Abstract
The prognosis of patients with relapsed/refractory (R/R) diffuse large B-cell lymphoma (DLBCL) remains unsatisfactory and, despite major advances in genomic studies, the biological mechanisms underlying chemoresistance are still poorly understood. We conducted for the first time a large-scale differential multi-omics investigation on DLBCL patient’s samples in order to identify new biomarkers that could early identify patients at risk of R/R disease and to identify new targets that could determine chemorefractoriness. We compared a well-characterized cohort of R/R versus chemosensitive DLBCL patients by combining label-free quantitative proteomics and targeted RNA sequencing performed on the same tissues samples. The cross-section of both data levels allowed extracting a sub-list of 22 transcripts/proteins pairs whose expression levels significantly differed between the two groups of patients. In particular, we identified significant targets related to tumor metabolism (Hexokinase 3), microenvironment (IDO1, CXCL13), cancer cells proliferation, migration and invasion (S100 proteins) or BCR signaling pathway (CD79B). Overall, this study revealed several extremely promising biomarker candidates related to DLBCL chemorefractoriness and highlighted some new potential therapeutic drug targets. The complete datasets have been made publically available and should constitute a valuable resource for the future research.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most frequent subtype of non-Hodgkin lymphoma (NHL) and is a clinically and biologically heterogeneous disease. The anthracycline-based regimen R-CHOP (rituximab, cyclophosphamide, doxorubicine, vincristine and prednisone) is still considered as the standard of care for first-line treatment with approximately 60% of the patients achieving a complete response. The prognosis of patients with primary refractory or early-relapsed (R/R) disease is particularly poor with a median overall survival below one year. Because of the acquisition of chemoresistance, only a fraction of R/R patients can be cured with salvage therapies1.
Recent advances in molecular biology, genetics and high throughput –omics technologies have led to a better understanding of the biology of this disease and the distinction of several subtypes of DLBCL2. Based on the cell-of-origin classification, the two major molecular subgroups are germinal center B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL that notably differ in their clinical outcomes3. Cytogenetic studies have highlighted the major importance of MYC, BCL2 and BCL6 rearrangements4. In parallel, the mutational landscape of DLBCL has been extensively studied, demonstrating the intratumoral heterogeneity and allowing the identification of recurrent somatic mutations, some of which provide promising opportunities for new drug developments5. However, the mechanisms underlying the resistance to treatment still remain poorly understood and robust biomarkers for the early identification of patients at risk of R/R disease are still lacking.
Mass spectrometry-based proteomics has benefited from an instrumental and methodological revolution over the last two decades. Today, global label-free quantitative proteomic studies enable the identification and quantification of thousands of proteins and provide new opportunities for an in-depth characterization of complex proteomes6. As a complement to the static picture revealed by genome sequencing, the comprehensive analysis of the proteome that is dynamic provides crucial information on protein expression to decipher complex biological processes. To date, no data are available in the literature focusing on the proteomic characterization of R/R DLBCL.
In this context, we conducted a large-scale differential proteomic investigation of R/R versus chemosensitive DLBCL patients in order to identify new potential biomarkers related to resistance to treatment and to better understand the biological mechanisms underlying chemoresistance. This proteomic investigation was combined with a quantitative transcriptomics experiment performed on the same samples to correlate genes expression and their impact at the proteomic level.
Results and Discussion
We performed for the first time a large-scale differential multi-omics study on DLBCL patient’s samples in order to search for new potential biomarkers that could help to early identify patients at risk of R/R disease and to better understand the biological mechanisms underlying chemorefractoriness. In the context of our current knowledge from the literature, a detailed study of some promising new biomarkers is provided below, demonstrating the high value of the present proteogenomic dataset.
Fresh-frozen tumour tissues were collected at the time of diagnosis, before any treatment, for 8 chemorefractory and 12 chemosensitive DLBCL patients who were uniformly treated in first-line with rituximab and an anthracycline-based chemotherapy regimen in a single institution. Patients were considered as chemorefractory if they had a stable or progressive disease after first-line (n = 6), or if they relapsed less than one year after having achieved a complete response (n = 2). Patients who achieved a complete response and did not relapse thereafter, with a minimal follow-up of at least 24 months after the end of treatment, were considered as chemosensitive. Chemorefractory patients were most likely to have an aggressive disease according to the age-adjusted International Prognostic Index (aaIPI) with 87% aaIPI 2–3 in the chemorefractory group and 42% in the chemosensitive group but the difference was not significant (p = 0.07). The two groups did not differ significantly regarding age (p = 0.58), sex (p = 0.64) and Ann Arbor stage (p = 0.16) (Table 1). RNA could be extracted from the same tissue samples that were used for proteomics analysis for 17 patients (7 chemorefractory and 10 chemosensitive). In both groups, the majority of patients were classified into Germinal Center B-Cell-like (GCB) molecular subtype (72% of the chemorefractory patients and 70% of the chemosensitive patients) as determined by rapid reverse transcriptase multiplex ligation-dependent probe amplification assay (RT-MLPA)7. The mean percentage of tumor-cells, determined by morphological examination and immunohistochemistry, was 76%, and was ≥70% in 18/20 samples. A single case had a low percentage of tumor cells (20%) but this sample corresponded to a particular subtype of DLBCL (T-cell/histiocyte-rich large B-cell lymphoma). Patient’s characteristics are summarized in Table 1 and a detailed description of the 20 patients is provided in Supplementary Table 1.
Table 1.
Patients’ characteristics.
| Chemosensitive | Chemorefractory | p value | |
|---|---|---|---|
| Age (years) | 0.58* | ||
| Median | 55 | 57 | |
| range | 18–79 | 31–73 | |
| Sex, n (%) | 0.64** | ||
| Male | 9/12 (75) | 5/8 (63) | |
| Female | 3/12 (25) | 3/8 (37) | |
| Ann Arbor Stage, n (%) | 0.16** | ||
| 1–2 | 6/12 (50) | 1/8 (13) | |
| 3–4 | 6/12 (50) | 7/8 (87) | |
| aaIPI, n (%) | 0.07** | ||
| 0–1 | 7/12 (58) | 1/8 (13) | |
| 2–3 | 5/12 (42) | 7/8 (87) | |
| Response to first-line, n (%) | <0.01** | ||
| Complete response | 12/12 (100) | 2/8 (25) | |
| Primary refractory | 0/12 | 6/8 (75) | |
| Number of treatment lines | <0.01*** | ||
| Median | 1 | 5 | |
| Range | 1–1 | 3–7 | |
| Cell of origin, n (%) | 0.99** | ||
| GC | 7/10 (70) | 5/7 (72) | |
| ABC | 1/10 (10) | 1/7 (14) | |
| Unclassifiable | 2/10 (20) | 1/7 (14) |
Abbreviations: aaPIP, age-adjusted International Prognostic Index; GC, Germinal Center B-Cell-Like; ABC, Activated B-Cell-Like. *Student’s t-test; **Fisher’s exact test; ***Mann-Whitney test.
Overall, the combined proteomics analysis of the 20 samples resulted in the identification of 4774 unique protein groups (proteins which cannot be unambiguously identified by unique peptides are grouped in one protein group and quantified together). Pairwise comparisons of all samples against each other resulted in a high Pearson coefficient correlation (average r = 0.89) demonstrating a high quantitative accuracy and a high similarity in the global proteomes. The statistical analysis with the peptide-level Robust Ridge Regression model (MSqRob) allowed the relative quantification of 3101 proteins between the two groups of patients, with 586/3101 (18.9%) being significantly differentially abundant with a false discovery rate <5%. Among these differentially abundant proteins, 246 were overexpressed in chemorefractory patients and 340 overexpressed in chemosensitive patients (Supplementary Table 2).
Transcriptomic analysis was performed on 17 samples (these 17 samples are indicated in Supplementary Table 1). It allowed the quantification of 17695 transcripts across the 17 samples. For the 4774 previously identified proteins, the transcript counterpart was also identified by RNAseq in 4338/4774 (90.8%). With an adjusted p-value < 0.1, 244 transcripts were differentially abundant between the two groups of patients (Supplementary Table 3).
Among the 3101 quantified proteins with MSqRob, 2965/3101 (95.6%) were also quantified at the transcriptomic level. The combination of transcriptomics and proteomics data thus resulted in 2965 transcripts/proteins commonly quantified at both levels. When focusing on the 246 proteins overexpressed in chemorefractory patients, only 7 were not identified at the transcriptomic level (Supplementary Table 4) and 24 had a high fold-change (FC) at the proteomic level (log2FC (R vs S) >1) and low at the transcriptomic level (log2FC (R vs S) <1) (Supplementary Table 5). Conversely, 16 proteins had a low fold-change at the proteomic level (log2FC (R vs S) <1) and high at the transcriptomic level (log2FC (R vs S) >1) (Supplementary Table 5). When focusing on the 340 proteins overexpressed in the chemosensitive patients, only 11 were not identified at the transcriptomic level (Supplementary Table 4) and 23 had a high fold-change at the proteomic level (log2FC (R vs S) <−1) and low at the transcriptomic level (log2FC (R vs S) >−1) (Supplementary Table 5). Conversely, only 5 proteins had a low fold-change at the proteomic level (log2FC (R vs S) >−1) and high at the transcriptomic level (log2FC (R vs S) <−1) (Supplementary Table 5).
By considering only the significantly differentially expressed proteins between the two groups of patients, only 22/586 (3.8%) were found to be also differentially expressed at the transcriptomic level. In all but one case, the variation direction was similar between the two methods with 16/22 transcripts and proteins overexpressed in chemorefractory patients (log2FC (R vs S) >0), and 5/22 transcripts and proteins overexpressed in chemosensitive patients (log2FC (R vs S) <0) (Table 2). Only one discordant case was observed with Complement C3 that was overexpressed in chemorefractory patient at the proteomics level but overexpressed in chemosensitive patients at the transcriptomics level. Among these 22 differentially expressed transcripts/proteins, 6 were selected and discussed below. The selection was based on a proteomic fold-change threshold (FC (R vs S) >1.5 or <−1.5) and a literature-based strong biological relevance in the context of treatment-resistance in DLBCL.
Table 2.
Sub-list of 22 differentially abundant transcripts/proteins pairs at both transcriptomics and proteomics levels.
| Protein name | Gene name | Proteomics | Transcriptomics | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of patients (n = 20) | log2FC (R vs S) | q-value | Number of patients (n = 17) | log2FC (R vs S) | adjusted p-value | ||||
| R (n = 8) | S (n = 12) | R (n = 7) | S (n = 10) | ||||||
| C-X-C motif chemokine 13 | CXCL13 | 3 | 2 | 2,9 | 0.006 | 7 | 10 | 1,6 | 0.019 |
| Indoleamine 2,3-dioxygenase 1 | IDO1 | 6 | 12 | 2,9 | <0.001 | 7 | 10 | 1,7 | 0.061 |
| Granzyme H | GZMH | 3 | 2 | 2,7 | 0.028 | 7 | 10 | 1,7 | 0.047 |
| Protein THEMIS2 | THEMIS2 | 7 | 9 | 1,2 | 0.015 | 7 | 10 | 1,3 | 0.067 |
| Granzyme K | GZMK | 8 | 11 | 0,6 | 0.017 | 7 | 10 | 1,5 | 0.089 |
| Complement C3 | C3 | 8 | 12 | 0,3 | <0.001 | 7 | 10 | −1,4 | 0.060 |
| Hexokinase-3 | HK3 | 8 | 12 | 1,7 | <0.001 | 7 | 10 | 1,9 | 0.028 |
| Superoxide dismutase [Mn], mitochondrial | SOD2 | 8 | 12 | 1,2 | <0.001 | 7 | 10 | 1,6 | 0.039 |
| Protein S100-A8 | S100A8 | 8 | 12 | 1,7 | <0.001 | 6 | 9 | 1,7 | 0.066 |
| Protein S100-A4 | S100A4 | 8 | 12 | 0,9 | 0.003 | 7 | 10 | 1,4 | 0.020 |
| PRA1 family protein 3 | ARL6IP5 | 8 | 12 | 0,5 | 0.009 | 7 | 10 | 0,9 | 0.089 |
| rRNA 2′-O-methyltransferase fibrillarin | FBL | 8 | 12 | −0,4 | 0.006 | 7 | 10 | −0,8 | 0.070 |
| 40 S ribosomal protein S18 | RPS18 | 8 | 12 | −0,3 | 0.011 | 7 | 10 | −1,1 | 0.076 |
| 40 S ribosomal protein S6 | RPS6 | 8 | 12 | −0,3 | 0.043 | 7 | 10 | −0,9 | 0.033 |
| 40 S ribosomal protein S12 | RPS12 | 8 | 12 | −0,4 | 0.009 | 7 | 10 | −0,9 | 0.089 |
| Alpha-1-antitrypsin | SERPINA1 | 8 | 12 | 1,1 | 0.007 | 7 | 9 | 1,8 | 0.039 |
| Serpin B6 | SERPINB6 | 8 | 12 | 1,2 | <0.001 | 7 | 10 | 1,5 | 0.001 |
| Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | PREX1 | 8 | 12 | 0,8 | 0.006 | 7 | 10 | 1,1 | 0.067 |
| Ceruloplasmin | CP | 8 | 12 | 0,6 | <0.001 | 7 | 8 | 1,7 | 0.083 |
| CD97 antigen | CD97 | 8 | 12 | 0,5 | 0.003 | 7 | 10 | 1,2 | 0.074 |
| B-cell antigen receptor complex-associated protein beta chain | CD79B | 5 | 10 | −1,7 | 0.013 | 7 | 10 | −1,3 | 0.024 |
| Syntaxin-11 | STX11 | 6 | 11 | 1,4 | <0.001 | 7 | 10 | 1,2 | 0.070 |
Abbreviations: Log2FC, log2 fold-change; R, chemorefractory patients; S, chemosensitive patients.
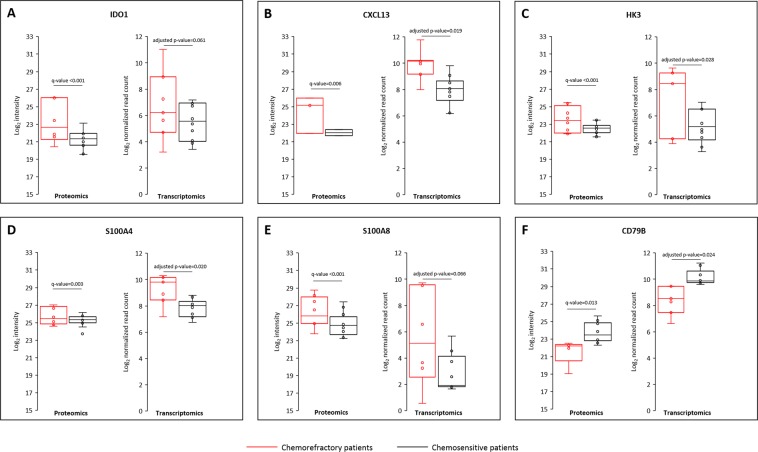
Indoleamine 2,3-dioxygenase 1 (IDO1) was overexpressed in chemorefractory patients (Fig. 1A). This enzyme is involved in the degradation of the amino acid tryptophan. L-kynurenine, one of the metabolites resulting from tryptophan degradation, has the ability to inhibit T-cell proliferation and to induce T-cell death, contributing to an immunosuppressive microenvironment8. Expression of IDO1 evaluated by immunohistochemistry was already found to be positive in one third of DLBCL cases and was associated with a worse response rate and a worse 3-year overall survival after first-line therapy with R-CHOP9. From a therapeutic point of view, IDO1 represents a novel immune checkpoint target. Several IDO1 inhibitors (epacadostat (INCB024360), indoximod, navoximod (GDC-0919) or BMS-986205) are now available but failed to demonstrate a therapeutic efficacy as a monotherapy. However, several ongoing trials in various solid tumors (ovarian cancer, pancreatic cancer, squamous cell carcinoma of the head and neck, non-small cell lung cancer, metastatic renal-cell carcinoma for example) are currently evaluating IDO1 inhibitors in combination with other agents such as PD1 or PD-L1 inhibitors10,11. Although results from phase 2 studies were encouraging, such as with the combination of epacadostat and anti-PD1 in melanoma patients12, recent results from phase 3 studies failed to confirm these results13, suggesting that further analysis are warranted to better define the subset of patients who are most likely to benefit from IDO1 inhibitors. No clinical trial is currently ongoing in order to evaluate the potential of IDO1 inhibitors in the context of R/R DLBCL.
Figure 1.
Top six differentially abundant proteins and genes between chemorefractory and chemosensitive patients. For each protein, dots represent the mean of the log2 intensities of all the peptides quantified for each patient in each group. For each gene, dots represent the log2 normalized read count distribution of the considered gene for each patient in each group.
We also demonstrated an overexpression of the chemokine C-X-C motif ligand 13 (CXCL13) in chemorefractory patients (Fig. 1B). CXCL13, the unique ligand of CXCR5, is an inflammatory chemokine that contributes to generate a pro-inflammatory microenvironment in angioimmunoblastic T-cell lymphoma14. Moreover, it has been shown to be an adverse prognosis factor in advanced colon cancer15. In colon cancer cells, the CXCL13-CXCR5 axis participates in tumour growth and invasiveness by activation of the PI3K/AKT signalling pathway16. Our results indicate that CXCL13 could also play a key role in the microenvironment of DLBCL.
Hexokinase 3 (HK3) is one of the four isoforms of hexokinase involved in the first step of the glycolysis pathway, converting glucose into glucose-6-phosphate. Glucose metabolism of cancer cells highly differs from that of normal cells. In cancer cells, pyruvate generated by glycolysis is converted into lactate via a phenomenon called “aerobic glycolysis” (Warburg effect)17. Overexpression of hexokinase is crucial for cancer cells to produce enough ATP by aerobic glycolysis. Recently, hexokinase 2 was shown to be overexpressed in rituximab-resistant cell lines and to be associated with inhibition of mitochondrial-mediated apoptosis18. In our study, hexokinase 2 was identified with a high degree of confidence (by 49 unique peptides covering almost 50% of its sequence) while its expression was not affected between the two groups of patients both at proteomic (log2FC = 0.4, q-value = 0.115) and transcriptomic (log2FC = 0.7, adjusted p-value = 0.58) levels. Interestingly, hexokinase 3 was also identified with a high degree of confidence (by 29 unique peptides covering 57% of its sequence), and its expression was significantly higher in chemorefractory patients at both proteomics (log2FC = 1.7, q-value < 0.001) and transcriptomics levels (log2FC = 1.9, adjusted p-value = 0.028) (Fig. 1C). These results suggest that hexokinase 3 could play a key role in DLBCL chemorefractoriness.
Proteins S100 are involved in the regulation of proliferation, migration and invasion of cancer cells, and their dysregulation has been demonstrated in the majority of human cancers19. Fifteen S100 family members were identified in our study by proteomic analysis (S100A2, S100A4, S100A6, S100A7, S100A7A, S100A8, S100A9, S100A10, S100A11, S100A12, S100A13, S100A14, S100A16, S100B and S100P) and 4 were found to be significantly overexpressed in chemorefractory patients: S100A4 (log2FC = 0.9, q-value = 0.003), S100A8 (log2FC = 1.7, q-value < 0.001), S100A9 (log2FC = 1.9, q-value < 0.001) and S100A11 (log2FC = 0.5, q-value = 0.007). Only S100A4 and S100A8 were significantly overexpressed at both transcriptomics and proteomics levels (Fig. 1D,E). S100A4 and S100A8 were already extensively studied in solid tumours and were found to be associated with tumour growth and metastasis20,21. However, few data are available in lymphoma with only S100A9 being described as associated with tumour growth and immune evasion22. Our data thus suggest that these proteins could be associated in DLBCL with a more aggressive disease and could participate in the development of resistance to treatment. Based on our results, targeting S100 proteins may represent a therapeutic potential for the treatment of R/R DLBCL.
Finally, the B-cell antigen receptor complex-associated protein beta chain (CD79B) was significantly under-expressed in chemorefractory patients both at proteomic (log2FC = −1.7, q-value = 0.013) and transcriptomic (log2FC = −1.3, adjusted p-value = 0.023) levels (Fig. 1F). CD79B is necessary for the function of the B-cell receptor and somatic genetic alterations in the CD79B gene participate in the constitutive activation of the NF-kB pathway, in particular for the ABC DLBCL subtypes. CD79B mutations have been reported in 23% of R/R ABC DLBCL23. However, few data are available which investigate the level of CD79B expression. This protein expression level could eventually affect the efficacy of the anti-CD79B antibody-drug conjugate (polatuzumab vedotin) that has recently emerged as a potential active drug in R/R DLBCL24.
We used the gene ontology (GO) (http://www.geneontology.org/) and PANTHER database25 to perform enrichment analysis based on the Gene Ontology – Biological Process (GO-BP) annotations in our proteomics dataset. This enrichment analysis indicated that proteins significantly overexpressed in chemorefractory patients were particularly enriched in GO-BP associated with inflammation and immune response, as well as the coagulation cascade. At the opposite, proteins significantly overexpressed in chemosensitive patients were enriched in GO-BP associated with ribosome biogenesis and ribosomal RNA (rRNA) processing (Table 3).
Table 3.
Gene Ontology-Biological Process enrichment among the differentially expressed proteins using the PANTHER database system.
| Gene Ontology-Biological Process | Fold enrichment | p value | Fisher’s Exact with FDR multiple test correction |
|---|---|---|---|
| Chemorefractory patients | |||
| Fibrinolysis | 8.5 | 0.0001 | <0.0001 |
| Regulation of complement activation | 4.1 | <0.0001 | 0.0065 |
| Platelet activation | 3.5 | 0.0002 | 0.0198 |
| Platelet degranulation | 3.2 | 0.0001 | 0.0154 |
| Inflammatory response | 3.2 | <0.0001 | 0.0001 |
| humoral immune response | 3.1 | <0.0001 | 0.0051 |
| Neutrophil degranulation | 2.5 | <0.0001 | <0.0001 |
| negative regulation of immune system process | 2.5 | 0.0004 | 0.0034 |
| activation of immune response | 2.4 | <0.0001 | 0.0018 |
| Chemosensitive patients | |||
| Ribosome biogenesis | 3.2 | <0.0001 | <0.0001 |
| rRNA processing | 3.4 | <0.0001 | <0.0001 |
Abbreviations: FDR, False Discovery Rate.
These results suggest that cancer-related inflammation and disturbed immune response may play an important role and contribute to chemorefractoriness in DLBCL. The role of cancer-related inflammation in the development and progression of tumors, as well as in patient outcomes, has been recognized since many years26. In addition to CXCL13 and S100A8 proteins, that were previously discussed, the other proteins from our dataset that were found to be significantly overexpressed at proteomics and transcriptomics level in chemorefractory patients and involved in inflammation or immune response are Granzyme H, Granzyme K, Complement C3 and Alpha-1-antitrypsin. Our results also suggest that a local activation of coagulation may contribute to treatment-resistance and tumor progression, and are consistent with previously published works having already highlighted the role of blood coagulation in cancer progression. In particular, it has been shown that blood coagulation enzymes in the tumor microenvironment played a role in solid-tumor progression and metastasis27,28. These results highlight the potential major role played by the microenvironment in tumor progression and drug-resistance in DLBCL. The strong interactions between cancer-cells and their surrounding microenvironment have been already largely studied, and targeting the microenvironment offers now novel therapeutic perspectives in cancer29,30. In this study, proteomic and transcriptomic analysis were performed on whole tissue sections, thus allowing the analysis of DLBCL-cells and their microenvironment. One major drawback of this approach is that these two compartments cannot be analysed separately, but this type of approach could be considered as an opportunity, in the context of a biomarker discovery study, to provide the most exhaustive list of potential new biomarkers. However, in this context, validation studies are a crucial need in order to determine more precisely the relative contribution of both compartments that are tumor-cells and microenvironment. To achieve such an objective, immunohistochemistry could be considered as a method of choice.
In this study, we present the first high-throughput multi-omics study in DLBCL. Over the past two decades, genomics and transcriptomics have largely dominated in cancer research, in particular with the advent of next-generation sequencing (NGS) technologies. In 2000, gene-expression profiling allowed the clear distinction of two molecular DLBCL subtypes, namely Activated B-Cell (ABC) and Germinal Center B cell-Like (GCB) subtypes3. The development of NGS technologies resulted in the recent publication of exome sequencing in 1001 DLBCL patients allowing to depict the nearly complete mutational landscape in DLBCL and the identification of 150 driver genes31. This in-depth and extensive molecular characterization of DLBCL at genomic and transcriptomic levels also recently led to the proposal of novel molecular classifications in DLBCL identifying subgroups of patients with distinct clinical behaviour and prognosis32,33. More recently, MS-based proteomics has emerged as an important tool for the characterization of DLBCL. Various methodological approaches have been used with an aim to exploring various aspects of the disease, such as pathogenesis, subtypes classification or therapeutic issues. Super-SILAC-based approaches have demonstrated the ability to distinguish DLBCL subtypes according to their cell of origin in patient-derived DLBCL cell lines as well as on tumor samples from patients34–36. So far, few proteomics studies have addressed the drug-resistance challenge in DLBCL. This issue has been first addressed by a proteomic study (two-dimensional gel electrophoresis with MALDI-TOF/TOF-MS analysis) aiming to identify differential proteins expressed by DLBCL cells with high or low sensitivity to chemotherapy after in vitro exposure to the CHOP regimen compounds. Nineteen differentially expressed proteins were identified between the two groups. Among these differentially expressed proteins, immunohistochemical analysis performed in DLBCL tissue samples from 98 patients confirmed a higher expression of Glutathione S-transferase (GSTP1) and Heat shock protein beta-1 (HSPB1), and a lower expression of Ezrin (EZR) and Pleckstrin (PLEK) in patients with relapse or progressive disease after CHOP chemotherapy37. In another study, by using a SILAC-based quantitative proteomic approach on 10 DLBCL patients selected according to their response to treatment (5 patients with primary refractory disease or early relapse, and 5 patients considered cured), 87 proteins, among a total of 3027 successfully quantified proteins, were differentially expressed between the two groups of patients with 21 overexpressed in refractory patients. The authors could demonstrate an up-regulation of proteins involved in the regulation of the actin cytoskeleton in chemosensitive patients38. This work was pursued by using a tandem mass tag (TMT)-based quantitative proteomic approach performed on microdissected samples obtained from formalin-fixed paraffin-embedded tissues. This study allowed identifying 102 DA proteins and the authors could confirm the up-regulation of proteins involved in actin regulation in chemosensitive patients. Interestingly, they managed to highlight a potential role for ribosomal proteins in treatment-resistance as these proteins were largely represented in those found to be overexpressed in chemorefractory patients39. In comparison with these previously published works, our study points out the potential role of the microenvironment in drug-resistance in DLBCL. However, we observed one discrepancy between our study and the study published by Bram Ednersson et al. regarding the potential role of ribosomal proteins. In our study, ribosomal proteins and ribosome biogenesis appeared over-represented in chemosensitive patients, while it was the opposite in the work of Bram Ednersson et al. There is no obvious explanation, but it remains hazardous to make a direct comparison between two exploratory studies that differ in several technical and methodological aspects such as the tissue used for protein extraction (fresh-frozen vs formalin-fixed paraffin-embedded, whole-tissue vs microdissection), quantitative proteomic workflow (label-free vs super-SILAC) and statistical analysis. Nevertheless, these two studies point out a potential role for ribosome proteins in drug-resistance and, therefore, warrant continued research to clarify the role of these proteins in treatment-resistance of DLBCL.
In conclusion, this study revealed several extremely promising biomarker candidates associated with chemorefractoriness, related to tumour metabolism, microenvironment, BCR signalling pathway, hence highlighting new potential therapeutic drug targets. The combination of multilevel –omics datasets is very useful to reduce lists of thousands of candidates to a subset of significant targets, as well as to cross-validate candidates by different techniques. Further studies will be necessary to validate these findings in a larger and independent cohort of patients. However, the present work already provides greater insights in the underlying mechanisms of chemoresistance in DLBCL, supported by a publically available dataset.
Materials and Methods
Patients selection
Patients were selected among the fresh-frozen tissue-sample collection available from the “Centre de ressources Biologiques des Hôpitaux Universitaires de Strasbourg”. We retrospectively analyzed the treatment and outcome of each patient. Only patients for whom a tissue-sample collected at the time of diagnosis and treated in first-line with the combination of anti-CD20 monoclonal antibody and an anthracycline-based regimen were selected. Patients were considered as chemorefractory if they had a stable or progressive disease after first-line, or if they relapsed less than one year after having achieved a complete response. Patients who achieved a complete response after first-line and did not relapse thereafter, with a minimal follow-up of at least 24 months after the end of treatment, were considered as chemosensitive.
Samples handling
All samples were obtained by a surgical resection or radiological-guided biopsy from a tumor mass. Proteomic and transcriptomic analysis were performed on the same specimens that were used for the diagnosis of the disease. After collection, all samples were stored at −80 °C, without conservative medium, until protein or RNA extraction. The neoplastic content in each sample was determined by a pathological review of all cases, based on morphology and immunohistochemistry for distinguishing B-cells and T-Cells.
Proteomics analysis
Sample preparation
Proteins were extracted from ~10 mg of fresh frozen tumor tissues in a lysis buffer containing 62.5 mM Tris HCl pH 6.8, 2% SDS and 10% glycerol. Protein concentration was determined with DCTM method (Bio-Rad) according to manufacturer’s instructions. For each sample, 20 µg of proteins were used for tube-gel preparation, as previously described40. Briefly, 7.5% acrylamide/Bis-acrylamide, and 0.25 µL TEMED were added for a final volume of 100 μL. Ammonium persulfate (2.50 µL) was added to initiate polymerization. After fixation with 50% ethanol/3% phosphoric acid, tube-gels were cut in 2 mm sections and each section in ~2 mm2 pieces. The gel pieces were washed and the cysteine residues were reduced by adding 10 mM DTT for 30 min at 60 °C and 30 min at room temperature, and alkylated by adding 55 mM IAA for 20 min in the dark. The gel pieces were then washed three times by adding 50/50 (v/v) 25 mM NH4HCO3/acetonitrile (ACN). After two dehydrations with ACN, the proteins were cleaved in an adequate volume to cover all gel pieces with a modified porcine trypsin (Promega) solution at a 1:80 (w/w) enzyme:protein ratio. Digestion was performed overnight at 37 °C. Tryptic peptides were extracted twice under agitation, first with 60% ACN in 0.1% FA for 1 h and then with 100% ACN for 1 h. The excess of ACN was vacuum dried, and the samples were resolubilized with H2O/ACN/FA (98/2/0.1 v/v/v).
NanoLC-MS/MS analysis
The nanoLC-MS/MS analysis was performed on a nanoAcquity UPLC device (Waters Corporation, Milford, USA) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Peptide separation was performed on an ACQUITY UPLC BEH130 C18 column (250 mm × 75 μm with 1.7 μm diameter particles) and a Symmetry C18 precolumn (20 mm × 180 μm with 5 μm diameter particles, Waters). The solvent system consisted of 0.1% FA in water (solvent A) and 0.1% FA in ACN (solvent B). Samples (equivalent to 800 ng of proteins) were loaded into the enrichment column over 3 min at 5 μL/min with 99% of solvent A and 1% of solvent B. The peptides were eluted at 450 nL/min with the following gradient of solvent B: from 1 to 35% over 120 min and 35 to 80% over 1 min. The 20 samples were injected in randomized order. The MS capillary voltage was set to 1.8 kV at 250 °C. The system was operated in Data Dependent Acquisition mode with automatic switching between MS (mass range 300–1800 m/z with R = 140,000, Automatic gain control (AGC) fixed at 3 × 106 ions and a maximum injection time set at 50 ms) and MS/MS (mass range 200–2000 m/z with R = 17,500, AGC fixed at 1 × 105 and the maximal injection time set to 100 ms) modes. The ten most abundant peptides were selected on each MS spectrum for further isolation and higher energy collision dissociation fragmentation, excluding unassigned and monocharged ions. The dynamic exclusion time was set to 60 s.
Data analysis
Raw data obtained for each sample were processed using MaxQuant (version 1.5.5.1). Peaks were assigned with the Andromeda search engine with full trypsin specificity. The isoform-containing human database used for the search was extracted from the UniProtKB-SwissProt database (26 sept 2016, 42,144 entries). The minimum peptide length required was seven amino acids and a maximum of one missed cleavage was allowed. Methionine oxidation was set as a variable modification and peptides with modified methionines, as well as their unmodified counterparts, were excluded from protein quantification. Cysteine carbamidomethylation was set as a variable modification to account for the potential propionamide modifications of cysteine residues. Cysteine propionamidation was thus also set as a variable modification. For protein quantification, the “match between runs” option was enabled. The maximum false discovery rate was 1% at peptide and protein levels with the use of a decoy strategy. We used the “peptides.txt” files exported from MaxQuant for further statistical analysis with the peptide-level Robust Ridge Regression model (MSqRob) R-package41.
Statistical analysis was performed for all quantified proteins and transcripts, independently of the number of patients in whom the proteins and/or transcripts were quantified.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium database with the identifier PXD00908942.
Transcriptomics analysis
RNA isolation and sequencing
Total RNA was isolated from fresh frozen tumor tissues with the with TRIzol reagent (Invitrogen, Carlsbad, California, USA). RNA extraction could not be performed for 3 patients (samples #5, #12 and #15 in supplementary Table 1) because of the low quantity of available tissue. RNA integrity was assessed with the Agilent total RNA Pico Kit on a 2100 Bioanalyzer instrument (Agilent Technologies, Paolo Alto, USA). The sequencing library was prepared with the Ion AmpliSeq Transcriptome Human Gene Expression Panel (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s protocol43. Briefly, after reverse transcription of total RNA, the cDNAs were amplified by multiplex PCR including a total of 20,812 amplicons. These amplicons were then partially digested, and after barcoded sequencing adapter ligation, the libraries were loaded at a concentration of 75 pM on an Ion PI IC 200 chip using the Ion Chef Instrument (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Finally, the sequencing took place on an Ion Proton sequencer with the Ion PI IC 200 Kit, according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Analysis of RNA-sequence reads
The raw reads were processed by the Torrent Suite analysis pipeline and mapped to the human genome assembly hg19 AmpliSeqTranscriptome. The Torrent AmpliSeqRNA Plugin was used to generate raw read counts which were further used for differential analysis. An average of 3.5 million reads were generated per sample with 93.25% reads on target and an average of 11,005 amplicons covered by at least 10 reads. We applied the R Bioconductor package DESeq2 to identify genes that were differentially expressed. The gene selection was based on the adjusted p-value. All genes with an adjusted p-value lower than 0.1 were selected as differentially expressed44.
RNAseq raw data have been deposited in fastaq format in the EMBL-EBI ArrayExpress archive (https://www.ebi.ac.uk/arrayexpress/) with the accession number E-MTAB-6597.
Ethical Committee
Sample collection for further research analysis was approved by an Ethical Committee (“Comité de Protection des Personnes Est IV”, Strasbourg, France) and all patients provided an informed consent according to the Declaration of Helsinki.
Supplementary information
Acknowledgements
The authors acknowledge “Alsace contre le Cancer” for research grant and the “Centre de Ressources Biologiques” (CRB) from the University Hospital of Strasbourg for providing the samples. This work was supported by the “Agence Nationale de la Recherche” (ANR) and the French Proteomic Infrastructure (ProFI; ANR-10-INBS-08-03). This work was further supported by the Laboratory of Excellence ANR-11-LABX-0070_TRANSPLANTEX, the INSERM (UMR_S 1109), and the Cancéropole Grand-Est, all to SB; and the European regional development fund (European Union) INTERREG V program (project n°3.2 TRIDIAG) to RC and SB.
Author Contributions
L.M.F., L.Mu, C.C. and R.C. conceived the project and designed the study. L.M.F., L.Mu and A.P. performed experiments. N.P. and F.B. performed statistical analysis. L.M.F., L.Mu, R.C. and C.C. wrote the manuscript. S.B., S.C., R.H., M.P.C., L.Ma and L.V. analysed data and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Luc-Matthieu Fornecker and Leslie Muller contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37273-4.
References
- 1.Crump M, et al. Outcomes in refractory diffuse large B-cell lymphoma: results from the international SCHOLAR-1 study. Blood. 2017;130:1800–1808. doi: 10.1182/blood-2017-03-769620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow SH, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Sarkozy C, Traverse-Glehen A, Coiffier B. Double-hit and double-protein-expression lymphomas: aggressive and refractory lymphomas. Lancet Oncol. 2015;16:e555–567. doi: 10.1016/S1470-2045(15)00005-4. [DOI] [PubMed] [Google Scholar]
- 5.Bohers E, et al. Activating somatic mutations in diffuse large B-cell lymphomas: lessons from next generation sequencing and key elements in the precision medicine era. Leukemia & lymphoma. 2015;56:1213–1222. doi: 10.3109/10428194.2014.941836. [DOI] [PubMed] [Google Scholar]
- 6.Doll S, et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun. 2017;8:1469. doi: 10.1038/s41467-017-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mareschal, S. et al. Accurate Classification of Germinal Center B-Cell-Like/Activated B-Cell-Like Diffuse Large B-Cell Lymphoma Using a Simple and Rapid Reverse Transcriptase-Multiplex Ligation-Dependent Probe Amplification Assay: A CALYM Study. The Journal of molecular diagnostics: JMD, 10.1016/j.jmoldx.2015.01.007 (2015). [DOI] [PubMed]
- 8.Frumento G, et al. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. The Journal of experimental medicine. 2002;196:459–468. doi: 10.1084/jem.20020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninomiya S, et al. Indoleamine 2,3-dioxygenase in tumor tissue indicates prognosis in patients with diffuse large B-cell lymphoma treated with R-CHOP. Annals of hematology. 2011;90:409–416. doi: 10.1007/s00277-010-1093-z. [DOI] [PubMed] [Google Scholar]
- 10.Zhai, L. et al. IDO1 in cancer: a Gemini of immune checkpoints. Cellular & molecular immunology, 10.1038/cmi.2017.143 (2018). [DOI] [PMC free article] [PubMed]
- 11.Vacchelli E, et al. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. 2014;3:e957994. doi: 10.4161/21624011.2014.957994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daud A, et al. Epacadostat plus nivolumab for advanced melanoma: Updated phase 2 results of the ECHO-204 study. Journal of Clinical Oncology. 2018;36:9511–9511. doi: 10.1200/JCO.2018.36.15_suppl.9511. [DOI] [Google Scholar]
- 13.Garber K. A new cancer immunotherapy suffers a setback. Science (New York, N.Y.) 2018;360:588. doi: 10.1126/science.360.6389.588. [DOI] [PubMed] [Google Scholar]
- 14.Tripodo C, et al. Mast cells and Th17 cells contribute to the lymphoma-associated pro-inflammatory microenvironment of angioimmunoblastic T-cell lymphoma. The American journal of pathology. 2010;177:792–802. doi: 10.2353/ajpath.2010.091286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi XW, et al. Expression features of CXCR5 and its ligand, CXCL13 associated with poor prognosis of advanced colorectal cancer. Eur Rev Med Pharmacol Sci. 2014;18:1916–1924. [PubMed] [Google Scholar]
- 16.Zhu Z, et al. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway. Molecular and cellular biochemistry. 2015;400:287–295. doi: 10.1007/s11010-014-2285-y. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, N.Y.) 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu JJ, et al. Up-regulation of hexokinase II contributes to rituximab-chemotherapy resistance and is a clinically relevant target for therapeutic development. Oncotarget. 2018;9:4020–4033. doi: 10.18632/oncotarget.23425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nature reviews. Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei F, Qu J, Zhang M, Li Y, Zhang S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget. 2017;8:73219–73239. doi: 10.18632/oncotarget.18016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zha H, et al. S100A8 facilitates the migration of colorectal cancer cells through regulating macrophages in the inflammatory microenvironment. Oncology reports. 2016;36:279–290. doi: 10.3892/or.2016.4790. [DOI] [PubMed] [Google Scholar]
- 22.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. The Journal of experimental medicine. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin RD, et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:2290–2300. doi: 10.1158/1078-0432.ccr-15-2123. [DOI] [PubMed] [Google Scholar]
- 24.Palanca-Wessels MC, et al. Safety and activity of the anti-CD79B antibody-drug conjugate polatuzumab vedotin in relapsed or refractory B-cell non-Hodgkin lymphoma and chronic lymphocytic leukaemia: a phase 1 study. The Lancet. Oncology. 2015;16:704–715. doi: 10.1016/s1470-2045(15)70128-2. [DOI] [PubMed] [Google Scholar]
- 25.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 27.Repetto O, De RV. Coagulation and fibrinolysis in gastric cancer. Ann N Y Acad Sci. 2017;1404:27–48. doi: 10.1111/nyas.13454. [DOI] [PubMed] [Google Scholar]
- 28.Lima, L. G. & Monteiro, R. Q. Activation of blood coagulation in cancer: implications for tumour progression. Biosci Rep33, 10.1042/BSR20130057 (2013). [DOI] [PMC free article] [PubMed]
- 29.Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015;368:7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 30.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 31.Reddy A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171:481–494 e415. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapuy B, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz R, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deeb SJ, D’Souza RC, Cox J, Schmidt-Supprian M, Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics. 2012;11:77–89. doi: 10.1074/mcp.M111.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deeb SJ, Cox J, Schmidt-Supprian M, Mann M. N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol Cell Proteomics. 2014;13:240–251. doi: 10.1074/mcp.M113.033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deeb SJ, et al. Machine Learning-based Classification of Diffuse Large B-cell Lymphoma Patients by Their Protein Expression Profiles. Mol Cell Proteomics. 2015;14:2947–2960. doi: 10.1074/mcp.M115.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. Identification of differentially expressed proteins in chemotherapy-sensitive and chemotherapy-resistant diffuse large B cell lymphoma by proteomic methods. Med Oncol. 2013;30:528. doi: 10.1007/s12032-013-0528-5. [DOI] [PubMed] [Google Scholar]
- 38.Ruetschi U, et al. SILAC-Based Quantitative Proteomic Analysis of Diffuse Large B-Cell Lymphoma Patients. Int J Proteomics. 2015;2015:841769. doi: 10.1155/2015/841769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bram Ednersson S, et al. Expression of ribosomal and actin network proteins and immunochemotherapy resistance in diffuse large B cell lymphoma patients. British journal of haematology. 2018;181:770–781. doi: 10.1111/bjh.15259. [DOI] [PubMed] [Google Scholar]
- 40.Muller L, Fornecker L, Van Dorsselaer A, Cianferani S, Carapito C. Benchmarking sample preparation/digestion protocols reveals tube-gel being a fast and repeatable method for quantitative proteomics. Proteomics. 2016;16:2953–2961. doi: 10.1002/pmic.201600288. [DOI] [PubMed] [Google Scholar]
- 41.Goeminne LJE, Gevaert K, Clement L. Experimental design and data-analysis in label-free quantitative LC/MS proteomics: A tutorial with MSqRob. J Proteomics. 2018;171:23–36. doi: 10.1016/j.jprot.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Deutsch EW, et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, et al. Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genomics. 2015;16:1069. doi: 10.1186/s12864-015-2270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.