Highlights
-
•
Viral, organelle and bacterial genomes can be synthesized.
-
•
Sc2.0 is on the way to generating the first synthetic designer eukaryote.
-
•
6 of 17 S. cerevisiae chromosomes are synthesized and published.
-
•
GP-write will open a new era of synthetic genomics.
Abstract
Since the first synthetic gene was synthesized in 1970s, the efficiency and the capacity of made-to-order DNA sequence synthesis has increased by several orders of magnitude. Advances in DNA synthesis and assembly over the past years has resulted in a steep drop in price for custom made DNA. Similar effects were observed in DNA sequencing technologies which underpin DNA-reading projects. Today, synthetic DNA sequences with more than 10 000 bps and turn-around times of a few weeks are commercially available. This enables researchers to perform large-scale projects to write synthetic chromosomes and characterize their functionalities in vivo. Synthetic genomics opens up new paradigms to study the genome fundamentals and engineer novel biological functions.
Current Opinion in Chemical Biology 2018, 46:56–62
This review comes from a themed issue on Synthetic biomolecules
Edited by Richard J Payne and Nicolas Winssinger
For a complete overview see the Issue and the Editorial
Available online 9th May 2018
https://doi.org/10.1016/j.cbpa.2018.04.002
1367-5931/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
One of the major challenges in biological sciences was the determination of DNA sequences. In the beginning, only single DNA fragments were sequenced using the chain termination sequencing technique [1]. However, the Human Genome Project (GP-Read) accelerated the evolution of new sequencing techniques by having the ambitious goal to sequence the human genome within 15 years. The development of Next Generation Sequencing techniques today allows sequencing of a human genome within days. However, most eukaryotic genomes are not fully sequenced and new sequencing techniques are still being developed. As exemplary achievement of this development, in 2017 sequencing of one of the highly repetitive human centromeres was achieved [2•]. Scientists are now performing well in reading genomes, a measurable output being the growing number of genome sequences in public databases. However, reading a book alone does not make a good writer, instead it requires one to start writing extensively and creatively to master the art and ultimately it leads to a better understanding of grammar and expression. In this case, one needs to write synthetic DNA sequences in order to better understand the grammar of life.
Writing DNA starts with short single-stranded fragments: the oligonucleotides. Since the development of the Polymerase Chain Reaction and the first complete synthesis of a gene, writing DNA in vitro has progressed impressively (Figure 1 ) [3, 4]. Recent drops in DNA synthesis costs and the improved capability of synthesizing longer stretches of DNA allow the design and construction of whole synthetic chromosomes in the mega-base range. Recent publications report the construction of viral and microbial synthetic genomes, and the Sc2.0 project aims to generate the first synthetic eukaryotic genome. It is an open discussion how to define whether a chromosome or genome is synthetic. In this review, chromosomes and genomes are defined as synthetic when all building blocks of the final DNA molecule are generated by chemical synthesis. Chromosomes and genomes which are not completely synthesized are considered ‘engineered’ or ‘modified’ and are outside the scope of this review. We define synthetic genomics to be a new field where biology is being engineered at the genome level, and it is an intersection of synthetic biology and systems biology. This review neither aims to discuss assembly methods nor the dual-use character of synthetic genomics. The authors are fully aware of the potential dual use character, especially for the synthesis of viral genomes. However, these issues are discussed and reviewed extensively elsewhere [5, 6, 7].
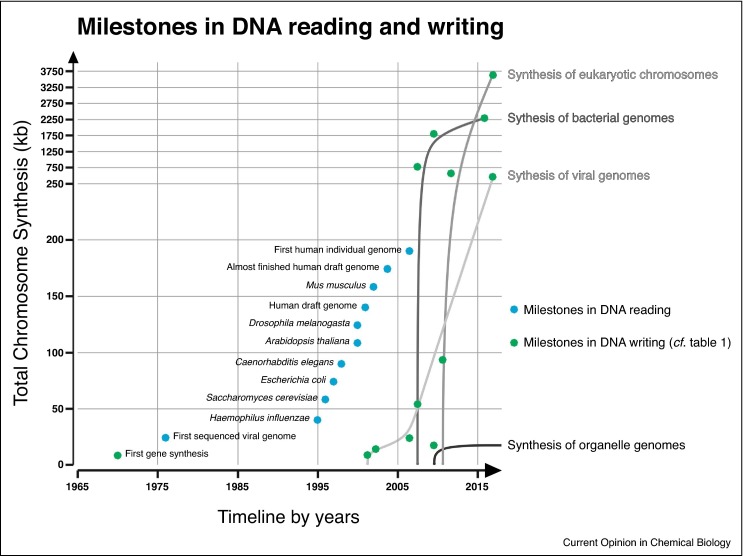
Figure 1.
Milestones in DNA reading and writing. Reading and writing technologies depend on technological breakthroughs. Large scale DNA-reading projects (examples in blue) were accomplished after development of Sanger Sequencing, PCR and Next Generation Sequencing. The number of studies utilizing new technologies grows quickly after a developmental lag phase. The number of genome sequences uploaded to databases is exploding and it is impossible to give a number which would be accurate and valid for some time. The knowledge gained by genome sequencing and advantages in gene synthesis is the dawn of writing chromosomes. By now the number of bases incorporated into completely synthetic chromosomes is: 6.1 mb. The cost by today would be roughly $425 000 assuming the current competitive price rate of 7 cents per base for non-clonal 1.8 kb DNA-fragments. The synthesis cost for a haploid human genome would by today be roughly $45 000 000. However, lowering DNA synthesis costs is one of the major goals of GP-write. In the future, the DNA synthesis cost of a human genome in will be less than the price of the Mycoplasma mycoides JCVI-syn1.0 project (estimated $40 000 000 [33]).
Design concepts and assembly strategies for synthetic chromosomes
Computer-assisted design software (CADs) have been developed to ensure efficient and consistent design of synthetic DNA sequences at the genome scale [8••, 9•]. The design space of synthetic DNA is enormous and many (if not infinite) design blueprints are possible, as long as they can result in the viability of the cell, to achieve the design intention [10]. Initial projects aiming to synthesize a whole genome were conservative in changes to the genetic content, but nonetheless resulted in the breakthrough in synthesizing, assembling and ultimately transplanting chromosome-scale synthetic DNA [11, 12•]. With increasing knowledge and progress in chromosome-scale DNA synthesis, the designs of synthetic sequences are becoming more complex and ambitious [8••, 13••]. Many genome synthesis projects utilize a hierarchical genome assembly strategy starting with small building blocks which are assembled, by the technique of choice, to larger building blocks of around 50–100 kb. These fragments are used to further assemble the synthetic chromosome in a heterologous host or to replace the corresponding wildtype sequence in a stepwise manner. Each of the techniques have advantages and disadvantages (Box 1 ), and should be chosen carefully based on the use cases.
Box 1. Advantages of heterologous host or native host for chromosome assembly.
Advantages of chromosome assembly in heterologous host:
-
•
Assembly in a heterologous host potentially prevents cellular burden to the targeted host.
-
•
Well-established assembly methods are readily available (e.g. homologous recombination in S. cerevisiae).
Disadvantages of chromosome assembly in heterologous host:
-
•
The size of synthetic chromosomes might be limited by the capacity of the heterologous host.
-
•
The synthetic chromosome needs to be transferred to the final host.
-
•
Difficult to debug design defects until the synthesized chromosome is transplanted to the target host.
Advantages of step-by-step chromosome replacement:
-
•
Replacing the wildtype sequence gives a real-time fitness monitoring.
-
•
No need to transfer the final chromosome to a different organism.
-
•
Multiplex replacement can be carried out simultaneously for more efficient assembly.
Disadvantages of step-by-step chromosome replacement:
-
•
Some organisms are not amenable to homologous recombination.
-
•
Might be very time consuming if the host is a slow growing organism.
-
•
It might be necessary to develop new tools to manipulate the target organism.
Synthesizing DNA goes viral
Although viruses and phages are not considered to be ‘alive’ they have a genome. They can reproduce themselves by leveraging the resources from a host. Viral genomes are rather small, with sizes between 1759 bps (Porcine circovirus [14]) and 1259 kb (Megavirus chilensis [15]) and can consist of DNA or RNA. The first complete synthesis of a viral RNA genome, the polio virus, was accomplished in 2002 [16••]. The 7.5 kb synthesized cDNA genome was in vitro transcribed by RNA polymerase and can generate infectious virus particles after transfer into a cell free extract. Further viral RNA and DNA genomes were synthesized up to a size of 212 kb in recent years (Table 1 ). Synthesizing, as well as engineering variations of viral genomes to produce genome libraries, has an enormous potential for therapeutic applications. Vaccines and drugs could be quickly generated in response to the emergency of a certain virus variant, which may help to prevent wider outbreaks [7].
Table 1.
Overview of finished synthesized chromosomes and genomes
| Year | Species | Size in kb (% of wt genome) | Highlights of the study | Reference |
|---|---|---|---|---|
| Synthetic viral genomes | ||||
| 2002 | Polio virus | 7.5 | First infectious viral particles, based on a fully synthetic genome. | [16••] |
| 2003 | φX174 bacteriophage | 5.4 | Whole workflow: design, oligo synthesis, genome assembly and generation of infectious viral particles in fourteen days. | [34] |
| 2007 | Human endogenous retrovirus (HERV-K) consensus | 9.5 | Generation of a consensus genome of a human endogenous retrovirus which can replicate and is infectious. | [35] |
| 2008 | Bat severe acute respiratory syndrome (SARS)-like coronavirus (Bat-SCoV) | 29.7 | Until 2010 the largest synthetic replicating life form. | [36] |
| 2017 | Horsepox | 212 | First de novo synthesis of an in nature extinct orthopoxvirus of which close relatives causes smallpox in humans. | [7] |
| Synthetic bacterial genomes | ||||
| 2008 | Mycoplasma genitalium | 583 | Watermark sequences were inserted to identify synthetic DNA; disruption of one gene (MG408) to prevent pathogenicity and to do antibiotic selection. | [11] |
| 2010 |
Mycoplasma mycoides JCVI-syn1.0 |
1079 | Watermark sequences were inserted to identify synthetic DNA; deletion or disruption of fourteen genes (one accidentally mediated by an IS1 E. coli transposon). | [12•] |
| 2016 |
Mycoplasma mycoides JCVI-syn3.0 |
531.6 (49.3%) | Reduction of Mycoplasma mycoides JCVI-syn1.0 in a design, build, test manner based on transposon mutagenesis to reduce the genome from 901 to 473 genes. | [13••] |
| Synthetic eukaryotic organelle genomes | ||||
| 2010 | Mus musculus mtDNA | 16.3 | Complete in vitro synthesis in four steps to produce a copy of the mouse mtDNA consensus genome. | [18] |
| Synthetic eukaryotic chromosomes | ||||
| 2011 | Saccharomyces cerevisiae synIXR | 91.0 (101.9%)a | Applied design rules (if possible): • Synonymous replacement of TAG stop codons with TAA • Synonymous recoding of tandem repeats in CDSs • Deletion of genomic instability causing elements: introns, transposons, subtelomeric repeats and tRNA genes • Insertion of loxPsym sites to ‘SCRaMbLE’ the genome • Insertion of PCRTag sequences to identify synthetic DNA |
[20] |
| 2012 | Saccharomyces cerevisiae synIII | 272.2 (86.2%) | [24••] | |
| 2017 | Saccharomyces cerevisiae synII | 770.1 (94.7%) | [25] | |
| 2017 | Saccharomyces cerevisiae synV | 536.0 (92.9%) | [28] | |
| 2017 | Saccharomyces cerevisiae synVI | 242.7 (89.9%) | [26] | |
| 2017 | Saccharomyces cerevisiae synX | 707.5 (94.9%) | [29] | |
| 2017 | Saccharomyces cerevisiae synXII | 999.4 (92.7%)b | [27] | |
The size of synIXR is slightly longer due to the insertion of 43 loxPsym sites (34 bps each).
Chromosome XII contains a cluster with >100 copies of the 9.1 kb rDNA operon which is not included in the shown size.
Synthesizing genomes of organelles
Mitochondria in general, and the plastids of plants, contain a genome. Their sizes are rather small but show a huge variation in size and content. Studying these organelles is very interesting but challenging. Transformation of organelles must be done by bio-ballistic transformation [17]. The efficiency of synthetic DNA transformation is rather low. Mitochondria are the only organelles for which a complete organelle genome has been synthesized so far. The synthesis of the 16.3 kb mouse mtDNA genome was achieved by using 600 60mer oligonucleotides in four consecutive assembly rounds [18]. This step was predominantly the proof of principle for a DNA assembly method. However, it is an intriguing question why organelles still contain genetic content and have not migrated all necessary genes to the nucleus. There are exceptions in nature where the mitochondria do not contain any DNA [19].
Synthesizing microbial genomes
The genome of Mycoplasma genitalium was the first completely chemically synthesized genome [11]. However, the genome could not be transferred to produce a viable synthetic M. genitalium strain, presumably because of an interruption of rnpB, a subunit of RNaseP. In 2010, the same group demonstrated, in a remarkable work, the complete chemical synthesis and transfer of the 1.08 Mb Mycoplasma mycoides genome into the close relative Mycoplasma capricolum [12•]. This is the first organism which is controlled by a synthetic genome, and is referred as M. mycoides JCVI-syn1.0. The genomic differences to M. mycoides are marginal and consist of designed ‘watermark’ sequences, 14 genes are deleted or disrupted and nineteen harmless polymorphisms were acquired during the building process.
This successful project was the starting point to generate a minimal Mycoplasma organism based on JCVI-syn1.0. Briefly, two independent teams failed to generate a viable cell, based on knowledge and genome synthesis, from scratch. However, multiple rounds of transposon mutagenesis and genome reduction finally generated M. mycoides JCVI-syn3.0 a minimal genome with a genome reduction of 50.8% in a design, build and test cycle manner [20, 13••]. The 901 genes of M. mycoides JCVI-syn1.0 were reduced to 473 genes of which 149 are of unknown function and will give deeper insights into essentiality of genes [21].
An interesting ongoing project is the generation of a synthetic Escherichia coli genome. The genome of a previously diminished E. coli strain is redesigned in 87 ca. 50 kb segments to eliminate 7 codons in the coding sequence in a stepwise manner [9•]. The 62 214 (5.4%) excluded codons are replaced by synonymous codons to maintain viability. The freed-up codons may be used to incorporate non-natural amino acids into proteins in the future. Absence of seven codons and corresponding tRNAs will, in addition, provide sufficient resistance to phages, rendering this strain of great general interest. Currently 55 of the 87 segments have been tested experimentally but the incorporation into a fully synthetic E. coli genome still needs to be proven functional.
Synthesizing eukaryotic genomes
As of today, there is no complete synthetic eukaryotic genome. However, the synthetic yeast genome — Sc2.0 project (www.syntheticyeast.org) aims to generate the first eukaryotic cell operated by a synthetic genome. The 16 chromosomes are synthesized in individual strains by teams of scientists within the Sc2.0 Consortium. The chromosomes are re-designed in a higher order of magnitude compared to any other existing write project [8••]. The major changes include the removal of most introns, transposons and repetitive elements. One central element of Sc2.0 design is the relocation of all tRNA genes to an independent 17th Saccharomyces cerevisiae chromosome, designated as the tRNA neochromosome. tRNA genes are heavily transcribed and therefore are hotspots of genomic instability caused by replication stress and transposon insertions. In addition, all non-essential genes are flanked by loxPsym sites which allow inducible large-scale genomic re-arrangements mediated by Cre-recombinase. This implemented genome rearrangement technique is therefore referred to as Synthetic Chromosome Rearrangement and Modification by LoxP-mediated Evolution (SCRaMbLE) and has already proven its functionality [20, 22•, 23•].
Recent publications report the synthesis and characterization of six Sc2.0 chromosomes and the right arm of synthetic chromosome IX (Table 1) which collectively correspond to 32% of the yeast genome [20, 24••, 25, 26, 27, 28, 29]. Strikingly, the individually synthesized chromosomes can be merged in a single cell by mating with a technique called endoreduplication intercross [8••]. Currently, the strain with the most synthetic chromosomes in one cell contains synIII, synVI and synIXR. With further progression of the Sc2.0 project more synthetic chromosomes will be finalised and ultimately merged to the final Sc2.0 strain.
GP-write: a sneak preview into the future of synthetic genomics
The successes in current genome synthesizing projects are leading to the next grand challenge in modern biological science: The Genome Project-write (GP-write). This project is a grand challenge using synthesis, gene editing and other technologies to understand, engineer and test living systems with the overarching goal to understand the blueprint for life provided by the Human Genome Project (HGP-read) [30••, 31, 32]. Therefore, a new international consortium was formed and first meetings were held in 2016 and 2017. The consortium is an open, interdisciplinary and international research group to focus efforts to realize GP-write.
GP-write has several goals, one being the development of new techniques and to accelerate the evolution of existing techniques with an overall goal to reduce synthesis costs by 1000-fold within ten years. Similar effects were achieved by HGP-read: today the cost of sequencing a human genome are magnitudes lower than the initial human genome sequence. The open nature of GP-write allows everyone to submit project proposals which will be evaluated by the Scientific Executive Committee. As of January 2018 there are 13 pilot projects approved (Table 2 ). The projects cover many aspects of synthetic genomics, two highlights are the projects dedicated to the Concepts and Ethics in GP-write as well as Anticipating and Understanding Governance Systems and the Publics Views on HGP-write, which shows the importance to consider ethics and the publics views within GP-write.
Table 2.
Approved GP-write pilot projects (by January 2018)
| Project title | Project goalsa | Project lead(s) |
|---|---|---|
| UltraSafe Cell Line | The project aims to generate an Ultrasafe cell line by altering roughly 1% of the human genome. Some key goals are: Virus and prion resistance, removal of transposable elements, recoding of triplet repeats, recoding to a human consensus sequence in regard to SNPs and indels, implementing the bespoke SCRaMbLE system beside further alterations. | Jef Boeke & George Church |
| High-throughput HAC Design to Test Connections Between Gene Expression, Location and Conformation | There is still a lack of understanding of the regulation of gene expression. The project will build two 1 mb regions of the human genome. The regions will be constructed as combinatorial libraries with different promoters and insulators to investigate ‘rules’ for optimal gene expression. | Pamela Silver & Jeffrey Way |
| Safety and Containment; Chromatin and Chromosome Structure | This project tries to solve two questions based on Sc2.0 strains and implement the results into GP-write. Firstly: how can hybridization with natural occurring strains or organisms be excluded? Second: what shapes the contact maps of chromosomes? The latter will be answered by analysing multiple SCRaMbLed Sc2.0 strains to investigate chromatin structure based on re-arranged chromosomes | Jasper Rine |
| Empirically Designing Genomically Recoded Human Cell Lines | Codon alteration is an important part of GP-write. The project aims to develop: Firstly, a rapid method for multiplex targeted genome modification; secondly, a respective rapid and robust screening system for living cells in 96-well format; thirdly, a strategy for rapid evaluation of heterogenic cell populations; and finally, a software to design the synthetic DNA fragments and evaluate viability of codon replacements. | Marc Lajoie |
| The Seven Signals Toolbox: Leveraging Synthetic Biology to Define the Logic of Stem-Cell Programming | Cell differentiation is mainly driven by seven signal types. This project aims to generate a toolbox which allows the in vitro differentiation of GP-write cell lines. This is a crucial step for future applications in the field of cell therapies, tissue replacement or transplantation of organs. | Liam Holt |
| Precision Human Genome Engineering of Disease-Associated Noncoding Variants | Efficient and precise engineering of the human genome is still a challenge. This project aims to create a complete pipeline for rapid engineering of human cells with an enrichment for homologous recombination repair. The project will also provide bioinformatic tools to optimize CRISPR based engineering. | Neville Sanjana |
| Synthesizing a Prototrophic Human Genome | This project postulates to introduce pathways for the nine amino acids and a variety of vitamins which cannot be synthesized by humans. These molecules derive from the diet. It investigates whether the milieu in the cell makes a prototrophic cell line feasible. If the project succeeds, further engineering would be performed and the first achievement would be a drastic cost reduction of cell line cultivation media. | Harris Wang |
| Through the Looking Glass: Anticipating and Understanding Governance Systems and the Public's Views on HGP-write | Including the publics view and governance systems into GP-write is an important step. This project will generate a dialogue between scientists and the public. Incorporation of the society will enable acceptance and support for GP-write. | Todd Kuiken & Gigi Gronvall |
| Synthetic Screening for Essential Introns and Retroelements in Human Cell and Animals | This project aims to perform systematic screenings of intron and retroelements in the genome. Combinatorial variants of chosen genes will be investigated in a diploid background. The outcome will indicate if the removal of these elements, like in Sc2.0, is feasible in GP-write. | Yasunori Aizawa |
| Isothermal Amplification Array | This project proposes a new method to synthesize DNA. It depends on two steps. Firstly: generation of short oligonucleotides by an isothermal amplification on an array. Secondly: the amplified oligonucleotides can anneal according to their design and nicks are sealed by a ligase. | Max Berry |
| Recombinase-Mediated Assembly | This project proposes a new method to assemble DNA fragments by utilizing a RecA-like recombinase (UvsX). The method should allow, with the Isothermal Amplification Array assembly from short oligos to chromosome-sized DNA, with a significant labour reduction. | Max Berry |
| Synthetic Regulatory Genomics | The project aims to study regulatory variations of non-coding regions. The project will use multi-edited regulatory DNA sequences and analyse their function with multiple techniques. This project will give deeper insights into non-coding regions of complex genomes. | Matt Maurano |
| Concepts & Ethics in GP-write: Understand, Question, Advance | This project aims to build a model for deep analysis of concepts and ethics in GP-write, and aims include the dynamics of science and society. It aims to expand collaborations between sciences and the humanities and provide proper education and training. | Jeantine Lunshof |
More detailed information can be found at http://www.engineeringbiologycenter.org/.
One major remaining question is: What can we learn from GP-write? On one hand, there will be the ad hoc advances in enabling technologies, on the other hand there will be an immense gain of knowledge in biological sciences. Our knowledge of complex genomes is still limited. For instance, roughly 1% of the genome is responsible for all proteins in the cell. The remaining 99% are often referred as the ‘dark matter’ of the genome. Stepwise replacement of these elements, like in the Sc2.0 project, will potentially help us decipher the functions of the dark matter in the genome. On the application front, the pilot project to engineer a stable and safe cell line, has a profound implication for biomanufacturing and bioproduction (Table 2). GP-write still has a long way to go. However, the scientific community is curious about the outcome of the first pilot projects in the GP-write framework.
Conclusion
The initial genome writing projects summarized here show that individual native chromosomes and whole genomes can be replaced by chemically synthesized genomes. So far, the changes to DNA sequences are relatively modest but with growing knowledge of biological systems, the design will become more aggressive and adventurous which will lead us into previously unexplored territories. The exciting field of synthetic genomics will give new insights in basic research and will open new possibilities in applied science.
Funding
The work in the UK is funded through a Biotechnology and Biological Sciences Research Council grant (BB/P02114X/1), and the University of Manchester President's Award for Research Excellence to YC. This work is also supported by the National Natural Science Foundation of China (31471254 and 31725002) and partially supported by the Bureau of International Cooperation, Chinese Academy of Sciences (172644KYSB20170042) to JD.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The authors are grateful to the Sc2.0 consortium and the GP-Write community for helpful discussions. We thank Sally Jones for proof-reading the manuscript, and the support from the Manchester Centre for Synthetic Biology of Fine and Speciality Chemicals (SYNBIOCHEM, grant BB/M017702/1).
References
- 1.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.Jain M., Olsen H.E., Turner D.J., Stoddart D., Bulazel K.V., Paten B., Haussler D., Willard H., Akeson M., Miga K.H. Linear assembly of a human Y centromere using nanopore long reads. bioRxiv. 2017 doi: 10.1038/nbt.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]; The human genome reference still contain gaps, especially in highly repetitive regions. This study shows how highly repetitive regions can be analysed using a targeted aproach and a new generation of sequencing devices.
- 3.Agarwal K.L., Buchi H., Caruthers M.H., Gupta N., Khorana H.G., Kleppe K., Kumar A., Ohtsuka E., Rajbhandary U.L., Van de Sande J.H. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature. 1970;227:27–34. doi: 10.1038/227027a0. [DOI] [PubMed] [Google Scholar]
- 4.Saiki R.K., Scharf S., Faloona F., Mullis K.B., Horn G.T., Erlich H.A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 5.Oldfield L.M., Grzesik P., Voorhies A.A., Alperovich N., MacMath D., Najera C.D., Chandra D.S., Prasad S., Noskov V.N., Montague M.G. Genome-wide engineering of an infectious clone of herpes simplex virus type 1 using synthetic genomics assembly methods. Proc Natl Acad Sci U S A. 2017;114:E8885–E8894. doi: 10.1073/pnas.1700534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell L.A., Ellis T. Synthetic genome engineering gets infectious. Proc Natl Acad Sci U S A. 2017;114:11006–11008. doi: 10.1073/pnas.1715365114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koblentz G.D. The de novo synthesis of horsepox virus: implications for biosecurity and recommendations for preventing the reemergence of smallpox. Health Secur. 2017;15:620–628. doi: 10.1089/hs.2017.0061. [DOI] [PubMed] [Google Scholar]
- 8••.Richardson S.M., Mitchell L.A., Stracquadanio G., Yang K., Dymond J.S., DiCarlo J.E., Lee D., Huang C.L., Chandrasegaran S., Cai Y. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]; Summary of the design and principle of the Sc2.0 project including how to construct the final Sc2.0 strain with all synthetic chromosomes merged in one cell.
- 9•.Ostrov N., Landon M., Guell M., Kuznetsov G., Teramoto J., Cervantes N., Zhou M., Singh K., Napolitano M.G., Moosburner M. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]; Ongoing study to generate a re-coded E. coli genome which has the greatest degree of re-coding of coding sequence so far. One result of this project will be a phage resistant E. coli strain.
- 10.Schindler D., Waldminghaus T. Synthetic chromosomes. FEMS Microbiol Rev. 2015;39:871–891. doi: 10.1093/femsre/fuv030. [DOI] [PubMed] [Google Scholar]
- 11.Gibson D.G., Benders G.A., Andrews-Pfannkoch C., Denisova E.A., Baden-Tillson H., Zaveri J., Stockwell T.B., Brownley A., Thomas D.W., Algire M.A. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 12•.Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]; The first chemically synthesized genome which can control a cell. This study is the dawn of synthetic genomics, it shows feasability of synthetic genomes.
- 13••.Hutchison C.A., 3rd, Chuang R.Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L. Design and synthesis of a minimal bacterial genome. Science. 2016;351 doi: 10.1126/science.aad6253. aad6253. [DOI] [PubMed] [Google Scholar]; Reduction of the first synthetic bacterial genome by 49.3% in multiple rounds with classification of the remaining genes into essential and quasi-essential (necessary for growth) genes.
- 14.Finsterbusch T., Mankertz A. Porcine circoviruses — small but powerful. Virus Res. 2009;143:177–183. doi: 10.1016/j.virusres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Arslan D., Legendre M., Seltzer V., Abergel C., Claverie J.M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A. 2011;108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Cello J., Paul A.V., Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]; The first chemically synthesized and assembled genome which is able to produce infectious viral particles.
- 17.Klein R.M., Wolf E.D., Wu R., Sanford J.C. High-velocity microprojectiles for delivering nucleic acids into living cells. Biotechnology. 1992;24:384–386. [PubMed] [Google Scholar]
- 18.Gibson D.G., Smith H.O., Hutchison C.A., 3rd, Venter J.C., Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 19.Henriquez F.L., Richards T.A., Roberts F., McLeod R., Roberts C.W. The unusual mitochondrial compartment of Cryptosporidium parvum. Trends Parasitol. 2005;21:68–74. doi: 10.1016/j.pt.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Dymond J.S., Richardson S.M., Coombes C.E., Babatz T., Muller H., Annaluru N., Blake W.J., Schwerzmann J.W., Dai J., Lindstrom D.L. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danchin A., Fang G. Unknown unknowns: essential genes in quest for function. Microb Biotechnol. 2016;9:530–540. doi: 10.1111/1751-7915.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Shen Y., Stracquadanio G., Wang Y., Yang K., Mitchell L.A., Xue Y., Cai Y., Chen T., Dymond J.S., Kang K. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2016;26:36–49. doi: 10.1101/gr.193433.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first study which shows a great diversification in synthetic chromosomes by SCRaMbLE. It highlights the need for better techniques to resolve the structure of the re-arranged chromosomes as some SCRaMbLEd chromosomes could not be resolved by short sequencing reads.
- 23•.Mercy G., Mozziconacci J., Scolari V.F., Yang K., Zhao G., Thierry A., Luo Y., Mitchell L.A., Shen M., Shen Y. 3D organization of synthetic and scrambled chromosomes. Science. 2017;355 doi: 10.1126/science.aaf4597. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study indicates that the re-design of the analyzed synthetic yeast chromosomes has no or very little effect on the 3D organization within the nucleus.
- 24••.Annaluru N., Muller H., Mitchell L.A., Ramalingam S., Stracquadanio G., Richardson S.M., Dymond J.S., Kuang Z., Scheifele L.Z., Cooper E.M. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first synthetic eukaryotic designer chromosome which produces viable cells. The study proves synthetic chromosomes are not restricted to viral and bacterial genomes.
- 25.Shen Y., Wang Y., Chen T., Gao F., Gong J., Abramczyk D., Walker R., Zhao H., Chen S., Liu W. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355 doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell L.A., Wang A., Stracquadanio G., Kuang Z., Wang X., Yang K., Richardson S., Martin J.A., Zhao Y., Walker R. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355 doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W., Zhao G., Luo Z., Lin Y., Wang L., Guo Y., Wang A., Jiang S., Jiang Q., Gong J. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355 doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 28.Xie Z.X., Li B.Z., Mitchell L.A., Wu Y., Qi X., Jin Z., Jia B., Wang X., Zeng B.X., Liu H.M. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355 doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y., Li B.Z., Zhao M., Mitchell L.A., Xie Z.X., Lin Q.H., Wang X., Xiao W.H., Wang Y., Zhou X. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355 doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Boeke J.D., Church G., Hessel A., Kelley N.J., Arkin A., Cai Y., Carlson R., Chakravarti A., Cornish V.W., Holt L. GENOME ENGINEERING. The Genome Project-Write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]; Publication which launch the GP-write and opens the discussion for the grand challenge to synthesize complex eukaryotic genomes.
- 31.Boeke J.D., Church G., Hessel A., Kelley N.C. Engineer and Test Living Systems. White Paper; 2016. The GP-write Consortium: Genome Project-write: A Grand Challenge. Using Synthesis, Gene Editing and Other Technologies to Understand. [Google Scholar]
- 32.http://www.engineeringbiologycenter.org/
- 33.Pennisi E. Genomics. Synthetic genome brings new life to bacterium. Science. 2010;328:958–959. doi: 10.1126/science.328.5981.958. [DOI] [PubMed] [Google Scholar]
- 34.Smith H.O., Hutchison C.A., 3rd, Pfannkoch C., Venter J.C. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y.N., Bieniasz P.D. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]