CovR/CovS is a two-component regulatory system in group A Streptococcus (GAS). The D53 residue of CovR is phosphorylated by CovS, and the phosphorylated CovRD53 binds to the rgg-speB intergenic region and acts as the transcriptional repressor. Nonetheless, the transcription of rgg and Rgg-controlled speB is upregulated in the covR mutant but inhibited in the covS mutant. The present study showed that nonphosphorylated CovRD53 protein inhibits rgg and speB transcription in the presence of the phosphorylated CovRD53 in vivo, indicating that nonphosphorylated CovRD53 has a dominant role in suppressing rgg transcription. These results reveal the roles of nonphosphorylated CovRD53 in regulating rgg transcription, which could contribute significantly to invasive phenotypes of covS mutants.
KEYWORDS: CovR/CovS, Rgg, SpeB, group A Streptococcus
ABSTRACT
CovR/CovS is a two-component regulatory system in group A Streptococcus and primarily acts as a transcriptional repressor. The D53 residue of CovR (CovRD53) is phosphorylated by the sensor kinase CovS, and the phosphorylated CovRD53 protein binds to the intergenic region of rgg-speB to inhibit speB transcription. Nonetheless, the transcription of rgg and speB is suppressed in covS mutants. The T65 residue of CovR is phosphorylated in a CovS-independent manner, and phosphorylation at the D53 and T65 residues of CovR is mutually exclusive. Therefore, how phosphorylation at the D53 and T65 residues of CovR contributes to the regulation of rgg and speB expression was elucidated. The transcription of rgg and speB was suppressed in the strain that cannot phosphorylate the D53 residue of CovR (CovRD53A mutant) but restored to levels similar to those of the wild-type strain in the CovRT65A mutant. Nonetheless, inactivation of the T65 residue phosphorylation in the CovRD53A mutant cannot derepress the rgg and speB transcription, indicating that phosphorylation at the T65 residue of CovR is not required for repressing rgg and speB transcription. Furthermore, trans complementation of the CovRD53A protein in the strain that expresses the phosphorylated CovRD53 resulted in the repression of rgg and speB transcription. Unlike the direct binding of the phosphorylated CovRD53 protein and its inhibition of speB transcription demonstrated previously, the present study showed that inactivation of phosphorylation at the D53 residue of CovR contributes dominantly in suppressing rgg and speB transcription.
IMPORTANCE CovR/CovS is a two-component regulatory system in group A Streptococcus (GAS). The D53 residue of CovR is phosphorylated by CovS, and the phosphorylated CovRD53 binds to the rgg-speB intergenic region and acts as the transcriptional repressor. Nonetheless, the transcription of rgg and Rgg-controlled speB is upregulated in the covR mutant but inhibited in the covS mutant. The present study showed that nonphosphorylated CovRD53 protein inhibits rgg and speB transcription in the presence of the phosphorylated CovRD53 in vivo, indicating that nonphosphorylated CovRD53 has a dominant role in suppressing rgg transcription. These results reveal the roles of nonphosphorylated CovRD53 in regulating rgg transcription, which could contribute significantly to invasive phenotypes of covS mutants.
INTRODUCTION
Streptococcus pyogenes (group A Streptococcus [GAS]) is a Gram-positive human pathogen causing diseases from mild pharyngitis and tonsillitis to severe cellulitis, necrotizing fasciitis, and toxic shock syndrome. An increase in invasive GAS infections has been reported globally (1–6) and is associated with high mortality and economic burden (7, 8). Therefore, there is an urgent need to understand the pathogenesis of invasive GAS infections.
Mutations in covS and covR genes are detected more frequently in strains isolated from patients with severe manifestations than from patients with mild symptoms (9–12). CovR/CovS is a two-component regulatory system of GAS (13). CovS acts as a membrane-associated sensor kinase/phosphatase to modulate the phosphorylation levels of the intracellular response regulator CovR (14, 15). There are two residues of CovR that can be phosphorylated. The phosphorylation level of the D53 residue of CovR (CovRD53) is modulated by CovS; however, the T65 residue of CovR (CovRT65) is phosphorylated by a CovS-independent mechanism (16). In addition, in vitro analysis showed that phosphorylation of these two residues in the recombinant CovR protein is mutually exclusive (16). Inactivation of CovS results in decreased levels of phosphorylated CovRD53 and the derepression of CovR-regulated virulence factors, including the hyaluronic acid capsule synthesis (has operon), streptolysin S (sls), streptolysin O (slo), NADase (nga), and DNase (sda1) (14, 17–19). In addition, although CovS is considered to regulate target genes solely through CovR (20), a strain that expresses nonphosphorylated CovRD53 is more virulent than the covR deletion strain in a mouse infection model (16), suggesting that nonphosphorylated CovRD53 in the covS mutant still has important roles in GAS pathogenesis. Nonetheless, how CovR contributes to GAS pathogenesis in the absence of CovS is still not clear.
Rgg is a positive regulator of the cysteine protease SpeB (21, 22). The emm3-type isolates with mutations in rgg express a higher level of virulence factors, such as slo, nga, and ska, than do isolates with intact rgg (11). In the emm49-type NZ131 strain, Rgg acts as a transcriptional repressor of genes that are involved in virulence, metabolism, and oxidative stress responses (23–25). Both rgg mutants of emm3- and emm49-type strains showed higher lethality in the mouse infection models than did wild-type strains (11, 23), indicating that inactivation of Rgg contributes to the increase of bacterial virulence.
The phosphorylated CovRD53 protein has better DNA-binding activity than the nonphosphorylated CovRD53 protein (26–28). The phosphorylated CovRD53 protein binds to the rgg-speB intergenic region to inhibit speB transcription (28–30). Nonetheless, the transcription of rgg is repressed in the covS mutant and therefore resulted in the inactivation of speB transcription (19, 30). These results indicate that the covS mutant and covR mutant are not identical. In addition, unlike the direct binding of the phosphorylated CovRD53 protein and its inhibition of speB transcription, which have been extensively demonstrated previously, whether nonphosphorylated CovRD53 is capable of repressing rgg transcription and subsequently preventing SpeB production has not been verified.
Phosphorylation of the CovR D53 and T65 residues is mutually exclusive (16). Inactivation of the CovR protein phosphorylation by a D53A or T65A amino acid substitution does not result in aberrantly folded protein in vitro (16). Nonetheless, the CovRT65A mutant was hypervirulent compared to the wild-type strain (16), suggesting that the phosphorylated CovRT65 protein contributes significantly to the regulatory networks of GAS. The present study aims to analyze how phosphorylated CovRD53 and CovRT65 proteins mediate the repression of rgg and speB transcription. The results showed that phosphorylation at the T65 residue of CovR was not required for suppressing rgg and speB transcription. In addition, the nonphosphorylated CovRD53 had the more dominant roles in inhibiting rgg and speB transcription than did the phosphorylated CovRD53 protein.
RESULTS
Transcription of rgg was inhibited in the CovRD53A mutant but derepressed in the CovRT65A mutant.
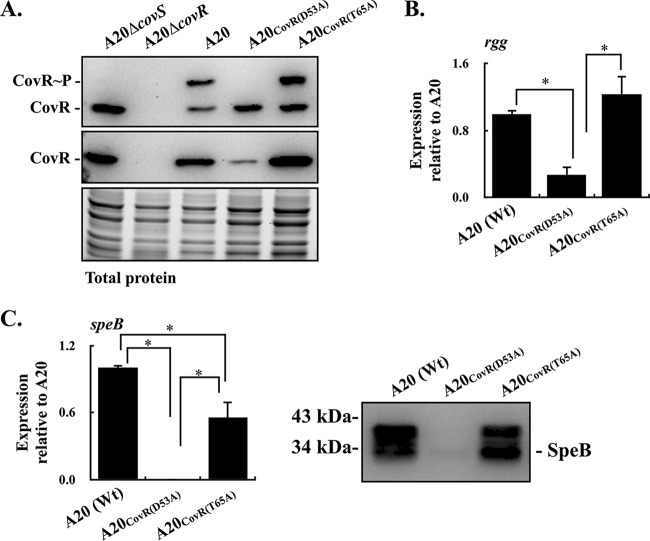
Phosphorylation of CovRD53 promotes its dimerization and increases its activity to bind to the rgg-speB intergenic region (28). CovR acts as a transcriptional repressor of rgg; however, the deletion of covS and inactivation of the D53 residue phosphorylation of CovR result in the repression of rgg transcription (19, 30), suggesting that the repression is unrelated to the phosphorylation at D53. In addition to the D53 residue, the T65 residue of CovR is phosphorylated by a CovS-independent mechanism (16). Horstmann et al. (16) showed that phosphorylation at the D53 and T65 residues of CovR is mutually exclusive; therefore, the phosphorylated CovRT65 could mediate the repression of rgg in the covS mutant. In order to elucidate whether the phosphorylated CovRT65 is involved in the repression of rgg transcription, the isoallelic GAS strains that were unable to be phosphorylated at D53 [A20CovR(D53A) mutant] and T65 [A20CovR(T65A) mutant] were constructed, and the levels of CovR protein expression and phosphorylation in these mutants were analyzed by Western blotting and Phos-tag Western blotting, respectively. The results showed that the expression of CovR protein in the CovRD53A mutant was decreased compared to those in the wild-type A20 strain and CovRT65A mutant (Fig. 1A). The phosphorylated CovRD53 protein was detected in the wild-type A20 strain and its CovRT65A mutant (Fig. 1A). Nonetheless, the phosphorylated CovRT65 protein cannot be observed in the covS and CovRD53A mutants (Fig. 1A), suggesting that low levels of phosphorylated CovRT65 were presented in those mutants or that the phosphorylated CovRT65 cannot be detected by Phos-tag Western blotting effectively. Next, the transcription levels of rgg in the wild-type A20 strain, A20CovR(D53A) mutant, and A20CovR(T65A) mutant were analyzed. Compared to the wild-type A20 strain, the transcription of rgg was inhibited in the A20CovR(D53A) mutant (Fig. 1B). Nonetheless, the transcription of rgg in the A20CovR(T65A) mutant was restored to a level similar to that in the wild-type A20 strain (Fig. 1B).
FIG 1.
Phosphorylation levels of CovR and the expression of CovR, rgg, and speB in the wild-type A20 strain (emm1 type) and its CovRD53A and CovRT65A mutants. Bacterial strains were cultured to the exponential (for detecting CovR) and stationary (for detecting rgg and speB) phases of growth. RNAs, culture supernatants, and total proteins were collected for quantitative real-time PCR (RT-qPCR), Western blotting, and Phos-tag Western blot analyses. (A) Levels of phosphorylation and expression of CovR protein in A20, CovRD53A mutant, and CovRT65A mutant. CovR∼P, phosphorylated CovR; CovR, nonphosphorylated CovR. Total protein serves as the loading control. (B) Transcription of rgg in A20, CovRD53A mutant, and CovRT65A mutant. (C) The transcription of speB and secretion of SpeB protein in A20, CovRD53A mutant, and CovRT65A mutant. Biological replicate experiments were performed using three independent preparations. The expression of rgg and speB was normalized to that of gyrA. Wt, wild-type strain; *, P < 0.05.
Rgg is the positive regulator of streptococcal pyrogenic exotoxin B (SpeB) (21). Therefore, whether the expression of SpeB was derepressed in the A20CovR(T65A) mutant was further analyzed with quantitative PCR (qPCR) and Western blotting. The results showed that the transcription of speB was partially restored in the A20CovR(T65A) mutant compared to the A20CovR(D53A) mutant (Fig. 1C). Furthermore, both the zymogen (42 kDa) and mature (28 kDa) forms of SpeB proteins were detected in the supernatants from the wild-type A20 strain and A20CovR(T65A) mutant (Fig. 1C). These results support the idea that the nonphosphorylated CovRD53 acts as the repressor for rgg transcription; in addition, the phosphorylated CovRT65 may also participate in the regulation of rgg transcription.
Inactivation of CovRD53 phosphorylation in the emm49-type NZ131 strain inhibits rgg and rgg-controlled speB transcription.
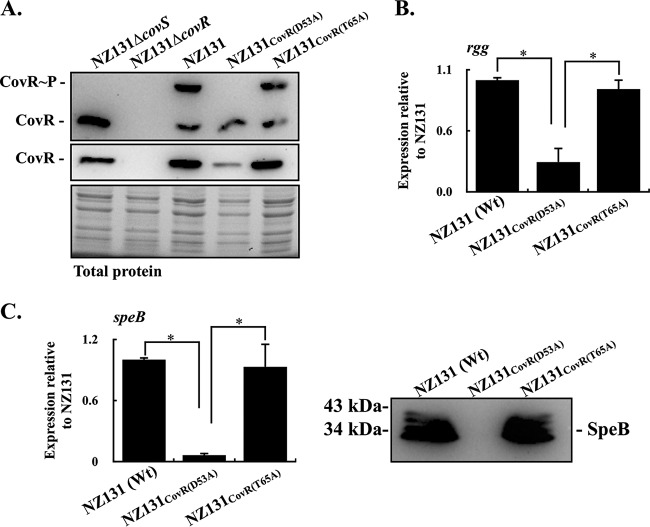
The results shown in Fig. 1 indicate that the transcription of speB was inhibited in the A20CovR(D53A) mutant but derepressed in the A20CovR(T65A) mutant. Nonetheless, in the emm3-type strain, the transcription of speB was derepressed both in CovRD53A and CovRT65A mutants (16). The emm3-type strain is rarely found in Taiwan. Therefore, to elucidate how CovRD53A and CovRT65A mutations influence the transcription of rgg and speB, the CovRD53A and CovRT65A mutants of another reference strain, NZ131 (emm49 type), were constructed, and the transcription of rgg and speB was analyzed. The levels of expression and phosphorylation of CovR proteins in NZ131 and its covR mutants were similar to those observed in A20 and its covR mutants (Fig. 2A). Compared to the wild-type NZ131 strain, the transcription of rgg and speB was repressed in the NZ131CovR(D53A) mutant but derepressed in the NZ131CovR(T65A) mutant (Fig. 2B and C). These results further support the idea that the nonphosphorylated CovRD53 or phosphorylated CovRT65 could mediate the repression of rgg and speB transcription.
FIG 2.
Phosphorylation levels of CovR and the expression of CovR, rgg, and speB in the wild-type NZ131 strain (emm49 type) and its CovRD53A and CovRT65A mutants. Bacterial strains were cultured to the exponential (for detecting CovR) and stationary (for detecting rgg and speB) phases of growth. RNAs, culture supernatants, and total proteins were collected for RT-qPCR, Western blotting, and Phos-tag Western blot analyses. (A) Levels of phosphorylation and expression of CovR protein in NZ131, CovRD53A mutant, and CovRT65A mutant. CovR∼P, phosphorylated CovR; CovR, nonphosphorylated CovR. Total protein serves as the loading control. (B) Transcription of rgg in NZ131, CovRD53A mutant, and CovRT65A mutant. (C) Transcription of speB and secretion of SpeB protein in NZ131, CovRD53A mutant, and CovRT65A mutant. Biological replicate experiments were performed using three independent preparations. The expression of rgg and speB was normalized to that of gyrA. Wt, wild-type strain; *, P < 0.05.
Inactivation of CovRT65 phosphorylation in the CovRD53A mutant cannot derepress rgg and speB transcription.
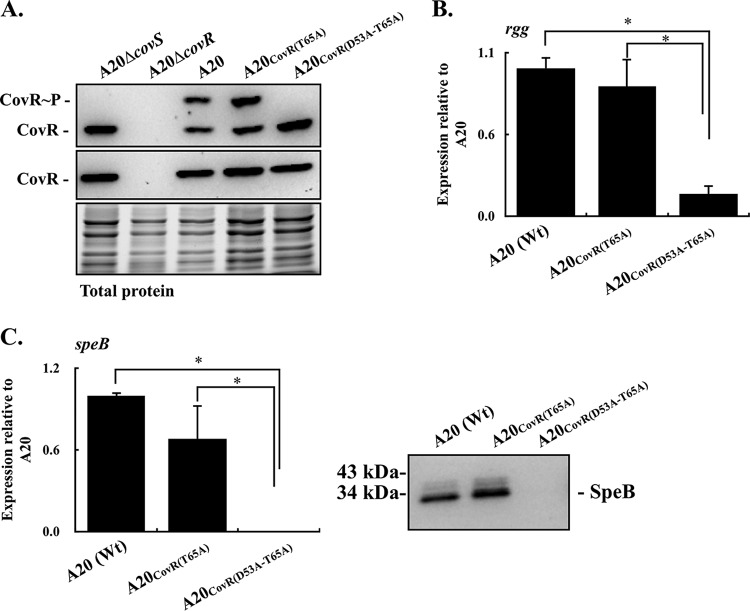
In order to further verify whether phosphorylation at the T65 residue of CovR is required for suppressing rgg and speB transcription, the T65A amino acid substitution of CovR in the A20CovR(D53A) mutant was constructed [A20CovR(D53A-T65A) mutant]. As expected, results from the Phos-tag Western blot analysis showed that the phosphorylated CovR proteins cannot be detected in the A20CovR(D53A-T65A) mutant (Fig. 3A). In addition, different from the A20 and its CovRD53A mutant, the expression level of CovR in CovRT65A and CovRD53A-T65A mutants was similar (Fig. 1A and 3A). Although the phosphorylated CovRT65 protein in the CovRD53A mutant cannot be detected by Phos-tag Western blotting, these results support the idea that the phosphorylated CovRT65 contributes to regulating CovR expression (16). In line with the results shown in Fig. 1B, the wild-type A20 strain and A20CovR(T65A) mutant had similar levels of rgg transcription (Fig. 3B). Nonetheless, the transcription of rgg was still suppressed in the A20CovR(D53A-T65A) mutant (Fig. 3B). In addition, the expression of SpeB was inhibited both in the transcriptional and translational levels in A20CovR(D53A-T65A) mutant compared to the wild-type A20 strain and A20CovR(T65A) mutant (Fig. 3C). These results indicate that phosphorylation at the T65 residue of CovR is not required for repression of rgg and speB transcription.
FIG 3.
Phosphorylation levels of CovR and the expression of CovR, rgg, and speB in emm1-type A20 and its CovRT65A and CovRD53A-T65A mutants. Bacterial strains were cultured to the exponential (for detecting CovR) and stationary (for detecting rgg and speB) phases of growth. RNAs, culture supernatants, and total proteins were collected for RT-qPCR, Western blotting, and Phos-tag Western blot analyses. (A) Levels of phosphorylation and expression of CovR protein in A20, CovRD53A mutant, and CovRD53A-T65A mutant. CovR∼P, phosphorylated CovR; CovR, nonphosphorylated CovR. Total protein serves as the loading control. (B) Transcription of rgg in A20, CovRT65A mutant, and CovRD53A-T65A mutant. (C) Transcription of speB and secretion of SpeB protein in A20, CovRT65A mutant, and CovRD53A-T65A mutant. Biological replicate experiments were performed using three independent preparations. The expression of rgg and speB was normalized to that of gyrA. Wt, wild-type strain; *, P < 0.05.
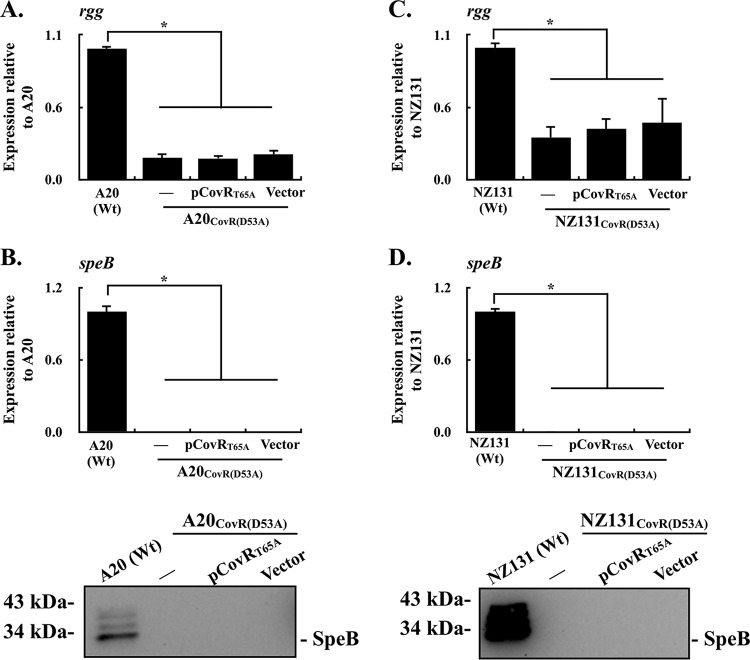
Expression of the CovRD53A protein in CovRT65A mutants inhibits rgg and speB transcription.
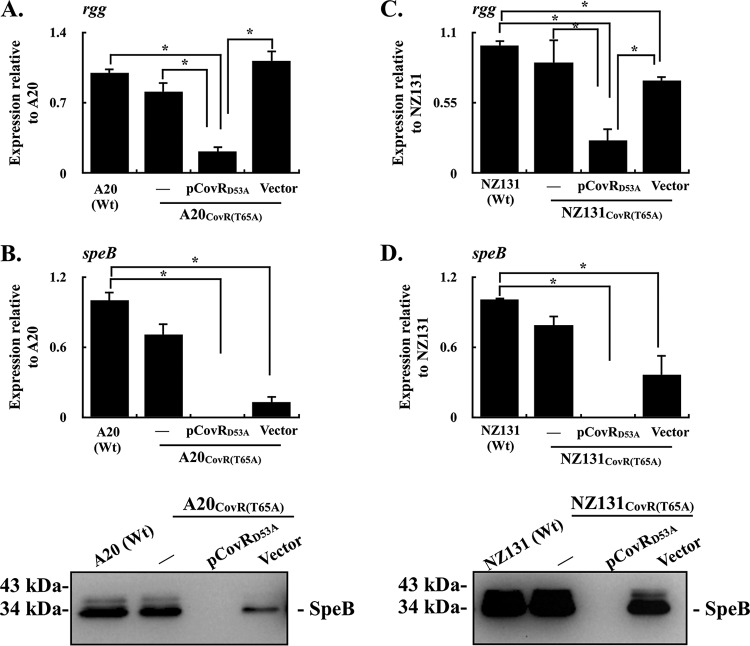
Phosphorylated CovRD53 has better DNA-binding activity than does the nonphosphorylated CovRD53 protein (28); therefore, whether the nonphosphorylated CovRD53 protein could inhibit rgg and speB transcription in the presence of phosphorylated CovRD53 was further verified. The covR gene encoding the D53A amino acid substitution was carried by the pTRKL2 vector and was electroporated into the strain that expresses the phosphorylated CovRD53 protein [A20CovR(T65A)] (see Fig. S1 in the supplemental material). The transcription of rgg in the A20CovR(T65A) mutant, its vector control strain, and the CovRD53A trans-complemented strain were further compared. The results showed that trans complementation of the CovRD53A protein in the A20CovR(T65A) mutant suppressed the transcription of rgg compared to the wild-type A20 and its vector control strains (Fig. 4A). Next, the expression levels of SpeB in the wild-type A20 strain, A20CovR(T65A) mutant, its vector control strain, and the CovRD53A trans-complemented strain were analyzed by qPCR and Western blotting. Compared to the wild-type strain, the expression of SpeB in both transcriptional and translational levels was decreased in the vector control strain (Fig. 4B). Nonetheless, the expression of SpeB cannot be detected in the CovRD53A trans-complemented strain (Fig. 4B). Furthermore, the transcription of rgg and speB was similarly analyzed in NZ131 strains. In line with results shown in the A20 strains, the transcription of rgg and speB was inhibited in the CovRD53A trans-complemented strain of NZ131CovR(T65A) (Fig. 4C and D). These results suggest that the nonphosphorylated CovRD53A protein has a more dominant role in repressing the transcription of rgg and speB than does the phosphorylated CovRD53 protein.
FIG 4.
Expression of rgg and speB in CovRT65A mutants and their vector control and CovRD53A trans-complementary strains. Bacterial strains were cultured to the stationary phase of growth. RNAs and culture supernatants were collected for RT-qPCR and Western blot analyses. (A and B) Transcription of rgg and the transcriptional and translational levels of SpeB expression in the emm1-type A20 strain, CovRT65A mutant, and its vector control (Vector) and CovRD53A trans-complementary (pCovRD53A) strains. (C and D) Transcription of rgg and the transcriptional and translational levels of SpeB expression in the emm49-type NZ131 strain, CovRT65A mutant, and its vector control and CovRD53A trans-complementary strains. Biological replicate experiments were performed using three independent preparations. The expression of rgg and speB was normalized to that of gyrA. Wt, wild-type strain; *, P < 0.05.
Expression of phosphorylated CovRD53 protein in CovRD53A mutants cannot restore rgg and speB transcription.
The transcription of speB is repressed in the covS mutant, suggesting that CovS acts as a positive regulator of speB (20, 31). In addition, it has been considered that CovS regulates downstream genes solely through CovR and is required for the CovR phosphorylation (20). Therefore, whether the phosphorylated CovRD53 could activate the rgg and speB transcription in A20CovR(D53A) was analyzed. The covR gene with the T65A amino acid substitution was carried by the pTRKL2 vector and was electroporated into CovRD53A mutants. The D53 residue of the CovRT65A protein can be phosphorylated by CovS, and expression of the phosphorylated CovRT65A protein in CovRD53A mutants was confirmed by Phos-tag Western blotting (Fig. 1; Fig. S1). Next, the transcription of rgg in the A20CovR(D53A) mutant, its vector-control strain, and the CovRT65A trans-complemented strain were further compared. The results showed that expression of the CovRT65A protein in the A20CovR(D53A) mutant had no effect on activating rgg and speB transcription (Fig. 5A and B). In addition, expression of the SpeB protein can only be detected in the wild-type A20 strain but not in A20CovR(D53A) and CovRT65A trans-complementary mutant strains (Fig. 5B). Furthermore, similar results were observed in NZ131 strains (Fig. 5C and D). These results indicate that the phosphorylated CovRD53 cannot attenuate the repressor activity of the CovRD53A protein to rgg and speB transcription.
FIG 5.
Expression of rgg and speB in CovRD53A mutants and their vector control and CovRT65A trans-complementary strains. Bacterial strains were cultured to the stationary phase of growth. RNAs and culture supernatants were collected for RT-qPCR and Western blot analyses. (A and B) Transcription of rgg and the transcriptional and translational levels of SpeB expression in the emm1-type A20 strain, CovRD53A mutant, and its vector control (Vector) and CovRT65A trans-complementary (pCovRT65A) strains. (C and D) Transcription of rgg and the transcriptional and translational levels of SpeB expression in the emm49-type NZ131 strain, CovRD53A mutant, and its vector control and CovRT65A trans-complementary strains. Biological replicate experiments were performed using three independent preparations. The expression of rgg and speB was normalized to that of gyrA. Wt, wild-type strain; *, P < 0.05.
DISCUSSION
The phosphorylated CovRD53 protein has better activity for binding to the rgg-speB intergenic region than does nonphosphorylated CovRD53 (28). In addition, the transcription of rgg and speB is upregulated in the covR mutant, suggesting that CovR inhibits rgg and speB transcription. Nonetheless, the transcription of speB is repressed in the covS mutant. In the present study, how phosphorylated and nonphosphorylated CovRD53 proteins contribute to regulating rgg and speB expression was elucidated. The results showed that the nonphosphorylated CovRD53 protein has a more dominant role in suppressing rgg and speB transcription than does the phosphorylated CovRD53 protein. Furthermore, inactivation of the phosphorylation at D53 but not the T65 residue of CovR is crucial for repressing rgg and speB transcription.
The D53 (CovRD53) and T65 (CovRT65) residues of CovR can be phosphorylated, and the phosphorylation at these two residues is mutually exclusive (16). CovRD53 is phosphorylated by CovS; however, CovRT65 is phosphorylated by the serine/threonine kinase (16). The strain unable to be phosphorylated at CovRT65 was hypervirulent compared to the wild-type strain (16), suggesting that the phosphorylated CovRT65 could have an important role in modulating the regulatory activity of CovR. The present study showed that the transcription of rgg and speB was repressed in strains unable to be phosphorylated at CovRD53 (CovRD53A mutants) but restored to levels similar to the wild-type strains in CovRT65A mutants (Fig. 1B and C and 2B and C), suggesting that phosphorylation at CovRT65 could contribute to the repression of rgg and speB transcription. Nonetheless, inactivation of CovRT65 phosphorylation in the CovRD53A mutant cannot derepress rgg and speB transcription (Fig. 3B and C). Although these results indicate that phosphorylation at the CovRT65 is not required for inhibiting rgg and speB transcription, whether the phosphorylated CovRT65 contributes to regulating other regulatory systems still needed to be verified.
The transcription of covR is negatively regulated by the CovR protein (28, 32). Compared to the wild-type A20 strain, the transcription of covR increased 2.95-fold ± 0.23-fold in its CovRD53A mutant. Nonetheless, the decreased expression of the CovR protein was observed in the CovRD53A mutants compared to that in the wild-type A20 and NZ131 strains (Fig. 1A and 2A). In addition, we also found that the expression level of CovR protein in the CovRD53A mutants was lower than that in the CovRT65A mutants (Fig. 1A and 2A). The underlying mechanisms of the decreased CovR protein in the CovRD53A mutants remain to be further studied; however, our results showed that the transcription of rgg and speB was not repressed in the CovRT65A mutants even though those mutants had higher levels of CovR protein than the CovRD53A mutants (Fig. 1 and 2). In addition, the present study showed that trans complementation of nonphosphorylated CovRD53 in strains that express phosphorylated CovRD53 resulted in the repression of rgg and speB transcription (Fig. 4). These results support the idea that the phosphorylated CovRD53 has minor effects on repressing rgg and speB transcription. Furthermore, it would be reasonable to suggest that the nonphosphorylated CovRD53 inhibits rgg and speB transcription through interacting with unknown regulatory systems but not through direct binding to the rgg-speB intergenic region.
In the emm3-type strain, the transcription levels of speB in covR deletion, CovRD53A, and CovRT65A mutants were all upregulated compared to those of the wild-type strain (16). The present study showed that trans complementation of phosphorylated CovRD53 in CovRD53A mutants cannot derepress rgg and speB transcription (Fig. 5), further supporting the notion that the CovR protein acts as the negative regulator for rgg and speB. Nonetheless, in the emm1- and emm49-type strains used in this study, the transcription of speB was repressed in the CovRD53A mutants but not in the CovRT65A mutants (Fig. 1C and 2C). In addition, trans complementation of the CovRD53A protein in strains that have the endogenous phosphorylated CovRD53 protein resulted in the repression of rgg and speB transcription (Fig. 4). These results indicate that the nonphosphorylated CovRD53 protein is crucial to mediate the repression of rgg and speB transcription. The different phenotypes observed in emm1-, emm49-, and emm3-type strains could be caused by the variations in the CovR/CovS phosphorelay signal among these strains (14). Nonetheless, how these variations influence the CovR/CovS regulatory activity to its target genes still needs to be further elucidated.
Rgg is required for triggering the expression of the cysteine protease SpeB (21, 33). In addition, inactivation of Rgg results in the increased resistance to oxidative stresses as well as the overproduction of bacterial virulence factors, such as DNase and streptolysin O (SLO) (11, 23, 34); these phenotypes may lead to the increase in bacterial virulence. Spontaneous mutations in rgg were also frequently found in clinical isolates from patients with invasive manifestations (11). Nonetheless, Friães et al. (17) showed that acquisition of the covS null allele but not the rgg null allele is associated with invasive infections. The present study showed that nonphosphorylated CovRD53 enhances its repression of rgg transcription (Fig. 1 to 4). These results suggest that inactivation of covS may not only derepress the expression of CovR-controlled genes but also upregulate the expression of Rgg-suppressed virulence genes. These phenomena may provide an explanation as to why the covS but not the rgg mutation is associated with invasive infections.
CovR/CovS is a two-component regulatory system; however, the covS and covR null mutants are not identical. The present study demonstrated that nonphosphorylated CovRD53 has a more dominant role in repressing rgg and speB transcription than does the phosphorylated CovRD53 protein. Whether the nonphosphorylated CovRD53 represses rgg transcription directly or indirectly through binding to the rgg-speB intergenic region still needs to be verified; however, these results reveal the potential role of CovR in regulating its controlled genes in invasive covS mutants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
GAS A20 (emm1 type) and NZ131 (emm49 type) were described previously (30). GAS strains (Table 1) were cultured on Trypticase soy agar with 5% sheep blood or in tryptic soy broth (Becton, Dickinson and Company, Sparks, MD) supplemented with 0.5% yeast extract (TSBY). Escherichia coli DH5α was purchased from Yeastern Biotech Co., Ltd. (Taipei, Taiwan) and was cultured in Luria-Bertani (LB) broth at 37°C with vigorous aeration. When appropriate, the antibiotics chloramphenicol (25 µg/ml and 3 µg/ml for E. coli and GAS, respectively), spectinomycin (100 µg/ml), and erythromycin (150 µg/ml and 5 µg/ml for E. coli and GAS, respectively) were used for selection.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Parent strain | Descriptiona | Reference or source |
|---|---|---|---|
| Plasmids | |||
| pTRKL2 | E. coli-Streptococcus shuttle vector | 30 | |
| pCN143 | Temperature-sensitive vector | 30 | |
| pMW506 | pSF152::covRΔcm | 30 | |
| pCN114 | pSF152::covSΔcm | This study | |
| pCN129 | pCN143::CovRD53A | 30 | |
| pCN160 | pCN143::covSΔcm | This study | |
| pCN167 | pCN143::CovRT65A (covR with A203G substitution) | This study | |
| pCN169 | pTRKL2::CovRD53A | This study | |
| pCN176 | pCN143::CovRD53A-T65A | This study | |
| pCN180 | pTRKL2::CovRT65A | This study | |
| Strains | |||
| A20 | emm1-type wild-type strain | 40 | |
| NZ131 | emm49-type wild-type strain | 40 | |
| A20 ΔcovS | A20 | A20 covS isogenic mutant | This study |
| A20 ΔcovR | A20 | A20 covR isogenic mutant | 30 |
| NZ131 ΔcovS | NZ131 | NZ131 covS isogenic mutant | This study |
| NZ131 ΔcovR | NZ131 | NZ131 covR isogenic mutant | This study |
| A20CovR(D53A) mutant | A20 | CovRD53A mutant strain | 30 |
| NZ131CovR(D53A) mutant | NZ131 | CovRD53A mutant strain | This study |
| A20CovR(T65A) mutant | A20 | CovRT65A mutant strain | This study |
| NZ131CovR(T65A) mutant | NZ131 | CovRT65A mutant strain | This study |
| A20CovR(D53A-T65A) mutant | A20 | CovRD53A and CovRT65A mutant strain | This study |
cm, chloramphenicol cassette.
DNA and RNA manipulations.
GAS genomic DNA extraction, RNA extraction, and reverse transcription were performed as described previously (35). Real-time PCR was performed in a 20-µl mixture containing 1 µl of cDNA, 0.8 µl of primers (10 µM), and 10 µl of SensiFAST SYBR Lo-ROX premixture (Bioline Ltd., London, UK), according to the manufacturer's instructions. Biological replicate experiments were performed from three independent RNA preparations in duplicate. The expression level of each target gene was normalized to that of gyrA and analyzed using the ΔΔCT method (7500 software version 2.0.5; Applied Biosystems, Thermo Fisher Scientific, Inc.). In addition, all values of control and experimental groups were divided by the mean of control samples before statistical analysis (36). Primers used for real-time PCR analysis (Table 2) were designed by Primer3 (version 0.4.0; http://frodo.wi.mit.edu) according to the MGAS5005 sequence (NCBI RefSeq accession no. NC_007297.1).
TABLE 2.
Primers used in this study
| Primer | Use | Sequence (5′ to 3′)a | Reference or source |
|---|---|---|---|
| covR-BamHI-F7 | Construction | GCGGGATCCAAATGACAAAGAAAATTTTA | This study |
| covR-BamHI-R6 | Construction | GCGGGATCCTAGGCACTTTCTTCTCAGAT | This study |
| covR-T65A-F | Construction | GATGGTTTTGAAGTGgCCCGTCGTTTGCAAACC | 16 |
| covR-T65A-R | Construction | GGTTTGCAAACGACGGGcCACTTCAAAACCATC | 16 |
| covS-F-2 | Construction | CGGGATCCTCGTGAAGGGTTAGAAACTG | This study |
| covS-R-2-2 | Construction | CCCCGGGCCATATGACTTATTTCTCACGAA | This study |
| covS-F-3-2 | Construction | CCCCGGGGGCCCAGTCTAAAGAGAGTTAGAG | This study |
| covS-R-3-2 | Construction | CCGGAATTCTTCGATTTCATCAACGTCCT | This study |
| Vec78_cat-F-1 | Construction | CGCCGCGTTAACGATAGATTTATGATATAG | This study |
| Vec78_cat-R-1 | Construction | CGCCGCGTTAACATTTATTCAGCAAGTCTT | This study |
| CovR/S-F-3 | Construction | GCGGATCCGCTTGCAAGGGTTGTTTGATG | 30 |
| CovR-R-5 | Construction | GCGGGATCCATGCAAGCCAGGAGATGATT | This study |
| Rgg-F-3 | qPCR | TTTGAATGCCGAAACATAGAAGGTT | 30 |
| Rgg-R-2 | qPCR | CTAATAACACCTTGACCAAGGCAAA | 30 |
| speB-F-2 | qPCR | TGCCTACAACAGCACTTTGG | 30 |
| speB-R-2 | qPCR | GGTAAAGTAGGCGGACATGC | 30 |
| gyrA-F-3 | qPCR | CGTCGTTTGACTGGTTTGG | 30 |
| gyrA-R-3 | qPCR | GGCGTGGGTTAGCGTATTTA | 30 |
Underlining indicates restriction enzyme sites; lowercase letters denote mutated nucleotides.
Construction of covR deletion, covS deletion, CovRD53A, CovRT65A, and CovRD53A-T65A mutants.
The covR gene of NZ131 was interrupted by pMW506 with a double-cross homologous recombination, as described previously (30). To construct the covS isogenic mutants, the 592-bp covS upstream region and 659-bp covS downstream region were amplified with primers covS-F-2, covS-R-2-2, covS-F-3-2, and covS-R-3-2 and ligated into the vector pSF152 (30) to generate pCN114 (Tables 1 and 2). The chloramphenicol cassette of vector 78 (37) was amplified using primers Vec78_cat-F-1 and Vec78_cat-R-1 and ligated into pCN114 with the SmaI site. The upstream and downstream regions of covS and the chloramphenicol cassette in pSF152 were amplified by primers covS-F-2 and covR-3-2 and ligated into TA Cloning vector (Yeastern Biotech Co., Ltd., Taipei, Taiwan) and then transferred to the temperature-sensitive vector pCN143 (30) with the BamHI site (pCN160, Table 1). The amino acid residue substitution of CovR protein was generated by overlap PCR, as described previously (30). The T65A amino acid substitution of CovR was generated by primers CovR-BamHI-F7, CovR-T65A-R, CovR-BamHI-R6, and CovR-T65A-F from the wild-type A20 strain and its CovRD53A mutant. PCR products were ligated into the TA Cloning vector (Yeastern Biotech Co., Ltd.), and the amino acid substitution was confirmed by Sanger sequencing. The mutated covR genes were transferred to pCN143 with the BamHI site. The constructed plasmids (pCN160, pCN167, and pCN176) were transfected into strain A20 or NZ131 by electroporation, as described previously (38), and cultured at 30°C with the spectinomycin selection. Transformants were then transferred to 37°C to force the plasmid integration. Finally, transformants in which plasmid excised from the chromosome via recombination were selected by chloramphenicol (for covS and covR mutants) or in the blood agar plate (for CovRD53A, CovRT65A, and CovRD53A-T65A mutants) at 30°C. The covS and covR deletion mutants and GAS strains with the D53A substitution or D53A and T65A substitutions of CovR were confirmed by Sanger sequencing. All primers used are shown in Table 2.
Construction of vector control and covR trans-complemented strains.
The E. coli-Streptococcus shuttle vector pTRKL2 was described previously (30). To construct covR trans-complemented strains, the genomic DNAs extracted from A20 CovRD53A and CovRT65A mutants served as the templates, and the covR gene, including its native promoter (1.3 kb), was amplified with primers CovR/S-F-3 and CovR-R-5 (Table 2). The PCR products were ligated into the BamHI site of pTRKL2, and constructed plasmids (pCN169 and pCN180) were transformed into CovRT65A or CovRD53A mutants with electroporation, as described previously (39).
Western blotting for SpeB and CovR proteins.
Bacteria were cultured in TSBY broth for 8 h, and the culture supernatant was collected and sterilized with a 0.22-µm-pore-size filter for detecting SpeB protein. Thirty microliters of bacterial culture supernatants was mixed with 6× protein loading dye and separated by 12% SDS-PAGE gels. In order to analyze the expression of CovR protein, bacteria that were grown to exponential phase (6 h of incubation) were collected, and total proteins were extracted by the bead beater (Mini-beadbeater; BioSpec Products, Inc., Bartlesville, OK) (30). Ten micrograms of total protein was subjected to analysis with 12% SDS-PAGE gels. Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA), and membranes were blocked by 5% skim milk in PBST buffer (PBS containing 0.2% Tween 20) at 37°C for 1 h. SpeB and CovR proteins were detected by the anti-SpeB antibody (Toxin Technology, Inc., Sarasota, FL) and anti-CovR serum (30), respectively. After hybridization, the membrane was washed with PBST buffer and hybridized with a secondary antibody, the peroxidase-conjugated goat anti-rabbit IgG (1:10,000 dilution; Cell Signaling Technology, Inc., Danvers, MA), at room temperature for 1 h. The blot was developed using Pierce ECL Western blotting substrate (Thermo Fisher Scientific, Inc., Rockford, IL), and the signal was detected using the Gel Doc XR+ system (Bio-Rad).
Phos-tag Western blotting.
Bacterial protein was extracted according to a previously described method (39). Ten micrograms of bacterial proteins was mixed with 6× protein loading dye (without boiling) and separated by 10% SDS-PAGE gels containing 10 µM Phos-tag (Wako Pure Chemical Industries Ltd., Richmond, VA) and 0.5 µM MnCl2. Phosphorylated and nonphosphorylated proteins were separated on Phos-tag SDS-PAGE gels for 120 to 140 min at 100 V at 4°C. Protein transfer, membrane blocking, hybridization, and signal detection were performed as described previously (39).
Statistical analysis.
Statistical analysis was performed by using the Prism software, version 5 (GraphPad, San Diego, CA). Significant differences in multiple groups were determined using analysis of variance (ANOVA). Posttest for ANOVA was analyzed by Tukey’s honestly significant difference test. A P value of <0.05 was taken as significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by parts of grants from Chang Gung Memorial Hospital, LinKou, Taiwan (grants CMRPD1G0381-3 and BMRPD19), and Ministry of Science and Technology, Taiwan (grant MOST 106-2320-B-182-023-MY3).
We appreciate the constructive comments from Shih-Tung Liu (Department of Microbiology and Immunology, Chang Gung University, Taiwan) for this work.
We affirm no conflicts of interest relative to any source of funding, sponsorship, or financial benefit.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00681-18.
REFERENCES
- 1.Seale AC, Davies MR, Anampiu K, Morpeth SC, Nyongesa S, Mwarumba S, Smeesters PR, Efstratiou A, Karugutu R, Mturi N, Williams TN, Scott JA, Kariuki S, Dougan G, Berkley JA. 2016. Invasive group A Streptococcus infection among children, rural Kenya. Emerg Infect Dis 22:224–232. doi: 10.3201/eid2202.151358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelthaler DM, Valentine M, Bowers J, Pistole J, Driebe EM, Terriquez J, Nienstadt L, Carroll M, Schumacher M, Ormsby ME, Brady S, Livar E, Yazzie D, Waddell V, Peoples M, Komatsu K, Keim P. 2016. Hypervirulent emm59 clone in invasive group A Streptococcus outbreak, southwestern United States. Emerg Infect Dis 22:734–738. doi: 10.3201/eid2204.151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athey TB, Teatero S, Sieswerda LE, Gubbay JB, Marchand-Austin A, Li A, Wasserscheid J, Dewar K, McGeer A, Williams D, Fittipaldi N. 2016. High incidence of invasive group A Streptococcus disease caused by strains of uncommon emm types in Thunder Bay, Ontario, Canada. J Clin Microbiol 54:83–92. doi: 10.1128/JCM.02201-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 5.Tse H, Bao JY, Davies MR, Maamary P, Tsoi HW, Tong AH, Ho TC, Lin CH, Gillen CM, Barnett TC, Chen JH, Lee M, Yam WC, Wong CK, Ong CL, Chan YW, Wu CW, Ng T, Lim WW, Tsang TH, Tse CW, Dougan G, Walker MJ, Lok S, Yuen KY. 2012. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J Infect Dis 206:341–351. doi: 10.1093/infdis/jis362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luk EY, Lo JY, Li AZ, Lau MC, Cheung TK, Wong AY, Wong MM, Wong CW, Chuang SK, Tsang T. 2012. Scarlet fever epidemic, Hong Kong, 2011. Emerg Infect Dis 18:1658–1661. doi: 10.3201/eid1810.111900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes GJ, Van Hoek AJ, Sriskandan S, Lamagni TL. 2015. The cost of hospital care for management of invasive group A streptococcal infections in England. Epidemiol Infect 143:1719–1730. doi: 10.1017/S0950268814002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens DL. 2000. Streptococcal toxic shock syndrome associated with necrotizing fasciitis. Annu Rev Med 51:271–288. doi: 10.1146/annurev.med.51.1.271. [DOI] [PubMed] [Google Scholar]
- 9.Lin JN, Chang LL, Lai CH, Lin HH, Chen YH. 2014. Association between polymorphisms in the csrRS two-component regulatory system and invasive group A streptococcal infection. Eur J Clin Microbiol Infect Dis 33:735–743. doi: 10.1007/s10096-013-2005-7. [DOI] [PubMed] [Google Scholar]
- 10.Flores AR, Sahasrabhojane P, Saldana M, Galloway-Pena J, Olsen RJ, Musser JM, Shelburne SA. 2014. Molecular characterization of an invasive phenotype of group A Streptococcus arising during human infection using whole genome sequencing of multiple isolates from the same patient. J Infect Dis 209:1520–1523. doi: 10.1093/infdis/jit674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. 2010. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog 6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ato M, Ikebe T, Kawabata H, Takemori T, Watanabe H. 2008. Incompetence of neutrophils to invasive group A Streptococcus is attributed to induction of plural virulence factors by dysfunction of a regulator. PLoS One 3:e3455. doi: 10.1371/journal.pone.0003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin JC, Wessels MR. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol 30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 14.Horstmann N, Sahasrabhojane P, Saldana M, Ajami NJ, Flores AR, Sumby P, Liu CG, Yao H, Su X, Thompson E, Shelburne SA. 2015. Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infect Immun 83:1068–1077. doi: 10.1128/IAI.02659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran-Winkler HJ, Love JF, Gryllos I, Wessels MR. 2011. Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog 7:e1002361. doi: 10.1371/journal.ppat.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horstmann N, Saldana M, Sahasrabhojane P, Yao H, Su X, Thompson E, Koller A, Shelburne SA. 2014. Dual-site phosphorylation of the control of virulence regulator impacts group A streptococcal global gene expression and pathogenesis. PLoS Pathog 10:e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friães A, Pato C, Melo-Cristino J, Ramirez M. 2015. Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci Rep 5:12057. doi: 10.1038/srep12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med 13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 19.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treviño J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely MN, Lyon WR, Runft DL, Caparon M. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol 185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaussee MS, Watson RO, Smoot JC, Musser JM. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect Immun 69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pulliainen AT, Hytonen J, Haataja S, Finne J. 2008. Deficiency of the Rgg regulator promotes H2O2 resistance, AhpCF-mediated H2O2 decomposition, and virulence in Streptococcus pyogenes. J Bacteriol 190:3225–3235. doi: 10.1128/JB.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaussee MA, Callegari EA, Chaussee MS. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J Bacteriol 186:7091–7099. doi: 10.1128/JB.186.21.7091-7099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaussee MS, Somerville GA, Reitzer L, Musser JM. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J Bacteriol 185:6016–6024. doi: 10.1128/JB.185.20.6016-6024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Federle MJ, Scott JR. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol Microbiol 43:1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Gusa AA, Scott JR, Churchward G. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J Biol Chem 280:38948–38956. doi: 10.1074/jbc.M506121200. [DOI] [PubMed] [Google Scholar]
- 28.Miller AA, Engleberg NC, DiRita VJ. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol Microbiol 40:976–990. doi: 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- 29.Heath A, DiRita VJ, Barg NL, Engleberg NC. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun 67:5298–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang-Ni C, Chu TP, Wu JJ, Chiu CH. 2016. Repression of Rgg but not upregulation of LacD.1 in emm1-type covS mutant mediates the SpeB repression in group A Streptococcus. Front Microbiol 7:1935. doi: 10.3389/fmicb.2016.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatsuno I, Okada R, Zhang Y, Isaka M, Hasegawa T. 2013. Partial loss of CovS function in Streptococcus pyogenes causes severe invasive disease. BMC Res Notes 6:126. doi: 10.1186/1756-0500-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gusa AA, Scott JR. 2005. The CovR response regulator of group A Streptococcus (GAS) acts directly to repress its own promoter. Mol Microbiol 56:1195–1207. doi: 10.1111/j.1365-2958.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 33.Lyon WR, Gibson CM, Caparon MG. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J 17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anbalagan S, Chaussee MS. 2013. Transcriptional regulation of a bacteriophage encoded extracellular DNase (Spd-3) by Rgg in Streptococcus pyogenes. PLoS One 8:e61312. doi: 10.1371/journal.pone.0061312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CH, Chiang-Ni C, Kuo HT, Zheng PX, Tsou CC, Wang S, Tsai PJ, Chuang WJ, Lin YS, Liu CC, Wu JJ. 2013. Peroxide responsive regulator PerR of group A Streptococcus is required for the expression of phage-associated DNase Sda1 under oxidative stress. PLoS One 8:e81882. doi: 10.1371/journal.pone.0081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valcu M, Valcu CM. 2011. Data transformation practices in biomedical sciences. Nat Methods 8:104–105. doi: 10.1038/nmeth0211-104. [DOI] [PubMed] [Google Scholar]
- 37.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2010. Oxidative stress and metal ions regulate a ferritin-like gene, dpr, in Streptococcus pyogenes. Int J Med Microbiol 300:259–264. doi: 10.1016/j.ijmm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Chiang-Ni C, Tsou CC, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2008. The transcriptional terminator sequences downstream of the covR gene terminate covR/S operon transcription to generate covR monocistronic transcripts in Streptococcus pyogenes. Gene 427:99–103. doi: 10.1016/j.gene.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Chiang-Ni C, Tseng HC, Hung CH, Chiu CH. 2017. Acidic stress enhances CovR/S-dependent gene repression through activation of the covR/S promoter in emm1-type group A Streptococcus. Int J Med Microbiol 307:329–339. doi: 10.1016/j.ijmm.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Chiang-Ni C, Zheng PX, Ho YR, Wu HM, Chuang WJ, Lin YS, Lin MT, Liu CC, Wu JJ. 2009. emm1/sequence type 28 strains of group A streptococci that express covR at early stationary phase are associated with increased growth and earlier SpeB secretion. J Clin Microbiol 47:3161–3169. doi: 10.1128/JCM.00202-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.