Abstract
In commercial research and development projects, public disclosure of new chemical compounds often takes place in patents. Only a small proportion of these compounds are published in journals, usually a few years after the patent. Patent authorities make available the patents but do not provide systematic continuous chemical annotations. Content databases such as Elsevier’s Reaxys provide such services mostly based on manual excerptions, which are time-consuming and costly. Automatic text-mining approaches help overcome some of the limitations of the manual process. Different text-mining approaches exist to extract chemical entities from patents. The majority of them have been developed using sub-sections of patent documents and focus on mentions of compounds. Less attention has been given to relevancy of a compound in a patent. Relevancy of a compound to a patent is based on the patent’s context. A relevant compound plays a major role within a patent. Identification of relevant compounds reduces the size of the extracted data and improves the usefulness of patent resources (e.g. supports identifying the main compounds). Annotators of databases like Reaxys only annotate relevant compounds. In this study, we design an automated system that extracts chemical entities from patents and classifies their relevance. The gold-standard set contained 18 789 chemical entity annotations. Of these, 10% were relevant compounds, 88% were irrelevant and 2% were equivocal. Our compound recognition system was based on proprietary tools. The performance (F-score) of the system on compound recognition was 84% on the development set and 86% on the test set. The relevancy classification system had an F-score of 86% on the development set and 82% on the test set. Our system can extract chemical compounds from patents and classify their relevance with high performance. This enables the extension of the Reaxys database by means of automation.
Background
The number of chemistry-related publications has massively increased in the past decade (1). These publications are mainly in the form of patent applications and scientific journal articles. A crucial step in the early stages of medicinal chemistry activities is the exploration of the chemical space covered by these sources (1–4). In commercial research and development projects, initial public disclosure of new chemical compounds often takes place in patent applications (4, 5). On average, it takes an additional 1 to 3 years for a small fraction of these chemical compounds to appear in journal publications (5). Therefore, a large selection of these chemical compounds is only available through patent documents (6). Additionally, chemical patent documents contain unique information such as reactions, experimental conditions, mode of action (7), bioactivity data and catalysts (1, 3). Analyzing such information becomes crucial (1, 4, 5, 8), as it allows the understanding of compound prior art, it provides a means for novelty checking and validation, and it points to starting points for chemical research in academia and industry (3, 7, 9, 10).
Patent data are freely available through different patent offices. Major patent authorities include the European Patent Office (EPO) (11), the United States Patent and Trademark Office (USPTO) (12) and the World Intellectual Property Organization (WIPO) (13). Depending on the patent authority, the data are made available in the form of XML, HTML, text PDF, Optical Character Recognition (OCR) PDF or image PDF. Patent documents usually follow a systematic structure consisting of title, bibliographic information [such as patent number, dates, inventors, assignees and International Patent Classification (IPC) classes], abstract, description and claims. Most of the chemical data are available in the experimental section of the description, while chemical compounds that are claimed (i.e. will become protected by the patent) are available in the claim section (4). Drawings, sequences or other additional information will normally be found at the very end of the patent.
While patent authorities make available the patent documents, they do not provide systematic continuous chemical annotations and full-text searching capabilities (3), so manual or automatic excerption processes have been considered (1, 5, 7, 14). Manual excerption processes result in high-quality content but are costly and time-consuming, and are therefore limited to commercial content providers (5). Examples of content databases are Elsevier Reaxys (15, 16), CAS SciFinder (17), and Thomson Reuters Pharma (18). These commercial resources provide high-quality content, such as compounds and their associated structures, facts associated to compounds, and reactions. Automatic approaches to extract information from patents have recently come into existence to overcome some of the aforementioned cost and time limitations. Examples of such resources include SureChEMBL (3), SCRIPDB (19), ChEBI database (20), IBM database (21), NextMove Software’s reaction database (22) and databases that combine data from different sources [e.g. PubChem (23)]. SureChEMBL provides continuous, up-to-date chemical annotations with structures derived from USPTO, EPO, WIPO and the Japanese Patent Office (JPO) (24). The information is extracted from full-text patents (except JPO), images and attachment files (3). This information is mostly derived by text mining and image mining. SCRIPDB is a chemical structure database from compounds and reactions. This information is built based on the digital chemical structure files provided by USPTO for a subset of its patents (grant patents, from 2001 until 2011) (19). The ChEBI database provides chemistry compounds and structures extracted from a subset of patent documents from the EPO office (20). The IBM database provides chemical compounds and structures derived from a subset of EPO, WIPO and USPTO patents (21). This information is derived by text-mining approaches. The reaction database of NextMove Software is also automatically generated by text mining the relevant experimental sections of patents covering the period 1976–2013 (22). It proves difficult to maintain public databases and many of the above have become outdated.
Some of the automatic resources mentioned above incorporate the textual data content supplied by the content providers to build their database (such as SCRIPDB). Others use image mining and text mining approaches to extract data from the patent full-text document (e.g. SureChEMBL and IBM). Image-mining approaches convert images attached to patents into structures using image-to-structure tools [e.g. CLiDE Pro (25) in SureChEMBL] (4). These tools have limitations in the interpretation of individual drawing features (such as chemical bonds) found in the structure diagrams of some images (25) and will not further be considered in this study. Text-mining approaches focus on the recognition of chemical compounds in patents (4). Each recognized small compound should also be associated with a chemical structure. Different text-mining approaches exist to extract chemical entities from patents. The approaches can be categorized as dictionary based, morphology based (or grammar based) or statistical (26–29). Dictionary-based approaches use matching methods to identify compounds mentioned in a dictionary (e.g. generic drug names) within patents. This approach is limited by the compounds contained in the dictionary. Addition of all systematic compound identifiers to a dictionary is almost impossible as they are algorithmically generated based on the structure of a compound and a set of rules (30). Grammar-based approaches use these rules to overcome this limitation and provide functionality to recognize systematic identifiers (26). Statistical approaches use supervised machine-learning techniques to recognize chemical compounds. These statistical-based recognizers are trained on manually annotated chemical terms (7). Among the three approaches, statistical approaches have been shown to perform the best (4, 31, 32) but they require a large annotated corpus for training (26, 33) and cannot associate structures with compounds (4, 27, 34). Correctness of the associated chemical structure to a recognized compound is essential in the field of chemistry (34, 35). Often a combination of the methods above in the form of an ensemble system is used for chemical compound recognition (31, 36). All systems require a gold-standard corpus for training, developing and testing performance (30). Producing such a corpus is laborious and expensive (7). It involves development of well-defined annotation guidelines, selection and training of domain experts for annotation, selection of the data, annotation of the data by multiple annotators and finally harmonization of the annotations (7).
Extracting information from patents automatically is fast but has limitations (7, 29, 37). The majority of patent text-mining systems have been developed, trained and tested using the title and abstract of the patent documents. Therefore, their usage is not evaluated on full-text documents (31, 36). More importantly, automatic extraction is mostly focused on extraction of all chemical compounds mentioned. In manually excerpted databases, the focus is on relevant compounds (5, 38). A compound is relevant to a patent when it plays a major role within the patent application (e.g. starting material or a product in a reaction specified in the claim section). Relevant compounds are a small fraction of all the compounds mentioned within the patent document (9, 39). Automatic identification of the relevant compounds would greatly reduce the amount of extracted data from patents and can improve the usefulness of patent resources. Furthermore, these compounds can be used in predictive analyses to identify the key compounds within the patent (key compounds are the main compounds protected by the patent application and are usually well-hidden within the context) (9, 39). To our knowledge, automatic identification of relevant compounds within patents has not yet been investigated.
The objective of this study is to identify relevant chemical compounds in patents using an automatic approach. To develop and evaluate our approach, a patent corpus with named-entity and relevancy annotations was built.
Materials and methods
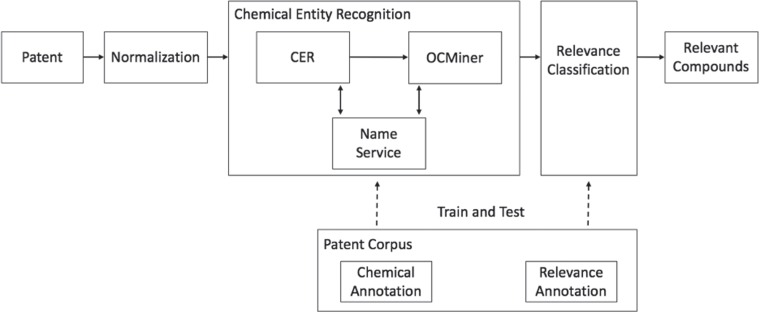
Figure 1 shows the relevancy classification workflow. The chemical patents are pulled through patent offices. The patent source documents are first normalized into a unified format. They are then fed into the chemical entity recognition system that consists of two different named-entity extraction systems, Chemical Entity Recognizer (CER; Elsevier, Frankfurt, Germany) (40) and OCMiner (OntoChem, Halle, Germany) (41). CER extracts chemical entities and tags them in the normalized input document. OCMiner further enriches the output of CER by extracting additional chemical entities and assigning confidence scores to all extracted entities of both systems. The associated structures of chemical compounds extracted by CER or OCMiner are generated, validated and standardized using the Reaxys Name Service (42). The chemical annotations in the patent corpus are used to train and test the chemical entity recognition system. The relevancy annotations in the corpus are used to train and test the relevancy classifier, which labels the chemical entities extracted by the chemical entity recognition system as relevant or irrelevant. Below we describe each of the components in more detail.
Figure 1.
Workflow of the relevancy classification.
Normalization
The variety of input sources and file types needs to be normalized into a unified text representation (4). The normalization step is performed by converting all input files (e.g. XML, HTML and PDF) into a unified XML representation format. Predefined XML tags corresponding to heuristic information such as document sections (title, abstract, claims, description and metadata) are used within this unified representation. The normalization also converts all character encodings into UTF-8 (8-bit Unicode Transformation Format).
During normalization, we store a one-to-one mapping between each character in the original text and the corresponding character in the normalized document. This provides us with the possibility to go back to the original document from the normalized text and vice versa. It also minimizes the efforts to update the annotations in the patent corpus in case of changes in normalization methodology (note that the documents in the corpus have also been normalized).
Patent corpus development
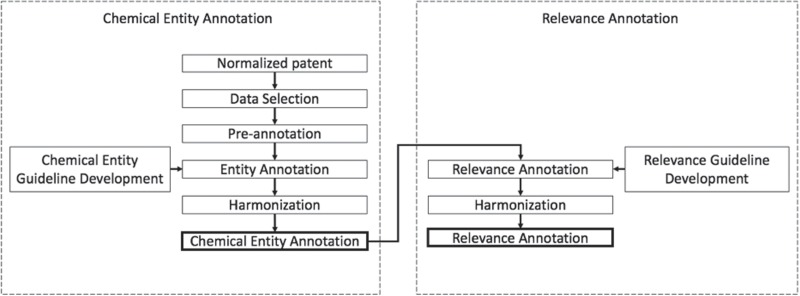
The development of the chemical patent corpus with chemical entity and relevancy annotations was done in two phases. Figure 2 illustrates the corpus creation process. The first phase focuses on building a corpus with chemical entity annotations. In phase 2 the corpus obtained from phase 1 is used to assign relevancy annotations to the entities annotated in phase 1. In this phase, annotators also flagged any compounds with spelling mistakes. For each phase, a set of well-defined guidelines was developed that helped achieve annotation consistency.
Figure 2.
Patent corpus development.
Chemical entity annotation guideline
The chemical entity annotation guideline was developed based on our previous patent corpus development guideline (7), previous work by other scholars (32, 43–46), and the help of subject matter experts in Elsevier. The guidelines define the entities to be annotated. For each entity, positive and negative examples were provided. Additionally, any exception was defined and illustrated through examples. The guideline also defined how the annotation should be performed within the brat rapid annotation tool (47, 48). Brat allows online annotation of text using pre-defined entity types. Annotators were asked to annotate chemical compounds (e.g. tetrahydrofuran), chemical classes (e.g. zirconium alkoxide) and suffixes or prefixes of these compounds (e.g. `stabilized’ as prefix in `stabilized zirconia’ and `nanoparticles’ as suffix in `silver nanoparticles’).
Chemical compounds could be annotated in three categories: mono-component compound (pure chemical compounds, e.g. systematic identifiers, trivial names, elements and chemical formulas), compound mixture part (e.g. `Magnesiaflux’, which scientifically is a mixture of 30% MgF2 and 70% MgO) or prophetic compound (specific compounds that are uncharacterized within the text and are mentioned in claims or descriptions only for intellectual property protection).
Compound classes could be annotated in six categories: chemical class (natural products or substructure names, e.g. heterocycle), biomolecules (e.g. insulin), polymers (e.g. polyethylene), mixture classes (e.g. opium), mixture part classes (e.g. quinupristin) or Markush (textual description of a Markush formula, e.g. HaXbC-C-H).
Relevancy annotation guideline
For the relevancy annotation, a new set of guidelines were developed, which defined how relevant compounds should be identified. The legal status of a compound (e.g. prophetic or claimed) and its characterization (e.g. NMR or MS measurement), properties (e.g. superconductivity), effects (e.g. toxicity) and transformation (e.g. reaction) were taken into consideration for defining the guidelines. The relevancy annotation did not include suffixes and prefixes of compounds. In brief, relevancy is assigned as follows:
Prophetic compounds and Markush classes are relevant.
Compound mixture parts, mixture part classes, mixture classes, polymers and biomolecules are irrelevant.
Mono-component compounds and chemical- classes are assigned relevance based on the context of the full patent text. They are considered relevant to the patent if (a) the entity is present in the title or abstract section of the patent, (b) the entity is part of a reaction context (e.g. product, intermediate product, catalyst or starting material used in synthetic procedures) or (c) the entity or its measured property belongs to the invention in the claim section and is connected to the core invention of the patent. The mono-component compounds and chemical classes are irrelevant if (a) the entity is only introduced for further explanation and is described beyond the invention, (b) the entity is described for reference or comparison or (c) the entity is involved in a chemical reaction but not a starting material, product or catalyst.
Data selection
Patent documents are long and extensive. Annotation of full-text documents is time-consuming and expensive. Complexity was reduced by selecting snippets of patent text from a large set of patent documents that represented the diversity of the data. We downloaded all EPO patents with IPC class A or C (corresponding to chemistry) from a 3-month period in 2016 (15, 16). This yielded 19 274 patents, which were divided into snippets as follows. First, each patent was divided into six snippets containing title, abstract, claims, description, metadata and non-English section of the patent. Second, since the performance of the brat toolkit drops on long files (7), snippets of more than 50 paragraphs were further divided into multiple snippets. From this set of snippets, a small set was selected for annotation. We performed random stratified sampling based on the sub-classes of IPC A and C (list available at https://www.wipo.int/classifications/ipc/en/). In addition, the following conditions were satisfied: 10% of the snippets were from titles, 10% from abstracts, 40% from claims and 40% from descriptions, and all snippets were from different patents.
We selected a total of 131 snippets, which constitute our patent corpus. The IPC sub-classes that occurred most frequently were A61K, A61B, C07D, A61F, A61M and C12N.
Chemical entity annotation process
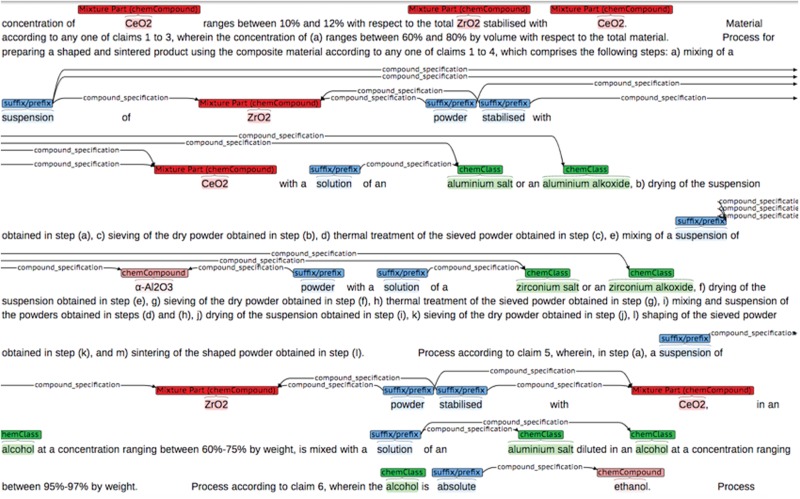
We selected 10 chemistry graduates as annotators. The annotators were located in different European countries. To train the annotators, 11 of the 131 patent snippets were distributed among the annotators using the brat annotation tool (47, 48). The snippets were pre-annotated with an untuned version of the chemical entity recognition software that is used in this study (only for categories mono-component compound and chemical class, see next section for the description of this software). The pre-annotations were displayed in brat, and annotators were asked to modify incorrect pre-annotated entities (wrong boundary or entity type) and add missing entities according to the guideline (see Figure 3).
Figure 3.
Annotations in a patent snippet with the brat annotation tool.
The 11 snippets were also annotated by two Elsevier subject-matter experts (SMEs) who defined the guidelines. The SMEs had PhDs in chemistry and ~15 years of professional experience in the field. Any discrepancies between the annotations of the two SMEs were resolved in consensus discussions. The resulting annotations (the training corpus) were used as a reference and compared to the annotations of each of the other annotators by inter-annotator agreement (IAA) scores. We used the F-score (harmonic mean of recall and precision) as a measure of IAA, similar to other studies (7, 43, 46). Several review sessions were held to compare annotations and resolve inconsistencies, and the annotation guideline was updated for clarity if needed. For each annotator, training continued until the IAA between the annotator and the SMEs was at least 85%.
After successful completion of the training, the remaining 120 snippets of the corpus were distributed between the annotators. Each snippet was annotated by three annotators, after which the annotations were harmonized. The harmonization was done for each entity as follows: if at least two annotators agreed on the entity boundaries and the entity type, that annotation was added to the gold-standard set, otherwise an SME adjudicated the disagreement.
Relevancy annotation process
The same training corpus of 11 snippets was also annotated for relevant compounds by the annotators and the SMEs. They were provided with the reference annotations of the chemical entities and had to indicate whether the annotations were relevant or not. For every snippet, we also delivered the corresponding full patent text to the annotators and the SMEs. This allowed them to determine relevance based on the complete document, which included title, abstract, description and claims. The relevancy annotations of the annotators and SMEs were compared, and questions were resolved.
After training, the 120 snippets of the chemical entity corpus created in the previous step were distributed between the annotators. Each snippet was annotated by five annotators. If more than three annotators annotated the chemical entity as relevant it was considered relevant. If three annotators annotated the chemical entity as relevant it was considered equivocal. If less than three annotators annotated the chemical entity as relevant, it was considered irrelevant. The equivocal category was introduced since relevance determination is sometimes complex and judged differently by different experts (as relevance is decided based on the full text). To capture this complexity, we did not try to resolve ambiguity by enforcing a decision by the SMEs. As per the guidelines, relevance is document based. As a result, if a compound is considered relevant at one occurrence in the snippet, it is marked automatically relevant at any other occurrence. Finally, the annotators were also asked to annotate any spelling errors. This annotation can be helpful for improvement of chemical entity recognition systems. As spelling errors can be hard to detect, we decided to accept each spelling-error annotation, irrespective of the number of annotators that made that annotation. The corpus was divided into a development and test set consisting of 50 and 70 snippets, respectively.
Chemical entity recognition
We focused on non-statistical approaches for chemical entity recognition as we wanted to associate a chemical structure to extracted chemical compounds. A dictionary-based approach was used in combination with a morphology-based approach to identify chemical entities. The structures of these compounds were produced, validated and standardized using Reaxys Name Service (42). Since the gold-standard annotations showed that only a small set of relevant entities are from compound class categories (see results), we decided to reduce our chemical entity recognition scope to the identification and classification of chemical compounds.
Name service
The Reaxys system uses a name-to-structure toolkit [Reaxys Name Service (42)] and a set of standardization rules (e.g. eliminate hydrogen bonds when constructing structures) when new compounds are inserted into the database. In this study, the Name Service was used to convert names to structures and standardize those structures as well as the structures in different dictionaries based on the Reaxys standardization rules, and to validate the structures assigned to chemical compounds.
Chemical entity recognizers
An ensemble system was used for chemical entity recognition. First, we used Elsevier’s CER software (40). CER identifies and tags chemical compounds and their physical properties (e.g. color, melting point and boiling point) within a text document and converts extracted compounds into a chemical structure (using Name Service). In addition, CER also identifies chemical reactions and chemical properties within the patent. The software uses a combination of dictionary-based and morphology-based approaches to extract chemical compounds from patents. CER was loaded with a dictionary derived from the manually curated compounds in the Reaxys database. Similar to previous studies (27, 28), an exclusion list was used to filter out any noise (e.g. frequent compounds such as oxygen) from the extracted compounds. The morphology-based approach in CER identifies different elements within a compound and combines them to create the final compound only if it can validate the compound based on its structural chemistry (e.g. can two elements bind with each other in this manner). This validation is done on the structural level and through a set of pre-defined rules processed by the Name Service. CER cannot assign the extracted compounds to the different compound groups that are defined in the guidelines.
Second, we used and improved OCMiner (41) to identify chemical entities. OCMiner also uses a dictionary-based approach along with a morphology-based approach to extract chemical compounds. The dictionary used for OCMiner was generated from a compound database built from various publicly available sources such as PubChem (23), DrugBank (49), ChEMBL (50) and ChEBI (20), among others (41). To improve the quality of the dictionary, frequent chemical identifiers that were associated to more than one structure were manually resolved and the name-to-structure mappings of the most-frequent identifiers were manually validated. OCMiner also used other resolution mechanisms to improve the quality of the dictionary [e.g. counting the number of stereocenters (41)]. The Name Service was used to standardize the compounds within these dictionaries based on the same standardization rules applied by CER and Reaxys. In comparison to CER, OCMiner has additional functionality, such as abbreviation expansion and spelling-error correction (41). The software also has post-dictionary modules to identify systematic names. In a separate module built for this study, OCMiner cleans up the chemical entities identified by both CER and OCMiner (e.g. overlapping annotations and combination of simple annotations to complex entities) and assigns compounds to the different compound groups. Finally, OCMiner generates a confidence score for all recognized chemical entities extracted by CER or OCMiner.
Relevancy classification
Relevance of a chemical compound is defined based on the context of the full patent. To identify the relevance of a specific entity, the complete patent should be analyzed for that entity. We therefore gathered statistical information for each unique entity (recognized in the snippet) from the whole patent text and used that information to classify the extracted entity. Relevancy classification was expressed as a scalar relevance score that after normalization can vary between zero (irrelevant) and one (relevant). We divided the corpus into a training set and a test set to experimentally find the best threshold for relevancy classification. The training set was used along with the relevance score to define the best cut-off point for the relevancy classification. The results were then tested on the test set.
Relevance score
Several features derived from the full text are used to calculate the relevance score. The relevancy score is a linear combination of these features, where the coefficients (or weights) are heuristically determined.
These features include the following:
Compound frequency: Frequency of the compound within the document. Usually compounds that occur frequently in a patent document are less relevant (due to the nature of patents), unless the compound is unique to the patent.
Compound section: Occurrence of the compound within specific sections of a patent (e.g. title and claim). A compound in a claim section is more relevant than a compound in a description section of a patent. If a compound appears in multiple sections we prioritize it in the following order: Title, Abstract, Claim and Description.
Compound length: Length of the extracted term. We have noticed that longer names are more likely to be International Union of Pure and Applied Chemistry (IUPAC) names and hence have a higher chance of being relevant.
Surrounding characters: Occurrence of the compound within special characters (e.g. `[’, `(’). Examples are usually mentioned between special characters and they will be less relevant.
Compound section uniqueness: Compound single occurrence within a section of the patent. If a compound is mentioned once in the claims and a few times in the description it has higher probability to be relevant than the other way around.
Compound without solvent: If the compound does not contain solvents or laboratory chemicals, there is a higher probability of the compound being relevant.
Compound wide usage: Presence of the compound in one of a number of predefined groups representing the frequency of compounds in a large set of chemistry patents. To create the groups, all chemical entities from a large set of patent documents (selection of chemical patents in 2015, excluding patents from the patent corpus) were extracted using OCMiner and ranked according to their frequency of occurrence. The resultant compound list was divided in 16 equally-sized groups (16 an arbitrary number). Note here that we are extending our calculation to data derived from a larger set of patents. If a compound is frequently mentioned in other patents, then there is a lower probability of it being relevant.
Performance evaluation
The performance of the system against the gold-standard annotations was evaluated using recall, precision and F-score, given the number of true positives (TP), false positives (FP) and false negatives (FN). For the entity recognition task, TP represents the total number of correctly identified chemical entities by the system (based on starting and ending position of the entity in text), FP represents the number of entities wrongly identified by the system and FN represents the number of entities that are missed by the system. Recall, precision and F-score metrics are calculated as follows: recall = TP/(TP + FN), precision = TP/(TP + FP) and F-score = 2*precision*recall/(precision + recall).
For the relevancy classification task, TP, FP and FN are determined at the document level and only take into account the unique entities identified in each of the documents. TP represents the number of compounds correctly classified as relevant, FP represents the number of compounds wrongly classified as relevant by the system and FN represents the number of relevant compounds missed by the system. The compounds in the corpus that were annotated as equivocal were disregarded from relevancy calculation. This pragmatic choice was made for those compounds where evidently human annotators could not agree on their relevance.
Results
Chemical entity annotation
The average IAA between the annotators on the 11 training documents initially was 72% and reached 92% after two rounds of training. On the gold-standard set of 120 snippets, the average IAA between the annotators and the harmonized annotations was 87%. This was higher than the IAA between pre-annotation and the gold-standard (77% for mono-component compound and 23% for chemical class) indicating that annotators considerably changed the pre-annotations. Table 1 provides the frequency of entities within the corpus. Overall, 18 789 chemical entities were annotated, of which 15 199 were chemical compounds and 3 590 were chemical classes. This resulted in an average of around 150 annotations per snippet. The majority of the annotations consisted of mono-component compounds (13 564). In addition, the corpus contains 1848 relationships from chemical compound or classes to 628 suffix or prefixes annotations (a suffix or prefix can have a relationship with one or more chemical compounds or classes).
Table 1.
Number of annotations in the gold-standard set
| Annotation type | Annotation subtype | Annotation | Relevant | Equivocal | Irrelevant |
|---|---|---|---|---|---|
| Compounds | Mono Component | 13 564 | 883 | 362 | 12 319 |
| Mixture part | 1010 | 0 | 0 | 1010 | |
| Prophetic | 625 | 625 | 0 | 0 | |
| Classes | Chemical class | 1848 | 249 | 30 | 1569 |
| Biomolecule | 1039 | 0 | 0 | 1039 | |
| Markush | 17 | 17 | 0 | 0 | |
| Mixture | 286 | 0 | 0 | 286 | |
| Mixture part | 174 | 0 | 0 | 174 | |
| Polymer | 226 | 0 | 0 | 226 | |
| Total chemical entities | 18 789 | 1774 | 392 | 16 623 | |
| Other | Suffix and prefix | 628 | — | — | — |
| Relation | 1848 | — | — | — | |
Relevancy annotation
All 18 789 chemical entities were annotated for relevance (Table 1). Of the 15 199 compounds, 1508 (9.9%) were considered relevant and 362 (2.4%) were equivocal. Of the 3590 chemical classes, 266 (7.4%) were relevant, while 30 (0.8%) were equivocal. Thus, the majority of entities were considered irrelevant (87.7% of the compounds and 91.8% of the classes).
Chemical entity recognition
The performance of the ensemble system on compound recognition is shown in Table 2 for different thresholds of the confidence score. On the development set, a threshold of 0.2 yielded the best F-score of 83.7% (precision, 89.1%, and recall, 78.9%). For this threshold, the best result was also obtained on the test set (F-score, 86.2%; precision, 90.1%; and recall, 82.3%). Error analysis of the results indicated that the performance of the system may further be improved by better recognizing prophetic compounds, reactants and products of synthesis procedures.
Table 2.
Performance of the ensemble system on compound recognition for different confidence score thresholds
| Confidence score threshold | Development | Test | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| 0.0 | 88.5 | 79.3 | 83.6 | 86.5 | 82.3 | 84.3 |
| 0.1 | 88.6 | 79.1 | 83.6 | 89.1 | 82.3 | 85.6 |
| 0.2 | 89.1 | 78.9 | 83.7 | 90.1 | 82.3 | 86.2 |
| 0.3 | 89.1 | 78.6 | 83.5 | 90.1 | 81.6 | 85.7 |
| 0.4 | 89.1 | 78.4 | 83.4 | 90.1 | 81.5 | 85.6 |
| 0.5 | 89.1 | 78.4 | 83.4 | 90.1 | 81.5 | 85.6 |
| 0.6 | 89.1 | 78.4 | 83.4 | 90.1 | 81.3 | 85.5 |
| 0.7 | 87.2 | 60.6 | 71.5 | 90.7 | 69.4 | 78.6 |
| 0.8 | 82.0 | 36.2 | 50.3 | 96.2 | 39.8 | 56.3 |
| 0.9 | 100.0 | 0.1 | 0.2 | 96.4 | 0.8 | 1.7 |
| 1.0 | 100.0 | 0.1 | 0.2 | 97.2 | 0.8 | 1.7 |
Relevancy classification
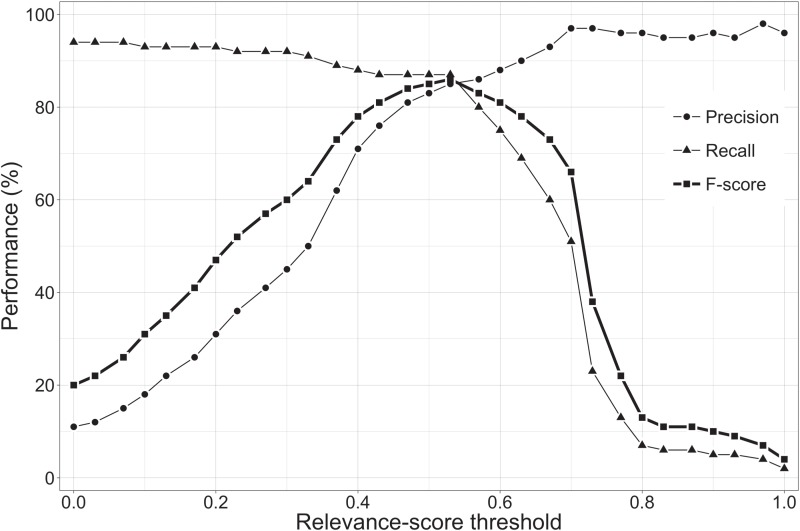
Figure 4 shows the performance of the relevance system for different relevance score thresholds on the training set. The best performance (in terms of F-score) was obtained for a relevance score threshold of 0.53, with a precision of 85%, a recall of 87% and an F-score of 86%. For the same threshold, the performance on the test set was slightly lower with 81% precision and 82% recall, resulting in an F-score of 82%. Further investigation into the compounds that the system classified as relevant showed that 97% of these compounds were annotated as chemical compounds in the chemical entity corpus. Therefore, only 3% of the compounds classified by the system as relevant were not chemical entities.
Figure 4.
The performance of the relevance system based on precision, recall and F-score.
The relevancy classification is dependent on the performance of the chemical entity recognition system in two ways. First, only compounds that are found by the CER can be classified as relevant. Second, the relevance-score features for a given chemical entity are based on the full patent text. The recognizer needs to correctly identify all occurrences of that entity in the full text. To assess the effect of the first dependency on the performance of the relevance system, we fed the gold-standard chemical entities as input to the relevance system (simulating a scenario where the chemical entity recognition system has a precision and recall of 100%). Apart from the patent snippet, all other parts of the full patent document were analyzed with the original system because gold-standard annotations were not available. When evaluated on our test set, the relevance classification system obtained 93% precision, 88% recall and 91% F-score. Further investigation into these scores indicated that the system could have performed better if we could also eliminate the second dependency.
We also investigated the contribution of individual relevancy features to the performance of the relevancy classification system. For this we removed each feature in turn from the relevance score and adjusted the relevance-score threshold for optimal performance. Table 3 shows that the length of the compound is a major indicator of the relevance of the compound (10 percentage points added value). Additionally, the patent section in which the compound was found and compound wide usage in other publications are also good indicators of the relevance of the compound (around 5 percentage points added value). The other features contribute between 1 and 2 percentage points to the relevancy classification performance.
Table 3.
The added value of individual features based on ``leave-one-out'' methodology
| Setting | Threshold | Precision | Recall | F-Score | Added value |
|---|---|---|---|---|---|
| All features | 0.53 | 84.8 | 86.8 | 85.8 | - |
| A—Compound frequency | 0.47 | 82.8 | 86.2 | 84.5 | 1.3 |
| B—Compound section | 0.40 | 95.5 | 70.0 | 80.8 | 5.0 |
| C—Compound length | 0.40 | 75.9 | 75.5 | 75.7 | 10.1 |
| D—Surrounding characters | 0.53 | 85.1 | 82.9 | 84.0 | 1.8 |
| E—Compound section uniqueness | 0.53 | 84.8 | 82.9 | 83.9 | 1.9 |
| F—Compound without solvent | 0.53 | 85.1 | 82.9 | 84.0 | 1.8 |
| G—Compound wide usage | 0.53 | 83.9 | 76.4 | 80.0 | 5.8 |
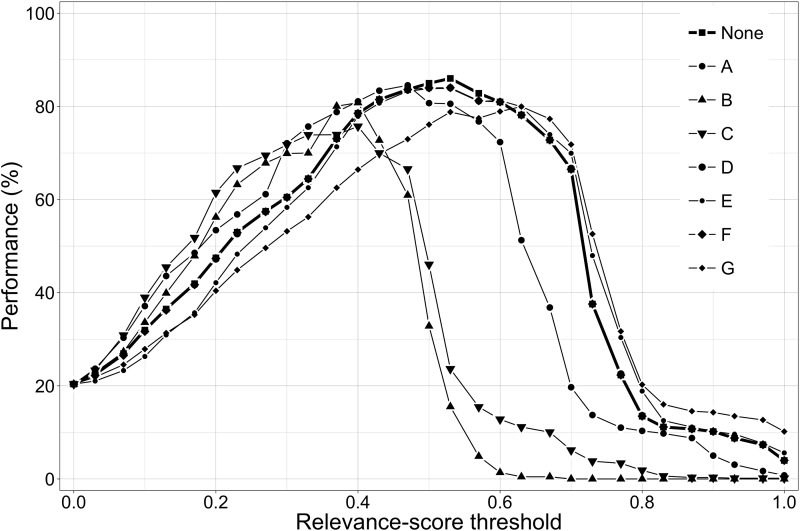
As can be seen from Table 3, leaving out a feature can affect the optimal value of the relevance-score threshold. Figure 5 shows the performance of the relevancy classification system as a function of the threshold value when a feature is left out.
Figure 5.
Performance of the relevancy classification system as a function of the relevance-score threshold when one of relevancy features A-G is removed (see Table 3 for feature legend).
Discussion
Extraction of chemical compounds from chemical-related patents has recently been studied, focusing on patent titles and abstracts (28, 31, 52) or full texts (3, 20, 21, 27, 51). The majority of these studies concentrated on identifying chemical compounds in text while disregarding the structures of the extracted compounds (31, 52). Some have also looked at associating structures to extracted compounds [e.g. (3, 20, 21)] and have resulted in products and databases of chemical compounds in patents (3, 20, 21). To our knowledge, this is the first attempt to narrow down the focus to relevant compounds and their structures within a chemical patent. Relevance of a chemical compound is based on the context of the full patent document. Generally, a relevant compound is a compound that plays a major role in the patent (e.g. a product of a reaction that is mentioned in the Claim section of a patent). We have shown that these compounds are a small subset (<10%) of all compounds mentioned in the textual part of a patent.
We have presented a two-step approach to identify relevant compounds in patent documents: compound identification (first step) followed by compound classification (second step). This approach allows the use of the output of the first step for additional purposes (such as indexing chemical compounds mentioned in patents) but at the same time introduces dependencies. Obtaining high precision and recall values in the first step is essential for the success of the second step. Based on the findings of our previous studies (27, 28), we used an ensemble approach combining dictionary-based and morphology-based approaches to obtain high precision and recall. These approaches require a small annotated corpus (26, 33) and can provide a structural representation of the extracted compounds. Associating correct chemical structures to compounds is essential when extracting chemical compounds. To reduce the possibility of associating a compound with the wrong structure (34, 35) we regenerated the structures of compounds in different databases with our name to structure toolkit (Name Service) and standardized the structures based on standardization rules used for Reaxys (15).
The structures of non-systematic identifiers associated with a compound within Reaxys are manually drawn by excerpters and are later validated and standardized using Name Service. Adding such structures to the Name Service database allowed us to generate structures for non-systematic identifiers. We used the same toolkit with the same standardization functionalities to validate compounds extracted using the grammar-based approach. This ensures high quality and consistency of the extracted compounds.
To build the chemical entity recognition and relevancy classifier system, a patent corpus annotated with chemical entities and their relevance was needed. To our knowledge, such a corpus did not exist (7). Currently available patent corpora either are limited to subsections of the patents, mostly title and abstract [e.g. the BioCreative corpus (36)], or had other limitations that prevented their use, such as different guideline definitions (focus on different entity types), harmonization approaches (manual using SMEs vs automation), low or unidentified IAA scores and limited scope of coverage (only one chemical IPC class or one section of a document) (7). We developed the corpus in two steps. First, we constructed a chemical entity corpus using random stratified sampling for content selection and manual harmonization to ensure high quality. Later we extended this corpus with relevancy annotations. We took into account the inherent difficulty of classifying relevance of some compounds by introducing `equivocal’ as a classification in the corpus. Chemical compounds identified as equivocal can be classified as both relevant and irrelevant. The system can assign relevant or irrelevant for compounds extracted in this area. Any compound identified as equivocal was disregarded from our evaluation. Using five annotators for relevancy annotation, we showed that the equivocal label is only limited to ~2% of the compounds.
Normalized patent documents were used to develop the corpus and the system. Any change in the normalization approach will lead to changes to the corpus and might result in a need for retraining the system. We reduced this dependency by finalizing the normalization before developing the corpus and the software. We also introduced a one-to-one mapping between the original patent document and the normalized patent document to allow possible changes to the corpus with limited efforts. The relevancy classification system has lower dependency to the normalization step as its performance is calculated on unique mentions of compounds within a patent. The dependency to the normalization step relies on the quality of the patent source file. Digital patents [e.g. from EPO (11) or USPTO (12)] have a higher quality than OCR patents [e.g. from WIPO (13)]. Therefore, the system is more dependable on the normalization when dealing with OCR patents.
The chemical entity recognition software showed a precision of 90.1% and a recall of 82.3% for compound recognition on EPO patents. The state-of-the-art statistical systems (tested on patent title and abstract) have obtained higher recall (precision of 87.5% and recall of 91.3%) (31). These systems do not generate structures for the identified chemical compounds. Error analysis of our system indicated that the loss in recall in our system is mainly due to the fact that reactants and products of synthesis procedures are not recognized, and prophetic compounds are missed. Identification of prophetic compounds may be improved by taking into account trigger phrases (e.g. `The compound of claim is:’, `A compound selected from’) or negative triggers for these compounds (e.g. `catalysts’).
Our current process only investigates the identification of relevant compounds in the textual part of non-OCR patents. Expanding this approach to chemical classes (such as Markush) can further improve the software. A large proportion of relevant compound information is only available through scaffolds, pictures and tables. Successful identification of these compounds can result in a higher coverage. Since 2001, some patent offices including the USPTO (12) are requesting applicants to submit chemical structures and reactions [as MDL Molfiles or ChemDraw CDX files (30)] when submitting their patent applications. Note that in many cases these are not drawn by authors or chemists and are presented usually with defects in the connection table of chemical structure. This can be a good starting point for future research.
We have successfully managed to identify relevant compounds in chemical-related patents. The resulting relevant compounds can be used to predict key compounds within a patent (9, 39, 53, 54). In future research, we want to extend this work to chemical classes, increase the coverage by dealing with OCR patents (that contain many spelling errors) and utilize data from tables, scaffolds and images.
Our system uses proprietary toolkits to extract chemical compounds. License to the tools can be requested from Elsevier. A demo of OCMiner is available through [http://www.ontochem.de/our-products/information-discovery.html]. A demo of CER can be requested from https://www.reaxys.com. The same methodology can be applied using non-proprietary toolkits [e.g. toolkits developed for BioCreative V, CHEMDNER track (55)]. The training corpus used to train the annotators is made available through Mendeley Data (56).
Declarations
Availability of data and material
This study uses proprietary toolkits to extract chemical compounds. License to the tools can be requested from Elsevier. A demo of OCMiner is available through [http://www.ontochem.de/our-products/information-discovery.html]. A demo of CER can be requested from https://www.reaxys.com. The same methodology can be applied using non-proprietary toolkits. The training corpus used to train the annotators is made available through Mendeley Data. The full gold set is a property of Elsevier. Access to the gold set can be requested from Elsevier. Please contact author for data requests.
Authors’ contributions
S.A.A., H.R., M. Schwörer, N.H., M.M., C.B. and M.I. conceived and designed the experiments. S.A.A., H.R., M. Schwörer, H.N. and C.B. developed the gold set. S.A., H.R., M. Schwörer, CB, M.I., H.N., and J.A.K. analyzed the data. S.A.A., H.N., G.I., J.T., M.D., M.G., M. Sheehan and J.A.K coordinated the project. S.A.A. drafted the manuscript, and J.A.K. revised it. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to acknowledge the patent annotation team for annotating the patent documents. The contents of this work have been included in a patent application at the US Patent and Trademark Office and the patent is pending.
Database URL: https://data.mendeley.com/datasets/6hykykmn65/1
Funding
Elsevier B.V.
Conflict of interest. S.A.A., M.D. and M.G. are employed by Elsevier B.V., Amsterdam, the Netherlands. H.R., G.I., M. Schwörer, M.M., M. Sheehan and H.N. are employed by Elsevier Information Systems GmbH, Frankfurt, Germany. J.T. is employed by Elsevier Limited, UK. Reaxys is a product of Elsevier. C.B. and M.I. are employed by OntoChem IT Solutions GmbH, Halle (Saale), Germany. OCMiner is a product of OntoChem. Elsevier and OntoChem jointly supported the changes made to OCMiner for the purpose of this project.
References
- 1. Muresan S., Petrov P., Southan C. et al. (2011) Making every SAR point count: the development of chemistry connect for the large-scale integration of structure and bioactivity data. Drug Discov. Today, 16, 1019–1030. [DOI] [PubMed] [Google Scholar]
- 2. Southan C., Boppana K., Jagarlapudi S.A. et al. (2011) Analysis of in vitro bioactivity data extracted from drug discovery literature and patents: ranking 1654 human protein targets by assayed compounds and molecular scaffolds. J. Cheminform., 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Papadatos G., Davies M., Dedman N. et al. (2016) SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res., 44, D1220–D1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krallinger M., Rabal O., Lourenço A. et al. (2017) Information retrieval and text mining technologies for chemistry. Chem. Rev., 117 (12),7673–7761. [DOI] [PubMed] [Google Scholar]
- 5. Senger S., Bartek L., Papadatos G. et al. (2015) Managing expectations: assessment of chemistry databases generated by automated extraction of chemical structures from patents. J. Cheminform., 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bregonje M. (2015) Patents: a unique source for scientific technical information in chemistry related industry? World Pat. Inf., 27, 309–315. [Google Scholar]
- 7. Akhondi S.A., Klenner A.G., Tyrchan C. et al. (2014) Annotated chemical patent corpus: a gold standard for text mining. PLoS One, 9, e107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Asche G. (2017) “80% of technical information found only in patents”—is there proof of this? World Pat. Inf., 48, 16–28. [Google Scholar]
- 9. Tyrchan C., Boström J., Giordanetto F. et al. (2012) Exploiting structural information in patent specifications for key compound prediction. J. Chem. Inf. Model., 52, 1480–1489. [DOI] [PubMed] [Google Scholar]
- 10. Benson C.L. and Magee C.L. (2015) Quantitative determination of technological improvement from patent data. PLoS One, 10(4), e0121635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. European Patent Office http://www.epo.org. 10 Jan 2019.
- 12. United States Patent and Trademark Office http://www.uspto.gov/.10 Jan 2019.
- 13. World Intellectual Property Organization http://www.wipo.int/.10 Jan 2019.
- 14. Zimmermann M., Fluck J., Thi Le T.B. et al. (2005) Information extraction in the life sciences: perspectives for medicinal chemistry, pharmacology and toxicology. Curr. Top. Med. Chem., 5, 785–796. [DOI] [PubMed] [Google Scholar]
- 15. Reaxys . https://www.reaxys.com. 10 Jan 2019.
- 16. Lawson A.J., Swienty-Busch J., Géoui T. et al. (2014) The making of Reaxys—towards unobstructed access to relevant chemistry information In: The Future of the History of Chemical Information, Washington D.C., USA: American Chemical Society, 127–148. [Google Scholar]
- 17. SciFinder https://scifinder.cas.org/scifinder/. 10 Jan 2019.
- 18. Thomson Reuters Pharma . http://lifesciences.thomsonreuters.com/. 10 Jan 2019.
- 19. Heifets A. and Jurisica I. (2012) SCRIPDB: a portal for easy access to syntheses, chemicals and reactions in patents. Nucleic Acids Res., 40, D428–D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matos P., Alcantara R., Dekker A. et al. (2010) Chemical entities of biological interest: an update. Nucleic Acids Res., 38:D249–D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. IBM IBM contributes data to the National Institutes of Health to speed drug discovery and cancer research innovation. http://www-03.ibm.com/press/us/en/pressrelease/36180.wss. 10 Jan 2019.
- 22. NextMove Software Unleashing over a million reactions into the wild. https://nextmovesoftware.com/blog/2014/02/27/unleashing-over-a-million-reactions-into-the-wild/. 10 Jan 2019.
- 23. Kim S., Thiessen P.A., Bolton E.E. et al. (2016) PubChem substance and compound databases. Nucleic Acids Res., 44, D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Japan Patent Office https://www.jpo.go.jp/. 10 Jan 2019.
- 25. Valko A.T. and Johnson A.P. (2009) CLiDE Pro: the latest generation of CLiDE, a tool for optical chemical structure recognition. J. Chem. Inf. Model., 49, 780–787. [DOI] [PubMed] [Google Scholar]
- 26. Vazquez M., Krallinger M., Leitner F. et al. (2011) Text mining for drugs and chemical compounds: methods, tools and applications. Molecular Informatics, 30, 506–519. [DOI] [PubMed] [Google Scholar]
- 27. Akhondi S.A., Hettne K.M., Horst E. et al. (2015) Recognition of chemical entities: combining dictionary-based and grammar-based approaches. J. Chem., 7, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akhondi S.A., Pons E., Afzal Z. et al. (2016) Chemical entity recognition in patents by combining dictionary-based and statistical approaches. Database (Oxford), 2016, baw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tseng Y.-H., Lin C.-J. and Lin Y.-I. (2007) Text mining techniques for patent analysis. Inf. Process. Manag., 43, 1216–1247. [Google Scholar]
- 30. Dalby A., Nourse J.G., Hounshell W.D. et al. (1992) Description of several chemical structure file formats used by computer programs developed at molecular design limited. J. Chem. Inf. Comput. Sci., 244–255. [Google Scholar]
- 31. Krallinger M., Rabal O., Lourenc O.A. et al. (2015) Overview of the CHEMDNER patents task. In: Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, 63–75.
- 32. Krallinger M., Leitner F., Rabal O. et al. (2015) CHEMDNER: the drugs and chemical names extraction challenge. J. Chem., 7, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eltyeb S. and Salim N. (2014) Chemical named entities recognition: a review on approaches and applications. J. Cheminform., 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Akhondi S.A., Kors J.A. and Muresan S. (2012) Consistency of systematic chemical identifiers within and between small-molecule databases. J. Cheminform., 4, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akhondi S.A., Muresan S., Williams A.J. et al. (2015) Ambiguity of non-systematic chemical identifiers within and between small-molecule databases. J. Cheminform., 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krallinger M., Rabal O., Leitner F. et al. (2015) The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminform., 7(suppl 1), S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jessop D.M., Adams S.E. and Murray-Rust P. (2011) Mining chemical information from open patents. J. Cheminform., 3, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ede M., Endacott J., Harper M. et al. (2016) Indexing chemical structures: exemplified compound indexing in patents by the vendors Thomson Reuters, Chemical Abstracts and Elsevier—a comparative study by the Patent Documentation Group (PDG). World Pat. Inf., 44, 48–52. [Google Scholar]
- 39. Hattori K., Wakabayashi H. and Tamaki K. (2008) Predicting key example compounds in “competitors” patent applications using structural information alone. J. Chem. Inf. Model., 48, 135–142. [DOI] [PubMed] [Google Scholar]
- 40. Lawson A., Roller S., Grotz H. et al. (2011) Method and software for extracting chemical data. Unites States Patent Office (USPTO). US7933763.
- 41. Irmer M., Weber L., Böhme T. et al. (2015) OCMiner for patents. extracting chemical information from patent texts. In: Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, pp. 119–123.
- 42. Roller S. Using Reaxys for searching chemistry in Patents. https://www.yumpu.com/en/document/read/25882921/using-reaxys-for-searching-chemistry-in-patents-stefan-roller/. 10 Jan 2019.
- 43. Kolárik C., Klinger R., Friedrich C.M.. et al. (2008) Chemical names: terminological resources and corpora annotation. In: Workshop on Building and Evaluating Resources for Biomedical Text Mining, 51–58.
- 44. Kulick S., Bies A., Liberman M. et al. (2004) Integrated annotation for biomedical information extraction. In Proceedings of the Human Language Technology Conference and the Annual Meeting of the North American Chapter of the Association for Computational Linguistics (HLT/NAACL), 61–68.
- 45. Kim J.-D., Ohta T., Tateisi Y. et al. (2003) GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics, 19, i180–i182. [DOI] [PubMed] [Google Scholar]
- 46. Corbett P., Batchelor C. and Teufel S. (2007) Annotation of chemical named entities. In: Proceedings of the Workshop on BioNLP 2007 Biological, Translational, and Clinical Language Processing—BioNLP “07”. Morristown, NJ, USA: Association for Computational Linguistics, 57–64. [Google Scholar]
- 47. The Brat Contributors . http://brat.nlplab.org/. 10 Jan 2019.
- 48. Stenetorp P., Pyysalo S., Topić G. et al. (2012) BRAT: a web-based tool for NLP-assisted text annotation. In: 13th Conference of the European Chapter of the Association for Computational Linguistics, Avignon, France, April 23-27, 2012. Association for Computational Linguistics, 102–107. [Google Scholar]
- 49. Law V., Knox C., Djoumbou Y. et al. (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res., 42, D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bento A.P., Gaulton A., Hersey A. et al. (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res., 42, D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lowe D. (2012) Extraction of chemical structures and reactions from the literature. Ph.D. Thesis University of Cambridge, https://doi.org/10.17863/CAM.16293. [Google Scholar]
- 52. Pérez-Pérez M., Pérez-Rodríguez G., Rabal O. et al. (2016) The Markyt visualisation, prediction and benchmark platform for chemical and gene entity recognition at at BioCreative/CHEMDNER challenge. Database (Oxford), 2016, baw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lepp Z., Huang C. and Okada T. (2009) Finding key members in compound libraries by analyzing networks of molecules assembled by structural similarity. J. Chem. Inf. Model., 49, 2429–2443. [DOI] [PubMed] [Google Scholar]
- 54. Kettle J.G., Ward R.A. and Griffen E. (2010) Data-mining patent literature for novel chemical reagents for use in medicinal chemistry design. Med. Chem. Commun., 1, 331. [Google Scholar]
- 55. Oxford Academic BioCreative Virtual Issue .https://academic.oup.com/database/pages/biocreative_virtual_issue. 10 Jan 2019.
- 56. Akhondi S.A., Rey H., Schwörer M. et al. (2018) Automatic identification of relevant chemical compounds from patents. The training corpus. Mendeley Data v1, 10.17632/6hykykmn65.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study uses proprietary toolkits to extract chemical compounds. License to the tools can be requested from Elsevier. A demo of OCMiner is available through [http://www.ontochem.de/our-products/information-discovery.html]. A demo of CER can be requested from https://www.reaxys.com. The same methodology can be applied using non-proprietary toolkits. The training corpus used to train the annotators is made available through Mendeley Data. The full gold set is a property of Elsevier. Access to the gold set can be requested from Elsevier. Please contact author for data requests.