Bacterial phospholipases act as intracellular and extracellular enzymes promoting the destruction of phospholipid barriers and inflammation during infections. Identifying enzymes with a common mechanism of activation is an initial step in understanding structural and functional properties. These properties serve as critical information for the design of specific inhibitors to reduce enzymatic activity and ameliorate host cell death. In this study, we identify and verify cytotoxic PLA2 enzymes from several bacterial pathogens. Similar to the founding member of the group, ExoU, these enzymes share the property of ubiquitin-mediated activation. The identification and validation of potential toxins from multiple bacterial species provide additional proteins from which to derive structural insights that could lead to paninhibitors useful for treating a variety of infections.
KEYWORDS: ExoU, orthologous enzymes, phospholipase, ubiquitin
ABSTRACT
ExoU is a potent type III secretion system effector that is injected directly into mammalian cells by the opportunistic pathogen Pseudomonas aeruginosa. As a ubiquitin-activated phospholipase A2 (PLA2), ExoU exhibits cytotoxicity by cleaving membrane phospholipids, resulting in lysis of the host cells and inhibition of the innate immune response. Recently, ExoU has been established as a model protein for a group of ubiquitin-activated PLA2 enzymes encoded by a variety of bacteria. Bioinformatic analyses of homologous proteins is a powerful approach that can complement and enhance the overall understanding of protein structure and function. To conduct homology studies, it is important to have efficient and effective tools to screen and to validate the putative homologs of interest. Here we make use of an Escherichia coli-based dual expression system to screen putative ubiquitin-activated PLA2 enzymes from a variety of bacteria that are known to colonize humans and to cause human infections. The screen effectively identified multiple ubiquitin-activated phospholipases, which were validated using both biological and biochemical techniques. In this study, two new ExoU orthologs were identified and the ubiquitin activation of the rickettsial enzyme RP534 was verified. Conversely, ubiquitin was not found to regulate the activity of several other tested enzymes. Based on structural homology analyses, functional properties were predicted for AxoU, a unique member of the group expressed by Achromobacter xylosoxidans.
IMPORTANCE Bacterial phospholipases act as intracellular and extracellular enzymes promoting the destruction of phospholipid barriers and inflammation during infections. Identifying enzymes with a common mechanism of activation is an initial step in understanding structural and functional properties. These properties serve as critical information for the design of specific inhibitors to reduce enzymatic activity and ameliorate host cell death. In this study, we identify and verify cytotoxic PLA2 enzymes from several bacterial pathogens. Similar to the founding member of the group, ExoU, these enzymes share the property of ubiquitin-mediated activation. The identification and validation of potential toxins from multiple bacterial species provide additional proteins from which to derive structural insights that could lead to paninhibitors useful for treating a variety of infections.
INTRODUCTION
Patatins are a large family of lipid acyl hydrolase proteins that are expressed by both prokaryotic and eukaryotic organisms (1). The enzymatic domain is characterized by an α/β-hydrolase fold and serine and aspartate catalytic residues. Serine is located in the conserved hydrolase motif GXSXG, and aspartate is located within a DGX motif. Importantly, the patatins are considered promiscuous enzymes that are capable of cleaving a variety of phospholipid substrates (2, 3). Based on the presence of a patatin domain and catalytic dyad motifs, over 4,400 potential enzymes have been bioinformatically identified in sequenced bacterial genomes (4). Conservation of the patatin motif suggests that these enzymes may serve a fundamental function in a broad array of bacteria. The biological function of the patatins in bacteria is poorly understood, however.
Early bioinformatic information suggested that genes encoding patatin-like enzymes are overrepresented in bacterial pathogens (5). A subclass of proteins, including ExoU from Pseudomonas aeruginosa, possess a patatin domain that is responsible for catalytic activity and C-terminal extensions that are predicted to have a variety of functional attributes (5). ExoU serves as a model enzyme for this subgroup of larger patatins; it is a type III secretion system (T3SS) substrate that is injected directly into the cytoplasm of host cells and is well characterized both biologically as a toxin and biochemically as an A2 phospholipase. The C-terminal extension of ExoU consists of a linker domain (amino acid residues 480 to 581) and a four-helix bundle (6, 7) and is essential for phospholipase activity, despite being distal from the patatin domain. Subsequent studies have demonstrated that the four-helix bundle is required for ubiquitylation of ExoU at K178 in host cells (8). It also serves as a membrane localization/stabilization domain (9–11), binding to phosphatidylinositol 4,5-bisphosphate (PIP2) with high affinity (6). Binding to and cleavage of PIP2 at focal adhesion complexes in eukaryotic cells is postulated to be responsible for the observed cytoskeletal collapse at early time points during ExoU intoxication (12). The linker domain was recently shown to be important for binding to monoubiquitin, a cofactor required for enzymatic activity (13, 14).
In previous studies, we identified patatin-like enzymes, from a variety of bacteria, that contain C-terminal extensions. Of the 17 potential candidates, 3 ExoU orthologous enzymes from bacteria with different ecological niches (Photorhabdus asymbiotica, Burkholderia thailandensis, and Pseudomonas fluorescens) were tested for ubiquitin-activated phospholipase activity (15). All tested enzymes were catalytically active only in the presence of ubiquitin, indicating that the activation mechanism is conserved across multiple species of Gram-negative bacteria. Comparisons of different orthologs for toxicity in eukaryotic and prokaryotic systems, enhancement of enzymatic activity by PIP2, or binding to different liposome compositions, however, yielded no consistent pattern that correlated with pathogenic versus nonpathogenic or intracellular versus extracellular lifestyles of the bacteria containing the ExoU orthologs (15). These results suggest that the ubiquitin-activated patatins may have multiple biological roles and may differ structurally.
This study focuses on patatin-like enzymes from bacteria associated with human hosts, as either pathogens or opportunists (Table 1). By limiting the producing bacteria to a specific lifestyle, it was anticipated that the enzymes might share common properties important for colonization of mammalian tissues. A series of primary and secondary screens was used to demonstrate that enzymes from Aeromonas diversa, Rickettsia prowazekii, and Achromobacter xylosoxidans possess phospholipase activity regulated by a noncovalent interaction with human monoubiquitin. A cloned ortholog from Legionella longbeachae (LlbU) was shown to possess minor phospholipase activity in the presence or absence of human monoubiquitin. While requiring ubiquitin for activity, the enzyme from Achromobacter xylosoxidans is predicted to possess a structural organization that differs significantly from that of ExoU. Ultimately, understanding both the similarities and the differences of this group of enzymes may provide insights into the activation mechanism and ideally will lead to the development of specific inhibitors that may be applicable to a diverse set of bacterial pathogens, as therapeutic agents.
TABLE 1.
Orthologs of P. aeruginosa ExoU used in this study
| Organism | No. of amino acid residues | % identitya | GenBank accession no. | Reference |
|---|---|---|---|---|
| Pseudomonas aeruginosa PA103 | 687 | WP_003134060.1 (ExoU) | Finck-Barbançon et al. (40) | |
| Achromobacter xylosoxidans MCW clinical isolate GN008 | 595 | 21.9 | MK248684 (AxoU) | NH44784-1996 is used as a reference sequence (41) |
| Aeromonas diversa ATCC 43946 | 669 | 47.8 | MK249729 (AdiU) | Farfan et al. (42) |
| Legionella longbeachae JV595 (ATCC 33462)b | 652 | 20.1 | WP_003634365.1 (LlbU) | Leggieri et al. (43) |
| Legionella pneumophila Philadelphia 1 LP02b | 665 | 20.1 | WP_010948114.1 (VpdA) | Chien et al. (44) |
| Rickettsia prowazekii Madrid E | 598 | 20.2 | WP_004599065.1 (RP534) | Housley et al. (29) |
| Vibrio vulnificus CMCP6c | 779 | 19 | WP_011079573.1 (VvuU) | Testa et al. (45) |
Biology WorkBench ALIGN, for optimal global alignment of two protein sequences.
Legionella genomic DNA was generously provided by Joseph Vogel, Washington University.
V. vulnificus strain CMCP6 was generously provided by James Oliver, University of North Carolina at Charlotte.
RESULTS
Ortholog sequence alignments and homology.
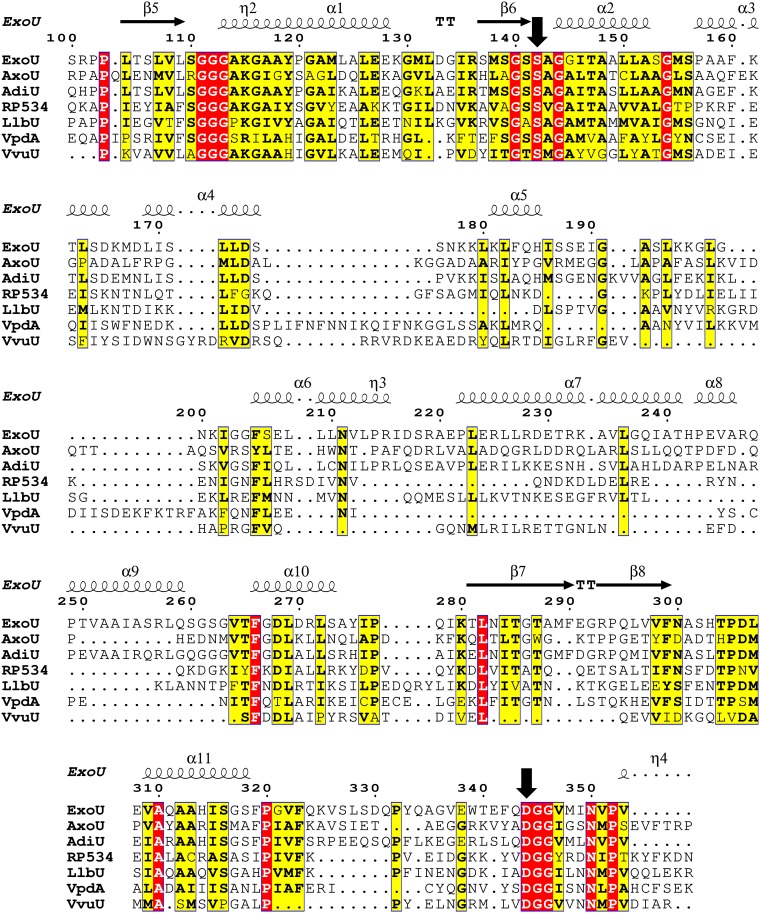
Previously studied enzymes from P. asymbiotica, B. thailandensis, and P. fluorescens were queried for ubiquitin activation, based on their significant sequence homology (∼35% to 50%) to ExoU (15). The open reading frames encode amino acid sequences that are highly homologous, but the bacteria potentially producing each ortholog inhabit different environments. While the enzymes shared ubiquitin as a common cofactor, they differed in a number of biochemical properties, suggesting that their biological roles might be divergent. In this study, candidate enzymes were queried based on the criteria that they might play roles in colonizing human hosts. Generally, the selected enzymes shared significantly less homology to ExoU (∼20% amino acid identity), except for a protein assembled from 2 contigs in Aeromonas diversa (∼48% identity) (Table 1). Despite the low sequence homology, alignments of the catalytic domains revealed the highly conserved GXSXG and DGX motifs that are present in all patatin-like phospholipases (Fig. 1). Additionally, there was a conserved GGGX(K/R) motif that provides the oxyanion hole necessary for catalysis. The only other region of significant homology mapped to a conserved β-hairpin and helix preceding the catalytic aspartate (residues 280 to 323) (Fig. 1). Outside these regions and the catalytic domain, sequence homology and alignment confidence decreased significantly. Identifying which, if any, of these orthologs are activated by ubiquitin may provide insight into the activation mechanism.
FIG 1.
Protein sequence alignment of the patatin domain for each ExoU ortholog. Red boxes indicate identical residues for all enzymes, while yellow boxes indicate similar or identical residues for a significant portion of the orthologous enzymes. Alignments reveal conserved motifs surrounding the catalytic amino acids (downward arrows) but show strong divergence elsewhere. Standard nomenclature for protein secondary structure is used; η indicates a 310 helix, α indicates an α-helix, β indicates a β-sheet, and TT indicates a tight turn.
Bacterial cytotoxicity as an indicator of ubiquitin interactions.
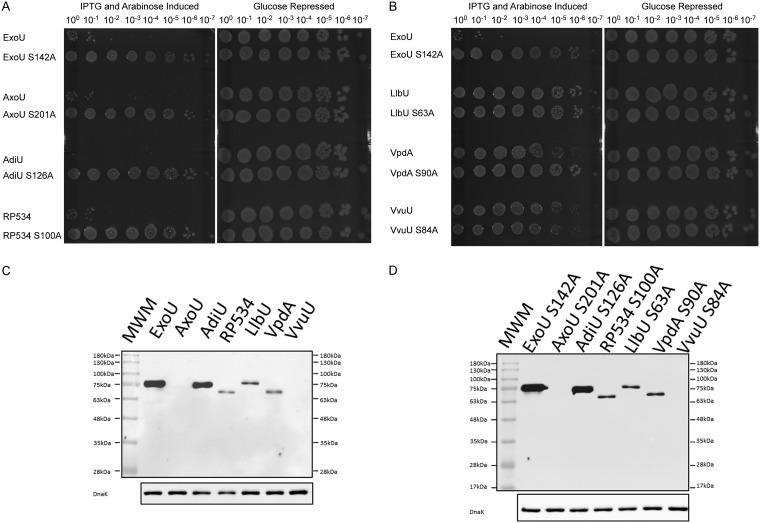
To determine whether ubiquitin plays a role in activation in the presence of biological membranes, we screened for toxicity in an expression system utilizing independent induction of the putative toxin and monoubiquitin in Escherichia coli. ExoU, ExoU orthologs, or predicted noncatalytic control constructs were expressed in E. coli BL21(DE3)/pJY2. Ubiquitin and the putative toxin genes were cloned in compatible plasmids (pCola-Duet1 and pJN105, respectively) and transformed individually into a single host. Strains containing both plasmids were grown in the presence of glucose or fucose (16) to repress gene expression and then were tested by transferring cultures to growth conditions with both arabinose and isopropyl-β-d-1-thiogalactopyranoside (IPTG) inducers. Bacterial cell death is a measure of activation of the candidate phospholipase by ubiquitin. As an initial screen, cultures were serially diluted and plated on agar plates with glucose (growth controls) or plates with IPTG and arabinose (test) (Fig. 2A and B).
FIG 2.
Screen for ubiquitin activation of orthologous phospholipases in bacteria and protein expression controls. (A and B) Strains expressing ExoU or an orthologous enzyme and human monoubiquitin were grown on agar plates containing glucose. Growth was emulsified, normalized to the same OD600, and diluted serially (100 to 10−7), and each dilution was spotted on control medium (glucose) or induction medium (arabinose and IPTG). AxoU, AdiU, and RP534 induced with arabinose and IPTG showed significant growth defects, compared to glucose-repressed controls, indicating cytotoxicity. (C and D) Relative protein expression of clones used in the bacterial surrogate toxicity assay. Cultures with BL21 pJY2 pJN105 His-tagged parental enzyme (C) or SA derivatives (D) were grown and lysates were prepared after induction with arabinose for 3 h. Samples were collected, subjected to Western blot analysis, and probed with anti-His MAb. Anti-DnaK antibody was used as a loading control. MWM, molecular weight markers.
LlbU (Legionella longbeachae), VpdA (Legionella pneumophila), and VvuU (Vibrio vulnificus) showed little to no cytotoxicity under both repressive and inducing conditions (Fig. 2B). In contrast, AxoU (Achromobacter xylosoxidans), AdiU (Aeromonas diversa), and RP534 (Rickettsia prowazekii) exhibited significant toxicity only under inducing conditions, suggesting that these enzymes are active and cytotoxic (Fig. 2A). As a control for enzymatic activity, we performed the same experiments with variants expressing bioinformatically predicted catalytic serine-to-alanine (SA) mutations (Fig. 2A and B). AxoU S201A, AdiU S126A, and RP534 S100A were not cytotoxic, suggesting that the cytotoxicity observed in the wild-type (WT) strains was due to phospholipase activity. To test whether cytotoxicity was ubiquitin dependent, we performed the same assay with strains containing only the pJN105 plasmid encoding a parental enzyme (see Fig. S1 in the supplemental material). Arabinose-induced constructs exhibited no detectable cytotoxicity, compared to glucose-repressed controls. BL21 strains with LlbU, VpdA, and VvuU with catalytic SA mutations grew similarly to control plates with glucose (Fig. 2B).
In previous studies, protein expression in the bacterial cytotoxicity assay was variable, likely due to differences in codon usage or protein stability. To ensure that each protein was expressed, Western blot analyses were performed with BL21 hosts containing JN105 clones with a parental enzyme in the absence of ubiquitin. All clones were constructed with the pET15b ribosome binding sequence and 6 histidines at the N terminus, to provide a tag for detection. ExoU and AdiU were expressed at comparable levels (Fig. 2C). RP534, LlbU, and VpdA were expressed but at lower levels than ExoU or AdiU. AxoU and VvuU were undetectable. To ensure that the differences in protein expression were inherent to the protein and not due to toxicity of the parental enzyme, SA derivatives were also subjected to protein expression studies (Fig. 2D). SA alleles reproduced a pattern of expression similar to that of the parental clones, indicating that there was no inherent toxicity reducing the amount of protein detected. To determine whether the levels of transcription were similar for each clone, RNA was extracted at time points ranging from 0.5 to 1.5 h after addition of arabinose (induced) or glucose (uninduced), and reverse transcription (RT)-PCR was performed with primers optimized and specific for ExoU and each ortholog. Results shown in Fig. S2 suggest that, during growth in arabinose, transcription of each clone was induced to a level similar to that of ExoU. Additionally, regions of the pJN105 plasmid encoding AraC and the pBAD promoter and operator regions were subjected to double-strand nucleotide sequence analysis. These regions were identical in each clone. Overall, our results suggest that the observed differences in protein expression may be due to differences in codon usage (translation) or posttranslational differences in peptide stability.
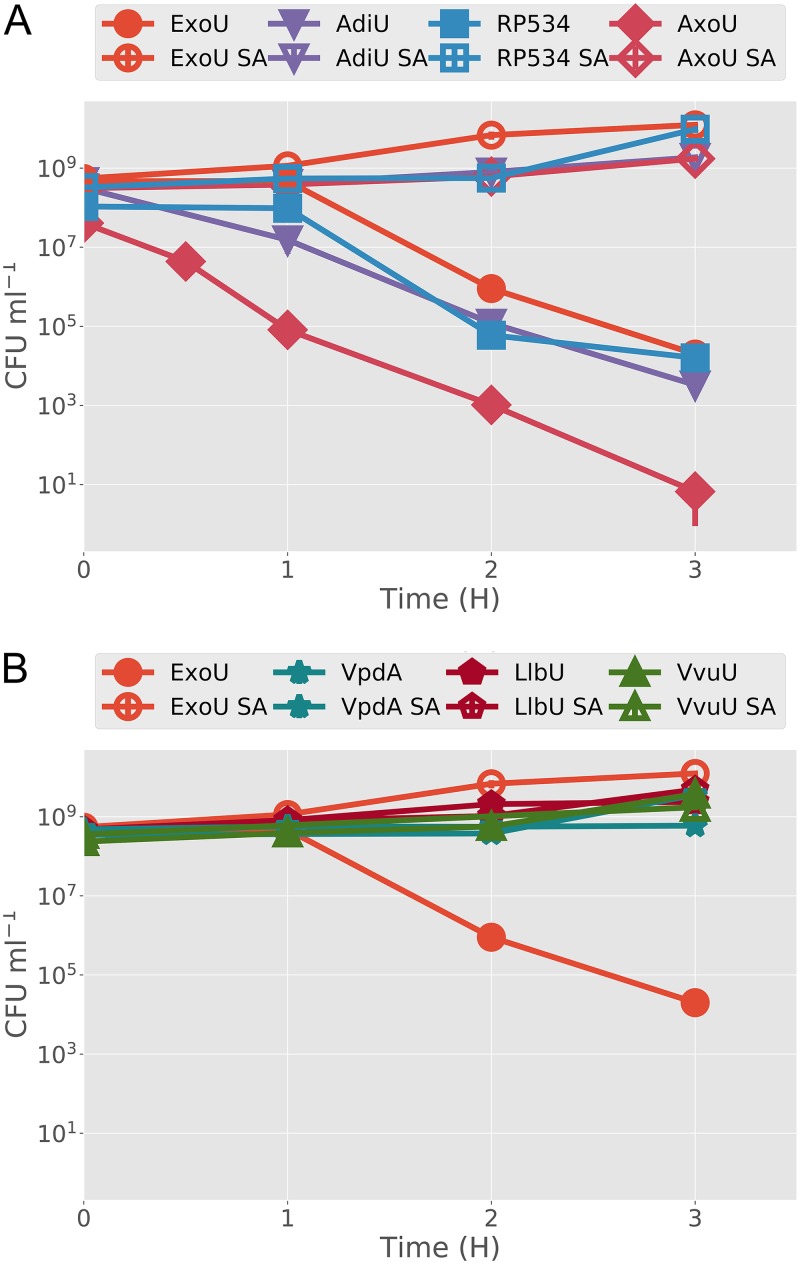
To perform a more quantitative assessment of toxicity, strains were grown under inducing conditions, and aliquots were plated, over a 3-h time course, on medium that rescues live cells retaining each plasmid (Luria-Bertani [LB] medium–glucose plates with chloramphenicol [pJY2], gentamicin [pJN105 with toxin gene], or kanamycin [pCola, encoding human ubiquitin]). The resulting colonies were counted, and the data were plotted as CFU per milliliter with respect to time postinduction. These analyses recapitulated the spot plate results, as ExoU, AdiU, AxoU, and RP534 exhibited significant reductions in CFU per milliliter over the time course of the experiment (Fig. 3A). Strains with VpdA, LlbU, and VvuU lacked detectable cytotoxicity (Fig. 3B). The lack of toxicity of VvuU may be due to a lack of protein expression under these conditions (Fig. 2C); however, AxoU was not detected in the expression assay but demonstrated enhanced cytotoxicity. Changing the catalytic serine to an alanine in each toxic clone eliminated detectable cytotoxicity. Conversely, there was no change in the growth of nontoxic orthologs with SA mutations. These results further support the hypothesis that toxicity is caused by phospholipase activity, and they predict that AdiU, RP534, and AxoU would be toxic to eukaryotic cells.
FIG 3.
Kill curves for each ortholog and SA control strain. BL21(DE3) pJY2 strains containing plasmids encoding ubiquitin (pCola Duet 1) and a parental or SA toxin (pJN105) were induced with both IPTG and arabinose for the indicated time periods (x axis). At each time point, samples were removed from the induced culture and plated on medium that rescues surviving cells. All assays were performed in triplicate, and the average values for CFU per milliliter were plotted. Vertical bars indicate standard deviations. (A) Assays with ExoU, AdiU, RP534, and AxoU. (B) Assays with ExoU, VpdA, LlbU, and VvuU.
AdiU and AxoU are toxic to HeLa cells.
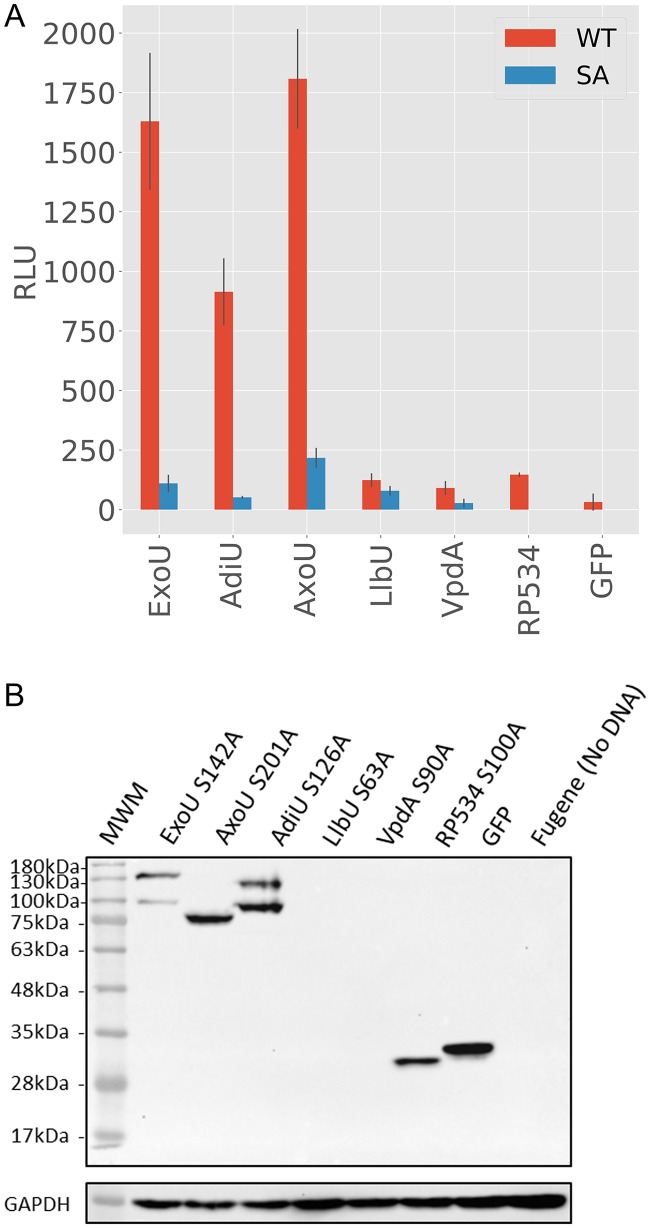
To test the cytotoxicity of the orthologs on mammalian membranes, HeLa cells were transfected with eukaryotic expression constructs encoding ExoU, ExoU orthologs, or SA variants as green fluorescent protein (GFP) fusion proteins. In contrast to the bacterial cytotoxicity system expressing only monoubiquitin, mammalian cells contain a variety of ubiquitin isoforms, including modified ubiquitin proteins. Importantly, the cellular environment also may contain cofactors, other than ubiquitin, that trigger activity. If activated, ExoU or orthologous enzymes should lyse the cells, resulting in the release of adenylate kinase (AK) into the supernatant. AK activity converts ADP to ATP, which can in turn be used by firefly luciferase, allowing toxicity to be measured as luminescence. Cell culture supernatants were collected 24 h posttransfection, subjected to centrifugation to remove any remaining cells, and assayed for AK activity. The resulting data were plotted to compare cytotoxicity with that of SA variants (Fig. 4A). In preliminary experiments, VvuU and VvuU SA were not toxic, were undetectable in protein expression assays regardless of conditions, and were not analyzed further in either prokaryotic or eukaryotic hosts (data not shown). Similar to the bacterial toxicity assays, LlbU and VpdA demonstrated little to no AK activity. Interestingly, RP534 also did not show cytotoxicity to eukaryotic cells, despite being toxic to prokaryotes. Both AdiU and AxoU exhibited significant cytotoxicity, comparable to that of the positive control, ExoU. Cytotoxicity was abrogated when SA variants were tested, supporting the hypothesis that the cytotoxicity of AdiU and AxoU is caused by phospholipase activity.
FIG 4.
AK release and protein expression. (A) AK release assay. Expression clones encoding ExoU orthologs and SA variants as eGFP fusion proteins were transfected into HeLa cells for 24 h. Supernatants were collected and tested for AK activity. An increase in RLU when SA variants are compared to parental enzymes indicates release of AK as a result of cellular lysis. AK release experiments were performed at least 3 times, each point in triplicate. Error bars are standard deviations, and the data bars are the means. (B) Analysis of protein expression. Western blot analysis of HeLa cells transfected for 24 h with eGFP fusion constructs encoding catalytically defective enzymes was performed. GAPDH serves as a loading control. Antiserum to GFP was used to detect GFP fusion proteins. MWM, molecular weight markers.
Protein expression from cells transfected with a SA derivative of each gene was assessed by Western blot analysis using anti-GFP antibodies (Fig. 4B). AdiU S126A and AxoU S201A were expressed as well as, or better than, the ExoU S142A control. LlbU and VpdA were undetectable (Fig. 4B), despite extensive efforts to optimize transfection conditions and reagents. Each clone, including VvuU, was reconstructed as a C-terminal fusion in pEGFPN1 (toxin-enhanced GFP [eGFP]) and used in transfection studies. Our results were identical, in that only ExoU, AdiU, and AxoU fusion proteins were detectable (data not shown). As a control for translation, we codon optimized the vpdA gene (Integrated DNA Technologies, Inc., Coralville, IA) for HeLa cell expression (Text S3). The optimized vpdA gene was transfected under a number of conditions but little to no protein was detectable, suggesting that VpdA is produced but is highly unstable (data not shown). Only a breakdown product was observed for RP534, likely explaining the lack of cytotoxicity in HeLa cells, compared to our bacterial toxicity assays. ExoU and AdiU were detected as two distinct bands, while AxoU appeared as a single fusion protein. These results suggest that, similar to ExoU (8), AdiU may be modified by ubiquitin after delivery into eukaryotic cells.
AdiU, RP534, and AxoU exhibit ubiquitin-dependent PLA2 activity in vitro.
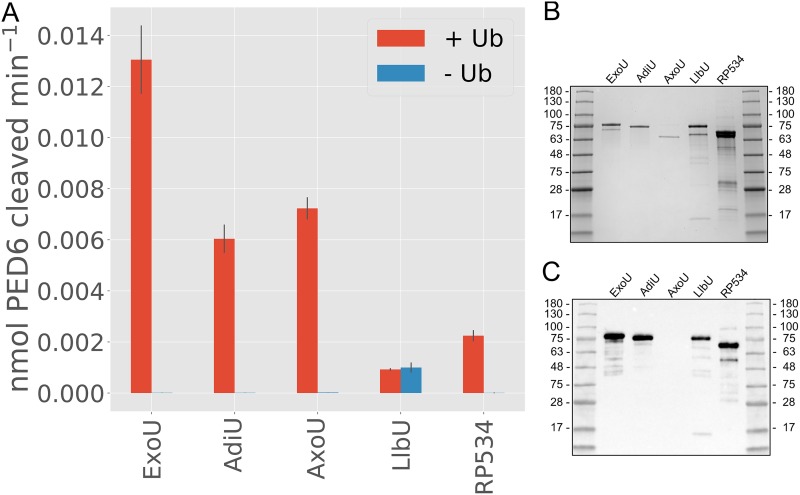
In vivo assays are limited by the composition of different membranes and variable levels of protein expression and stability. To determine whether the orthologous proteins possess ubiquitin-dependent activity, each gene was cloned into pET15b, expressed, and purified. For each ortholog, protein expression was optimized regarding temperature and length of time for induction, as well as medium composition. Different buffer conditions were used to optimize both protein purification efforts and enzyme activity measurements. These experiments resulted in separate purification protocols for AxoU and RP534, as described in Materials and Methods. Unfortunately, we were unable to purify VpdA in large enough quantities to perform activity assays. Protein quantities were determined using absorbance at 280 nm. Purity was assessed by detection of protein bands in Coomassie-stained SDS-polyacrylamide gels and by Western blot analysis with an anti-His antibody (Fig. 5B and C). Notably, AxoU was purified without an N-terminal His tag and therefore was undetectable in the Western blot analysis. Due to variability in the purity of the enzymes, we consider the activity assays semiquantitative. Despite this limitation, we can confidently assess both ubiquitin dependence and phospholipase A2 (PLA2) specificity of these enzymes. Activity assays were performed in 50-µl reaction volumes with approximately 10 nM ExoU or orthologous enzyme, in the presence or absence of 50 µM monoubiquitin. All enzymes tested exhibited at least some phospholipase activity (Fig. 5A). LlbU had the lowest activity, which was the same in both the presence and the absence of ubiquitin, suggesting that LlbU phospholipase activity is not ubiquitin dependent or that the PLA2 substrate PED6 [N-([6-(2,4-dinitrophenyl)amino]hexanoyl)-2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt] is not optimal. RP534 exhibited a small amount of activity that was ubiquitin dependent under our assay conditions. AxoU and AdiU exhibited moderate activity, compared to the ExoU control, and both activities were ubiquitin dependent.
FIG 5.
(A) In vitro PLA2 activity assays. ExoU and orthologs were purified and tested for in vitro PLA2 activity using PED6 as a substrate. All tested constructs exhibited at least some PLA2 activity, and all except LlbU were regulated by the presence of ubiquitin (Ub). LlbU exhibited equal activity in both the presence and the absence of ubiquitin. Assays were performed in triplicate. Error bars are standard deviations, and the data bars are the means. (B) Coomassie-stained SDS-polyacrylamide gel analysis of each protein used in the enzyme assay. (C) Western blot analysis of purified proteins probed with antihistidine antibody at a 1:10,000 dilution. AxoU was purified without a tag and is not detectable in this analysis.
Modeling and structural analysis of cytotoxic ExoU orthologs.
Since only AxoU and AdiU were ubiquitin dependent and cytotoxic in all of the aforementioned assays, we attempted to model these protein structures, to look for similarities that may better indicate the ExoU activation mechanism. Based on amino acid sequence alignment, AdiU shares over 40% identity to P. aeruginosa ExoU (Table 1 and crystal structure under PDB ID 3TU3 [7]) and Pseudomonas fluorescens ExoU (crystal structure under PDB ID 4QMK [17]). RosettaCM (18) was used to perform multitemplate modeling with the 4QMK (17) and 3TU3 (7) structures. Approximately 2,000 decoys were generated, as described in Materials and Methods. The top 10 scoring models were aligned, and all were within 5 Å root mean square deviation (RMSD) of the top scoring structure. Most of the observed variance came from regions in which residues are not resolved in both the 3TU3 and 4QMK structures. Excluding these regions, the top 10 scoring structures were all less than 2.0 Å RMSD apart (Fig. S3A). Additional variation likely derives from flexible regions of the protein; for example, the L3 loop in the membrane localization domain (MLD) is flexible and exhibits variation. The models align to the crystal structure of ExoU with less than 2.2 Å RMSD (Fig. S3B). Combined with the biological and enzymatic analyses, modeling data support the hypothesis that AdiU is similar structurally and functionally to ExoU.
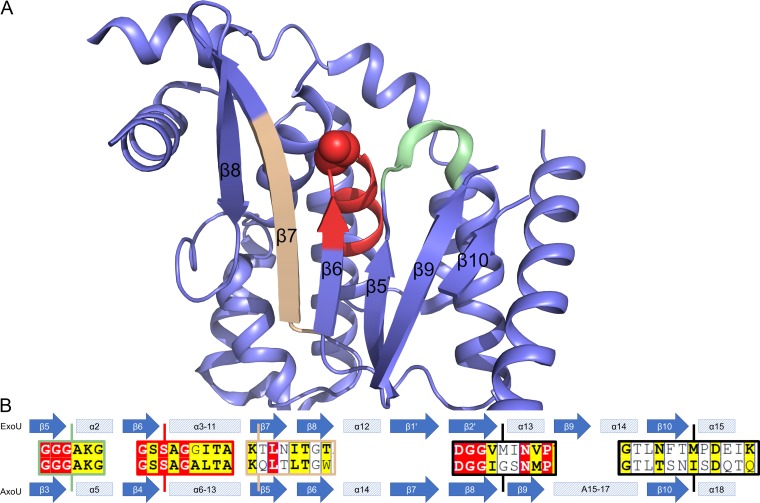
AxoU possesses low sequence homology (∼22%) to P. aeruginosa ExoU (3TU3 [7]) and P. fluorescens ExoU (4QMK [17]) despite being ubiquitin activated and cytotoxic to both prokaryotes and eukaryotes. Due to the low sequence homology, we were unable to obtain quality homology models of AxoU. Using conserved motifs as reference points, we were able to compare secondary structure predictions (19) for the AxoU catalytic domain with the secondary structure of ExoU (Fig. 6). These comparisons revealed the conserved α/β-hydrolase fold of AxoU. The most apparent alignments correspond to the six β-strands forming a primarily parallel β-sheet (ExoU β5 to β7 and β9 to β10) with a single antiparallel strand on one end (ExoU β8) (Fig. 6A). Alignment of the AxoU predicted secondary structure with these elements of ExoU (Fig. 6B) is consistent with the sequence alignments (Fig. 1). Since this fold is highly conserved among all patatin-like phospholipases, it is likely that AxoU takes on a similar fold. Additionally, we can observe two β-strands preceding the catalytic aspartate (ExoU β1ʹ and β2ʹ). These β strands are hypothesized to fold into a β-hairpin, as observed in most other patatin-like phospholipases (patatin, VipD, cytosolic PLA2, and PlpD) (1, 20–23). While these residues are not resolved in the ExoU crystal structure, PSIPRED also predicts that these residues form β-strands in ExoU. The folding of this β-hairpin is likely a major mechanism for regulation of ExoU and ubiquitin-activated orthologs. Outside the catalytic domain, we were unable to make reasonable secondary structure comparisons. Considering the patatin domain as being central to the protein, AxoU appears to be composed of approximately 148 N-terminal residues and 56 C-terminal residues. Accordingly, we separated AxoU into three domains, i.e., N-terminal, catalytic, and C-terminal. Each domain was searched against the Protein Data Bank independently using BLAST, and these searches yielded no significant matches.
FIG 6.
Structural comparison of ExoU and AxoU. (A) Cartoon representation of the ExoU catalytic domain. The structure (PDB ID 3TU3) and annotations were obtained from the report by Halavaty et al. (7). Conserved regions are highlighted and correspond to the alignments. Green, conserved GGGX(K/R) motif (ExoU β5-α2 and AxoU β3-α5); red, catalytic serine and conserved GXSXG motif (ExoU β6 and AxoU β4); tan, conserved residues in ExoU β7 and AxoU β5. (B) Secondary structure alignment of ExoU (top) and AxoU (bottom). AxoU secondary structure was predicted using the PSIPRED server and compared to the known structure of ExoU (PDB ID 3TU3) (7). ExoU secondary structure annotation is the same as used by Halavaty et al. (7) except for β1ʹ and β2ʹ, which are not resolved in the crystal structure but are predicted by PSIPRED. Alignments in the central area are reference locations from Fig. 1. Vertical lines indicate the approximate locations of these sequences. The colors of the boxes correspond to structural elements highlighted above. Black boxes were not resolved in the crystal structure.
To better assess the structure of the N-terminal domain, we attempted to perform ab initio modeling using several applications, including Rosetta, BCL::Fold, QUARK, and I-TASSER (24–28). All applications yielded low confidence results, with significantly different folds. Secondary structure predictions suggest that the N terminus consists of four α-helix segments. This is significantly different from the N terminus of ExoU, which is much shorter (∼70 amino acids) and interacts primarily with its chaperone, SpcU (6, 7).
The C-terminal domain of AxoU is far shorter than that of ExoU. In ExoU, this region consists of a bridging domain that has been shown to interact with ubiquitin (14) and a four-helix bundle that serves as a MLD (9–11). We utilized Rosetta to perform ab initio modeling of the C-terminal domain of AxoU (27, 28). The top scoring model has a globular fold of small helices. The next nine models have RMSDs between 1.6 and 5.9 Å, with respect to the top model (Fig. S4). Notably, the first 10 residues of the top models do not align well. It is possible that these residues form a flexible linker between the catalytic domain and the C-terminal domain of AxoU or they are normally stabilized by the catalytic domain. PSIPRED predicts, although with relatively low confidence, that these residues take on a coil conformation. The analogous region of ExoU is a β-strand that is part of a β-sheet formed with N-proximal residues of ExoU. If the first 10 residues are excluded from the alignment (RMSD11–56), then the structures align with RMSD11–56 values of 0.71 to 3.6 Å. These structures do not yield significant similarity to that of the ExoU C terminus, despite the fact that AxoU is ubiquitin activated. A major caveat of predictive modeling is that the models may differ significantly from the actual structures. This would be particularly true if the AxoU C-terminal domain makes significant tertiary contacts with the rest of the AxoU structure. Interestingly, AxoU is predicted to have two β-strands preceding the catalytic domain. It is possible that these strands form a sheet with residues directly following the AxoU catalytic domain, as seen in ExoU and VipD (7, 20). If so, this places the N- and C-terminal domains of AxoU in close proximity.
DISCUSSION
Identifying members of a protein family is generally accomplished bioinformatically by searching for conserved domains. Patatin and patatin-like enzymes are expressed by many microorganisms. By definition, they display a patatin domain, which is responsible for lipase activity, but they can differ dramatically in their biochemical properties and amino acid sequences outside the catalytic domain. These data suggest that the patatin enzyme family is likely composed of different subgroups that may possess diverse biological activities, including facilitating replication and transmission of pathogenic species of bacteria.
A model patatin-like enzyme that plays a prominent role in the pathogenesis of P. aeruginosa is ExoU. ExoU is composed of several functional domains that bind its chaperone SpcU, stabilize membrane interactions by binding PIP2, and regulate phospholipase activity such that the enzyme is expressed only within eukaryotic cells. Activation of ExoU depends on a noncovalent interaction with ubiquitin, a molecule that is present only in eukaryotic cells. In this study, the ubiquitin-mediated activation of ExoU is exploited in a surrogate toxicity assay to identify other members of this subgroup that may be associated with pathogenesis. Importantly, we postulated that the members of the ubiquitin-activated patatin subgroup may share structural properties that will help define the mechanism of activation, a key step in the rational design of therapeutically useful inhibitors.
The method for identifying ubiquitin-activated enzymes involves simple cloning and assays for bacterial or eukaryotic cell viability postinduction or posttransfection. Screens are performed in vivo to provide native membrane substrates. Verification assays include protein expression coupled with protein purification and in vitro enzyme assays with a phospholipid mimetic, PED6. With this work flow and mutagenesis analyses of predicted catalytic residues, we identified three enzymes that can be integrated into the ExoU subgroup. Ubiquitin-activated molecules include AxoU (Achromobacter xylosoxidans), AdiU (Aeromonas diversa), and RP534 (Rickettsia prowazekii). RP534 was shown previously to exhibit PLA1, PLA2, and lyso-PLA2 activities independent of a eukaryotic cofactor (29). Activity was stimulated by the addition of bovine liver superoxide dismutase 1 (SOD1), a cofactor that was initially identified by Sato et al. as being required for ExoU activation (30). We subsequently demonstrated that SOD1 was modified by ubiquitin and that the addition of ubiquitin to the ExoU enzyme reaction mixture allowed detection of activity when PED6 was used as a substrate (31). Our results verify that ubiquitin enhances the PLA2 activity of RP534; however, the enzyme differs from the ExoU subgroup in its biochemical profile.
Enzymes from Legionella (VpdA [L. pneumophila] and LlbU [L. longbeachae]) and Vibrio (VvuU) were also tested. VvuU was undetectable in any expression system and could not be characterized further. VpdA and LlbU were noncytotoxic in bacterial and eukaryotic toxicity assays and were detectable as recombinant proteins in bacteria but were undetectable after transfection. Originally identified as a VipD homolog, VpdA does not appear to bind Rab5 (32), a required cofactor for VipD phospholipase activity. VpdA is conserved, is expressed, and appears to be translocated by L. pneumophila (33), suggesting that it serves a biological purpose. Expression of VpdA in yeast results in significant cytotoxicity (34). In our hands, however, VpdA was not stable enough to be purified using standard techniques, and the lack of cytotoxicity in both eukaryotic and prokaryotic systems indicates that ubiquitin likely is not involved in regulating enzymatic activity. LlbU was stable enough for purification and demonstrated minor phospholipase activity, independent of ubiquitin. The limitations of our work flow include failure to express protein in one or more systems, which is likely related to the inherent stability of the enzyme. Our analyses suggest that the biological screens are sensitive and specific for identifying potential members of the ExoU-like subgroup of phospholipases that are likely involved in bacterial pathology versus bacterial physiology. These enzymes would be defined by their ability to kill prokaryotes (supplemented with the correct cofactor) and eukaryotes and likely would be able to degrade a broad range of lipid substrates.
To overcome the biochemical limitations of protein stability in different hosts, it will be imperative to develop better predictive algorithms both to identify members of specific protein subgroups and to predict structural and functional properties. We utilized secondary structure predictions and computational modeling techniques to perform a structural analysis of the two ubiquitin-activated enzymes that most closely fall into the ExoU subgroup. AdiU, similar to previously studied orthologs (15), shares significant sequence and structural homology to ExoU. The comparative models generated using RosettaCM are likely very close to the native AdiU structure.
In contrast, AxoU has very low sequence homology to ExoU, even within the catalytic domain. Despite this major difference, AxoU has been shown to be a ubiquitin-activated phospholipase. Secondary structure analysis of AxoU in comparison to ExoU allowed us to identify the boundaries of the AxoU catalytic domain, revealing a significantly longer N terminus and shorter C terminus. The N terminus of ExoU functions primarily as a chaperone binding domain (6, 7, 35). Truncation of the N terminus (to residue 52) has little to no effect on the toxic activity of ExoU (36). Amino acid 52 is located a few residues from the start of the catalytic domain. The N-terminal region of AxoU is significantly longer. Taken together with the dramatically truncated C-terminal domain, the N terminus of AxoU may have additional functional roles. Unfortunately, we were unable to model the N terminus of AxoU with any confidence. Secondary structure predictions suggest an entirely helical domain.
The C terminus of ExoU contains two important functional domains that mediate ubiquitin binding and membrane localization (MLD). Ab initio predictions of the C-terminal domain of AxoU, however, could not be confidently aligned with either the MLD or ubiquitin binding domain of ExoU. ExoU catalytic activity is significantly impaired when the C terminus is truncated by only 9 residues (8). Due to the lack of an identifiable MLD or ubiquitin binding domain within the C terminus of AxoU, the N-terminal region may have one or both functions. Secondary structure predictions are consistent with the presence of a β-sheet that brings the N- and C-terminal domains of AxoU together, as seen in ExoU and VipD (7, 20). The analogous β-sheet is thought to play a crucial role in the activation of VipD and may be a conserved mechanism in bacterial phospholipases activated by eukaryotic cofactors (21). The close proximity of these two domains may have allowed a domain rearrangement that transferred the ubiquitin binding domain, the MLD, or both, to the N-terminal domain of AxoU. Identifying the location of ubiquitin binding to AxoU should shed light on the activation mechanisms of both AxoU and ExoU. Future studies will seek to establish the significance of AxoU during infection, as well as the structural factors shared between AxoU and ExoU that allow ubiquitin-mediated activation.
MATERIALS AND METHODS
Reagents.
The antibodies used for Western blot detection are as follows: mouse antiubiquitin (product no. sc-271289; Santa Cruz Biotechnology), mouse anti-His (product no. 27-4701-01; GE Healthcare), anti-GFP (product no. MMS-118R; Covance), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (product no. SC-32233; Santa Cruz Biotechnology), anti-DnaK (product no. 8E2/2; Enzo), and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (product no. F-21453; Invitrogen). Each ExoU ortholog was amplified from chromosomal DNA purchased from ATCC, obtained as a gift, or purified using Qiagen 100 genomic tips (Table 1). Primers were designed and synthesized by Integrated DNA Technologies, Inc. Platinum Pfx DNA polymerase was used to amplify each gene (Invitrogen). For each vector used (pEGFPN1, pEGFPC1, pET15b, or pJN105), plasmid DNA was purified using a midiprep protocol (PureYield; Promega) and digested with restriction endonucleases to generate an oriented insertion. Gene amplifications were designed for Gibson assembly using the NEB Builder HiFi DNA assembly master mix. All resulting clones were subjected to double-stranded DNA sequence analysis to ensure that no other mutations were introduced as a result of PCR amplifications. The clone encoding human monoubiquitin was described previously (15) and was expressed from pCola Duet1 (50 µg/ml kanamycin). Mutations in each gene were introduced using a Q5 mutagenesis kit from New England Biolabs.
In vivo toxicity assays.
The bacterial dual expression assay was performed as described previously (15) except that the ubiquitin clones were constructed in pCola Duet-1. Briefly, BL21(DE3)/pJY2 strains of E. coli (30 µg/ml chloramphenicol) were transformed with each expression plasmid. pJN105 plasmids contained histidine-tagged ExoU (selected with 5 µg/ml gentamicin), an orthologous gene, or derivatives constructed to be defective in phospholipase activity and were selected on medium with 5 µg/ml gentamicin and 30 µg/ml chloramphenicol. Toxin genes or their noncatalytic derivatives were induced with 0.5% arabinose. pCola Duet-1 plasmids contained a human monoubiquitin gene and were retained by selection on 50 µg/ml kanamycin. IPTG was added to the medium at 0.5 mM as the inducer. Strains containing both toxin and ubiquitin expression constructs were grown overnight at 30°C on agar plates containing 5 µg/ml gentamicin, 30 µg/ml chloramphenicol, 50 µg/ml kanamycin (triple antibiotics), and 0.5% glucose. Several colonies were harvested, and an emulsion was standardized to an optical density at 600 nm (OD600) of 3.0. The emulsion was rested statically at 30°C for 40 min and then was diluted in broth with triple antibiotics, IPTG, and arabinose to measure CFU over time (kill curves) or was diluted serially, with each dilution being applied directly to agar medium containing inducers (IPTG and arabinose) or glucose (spot plates).
Protein expression from bacterial cells was tested in BL21(DE3)/pJY2 strains containing only the pJN105 ExoU ortholog constructs. Bacterial cells were grown overnight at 30°C on agar plates (5 µg/ml gentamicin, 30 µg/ml chloramphenicol, and 0.5% glucose), and an emulsion was made and standardized as described for kill curve analyses. The emulsion was diluted to an OD600 of 0.25 and induced with 0.5% arabinose. Samples containing cells and growth medium were removed after 3 h, diluted into SDS sample buffer, boiled, and subjected to Western blot analysis with antiserum to the histidine tag.
To test toxicity in eukaryotic cells, each ortholog or noncatalytic derivative was subcloned into pEGFPC1, encoding a C-terminal (eGFP) fusion to the putative toxin N terminus (eGFP-toxin). HeLa cells (CCL-2 from ATCC) were seeded at a density of 5.75 × 104 cells in a 24-well plate and were grown overnight at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), with 5% CO2. Cells were washed two times with Hanks’ balanced salt solution (HBSS) and transfected with the FuGENE HD transfection reagent (Promega) mixed with 400 ng DNA/well, in triplicate, at the ratio of 3.2 µl FuGENE/1 µg DNA, according to manufacturer’s instructions. The eGFP control plasmid was transfected under the same conditions. After 24 h, the contents of each well were mixed, and 100 µl of sample was removed and subjected to a 10-min centrifugation at 2,300 × g at 4°C. Approximately 50 µl of supernatant was added to 50 µl of Toxilight substrate to measure AK release from lysed cells. Relative light units (RLU) were measured during a 1-s interval.
Western blot detection of eGFP-ortholog chimeric proteins was conducted using cells from 60-mm plates seeded with 6.8 × 105 cells. Cells were transfected with 4.0 µg of plasmid DNA at a 3.0 µl FuGENE/1 µg DNA ratio. After 24 h of incubation, cells were scraped in 900 µl sucrose buffer (250 mM sucrose, 3 mM imidazole, and cOmplete mini protease inhibitors [EDTA-free; Roche] in phosphate-buffered saline [PBS]), transferred to a microcentrifuge tube, and collected by centrifugation at 5,000 × g for 15 min at 40C. Floating cells in the culture medium were also collected by centrifugation at 680 × g for 10 min and were combined with the harvested cells. Cell pellets were combined with 50 µl of 2× lysis buffer (6 mM imidazole, 2× cOmplete mini protease inhibitors [EDTA-free], 100 µg/ml DNase, and 100 µg/ml RNase in PBS), and 50 µl of 5× protein sample buffer was added. After 15 min of incubation at room temperature, samples were boiled for 5 min prior to analysis by SDS-PAGE and Western blotting. GFP-tagged proteins were probed with an anti-GFP monoclonal antibody (MAb) (1:5,000 dilution), followed by HRP-conjugated anti-mouse IgG (1:10,000 dilution). As a loading control, GAPDH was probed with an anti-GAPDH MAb (1:2,000 dilution) and HRP-conjugated anti-mouse IgG.
mRNA assessment.
Total RNA was isolated from 1 × 108 cells from each time point, utilizing TRIzol Max bacterial RNA isolation reagents (Ambion). RNA was further purified using RNAeasy columns, according to the manufacturer’s instructions (Qiagen). RNAs were quantified using Qubit reagents and fluorometer (Thermo Fisher Scientific) and were diluted to 25 ng/µl for DNase treatment with ezDNase (Invitrogen). Approximately 200 ng of RNA was incubated with DNase at 37°C for 15 min. DNase-treated RNA was diluted to 1 ng/µl and reverse transcribed with SuperScript IV VILO master mix (Invitrogen). Controls included no-reverse transcriptase mixtures. PCRs were performed with SSo Advanced Universal IT Sybr green SMx reagents (Bio-Rad). Plasmid DNA with each ortholog gene was quantified with Qubit reagents. For each experiment, plasmid DNA was titrated in triplicate (107 to 101 copies). Copy number was calculated and plotted for each time point over a period of 1.5 h. Each time point for each growth condition was performed in triplicate. Primers were designed using the Integrated DNA Technologies website and verified in melt curve analyses as containing a single predominant peak.
Protein purification and enrichment.
All orthologous enzymes were expressed from E. coli strain BL21(DE3) containing the pLysS plasmid. BL21(DE3)/pLysS/pET15b ExoU, -pET15b AdiU, and -pET15b LlbU cells were grown in LB medium (30 µg/ml chloramphenicol and 100 µg/ml ampicillin) at 37°C to an OD600 of 0.5 and were induced with 0.5 mM IPTG for 3 h. Cultures were subjected to centrifugation (12,000 × g at 4°C) to harvest bacterial cells after induction. Pellets were stored at −80°C until purification. ExoU, AdiU, and LlbU were purified using metal affinity chromatography, as described previously (15). Histidine-tagged RP534 was expressed from pET15b and purified as described by Housley et al. (29).
AxoU protein expression and purification.
Untagged AxoU was coexpressed with its chaperone SacU, containing an N-terminal histidine tag, from pET15b in E. coli BL21(DE3)/pLysS. Cells were grown at 37°C in yeast extract nutrient broth with chloramphenicol (30 µg/ml) and ampicillin (100 µg/ml) to an OD600 of 0.5 and then were induced with 0.25 mM IPTG for 3 h at 37°C. Induced cells were harvested by centrifugation at 12,000 × g at 4°C and were stored at −80°C.
Frozen pellets were suspended in lysis buffer containing protease inhibitors (32 µg/ml benzamidine, 64 µg/ml leupeptin, 3.2 µg/ml pepstatin, and 32 µg/ml aprotinin) and 32 µg/ml DNase I. Suspensions were lysed by passage through a cold French pressure cell. Cell lysates were subjected to centrifugation (30,000 × g at 4°C for 20 min) to remove cell debris. The supernatant fraction was then applied to a gravity flow column containing 3 ml of Talon metal affinity resin charged with cobalt. The column was washed according to the manufacturer’s recommendations and eluted with 50 mM sodium phosphate-300 mM NaCl (pH 7.0) with 0.02% EDTA. The chaperone-toxin complex was then concentrated to 500 µl using an Amicon Ultra-15 centrifugal filter with a 30-kDa molecular weight cutoff (MWCO). Size exclusion chromatography was performed on the concentrated samples using a HiLoad 16/600 Superdex 200-pg column (GE) on an AKTA Explorer 10 fast protein liquid chromatography (FPLC) system. Fractions containing the chaperone-toxin complex were then applied to an additional gravity flow Talon metal affinity column, as described previously. AxoU was eluted in 25 mM HEPES-300 mM NaCl-6 M urea with 20% glycerol (pH 7.5). Urea was removed from the eluent by dialysis overnight at 4°C, using a Spectra/Por 7 regenerated cellulose dialysis membrane with a 10-kDa MWCO, in 25 mM HEPES-300 mM NaCl with 20% glycerol (pH 7.5). The dialyzed product was quantified using absorbance at 280 nm and was stored at −80°C.
Purification of recombinant monoubiquitin.
Monoubiquitin was expressed, tag free, from E. coli AR58(DE3)/pJY2 from pET15b. These strains were grown in Terrific broth at 37°C, to an OD600 of 0.5 to 0.7. Protein expression was induced with 0.25 mM IPTG for 3 to 4 h. Cultures were harvested by centrifugation, as described previously, and bacterial pellets were stored at −80°C until purification. Frozen pellets were suspended in 20 ml 50 mM sodium acetate buffer (pH 7.3) with 32 µg/ml benzamidine, 64 µg/ml leupeptin, 3.2 µg/ml pepstatin, and 32 µg/ml aprotinin and were lysed by three passes through a French pressure cell. Lysates were cleared by centrifugation at 30,000 × g at 4°C for 20 min. The supernatant was collected, acidified to pH 4.5 using glacial acetic acid, and diluted to 40 ml with 50 mM sodium acetate (pH 4.5). Precipitated protein was removed by centrifugation at 30,000 × g at 4°C for 20 min. The supernatant was collected and filter sterilized. Cation exchange chromatography was performed on the filtered supernatant using a HiPrep SP FF 16/10 cation exchange chromatography column (GE) on an AKTA Explorer 10 FPLC system. Samples were loaded onto the column at 0.5 ml/min and washed with 5 column volumes. Ubiquitin was eluted over 10 column volumes by a linear gradient of 50 mM sodium acetate (pH 4.5) with 0 to 0.5 M NaCl. Eluted fractions were analyzed by SDS-PAGE. Fractions containing ubiquitin were identified after SDS-PAGE and Coomassie blue staining. Fractions containing ubiquitin were pooled and concentrated using an Amicon Ultra-15 centrifugal concentrator with a 3-kDa cutoff. Size exclusion chromatography was performed on concentrated ubiquitin using the same FPLC system and a HiPrep 26/60 Sephacryl S-200 column with 50 mM phosphate-150 mM NaCl (pH 7.0). Ubiquitin fractions were pooled and concentrated.
All purified proteins were quantified by absorbance at 280 nm. Extinction coefficients were calculated using the ExPASy ProtParam tool (37). Protein was aliquoted and stored at −80°C until used in activity assays.
In vitro enzyme activity assays.
Activity assays were performed in triplicate as described previously (38), with the following modifications. Reaction mixtures (50 µl) contained 50 mM morpholineethanesulfonic acid (MES) buffer, 750 mM monosodium glutamate, 50 µM PED6, and 50 µM monoubiquitin. Reactions were initiated by adding 5 µl of 100 nM enzyme, resulting in 10 nM enzyme per reaction. The PED6 fluorophore was excited with 488-nm light, and fluorescence readings were taken at 530 nm every minute for 60 min. Linear ranges were fit to determine slopes in relative fluorescence units (RFU) per minute and were normalized to the RFU of wells containing 50 µl of 10 µM boron-dipyrromethene (BODIPY) to obtain an enzymatic rate.
Molecular modeling.
Comparative modeling of AdiU was conducted using RosettaCM from Rosette 3.8. Initial threaded structures were prepared for the 3TU3 and 4QMK structures using the setup_RosettaCM.py script distributed with Rosetta; 3TU3 and 4QMK structures were weighted equally. Rosetta scripts were used to run the RosettaCM protocol to generate 2,000 decoys, using the options described in Protocol S1 in the supplemental material. Decoys were sorted by score, and the top 10 results were used for structural analysis.
Ab initio modeling for the C terminus of AxoU was conducted using the Rosetta AbinitioRelax application. Three-mer and 9-mer fragment files were generated using the Robetta server (39). The AbinitoRelax application was used with the options described in Protocol S2, to generate 50,000 decoys. Decoys were sorted by score, and the top 10 results were used for analysis. Structural alignments and figure making were performed with open-source PyMol version 2.1.0.
Accession number(s).
Nucleotide sequences for A. diversa ATCC 43946 adiU and A. xylosoxidans GN008 axoU have been submitted to GenBank under nucleotide accession numbers MK249729 and MK248684, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Allergy and Infectious Disease grant 1R01 AI104922 (D.W.F.).
This research was completed in part with computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00623-18.
REFERENCES
- 1.Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 2.Leslie CC, Voelker DR, Channon JY, Wall MM, Zelarney PT. 1988. Properties and purification of an arachidonoyl-hydrolyzing phospholipase A2 from a macrophage cell line, RAW 264.7. Biochim Biophys Acta 963:476–492. doi: 10.1016/0005-2760(88)90316-5. [DOI] [PubMed] [Google Scholar]
- 3.Strickland JA, Orr GL, Walsh TA. 1995. Inhibition of Diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiol 109:667–674. doi: 10.1104/pp.109.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang C, Flieger A. 2011. Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur J Cell Biol 90:903–912. doi: 10.1016/j.ejcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Banerji S, Flieger A. 2004. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150:522–525. doi: 10.1099/mic.0.26957-0. [DOI] [PubMed] [Google Scholar]
- 6.Gendrin C, Contreras-Martel C, Bouillot S, Elsen S, Lemaire D, Skoufias DA, Huber P, Attree I, Dessen A. 2012. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog 8:e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halavaty AS, Borek D, Tyson GH, Veesenmeyer JL, Shuvalova L, Minasov G, Otwinowski Z, Hauser AR, Anderson WF. 2012. Structure of the type III secretion effector protein ExoU in complex with its chaperone SpcU. PLoS One 7:e49388. doi: 10.1371/journal.pone.0049388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stirling FR, Cuzick A, Kelly SM, Oxley D, Evans TJ. 2006. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol 8:1294–1309. doi: 10.1111/j.1462-5822.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 9.Rabin SD, Veesenmeyer JL, Bieging KT, Hauser AR. 2006. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect Immun 74:2552–2561. doi: 10.1128/IAI.74.5.2552-2561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veesenmeyer JL, Howell H, Halavaty AS, Ahrens S, Anderson WF, Hauser AR. 2010. Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect Immun 78:3346–3357. doi: 10.1128/IAI.00223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessmer MH, Anderson DM, Buchaklian A, Frank DW, Feix JB. 2017. Cooperative substrate-cofactor interactions and membrane localization of the bacterial phospholipase A2 (PLA2) enzyme, ExoU. J Biol Chem 292:3411–3419. doi: 10.1074/jbc.M116.760074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato H, Frank DW. 2014. Intoxication of host cells by the T3SS phospholipase ExoU: PI(4,5)P2-associated, cytoskeletal collapse and late phase membrane blebbing. PLoS One 9:e103127. doi: 10.1371/journal.pone.0103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson DM, Feix JB, Monroe AL, Peterson FC, Volkman BF, Haas AL, Frank DW. 2013. Identification of the major ubiquitin-binding domain of the Pseudomonas aeruginosa ExoU A2 phospholipase. J Biol Chem 288:26741–26752. doi: 10.1074/jbc.M113.478529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tessmer MH, Anderson DM, Pickrum AM, Riegert MO, Moretti R, Meiler J, Feix JB, Frank DW. 2018. Identification of a ubiquitin-binding interface using Rosetta and DEER. Proc Natl Acad Sci U S A 115:525–530. doi: 10.1073/pnas.1716861115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson DM, Sato H, Dirck AT, Feix JB, Frank DW. 2015. Ubiquitin activates patatin-like phospholipases from multiple bacterial species. J Bacteriol 197:529–541. doi: 10.1128/JB.02402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 17.Tyson GH, Halavaty AS, Kim H, Geissler B, Agard M, Satchell KJ, Cho W, Anderson WF, Hauser AR. 2015. A novel phosphatidylinositol 4,5-bisphosphate binding domain mediates plasma membrane localization of ExoU and other patatin-like phospholipases. J Biol Chem 290:2919–2937. doi: 10.1074/jbc.M114.611251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, DiMaio F, Wang R, Kim D, Miles C, Brunette TJ, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku B, Lee K-H, Park W, Yang C-S, Ge J, Lee S-G, Cha S-S, Shao F, Heo W, Jung JU, Oh B-H. 2012. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog 8:e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas M, Gaspar AH, Pallara C, Rojas A, Fernández-Recio J, Machner MP, Hierro A. 2014. Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc Natl Acad Sci U S A 111:3514–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. 1999. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97:349–360. doi: 10.1016/S0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 23.da Mata Madeira PV, Zouhir S, Basso P, Neves D, Laubier A, Salacha R, Bleves S, Faudry E, Contreras-Martel C, Dessen A. 2016. Structural basis of lipid targeting and destruction by the type V secretion system of Pseudomonas aeruginosa. J Mol Biol 428:1790–1803. doi: 10.1016/j.jmb.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Karakaş M, Woetzel N, Staritzbichler R, Alexander N, Weiner BE, Meiler J. 2012. BCL::Fold: de novo prediction of complex and large protein topologies by assembly of secondary structure elements. PLoS One 7:e49240. doi: 10.1371/journal.pone.0049240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Zhang Y. 2012. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender BJ, Cisneros A, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, DiMaio F, Meiler J, Moretti R. 2016. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry 55:4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban Y-EE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Housley NA, Winkler HH, Audia JP. 2011. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. J Bacteriol 193:4634–4642. doi: 10.1128/JB.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato H, Feix JB, Frank DW. 2006. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry 45:10368–10375. doi: 10.1021/bi060788j. [DOI] [PubMed] [Google Scholar]
- 31.Anderson DM, Schmalzer KM, Sato H, Casey M, Terhune SS, Haas AL, Feix JB, Frank DW. 2011. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol 82:1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaspar AH, Machner MP. 2014. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci U S A 111:4560–4565. doi: 10.1073/pnas.1316376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanRheenen SM, Luo Z-QQ, O’Connor T, Isberg RR. 2006. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun 74:3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viner R, Chetrit D, Ehrlich M, Segal G. 2012. Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog 8:e1002988. doi: 10.1371/journal.ppat.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finck-Barbançon V, Yahr TL, Frank DW. 1998. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J Bacteriol 180:6224–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck-Barbançon V, Frank DW. 2001. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol 183:4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. [DOI] [PubMed] [Google Scholar]
- 38.Benson MA, Schmalzer KM, Frank DW. 2010. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA2 activity. Clin Chim Acta 411:190–197. doi: 10.1016/j.cca.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DE, Chivian D, Baker D. 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 41.Jakobsen TH, Hansen MA, Jensen PØ, Hansen L, Riber L, Cockburn A, Kolpen M, Rønne Hansen C, Ridderberg W, Eickhardt S, Hansen M, Kerpedjiev P, Alhede M, Qvortrup K, Burmølle M, Moser C, Kühl M, Ciofu O, Givskov M, Sørensen SJJ, Høiby N, Bjarnsholt T. 2013. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS One 8:e68484. doi: 10.1371/journal.pone.0068484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farfán M, Spataro N, Sanglas A, Albarral V, Lorén JG, Bosch E, Fusté MC. 2013. Draft genome sequence of the Aeromonas diversa type strain. Genome Announc 1:e00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leggieri N, Gouriet F, Thuny F, Habib G, Raoult D, Casalta J-PP. 2012. Legionella longbeachae and endocarditis. Emerg Infect Dis 18:95–97. doi: 10.3201/eid1801.110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou ICC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 45.Testa J, Daniel LW, Kreger AS. 1984. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect Immun 45:458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.