upregulated JMJD1A then directly binds to the MALAT1 promoter to demethylate H3K9, to activate MALAT1 expression (Tee et al., 2014; Peng et al., 2018). Furthermore, H19 and mir-675 were found to participate in the adipogenesis through mir-675, targeting the histone deacetylase (HDAC) 4–6 3′ untranslated regions and inducing HDACs 4–6 down-regulation. The reduced HDAC 4–6 then reduced H19 expression, possibly by reducing the levels of CTCF occupancy in the H19 imprinting control region. H19 inhibition then facilitates the bone marrow mesenchymal stem cells differentiating into adipocytes (Huang et al., 2016).
miRNA Sponges
A handful of microRNAs have been reported to influence the mRNA stability of protein-coding genes on post-transcriptional level. Recent studies on lncRNAs discovered several miRNA-lncRNA interactions based on in silico and experimental analyses (Paraskevopoulou and Hatzigeorgiou, 2016). One of the well-studied lncRNAs which act as miRNA sponges, is lincRNA-ROR, which decoys miR-145 in self-renewing human ESCs. A regulatory feedback loop formed by lincRNA-RoR, miR-145 and the core transcription factors OCT4, SOX2, and NANOG is closely related to the ESCs pluripotency (Wang et al., 2013). MALAT1 not only interacts with several splicing factors, but also binds to miRNAs including miR-101, miR-9, miR-125b, and miR217 to regulate the interactions between miRNA and mRNAs (Paraskevopoulou and Hatzigeorgiou, 2016).
Other Mechanisms
Some lncRNAs exert their function by maintaining DNA looping between enhancer and promoter regions, or by recruiting chromatin regulatory proteins to establish high affinity interactions between different regions of the DNA, resulting in closely positioned promoters and enhancers (Yang et al., 2013). For example, linRNA-p21 regulates the expression of its neighboring gene p21, by cis-regulatory enhancer-like DNA elements, which embed within the p21 gene body (Dimitrova et al., 2014). lncRNAs can also interact with DNA directly through nucleic-acid hybridization, and regulate nearby gene expression, such as Airn (antisense of IGF2R non-protein coding RNA), which produces transcription interference on paternal allele, by spanning the Igf2 gene promoter (Latos et al., 2012). Additionally, lncRNAs may influence the three-dimensional organization of the mammalian nucleus. FIRRE (firre intergenic repeating RNA element) is transcribed from the X chromosome, and interacts with hnRNPU in the process of nuclear organization (Yang et al., 2015). Trans-acting lncRNAs also modulate the protein activity and RNA stability by directly binding in the nucleus or cytoplasm. lncRNA NORAD functions as a negative regulator of the RNA binding protein PUMILIO1 and 2 (PUM1 and 2) in the cytoplasm. The knockdown of NORAD leads to the degradation of PUM1/2 targeted mRNAs (Lee et al., 2016).
Perspectives
Previous studies have revealed that lncRNAs play an important role in neuronal development and function through differential mechanisms. The dysregulation of lncRNAs could result in neurological diseases. Advances in sequencing technologies and their applications will contribute substantially to uncovering and investigating novel lncRNAs and their functions.
One challenge in the lncRNA field is whether lncRNAs can be used as diagnostic biomarkers or therapeutic targets for diseases. Considering that the down-stream targets of lncRNAs could be broad, it is hard to use it as a specific “key” to one “lock.” Although the validation of their functions could be performed in vitro and in vivo, it is difficult to claim a specific target. Another challenge is to better understand the mechanisms of lncRNAs functions. It is highly necessary to develop proper genetic tools and to establish animal models to dissect the regulatory networks of lncRNAs, and their interaction with other epigenetic modifications.
Author Contributions
LL, YZ, XZ, and XL wrote the manuscript. All authors commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. LL was supported by the Natural Science Foundation of Zhejiang Province (Grant No. LQ18C060001). XL was supported in part by the National Key Research and Development Program of China (Grant No. 2016YFC0900400) and the National Natural Science Foundation of China (Grant Nos. 31771395 and 31571518).
References
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. P., Bartolomei M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6:a018382. 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard T. G., Marques A. C., Oliver P. L., Abaan H. O., Sirey T. M., Hoerder-Suabedissen A., et al. (2011). A transcriptomic atlas of mouse neocortical layers. Neuron 71 605–616. 10.1016/j.neuron.2011.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff E. G., Clark M. F., Chen S., Cajigas I., Leib D. E., Kohtz J. D. (2013). Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140 4407–4416. 10.1242/dev.099390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes De Jesus B., Marinho S. P., Barros S., Sousa-Franco A., Alves-Vale C., et al. (2018). Silencing of the lncRNA Zeb2-NAT facilitates reprogramming of aged fibroblasts and safeguards stem cell pluripotency. Nat. Commun. 9:94. 10.1038/s41467-017-01921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A., Deb P., Shihabeddin N., Ansari K. I., Brotto M., Mandal S. S. (2017). Histone methylase MLL1 coordinates with HIF and regulate lncRNA HOTAIR expression under hypoxia. Gene 629 16–28. 10.1016/j.gene.2017.07.069 [DOI] [PubMed] [Google Scholar]
- Bian S., Sun T. (2011). Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 44 359–373. 10.1007/s12035-011-8211-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmdorfer G., Wierzbicki A. T. (2015). Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 25 623–632. 10.1016/j.tcb.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A. M., Vangompel M. J. W., Sametsky E. A., Clark M. F., Savage J. C., Disterhoft J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12:1020. 10.1038/nn.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Song X. Q., Cai J. P., Zhang S. (2014). HOTAIR: a cancer-related long non-coding RNA. Neoplasma 61 379–391. 10.4149/neo_2014_075 [DOI] [PubMed] [Google Scholar]
- Cai X. Z., Cullen B. R. (2007). The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13 313–316. 10.1261/.rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas I., Chakraborty A., Swyter K. R., Luo H., Bastidas M., Nigro M., et al. (2018). The Evf2 ultraconserved enhancer lncRNA functionally and spatially organizes megabase distant genes in the developing forebrain. Mol. Cell. 71:e959. 10.1016/j.molcel.2018.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Wahlestedt C., Kapranov P. (2018). Strategies to annotate and characterize long noncoding RNAs: advantages and pitfalls. Trends Genet. 34 704–721. 10.1016/j.tig.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491 454–457. 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Carrieri C., Forrest A. R. R., Santoro C., Persichetti F., Carninci P., Zucchelli S., et al. (2015). Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 9:114. 10.3389/Fncel.2015.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayre A., Rossignol F., Clottes E., Penault-Llorca F. (2003). aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 5 R223–R230. 10.1186/bcr652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A., Pintacuda G., Tattermusch A., Avner P. (2015). Xist localization and function: new insights from multiple levels. Genome Biol. 16:166. 10.1186/s13059-015-0733-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalei V., Sansom S. N., Kong L. S., Lee S., Montiel J., Vance K. W., et al. (2014). The long non-coding RNA dali is an epigenetic regulator of neural differentiation. Elife 3:e04530. 10.7554/eLife.04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Yuan Y., Li C., Guo T., Qi H., Xiao Y., et al. (2016). Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 383 183–194. 10.1016/j.canlet.2016.09.034 [DOI] [PubMed] [Google Scholar]
- Chen L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41 761–772. 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Chen S. D., Sun X. Y., Niu W., Kong L. M., He M. J., Li W. S., et al. (2016). Aberrant expression of long non-coding RNAs in schizophrenia patients. Med. Sci. Monitor 22 3340–3351. 10.12659/Msm.896927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chery J., Larschan E. (2014). X-marks the spot: X-chromosome identification during dosage compensation. Biochim. Biophys. Acta 1839 234–240. 10.1016/j.bbagrm.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa N., De Crescenzo A., Mishra K., Perone L., Carella M., Palumbo O., et al. (2012). The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of beckwith-wiedemann syndrome and silver-russell syndrome cases. Hum. Mol. Genet. 21 10–25. 10.1093/hmg/ddr419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. P., Cifuentes-Rojas C., Kesner B., Aeby E., Lee H. G., Wei C., et al. (2017). TERRA RNA antagonizes ATRX and protects telomeres. Cell 170:e116. 10.1016/j.cell.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B. S., Blackshaw S. (2014). Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front. Genet. 5:164. 10.3389/fgene.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., Chess A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33 717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congrains A., Kamide K., Ohishi M., Rakugi H. (2013). ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci. 14 1278–1292. 10.3390/ijms14011278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diman A., Decottignies A. (2018). Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: from Darkness to Dawn. FEBS J. 285 1389–1398. 10.1111/febs.14363 [DOI] [PubMed] [Google Scholar]
- Dimitrova N., Zamudio J. R., Jong R. M., Soukup D., Resnick R., Sarma K., et al. (2014). LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 54 777–790. 10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. (2012). Landscape of transcription in human cells. Nature 489 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes I. M., Emanueli C. (2017). Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform. 15 177–186. 10.1016/j.gpb.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G., Morgan T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14 723–730. 10.1038/nm1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Zhang M., Huang J., Modarresi F., Van Der Brug M. P., Nalls M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11:R56. 10.1186/gb-2010-11-5-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y. W., Fullwood M. J. (2016). Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 14 42–54. 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi R. P., Salah-Uddin S., Modarresi F., Khoury N., Wahlestedt C., Faghihi M. A. (2015). Screening for small-molecule modulators of long noncoding RNA-protein interactions using alphascreen. J. Biomol. Screen. 20 1132–1141. 10.1177/1087057115594187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Coller J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14 699–712. 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Heard E. (2014). Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu. Rev. Cell Dev. Biol. 30 561–580. 10.1146/annurev-cellbio-101512-122415 [DOI] [PubMed] [Google Scholar]
- Ghosal S., Das S., Chakrabarti J. (2013). Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 22 2240–2253. 10.1089/scd.2013.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C. G., Maquat L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., et al. (2013). The tissue-specific IncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Chen X., Xie R., Han J., Xie W., Wang B., et al. (2017). lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol. Ther. J. Am. Soc. Gene Ther. 25 1959–1973. 10.1016/j.ymthe.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Hammerle M., Diederichs S. (2013). MALAT1 – a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 91 791–801. 10.1007/s00109-013-1028-y [DOI] [PubMed] [Google Scholar]
- Haladyna J. N., Yamauchi T., Neff T., Bernt K. M. (2015). Epigenetic modifiers in normal and malignant hematopoiesis. Epigenomics-UK 7 301–320. 10.2217/Epi.14.88 [DOI] [PubMed] [Google Scholar]
- Huang Y., Zheng Y., Jin C., Li X., Jia L., Li W. (2016). Long Non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci. Rep. 6:28897. 10.1038/srep28897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21 1253–1261. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- Hwang J. Y., Zukin R. S. (2018). REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 48 193–200. 10.1016/j.conb.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47 199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. (2012). Long non-coding RNAs in huntington’s disease neurodegeneration. Neurobiol. Dis. 46 245–254. 10.1016/j.nbd.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Johnson R., Richter N., Jauch R., Gaughwin P. M., Zuccato C., Cattaneo E., et al. (2010). Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genomics 41 269–274. 10.1152/physiolgenomics.00019.2010 [DOI] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., et al. (2005). Antisense transcription in the mammalian transcriptome. Science 309 1564–1566. 10.1126/science.1112009 [DOI] [PubMed] [Google Scholar]
- Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., et al. (2012). The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and lgf1r. Nat. Cell Biol. 14 659–665. 10.1038/ncb2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8. 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A., Steinhauser M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F., Mendell J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus T. F. J., Haider M., Spanner J., Steinmaurer M., Dietinger V., Kretzschmar H. A. (2017). Altered long noncoding RNA expression precedes the course of parkinson’s disease-a preliminary report. Mol. Neurobiol. 54 2869–2877. 10.1007/s12035-016-9854-x [DOI] [PubMed] [Google Scholar]
- Kretz M. (2013). TINCR, staufen1, and cellular differentiation. RNA Biol. 10 1597–1601. 10.4161/rna.26249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P. A., Pauler F. M., Koerner M. V., Senergin H. B., Hudson Q. J., Stocsits R. R., et al. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338 1469–1472. 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- Lee J. T., Bartolomei M. S. (2013). X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152 1308–1323. 10.1016/j.cell.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Lee S., Kopp F., Chang T. C., Sataluri A., Chen B. B., Sivakumar S., et al. (2016). Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164 69–80. 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekka E., Hall J. (2018). Noncoding RNAs in disease. FEBS Lett. 592 2884–2900. 10.1002/1873-3468.13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Hu C. P. (2015). Long non-coding RNA urothelial carcinoma associated 1 (UCA1): insight into its role in human diseases. Crit. Rev. Eukaryot. Gene Expr. 25 191–197. 10.1615/CritRevEukaryotGeneExpr.2015013770 [DOI] [PubMed] [Google Scholar]
- Li J., Li Z., Zheng W., Li X., Wang Z., Cui Y., et al. (2017). LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 50:e12381. 10.1111/cpr.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Shen J., Chan M. T., Wu W. K. (2016). TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 49 471–475. 10.1111/cpr.12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y. F., Cai Z. L., Gong H. B., Xue S. L., Wu D. D., Wang K. M. (2016). HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol. Biosyst. 12 3247–3253. 10.1039/c6mb00475j [DOI] [PubMed] [Google Scholar]
- Lipovich L., Tarca A. L., Cai J., Jia H., Chugani H. T., Sterner K. N., et al. (2014). Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb. Cortex 24 1451–1459. 10.1093/cercor/bhs414 [DOI] [PubMed] [Google Scholar]
- Liu B., Sun L., Liu Q., Gong C., Yao Y., Lv X., et al. (2015). A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27 370–381. 10.1016/j.ccell.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Liu Y. S., Rao S. Q., Xu Y., Zhang F. Q., Wang Z. Q., Zhao X. D. (2018). Changes in the level of long non-coding RNA gomafu gene expression in schizophrenia patients before and after antipsychotic medication. Schizophr. Res. 195 318–319. 10.1016/j.schres.2017.08.025 [DOI] [PubMed] [Google Scholar]
- Lloret-Llinares M., Mapendano C. K., Martlev L. H., Lykke-Andersen S., Jensen T. H. (2016). Relationships between PROMPT and gene expression. RNA Biol. 13 6–14. 10.1080/15476286.2015.1109769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S., Cabili M. N., Guttman M., Loh Y. H., Thomas K., Park I. H., et al. (2010). Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42 1113–1117. 10.1038/ng.710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Lingner J. (2009). TERRA: telomeric repeat-containing RNA. EMBO J. 28 2503–2510. 10.1038/emboj.2009.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F. Q., Sun Q. W., Dean C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54 156–165. 10.1016/j.molcel.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Qureshi I. A., Gokhan S., Dinger M. E., Li G., Mattick J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. 10.1186/1471-2202-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F., Faghihi M. A., Lopez-Toledano M. A., Fatemi R. P., Magistri M., Brothers S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30 453–459. 10.1038/nbt.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F., Faghihi M. A., Patel N. S., Sahagan B. G., Wahlestedt C., Lopez-Toledano M. A. (2011). Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimer’s Dis. 2011:929042. 10.4061/2011/929042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Fraser P. (2011). No-nonsense functions for long noncoding RNAs. Cell 145 178–181. 10.1016/j.cell.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Nanda J. S., Kumar R., Raghava G. P. S. (2016). dbEM: a database of epigenetic modifiers curated from cancerous and normal genomes. Sci. Rep. UK 6:19340. 10.1038/Srep19340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M., Wongpalee S. P., Sarkar S., Park J., Lee K. Y., Shibata Y., et al. (2014). A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One 9:e95216. 10.1371/journal.pone.0095216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. Y., Bogu G. K., Soh B. S., Stanton L. W. (2013a). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 51 349–359. 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Ng S. Y., Lin L., Soh B. S., Stanton L. W. (2013b). Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 29 461–468. 10.1016/j.tig.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Ng S. Y., Johnson R., Stanton L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31 522–533. 10.1038/emboj.2011.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y. H., Huang H., Chen Y. Q., Cao M. H., Zhou H. Z., Zhang Y. Y. (2017). Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell Mol. Neurobiol. 37 329–338. 10.1007/s10571-016-0373-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’leary V. B., Hain S., Maugg D., Smida J., Azimzadeh O., Tapio S., et al. (2017). Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. UK 7:1790. 10.1038/S41598-017-01875-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou M. D., Hatzigeorgiou A. G. (2016). Analyzing MiRNA-LncRNA interactions. Methods Mol. Biol. 1402 271–286. 10.1007/978-1-4939-3378-5_21 [DOI] [PubMed] [Google Scholar]
- Peng K., Su G., Ji J., Yang X., Miao M., Mo P., et al. (2018). Histone demethylase JMJD1A promotes colorectal cancer growth and metastasis by enhancing Wnt/beta-catenin signaling. J. Biol. Chem. 293 10606–10619. 10.1074/jbc.RA118.001730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. B., Ulitsky I. (2016). The functions of long noncoding RNAs in development and stem cells. Development 143 3882–3894. 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- Pnueli L., Rudnizky S., Yosefzon Y., Melamed P. (2015). RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proc. Natl. Acad. Sci. U.S.A. 112 4369–4374. 10.1073/pnas.1414841112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell 136 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Preker P., Almvig K., Christensen M. S., Valen E., Mapendano C. K., Sandelin A., et al. (2011). PROMoter uPstream transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 39 7179–7193. 10.1093/nar/gkr370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvvula P. K., Desetty R. D., Pineau P., Marchio A., Moon A., Dejean A., et al. (2014). Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun. 5:5323. 10.1038/ncomms6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. D., Diaz A., Nellore A., Delgado R. N., Park K. Y., Gonzales-Roybal G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem. Cell 12 616–628. 10.1016/j.stem.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli N. A., Poth E. M., Zhu H., Blackshaw S. (2011). The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 6:32. 10.1186/1749-8104-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. C., Morris K. V., Wood M. J. (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130507. 10.1098/rstb.2013.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T., Hoki Y., Sasaki H. (2005). Tsix silences Xist through modification of chromatin structure. Dev. Cell 9 159–165. 10.1016/j.devcel.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Salameh A., Lee A. K., Cardo-Vila M., Nunes D. N., Efstathiou E., Staquicini F. I., et al. (2015). PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. U.S.A. 112 8403–8408. 10.1073/pnas.1507882112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarola M., Comisso E., Pascolo R., Chiaradia R., Marion R. M., Schneider C., et al. (2015). Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 6:7631. 10.1038/ncomms8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C. J., Jacob C. P., Buttenschon H. N., Kittel-Schneider S., Boreatti-Hummer A., Zimmer M., et al. (2010). Functional variants of TSPAN8 are associated with bipolar disorder and schizophrenia. Am. J. Med. Genet. 153B, 967–972. 10.1002/ajmg.b.31057 [DOI] [PubMed] [Google Scholar]
- Seidman L. J., Mirsky A. F. (2017). Evolving notions of schizophrenia as a developmental neurocognitive disorder. J. Int. Neuropsychol. Soc. 23 881–892. 10.1017/S1355617717001114 [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J., Gaughwin P. M., Lim B., Robson P., Lipovich L. (2010). Conserved long noncoding RNAs transcriptionally regulated by Oct4 and nanog modulate pluripotency in mouse embryonic stem cells. RNA 16 324–337. 10.1261/rna.1441510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C. H., Zhang L., Qin C. (2017). Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 132 160–169. 10.1016/j.brainresbull.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Song Y., Xuan A., Bu C., Ci D., Tian M., Zhang D. (2018). Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. J. 10.1111/pbi.12955 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G., Wahlestedt C., Kapranov P. (2015). The landscape of long noncoding RNA classification. Trends Genet. 31 239–251. 10.1016/j.tig.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Huang L., Li Z., Leng K., Xu Y., Jiang X., et al. (2018). Long non-coding RNA MIAT in development and disease: a new player in an old game. J. Biomed. Sci. 25:23. 10.1186/s12929-018-0427-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Del Rosario B. C., Szanto A., Ogawa Y., Jeon Y., Lee J. T. (2013). Jpx RNA activates xist by evicting CTCF. Cell 153 1537–1551. 10.1016/j.cell.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo J. S., Lee S. T., Im W., Lee M., Byun J. I., Jung K. H., et al. (2017). Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol. Neurobiol. 54 1577–1586. 10.1007/s12035-016-9928-9 [DOI] [PubMed] [Google Scholar]
- Tang J., Yu Y. Z., Yang W. (2017). Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci. Ther. 23 645–656. 10.1111/cns.12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. E., Ling D., Nelson C., Atmadibrata B., Dinger M. E., Xu N., et al. (2014). The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget 5 1793–1804. 10.18632/oncotarget.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K. W., Sansom S. N., Lee S., Chalei V., Kong L. S., Cooper S. E., et al. (2014). The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 33 296–311. 10.1002/embj.201386225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P. X., Su W. R., Zhuo Y. H. (2017). The role of long noncoding RNAs in neurodegenerative diseases. Mol. Neurobiol. 54 2012–2021. 10.1007/s12035-016-9793-6 [DOI] [PubMed] [Google Scholar]
- Wang K. C., Chang H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhao Y., Bao X., Zhu X., Kwok Y. K., Sun K., et al. (2015a). LncRNA dum interacts with dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25 335–350. 10.1038/cr.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. H., Lin Y. S., Chen Y., Yeh C. T., Huang Y. L., Hsieh T. H., et al. (2015b). Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget 6 23342–23357. 10.18632/oncotarget.4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao X., Ju W., Flory M., Zhong J., Jiang S., et al. (2015c). Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Trans. Psychiatry 5:e660. 10.1038/tp.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pang W. J., Wei N., Xiong Y., Wu W. J., Zhao C. Z., et al. (2014). Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene 539 117–124. 10.1016/j.gene.2014.01.037 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., et al. (2013). Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25 69–80. 10.1016/j.devcel.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Wilkinson B., Campbell D. B. (2013). Contribution of long noncoding RNAs to autism spectrum disorder risk. Int. Rev. Neurobiol. 113 35–59. 10.1016/B978-0-12-418700-9.00002-2 [DOI] [PubMed] [Google Scholar]
- Wu B., Chen M., Gao M., Cong Y., Jiang L., Wei J., et al. (2018). Down-regulation of lncTCF7 inhibits cell migration and invasion in colorectal cancer via inhibiting TCF7 expression. Hum. Cell 32 31–40. 10.1007/s13577-018-0217-y [DOI] [PubMed] [Google Scholar]
- Wu D., Zhao B., Cao X., Wan J. (2017). Long non-coding RNA LINK-A promotes glioma cell growth and invasion via lactate dehydrogenase A. Oncol. Rep. 38 1525–1532. 10.3892/or.2017.5806 [DOI] [PubMed] [Google Scholar]
- Wu H., Yang L., Chen L. L. (2017). The diversity of long noncoding RNAs and their generation. Trends Genet. 33 540–552. 10.1016/j.tig.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Wu Y. T., Huang C., Meng X. M., Li J. (2015). Long noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr. Pharm. Design 21 5017–5028. 10.2174/1381612821666150724115625 [DOI] [PubMed] [Google Scholar]
- Xin Y., Li Z., Zheng H. Y., Chan M. T. V., Wu W. K. K. (2017). CCAT2: a novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 50:e12342. 10.1111/cpr.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z., Lin A., Li C., Liang K., Wang S., Liu Y., et al. (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159 1110–1125. 10.1016/j.cell.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Deng X., Ma W., Berletch J. B., Rabaia N., Wei G., et al. (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16:52. 10.1186/s13059-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lin C., Jin C., Yang J. C., Tanasa B., Li W., et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500 598–602. 10.1038/nature12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. F., Yan P. X., Lu J. L., Song G., Zhu Y. Y., Li Z. H., et al. (2015). Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16 504–516. 10.1016/j.stem.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Young T. L., Matsuda T., Cepko C. L. (2005). The noncoding RNA Taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 15 501–512. 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- Yu C. Y., Kuo H. C. (2016). The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells 34 2052–2062. 10.1002/stem.2386 [DOI] [PubMed] [Google Scholar]
- Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. (2016). The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 11:e0158347. 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang C., Gao Z., Wu H., Gu R., Jiang R. (2018). Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration, and invasion by regulating microRNA-21. J. Cell. Biochem. 119 6461–6469. 10.1002/jcb.26671 [DOI] [PubMed] [Google Scholar]
- Zhao Z. L., Senturk N., Song C. L., Grummt I. (2018). lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Gene Dev. 32 836–848. 10.1101/gad.311688.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Guo H., Zhou B., Feng J., Li Y., Han T., et al. (2016). Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene 35 5770–5780. 10.1038/onc.2016.110 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D. L., Gejman R., et al. (2007). Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282 24731–24742. 10.1074/jbc.M702029200 [DOI] [PubMed] [Google Scholar]
- Ziats M. N., Rennert O. M. (2013). Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 49 589–593. 10.1007/s12031-012-9880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]