Abstract
Immunomodulatory drugs (IMIDs) are very effective in the treatment of multiple myeloma (MM). The description of their cereblon-mediated mechanism of action was a hallmark in MM research. Although the importance of IMID-induced degradation of cereblon-binding proteins is well described in vitro, the prognostic value of their expression levels in MM cells is less clear. Based on recently published data showing somewhat conflicting RNA levels, we analyzed the association between the levels of the Ikaros family zinc finger protein 1 (IKZF1), IKZF3, and karyopherin subunit alpha 2 (KPNA2) proteins measured by flow cytometry and prognostic parameters in 214 newly diagnosed MM patients who were randomized in the GMMG HD6 trial. No statistically significant associations between the expression levels and age, gender, light chain type, International Staging System (ISS) stage or cytogenetic high- and normal risk groups could be identified. Hyperdiploid MM cells expressed significantly higher levels of IKZF1, IKZF3 and KPNA2 than nonhyperdiploid cells. In contrast, translocation t(11;14) was associated with significantly lower expression levels. In conclusion, the observed overexpression of cereblon-binding proteins in MM cells with gain of chromosomes 5, 9, 11, 15, and 19 is consistent with the previously proposed positive regulation of MYC by IKZF1 and IKZF3, as well as MYC activation in hyperdiploid MM cells.
Introduction
Lenalidomide, an immunomodulatory drug (IMID), is a highly effective treatment option for patients with newly diagnosed and relapsed multiple myeloma (MM) and achieves response rates of up to 70% in combination with dexamethasone1–3.
In terms of its mechanism of action, lenalidomide binds to cereblon (CRBN), the substrate adaptor of the CRL4CRBN E3 ubiquitin ligase complex4. By modulating the substrate specificity of the CRL4CRBN E3 ubiquitin ligase, lenalidomide induces the selective ubiquitination and subsequent degradation of Ikaros family zinc finger protein (IKZF) 1 and IKZF3 in MM cells5,6. IKZF3 induces the expression of interferon regulatory factor 4 (IRF4) and IRF4 is essentially involved in the positive feedback regulation of the MYC oncogene7. Consequently, proteasomal degradation of the transcription factors IKZF1 and IKZF3 leads to MM cell death.
The expression levels of several CRBN-binding proteins, including IKZF1, IKZF3, and karyopherin subunit alpha 2 (KPNA2), are associated with clinical variables and cytogenetic aberrations and have predictive and prognostic relevance in IMID-treated patients with relapsed or newly diagnosed MM8–10. Notably, in the study by Krönke et al., IKZF1 mRNA was expressed at higher levels in patients with newly diagnosed International Staging System (ISS) stage III MM than in patients diagnosed with ISS stage I and II MM; lower IKZF1 and IKZF3 expression was detected in patients with gains of 1q21. In this study, a high pretreatment IKZF1 RNA level was identified as an adverse prognostic factor for progression-free survival (PFS). IKZF1 expression was not correlated with the response to induction therapy in patients who received lenalidomide and intensive chemotherapy8. In contrast, Zhu et al. have shown that low IKZF1 expression levels are an adverse prognostic factor for the response and survival of patients with relapsed/refractory MM treated with an IMID-based therapy using gene expression profiling10. Similarly, in the study by Pourabdollah et al., low IKZF1 and IKZF3 levels correlated with shorter PFS and overall survival (OS), as evidenced by immunohistochemistry of bone marrow from patients with relapsed/refractory MM who were treated with lenalidomide9. Sehgal et al. did not identify correlations between baseline immunohistochemical staining for the IKZF1 and IKZF3 proteins in tumor cells and the response to IMIDS or survival11. Moreover, Zhu et al. noted the prognostic relevance of mRNA levels of KPNA2, a nuclear transport protein that has been associated with B-cell development, in plasma cells (PCs)12. In patients with MM treated with IMIDs, a high KPNA2 expression level was associated with shorter OS10.
These current studies have therefore yielded contradictory results regarding the prognostic value of IKZF1 and IKZF3 expression in MM cells, possibly because they were performed in relatively small patient cohorts. The aim of our current study was therefore to determine the expression levels of the CRBN-binding proteins IKZF1, IKZF3, and KPNA2 in MM cells by flow cytometry and to assess correlations of their expression levels with clinical and prognostic factors measured at the time of diagnosis in a large patient cohort.
Material and methods
Patient selection and data matching
An analysis of 214 patients who were newly diagnosed with MM and randomized to participate in the multicenter prospective phase III trial GMMG-HD6 on the “Effect of Elotuzumab in VRD (Velcade, Revlimid, and Dexamethasone) Induction/Consolidation and Lenalidomide Maintenance” conducted by the German-Speaking Myeloma Multicenter Group (GMMG) was performed. Clinical parameters, including age, gender, and serum heavy and light chain type by immunofixation, and ISS were assessed as a part of the routine clinical examination. The cytogenetic examinations were performed in a central lab. All patients provided written informed consent before participating in the study. Approval was obtained by the ethics committee of the University of Heidelberg in collaboration with the participating ethics committees.
Molecular cytogenetic testing
Molecular cytogenetic testing was performed using a previously described method13. Briefly, CD138+ bone marrow PCs were purified using auto-magnetic-activated cell sorting with anti-CD138 immunobeads as published14. For interphase fluorescence in situ hybridization analyses, a panel of two-color probe sets was used to detect numerical changes at the chromosomal loci 1q21/13q14, 5p15/5q35, 8p21/19q13, 9q34/15q22, and 11q22.3/17p13 as well as the IgH-translocations t(11;14)(q13;q32), t(4;14)(p16;q32), t(14;16)(q32;q23), or any other IgH-rearrangement. Hybridization was performed according to the manufacturer’s instructions (MetaSystems, Altlussheim, Germany and Cytocell, Cambridge, UK) and a minimum of 100 interphase nuclei per probe were evaluated using an automated spot counting system (Applied Spectral Imaging, Edingen-Neckarhausen, Germany). Hybridization efficiency was validated using interphase nuclei obtained from the bone marrow of a healthy donor, and the thresholds for gains, deletions, and translocations were set at 10%. Hyperdiploidy was defined according to the criteria described by Wuilleme et al., which require trisomy of at least two of three chromosomes: 5, 9, and 1515. Two high-risk cytogenetic definitions were applied to facilitate comparisons with the results from other research groups: Neben et al.—the presence of deletion 17p13 and/or translocation t(4;14), gain of 1q21 (>3 copies)16, and Krönke et al.—the presence of deletion 17p and/or t(4;14) or t(14;16)8.
Plasma cell immunophenotyping
Plasma cell immunophenotyping was performed using cytoplasmic anti-κ-allophycocyanin (APC, TB28-2), cytoplasmic anti-λ-APC-H7 (1-155-2), CD19-phycoerythrin (PE)-Cy7 (HIB19), CD27-PerCP-Cy5.5 (L128), CD38-PE (HB7), CD45-V450 (2D1), CD56-fluorescein (FITC, NCAM16.2), CD81-APC-H7 (JS-81), and CD138-V500-C (MI15) antibodies (all obtained from BD Biosciences, Heidelberg, Germany). Fixation and permeabilization (FIX&PERM) solutions A and B (Biozol, Eching, Germany) were used for cytoplasmic anti-κ and anti-λ staining, and BD FACSTM Lysing Solution was used to lyse red blood cells (BD, Heidelberg, Germany). Before measurements were obtained, the cells were washed twice and resuspended in phosphate-buffered saline (Life Technologies, Carlsbad, USA). Measurements were performed on a FACSCanto™ II cell analyzer (BD, Heidelberg, Germany). Data were analyzed using the BD FACSDiva™ (BD, Heidelberg, Germany) and Infinicyt™ software packages (Cytognos, Salamanca, Spain). PCs were identified based on the coexpression of CD38 and CD138 antigens. An aberrant PC expression profile was defined as CD45-low/negative, CD56-positive, CD19-negative, and light chain-restricted17,18. CD27 and CD81 were evaluated separately as prognostic markers. The cutoff for the level of positive CD27 and CD81 antigen expression was defined as 20% compared to negative controls.
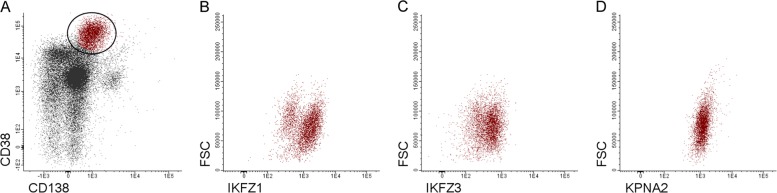
Similarly, the expression of CRBN-binding proteins was detected by flow cytometry according to the antibody combinations and the protocol for cytoplasmic staining listed in Table 1. IKZF1, IKZF3, and KPNA2 expression in PCs (CD138+, CD38+) was determined by measuring the fluorescence intensity (FI, Fig. 1).
Table 1.
Antibody panel used in flow cytometry assays to identify cereblon-binding proteins in myeloma cells
| Fluorochrome | V450 | V500-C | Alexa Fluor488/FITC | PE | PerCP | APC |
|---|---|---|---|---|---|---|
| Control tube (clone, company) |
/ | CD138 (MI15, BD) |
/ | / | / | CD38 (MHCD3805, Life Technologies) |
| Tube 1 (clone, company) |
CD45 (2D1, BD) |
CD138 (MI15, BD) |
IKZF1 (R32–1149, BD Pharmingen) |
IKZF3 (S50–895, BD Pharmingen) |
CD56 (5.1H11, BioLegend) |
CD38 (MHCD3805, Life Technologies) |
| Tube 2 (clone, company) |
CD45 (2D1, BD) |
CD138 (MI15, BD) |
KPNA2 (orb188808, Biorbyt) |
/ | CD56 (5.1H11, BioLegend) |
CD38 (MHCD3805, Life Technologies) |
Fig. 1. Gating strategy used to identify CRBN-binding proteins in MM cells.
Plasma cells (PCs) were identified as CD38/CD138-positve cells among leukocytes (a). The expression levels of IKZF1, IKZF3 and KPNA2 were determined by measuring the absolute median fluorescence intensity of the whole PC population (b, c, d; FSC, forward scatter)
Statistical analysis
For descriptive statistics, categorical data are presented as absolute numbers and percentages, and continuous data are presented as medians and ranges. The nonparametric Kruskal−Wallis rank sum test was used to test for differences in location of the distribution of continuous variables (IKZF1, IKZF3, and KPNA2 FI in PCs) with respect to categorical variables with two or more categories (gender, light chain type, ISS stage, cytogenetic risk category, CD27 and CD81 expression, and single cytogenetic abnormalities). Age was considered as a continuous variable and a univariate linear regression model was used to investigate the impact of age on log-transformed median FI values variables (IKFZ1, IKZF3, and KPNA2). All investigations were provided in an exploratory manner. Therefore, no correction for multiple testing was done. A two-sided significance level of 5% was used for all statistical analyses. All statistical analyses were performed with the statistical software environment R, version 3.3.2 (R Development Core Team).
Results
Patient characteristics
Bone marrow samples from 214 patients (129 males and 85 females) with newly diagnosed MM patients, randomized in the GMMG HD6 trial, were analyzed. The median age was 60 (range 37–70) years. ISS stage I, II, and III were observed in 80 (37.4%), 81 (37.9%), and 53 (24.8%) patients, respectively. Depending on the definition (Neben et al. or Krönke et al.), high-risk cytogenetic abnormalities were detected in 40 (20.5%) and 46 (23.6%) patients, respectively. MM cells from 7 patients (3.4%) were negative for CD27, whereas CD81-positive MM cells were detected in 140 patients (69.3%). The demographic and clinical parameters are summarized in Table 2.
Table 2.
Clinical characteristics and prognostic factors
| Patient number, n | 214 |
| Age, years | 60 (37–70) |
| Gender, n (%) | |
| Male | 129 (60.3) |
| Female | 85 (39.7) |
| Heavy chain type, n (%) | |
| IgG | 136 (63.6) |
| IgA | 30 (14.0) |
| Other (IgM, IgD, and IgE) | 4 (1.9) |
| None | 44 (20.6) |
| Light chain type, n (%) | |
| κ | 146 (68.2) |
| λ | 68 (31.8) |
| β2-microglobulin (mg/l) | 3.4 (1.0–17.4) |
| Albumin (g/l) | 37 (20.0–56.0) |
| ISS, n (%) | |
| I | 80 (37.4) |
| II | 81 (37.9) |
| III | 53 (24.8) |
| Cytogenetic risk groupa, n (%) | |
| Standard risk | 155 (79.5) |
| High risk | 40 (20.5) |
| Missing | 19 |
| Cytogenetic risk groupb, n (%) | |
| Standard risk | 149 (76.4) |
| High risk | 46 (23.6) |
| Missing | 19 |
| Immonophenotypic prognostic markers, n (%) | |
| CD27-positive | 196 (96.5) |
| CD27-negative | 7 (3.4) |
| Missing | 11 |
| CD81-positive | 140 (69.3) |
| CD81-negative | 62 (30.7) |
| Missing | 12 |
Unless indicated otherwise, data are presented as medians (ranges)
ISS International Staging System
aCytogenetic high-risk group: presence of del17p and/or t(4;14) or a gain at 1q of >3 copies
bCytogenetic high-risk group: presence of del17p and/or t(4;14) or t(14;16)
IKZF1, IKZF3, and KPNA2 expression in MM cells
The levels of IKZF1, IKZF3, and KPNA2 expression were determined by flow cytometry. The distribution of the expression levels in myeloma cells for the whole cohort is shown in Fig. 2.
Fig. 2. Distribution of IKZF1, IKZF3, and KPNA2 expression in MM cells.
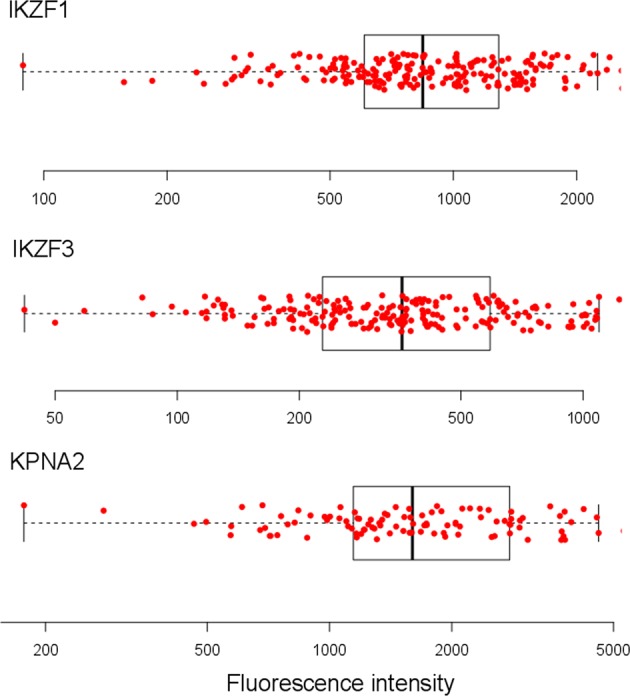
IKZF1 (n = 214), IKZF3 (n = 214), and KPNA2 (n = 108) fluorescence intensity was determined on the whole plasma cell population in all patients. Each dot represents a single patient. The boxplots in the background show the median, the first and the third quartiles as well as the lower and upper whiskers
The main focus of the study was to assess whether the expression levels of the individual CRBN-binding proteins differ between clinical and prognostic subgroups (Table 3). No statistically significant differences in IKZF1, IKZF3, and KPNA2 expression levels were identified with regard to age, gender, light chain type, ISS stage, and cytogenetic risk groups.
Table 3.
IKZF1, IKZF3, and KPNA2 expression levels in MM cells according to clinical and prognostic subgroups
| Variable | IKZF1 expression level | IKZF3 expression level | KPNA2 expression level | |||||
|---|---|---|---|---|---|---|---|---|
| n | Median | P value | Median | P value | Median | P value | ||
| Age, 10-year increase | 214 | 60 | 0.06 | 60 | 0.32 | 60 | 0.98 | |
| Gender | ||||||||
| Male | 129 | 844 | 0.94 | 341 | 0.03 | 1582 | 0.8 | |
| Female | 85 | 841 | 380 | 1678 | ||||
| Light chain type | ||||||||
| κ | 146 | 854 | 0.26 | 367 | 0.097 | 1647 | 0.33 | |
| λ | 68 | 799 | 337 | 1600 | ||||
| ISS | ||||||||
| I | 80 | 797 | 341 | 1368 | ||||
| II | 81 | 832 | 0.69 | 355 | 0.11 | 1594 | 0.5 | |
| III | 53 | 886 | 406 | 1809 | ||||
| Cytogenetic risk groupa | ||||||||
| Standard risk | 155 | 850 | 0.53 | 357 | 0.8 | 1588 | 0.99 | |
| High risk | 40 | 872 | 371 | 1695 | ||||
| Cytogenetic risk groupb | ||||||||
| Standard risk | 149 | 844 | 0.66 | 355 | 0.5 | 1577 | 0.79 | |
| High risk | 46 | 914 | 379 | 1702 | ||||
| Cytogenetic aberrations | ||||||||
| gain 1q21 | Yes | 66 | 866 | 0.54 | 382 | 0.33 | 1702 | 0.87 |
| No | 132 | 842 | 358 | 1579 | ||||
| gain 1q21 > 3 | Yes | 16 | 929 | 0.65 | 505 | 0.07 | 1691 | 0.77 |
| No | 182 | 847 | 356 | 1582 | ||||
| gain 5p15 | Yes | 87 | 1057 | <0.001 | 403 | 0.0013 | 1881 | 0.0026 |
| No | 98 | 745 | 321 | 1371 | ||||
| gain 5q35 | Yes | 85 | 1075 | <0.001 | 403 | 0.0013 | 1798 | 0.026 |
| No | 100 | 745 | 327 | 1458 | ||||
| gain 5 | Yes | 82 | 1096 | <0.001 | 404 | <0.001 | 1810 | 0.01 |
| No | 103 | 748 | 321 | 1456 | ||||
| del 8p21 | Yes | 48 | 1052 | 0.14 | 417 | 0.074 | 1939 | 0.085 |
| No | 138 | 802 | 336 | 1520 | ||||
| gain 9q34 | Yes | 109 | 1018 | <0.001 | 366 | 0.057 | 1735 | 0.035 |
| No | 78 | 745 | 337 | 1308 | ||||
| gain 11q23 | Yes | 111 | 998 | 0.015 | 366 | 0.34 | 1551 | 0.7 |
| No | 87 | 798 | 359 | 1702 | ||||
| del 13q14 | Yes | 100 | 811 | 0.14 | 358 | 0.83 | 1577 | 0.18 |
| No | 98 | 976 | 378 | 1798 | ||||
| gain 15q22 | Yes | 100 | 1015 | <0.001 | 372 | 0.041 | 1761 | 0.044 |
| No | 87 | 762 | 334 | 1492 | ||||
| gain 19q13 | Yes | 100 | 1037 | <0.001 | 402 | 0.0023 | 1881 | 0.01 |
| No | 86 | 725 | 304 | 1408 | ||||
| del 17p13 | Yes | 24 | 823 | 0.38 | 362 | 0.77 | 2044 | 0.37 |
| No | 174 | 854 | 359 | 1582 | ||||
| t(4;14) | Yes | 19 | 858 | 0.48 | 377 | 0.8 | 1459 | 0.21 |
| No | 177 | 850 | 359 | 1606 | ||||
| t(6:14) | Yes | 3 | 646 | 0.26 | 325 | 0.85 | 2345 | 0.49 |
| No | 184 | 847 | 357 | 1575 | ||||
| t(11;14) | Yes | 46 | 646 | <0.001 | 225 | <0.001 | 1064 | 0.0062 |
| No | 144 | 1019 | 388 | 1712 | ||||
| t(14;16) | Yes | 7 | 1075 | 0.51 | 568 | 0.21 | 1789 | 0.58 |
| No | 190 | 850 | 359 | 1588 | ||||
| Hyperdiploidy | Yes | 94 | 1033 | <0.001 | 384 | 0.0049 | 1951 | 0.0023 |
| No | 93 | 762 | 321 | 1334 | ||||
| Immunophenotypic prognostic markers | ||||||||
| CD27 | Positive | 196 | 807 | 0.5 | 357 | 0.39 | 1579 | 0.42 |
| Negative | 7 | 959 | 307 | 1215 | ||||
| CD81 | Positive | 140 | 801 | 0.59 | 357 | 0.97 | 1432 | 0.046 |
| Negative | 62 | 997 | 364 | 1919 | ||||
Median expression levels and P values are presented for clinical and prognostic subgroups. The P values for categorical prognostic factors are calculated by the Kruskal−Wallis test. For the continuous values of age the linear regression was used. Bold P values indicate statistical significance. KPNA2 expression levels were measured in 108 patients
A gain at chromosome 5 was considered positive when a gain at both 5p15 and 5q35 was present
Hyperdiploidy was defined as trisomy of at least two of three chromosomes: 5, 9, and 15
del deletion, ISS International Staging System
aCytogenetic high-risk group: presence of del17p and/or t(4;14) and/or a gain at 1q of >3 copies
b Cytogenetic high-risk group: presence of del17p and/or t(4;14) and/or t(14;16)
However, we observed a significant increase in IKZF1, IKZF3, and KPNA2 expression in patients with gains at chromosomes 5, 9 and 15, all of which exhibited gain (a borderline significant difference in IKZF3 expression was observed in MM patients with gain of chromosome 9, P = 0.057). Accordingly, patients with hyperdiploidy expressed significantly higher levels of IKZF1, IKZF3, and KPNA2 in MM cells compared to patients without hyperdiploidy (Fig. 3). Additionally, patients with gain 19 also expressed significantly higher levels of all three CRBN-binding proteins in MM cells.
Fig. 3. Distribution of IKZF1, IKZF3, and KPNA2 expression in MM cells among cytogenetic aberration subgroups.
The figure shows the fluorescence intensity of IKZF1, IKZF3, and KPNA2 on the whole plasma cell population in MM patients with (n = 46) and without (n = 144) translocation t(11;14) and with (n = 94) and without (n = 93) hyperdiploidy. Each dot represents a single patient. The boxplots in the background show the median, the first and the third quartiles as well as the lower and upper whiskers. MM multiple myeloma
Interestingly, in contrast to hyperdiploidy, t(11;14) was associated with significantly lower expression of IKZF1, IKZF3, and KPNA2 in MM cells (Fig. 3).
Regarding the prognostic factors CD27 and CD81, significant differences in KPNA2, but not IKZF1 or IKZF3, expression levels were detected between CD81-positive and -negative MM cells. Compared to CD81-positive MM cells, CD81-negative MM cells showed a higher KPNA2 FI (1919 vs. 1432, P = 0.046). No statistically significant differences in the levels of CRBN-binding proteins were observed between patients with CD27-positive and -negative MM cells.
Discussion
The expression levels of IKZF1, IKZF3, and KPNA2 have predictive and prognostic value in determining the response of patients with MM to IMID treatment. Moreover, an association with clinical variables and cytogenetic aberrations has been proposed8–10. In the current study, we determined the expression levels of the CRBN-binding proteins IKZF1, IKZF3, and KPNA2 in MM cells in a cohort of 214 patients with newly diagnosed MM before treatment with an induction therapy containing lenalidomide. A comparison of the protein expression levels in different clinical and prognostic subgroups was performed.
We did not identify any significant differences in IKZF1 and IKZF3 expression with regard to age, gender, ISS stage, and light chain type. These results are consistent with the results reported by Krönke et al., who assessed IKZF1 and IKZF3 mRNA expression levels in PCs from 60 patients with newly diagnosed MM8. A high-risk cytogenetic status was defined using the criteria reported by Krönke et al. (presence of del17p and/or t(4;14) or t(14;16)8) and criteria applied at our institution (presence of del17p and/or t(4;14) or a gain at 1q21 of >3 copies16) to ensure comparability. However, no differences in IKZF1 and IKZF3 expression were identified between cytogenetic standard and high-risk groups using either definition, which also confirms the data reported by Krönke et al. Moreover, we found that del 17p13, del 13q14 and t(4;14) were not associated with higher or lower IKZF1 and IKZF3 expression at the protein level. In contrast, lower IKZF1 expression in patients with a gain at 1q21 was not confirmed in the current study, as similar levels of the IKZF1 protein were detected in MM cells from patients with and without a gain at 1q21.
Analogous results were obtained for the levels of the KPNA2 protein. Although Zhu et al. reported an association between a high KPNA2 RNA expression level and shorter OS in patients with relapsed/refractory MM who were treated with IMIDs10, we did not identify an association between the expression of the KPNA2 protein and clinical/prognostic variables.
Genetically, two major groups of MM are distinguished according to ploidy and chromosome 14q32 translocations, hyperdiploid and nonhyperdiploid groups, both of which are assumed to be early mutagenic events19. Hyperdiploidy in MM is defined as gain of certain odd-numbered chromosomes and is observed in approximately one-half of patients and, with some exceptions, was described to have a positive prognostic value20. A major new finding of the current study is the significant increase in IKZF1, IKZF3, and KPNA2 expression observed in patients with hyperdiploidy defined as trisomy of chromosomes 5, 9, and 15. Similarly, patients with gains at 19q13 expressed significantly higher levels of these proteins, and patients with gain of 11q22.3 expressed higher levels of IKZF1, but not IKZF3 or KPNA2. As described in the study by Wuilleme et al. reviewing large published studies constituting 173 hyperdiploid karyotypes, the most frequent gained chromosomes are the odd-numbered chromosomes: 3, 5, 7, 9, 11, 15, 19, and 21. The use of the combination of chromosomes 5, 9, and 15 is generally the best compromise between specificity and sensitivity for the detection of hyperdiploidy15. Therefore, a common underlying mechanism of hyperdiploidy was postulated and was reflected by uniformly increased IKZF1, IKZF3, and KPNA2 expression levels in MM cells with gains at all chromosomes analyzed in the current study (5p15/5q35, 9q34, 11q22.3, 15q22, and 19q13).
MYC has recently been shown to be activated in 67% of myeloma samples and is associated with hyperdiploidy21,22. Moreover, kinetic analyses have revealed that IKZF1 and IKZF3 may act as positive regulators of IRF4 and MYC in MM cells, thus providing new understanding of the mechanism of action of IMIDs. As shown in the study by Bjorklund et al., downregulation of MYC led to the sequential downregulation or proteasomal degradation of IKZF1, IKZF3 and IRF4, followed by growth inhibition and apoptosis of MM cells23. Our results complement these studies, as we observe increased levels of the IKZF1 and IKZF3 proteins in hyperdiploid MM cells, consistent with the positive regulation and activation of MYC by IKZF1 and IKZF3.
Nonhyperdiploid MM involves translocations of the immunoglobulin heavy chain alleles at chromosome 14q32 with different chromosomes, namely chromosomes 4, 6, 11, 16, and 2015,24. In the current study, we observed a significant decrease in the IKZF1, IKZF3, and KPNA2 levels in MM cells with translocation t(11;14), but no differences were observed in cells with other chromosome 14 translocations. Therefore, our current findings differ from the results published by Krönke et al., who did not observe differences in IKZF1 or IKZF3 RNA expression levels in the presence or absence of t(11;14). A prognostic value for t(11;14), which is detected in approximately 15–20% of patients with MM, has not yet been determined19, and the obtained results are difficult to interpret in terms of the prognosis or underlying biological mechanism. However, IKZF1, IKZF3, and KPNA2 expression differed in hyperdiploid MM (high protein levels) and t(11;14) MM (low protein levels), highlighting the two major genetically distinguished groups.
Several studies have reported the predictive value of the levels of CRBN-binding proteins regarding the response to IMID therapy and survival outcomes in patients with MM, indicating their considerable potential for use as biomarkers. For example, Zhu et al. reported a lack of clinical response in pomalidomide-treated patients with relapsed/refractory MM who exhibited the lowest quartile range of IKZF1 RNA expression in their MM cells. In this study, cohorts with low IKZF1 and high KPNA2 RNA expression levels showed shorter OS. Moreover, in terms of potential clinical use, the assessment of IKZF1 expression using flow cytometry achieved results consistent with western blot analyses10. Bolomsky et al. analyzed IKZF1 expression in MM cells from a cohort of patients with MM who were treated with lenalidomide using flow cytometry and determined correlations with outcomes. High levels of IKZF1 expression on MM cells in patients before treatment were associated with prolonged PFS25. Contrary, Dimopoulos et al. could not identify any predictive value of CRBN, IKZF1, and IKZF3 for IMID response in 23 MM patients treated with a lenalidomide-containing regimen and argue against the use of these proteins as predictive biomarkers. In this study, the expression levels of CRBN, IKZF1, and IKZF3 were determined by standard immunohistochemistry in formalin-fixed paraffin embedded bone marrow samples26. A correlation between the levels of CRBN-binding proteins in MM cells assessed using flow cytometry and the response of patients with MM to lenalidomide-based induction therapy is the subject of current investigations by our group.
Low CD27 expression was associated with high-risk disease and CD81 positivity was described as an adverse prognostic marker in patients with MM27–29. No differences in IKZF1, IKZF3 and KPNA2 expression were observed between CD27-positive and -negative MM cells. On the contrary, KPNA2 expression was increased in CD81-negative MM cells. However, the functional and prognostic importance of this finding remains unclear.
The current study provides the largest scale effort to date, in terms of patient samples analyzed, to characterize the association between CRBN-binding proteins in MM cells and clinical characteristics at first diagnosis. Nonetheless, the present analysis shows several limitations. Compared to other techniques, flow cytometry is a relatively easy-to-establish and fast method to assess protein expression levels and is feasible in large numbers of patients in clinical routine diagnostics. Combined with further antigens it also allows to determine the expression levels of proteins in selected subpopulations of cells (e.g. monoclonal MM cells characterized by light chain restriction). However, no uniform IKZF1, IKZF3 and KPNA2 flow cytometric protocols are available and standardization is warranted. Also, further studies on CRBN-binding proteins discussed in this manuscript relay on other techniques than flow cytometry for expression level measurement. Exemplarily, Dimopoulos et al. use standard immunohistochemistry in formalin-fixed paraffin-embedded bone marrow samples26. Krönke et al. and Zhu et al. assess IKZF1, IKZF3 and KPNA2 expression on RNA but not protein level8,10. Therefore, comparability of results might be hampered and further studies are required to prove the correlation between different methods.
The current study gives detailed information on the CRBN-binding proteins expression levels in different clinical and prognostic subgroups providing a functional link between previously reported positive regulation of MYC by IKZF1 and IKZF3 and MYC activation in hyperdiploid MM. One could hypothesize that high CRBN-binding proteins expression levels—and therefore hyperdiploidy—might indicate higher dependency on this pathway and IMID treatment might be associated with better response. However, the correlation results and subgroup analyses on CRBN-binding protein expression levels and response to lenalidomide-based induction therapy are still pending and will provide more insights into the predictive value of CRBN-binding proteins levels for IMID treatment.
Moreover, the assessment of cereblon-binding proteins expression in MM cells post lenalidomide treatment would provide additional information on IMID mechanism of action. However, after induction treatment the majority of patients reaches excellent response rates and only few or no MM cells can be detected in the bone marrow of MM patients corresponding to minimal residual disease negativity (MRD). Also, bone marrow assessment post induction (i.e. post lenalidomide) treatment is usually performed in case of serological CR in order to prove MRD status. Therefore, no accessible bone marrow sample with a sufficient number of MM cell to analyze the level of cereblon-binding proteins expression is available post lenalidomide treatment. Hence, cereblon-binding proteins expression levels in MM cells can only be measured at first diagnosis. Future analyses of our group will focus on cereblon-binding proteins expression levels at relapse, therefore providing more insights into resistance mechanism after IMID treatment.
In summary, we demonstrate the feasibility of large-scale IKZF1, IKZF3 and KPNA2 expression level assessment in MM cells by flow cytometry. Regarding cytogenetic abnormalities, high IKZF1, IKZF3, and KPNA2 expression were observed in hyperdiploid MM cells with gain of chromosomes 5, 9, 15, and 19. These results are consistent with the previously proposed positive regulation of MYC by IKZF1 and IKZF3 and MYC activation in hyperdiploid MM. Future correlation analysis of IKZF1, IKZF3, and KPNA2 expression levels and response to lenalidomide-based induction therapy are warranted and will establish the predictive value of CRBN-binding proteins levels.
Acknowledgements
The authors thank the participating patients and the study teams at the GMMG trial sites. We also thank Larissa Schönhoff for carrying out the flow cytometry analyses; Annekathrin Borowski, Michaela Brough, Michelle Ebentheuer, Marie-Christine Meffert and Stephanie Pschowski-Zuck for technical assistance in interphase-FISH analysis. This work was supported by Celgene GmbH and BMS GmbH & Co. KGaA.
Authors’ contributions
K.K., M.H.S.A., D.H., A.S., and A.J. performed experiments. K.K., M.-A.B., N.B., and A.B. analyzed results and made the figures. K.K., H.G., and M.H. designed the research and wrote the paper. M.Merz, M.S.R., J.H., U.B., K.W., H.J.S., M.H., R.F., M.Munder, J.D., and C.M.-T. recruited the patients and critically reviewed the manuscript.
Conflict of interest
K.K.: Research Support BMS, Celgene, Sanofi, Morphosys. M.-A.B.: Travel grants: Amgen, Novartis, Bristol-Myers Squibb, Janssen, Sanofi; Honoraria: Novartis, Takeda. M.Merz: Travel grants: Celgene, Takeda, AMGEN, Janssen; Advisory Board: AMGEN; Research grant: Takeda. M.R.: research funding from Novartis, Amgen, Morphosys. J.H.: honoraria from Amgen, BMS, Celgene, Janssen; research support from Sanofi; advisory boards: Amgen, Celegene, Janssen, Novartis. K.W.: advisory board and honoraria: AMGEN, Bristol-Myers Squibb, Celgene, Janssen, Novartis, Takeda, Onyx; research funding: Janssen, Celgene. H.J.S.: honoraria and travel support: Janssen Cilag, Celgene, Bristol Meyer Squibb. M.H.: honoraria Roche, Novartis. R.F.: Honoraria and travel grants: Celgene, Janssen, BMS; research funding: Celgene. M.Munder: Consultancy: Janssen Cilag, BMS, Takeda, Amgen, Celgene. J.D.: advisory Board: Bristol-Myers Squibb, Celgene, Janssen; speakers bureau: Celgene, Janssen; research funding: Celgene. H.G.: Research support: Celgene, Janssen, Chugai, Novartis, BMS; Advisory boards: Janssen, Celgene, Novartis, Amgen, Takeda, BMS; Honoraria: Celgene, Janssen, Novartis, Chugai, BMS. M.H.: Research support: BMS, Celgene, Sanofi, Morphosys. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palumbo A, et al. Continuous lenalidomide treatment for newly diagnosed multiple myeloma. N. Engl. J. Med. 2012;366:1759–1769. doi: 10.1056/NEJMoa1112704. [DOI] [PubMed] [Google Scholar]
- 2.Weber DM, et al. Lenalidomide plus dexamethasone for relapsed multiple myeloma in North America. N. Engl. J. Med. 2007;357:2133–2142. doi: 10.1056/NEJMoa070596. [DOI] [PubMed] [Google Scholar]
- 3.Rajkumar SV, et al. Combination therapy with lenalidomide plus dexamethasone (Rev/Dex) for newly diagnosed myeloma. Blood. 2005;106:4050–4053. doi: 10.1182/blood-2005-07-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fink EC, Ebert BL. The novel mechanism of lenalidomide activity. Blood. 2015;126:2366–2369. doi: 10.1182/blood-2015-07-567958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kronke, J. et al. IKZF1 expression is a prognostic marker in newly diagnosed standard-risk multiple myeloma treated with lenalidomide and intensive chemotherapy: a study of the German Myeloma Study Group (DSMM). Leukemia10.1038/leu.2016.384 (2017). [DOI] [PubMed]
- 9.Pourabdollah M, et al. High IKZF1/3 protein expression is a favorable prognostic factor for survival of relapsed/refractory multiple myeloma patients treated with lenalidomide. J. Hematol. Oncol. 2016;9:123. doi: 10.1186/s13045-016-0354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu YX, et al. Identification of cereblon-binding proteins and relationship with response and survival after IMiDs in multiple myeloma. Blood. 2014;124:536–545. doi: 10.1182/blood-2014-02-557819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehgal K, et al. Clinical and pharmacodynamic analysis of pomalidomide dosing strategies in myeloma: impact of immune activation and cereblon targets. Blood. 2015;125:4042–4051. doi: 10.1182/blood-2014-11-611426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovac CR, Emelyanov A, Singh M, Ashouian N, Birshtein BK. BSAP (Pax5)-importin alpha 1 (Rch1) interaction identifies a nuclear localization sequence. J. Biol. Chem. 2000;275:16752–16757. doi: 10.1074/jbc.M001551200. [DOI] [PubMed] [Google Scholar]
- 13.Merz M, et al. Baseline characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am. J. Hematol. 2016;91:E473–E477. doi: 10.1002/ajh.24533. [DOI] [PubMed] [Google Scholar]
- 14.Granzow, M. et al. Novel recurrent chromosomal aberrations detected in clonal plasma cells of light chain amyloidosis patients show potential adverse prognostic effect: first results from a genome-wide copy number array analysis. Haematologica10.3324/haematol.2016.160721 (2017). [DOI] [PMC free article] [PubMed]
- 15.Wuilleme S, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19:275–278. doi: 10.1038/sj.leu.2403586. [DOI] [PubMed] [Google Scholar]
- 16.Neben K, et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion17p. Blood. 2012;119:940–948. doi: 10.1182/blood-2011-09-379164. [DOI] [PubMed] [Google Scholar]
- 17.Robillard N, Wuilleme S, Moreau P, Bene MC. Immunophenotype of normal and myelomatous plasma-cell subsets. Front. Immunol. 2014;5:137. doi: 10.3389/fimmu.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawstron AC, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93:431–438. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 19.Robiou du Pont S, et al. Genomics of multiple myeloma. J. Clin. Oncol. 2017;35:963–967. doi: 10.1200/JCO.2016.70.6705. [DOI] [PubMed] [Google Scholar]
- 20.Chretien ML, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015;126:2713–2719. doi: 10.1182/blood-2015-06-650242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chng WJ, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misund K, et al. Complementary activation of Ccnd, MYC, RAS and NFkB by mutations in multiple myeloma. Blood. 2016;128:355–355. [Google Scholar]
- 23.Bjorklund CC, et al. Rate of CRL4(CRBN) substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J. 2015;5:e354. doi: 10.1038/bcj.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehl, W. M. & Bergsagel, P. L. Early genetic events provide the basis for a clinical classification of multiple myeloma. Hematol. Educ. Prog. 346−352 10.1182/asheducation-2005.1.346 (2005). [DOI] [PubMed]
- 25.Bolomsky A, et al. IKAROS expression in distinct bone marrow cell populations as a candidate biomarker for outcome with lenalidomide-dexamethasone therapy in multiple myeloma. Am. J. Hematol. 2017;92:269–278. doi: 10.1002/ajh.24634. [DOI] [PubMed] [Google Scholar]
- 26.Dimopoulos, K. et al. Expression of CRBN, IKZF1, and IKZF3 does not predict lenalidomide sensitivity and mutations in the cereblon pathway are infrequent in multiple myeloma. Leuk. Lymphoma 1−9. 10.1080/10428194.2018.1466290 (2018). [DOI] [PubMed]
- 27.Paiva B, et al. Clinical significance of CD81 expression by clonal plasma cells in high-risk smoldering and symptomatic multiple myeloma patients. Leukemia. 2012;26:1862–1869. doi: 10.1038/leu.2012.42. [DOI] [PubMed] [Google Scholar]
- 28.Moreau P, et al. Lack of CD27 in myeloma delineates different presentation and outcome. Br. J. Haematol. 2006;132:168–170. doi: 10.1111/j.1365-2141.2005.05849.x. [DOI] [PubMed] [Google Scholar]
- 29.Guikema JE, et al. CD27 is heterogeneously expressed in multiple myeloma: low CD27 expression in patients with high-risk disease. Br. J. Haematol. 2003;121:36–43. doi: 10.1046/j.1365-2141.2003.04260.x. [DOI] [PubMed] [Google Scholar]