Abstract
At present, the pathophysiology of autism spectrum disorder (ASD) remains unclear. Increasing evidence suggested that gut microbiota plays a critical role in gastrointestinal symptoms and behavioral impairment in ASD patients. The primary aim of this systematic review is to investigate potential evidence for the characteristic dysbiosis of gut microbiota in ASD patients compared with healthy controls (HCs). The MEDLINE, EMBASE, Web of Science and Scopus were systematically searched before March 2018. Human studies that compared the composition of gut microbiota in ASD patients and HCs using culture-independent techniques were included. Independent data extraction and quality assessment of studies were conducted according to PRISMA statement and Newcastle-Ottawa Scale. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to infer biological functional changes of the shifted microbiota with the available data in four studies. Sixteen studies with a total sample size of 381 ASD patients and 283 HCs were included in this systematic review. The quality of the studies was evaluated as medium to high. The overall changing of gut bacterial community in terms of β-diversity was consistently observed in ASD patients compared with HCs. Furthermore, Bifidobacterium, Blautia, Dialister, Prevotella, Veillonella, and Turicibacter were consistently decreased, while Lactobacillus, Bacteroides, Desulfovibrio, and Clostridium were increased in patients with ASD relative to HCs in certain studies. This systematic review demonstrated significant alterations of gut microbiota in ASD patients compared with HCs, strengthen the evidence that dysbiosis of gut microbiota may correlate with behavioral abnormality in ASD patients. However, results of inconsistent changing also existed and further big-sampled well-designed studies are needed. Generally, as a potential mediator of risk factors, the gut microbiota could be a novel target for ASD patients in the future.
Introduction
Autism spectrum disorder (ASD) is a complex, pervasive neurobiological disorder, characterized by impaired social and communication skills, as well as stereotyped behaviors and restricted patterns of interests1. ASD includes autism (AD), Asperger’s Syndrome, and Pervasive Development Disorder Not Otherwise Specified (PDD-NOS). According to recent estimate, the prevalence of ASD is elevating with 1–2% of children currently diagnosed worldwide2. The etiology of ASD remains unclear and appears to involve a complicated interaction of genetic and environmental factors3,4. By estimate, the heritability including de novo mutations, common variants, and short nucleotide polymorphisms identified in ASD cases altogether accounts for approximately 50% of the disorder5,6. As well, the possibility for environmental risk factors and related medical comorbidities which contribute to core neurobehavioral symptoms of the disorder has been highlighted by many studies. Among the comorbidities in ASD, gastrointestinal (GI) symptoms are quite common, such as diarrhea, constipation, and commutative diarrhea/constipation, they are also correlated with the severity of the neurobehavioral disorder7. The association of ASD with great prevalence of GI symptoms is spurring an intensive search of the ASD gut microbiota. There is growing evidence demonstrating that disturbances in the pathway underlying the microbiota-gut-brain axis, especially the disordered gut microbiota, may result in neurobehavioral and intestinal dysfunction in ASD patients8,9. Gut microbiota makes critical contribution to maintaining the integrity of intestinal epithelia, protecting intestinal barrier and preventing bacterial LPS and other toxins into bloodstream. It has been confirmed that systematic inflammation by LPS induced behavioral impairment and damaged the blood-brain barrier in animal models10,11. Conversely, gut microbiota reconstitution with probiotics could alter blood metabolic profiles, remediate gut permeability and improve ASD-related behaviors in mice model12. Moreover, gut microbiota may regulate the central nervous system (CNS) activities through neural, immune and endocrine pathways. For example, gut microbiota can regulate the hypothalamic-pituitary-adrenal (HPA) axis13, and produce many chemicals affecting brain function (e.g., serotonin, dopamine, r-aminobutyric acid, SCFAs, and p-cresol)14,15.
ASD individuals vary widely in clinical presentation, severity and treatment response. The complexity is motivating an exploration to identify biological factor helps to achieve earlier diagnoses and predict clinical prognosis. Thus, the gut microbiota in ASD patients has gained growing attention as a potential mediator of risk factors. So far, several case-control studies aiming to observe the aberrant gut microbiota have been performed16–32. Moreover, a recent open-label study has indicated that fecal microbiota transfer (FMT) therapy alters gut ecosystem and improves GI and neurobehavioral symptoms in ASD patients19. All these results have gained an insight into the potential mechanisms of gut microbiota in ASD. At present, no systematic review has addressed the evidence of the altered gut microbiota in ASD patients focusing on culture-independent methods especially the high-throughput sequencing techniques. These techniques enable the identification of previously unknown bacterial species, thereby provide novel insights into the compositional diversity and the functional capacity of gut microbiota.
The aim of this systematic review is to explore the current evidence for the alteration of gut microbiota in ASD patients compared with HCs using culture-independent techniques.
Materials and methods
Protocol
We conducted the systematic review to evaluate the altered gut microbiota in ASD patients compared with HCs. Available literatures were identified and examined as a systematic review but not a meta-analysis due to the heterogeneity of methods and results. To report this systematic review, the method was consistent with the PRISMA statement guidelines (Preferred Reporting Items for Systematic Reviews and Meta-Analysis), and the protocol was registered at PROSPERO (registration number: CRD42017060769).
Selection criteria
Studies compared gut microbiota in ASD patients and HCs were included. The inclusion criteria were as follows: (1) ASD diagnosis with definite criteria; (2) the age of participants ranged from 2 to 18; (3) detection of gut microbiota with gut biopsy or fecal samples; (4) metagenomic sequencing, 16S rDNA sequencing, quantitative real-time PCR techniques (qPCR) or FISH. The exclusion criteria were as follows: (1) medicated participants; (2) failure to provide data for the microbiota; (3) culture-dependent methods; (4) intervention studies without initial data or reviews; (5) duplicate publications.
Search strategy and study selection
A systematic search was conducted using MEDLINE, EMBASE, Web of Science and Scopus for the studies published before March 2018. The reference lists of all identified studies that matched the key search terms were manually searched for relevant trials. The specific search strategy was: (Autism OR autistic OR ASD) AND (microbiome OR microbiota OR microflora OR flora). At the beginning of study selection, irrelevant articles were excluded via an assessment of the tittle, abstract and keywords. The full-text of potentially relevant studies was then retrieved. Following the elimination of duplicates, two independent authors (FL and JL) assessed the articles for eligibility considering established criteria detailed above. Any disagreement between authors were resolved by discussion until consensus was achieved.
Data extraction
Data for gut microbiota in eligible studies were extracted to the Excel spreadsheet. The following information was extracted: author; publication year; country of origin; the characteristics of case and control (including sample size, mean age, sex ratio, GI symptom and the diagnosis criteria for ASD); the method for microbiota analysis (including sample source, DNA extraction, the information of PCR or FISH or sequencing and the referred database); outcomes (including the differences of overall microbiota structure and the specific bacteria). Besides, the raw sequencing data and biom files were also collected in studies using high-throughput sequencing methods. Two authors (FL and JL) independently extracted data from the selected articles, and the data was then cross-checked for accuracy (FW).
Quality assessment
Studies included in this systematic review were carefully evaluated for the methodological quality and the risk of bias by two authors (FL and JL). Study quality was assessed on the basis of Newcastle-Ottawa Scale (NOS) for case-control studies. NOS included three domains: selection, comparability and exposure criteria. The selection criteria included four aspects: (1) adequate definition of the cases; (2) representativeness of the cases; (3) selection of controls; (4) definition of controls. The comparability criteria included comparability of case and controls according to the design and analysis. The exposure criteria included three aspects: (1) ascertainment of exposure; (2) same method of ascertainment for cases and controls; (3) non-response rate.
Summary measures and secondary bioinformatics analysis
The overall microbiota structure, including α-diversity, β-diversity (compositional dissimilarity), the relative abundance of the specific genus and the quantity of specific bacteria by q-PCR or FISH were the primary outcomes. Moreover, to explore the potential function of gut microbiota in ASD patients, the Linear Discriminant Analysis (LDA) effect size (LEfSe) was applied to the relative abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways predicted by using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)33. As for the study provided with raw sequencing data, we re-clustered the available data in QIIME with the command of pick_closed_reference_otus.py with the Greengenes reference version 13_8. The operational taxonomic unit (OTU) picking method is Usearch61. As for the study provided OTU biome file, we directly used the biome file to analysis.
Results
Study selection
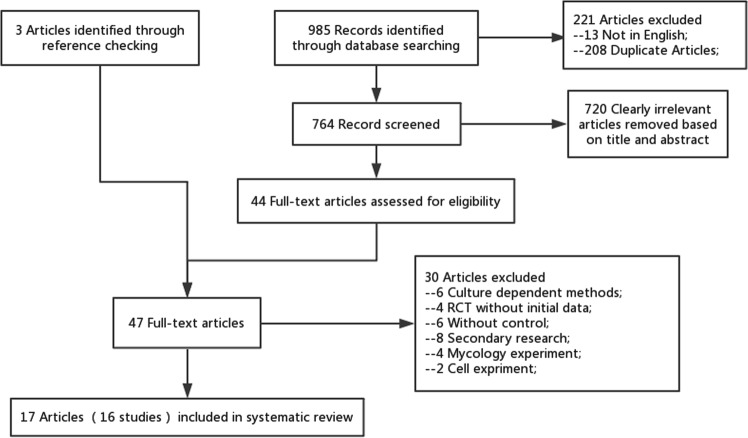
In total, 985 records were identified through the electronic search. 208 duplicate articles and 13 articles that were not published in English were discarded. 44 full-text articles were retrieved for eligibility following the exclusion of tittles and abstracts that were not relevant to the research. Three additional papers were identified via checking the references of relevant articles. The remaining 47 full-text papers were further assessed according to the fore-mentioned criteria, resulting in the exclusion of 30 papers due to the following reasons: culture-dependent method, randomized controlled trials (RCTs) without initial data, without control group, secondary research; mycology and cell experiment. Following the selection process (Fig. 1), 17 articles remained and were included in the systematic review16–32. Among them, William et al.24,25 reported one study in two different articles, so did Wang et al.29,30. It is also noteworthy that one article23 included two group of ASD simultaneously: AD and PDD-NOS, actually counted as two case-control studies. Thus, a total of 17 articles (16 studies) were included.
Fig. 1. Flow chart of identification, exclusion and inclusion of eligible studies.
Flow chart indicates the progression of trials through each stage of the selection process
Study characteristics
Sixteen studies were published in English journals between 2005 and 2018. Eight studies were conducted in USA16,17,19,21,22,24,27,31, three in Italy18,23, two in Australia26,29, one in UK32, one in Japan20 and one in Slovakia28. The characteristics and main results of studies were outlined in Table 1. A total of 381 ASD patients and 283 HCs were included, with sample size ranged from 12 to 104. Of 283 HCs, 107 subjects were healthy siblings21,23,26. The age of ASD patients ranged from 2–18 and the proportion of male ranged from 77.5–100%. Concerning about GI symptoms, five studies17,19,22,24,27 reported all of the ASD patients had GI symptoms, seven studies16,18,21,26,28,29,32 reported that a part of ASD patients had GI symptoms. And three studies20,23 conducted in ASD patients without any GI symptoms. Different diagnostic criteria were used: ADOS, DSM-5, ABD, ADI-R, and CARS. However, two studies31,32 did not report the specific diagnostic criteria. As restricted diet is very common in ASD patients, we tried to extract the information of eating habit in ASD and control group. Whereas, only six studies reported dietary information. Strati et al.18 reported that all subjects of the study went through a Mediterranean diet. Angelis et al.23 reported that the major dietary differences were excluded since each of these pairs of children belonged to the same family unit. The remaining four studies consistently reported that some of the ASD patients had special diets, such as casein-free (CF) or gluten-free (GF) diet21,22,27,32. In addition, Son et al.21 conducted further nutritional analysis of one-week food diary, no significant difference between two group with respect to daily intake of macronutrients (calories, protein, fats, carbohydrates, sugars or dietary fiber) were noted.
Table 1.
Characteristics of the trials included
| References | Case/Control participants | ↔Microbiology assessment | Outcomes (compared with control) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Study country | Sample size | Mean age (Year) | Male ratio (%) | Symptom of GI (%) | Diagnose of ASD | Sample source DNA extraction | PCR or FISH Or Sequencing | Referred Database | Outcomes (compared with control) |
| Kang16 | ASD:21 | 10.1 ± 4.1 | 15/21 | GI symptom were more severe in ASD | ATEC | Fecal sample; | 16S rRNA V2-3 region; Genome Sequencer FLX-Titanium System | Greengenes database | ↓α diversity; |
| 2018 | CON:23 | 8.4 ± 3.4 | 22/23 | PDD-BI | −80 °C; | β diversity (P < 0.05); | |||
| USA | PowerSoil® DNA Isolation Kit | Genus:↓Prevotella and Coprococcus | |||||||
| ↓Faecalibacterium, Haemophilus | |||||||||
| Luna17 | ASD:14 | Age:4-13 | 14/14 | 14/14 | ADOS | Rectal biopsy; | 16S rRNA V1V3, V4 region; MiSeq Illumina platform | Silva database | β diversity (P < 0.05); |
| 2017 | CON:15 | Age:3-18 | 12/15 | 15/15 | −80 °C; | ↑Clostridiales; Clostridium; Lachnoclostridium; Flavonifractor; | |||
| USA | PowerSoil® DNA Isolation Kit | ↓Dorea, Blautia, Sutterella; | |||||||
| Other findings: ↓tryptophan in rectal; | |||||||||
| Strati18 | ASD: 40 | 11.1 ± 6.8 | 31/40 | 5/50 | DSM-5 | Fecal sample; | V3-V5 | Greengenes database | ↔ α diversity;β diversity (P < 0.05); |
| 2017 | CON:40 | 9.2 ± 7.9 | 28/40 | 11/40 | ADOS | −80 °C; | 454 | Phylum:↑Firmicutes/Bacteroidetes ratio;↓Bacteroidetes; | |
| Italy | ABD | FastDNA™ SPIN Kit | pyrosequence | Genus:↑Collinsella, Corynebacterium, Dorea, and Lactobacillus; | |||||
| ↓Alistipes, Bilophila, Dialister, Parabacteroides and Veillonella; | |||||||||
| Other findings: Candida was more than double in the autistic; | |||||||||
| Kang19 | ASD:18 | 10.8 ± 1.6 | 16/18 | 18/18 | ADI-R | Stool samples | 16S rRNA V4 region; MiSeq | Greengenes database | ↓α diversity; |
| 2017 | CON:20 | 11.4 ± 2.5 | 18/20 | 0/20 | PowerSoil® DNA Isolation Kit | Illumina platform | Genus:↓Bifidobacterium; | ||
| USA | Other findings: ASD had lower fiber consumption; | ||||||||
| ASD were breastfed significantly shorter time; | |||||||||
| Inoue20 | ASD:6 | age 3–5 | NA | 0/6 | DSM-5 | Fecal sample; | 16S rRNA V3-4 region; MiSeq | Greengenes database | Genus:↑Faecalibacterium; ↓Blautia; |
| 2016 | CON:6 | age 3–5 | NA | 0/6 | PARS | −80 °C; | Illumina platform | Other findings: number of GO biological processes associated with response to viruses were enriched: IFN-γ and type-I IFN signaling pathways. | |
| Japan | M-CHAT | e QuickGene DNA Tissue kit | |||||||
| Son21 | ASD:59 | 10.3 ± 1.8 | 52/59 | 25/59 | ADOS | Fecal sample; | 16S rRNA V1V2, V1V3 region; MiSeq Illumina platform | Silva database | ↔α diversity; ↔β diversity; |
| 2015 | SIB:44 | 10.0 ± 1.8 (age 7–14) | 21/44 | 13/44 | ADI-R | -80 °C; | ↔ the relative abundance of any phylum and genus; | ||
| USA | ZR Fecal DNA MiniPre | ||||||||
| Kang22 | ASD:20 | 6.7 ± 2.7 | 18/20 | 20/20 | ADOS | Fecal sample; | 16S rRNA V2-3 region; bTEFAP using a 454 FLX Sequencer; | SSURef database | ↓α diversity; |
| 2013 | CON:20 | 8.3 ± 4.4(age 3–16) | 17/20 | 0/20 | ADI-R | −80 °C; QIAamp | Genus:↓Prevotella, Coprococcus, unclassified Veillonellaceae; | ||
| USA | ATEC | DNA Stool Mini Kit | Akkermansia was very high in several autism subjects; | ||||||
| PDD-BI | |||||||||
| De Angelis23 | AD:10 | age 4–10 | NA | 0/10 | ADI-R | Fecal sample; | 16S rRNA V1-3 region; bTEFAP using a 454 FLX Sequencer; | GenBank databases | ↑α diversity;β diversity(P < 0.05); |
| 2013 | SIB:10 | age 4–10 | NA | 0/10 | ADOS | −80 °C; | Phylum:↓Firmicutes; Fusobacteria and Verrucomicrobia; Firmicutes/Bacteroidetes ratio; | ||
| Italy | CARS | FastDNA Pro Soil-Direct Kit | ↑Bacteroidetes; | ||||||
| Genus: ↓Faecalibacterium, Oscillospira, Bifidobacterium, Fusobacterium, Escherichia, Turicibacter, Eubacterium; | |||||||||
| ↑Caloramator, Sarcina, Clostridium, Roseburia, Akkermansia, Shigella, Enterobacter, Dorea; | |||||||||
| Other findings: AD fecal samples contained higher FFA; phenol, 4-(1,1,3,3-tetramethylbutyl)-phenol, p-cresol; SCFAs was lower; | |||||||||
| De Angelis23 | PDD-NOS:10 | age 4–10 | NA | 0/10 | ADI-R | Fecal sample; | 16S rRNA V1-3 region; bTEFAP using a 454 FLX Sequencer; | GenBank databases | ↑α diversity; β diversity(P < 0.05); |
| 2013 | SIB:10 | age 4–10 | NA | 0/10 | ADOS | −80 °C; | Phylum:↓Fusobacteria and Verrucomicrobia; | ||
| Italy | CARS | FastDNA Pro Soil-Direct Kit | Genus: ↓Oscillospira, Bacteroides, Fusobacterium, Escherichia, Prevotella, Turicibacter; Clostridium; | ||||||
| ↑Faecalibacterium, Ruminococcus, Roseburia, Alistipes, Dorea; | |||||||||
| Other findings: AD fecal samples contained higher phenol, 4-(1,1,3,3-tetramethylbutyl)-phenol, p-cresol; SCFAs was lower; | |||||||||
| William24,25 | ASD:15 | 4.5 ± 1.3 | 15/15 | 15/15 | DSM-5 | ileal and cecal biopsies; −80 °C | 16S rRNA V2 region; bTEFAP using a 454 FLX Sequencer; | Greengenes database | Phylum:↑Firmicutes/Bacteroidetes ratio; Firmicutes, Betaproteobacteria; ↓Bacteroidetes; |
| 2011,2012 | CON:7 | 4.0 ± 1.1 | 7/7 | 7/7 | ADI-R | Quantitative Real-time PCR | Family: ↑ Lachnospiraceae and Ruminococcaceae; | ||
| USA | Genus:↑Sutterella, Faecalibacterium;Other findings: Presence of Alcaligenaceae in some ASD children but absence in controls. | ||||||||
| Gondalia26 | ASD:51 | age 2–12 | 42/51 | 28/51 | CARS | Stool samples | bTEFAP using a 454 FLX Sequencer; | NA | ↔α diversity; ↔β diversity; |
| 2012 | SIB:53 | age 2–12 | 19/34 | 4/53 | −20 °C; QIAamp DNA stool kit | ↔ the relative abundance of any phylum and genus; | |||
| Australia | |||||||||
| Finegold27 | ASD:11 | age 2–13 | NA | 11/11 | CARS | Fecal sample; | bTEFAP using a 454 FLX Sequencer; | RDP-II database | ↔ α diversity;β diversity (P < 0.05); |
| 2010 | CON:8 | age 2–13 | 5/8 | NA | -80 °C;QIAamp DNA stool mini kit | Phylum:↑Bacteroidetes, Proteobacteria; | |||
| USA | ↓Firmicutes, Actinobacteira; | ||||||||
| Genus:↑Alkaliflexus, Desulfovibrio, Acetanaerobacterium, | |||||||||
| Parabacteroides, Bacteroides; | |||||||||
| ↓Weissella, Turicibacter, Clostridium, Anaerofilum, Dialister, Pseudoramibacter, Ruminococcus, Streptococcus, Anaerovorax; | |||||||||
| Species:↑ Desulfovibrio spp. and Bacteroides vulgatus; | |||||||||
| ↓Bifidobacterium longum, Dialister invisus, Clostridium leptum; | |||||||||
| Tomova28 | ASD:10 | age 2–9 | 9/10 | 9/10 | ICD-10 | Stool samples | Quantitative Real-time PCR | NA | ↑Lactobacillus; |
| 2015 | CON:10 | age 2–11 | 10/10 | 6/10 | CARS | −80 °C; QIAamp DNA stool kit | ↑Clostridia cluster l and Desulfovibrio (not significant); | ||
| Slovakia | ADI | ↓Bacteroidetes/Firmicutes ratio; | |||||||
| Wang29,30 | ASD:23 | 10.3 ± 0.8 | 21/23 | 9/23 | CARS | Fecal sample; | Quantitative Real-time PCR | NA | ↑Sutterella spp, Ruminococcus torques (p = 0.08); |
| 2011 | CON:9 | 12 ± 1.3 | 4/9 | 1/9 | DSM-5 | −80 °C; A repeat bead beating plus column; | ↓Bifidobacterium spp; Akkermansia.muciniphila; | ||
| Australia | Other findings: No differences between groups in levels of | ||||||||
| Faecalibacterium prausnitzii; | |||||||||
| Song31 | ASD:15 | Age matched | Gender matched | NA | NA | Stool samples | TaqMan | NA | ↑Clostridia bolteae, Clostridia and clusters I and XI; |
| 2004 | CON:8 | −80 °C; QIAamp DNA stool kit | Real-time PCR | ||||||
| USA | |||||||||
| Helena32 | ASD:58 | 7 ± 3.76 | 48/58 | 53/58 | NA | Fecal sample; | FISH | NA | ↑Clostridium histolyticum group (Clostridium clusters I and II); |
| 2005 | CON:10 | 6 ± 2.88 | 6/10 | 0/10 | −20 °C; | ||||
| UK | |||||||||
The alteration of gut microbiota composition was assessed with fecal samples16,18–22,25–31 or gut biopsy16,23. Ten studies16–23,26,27 assessed gut microbiota by high-throughput molecular approaches: Illumina MiSeq platforms, 454 pyrosequencing or bTEFAP using a 454 FLX Sequencer. Three studies28–31 detected gut microbiota with qPCR and one32 with FISH approach. One study24,25 used both sequencing and qPCR method. Of the studies adopting 16S rDNA-based method, two studies26,27 did not report which hypervariable region of the 16S rDNA was targeted and the remaining studies targeted varied sets of regions. Meanwhile, the database used for mapping the sequences were GreenGenes16,18–20,24, RDP-227, Silva17,21, SSURef22 and GenBank23, while one study26 did not report the database used.
Risk of bias
16 studies were identified and assessed as medium (6–7) to high (8) quality by the NOS as presented in Table 2. When assessing the quality of selection, the studies of Song et al.31 and Helena et al.32 lacked an adequate definition of the ASD patients, thus these two studies achieved 3 points in the selection assessment. As for comparability, Luna et al.17, Inoue et al.20, De Angelis et al.23 and William et al.24,25 included case and control with better design, such as all participants has consistent GI symptoms at baseline, thus these studies achieved 2 points in the comparability assessment.
Table 2.
Quality assessment of included studies
| First author | Year | Selection | Comparability | Exposure | Total |
|---|---|---|---|---|---|
| Kang16 | 2018 | 4 | 1 | 2 | 7 |
| Luna17 | 2017 | 4 | 2 | 2 | 8 |
| Strati18 | 2017 | 4 | 1 | 2 | 7 |
| Kang19 | 2017 | 4 | 1 | 2 | 7 |
| Inoue20 | 2016 | 4 | 2 | 2 | 8 |
| Son21 | 2015 | 4 | 1 | 2 | 7 |
| Kang22 | 2013 | 4 | 1 | 2 | 7 |
| De Angelis23 | 2013 | 4 | 2 | 2 | 8 |
| De Angelis23 | 2013 | 4 | 2 | 2 | 8 |
| William24,25 | 2012 | 4 | 2 | 2 | 8 |
| Gondalia26 | 2012 | 4 | 1 | 2 | 7 |
| Finegold27 | 2010 | 4 | 1 | 2 | 7 |
| Tomova28 | 2015 | 4 | 1 | 2 | 7 |
| Wang29,30 | 2011 | 4 | 1 | 2 | 7 |
| Song31 | 2004 | 3 | 1 | 2 | 6 |
| Helena32 | 2005 | 3 | 1 | 2 | 6 |
Heterogeneity
Methodological sources of heterogeneity included the type of sample (fecal or biopsy), the temperature samples stored, the methods of DNA extraction and the primer used to PCR. Although the included studies had slightly differences in primer selection, there was significant overlap in the key variable and constant regions of the 16S gene. The bacterial identification platform was also a possible source of heterogeneity. 16S rDNA sequencing can quantitatively identify all bacteria present in one sample. However, qPCR and FISH only detected the specific bacteria, which lacked the evaluation of the whole community.
Possible clinical heterogeneity included age, gender, type of control (sibling vs non-sibling) and whether had GI symptoms. Although age structure varied greatly among studies, extensive overlap was found that age of all participants ranged from two to eighteen. In terms of gender, Luna et al.17 and William et al.24 only included male individuals, the remaining studies included both male and female. Besides, Son et al.21, De Angelis et al.23 and Gondalia et al.26 recruited healthy siblings as control, whereas other studies recruited normal control from the whole population. Considering the impact of GI symptoms on composition of gut microbiota, a potential source of heterogeneity may come from the varied baseline of GI symptoms.
The altered composition of gut microbiota in ASD patients
In the studies using high-throughput sequencing method, the diversity of species in the samples can be expressed in many ways, including richness, evenness, and α-diversity. α-diversity is the number of species and their proportion within one sampling site. Of the nine studies compared α-diversity, three studies demonstrated that α-diversity has a significant reduction in ASD patients16,19,22. In contrast, De Angelis et al.23 reported the increased α-diversity was found in both AD and PDD-NOS patients compared with their sibling control. β-diversity means the dissimilarity between communities of two sites or two samples. In this systematic review, ten studies analyzed β-diversity (unweighted UniFrac distance, weighted UniFrac distances, and Bray-Curtis), six of them consistently reported that the microbiota of ASD patients clustered significantly apart from that of HCs16–18,23,27.
To assess the specific changing of bacteria in ASD patients, we analyzed the phyla grouping at first. Three studies indicated a clear alteration of the gut bacterial community in ASD patients characterized by a higher Firmicutes/Bacteroidetes ratio in ASD than that in HCs due to a significant increase of Firmicutes and/or a reduction of Bacteroidetes18,24,28. Whereas, De Angelis et al.23 and Finegold et al.27 reported totally opposite results. Phylum of Fusobacteria and Verrucomicrobia were significantly decreased in patients of ASD (AD and PDD-NOS) in the data of De Angelis et al.23 Consistently, William et al.24 and Finegold et al.27 both reported that the Proteobacteria was elevated in ASD patients rather than HCs.
Further analysis of the alterations of genus and species in ASD, four studies consistently demonstrated a significantly decrease of Bifidobacterium in patients with ASD relative to HCs19,23,27,30. Consistent with this, the abundance of Blautia17,20, Dialister18,27, Prevotella16,22,23, Turicibacter23,27 and Veillonella18,22 were all decreased. In contrast, Lactobacillus18,28, Bacteroides23,27, and Desulfovibrio27,28 were all increased in ASD patients rather than controls. Five studies17,23,28,31,32 all revealed that there was a significant increase of Clostridium in ASD. In addition, De Angelis et al.23 indicated that Oscillopira decreased and Roseburia increased in both AD and PDD-NOS patients. Meanwhile, they also found that some opportunistic pathogen such as Enterobacter and Shigella were elevated in ASD patients23. Whereas, conflicting results were also reported. Two sequencing studies reported that the abundance of Akkermansia was elevated in ASD patients22,23. Inconsistently, one qPCR study30 indicated that lower amount of Akkermansia muciniphila was found in feces of ASD patients than HCs. Dorea18,23 and Sutterella25,29 were reported increased significantly in ASD patients, but they were reported decreased in another study17. Faecalibacterium20,23,24 and Ruminococcus23,24,29 were reported increased in ASD patients by three studies, but they were reported reduced in another study27.
The altered function of gut microbiota in ASD patients
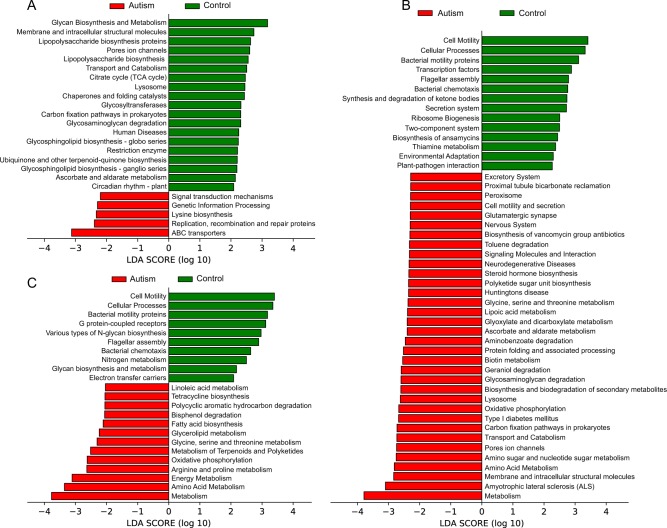
Functional profiles of microbial communities with high-throughput sequencing were predicted by PICRUSt. Raw sequence data was provided in three studies18,21,22 and the OTU biome file was provided in one study19. Among them, three studies18,19,22 indicated the functional modules of gut microbiota in ASD patients were substantially different from that in HCs (Fig. 2). In the study of Strati et al.18 (Fig. 2a), the pathways with the highest five discriminative power in HCs were “Glycan Biosynthesis and Metabolism”, “Membrane and intracellular structural molecules”, “Lipopolysaccharide biosynthesis proteins”, “Pores ion channels” and “Lipopolysaccharide biosynthesis”. The pathways with the highest discriminative power in ASD patients were “ABC transporters” under Membrane Transport category, followed by pathways of “Replication, recombination and repair proteins”, “Lysine biosynthesis”, “Genetic Information Processing” and “Signal transduction mechanisms”. In two studies of Kang et al. (Figs. 2b, c)19,22, the pathways of “Cell Motility”, “Cellular Processes” and “Bacterial motility proteins” had consistent significant discriminative power in HCs. In addition, “Bacterial chemotaxis” and “Flagellar assembly” were also noted. In ASD patients, the functional modules of metabolism were higher than that in HCs, such as “Oxidative phosphorylation” under Energy Metabolism category and “Glycine, serine and threonine metabolism” under Amino Acid Metabolism. It should be noted that pathways of “Huntingtons disease” and “Amyotrophic lateral sclerosis (ALS)” under Neurodegenerative Diseases category as well as “Glutamatergic synapse” under Nervous System category were also increased in ASD patients. Whereas, in the study of Son et al.21, there was no difference of functional modules between ASD patients and HCs, which was accordance with the result of gut microbiota composition.
Fig. 2. Linear discriminative analysis effect size (LEfSe) of statistically significantKEGG pathways between autism and control in the studies.
a Strati et al., b, c two studies of Kang et al. Positive LDA scores (green) are enriched in control while negative LDA scores (red) are enriched in autism
Discussion
This systematic review demonstrated that there was consistent evidence for the alterations of gut microbiota in ASD patients compared with HCs. Novel culture-independent techniques that analyzed bacterial DNA offered a unique, more in-depth look into the gut microbiome. Overall, the changed structure of gut bacterial community in terms of β-diversity was observed coherently in ASD patients compared with HCs. Consistently, ASD patients had elevated abundance of Proteobacteria rather than HCs. In addition, Bifidobacterium, Blautia, Dialister, Prevotella, Veillonella, and Turicibacter were consistently decreased, while Lactobacillus, Bacteroides, Desulfovibrio, and Clostridium were increased in ASD patients relative to HCs.
Several included studies reported the Firmicutes/Bacteroidetes ratio due to the alteration of Firmicutes and Bacteroidetes, but has not reached to any concordant conclusions. Some studies indicated that elevated Firmicutes /Bacteroidetes ratio was correlated with inflammatory conditions such as inflammation bowel diseases (IBDs)34 and obesity35. The elevated Proteobacteria phylum was found in ASD patients. It is noted that Proteobacteria is a major of Gram-negative bacteria and includes a variety of opportunistic pathogens. Meanwhile, as microbial signature of dysbiosis in gut microbiota, Proteobacteria is associated with host inflammation36. It is also remarkable that these Gram-negative bacteria produce a potent toxic factor LPS37. An animal study indicated that prenatal LPS exposure reduced the level of glutathione in the brain38. Glutathione is a significant antioxidant and closely related to detoxification in the brain39. Thus, the elevated abundance of Proteobacteria in ASD patients need more attention in future studies. As mentioned, the level of some genera (Bifidobacterium, Blautia, Veillonella and Prevotella) was decreased in ASD patients. Notably, these particular species are known to be versatile carbohydrate metabolizers40. Bifidobacterium is among the first colonizers of human intestinal and one of the dominant groups in the gut microbiota of breast-fed infants41. It can ferment complex polysaccharides to regulate host function and promote health42,43, encouraging interest in its use as probiotics. Blautia plays an important role in nutrient assimilation44 and gut maturation in children45. The reduction of these beneficial bacteria in ASD patients may be implicated in the pathogenesis of the disease. On the other hand, overgrowth of Bacteroides, Desulfovibrio, and Clostridium were also found in ASD patients. Indeed, Bacteroides is an abundant genus at all ages, from infants to adults46. It is the main producer of propionate in the gut, and the abundance of propionate in feces correlates strongly with the abundance of Bacteroides47. Propionate produced by microbiota is used for gluconeogenesis in liver and represents a source of glucose level for the host48. Whereas, a study indicated that neurodevelopmental abnormality in ASD patients accompanied with impaired propionic acid metabolism49, which may relate to the changing of propionate-producing bacteria. Desulfovibrio produces LPS as well as hydrogen sulfide which could be toxic to intestinal cells under certain circumstance27,50. Clostridium has been extensively studied in ASD51,52 due to its characteristic of producing exotoxins and propionate, which may aggravate the symptoms of ASD53. In addition, some species belonging to the Clostridium produce p-cresol. This chemical metabolite could cause the reduction of glutathione and reported to be a possible urinary biomarker for autism54,55. De Angelis et al.23 indicated that Enterobacter and Shigella were increased in ASD patients, which were positively correlated with the GI symptoms in autism18. These opportunistic pathogens have been previously reported to cause or underlie human infections such as bacteremia and intra-abdominal infection56. However, the high population of Lactobacillus in ASD patients was not expected. Lactobacillus has the ability of fermenting a series of carbon sources primarily to lactic acid and is widely recognized as probiotics57. Nonetheless, a recent research indicated that Lactobacillus was more abundant in T2DM patients than in HCs58.
There were still some conflicting results about the alterations of Akkermansia, Ruminococcus, Sutterella, and Faecalibacterium in ASD patients. Akkermansia and Ruminococcus are mucin-degrading bacterium and associated with the gut permeability59,60. Sutterella can regulate mucosal metabolism and intestinal epithelial integrity25,29. Changes of mucus-degrading microbes may be related with mucus producing, which could impact the mucosal barrier in the gut. Faecalibacterium is regarded as commensal or even beneficial due to its function of producing anti-inflammation butyrate61. Thus, the variation of these genera needs to be further explored with the pathogenesis of ASD in future studies.
The predicted biological functions of the observed microbial community in ASD patients were significantly different from that in HCs, but varied among studies. In two accordant studies19,22, functional modules of microbiota in HCs mostly involved in cell motility, cellular processes, bacterial motility proteins, bacterial chemotaxis, and flagellar assembly, which indicated the basic physiological maintenance under normal condition. Whereas, in ASD patients, the most enhanced functional module of microbiota was metabolism, including amino acid metabolism, lipid metabolism, carbohydrate metabolism, energy metabolism, cofactors and vitamins metabolism and tetracycline biosynthesis. Functions of xenobiotics degradation such as toluene, aminobenzoate, polycyclic aromatic hydrocarbon, and bisphenol were also enhanced. Moreover, some functional modules involved in neurodegenerative diseases in human, such as Huntington disease and ALS were also raised in ASD patients. So far, a variety of mechanism has been proposed in ASD associated with the function of gut microbiota, including immune activation/dysfunction, bacterial-derived toxin (e.g., LPS, phenols, p-cresol, 4-EPS), metabolites aberrations in fermentation process or products, such as propionic acid (PPA) and other SCFAs, and the dysregulated metabolism of free amino acids61. Thus, the functional analysis of gut microbiota may give some hints to these underlying mechanisms and needs to be studied in the future.
Microbiome reconstitution could be a potential therapy to ASD patients in future
Since gut microbiota appears strongly associated with ASD, the interests in remodeling gut microbiota with diet, antibiotics, prebiotics, probiotics, and FMT are advancing19,62–65. In ASD children, an open-label study indicated that treatment with 8 weeks oral vancomycin greatly improved GI symptoms and ASD disorders64. A randomized double-blind crossover trial showed that treatment with probiotics resulted in significant differences in the stool consistency compared to placebo and behavior scores compared to baseline65. In addition, Buffington et al. reported that maternal high fat diet (MHFD) can induce abnormal social behavior through mediating the dysbiosis of gut microbiota, but reconstituting microbiota with probiotics can correct social deficits in MHFD offspring66. Thus, targeting the gut microbiota could be a possible treatment for ASD patients in the future.
The possible factors for contradictory findings
There are numerous confounding factors that may limit the consensus of the studies analyzed in this systematic review. Geography and dietary habits are main factors that play a great role in microbiome composition. The studies included in this systematic review were conducted in different countries. Besides, implementation of restricted diet is very common in ASD subjects, such as GF and CF diet. Thus, the enormous variation of dietary habit and its effect on the gut microbiota may mask the true picture of the differences. Secondly, the selection of control (that is, sibling vs non-sibling) may also have subtle influence on gut microbiota. Krajmalnik et al. reported that neurotypical (NT) siblings of ASD children have altered microbiome compared to that of unrelated children67. It can be an alternative explanation that why studies21,26 using NT sibling controls did not find any significant difference, while other studies using non-sibling controls did. Thirdly, the frequent occurrence of GI symptoms in ASD patients may affect the composition of gut microbiota. Hence, the baseline comparability of GI symptoms in case and control, can facilitate a more robust interpretation of the results. Another important consideration is that biogeographic variation of gut microbiota. The samples were collected from different anatomic subsites within the gut tract or feces. Zoetendal et al. reported that using biopsies other than feces allowed to assess the mucosa-epithelia associated microbiota. As biopsies likely established more intimate interplay with the human intestinal epithelium and immune cells68. Besides, the different results of gut microbiota may also arise from distinct techniques for DNA isolation, PCR and sequencing, and small-sample sized studies.
Limitations
Generally, the analyses and results of gut microbiota were relatively difficult to evaluated. It is probably because that the gut microbiota is a new research field that currently depends much on non-parametric statistics and lacks generally accepted standard methods of reporting results69. As stated previously, methodological and clinical heterogeneity made it impossible to combine the results of different studies into a meta-analysis. Publication bias is a common challenge within the area of systematic reviews. However, two studies included in the present review provided null findings, showing that the concern about publication bias may be solved in some extent. In addition, only four studies provided the raw sequencing data or biome file for doing PICRUSt analysis, which may result in the loss of bias. Language bias cannot be excluded because our search strategy exclusively based on the English language dominated databases.
Conclusions
In summary, this systematic review demonstrated there were consistently alterations of gut microbiota in ASD patients compared with HCs, as assessed by culture-independent techniques. It strengthened evidence that dysbiosis of gut microbiota may correlate with behavioral abnormality in ASD. A number of issues resulted in heterogeneity has be drawn, including dietary factors, the selection of controls, frequent occurrence of GI symptoms and sample sources. Thus, more well-designed studies with available sequencing data are needed to better understand the significance of the host interaction with gut microbiota in ASD. Furthermore, as a potential risk factor, the gut microbiome could be a novel target for ASD patients in the future.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number: NSFC313220143).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Feitong Liu, Jie Li
References
- 1.Association A. P. Diagnostic and statistical manual of mental disorders (DSM-5th). American Psychiatric Pub 2013. [DOI] [PubMed]
- 2.Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 3.Kim YS, Leventhal BL. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol. Psychiatry. 2015;77:66–74. doi: 10.1016/j.biopsych.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colvert E, et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry. 2015;72:415–423. doi: 10.1001/jamapsychiatry.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaugler T, et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc. Natl Acad. Sci. USA. 2015;112:e5600–5607. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McElhanon BO, McCracken C, Karpen S, Sharp WG. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133:872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 8.Croen LA, et al. The health status of adults on the autism spectrum. Autism. 2015;19:814–823. doi: 10.1177/1362361315577517. [DOI] [PubMed] [Google Scholar]
- 9.de Theije CGM, et al. Pathways underlying the gut-to-brain connection in autism spectrum disorders as future targets for disease management. Eur. J. Pharmacol. 2011;668:S70–S80. doi: 10.1016/j.ejphar.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Stolp HB, Dziegielewska KM, Ek CJ, Potter AM, Saunders NR. Long-term changes in blood-brain barrier permeability and white matter following prolonged systemic inflammation in early development in the rat. Eur. J. Neurosci. 2005;22:2805–2816. doi: 10.1111/j.1460-9568.2005.04483.x. [DOI] [PubMed] [Google Scholar]
- 11.Stolp HB, et al. Effects of neonatal systemic inflammation on blood-brain barrier permeability and behaviour in juvenile and adult rats. Cardiovasc. Psychiatry Neurol. 2011;2011:469046. doi: 10.1155/2011/469046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudo N. Role of microbiome in regulating the HPA axis and its relevance to allergy. Chem. Immunol. Allergy. 2012;98:163–175. doi: 10.1159/000336510. [DOI] [PubMed] [Google Scholar]
- 14.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Ray K. Gut microbiota: microbial metabolites feed into the gut-brain-gut circuit during host metabolism. Nat. Rev. Gastroenterol. Hepatol. 2014;11:76. doi: 10.1038/nrgastro.2014.8. [DOI] [PubMed] [Google Scholar]
- 16.Kang DW, et al. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe. 2018;49:121–131. doi: 10.1016/j.anaerobe.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Luna RA, et al. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children with Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2017;3:218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strati F, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang DW, et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5:10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue R, et al. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci. Biotechnol. Biochem. 2016;80:2450–2458. doi: 10.1080/09168451.2016.1222267. [DOI] [PubMed] [Google Scholar]
- 21.Son JS, et al. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS. ONE. 2015;10:e0137725. doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang DW, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS. ONE. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Angelis M, Francavilla R, Piccolo M, De Giacomo A, Gobbetti M. Autism spectrum disorders and intestinal microbiota. Gut Microbes. 2015;6:207–213. doi: 10.1080/19490976.2015.1035855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams BL, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS. ONE. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio. 2012;3:e00261–11. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gondalia SV, et al. Molecular characterization of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012;5:419–427. doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- 27.Finegold SM, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Tomova A, et al. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015;138:179–187. doi: 10.1016/j.physbeh.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, et al. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, et al. Low Relative Abundances of the Mucolytic Bacterium Akkermansia muciniphila and Bifidobacterium spp. in Feces of Children with Autism. Appl. Environ. Microbiol. 2011;77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J. Med. Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 33.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell. Host. Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Finegold SM, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Carvey PM, Ling Z. Altered glutathione homeostasis in animals prenatally exposed to lipopolysaccharide. Neurochem. Int. 2007;50:671–680. doi: 10.1016/j.neuint.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Navarro F, Liu Y, Rhoads JM. Can probiotics benefit children with autism spectrum disorders? World J. Gastroenterol. 2016;22:10093–10102. doi: 10.3748/wjg.v22.i46.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012;3:S415–S421. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menard S, et al. Bifidobacterium breve and Streptococcus thermophilus secretion products enhance T helper 1 immune response and intestinal barrier in mice. Exp. Biol. Med. (Maywood). 2005;230:749–756. doi: 10.1177/153537020523001008. [DOI] [PubMed] [Google Scholar]
- 43.Furusawa Y, Obata Y, Hase K. Commensal microbiota regulates T cell fate decision in the gut. Semin. Immunopathol. 2015;37:17–25. doi: 10.1007/s00281-014-0455-3. [DOI] [PubMed] [Google Scholar]
- 44.Eren AM, et al. A single genus in the gut microbiome reflects host preference and specificity. Isme. J. 2015;9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacFabe DF, Cain NE, Boon F, Ossenkopp KP, Cain DP. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: Relevance to autism spectrum disorder. Behav. Brain. Res. 2011;217:47–54. doi: 10.1016/j.bbr.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, et al. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 2012;57:2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 48.Wolever TM, Spadafora P, Eshuis H. Interaction between colonic acetate and propionate in humans. Am. J. Clin. Nutr. 1991;53:681–687. doi: 10.1093/ajcn/53.3.681. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, et al. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 2012;57:2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 50.Hughes R, Magee E, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr. Issues Intest. Microbiol. 2000;1:51–58. [PubMed] [Google Scholar]
- 51.Finegold SM, et al. Gastrointestinal microflora studies in late-onset autism. Clin. Infect. Dis. 2002;35:S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 52.Williams BL, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS. ONE. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frye RE, Rose S, Slattery J, MacFabe DF. Gastrointestinal dysfunction in autism spectrum disorder: the role of the mitochondria and the enteric microbiome. Microb. Ecol. Health Dis. 2015;26:27458. doi: 10.3402/mehd.v26.27458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altieri L, et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers. 2011;16:252–260. doi: 10.3109/1354750X.2010.548010. [DOI] [PubMed] [Google Scholar]
- 55.Persico AM, Napolioni V. Urinary p-cresol in autism spectrum disorder. Neurotoxicol. Teratol. 2013;36:82–90. doi: 10.1016/j.ntt.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 56.McClean KL, Sheehan GJ, Harding GK. Intraabdominal infection: a review. Clin. Infect. Dis. 1994;19:100–116. doi: 10.1093/clinids/19.1.100. [DOI] [PubMed] [Google Scholar]
- 57.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 58.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 59.Kang DW, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS. ONE. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quevrain E, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding HT, Taur Y, Walkup JT. Gut microbiota and autism: Key concepts and findings. J. Autism Dev. Disord. 2017;47:480–489. doi: 10.1007/s10803-016-2960-9. [DOI] [PubMed] [Google Scholar]
- 62.Tillisch K, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–1401. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moayyedi P, et al. The effect of fiber supplementation on irritable bowel syndrome: a systematic review and meta-analysis. Am. J. Gastroenterol. 2014;109:1367–1374. doi: 10.1038/ajg.2014.195. [DOI] [PubMed] [Google Scholar]
- 64.Sandler RH, et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J. Child Neurol. 2000;15:429–435. doi: 10.1177/088307380001500701. [DOI] [PubMed] [Google Scholar]
- 65.Parracho HM, Gibson GR, Bosscher D, Kleerebezem M, McCartney AL. A double-blind, placebo-controlled, crossover-designed probiotic feeding study in children diagnosed with autistic spectrum disorders. Int J. Probiotics Prebiotics. 2010;5:69–74. [Google Scholar]
- 66.Buffington SA, et al. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krajmalnik-Brown R, Lozupone C, Kang DW, Adams JB. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Health Dis. 2015;26:26914. doi: 10.3402/mehd.v26.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kristensen NB, et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]