Abstract
Histone methylation is thought to control the regulation of genetic program and the dysregulation of it has been found to be closely associated with cancer. JMJD3 has been identified as an H3K27 demethylase and its role in cancer development is context specific. The role of JMJD3 in gastric cancer (GC) has not been examined. In this study, JMJD3 expression was determined. The prognostic significance of JMJD3 and its association with clinical parameters were evaluated. JMJD3 dysregulation mechanism and targets were analyzed. The effect of JMJD3 mutation was determined by functional study. Results showed that JMJD3 was overexpressed in different patient cohorts and also by bioinformatics analysis. High JMJD3 expression was correlated with shortened overall survival in patients with GC and was an independent prognosis predictor. Genetic aberration and DNA methylation might be involved in the deregulation of JMJD3 in GC. Downstream network of JMJD3 was analyzed and several novel potential targets were identified. Furthermore, functional study discovered that both demethylase-dependent and demethylase-independent mechanisms were involved in the oncogenic role of JMJD3 in GC. Importantly, histone demethylase inhibitor GSK-J4 could reverse the oncogenic effect of JMJD3 overexpression. In conclusion, our study report the oncogenic role of JMJD3 in GC for the first time. JMJD3 might serve as an important epigenetic therapeutic target and/or prognostic predictor in GC.
Introduction
Epigenetic modifications play an important role in cancer initiation and progression1. Histone methylation is an essential epigenetic phenomenon and the dysregulation of it is associated with the processes of cancer occurrence/progression2. The most common histone modifications are acetylation and methylation, which result in target gene expression or repression3. The Jumonji domain containing-3 (JMJD3), also known as lysine (K)-specific demethylase 6B (KDM6B) can demethylate H3K27me3 to H3K27me2 or H3K27me1, and dissociate polycomb group complexes4.
Many studies have demonstrated that JMJD3 is involved in cancer progression via regulation of several cellular processes, such as proliferation, senescence, and apoptosis1,3,5. However, there is controversy regarding the expression pattern of JMJD3 in different cancers. Based on analysis of JMJD3 expression in diverse tumor tissues from the oncomine database, Agger et al. found that the expression of JMJD3 was significantly decreased in various cancers, including lung and liver carcinomas, as well as various hematopoietic malignancies6. Moreover, JMJD3 expression levels are lower in colorectal cancer relative to normal tissues7. Conversely, Shen et al. reported that both JMJD3 mRNA and protein levels were significantly elevated in renal carcinoma compared to adjacent normal tissue8. Moreover, JMJD3 expression was found to be significantly increased in prostate cancer, and increased further during metastasis9,10. Similarly, high expression levels of JMJD3 were found in gliomas11.
To our knowledge, there is no published study on the role of JMJD3 in gastric cancer (GC). In the current study, JMJD3 transcripts and JMJD3 protein expression were measured in different patient cohorts. The clinicalpathological and prognostic significance of JMJD3 expression were evaluated and the upstream regulating mechanism and downstream targets were identified. Elucidation of the role of JMJD3 in GC may lead to new therapeutic approach for the treatment of this disease.
Materials and Methods
Gastric clinical tissues
Clinical microarray tissues from 128 gastric cancer patients were retrieved from the tissue bank of the Prince of Wales Hospital (Hong Kong). Use of these tissues had been approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee. A total of 41 fresh gastric cancer and adjacent non-cancerous tumor tissue samples were collected from the tissue bank of Yijishan Hospital of Wannan Medical College (Wuhu, Anhui Province, China). All procedures using human tissue samples were performed in accordance with the relevant guidelines and regulations of the above institutions and informed consent for study participation were obtained from all patients involved.
RT-PCR and real-time quantitative PCR
Total RNA was extracted from tissues using TRIReagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. DNase I-treated RNA samples were reverse transcribed using M-MLV reverse transcriptase (Takara) plus a mixture of oligo (dT)12–18 and random primers. cDNA samples (1 µl) were used for conventional PCR amplification, using JMJD3-specific primer pairs. For real-time quantitative PCR analysis, the PCR reaction was performed in a real-time PCR system (Takara) and the expression levels of target gene relative to β-actin were determined using an SYBR Green-based comparative CT method (relative fold change = 2−ΔΔCT). Primers used are as follows: JMJD3: forward primer: 5′-GGAGGCCACACGCTGCTAC-3′, reverse primer: 5′-GCCAGTATGAAAGTTCCAGAGCTG-3′, β-actin: forward primer: 5′-CATGTACGTTGCTATCCAGGC-3′, reverse primer: 5′-CTCCTTAATGTCACGCACGAT-3′.
Immunohistochemistry
Immunohistochemistry of JMJD3 was conducted on a gastric cancer tissue microarray consisting of 128 tumor tissues. Tissue sections were deparaffinized, rehydrated and rinsed in distilled water. Antigen retrieval was done with sodium citrate buffer (pH 6.0), in a microwave oven for 5 min. The endogenous peroxidase activity was blocked using 3% (v/v) hydrogen peroxide for 10 minutes. Immunohistochemical staining for JMJD3 was performed using anti-JMJD3 antibodies (BD Biosciences) via the standard avidin-biotin method. Measurement of immunohistochemical staining was based on a semi quantitative scoring method. For the intensity of staining, 0 = negative (<5%), 1 = very weak (5~20%), 2 = weak (21~40%), 3 = moderate (41~60%), 4 = strong (61~80%), 5 = very strong (>80%). JMJD3 scores in gastric cancer tissues were further subdivided into high-expression (3, 4, 5) and low-expression groups (0, 1, 2).
Cell lines and transfection
Gastric cancer cell lines TMK-1 and MKN-45 were used in this study. TMK1 was obtained from Dr. Eiichi Tahara (University of Hiroshima, Hiroshima, Japan). MKN-45 was purchased from American Type Culture Collection (Manassas, VA, USA). Transient transfection of pMSCV-wt JMJD3 or pMSCV-mutant JMJD3 was mediated by Lipofectamine 2000.
MTT assay
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenylte–trazolium bromide (MTT) assay. After transfection for different time periods, MTT solution at the final concentration of 0.5 mg/ml was added to each well and the plates were incubated for another 3 h. Dimethyl sulfoxide (DMSO) (150 μl) was then added to solubilize MTT tetrazolium crystal. Finally, the optical density was determined at 570 nm.
BrdU assay
The colorimetric cell proliferation ELISA kit using Bradykinine U (BrdU) incorporation (Roche, Indianapolis, IN, USA) was used for cell proliferation assay. Experiment was carried out according to manufacturer’s instruction.
Bioinformatics analysis
The relationship of JMJD3 expression and overall survival was determined using TCGA data. The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) provides large scale cancer genomic data and cancer clinical sample information. The association of JMJD3 expression with clinical parameters and DNA methylation, the JMJD3 genetic alteration and downstream targets were analyzed by cBioportal. The comparison of expression level of different JMJD3 transcript variants in GC and normal tissue was carried out in Human Protein Atlas (http://www.proteinatlas.org/). Proteins that were positively co-expressed with JMJD3 and negatively co-expressed with EZH2 were identified by online program Venny using TCGA data.
Statistical analysis
GraphPad Prism 5 (GraphPad Software) was used for statistical analysis. All results were expressed as mean ± SD from at least three independent experiments. Student t-test was used to compare the discrepancy of two groups and one-way ANOVA was used to compare multiple groups. Overall survival was shown as Kaplan–Meier curve with p values calculated using the log-rank test. P values of less than 0.05 were considered statistically significant.
Results
JMJD3 was upregulated in GC and its high expression predicts poor survival
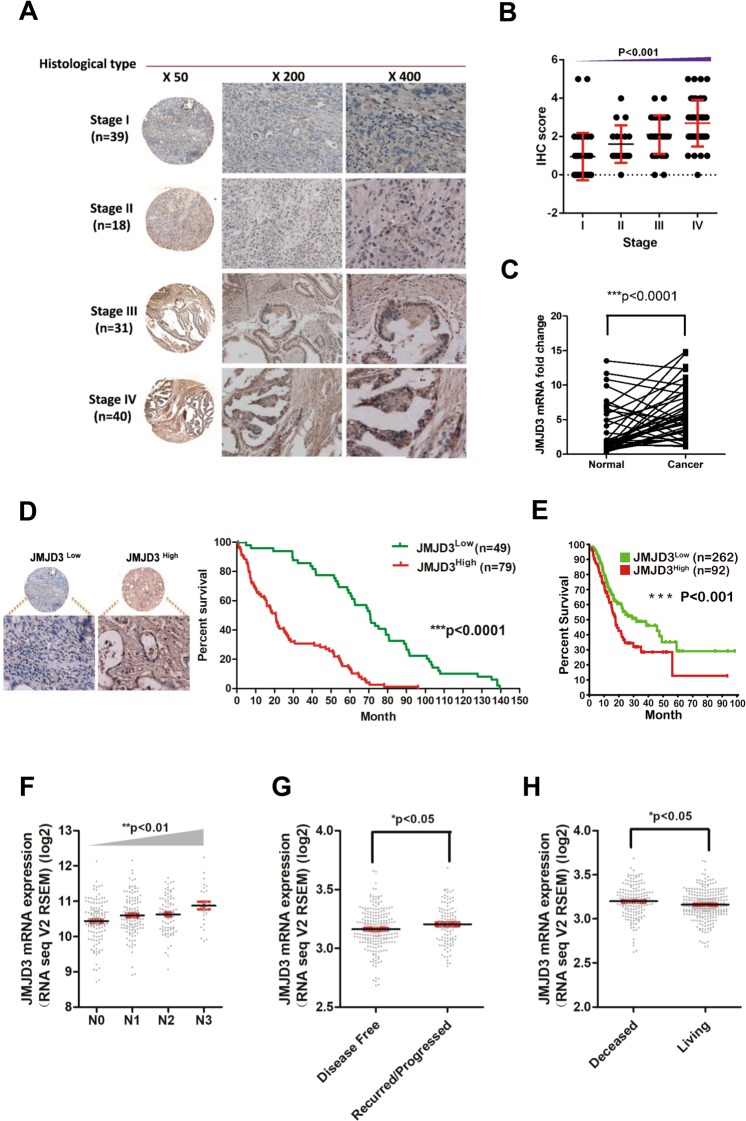
The expression level of JMJD3 was determined by IHC analysis in tissue microarray containing paraffin-embedded GC tissue samples from four different histological stages (Stage I n = 39; Stage II n = 18; Stage III n = 31; Stage IV n = 40). JMJD3 protein expression level significantly escalated (p < 0.001) with tumor stage (Fig. 1A,B). A total of 41 GC tissue and paired adjacent normal tissue samples were available from patients with GC who had undergone surgery. All patients were treated by radical gastrectomy and received no preoperative radiation or chemotherapy. Most patients (35/41) were at an early stage (stages 1 and 2) and no lymph node metastasis was present in any patient. The overall 5-year survival rate was 100%, suggesting that early diagnosis and surgical removal of the cancer tissue resulted in a good prognosis. Realtime PCR result of this cohort of patient sample demonstrated that JMJD3 level was significantly elevated in tumor compared with adjacent normal tissue (p < 0.001) (Fig. 1C). JMJD3 protein expression in gastric cancer specimens from 128 patients (Supplementary Table 1), including 89 male (69.5%) and 39 female (30.5%) (mean age was 59.3 years) was evaluated. According to JMJD3 expression level patients were categorized into two groups: low JMJD3 (n = 49) and high JMJD3 (n = 79) (Fig. 1D). The overall survival rate of patients with high JMJD3 expression was significantly lower compared to patients with low JMJD3 expression (p < 0.001) (Fig. 1D). We also analyzed patient survival according to JMJD3 expression level using TCGA data and obtained similar result (p < 0.001) (Fig. 1E). Furthermore, in the univariate Cox regression analysis, high JMJD3 expression was associated with a significantly increased risk of death (p < 0.001). Age (p < 0.05), tumor–node–metastasis (TNM) stage (p < 0.01) and lymph node metastasis (p < 0.001) were also significant predictors of overall survival (Table 1). After the adjustment for potential confounding factors, high JMJD3 expression (p < 0.01), age (p < 0.01), and TNM stage (p < 0.01) but not lymph node metastasis (p = 0.982) still predicted poorer survival in the multivariate model (Table 1). Moreover, we also evaluated the association of JMJD3 expression and clinical parameters by cBioportal. JMJD3 mRNA expression escalated with tumor stage (p < 0.01) and was higher in recurred/progressed disease than disease free (p < 0.05) and also higher in dead patients than living patients (p < 0.05) (Fig. 1F–H).
Figure 1.
Expression level of JMJD3 in GC and its association with patient survival and clinical parameters. (A) Expression level of JMJD3 was determined by IHC staining in tissue microarray containing 128 GC patient samples. (B) Statistical analysis of IHC result in different tumor stage. (C) JMJD3 was upregulated in 41 GC patient cohort as determined by realtime PCR. (D) High JMJD3 expression predicts poor patient survival in the 128 GC patient cohort. (E) High JMJD3 expression predicts poor patient survival in TCGA dataset. (F) JMJD3 expression escalated with tumor stage as analyzed by cBioportal using TCGA data. (G) Disease free patient had lower level of JMJD3 compared with recurred/progressed patient as analyzed by cBioportal. (H) Living patient had lower level of JMJD3 compared with deceased patient as analyzed by cBioportal. *p < 0.05, **p < 0.01 and ***p < 0.001.
Table 1.
Univariate and multivariate analysis of overall survival in GC patients (n = 128).
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | RR (95% CI) | P value | RR (95% CI) | P value |
| Age, y | 1.03 (1.00–1.05) | 0.021 | 1.05 (1.02–1.0) | 0.001 |
| Sex | ||||
| M | 1.55 (0.84–2.85) | 0.158 | 1.48 (0.77–2.84) | 0.236 |
| F | 1 | 1 | ||
| H. pylori infection | ||||
| Negative | 1.45 (0.82–2.56) | 0.201 | ||
| Positive | 1 | |||
| Lauren type | ||||
| Intestinal | 0.54 (0.22–1.35) | 0.188 | ||
| Diffuse | 1.33 (0.55–3.21) | 0.532 | ||
| Mix | 1 | |||
| Stage | ||||
| I | 0.13 (0.06–0.29) | <0.001 | 0.11 (0.03–0.36) | <0.001 |
| II | 0.17 (0.05–0.55) | 0.003 | 0.14 (0.04–0.50) | 0.002 |
| III | 0.39 (0.20–0.75) | 0.005 | 0.32 (0.16–0.63) | 0.001 |
| IV | 1 | 1 | ||
| Lymph node metastasis | ||||
| No | 0.24 (0.11–0.51) | <0.001 | 0.99 (0.30–3.22) | 0.982 |
| Yes | 1 | 1 | ||
| JMJD3 | ||||
| Low | 0.27 (0.14–0.50) | <0.001 | 0.36 (0.18–0.70) | 0.003 |
| High | 1 | 1 | ||
The underlying mechanism of JMJD3 dysregulation in GC
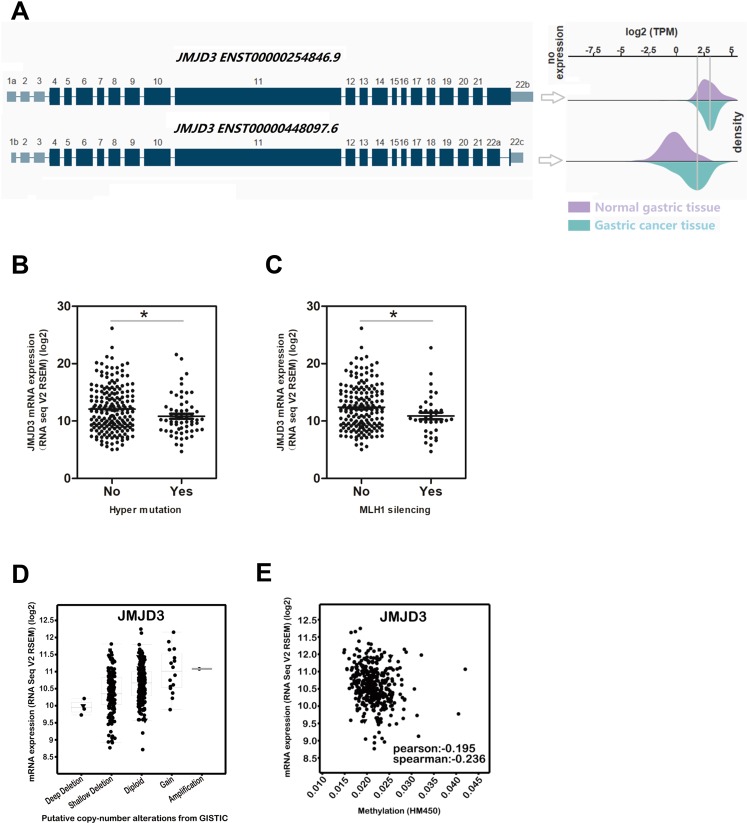
There are two transcript variants of JMJD3 from NCBI (Fig. 2A). Our result showed that the average expression level of both variants were higher in GC than in normal gastric tissue (Fig. 2A). Research evidence has shown that a proportion of colorectal cancer (CRC) are hypermutated, exhibit frequent epigenetic silencing of the MLH1 and have a better clinical prognosis12. Our result demonstrated that hyper mutation (Fig. 2B) and MLH1 silencing (Fig. 2C) were associated with lower JMJD3 expression (p < 0.05), supporting the oncogenic role of JMJD3. Genomic alteration analysis revealed that there were deep deletion, shallow deletion, diploid, copy number gain and amplification (Fig. 2D). JMJD3 mRNA expression was also negatively correlated with DNA methylation, although the correlation was weak (Fig. 2E).
Figure 2.
Deregulation of JMJD3 and its underlying mechanism by bioinformatics study. (A) The two transcript variants of JMJD3 were both upregulated in GC as analyzed by The Human Protein Atlas. (B) Patients with hypermutation had significantly lower level of JMJD3 expression as analyzed by cBioportal using TCGA data. (C) Patients with MLH1 silencing had significantly lower level of JMJD3 expression as analyzed by cBioportal using TCGA data. (D) Deletion, diploid, copy number gain and amplification were involved in the deregulation of JMJD3 expression as analyzed by cBioportal using TCGA data. (E) DNA methylation was also involved in JMJD3 deregulation stage as analyzed by cBioportal using TCGA data.*p < 0.05.
JMJD3 mutation and its influence on cell proliferation
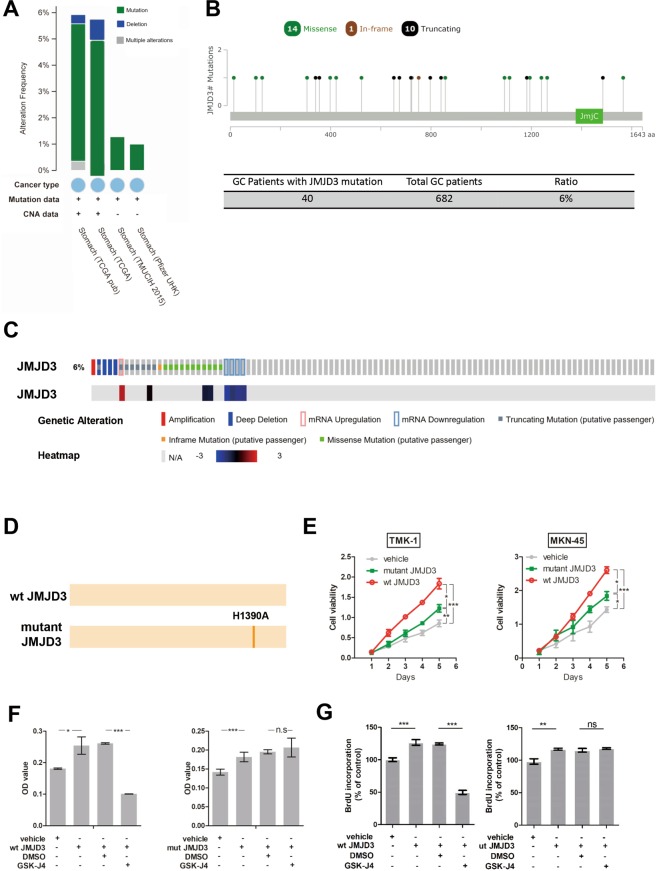
We analyzed JMJD3 genetic aberration and found mutation accounts for the majority of genetic alteration of JMJD3 (Fig. 3A). Further analysis revealed that in a cohort of 682 GC patients, approximately 6% patients have JMJD3 mutation including missense mutation, in-frame mutation and truncating mutation. Mutation distributed in areas other than the JmjC domain (Fig. 3B), which is a catalytic domain required for the demethylase activity of JMJD3. Representative proportion for each type of genetic alteration including amplification, deletion, mRNA upregulation and downregulation and mutation in JMJD3 was shown in Fig. 3C. JMJD3 expression level was changed accompanying these alterations (Fig. 3C). To confirm the function of JMJD3 mutation we generated two pMSCV plasmids expressing wildtype JMJD3 (wt JMJD3) or mutant JMJD3 which has lost the histone demethylase function (Fig. 3D). MTT result revealed that overexpression of wildtype and mutant JMJD3 significantly promoted cell viability in two GC cell lines. However, the promoting effect for mutant JMJD3 was significantly weaker than that of wildtype JMJD3 (Fig. 3E), suggesting that there was other mechanism involved in the oncogenic function of JMJD3 other than its histone demethylation function. To further confirm the result, we used a JMJD3 histone demethylation inhibitor GSK-J4 for MTT, BrdU and transwell experiments. Result showed that the proliferation promoting effect of wildtype JMJD3 could be partially reversed by GSK-J4 while GSK-J4 could not reverse the promoting effect of mutant JMJD3 (Fig. 3F,G). Similar result was found for cell migration (Supplementary Fig. 1). These results suggest that both demethylase-dependent and demethylase-independent mechanisms were involved in the oncogenic function of JMJD3.
Figure 3.
Mutation accounts for the majority of genetic aberration in JMJD3. (A) The percentage of JMJD3 genetic alteration in different GC patient cohorts analyzed by cBioportal. (B) The position of JMJD3 mutation in the four patient cohorts combined together cohorts analyzed by cBioportal. (C) Representative percentage of different genetic alteration in JMJD3 and the accompanying expression change cohorts analyzed by cBioportal. (D) Representative diagram of the two pMSCV plasmids established for functional study. (E) MTT result after transfection of wildtype or mutant JMJD3 plasmids in two GC cell lines. (F) MTT result after transfection of JMJD3 plasmids with or without GSK-J4. (G) BrdU result after transfection of JMJD3 plasmids with or without GSK-J4. *p < 0.05, **p < 0.01 and ***p < 0.001.
Potential targets of JMJD3 in GC
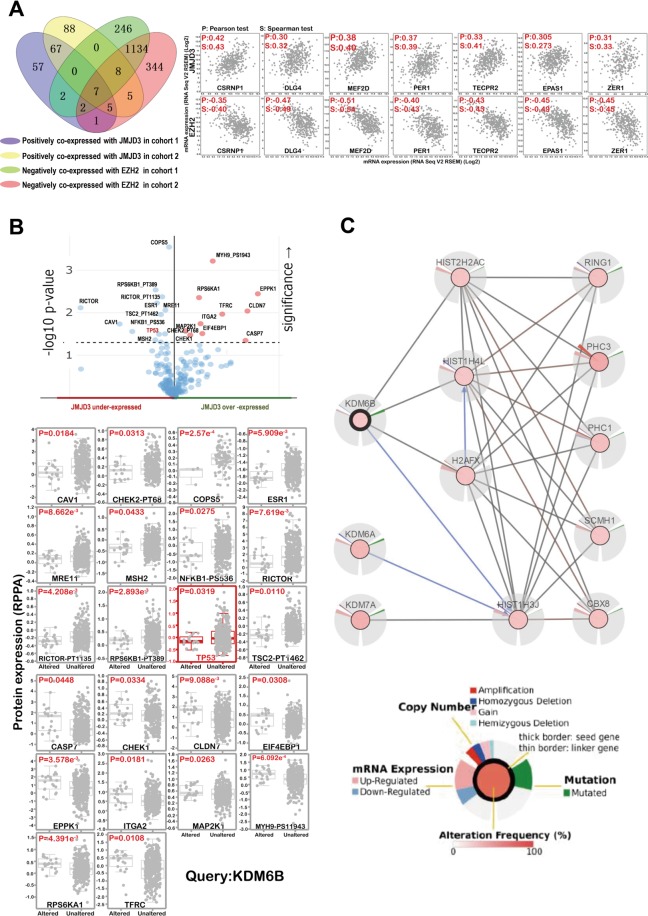
To identify proteins that are potentially regulated by JMJD3, we performed different analysis. H3K27me3 levels are determined by the balance between activities of histone methyltransferase EZH2 and histone demethylase JMJD39. We analyzed proteins that are positively co-expressed with JMJD3 (2 cohorts) and at the same time negatively co-expressed with EZH2 (2 cohorts) and found 7 potential targets (Fig. 4A). When validating these potential targets in patients samples, we found that three targets named CSRNP1, MEF2D and EPAS1 are significantly up-regulated in tumor tissues when compared with normal tissues (Supplementary Fig. 1). This is consistent with the previous result that they are positively co-expressed with JMJD3 which is overexpressed in GC. We also analyzed proteins that are significantly upregulated accompanying JMJD3 overexpression or downregulated accompanying JMJD3 underexpression. Twenty two potential targets were discovered for JMJD3 by this means including TP53, which has already been reported to be JMJD3 target13 (Fig. 4B). Furthermore, we investigated interacting proteins for JMJD3 using cBioportal network analysis. We identified 4 proteins that are directly regulated by JMJD3 and 5 proteins that are indirectly affected by JMJD3 (Fig. 4C).
Figure 4.
The downstream targets of JMJD3 by bioinformatics study. (A) Genes that were positively co-expressed with JMJD3 and at the same time negatively co-expressed with EZH2 were analyzed by Venny using TCGA data. (B) Genes that were significantly influenced by JMJD3 expression alteration were analyzed by cBioportal. (C) Net work analysis was performed in cBioportal and four direct and five indirect targets of JMJD3 were found.
Discussion
Gastric cancer (GC) is one of the leading causes of cancer deaths worldwide and the incidence and mortality rates are higher in Asian compared with Western countries2. In China, GC is the second most prevalent type of cancer, with 679,100 new cases and 498,000 deaths reported in 201514. Thus, investigating promising diagnostic and/or prognostic markers for GC is urgent and vitally important. Recently, JMJD3 has been highlighted in a variety of studies of human malignancies due to the increasing focus on histone methylation in cancer development15. To the best of our knowledge, this is the first study to determine the dysregulation of JMJD3 in GC and to evaluate the prognostic implications of overexpressed JMJD3 protein.
Result of IHC analysis of 128 gastric cancer tissues from four different histological stages indicated that JMJD3 protein expression is increasingly elevated with histological stage (Fig. 1A,B). Moreover, this study showed that JMJD3 transcripts were significantly increased in gastric cancer tissues from 41 cases when compared with paired adjacent normal tissues (Fig. 1C). We also found that high JMJD3 expression predicted unfavorable survival both in the 128 patients cohort (Fig. 1D) and in TCGA data (Fig. 1E). Multivariate analysis of the 128 patient cohort showed that JMJD3 was an independent prognosis predictor in GC (Table 1). Consistent with our result, bioinformatics analysis in cBioportal also revealed that JMJD3 expression increased along with tumor stage (Fig. 1F) and high expression of JMJD3 was associated with poor disease progression (Fig. 1G,H). The two JMJD3 transcript variants were also found to be upregulated in GC compared with normal tissue from The Human Protein Atlas (Fig. 2A). In accordance with our findings, Shen et al. observed that JMJD3 transcripts were significantly elevated in renal carcinoma and that JMJD3 expression was higher in cancer tissue compared to adjacent normal tissue8. JMJD3 expression was also found to be significantly higher in prostate cancer and further increased during metastasis9,10. Similarly, Sui et al. reported that JMJD3 expression was upregulated in glioma11. Conversely, Agger et al. found a significant decrease in JMJD3 expression in various cancers, including lung and liver carcinomas, as well as various hematopoietic malignancies6. Moreover, JMJD3 expression levels are lower in colorectal cancer relative to normal tissues7. Based on the above reports, we conclude that the expression patterns of JMJD3 are context and cancer type specific. For example, JMJD3 can promote breast cancer cell invasion through upregulation of the EMT-specific gene-SNAI116. However, in colorectal cancer cells, a decrease in JMJD3 can significantly increase cell proliferation through cell cycle progression and apoptosis suppression7.
Previous study has shown that a better clinical prognosis was found in some CRC patients in which there were hypermutation and epigenetic silencing of the MLH112. We analyzed JMJD3 expression, mutation status and MLH1 silencing in GC and our result demonstrated that hyper mutation (Fig. 2B) and MLH1 silencing (Fig. 2C) were associated with lower JMJD3 expression. Hyper mutation and MLH1 silencing were also found to be associated with longer overall survival in GC17,18. This is consistent with our finding that lower JMJD3 level predicts better survival in GC.
Our bioinformatics study also demonstrated that both genetic alteration and DNA methylation might participate in the dysregulation of JMJD3 in GC (Fig. 2D,E). Up to now, there has been few reports about mutation of JMJD3 in cancer. We further comprehensively investigated JMJD3 mutation in cBioportal (Fig. 3A–C). Results showed that in a total of 682 GC patients around 6% patients have JMJD3 mutation and the mutation was not in the catalytic domain. The functional impact of JMJD3 mutation (loss of demethylase function) was also studied by overexpressing mutant JMJD3 plasmids or using pharmacological inhibitor (Fig. 3D–G and Supplementary Fig. 1) and we found that JMJD3 promotes GC cell proliferation and migration by both demethylase-dependent and demethylase-independent mechanisms. The demethylase-independent function of JMJD3 has been reported but not in cancer19,20. The exact mechanism underlying the oncogenic function of JMJD3 needs further elucidation.
We also investigated the potential targets of JMJD3 by different bioinformatics methods. Totally 29 proteins were found significantly affected by JMJD3 and another 4 targets were found directly regulated by JMJD3 by network analysis (Fig. 4A–C). We validated the expression of potential targets that are positively co-expressed with JMJD3 and negatively co-expressed with EZH2 by realtime PCR. Results showed that CSRNP1, MEF2D and EPAS1 are significantly up-regulated in tumor tissues compared with normal tissues (Supplementary Fig. 2). Among them, the oncogenic role of MEF2D, EPAS1 has been well established21,22. MEF2D was upregulated in GC and was significantly associated with the clinical parameters. Its high expression predicts shorter overall survival23. EPAS1 also called hypoxia-inducible factor-2α (HIF-2α), has been reported to play an oncogenic role in various cancers including gastric cancer, colon cancer, lung cancer, pancreatic cancer, renal cancer, etc24–28. It is also a promising target for cancer therapy29,30. In contrary to our finding, CSRNP1 (AXUD1) has been reported to be decreased in cancers31 and play a pro-apoptotic role32. Besides, we also discovered twenty two potential targets that showed similar trend of expression change with JMJD3 in GC (Fig. 4B). The role of CAV1 in gastric cancer remains controversial33. The oncogenic role of COPS534, MRE1135, RICTOR36, CHEK137, CLDN738, EIF4EBP139, EPPK140, ITGA241, MAP2K142, RPS6KA143, TFRC44 have been reported in different cancer types. Specifically EIF4EBP1 were overexpressed in GC45 and MAP2K1 has been identified to be associated with GC by different studies46. Furthermore, high expression of MRE11 complex47, RICTOR48, ITGA249, MAP2K146 predicts poor survival in GC. ITGA2 has been reported to be a target of EZH250, suggesting it is very probably a JMJD3 target. Among the four genes that were found directly regulated by JMJD3 by network analysis (Fig. 4C), HIST2H2AC was reported increased in breast cancer51 and HIST1H4L was increased in prostate cancer52. Altogether, our findings demonstrated that it is highly possible that some of these potential targets are regulated by JMJD3 in GC. However, the direct interaction of these novel targets with JMJD3 awaits further validation.
In summary, our study report for the first time the oncogenic role of JMJD3 in GC and inhibition of JMJD3 might be an effective therapeutic approach for GC. Some inhibitors that suppress the histone demethylase activity of JMJD3 have been identified, such as JIB-04 and GSK-J1/4. They have been reported to exert tumor suppressive activity in several malignances, including breast cancer53, osteoblast cancer54, leukemia55, ovarian cancer stem cells56 and pediatric brainstem glioma57. These inhibitors may have therapeutic potential for GC via targeting JMJD3. Although the functional role of JMJD3 in GC is unclear, it is an emerging therapeutic epigenetic target for cancer treatment and might be associated with the progression of GC and serves as a prognostic indicator.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No.81503093, 81473269, 81503093, 81602166, 81672444 and 81602240),the Joint Funds of the Southwest Medical University & Luzhou (2016LZXNYD-T01, 2017LZXNYD-Z05 and 2017LZXNYD-J09), the Natural Science Foundation of Jiangsu Province (BK20140286), the University Natural Science Research Project of Anhui Province (KJ2016A732 and KJ2017A272) and the Talent Scientific Research Foundation of Yijishan Affiliated Hospital of Wannan Medical College (YR201504), the Basic Research Project of Knowledge Innovation Program in Shenzhen (JCYJ20150402152130696) .
Author Contributions
Z.Y.X., Y.X., Z.X. conducted the experiments, Y.J., L.N.L., Y. J. collected and processed the data, Q.Z., L.W., T.Y. prepared the figures, Y.Y., Y.Z. wrote manuscript, Q.W., B.Q., F.Z., J.S. managed the project, provided the funding and revised the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenyu Xu, Yabin Xia and Zhangang Xiao contributed equally.
Contributor Information
Bo Qin, Email: qinbozf@163.com.
Fan Zhang, Email: zhangfan401401@aliyun.com.
Jing Shen, Email: crystal_stray@126.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37340-w.
References
- 1.Mamta P, Puja S, Manisha S. Epigenetic modifications as prognostic marker in cancer. Res J Biotechnol. 2013;8:66–75. [Google Scholar]
- 2.Balakrishnan M, George R, Sharma A, Graham DY. Changing Trends in Stomach Cancer Throughout the World. Current gastroenterology reports. 2017;19:36. doi: 10.1007/s11894-017-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacology & therapeutics. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes & development. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokunaga R, et al. The Prognostic Significance of Histone Lysine Demethylase JMJD3/KDM6B in Colorectal Cancer. Ann Surg Oncol. 2016;23:678–685. doi: 10.1245/s10434-015-4879-3. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, et al. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. Bmc Cancer. 2012;12:470. doi: 10.1186/1471-2407-12-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daures M, et al. The JMJD3 Histone Demethylase and the EZH2 Histone Methyltransferase in Prostate Cancer. Omics. 2016;20:123–125. doi: 10.1089/omi.2015.0113. [DOI] [PubMed] [Google Scholar]
- 10.Xiang Y, et al. JMJD3 is a histone H3K27 demethylase. Cell research. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 11.Sui A, et al. The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget. 2017;8:68591–68598. doi: 10.18632/oncotarget.19793.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donehower LA, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. The Journal of pathology. 2013;229:99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ene CI, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PloS one. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 15.Burchfield JS, Li QT, Wang HY, Wang RF. JMJD3 as an epigenetic regulator in development and disease. Int J Biochem Cell B. 2015;67:148–157. doi: 10.1016/j.biocel.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigeyasu K, et al. Clinical Significance of MLH1 Methylation and CpG Island Methylator Phenotype as Prognostic Markers in Patients with Gastric Cancer. Plos One. 2015;10:e0130409. doi: 10.1371/journal.pone.0130409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristescu R, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature medicine. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 19.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS genetics. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W, et al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su L, et al. MEF2D Transduces Microenvironment Stimuli to ZEB1 to Promote Epithelial-Mesenchymal Transition and Metastasis in Colorectal Cancer. Cancer research. 2016;76:5054–5067. doi: 10.1158/0008-5472.CAN-16-0246. [DOI] [PubMed] [Google Scholar]
- 22.Ma L, et al. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer research. 2014;74:1452–1462. doi: 10.1158/0008-5472.CAN-13-2171. [DOI] [PubMed] [Google Scholar]
- 23.Xu K, Zhao YC. MEF2D/Wnt/beta-catenin pathway regulates the proliferation of gastric cancer cells and is regulated by microRNA-19. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:9059–9069. doi: 10.1007/s13277-015-4766-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Hypoxia-inducible factor-2alpha promotes tumor progression and has crosstalk with Wnt/beta-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16:119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Zhang H, Xue X, Shah YM. Hypoxia-inducible factor 2alpha (HIF-2alpha) promotes colon cancer growth by potentiating Yes-associated protein 1 (YAP1) activity. J Biol Chem. 2017;292:17046–17056. doi: 10.1074/jbc.M117.805655.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putra AC, et al. The A Allele at rs13419896 of EPAS1 Is Associated with Enhanced Expression and Poor Prognosis for Non-Small Cell Lung Cancer. Plos One. 2015;10:e0134496. doi: 10.1371/journal.pone.0134496.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong WW, Tong GH, Kong H, Liu Y. The tumor promoting roles of HSP60 and HIF2alpha in gastric cancer cells. Tumour Biol. 2016;37:9849–9854. doi: 10.1007/s13277-015-4783-2. [DOI] [PubMed] [Google Scholar]
- 28.Schodel J, et al. Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer. Eur Urol. 2016;69:646–657. doi: 10.1016/j.eururo.2015.08.007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoning JP, Monteiro M, Gu W. Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1alpha and HIF2alpha. Clin Exp Pharmacol Physiol. 2017;44:153–161. doi: 10.1111/1440-1681.12693. [DOI] [PubMed] [Google Scholar]
- 30.Pan X, et al. PLGA/poloxamer nanoparticles loaded with EPAS1 siRNA for the treatment of pancreatic cancer in vitro and in vivo. Int J Mol Med. 2015;35:995–1002. doi: 10.3892/ijmm.2015.2096. [DOI] [PubMed] [Google Scholar]
- 31.Ishiguro H, et al. Identification of AXUD1, a novel human gene induced by AXIN1 and its reduced expression in human carcinomas of the lung, liver, colon and kidney. Oncogene. 2001;20:5062–5066. doi: 10.1038/sj.onc.1204603. [DOI] [PubMed] [Google Scholar]
- 32.Glavic A, Molnar C, Cotoras D, de Celis JF. Drosophila Axud1 is involved in the control of proliferation and displays pro-apoptotic activity. Mech Dev. 2009;126:184–197. doi: 10.1016/j.mod.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Fu P, et al. The different functions and clinical significances of caveolin-1 in human adenocarcinoma and squamous cell carcinoma. OncoTargets and therapy. 2017;10:819–835. doi: 10.2147/OTT.S123912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou J, et al. Increased Jab1/COPS5 is associated with therapeutic response and adverse outcome in lung cancer and breast cancer patients. Oncotarget. 2017;8:97504–97515. doi: 10.18632/oncotarget.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho V, et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PloS one. 2016;11:e0167675. doi: 10.1371/journal.pone.0167675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuhua W, et al. Autophagy-related genes Raptor, Rictor, and Beclin1 expression and relationship with multidrug resistance in colorectal carcinoma. Human pathology. 2015;46:1752–1759. doi: 10.1016/j.humpath.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 37.Sankunny M, et al. Targeted inhibition of ATR or CHEK1 reverses radioresistance in oral squamous cell carcinoma cells with distal chromosome arm 11q loss. Genes, chromosomes & cancer. 2014;53:129–143. doi: 10.1002/gcc.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahiya N, Becker KG, Wood WH, III., Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PloS one. 2011;6:e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M, Kim EJ, Jeon MJ. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget. 2016;7:8726–8742. doi: 10.18632/oncotarget.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kudo I, Esumi M, Kusumi Y, Furusaka T, Oshima T. Particular gene upregulation and p53 heterogeneous expression in TP53-mutated maxillary carcinoma. Oncology letters. 2017;14:4633–4640. doi: 10.3892/ol.2017.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nones K, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. International journal of cancer. 2014;135:1110–1118. doi: 10.1002/ijc.28765. [DOI] [PubMed] [Google Scholar]
- 42.Waterfall JJ, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nature genetics. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salhi A, et al. RSK1 activation promotes invasion in nodular melanoma. The American journal of pathology. 2015;185:704–716. doi: 10.1016/j.ajpath.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wada S, Noguchi T, Takeno S, Kawahara K. PIK3CA and TFRC located in 3q are new prognostic factors in esophageal squamous cell carcinoma. Annals of surgical oncology. 2006;13:961–966. doi: 10.1245/ASO.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Riquelme I, et al. The Gene Expression Status of the PI3K/AKT/mTOR Pathway in Gastric Cancer Tissues and Cell Lines. Pathology oncology research: POR. 2016;22:797–805. doi: 10.1007/s12253-016-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu ZY, Chen JS, Shu YQ. Gene expression profile towards the prediction of patient survival of gastric cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2010;64:133–139. doi: 10.1016/j.biopha.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Altan B, et al. High Expression of MRE11-RAD50-NBS1 Is Associated with Poor Prognosis and Chemoresistance in Gastric Cancer. Anticancer research. 2016;36:5237–5247. doi: 10.21873/anticanres.11094. [DOI] [PubMed] [Google Scholar]
- 48.Bian Y, et al. Elevated Rictor expression is associated with tumor progression and poor prognosis in patients with gastric cancer. Biochemical and biophysical research communications. 2015;464:534–540. doi: 10.1016/j.bbrc.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Dong J, et al. HMGA2-FOXL2 Axis Regulates Metastases and Epithelial-to-Mesenchymal Transition of Chemoresistant Gastric Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:3461–3473. doi: 10.1158/1078-0432.CCR-16-2180. [DOI] [PubMed] [Google Scholar]
- 50.Ferraro A, Boni T, Pintzas A. EZH2 regulates cofilin activity and colon cancer cell migration by targeting ITGA2 gene. PloS one. 2014;9:e115276. doi: 10.1371/journal.pone.0115276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monteiro FL, et al. The histone H2A isoform Hist2h2ac is a novel regulator of proliferation and epithelial-mesenchymal transition in mammary epithelial and in breast cancer cells. Cancer letters. 2017;396:42–52. doi: 10.1016/j.canlet.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Camoes MJ, et al. Potential downstream target genes of aberrant ETS transcription factors are differentially affected in Ewing’s sarcoma and prostate carcinoma. PloS one. 2012;7:e49819. doi: 10.1371/journal.pone.0049819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, et al. A small molecule modulates Jumonji histone demethylase activity anselectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D, Okamura H, Teramachi J, Haneji T. Histone demethylase Utx regulates differentiation and mineralization in osteoblasts. Journal of cellular biochemistry. 2015;116:2628–2636. doi: 10.1002/jcb.25210. [DOI] [PubMed] [Google Scholar]
- 55.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakaki H, et al. GSKJ4, A Selective Jumonji H3K27 Demethylase Inhibitor, Effectively Targets Ovarian Cancer Stem Cells. Anticancer Res. 2015;35:6607–6614. [PubMed] [Google Scholar]
- 57.Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nature medicine. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.