Abstract
Non-small cell lung cancer (NSCLC) is a type of lung cancer which has a high mortality and low survival rate. Previous studies have revealed that long non-coding RNAs participate in tumorigenesis and metastasis in NSCLC. In the present study, the function of small nucleolar RNA host gene 12 (SNHG12) was investigated in NSCLC. Using reverse transcription-quantitative polymerase chain reaction analysis, it was identified that SNHG12 was significantly overexpressed in NSCLC specimens. Furthermore, overexpression of SNHG12 was identified to be associated with tumor progression and poor overall survival rates. Knockdown of SNHG12 in NSCLC cells could effectively induce cell apoptosis and suppress cell viability, proliferation, migration and invasion via inhibition of the epithelial-mesenchymal transition process. Furthermore, a direct interaction between microRNA (miR)-218 and the binding site of SNHG12 was identified. SNHG12 acted as an endogenous sponge for miR-218. Knockdown of SNHG12 upregulated the expression level of miR-218 as well as downregulating the Slug/zinc finger E-box-binding homeobox 2 EMT signaling pathway, and thus inhibited cell migration and invasion. Therefore, SNHG12 may serve as a key biomarker and a potential therapeutic target for the treatment of NSCLC.
Keywords: non-small cell lung cancer, small nucleolar RNA host gene 12, microRNA-218, metastasis, epithelial-mesenchymal transition, Slug/zinc finger E-box-binding homeobox 2
Introduction
Lung cancer, which results in an estimated 1.59 million mortalities worldwide annually is one of the most frequent malignancies (1). Non-small cell lung cancer (NSCLC), which exhibits high mortality and a low 5-year survival rate, accounts for between 80 and 85% of lung cancer cases (2–4). At present, cisplatin-, paclitaxel- and gefitinib-based chemotherapies are widely used as the first-line chemotherapy for advanced NSCLC (5). However, differences in the therapeutic effect among individuals and the development of multidrug resistance present challenges to successful chemotherapy in clinical application (6–8). More seriously, invasion and metastasis, which account for the principal causes of mortality in patients with cancer, are common in NSCLC following initial diagnosis (9,10). Therefore, further elucidation of the molecular mechanisms underlying tumorigenesis, invasion and metastasis is required to develop an efficient strategy for the therapy of NSCLC.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA molecule which are >200 nt in length and participate in the regulation of gene expression (11). Previous studies have demonstrated that lncRNAs were able to regulate various biological progresses such as tumorigenesis, differentiation and inflammation (12,13). In addition, overexpression or downregulation of lncRNAs is associated with processes in tumors including proliferation, apoptosis, invasion and metastasis in numerous types of cancer (14,15). For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1, which was overexpressed in various cancer cells, was identified as a key regulator of cell proliferation and the cell cycle (16). Additionally, it has been reported that the small nucleolar RNA host gene 12 (SNHG12) lncRNA enhances cell proliferation, invasion and migration by increasing the angiomotin gene expression level in human osteosarcoma (17). However, the biological function and mechanism of SNHG12 in NSCLC remain unknown. Previous studies have indicated that all RNA transcripts that possess microRNA-binding sites are able to interact with each other and regulate expression levels, suggesting interactions between microRNAs and lncRNAs in tumorigenesis (18–22).
In the present study, the expression level and function were investigated in NSCLC, and it was identified that SNHG12 was markedly overexpressed in NSCLC tissues and was associated with poor prognosis and survival of patients with NSCLC. Furthermore, further mechanical analysis revealed that SNHG12 suppresses NSCLC cell proliferation, invasion and migration by targeting microRNA-218 (miR-218) and then influences the Slug/zinc finger E-box-binding homeobox 2 (ZEB2) signaling pathway.
Patients and methods
Tissue specimens and cell lines
In total, 40 NSCLC tissue samples and adjacent normal tissue samples (3×2×1 cm) were obtained from patients with NSCLC who were undergoing surgery and without radiotherapy or chemotherapy. The patients' sex distribution (male, n=21; female, n=19) and age distribution in years (≥60, n=18; <60, n=22; range, 28–77; mean ± standard deviation, 58.88±12.65) were recorded. The present study was approved by the Ethic Review Committees of The Affiliated Hospital of Hebei University (Baoding, China) and written informed consent was obtained from all patients. A 48-month follow-up survival survey based on the medical records of the patients was performed. Overall survival (OS) time was defined as the interval between resection and mortality or the last follow-up visit. Pathological evaluations of tissues were performed by pathologists at the Department of Pathology of The Affiliated Hospital of Hebei University. The tissues obtained were stored at −80°C until further use.
NSCLC cell lines A549 (cat. no. CCL-185) and H1299 (cat. no. CRL-5803) were purchased from the American Type Culture Collection (Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and antibiotics at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was obtained from tumor tissue samples or cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT was performed using a TaqMan™ MicroRNA Reverse Transcription kit (cat. no. 4366596; Thermo Fisher Scientific, Inc.). qPCR was performed using IQ SYBR-Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) on an iCycler IQ Multi-Color Detection system (Bio-Rad Laboratories, Inc.). The thermocycling conditions for PCR were as follows: 95°C for 30 sec; 40 cycles of 95°C for 5 sec and 60°C for 34 sec; followed by dissociation. Stem-loop primers used to detect microRNAs were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). U6 small nucleolar RNA and GAPDH were used for normalization. Primers sequences were as follows: SNHG12, 5′-TCTGGTGATCGAGGACTTCC-3′ (forward) and 5′-ACCTCCTCAGTATCACACACT-3′ (reverse); human (hsa)-miR-218, 5′-GGAGTGGCGAATGGTAGTGGAGT-3′ (forward) and 5′-ACCAGGCTGGACAGTAGAGCG-3′ (reverse); GAPDH, 5′-TCTCTGCTCCTCCTGTTC-3′ (forward) and 5′-GGTTGAGCACAGGGTACTTTATTGA-3′ (reverse); and U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). Relative expression levels of RNA were calculated using the 2−ΔΔCq method (23) with CFX Manager software (version 3.1; Bio-Rad Laboratories, Inc.).
In situ hybridization (ISH)
ISH was used to detect SNHG12 in 40 paired NSCLC and adjacent normal samples. A digoxigenin (DIG)-UTP-labeled antisense RNA probe (Beijing View Solid Biotechnology, Beijing, China) which was derived from nucleotides 28–257 of the SNHG12 coding sequence was obtained by in vitro transcription using a DIG RNA Labeling kit (Roche Diagnostics, Basel, Switzerland). The sense RNA probe derived from nucleotides 28–257 of the SNHG12 coding sequence was labeled with DIG-UTP and used as a negative control. ISH was performed using the ISH kit (Boster Biological Technology, Pleasanton, CA, USA), according to the manufacturer's protocol.
Cell transfection
The plasmid for SNHG12 inhibitor (SNHG12-inhi) and the plasmid for hsa-miR-218 mimic (miR-218-mimic) were constructed by OBiO Technology (Shanghai) Corp., Ltd. (Shanghai, China). SNHG12-inhi or miR-218-mimic was transfected into A549 and H1299 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Cell viability analysis
Cell viability was determined using an MTT assay. First, A549 and H1299 cells were seeded into a 96-well plate at a density of 2,000 cells/well. After 12 h incubation at 37°C, cells were transfected with SNHG12-inhi or negative control-inhi (NC-inhi; OBiO Technology (Shanghai) Corp., Ltd.). At various times (24, 48, 72 and 96 h), cells from each well were treated with 20 µl MTT solution (5 mg/ml; Amresco, LLC, Solon, OH, USA). Following incubation for 4 h at 37°C, 200 µl dimethylsulfoxide was added to each well to dissolve the precipitated formazan product. Finally, the absorbance of each well was determined at 570 nm. All experiments were performed three times.
Colony formation assay
Cells were seeded in a 6-well plate at a density of 500 cells/well and incubated for 12 h at 37°C. Following transfection with SNHG12-inhi or NC-inhi, cells were incubated for a further 6–8 days. Finally, cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min at 25°C and stained with 0.1% crystal violet for 30 min at room temperature, and images were captured. Colonies were counted by eye and colonies containing ≥50 cells were scored.
Cell apoptosis evaluation
Cells were seeded in a 6-well plate and transfected with SNHG12-inhi or NC-inhi. Cells were trypsinized, washed and collected for further staining. In total, 2×105 cells/well, suspended in 400 µl 1X binding buffer, were stained with 5 µl fluorescein isothiocyanate-Annexin V and 5 µl propidium iodide in the dark at room temperature for 15 min. Cells were analyzed immediately using flow cytometry (FACScan®; BD Biosciences, San Jose, CA, USA).
Transwell assay
Transwell chambers assays were used to determine cell migration and invasion. For the determination of cell migration, 4×104 cells suspended in 200 µl serum-free DMEM were seeded into the upper chamber. For cell invasion assessment, 200 µl cell suspension (1×105 cells, without serum) were added into the upper chamber which was pre-coated with Matrigel (BD Biosciences). The lower chamber was filled with 800 µl DMEM containing 10% FBS. Cells were incubated for 24–48 h at 37°C. The non-invasive cells which remained in the upper chamber were removed with a cotton swab, whereas the invasive cells which were located in the bottom of membrane were fixed with methanol and stained with 0.1% crystal violet. Finally, images of invasive cells from each well were captured under a phase contrast microscope (CX43; Olympus Corporation, Tokyo, Japan) in three independent fields.
Wound healing assay
In total, 5×105 cells were seeded in a 6-well plate and incubated for 12 h at 37°C. When the cell density reached ~90%, cells were scratched with a 10 µl pipette tip. Cells were washed with PBS to remove the free-floating cells and transfected with SNHG12-inhi or NC-inhi. Cells were incubated for a further 24 h at 37°C in DMEM with 1% FBS and images were captured under an inverted light microscope (CKX53; Olympus, Japan) for migration analysis.
Western blot analysis
Cells were transfected with different plasmids (NC-inhi, SNHG12-inhi, miR-NC, miR-218-mimic) for 48 h at 37°C and collected for total protein extraction. Total protein was obtained from the cells by lysing with radioimmunoprecipitation buffer (Beyotime Institute of Biotechnology, Haimen, China). The concentration of total protein was determined using a BCA protein assay kit (cat. no. 23225; Thermo Fisher Scientific, Inc.). The protein (20 µg) was separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking with 5% non-fat milk powder suspended in TBS containing 0.25% Tween-20 for 2 h at room temperature, membranes were incubated with primary antibodies against cleaved caspase 3 (cat. no. 9661) cleaved caspase 9 (cat. no. 52873), matrix metalloproteinase 9 (MMP-9; cat. no. 13667S), epithelial (E-) cadherin (cat. no. 14472), vimentin (cat. no. 5741), Slug (cat. no. 9585), ZEB2 (cat. no. 3396) and β-actin (cat. no. 3700; all Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C for 8 h. The dilution for cleaved caspase-3, cleaved caspase-9, MMP-9, E-cadherin, vimentin, Slug and ZEB2 was 1:100, and the dilution for β-actin was 1:1,000. Subsequently, membranes were incubated further with horseradish peroxidase-labeled anti-rabbit IgG secondary antibodies (cat. no. 7047; 1:1,000; Cell Signaling Technology, Inc.) and protein bands were imaged and quantified using an enhanced chemiluminescence detection system.
Dual-luciferase assay
In total, 50 genes were simultaneously predicted by the miRcode database (http://www.mircode.org/), and small nuclear RNA host gene 12 (SNHG12) was detected as a candidate gene associated with NSCLC based on its associated Gene Ontology (GO; http://www.geneontology.org/) terms and because it harbors miR-218 binding sites. The wild-type (WT) and mutant 3′-untranslated region of human SNHG12 were the amplified from human genomic DNA and individually cloned into the luciferase vector of pmiR-RB-REPORT™ [OBiO Technology (Shanghai) Corp., Ltd.]. A549 cells were co-transfected with 200 ng mutant or WT pmiR-RB-REPORT™-SNHG12 plasmid, and 100 ng miR-218-mimic or miR-NC plasmid using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. pRL-SV40 plasmid (Promega Corporation, Madison, WI, USA) carrying Renilla luciferase was also co-transfected into the cells for standardizing transfection efficiency. After 48 h, cells were collected for luciferase activity analysis using a Dual-Luciferase Reporter assay system (Promega Corporation), according to the manufacturer's protocol. The firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
All results are presented as the mean ± standard deviations. The statistical analysis was performed using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). The significance among groups was determined using one-way analysis of variance. Kaplan-Meier analysis was performed to compare overall survival rates based on the expression levels of SNHG12 in 40 patients with NSCLC. Survival curve was assessed by a log-rank test. The median level of SNHG12 was used as the cut-off values. Patients with HCC were divided into a high SNHG12 expression group and a low SNHG12 expression group. P<0.05 was considered to indicate a statistically significant difference.
Results and Discussion
SNHG12 is significantly overexpressed, and is markedly associated with progression and prognosis of NSCLC
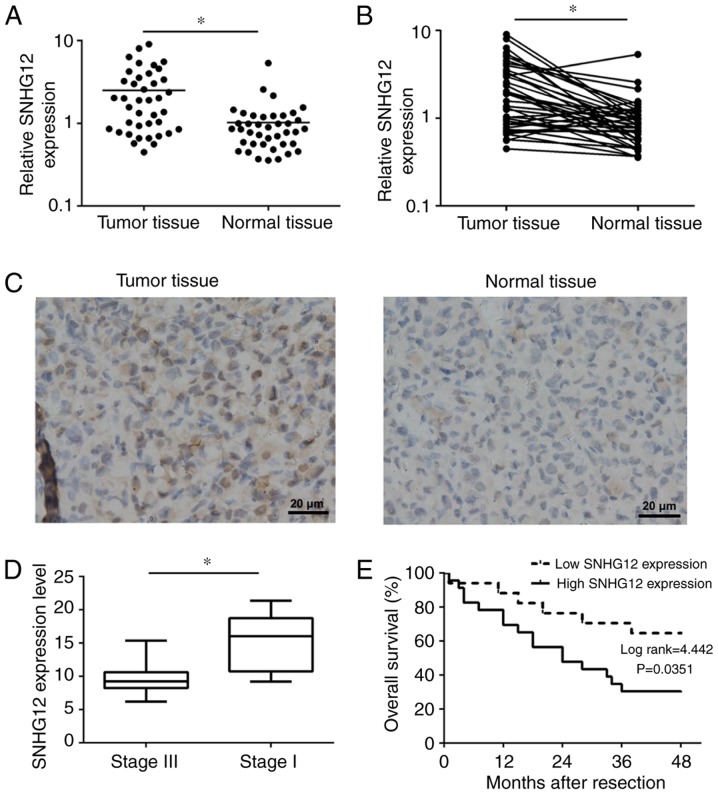
Previous studies have identified that lncRNA was regulated in a wide range of pathological processes such as tumorigenesis, cell death, invasion and inflammation, but also dysregulated in a number of types of cancer (24,25). To investigate whether there was a difference in SNHG12 expression between NSCLC tissues and adjacent normal tissues, RT-qPCR was used to determine the SNHG12 expression levels in 40 paired specimens. As presented in Fig. 1A and B, SNHG12 was significantly overexpressed in NSCLC tissues (P<0.01). Furthermore, an ISH assay was also performed to determine SNHG12 expression in tumor tissue sections and adjacent normal tissue sections. As presented in Fig. 1C, SNHG12, which was located in the cytoplasm, was markedly overexpressed in tumor tissue compared with normal tissue. In addition, the expression level of SNHG12 in advanced NSCLC (stage III, n=19) was significantly lower compared with that in early stages (stage I, n=21; P<0.01) (Fig. 1D). Furthermore, for the purpose of determining the association between SNHG12 expression and OS rate, patients with NSCLC were divided into a high-expression group (n=23) and a low-expression group (n=17), according to the median expression level of all tumor tissues. Compared with the low-expression group, NSCLC patients from the high-expression group have a significantly lower OS rate (P=0.0351; Fig. 1E). These results indicated that SNHG12 was overexpressed in NSCLC and may be associated with tumor progression and poor prognosis of NSCLC.
Figure 1.
SNHG12 is overexpressed in NSCLC, and is associated with tumor progression and poor overall survival. (A) The expression level of SNHG12 in tumor tissues and normal tissues. (B) The expression level of SNHG12 in tumor tissues was higher compared with that in adjacent normal tissues. (C) Images of in situ hybridization analysis of SNHG12 in tumor tissues and adjacent normal tissues. (D) Relative expression levels of SNHG12 in different stages of tumor tissues. (E) Kaplan-Meier analysis of overall survival rates based on expression levels of SNHG12 in 40 patients with NSCLC. *P<0.05. SNHG12, small nucleolar RNA host gene 12; NSCLC, non-small cell lung cancer.
Knockdown of SNHG12 inhibits NSCLC cell proliferation and induces cell apoptosis
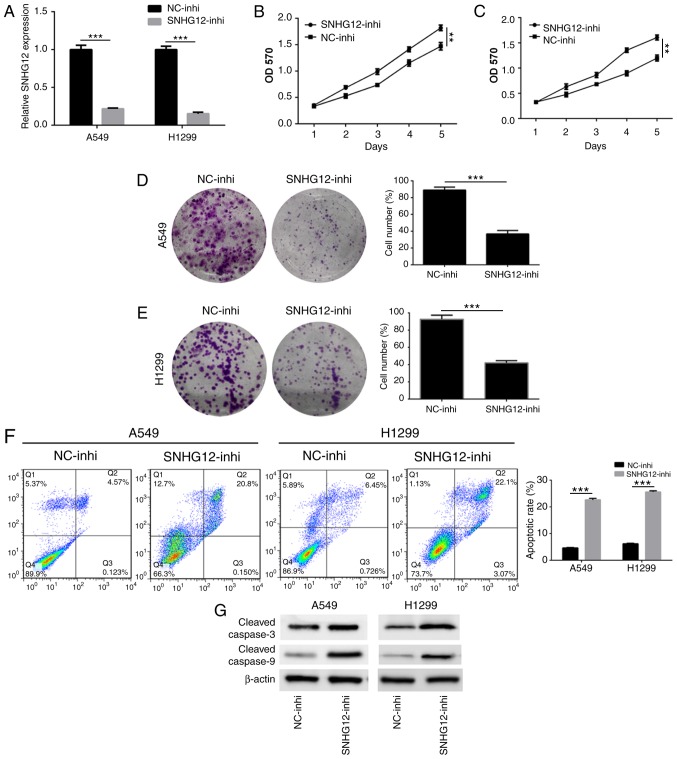
In order to investigate the action of SNHG12 on NSCLC cell proliferation and apoptosis, A549 and H1299 cells were transfected with SNHG12-inhi and NC-inhi. An RT-qPCR assay was used to assess the effect of SNHG12 knockdown. As presented in Fig. 2A, the expression level of SNHG12 in A549 and H1299 cells was significantly downregulated following treatment with SNHG12-inhi (P<0.001), indicating that SNHG12 was successfully and effectively knocked down. Results of the MTT assay showed that the viability of A549 and H1299 cells was significantly decreased following knockdown of SNHG12 (Fig. 2B and C; P<0.001). In addition, a colony-formation assay was performed to determine cell proliferation. As presented in Fig. 2D and E, compared with the control group, the colony-forming abilities of A549 and H1299 cells were significantly suppressed following knockdown of SNHG12 (P<0.001). Furthermore, cell apoptosis induced by knockdown of SNHG12 was also assessed. As presented in Fig. 2F, the apoptosis rate of A549 cells which was induced by SNHG12-inhi was 22.4±1.3%, which was significantly higher compared with that of the control group (4.2±0.9%; P<0.001). Compared with the control group (5.1±0.8%), apoptosis of H1299 cells which induced by SNHG12-inhi was 26.3±1.2% (P<0.001). To further elucidate the underlying molecular mechanisms of apoptosis induced by SNHG12, western blotting was used to investigate the caspase-mediated apoptotic pathway. As presented in Fig. 2G, knockdown of SNHG12 significantly enhanced expression of cleaved caspase 3 and cleaved caspase 9, indicating that knockdown of SNHG12 induced apoptosis through the caspase signaling pathway. These results indicated that downregulation of SNHG12 significantly suppressed proliferation and induced apoptosis in NSCLC cells.
Figure 2.
Knockdown of SNHG12 suppresses proliferation and induces apoptosis in non-small cell lung cancer cells. (A) Reverse transcription-polymerase chain reaction analysis of the expression level of SNHG12 in A549 and H1299 cells following treatment with SNHG12-inhi. Viability of (B) A549 and (C) H1299 cells was determined using an MTT assay following treatment with SNHG12-inhi. Proliferation of (D) A549 and (E) H1299 cells was assessed using a colony formation assay. (F) Apoptosis of A549 and H1299 cells induced by SNHG12 was detected using Annexin V/propidium iodide staining and flow cytometry. (G) The expression levels of caspase-associated proteins were analyzed by western blot analysis. **P<0.01, ***P<0.001. SNHG12, small nucleolar RNA host gene 12; inhi, inhibitor; NC, negative control.
Downregulation of SNHG12 strongly suppresses NSCLC cell migration and invasion by inhibiting the EMT process
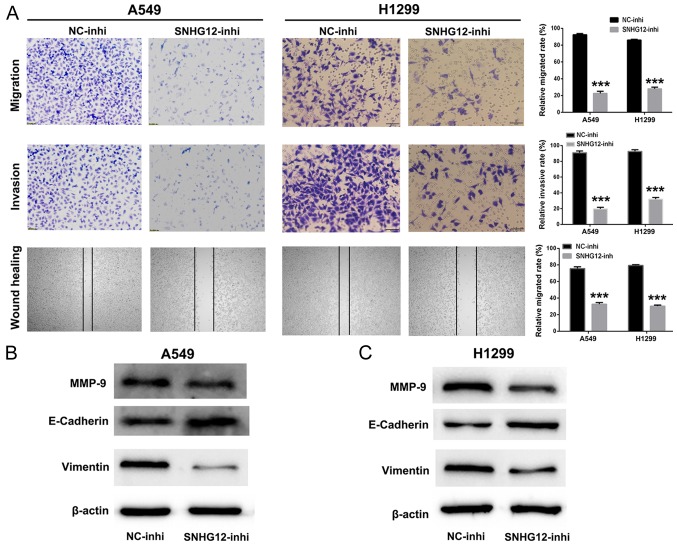
It was identified previously that SNHG12 participates in various biological processes including cell migration and invasion (26,27). Therefore, cell migration and invasion assays were used to investigate the function of SNHG12 in NSCLC cells. As presented in Fig. 3A, compared with control group, the numbers of migratory and invasive A549 and H1299 cells were significantly decreased in the SNHG12-inhi group (P<0.001). Additionally, the wound healing assay indicated that the mobility of A549 and H1299 cells treated with SNHG12-inhi was notably inhibited in comparison with the control group (P<0.001). These results indicated that knockdown of SNHG12 exhibited marked suppression of migration and invasion in NSCLC cells.
Figure 3.
Knockdown of SNHG12 inhibits migration and invasion by suppressing the EMT process in non-small cell lung cancer cells. (A) Migratory, invasive and mobility abilities of A549 and H1299 cells were evaluated using Transwell chambers assays and wound healing assays. The levels of EMT associated proteins in (B) A549 and (C) H1299 cells were analyzed by western blot assays. ***P<0.001 vs. NC-inhi. SNHG12, small nucleolar RNA host gene 12; inhi, inhibitor; NC, negative control; MMP, matrix metalloproteinase; E-cadherin, epithelial cadherin.
Furthermore, western blotting was performed to investigate further the anti-metastatic mechanism of SNHG12-inhi. MMPs are associated with the invasive behavior of tumor cells (28,29). As presented in Fig. 3B and C, the expression level of MMP-9 was decreased in A549 and H1299 cells following treatment with SNHG12-inhi. EMT has been identified to be an important process for tumor metastasis (30,31). As presented in Fig. 3B and C, when expression levels of SNHG12 in A549 and H1299 cells were downregulated by SNHG12-inhi, the expression level of the mesenchymal marker vimentin was decreased; in addition, the expression level of the epithelial marker E-cadherin was upregulated. Collectively, these results indicated that knockdown of SNHG12 could effectively inhibit metastasis in NSCLC cells.
SNHG12 interacts directly with miR-218 and serves as its sponge
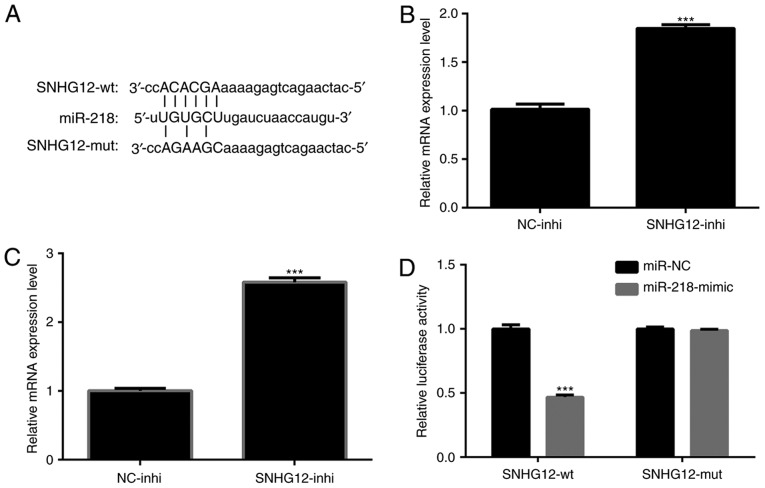
To further investigate the regulatory mechanism of SNHG12 in NSCLC cells, a bioinformatics prediction using miRcode software (www.mircode.org) was performed. Putative binding sites for miR-218 on SNHG12 were identified (Fig. 4A). It was determined whether the expression level of miR-218 was regulated by SNHG12. As presented in Fig. 4B and C, expression levels of miR-218 in A549 and H1299 cells were significantly upregulated by treatment of SNHG12-inhi (P<0.001). To verify that overexpression of miR-218 was indeed directly regulated by SNHG12, a dual-luciferase reporter assay was performed. pmiR-RB-REPORT™-SNHG12-WT and pmiR-RB-REPORT™-SNHG12-MUT were respectively transfected into A549 cells together with miR-218-mimic or miR-NC. The results indicated that the luciferase activity of pmiR-RB-REPORT™-SNHG12-WT was significantly decreased by miR-218-mimic (Fig. 4D; P<0.001). In contrast, the luciferase activity of pmiR-RB-REPORT™-SNHG12-MUT was not affected (Fig. 4D). These results indicated interactions between miR-218 and the binding sites of SNHG12.
Figure 4.
SNHG12 interacts directly with miR-218 and acts as a sponge. (A) Predicted binding sites between SNHG12 and miR-218. Expression levels of miR-218 in (B) A549 and (C) H1299 cells following treatment with SNHG12-inhi. ***P<0.001 vs. NC-inhi. (D) Luciferase activity in A549 cells co-transfected with miR-218-mimic or miR-NC or pmiR-RB-REPORT™-SNHG12-WT or pmiR-RB-REPORT™-SNHG12-MUT. ***P<0.001 vs. SNHG12-wt. SNHG12, small nucleolar RNA host gene 12; inhi, inhibitor; NC, negative control; wt, wild-type; miR, microRNA; mut, mutant.
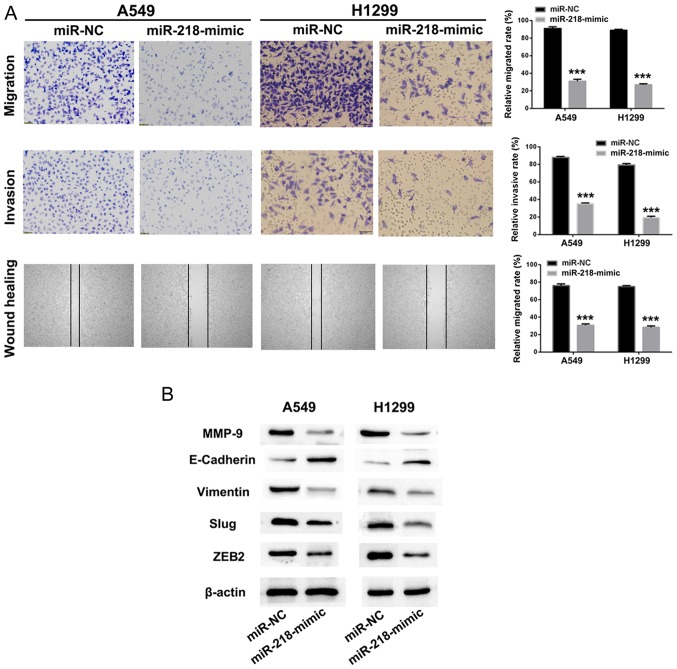
Knockdown of SNHG12 upregulates the expression level of miR-218, and suppresses NSCLC cells migration and invasion via the Slug/ZEB2 signaling pathway
As presented in Fig. 4B and C, knockdown of SNHG12 upregulated the expression level of miR-218 in A549 and H1299 cells. Knockdown of SNHG12 led to effective suppression of migration and invasion of NSCLC cells. Thus, we hypothesized that overexpression of miR-218 could also impair NSCLC cell migration and invasion. miR-218-mimic or miR-NC was transfected into A549 and H1299 cells, and cell migration, invasion and mobility of A549 and H1299 cells were identified to be significantly inhibited by treatment with miR-218-mimic, in comparison with the miR-NC group (Fig. 5A; P<0.001).
Figure 5.
miR-218 regulates non-small cell lung cancer cell metastasis via the Slug/ZEB2 signaling pathway. (A) Migratory, invasive and mobility abilities in A549 and H1299 cells were assessed using Transwell chambers assays and wound healing assays following treatment with miR-218-mimic. (B) Endothelial-mesenchymal transition-associated proteins and key proteins in the Slug/ZEB2 signaling pathway were detected using western blot. ***P<0.001 vs. miR-NC. ZEB2, zinc finger E-box-binding homeobox 2; miR, microRNA; NC, negative control; MMP, matrix metalloproteinase; E-cadherin, epithelial cadherin.
EMT is a progressive process in which epithelial cells gradually obtain a mesenchymal (fibroblast-like) cell phenotype, leading to increased metastasis and invasion (32). It has been identified that EMT is associated with the progression of lung cancer towards metastasis and invasion (33). Downregulation of E-cadherin is a hallmark of the EMT process, and transcription factors such as Slug, ZEB1 and ZEB2, which are regarded as regulators of E-cadherin, are the focus of investigation in tumor metastasis (34–36). Therefore, western blot assays were used to investigate whether the suppression of metastasis by SNHG12/miR-218 involves the Slug/ZEB2 signaling pathway. As presented in Fig. 5B, the metastasis-associated protein MMP-9 in A549 and H1299 cells was downregulated following treatment with miR-218-mimic. When treated with miR-218-mimic, the expression level of the mesenchymal marker vimentin in A549 and H1299 cells was decreased. Furthermore, the expression level of E-cadherin, a transcriptional regulator of Slug and ZEB2, was upregulated following treatment with miR-218-mimic, resulting in suppression of EMT. These results implied that knockdown of SNHG12 promoted expression of miR-218 and inhibited cell migration and invasion via the Slug/ZEB2 EMT signaling pathway.
Conclusion
In conclusion, the results of the present study indicated that SNHG12 was overexpressed in NSCLC, and is associated with tumor progression and poor prognosis. Knockdown of SNHG12 could directly increase the expression level of miR-218 and thus effectively suppressed NSCLC cell proliferation, migration and invasion by inhibiting the EMT process via the Slug/ZEB2 signaling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HZ and YW conceived and designed the study. SL and YY acquired and analyzed the data. YW and YS interpreted the data and wrote the manuscript. HZ and YW critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethic Review Committees of The Affiliated Hospital of Hebei University and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Grønberg BH, Lund-Iversen M, Strøm EH, Brustugun OT, Scott H. Associations between TS, TTF-1, FR-α, FPGS, and overall survival in patients with advanced non-small-cell lung cancer receiving pemetrexed plus carboplatin or gemcitabine plus carboplatin as first-line chemotherapy. J Thorac Oncol. 2013;8:1255–1264. doi: 10.1097/JTO.0b013e3182a406a3. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heist RS, Engelman JA. SnapShot: Non-small cell lung cancer. Cancer Cell. 2012;21:448.e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Song L, Liu X, Yang X, Li X, He T, Wang N, Yang S, Yu C, Yin T, et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 7.Mohammad IS, He W, Yin L. Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed Pharmacother. 2018;100:335–348. doi: 10.1016/j.biopha.2018.02.038. [DOI] [PubMed] [Google Scholar]
- 8.Song L, Liang X, Yang S, Wang N, He T, Wang Y, Zhang L, Wu Q, Gong C. Novel polyethyleneimine-R8-heparin nanogel for high-efficiency gene delivery in vitro and in vivo. Drug Deliv. 2018;25:122–131. doi: 10.1080/10717544.2017.1417512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrini D, Panagiotopoulos N, Bedetti B, Mitsos S, Crisci R, Solli P, Bertolaccini L, Scarci M. Surgical approach in oligometastatic non-small cell lung cancer. Ann Transl Med. 2018;6:93. doi: 10.21037/atm.2018.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng H, Perez-Soler R. Leptomeningeal metastases in non-small-cell lung cancer. Lancet Oncol. 2018;19:e43–e55. doi: 10.1016/S1470-2045(17)30689-7. [DOI] [PubMed] [Google Scholar]
- 11.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 13.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 14.Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925–1936. doi: 10.18632/oncotarget.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016;37:4065–4073. doi: 10.1007/s13277-015-4256-7. [DOI] [PubMed] [Google Scholar]
- 18.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 20.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Ji G, Ke X, Gu H, Jin W, Zhang G. MiR-141 inhibits gastric cancer PROLIFERATION BY interacting with long noncoding RNA MEG3 and down-regulating E2F3 expression. Dig Dis Sci. 2015;60:3271–3282. doi: 10.1007/s10620-015-3782-x. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Li J, Jia S, Wu M, An J, Zheng Q, Zhang W, Lu D. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget. 2015;6:31958–31984. doi: 10.18632/oncotarget.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8:84086–84101. doi: 10.18632/oncotarget.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T, Li J, Dong Y, Zhai D, Lai L, Dai F, Deng H, Chen Y, Liu M, Yi Z. Cucurbitacin E inhibits breast tumor metastasis by suppressing cell migration and invasion. Breast Cancer Res Treat. 2012;135:445–458. doi: 10.1007/s10549-012-2175-5. [DOI] [PubMed] [Google Scholar]
- 29.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan X, Banerjee P, Guo HF, Ireland S, Pankova D, Ahn YH, Nikolaidis IM, Liu X, Zhao Y, Xue Y, et al. Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J Clin Invest. 2017;127:117–131. doi: 10.1172/JCI88736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 32.Kraljevic Pavelic S, Sedic M, Bosnjak H, Spaventi S, Pavelic K. Metastasis: New perspectives on an old problem. Mol Cancer. 2011;10:22. doi: 10.1186/1476-4598-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Li F, Zheng C, Wang Y, Fang Z, Guo C, Wang X, Liu H, Deng L, Li C, et al. NEDD9 promotes lung cancer metastasis through epithelial-mesenchymal transition. Int J Cancer. 2014;134:2294–2304. doi: 10.1002/ijc.28568. [DOI] [PubMed] [Google Scholar]
- 34.Mallini P, Lennard T, Kirby J, Meeson A. Epithelial-to-mesenchymal transition: What is the impact on breast cancer stem cells and drug resistance. Cancer Treat Rev. 2014;40:341–348. doi: 10.1016/j.ctrv.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Nagaraju GP, Long TE, Park W, Landry JC, Taliaferro-Smith L, Farris AB, Diaz R, El-Rayes BF. Heat shock protein 90 promotes epithelial to mesenchymal transition, invasion, and migration in colorectal cancer. Mol Carcinog. 2015;54:1147–1158. doi: 10.1002/mc.22185. [DOI] [PubMed] [Google Scholar]
- 36.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.