The molecular mechanisms responsible for selective motor neuron loss in SMA remain elusive. Rizzo et al. show that deregulated transcripts in SMA-motor neurons share motif 7, targeted by SYNCRIP, which binds SMN. Impaired SYNCRIP-SMN interaction leads to dysregulation of downstream genes, such as NEUREXIN2, which could represent therapeutic targets.
Keywords: motor neurons, RNA sequencing, NRXN2, SYNCRIP, SMN1
Abstract
Spinal muscular atrophy is a motor neuron disorder caused by mutations in SMN1. The reasons for the selective vulnerability of motor neurons linked to SMN (encoded by SMN1) reduction remain unclear. Therefore, we performed deep RNA sequencing on human spinal muscular atrophy motor neurons to detect specific altered gene splicing/expression and to identify the presence of a common sequence motif in these genes. Many deregulated genes, such as the neurexin and synaptotagmin families, are implicated in critical motor neuron functions. Motif-enrichment analyses of differentially expressed/spliced genes, including neurexin2 (NRXN2), revealed a common motif, motif 7, which is a target of SYNCRIP. Interestingly, SYNCRIP interacts only with full-length SMN, binding and modulating several motor neuron transcripts, including SMN itself. SYNCRIP overexpression rescued spinal muscular atrophy motor neurons, due to the subsequent increase in SMN and their downstream target NRXN2 through a positive loop mechanism and ameliorated SMN-loss-related pathological phenotypes in Caenorhabditis elegans and mouse models. SMN/SYNCRIP complex through motif 7 may account for selective motor neuron degeneration and represent a potential therapeutic target.
Introduction
Spinal muscular atrophy (SMA) is a severe genetic neuromuscular disease with early onset and represents the most common genetic cause of infant mortality (Pellizzoni et al., 1998; Faravelli et al., 2015). Affected children present atrophic muscular masses, weak and hypotonic limbs, and life-threatening respiratory complications (Pellizzoni et al., 1998; Faravelli et al., 2015). SMA is caused by mutations in the survival motor neuron 1 gene (SMN1), which impairs the function and survival of lower motor neurons in the spinal cord (Lefebvre et al., 1995), but why deficiency in this ubiquitously expressed protein primarily affects motor neurons is unclear.
The majority of current SMA therapeutic approaches, such as nusinersen, the only compound recently approved by the FDA/EMA (www.curesma.it), are focused on increasing the levels of full-length SMN protein (Pellizzoni et al., 1998; Foust et al., 2010; Faravelli et al., 2015). However, SMN-independent approaches to target downstream pathological events can be valuable as complementary strategies, particularly in the symptomatic phase of the disease. Furthermore, understanding the complex series of mechanisms and pathways responsible for the effect of SMN deficiency is crucial for monitoring therapeutic efficacy and allowing further therapeutic progress, including the development of novel effective cures.
SMN plays a key role in RNA splicing (Pellizzoni et al., 1998), but its precise function is not completely clear. The absence of SMN causes improper assembly of spliceosomal small nuclear ribonucleoproteins (snRNPs), leading to impaired splicing. SMN may also play a direct role in splicing, interacting transiently with components of the splicing machinery, acting prior to their assembly into the spliceosome (Mourelatos et al., 2001), suggesting a direct molecular link between SMN, pre-mRNA splicing, and other RNA processing events that could be impaired when levels of full-length SMN are reduced.
Many studies have documented the association between SMN and several RNA binding proteins (RBPs) that are not part of the SMN complex, such as HuD or heterogeneous nuclear ribonucleoprotein (hnRNP) R, fragile X mental retardation protein (FMRP), and amyotrophic lateral sclerosis-related proteins FUS and TDP43. These factors are involved in the splicing, transport, stability, and translation of mRNAs, among other aspects of RNA metabolism (Pellizzoni et al., 1998).
Alterations in gene expression and splicing have been shown in tissues derived from SMA mice (Zhang et al., 2008, 2013; Lotti et al., 2012; Maeda et al., 2014) and patient samples (Corti et al., 2012; Ng et al., 2015). However, analyses of the correlation between SMN defects, specific gene expression, and splicing alterations in human SMA motor neurons, and studies of common motif sequences present in these altered genes are still lacking. This study could suggest a mechanistic link between aberrant transcripts and the motor neuron selective dysfunction observed in SMA.
To investigate these aspects, we performed RNA sequencing (RNA-Seq) of SMA motor neurons derived from patients’ induced pluripotent stem cells (iPSCs) and healthy lines and we identified SMA-specific molecular changes, including gene expression and splicing alterations in transcripts related to cytoskeletal, axonal, and synaptic functions, such as neurexin 2 (NRXN2), a neuron-specific gene encoding a presynaptic cell adhesion protein involved in motor neuron survival and function. Other groups have already observed reduced expression and altered alternative splicing of NRXN2 in transgenic SMA mice (See et al., 2014). In the present study, the overexpression of NRXN2 was shown to improve human SMA motor neuron survival and increase motor axon length, suggesting a role as a modifier gene.
Motif enrichment analysis of differentially expressed/spliced genes in SMA motor neurons identified motif 7 as one of the most significantly enriched. Notably NRXN2 also possesses motif 7, which is a common target sequence for RBPs, particularly hnRNPs. Among these, hnRNPQ (SYNCRIP), which is highly homologous to hnRNPR, recognizes a large number of key motor neuron genes and is likely implicated in misregulation of their gene expression/splicing (Mourelatos et al., 2001; Rossoll et al., 2002). This observation goes along with the finding that SYNCRIP protein directly interacts only with full-length SMN and not with the non-functional Δ7 truncated form, the most prevalent SMN mutant found in SMA patients (Mourelatos et al., 2001; Rossoll et al., 2002). Importantly, SYNCRIP has been studied as a splicing modulator of SMN, promoting the inclusion of exon 7 in SMN2, probably by activating the upstream 3′ splice site (Chen et al., 2008). Overall, this suggests that the disruption of SMN-SYNCRIP-dependent RNA pathways causes specific downstream molecular defects, including deregulation of NRXN2, which can contribute to motor neuron death and axonal impairment. Interestingly, the overexpression of SYNCRIP rescued the phenotype in our SMA cells due to the subsequent increase in SMN and NRXN2, demonstrating the presence of a positive loop among SYNCRIP, SMN, and NRXN2. Noteworthy, the overexpression of SYNCRIP also rescued the neurodegenerative phenotype observed in vivo in a C. elegans model (Gallotta et al., 2016). This study produced novel insights into the role of SMN loss in mRNA processing and its selective effects on human SMA-motor neurons, highlighting novel targets for therapeutic strategies.
Materials and methods
Induced pluripotent stem cell lines
The studies involving human samples were conducted in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and with national legislation and institutional guidelines. After obtaining informed consent, fibroblasts were generated from dermal biopsies (Eurobiobank) as described previously (ethical committee approval at the IRCCS Foundation Ca’ Granda Ospedale Maggiore Policlinico) (Corti et al., 2012; Rizzo et al., 2016, 2017). The iPSCs were generated by non-viral transduction with six reprogramming factors: OCT4, SOX2, NANOG, LIN28, c-Myc, and KLF4 (Supplementary Table 1; Corti et al., 2012). IPSC colonies with embryonic stem cell-like morphology were expanded and detached for analysis. IPSCs were grown on Matrigel® (BD Biosciences) with E8 media (Life Technologies). All cell cultures were maintained at 37°C in 5% CO2.
Differentiation of iPSCs into motor neurons
We generated spinal motor neurons using a multistage differentiation protocol (Corti et al., 2012; Rizzo et al., 2016, 2017). Wild-type and SMA iPSCs were plated with neuronal medium (DMEM/F12; Gibco, Invitrogen) supplemented with MEM non-essential amino acids, N2, and heparin (2 μg/ml, Sigma-Aldrich). After 10 days, we added retinoic acid (0.1 μM, Sigma-Aldrich) for neural caudalization. On Day 17, we collected the posteriorized neuroectodermal cells. These clusters were suspended for 1 week in the same medium with retinoic acid (0.1 μM) and sonic hedgehog (SHH; 100–200 ng/ml, R&D Systems, Inc.). On Day 24, we supplemented the medium with other growth factors e.g. brain-derived neurotrophic factor (BDNF), glial-derived neurotrophic factor (GDNF), and IGF1 (10 ng/ml, Peprotech). To achieve greater cell purity, motor neurons were enriched using a gradient centrifugation method (Corti et al., 2012). To monitor the proper acquisition of a motor neuron phenotype, cells were transduced with a lenti-Hb9::GFP construct (Marchetto et al., 2008), fixed, and stained for quantification using established neuronal and motor neuron markers. For genetic modification, cultures were infected with lentivirus-NRXN2 (Origene RC219788, Accession number NM_015080), lentivirus-SYNCRIP, lentivirus-SMN, or null vector, which was used as a negative control (Simone et al., 2014).
SH-SY5Y transfection
The human SH-SY5Y neural cell line was cultured in DMEM high glucose medium containing 10% foetal bovine serum (FBS), 2.5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all purchased from Euroclone). The cells were transfected with 15 nM SYNCRIP siRNA (Thermofisher, ID s20564) or with select negative control (Thermofisher, ID 4390843), which was used as a negative control, and harvested after 48 h.
Immunocytochemistry and phenotypic analysis of iPSCs and motor neurons
Cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with Triton™ 0.25%, and then blocked with 10% bovine serum albumin in phosphate-buffered saline and 0.3% Triton™ X-100 for 1 h at room temperature. We incubated the cells with primary antibodies to NANOG (1:100, Abcam), SSEA-3 (1.100, Covance), ChAT (1:200, Chemicon and Millipore), SMI32 (1:500, Covance), hnRNP-Q (1:100, Sigma), and NRXN 2α (1:100, Abcam) overnight, and then with anti-rabbit, anti-mouse, or anti-goat Alexa Fluor® 488 or 594 (1:400; Life Technologies) secondary antibody for 1.5 h at room temperature. Negative controls were performed for all stains. Microphotographs were taken with a LEICA LCS2 confocal microscope.
Motor neuron morphometric analysis
Phenotypic analysis was performed as described previously (Corti et al., 2012; Nizzardo et al., 2015; Allodi et al., 2016). We quantified the motor neurons by determining cells positive for motor neuron marker and counting 10 randomly selected fields/well. Morphometric and axon length analyses were performed by measuring soma diameter and length distance between two points (one point from the soma and one on the distal axon) (Corti et al., 2012). All quantification analyses were carried out in a double-blind fashion. For all imaging, we used a confocal LEICA SP2.
RNA analysis
Selected cells were treated with proteinase K for 1 h at 50°C before the addition of TRIzol® LS (Life Technologies) for RNA extraction. DNase treatment using Turbo DNA-free™ (Ambion) was performed to remove contaminating genomic DNA. The RNA analysis was processed by DNA vision as follows. A sample quality control was performed on an Agilent 2100 Bioanalyzer and quantity checked by Nanodrop. Next, the TrueSeq mRNA library was constructed, followed by Library Quality Control on the Agilent 2100 Bioanalyzer. Sequencing was performed on a HiSeq 2000, two samples per lane with a 2 × 100 cycles strategy.
RNA-Seq reads mapping
The raw data generated by sequencing were quality-checked using the FastQC tool and cleaned of low quality reads, adaptor sequences, and other contaminants. Quality was judged in terms of read length distribution, Phred score distribution, and nucleotide frequencies obtained from FastQC statistics. STAR software (Dobin et al., 2013) was then used to align the reads to the reference Homo sapiens Ensembl GRCh37 genome (Flicek et al., 2014). Parameters included the removal of non-canonical junctions, the generation of the XS strand attribute for all alignments that contain splice junctions, and the sorting of the output files in bam format. The annotation provided in Illumina iGenomes repository was given as an input to the STAR routine, as well as all other routines requiring an annotation file.
Alternative splicing differential analysis
To detect the differential usage of exons between patients and controls, data were processed using the rMATS tool (Shen et al., 2014). The tool was run in paired-end data processing mode, with declared length of reads of 101 bp and with a junction anchor length of 4. The statistical model of rMATS calculated the inclusion level difference (ILD), P-value, and false discovery rate (FDR) for each skipping exon from a list obtained from both data and annotation. Exons were then separated on the basis of three conditions: (i) enhanced exons (ILD > 0, |ILD| > median|ILD|, FDR < 0.05); (ii) silenced exons (ILD < 0, |ILD| > median|ILD|, FDR < 0.05); and (iii) invariant exons (|ILD| < median|ILD|, FDR > 0.50).
Motif discovery
The enhanced and silenced cassette exons were analysed to identify sequence patterns inside both exons and flanking sequences that discriminate cassette exons differentially included in patients. The genomic sequences were retrieved for enhanced, silenced, and control exons and their flanking upstream and downstream introns using GeCo++ library (Cereda et al., 2011). The flanking intronic regions were selected for 150 bp fixed length and location −10/−160 bp upstream and +10/+160 bp downstream from the exon. For each sequence, the frequency distribution of all possible tetramers was estimated and collected in a matrix. Projective non-negative matrix factorization was then applied to identify a small set of distributions (motifs) that approximate the full matrix. The factorization rank r corresponds to the number of desired motifs. For each sequence, we then calculated the Battacharyya coefficient between the corresponding distribution and each motif representing a set of r features. At this point, each cassette exon was characterized by a set of 3r features comprising upstream intronic, exonic, and downstream intronic sequence features.
This set of features was then used as variables in logistic regressions to identify the parameters of two distinct models and distinguish controls from enhanced and silenced exons. We evaluated the performance of the models by 10-fold cross-validation AUROC (area under the receiver operating characteristic). The entire procedure was repeated for increasing number of motifs (r) until no AUROC improvement. A manuscript with a full description of this method is in preparation.
The obtained motifs were then compared to a set of 119 vertebrate RBP motifs selected from a publicly available database (Ray et al., 2013). Again, we used Battacharyya coefficients to measure the similarity between one of our motifs and the tetramer distributions obtained for each of the Ray dataset position weight matrices. Significant similarities were obtained through 200 000 random permutations of the tetramer distributions. The proportion of permutations, resulting in a higher Battacharyya coefficient than the observed one, were used as empirical P-value for the comparison. Bonferroni correction was applied to take into account the fact that we performed 119 (Ray dataset) ×15 (our motifs) comparisons.
Gene and transcript quantification and differential expression
The files containing data alignments to genome assembly were processed by Cufflinks tools (Trapnell et al., 2013) to compute the gene and transcript expression levels. Cuffquant routine was run first on each individual sample with the ‘-u’ flag, supporting correction of multiple reads (i.e. reads mapped at multiple loci within the genome). Next, the Cuffnorm routine was run to normalize the quantified expression with respect to possible differences in library size, sequencing depth, and/or transcript length. The resulting normalized abundances, measured as fragments per kilobase of transcript per million mapped reads (FPKM), were further used to build a dendrogram plot of all samples and replicates. The results of the Cuffquant routine were further employed for the differential expression analysis between patients with SMA and controls. Thus, the Cuffdiff2 routine was run with the option of multiple reads correction. Differentially expressed genes were then separated into three sets on the basis of three conditions: (i) significant upregulation in patients with SMA (log2fold-change > 0, q < 0.05); (ii) statistically significant downregulation in patients with SMA (log2fold-change < 0, q < 0.05); and (iii) lack of significance between patients and controls.
Functional annotation analysis
The web-based functional annotation analysis tool Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang da et al., 2009) was applied to evaluate the list of significantly differentially used exons, and to obtain a set of enriched functional annotations in three domains: biological process (BP), cellular component (CC), and molecular function (MF). For each exon, the corresponding gene was considered. The complete set of genes, inclusive of all significantly up- and downregulated exons, was provided as the target (Nt = 4175) and tested against all genes, named background (Nb = 29 237).
The analysis was repeated for the differentially expressed genes (P < 0.05). The complete set of significantly up- and downregulated genes was provided as the target (Nt = 1795) and tested against all genes, named background (Nb = 16 122).
In all analyses, the classification stringency was set to high. We also used gene set enrichment analysis (GSEA) as a bioinformatics tool (Subramanian et al., 2005). Enrichment for up- or downregulated sets of genes from the REACTOME pathway and GO (gene ontology) term database was performed by running GSEA against the test statistic-ranked list of genes in the experiment. Ranking was based on the Cuffdiff 2-derived test statistic. All REACTOME and GO term gene sets with >15 members in the MSigDB package ‘c2.all.v3.0.symbols.gmt’ and ‘c5.all.v4.0.symbols.gmt’, respectively, were downloaded from ftp://gseaftp.broadinstitute.org/.
RBP motif enrichment of differentially expressed genes
Among the 29 237 genes analysed, we only kept those for which the most representative transcript (i.e. the one with the highest Cuffquant estimated mean FPKM) was fully annotated as coding [i.e. both the 3′ and 5′ untranslated region (UTR) and coding sequence (CDS) were present]. A subset of 13 853 genes was obtained with the most representative coding transcript for each, 1385 from differentially expressed genes.
Motif enrichment was performed in differentially expressed genes as follows. For each transcript and motif, a profile was calculated separately for the 5′ UTR, CDS, and 3′ UTR sequences. Profiles were obtained by evaluating the tetramer frequency distribution in 50-bp sliding windows at each position along each sequence and by assigning each position its similarity with the motif (Bhattacharyya coefficient). A motif was then considered present in a profile if its maximum window similarity was >0.7.
Considering the set of 5′ UTR profiles derived from differentially expressed genes (D5′UTR), we merged all of the 5′ UTR profiles into a unique profile, then starting from a random position we mapped a number of consecutive intervals equal to the number of D5′UTRs of matching length. We counted the number of motifs containing intervals and compared it to the number of motifs containing D5′UTRs. We repeated this step 10 000 times and obtained the empirical enrichment P-value as the proportion of iterations that gave a number of motifs containing intervals equal or greater to the number of motifs containing D5′UTR. The entire procedure was repeated for CDS and 3′ UTR profiles derived from differentially expressed genes.
Other bioinformatics analysis
For RNA-Seq data analysis, we used two bioinformatics tools that query for enriched GO terms: gene-annotation enrichment analysis with DAVID (Huang da et al., 2009) and GSEA (Subramanian et al., 2005). Enrichment for sets of genes from the REACTOME pathway and GO term database was performed by running GSEA against the test statistic-ranked list of genes in the experiment. Ranking was based on the Cuffdiff 2-derived test statistic. All REACTOME and GO term gene sets with >15 members in the MSigDB package ‘c2.all.v3.0.symbols.gmt’ and ‘c5.all.v4.0.symbols.gmt’, respectively, were downloaded from ftp://gseaftp.broadinstitute.org/.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells using the RNeasy® Mini Kit (Qiagen) and concentrations measured by a Nanodrop spectrophotometer. Only samples with ratios between 1.8 and 2.0 were analysed further. A total of 1.5 μg of total RNA was reverse-transcribed using the Ready-To-Go™ kit (GE Healthcare). Reverse-transcribed material (5 ng for each sample) was amplified using the TaqMan® Universal PCR Master Mix (Applied Biosystems) and appropriate probes to evaluate gene expression and exons (probe ID available upon request, Supplementary Table 2) in the 7500 Real Time PCR System (Software 2.01, Applied Biosystems). Expression levels were normalized to the average level of housekeeping gene 18S and referred to the relevant control samples.
Western blot analysis
Western blot analysis was performed as described previously (Nizzardo et al., 2014a; Rizzo et al., 2016). Briefly, cells were sonicated on ice for 10 min in buffer supplemented with protease and phosphatase inhibitor cocktail (Pierce) (Nizzardo et al., 2014a; Rizzo et al., 2016). Alternatively, 20 mg of frozen brain were homogenized in 0.4 ml of protein sample buffer containing 2% (w/v) SDS, 10% (v/v) glycerol, 50 mM Tris-HCl (pH 6.8), and 0.1 M DTT. A total of 50 µg was separated via 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were transferred to a nitrocellulose membrane and incubated with primary antibodies overnight at 4°C. The primary antibodies used in these experiments were: NFs (1:1000, Sigma), MMP14 (Thermofisher, 1:1000), SYT13 (1:1300, Proteintech), NRXN1 (1:2000, Abcam), NRXN2α (1:500, Abcam), SMN (1:1.000, BD), STMN2 (1:2000, Proteintech), PLP1(1:800, Sigma), and hnRNP-Q (1:1000, Sigma). The blots were then incubated in secondary antibody: polyclonal anti-rabbit (1:2700, Dako) and polyclonal anti-mouse (1:3200, Dako). The immune complexes were revealed by chemiluminescence assay (Amersham). The nitrocellulose membrane was stripped and reprobed with anti-actin (1:1000, Sigma) as a loading control. Densitometry was performed using ImageJ software.
In vivo experiments in the SMA C. elegans model
Nematodes were grown and handled following standard procedures in uncrowded conditions at 20°C on nematode growth medium (NGM) agar plates seeded with Escherichia coli strain OP50 (Brenner, 1974). The wild-type animals were strain N2, variety Bristol. The transgenic strains were: NA1330 gbIs4 [GBF109 punc-25::smn-1(RNAi); GB301 pchs-2::GFP] III; NA1678 gbEx612a [GB301 pchs-2::GFP]; NA1252 gbEx540a [GBF322 punc-119::dsRED; pelt-2::RFP]; EG1285 oxIs12 [punc-47::GFP; lin-15(+)] X; NA1355 gbIs4 III, oxIs12 III (Gallotta et al., 2016). EG2185 and N2 were provided by the Caenorhabditis Genetics Center (CGC) funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The following strains were obtained by genetic crosses: NA2052 gbIs4 III, gbEx540a, oxIs12 X; NA2017 gbIs4 III, gbEx540a. The rescue construct (GBF362 punc-119::HRP-2) for pan-neuronal expression of hrp-2 was created by PCR fusion (Hobert, 2002) of unc-119 promoter with hrp-2, both amplified from genomic sequences. All primer sequences are available upon request. Germline transformation was accomplished as described by Mello et al. (1991) by injecting a DNA mixture containing the transgenic construct at 2 ng/∝l (lower concentration, LC) and 20 ng/∝l (higher concentration, HC) into the gonad of NA1330 gbIs4 III adult animals together with a phenotypic marker for selection of the transgenic progeny. We used pJM371 pelt-2::RFP (RFP expression in the nuclei of intestinal cells) as the co-injection marker at 30 ng/∝l, a kind gift of Prof. J.D. McGhee (University of Calgary) (McGhee et al., 2009). Two independent transgenic lines were examined for each concentration: NA2078 gbIs4 III, gbEx656a [GBF362 punc-119::hrp-2 LC; pelt-2::RFP]; NA2079 gbIs4 III, gbEx656b [GBF362 punc-119::hrp-2 LC; pelt-2::RFP]; NA2075 gbIs4 III, gbEx655a [GBF362 punc-119::hrp-2 HC; pelt-2::RFP]; NA2076 gbIs4 III, gbEx655b [GBF362 punc-119::hrp-2 HC; pelt-2::RFP]. Genetic crosses were made to transfer transgenes to the appropriate genetic background to obtain NA2082 gbIs4 III, gbEx655a, oxIs12 X: NA2080/2081 gbIs4 III, gbEx656a/b, oxIs12 X. In all cases, the presence of the desired transgenes was verified by phenotyping the clonal F2s using a dissecting microscope equipped with epifluorescence. Two independent clones with the same genotype were examined after each cross, and the mean of the two clones has been reported in the results. Animals were immobilized in 0.01% tetramisole hydrochloride (Sigma-Aldrich) on 4% agar pads and visualized using a Zeiss Axioskop microscope. The phenotypes of dying motor neurons were scored by counting the number of ventrally located D-type motor neurons acquiring fluorescence, which we previously demonstrated to be specific for smn-1 silencing, a late sign of apoptosis, visible in the absence of any motor neuron-specific expression of GFP, different from endogenous autofluorescence in the intestine, and never observed in control animals (Gallotta et al., 2016). The degenerative phenotype was scored by counting the number of visible, and hence viable, D-type motor neurons expressing GFP due to a motor neuron-specific promoter (punc-47::GFP) (McIntire et al., 1997). Microscopes used for scoring and imaging were equipped with epifluorescence and DIC Nomarski optics. Epifluorescence and confocal images were collected with a Leica TCS SP8 AOBS microscope, using 40× and 20× objectives, respectively. Epifluorescence images were reconstructed using the Tile Scan function of the Leica LAS X program, which automatically assembles a picture by taking a number of adjoining images of the sample and merges them digitally to obtain a larger view. Confocal images were collected using a 20× objective on a Leica TCS SP8 AOBS confocal microscope. Well-fed, young adult animals were used for the backward movement assay (McIntire et al., 1993) to test D-type motor neuron function. The assay was performed blindly on 6-cm diameter NGM plates seeded with bacteria. Using an eyelash, the animal was touched first on the tail to induce forward movement and then on the head to test for backward movement. Defective movement was scored when animals were unable to fully move backward. For each dataset, the percentage of defective animals among the total number of tested animals was calculated.
In vivo experiments in the SMA mouse model
The SMAΔ7 mouse transgenic model was used. Heterozygous mice (Smn+/−, hSMN2+/+, SMNΔ7+/+, Jackson Laboratories) were bred and pups identified by genotyping (Le et al., 2005). All animal experiments were approved by the University of Milan and Italian Ministry of Health review boards. Homozygous affected pups were cryoanaesthetized and injected intracerebroventricularly as described previously (Nizzardo et al., 2014b) on postnatal Day 1 (P1) with 1011 particles of adenovirus-associated vector serotype 9 (AAV9) vector expressing Syncrip or null vector (SignaGen Laboratories). The brain (n = 3/group) was collected and harvested at P7 for western blot analysis. Disease onset, progression, survival, and motor function (righting test) were monitored after treatment (n = 5/group) as described previously (Nizzardo et al., 2014b). All tests were performed blinded to the mouse genotype and treatment. Intercostal muscles were collected, frozen on dry ice at P10 (n = 3/group), cryosectioned (20 µm), and stained for neuromuscular junction (NMJ) detection and counting. All sections were saturated with 10% bovine serum albumin and 0.3% Triton™ X-100 for 1 h at room temperature before incubation with rabbit Neurofilament Medium (NF-M, Millipore 1:250) overnight at 4°C. The next day, the slides were incubated with Alexa Fluor® 488 (1:1000; Life Technologies) and α-bungarotoxin 555 (1:500, Life Technologies). A minimum of 100 NMJs from each muscle were randomly selected and the number of denervated/degenerated NMJs was determined for each muscle group in each animal based on NF-M/α-BTX staining.
Statistical analysis
Statistical analyses were carried out in StatsDirect for Windows (version 2.6.4) or GraphPad Prism 5 software. Two-tailed, unpaired Student’s t-test was used to compare two groups. Differences in axonal length were investigated by the Kolmogorov-Smirnov test (http://www.physics.csbsju.edu/stats/KS-test.n.plot_form.html). The Kaplan–Meier log rank test and logistic regression analysis were used to compare lifespans. Contingency and chi-squared tests were used to determine differences in the righting test and NMJ innervation. All experiments were carried out in at least triplicate. One-way ANOVA with Kruskal-Wallis test was used for C. elegans data related to statistical analyses.
The experimental results are provided as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). The null hypothesis was rejected at the 0.05 level.
Data availability
The data that support the findings of this study are openly available in GEO at https://www.ncbi.nlm.nih.gov/geo/ reference number GSE108094.
Results
Motor neurons generated from SMA patient iPSCs present reduced cell survival and axonal length in culture
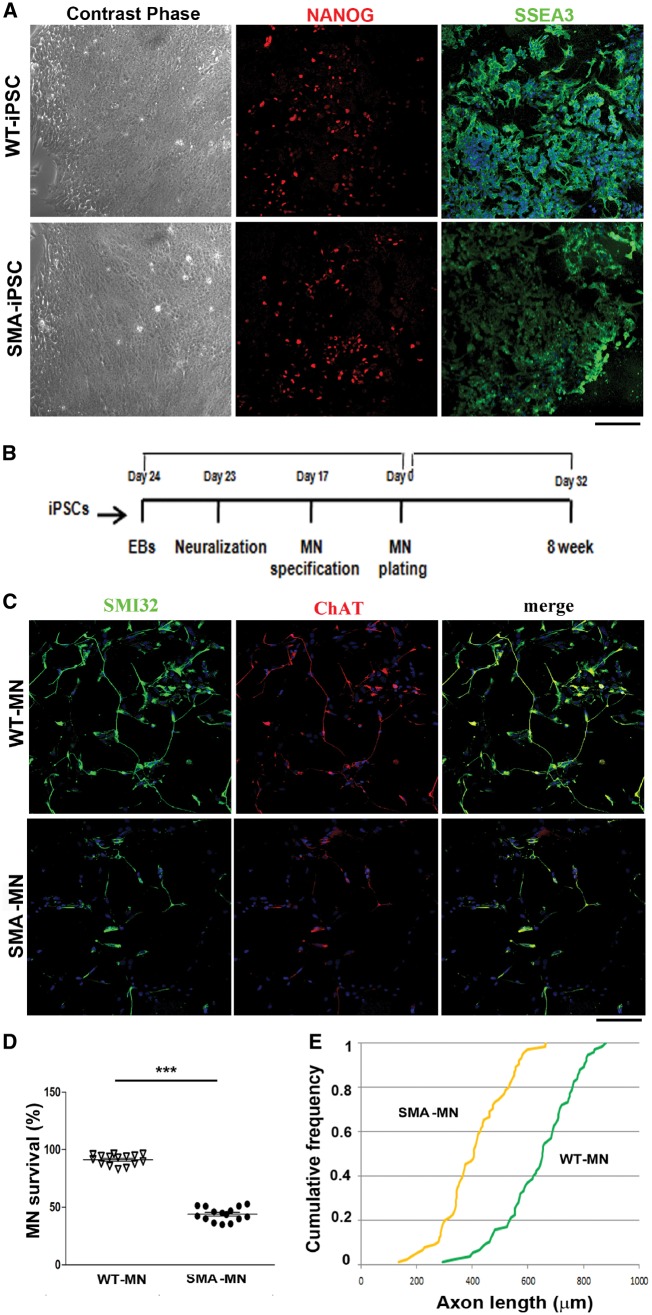
We previously generated iPSCs from type 1 SMA patients and healthy subject fibroblasts using a non-viral, non-integrating method (Supplementary Table 1; Corti et al., 2012; Nizzardo et al., 2014a). Obtained iPSCs exhibited markers of pluripotency (Fig. 1A) and were able to differentiate into motor neurons using established protocols (Fig. 1B) (Corti et al., 2012; Nizzardo et al., 2015).
Figure 1.
SMA iPSCs and the derived motor neurons express typical specific cell type markers. (A) Left: Wild-type and SMA iPSCs showed the typical pluripotent stem cell colony morphology under the contrast phase microscope. Right: The cells express the pluripotency markers NANOG (red) and SSEA3 (green). Nuclei are labelled with DAPI (blue). Scale bar = 75 µm. (B) Experimental outline for wild-type and SMA motor neuron differentiation. (C) Immunocytochemistry of wild-type and SMA motor neurons. The cells were positive for typical motor neuron markers SMI32 (green) and ChAT (red) (merge, yellow signal). Nuclei are labelled with DAPI (blue). Scale bar = 75 μm. (D) Eight weeks after differentiation, SMA motor neuron survival was significantly reduced compared to wild-type. ***P < 0.0001, Student t-test. Error bars represent SEM. (E) After 8 weeks of culture, SMA motor neurons exhibited significantly reduced axon elongation with respect to wild-type. P < 0.001, Kolmogorov-Smirnov test, five independent experiments.
Differentiated cells expressed motor neuron-specific transcription factors, such as spinal cord progenitor markers HB9, ISLET1, and OLIG2 and pan-neuronal markers TuJ1, neurofilament, and MAP2. The majority of these HB9/ISLET1-positive neurons expressed choline acetyl transferase (ChAT) and were positive for motor neuron marker SMI-32, demonstrating a mature motor neuron phenotype (Fig. 1C). The in vitro differentiation protocol yielded a mixed cell population that included non-motor neuron cells. Given the limited availability of surface markers to isolate and purify motor neurons, we applied a physical strategy based on gradient centrifugation. After selection, immunocytochemistry revealed that the percentage of ChAT+ SMI32+ cells was 89.6 ± 8.4% for cells derived from wild-type iPSCs and 87.6 ± 7.7% for cells derived from SMA-iPSCs. Less than 1% of cells differentiated from iPSCs expressed the astrocyte marker GFAP (data not shown). At 8 weeks, we observed a reduction in the number of motor neurons and axonal length in the SMA-iPSC cultures compared to wild-type-iPSCs (P < 0.0001, Fig. 1D; P < 0.001, Fig. 1E), which is in line with previous reports that SMA iPSC-derived motor neurons exhibit reduced survival in long-term culture (Ebert et al., 2009; Nizzardo et al., 2015).
RNA sequencing of SMA motor neurons shows specific changes in key motor neuron function
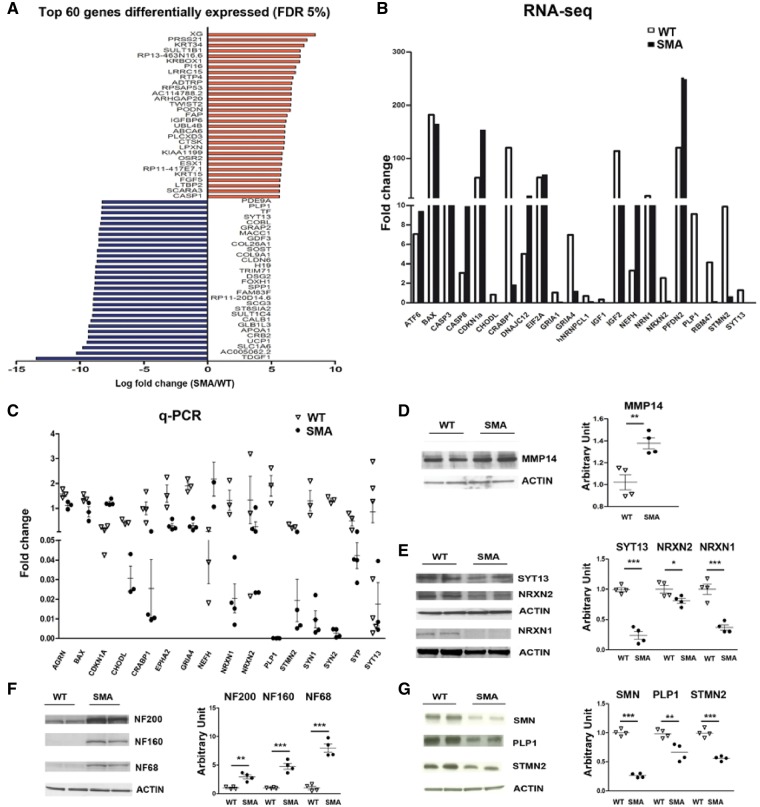
We performed massively parallel deep RNA sequencing targeting 40M clusters per sample to evaluate global gene expression in SMA type I and wild-type motor neurons to identify differentially expressed/spliced transcripts in SMA motor neurons. The analysis revealed a downregulation of 1084 genes and upregulation of 808 genes out of the whole set of 24 691 genes tested (fold change cut-off = 1.5; P < 0.01; Supplementary Table 3). We identified the 30 transcripts most increased and decreased in abundance at an FDR of 5% (Fig. 2A). The web-based functional annotation analysis tool DAVID was applied to evaluate the RNA-Seq data and obtain a set of enriched GO terms (Supplementary Tables 4 and 5). The most relevant GO terms are provided in Supplementary Fig. 1.
Figure 2.
Transcriptional analysis of SMA versus wild-type-motor neurons by RNA-Seq analysis shows deregulation of specific mRNAs involved in motor neuron functions. (A) Top 60 genes (based on fold change) differentially expressed in SMA versus wild-type motor neurons with an FDR of 5%. (B and C) Comparison of RNA-Seq and quantitative RT-PCR data or a selected number of transcripts in wild-type and SMA motor neurons. STMN2 (**P < 0.001), PLP1 (*P < 0.05), NEFH (*P < 0.05), NRXN1 (*P < 0.05), SYN1 (*P < 0.05), SYN2 (***P < 0.0001), EPH2A (*P < 0.05), SYP (*P < 0.05), CRABP1 (**P < 0.01), CDKN1A (***P < 0.0001), CHODL (*P < 0.05), GRIA4 (**P < 0.01). Student’s t-test. Error bars represent ± SEM from three independent experiments. (D–G) Representative image of western blot of selected proteins in SMA motor neurons compared to wild-type motor neurons: MMP14: **P < 0.01; SYT13: ***P < 0.0001; NRXN2: *P < 0.05; NRXN1: ***P < 0.001; NF200: **P < 0.01; NF160: ***P < 0.001; NF68: ***P < 0.001; SMN: ***P < 0.0001, PLP1: **P < 0.01; STMN2: ***P < 0.001, Student’s t-test. Data are presented as mean ± SEM from three independent experiments.
Interestingly, in the Reactome analysis we observed a significant reduction in transcripts related to specific neural assets in SMA motor neurons, including axon-related proteins [i.e. stathmin 2 (STMN2), proteolipid protein 1 (PLP1)], ion channels, particularly potassium channel, and synapses [i.e. synaptotagmin 13 (SYT13), and neurexin (NRXN) 1, 2, and 3] (Fig. 2B). An upregulation of neurofilament heavy polypeptide (NEFH) was detected (Table 1).
Table 1.
Selected enriched gene sets obtained with Reactome analysis that included terms related to axon-related proteins, synapses, and potassium channels
| Category | Gene | Expression |
|---|---|---|
| Axon-related protein | NEFH | Up |
| STMN2 | Down | |
| PLP1 | Down | |
| NCAN | Down | |
| RGMA | Down | |
| UNC5A | Down | |
| Synapsis | NRXN1 | Down |
| NRXN2 | Down | |
| NRXN3 | Down | |
| SYT7 | Up | |
| SYT9 | Down | |
| SYT11 | Up | |
| SYT13 | Down | |
| VAMP8 | Down | |
| SYCP2 | Down | |
| RIMS4 | Down | |
| BSN | Down | |
| SNAP91 | Down | |
| STXBP1 | Up | |
| Potassium channels | KCNA3 | Down |
| KCNA5 | Down | |
| KCNC1 | Down | |
| KCND2 | Down | |
| KCNF1 | Down | |
| KCNG1 | Down | |
| KCNJ10 | Down | |
| KCNK3 | Down | |
| KCNN1 | Down | |
| KCNN2 | Down | |
| KCNQ3 | Up | |
| KCNC4 | Up | |
| KCNN4 | Up |
The selected genes upregulated and downregulated in SMA motor neurons are shown in bold and italics, respectively.
Cross-checking our RNA-Seq data (Fig. 2B) with data published in the literature, we found sets of genes differentially expressed between SMA and wild-type motor neurons that are likely involved in SMA vulnerability. We validated some of the genes by western blot and quantitative PCR (qPCR) analysis (Fig. 2C–G). We detected a reduction in glutamate ionotropic receptor AMPA-type subunit 1 (GRIA1) and, even though not significant, GRIA2 and GRIA4, which encode core subunits of AMPA-type glutamate receptors that are critical for glutamatergic excitatory synapses (Zhang et al., 2013). We also found dysregulation of IGF-related pathways, detecting the downregulation of insulin-like growth factor 1 (IGF1) and one of its substrates, insulin receptor substrate 4 (IRS4), in motor neurons, as well as the downregulation of IGF2. Both IGF1 and IGF2 play a significant role in SMA motor neuron survival, as we recently demonstrated (Allodi et al., 2016). The pathology of SMA has been linked to the endoplasmic reticulum (ER) stress pathway and apoptosis (Ng et al., 2015), and we evaluated whether some of these genes were deregulated. Some heat shock proteins, unfolded protein response (UPR), and apoptosis genes were upregulated, suggesting that activation of the UPR pathway in SMA motor neurons leads to hyperactivation of the pro-apoptotic branches of the UPR pathway. In terms of mRNA metabolism, we detected the dysregulation of some proteins involved in mRNA splicing and processing, including the upregulation of muscle blind-like protein 1 (MBLN1), the key gene involved in splicing dysregulation in myotonic dystrophy, and the downregulation of heterogeneous nuclear ribonucleoprotein c-like 1 (hNRNPCL1), RNA binding motif protein 47 (RBM47), and epithelial splicing regulatory protein 2 (ESRP2). This alteration can further promote transcriptional/splicing alterations. We also confirmed the downregulation of some key transcripts altered in other SMA models, such as chondrolectin (CHODL1) (Gavrilina et al., 2008; Zhang et al., 2008; Baumer et al., 2009), neuritin (NRN1) (Akten et al., 2011), and cellular retinoic acid-binding protein 1 (CRABP1), further supporting the potential role of these genes as downstream targets of RNA processing dysfunction induced by SMN deficiency.
Differential splicing analysis
SMN plays a crucial role in the biogenesis of spliceosomal snRNPs; thus, its deficiency is expected to impact splicing (Pellizzoni, 2007; Neuenkirchen et al., 2008). Therefore, we started our analysis by searching for cassette exons that demonstrate a significant difference in the level of inclusion between SMA and wild-type motor neurons (alternative splicing differential analysis). A total of 12 144 cassette exons were deregulated in terms of the inclusion level; 2402 and 1035 exhibited a significant decrease and increase in inclusion, respectively, in patient samples with respect to the controls (Supplementary Table 6). Functional annotation analysis of genes containing both significantly silenced and enhanced cassette exons revealed that the most represented GO terms were in the categories ‘Molecular Function’, ‘Biological Process’, and ‘Cellular Component’ (Table 2, Supplementary Tables 4 and 7, and Supplementary Fig. 2).
Table 2.
GO terms of differentially spliced genes in SMA-motor neurons versus wild-type motor neurons (P < 0.05) using DAVID analysis
| GO category | GO terms |
|---|---|
| Molecular function | RNA binding |
| Protein binding | |
| Cytoskeletal protein binding | |
| Actin binding | |
| Tubulin binding | |
| Microtubule plus-end binding | |
| Protein serine/threonine kinase activity | |
| Rab GTPase activator activity | |
| RNA methyltransferase activity | |
| Biological process | Cell division |
| Cell cycle, differentiation, secretion and proliferation | |
| Regulation of translation and signal transduction | |
| RNA and snRNA | |
| Synaptosome | |
| Flotillin complex | |
| Cellular component | Membranes |
| Intracellular organelles | |
| Mitochondrion and mitochondrial membrane | |
| Cell junctions | |
| Microtubules | |
| Endoplasmic reticulum | |
| Endocytic vesicle |
Interestingly, the genes with the greatest degree of differential splicing, analysed with Reactome, belonged to functional classes considered responsible for SMA pathogenesis, including axon guidance and synapses (Supplementary Table 9). In the vast majority of cases, there was reduced inclusion of an exon.
Among differentially spliced genes, we identified genes involved in motor neuron diseases and function. In particular, PIP5K1C is the causative gene of Lethal Congenital Contracture Syndrome type 3 (LCCS3), a neurological syndrome in which patients present with rather selective atrophy of the spinal cord anterior horn, similar to SMA patients (Narkis et al., 2007). RAC1 has been implicated in motor neuron disease pathogenesis (D’Ambrosi et al., 2014). Furthermore, SINJ1 and RIMS1 were found as alternative spliced genes specifically in motor neurons in a pathway dependent from FUS protein, a key gene in amyotrophic lateral sclerosis (ALS) (Honda et al., 2013).
We also identified genes that were previously detected as mispliced in rodent SMA-motor neurons (Zhang et al., 2013), such as microtubule affinity regulating kinase 2 (MARK2), calcium/calmodulin dependent protein kinase II delta (CAMK2D), agrin (AGRN) (Zhang et al., 2013), which is critical for NMJ maintenance, and M-phase phosphoprotein 9 (MPHOSPH9) (Supplementary Table 9). Some of these genes with altered splicing were validated by RT-PCR in our SMA motor neurons (Supplementary Fig. 7A).
Overall, our data confirm the role of SMN in splicing modulation, particularly for genes related to axon guidance and synapses.
RNA binding motif analysis
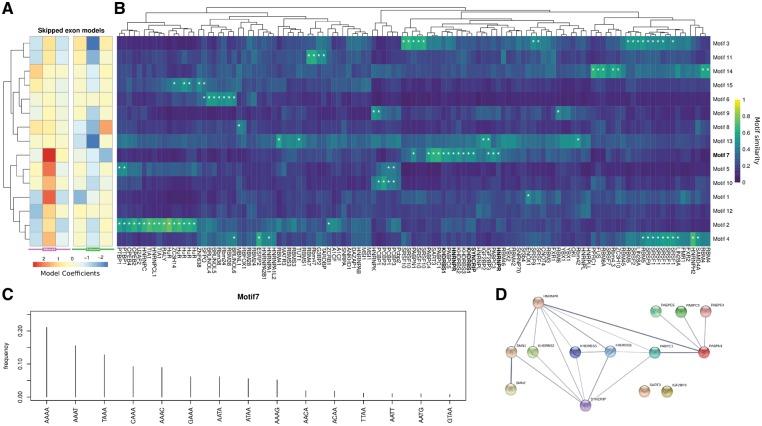
We searched for common RNA sequence motifs (i.e. tetramer distributions) that could predict either silenced or enhanced exons in the whole set of analysed cassette exons. In logistic regression, we found 15 motifs (Supplementary Fig. 3) with an area under the curve (AUC) of 0.629 and 0.623 for enhanced and silenced cassette exons, respectively. Each model assigned three different weights to each motif in exonic, upstream, and downstream intronic regions (Fig. 3A and B). In particular, motif 7 (Fig. 3C) in the silenced exonic region had the highest positive weight, suggesting an enrichment in these regions. We hypothesized that the altered expression of transcript variants among multiple genes is dependent on the interaction of common RBPs with SMN. To examine this possibility, the 15 motifs were compared to a set of 119 vertebrate RNA binding protein motifs derived from a previous work (Ray et al., 2013) using a similarity measure. We completed an inventory of the common motifs and their interacting RBPs (Fig. 3B). Significant similarities were observed, particularly for motif 7, to a group of polyA binding proteins involved in splicing and other mRNA processing, transport, and transcription (Glisovic et al., 2008). Among these, hnRNPQ (SYNCRIP), HNRNPR, and KHDRBS1 have been described as interacting with the full-length SMN protein (Rossoll et al., 2002; Pagliarini et al., 2015; Geuens et al., 2016). The interaction between these polyA binding proteins and SMN are illustrated in Fig. 3D. SYNCRIP emerged as one of the top RPBs regulating the splicing of dysregulated transcripts in SMA cells. SYNCRIP is an SMN-interacting protein partner highly related to hnRNPR (Rossoll et al., 2002). In our experiments, one of the most frequent motifs with highly probable interaction with SYNCRIP is the motif identified as 7 (Fig. 3C). This finding indicates a possible link between the loss of the interaction between SYNCRIP and full-length SMN and inefficient pre-mRNA splicing downstream. Given the possible role of RBPs in transcription and post-transcriptional regulation other than splicing, we wanted to test whether enrichment of these motifs can be detected in differentially expressed genes. Thus, we devised a motif enrichment procedure to test the coding, 5′, and 3′ UTR sequences of differentially expressed genes for motif enrichment (Supplementary Table 8). We found one motif enriched in 5′ UTRs, none in CDSs, and six in 3′ UTRs (including motif 7). No motif was depleted in 5′ UTRs, four in CDS, and three in 3′ UTRs. Remarkably, some genes with high motif 7 scores in 3′ UTRs exhibited axonal and synaptic functions, including NRXN1, NRXN2, and SYT13.
Figure 3.
Differentially spliced cassette exons in RNA motif analysis. (A) Heat map of the logistic regression model coefficients for silenced (violet) and enhanced (green) exons. Both model coefficients are organized according to the region to which they refer (i.e. intronic upstream, exonic and intronic downstream). The 15 motifs are ordered by hierarchical clustering of the corresponding coefficients. (B) Heatmap of the comparison between the 15 motifs and a set of known vertebrate RNA binding protein motifs derived from Ray et al. (2013). Motifs are ordered as in A, whereas RBPs are ordered by hierarchical cluster. White asterisks correspond to significant similarities (Bonferroni corrected empirical P < 0.01). (C) Motif 7 described probability distribution as a tetramer. Probability accounting for 99% is reported. (D) Known interactions between proteins significantly similar to motif 7 and SMN1/2 according to the STRING database.
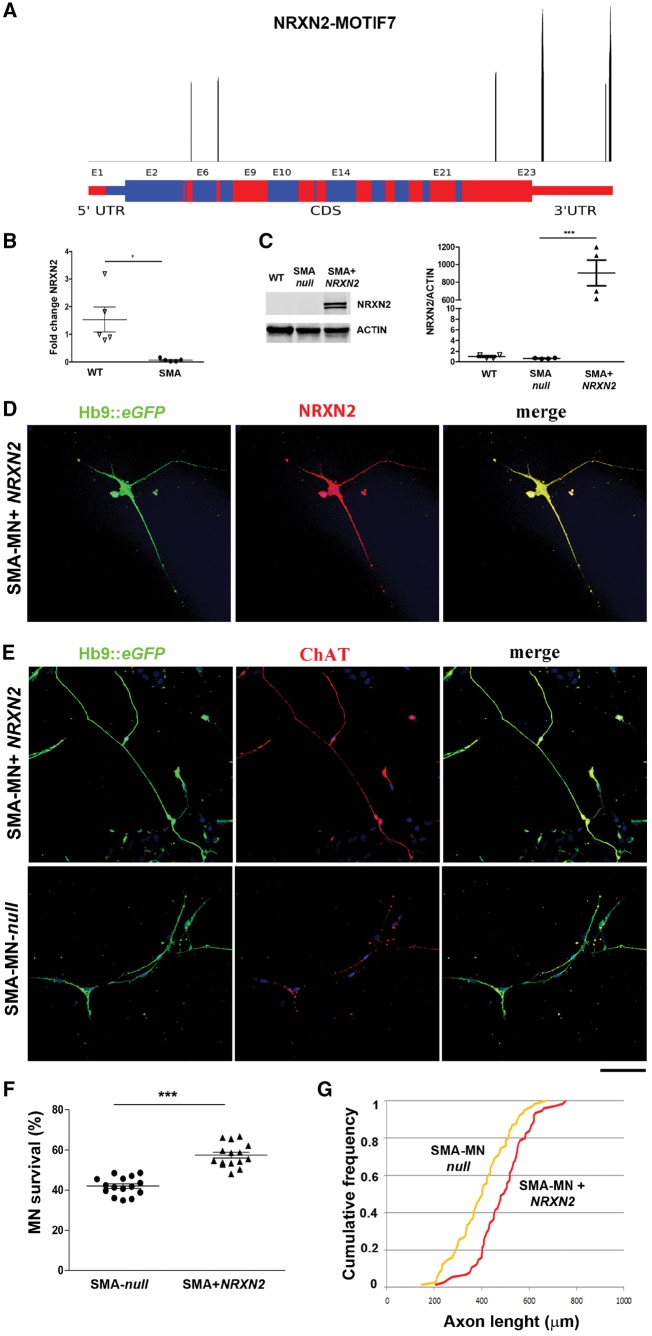
NRXN2 protects vulnerable SMA patient motor neurons from degeneration
Among the deregulated genes with motif 7, we focused our attention on NRXN2 (Fig. 5A), which has already been associated with SMA in animal models and is relevant for motor neuron survival and function (See et al., 2014). The reduction in NRXN2 was further confirmed by qPCR (P < 0.05, Fig. 4B).
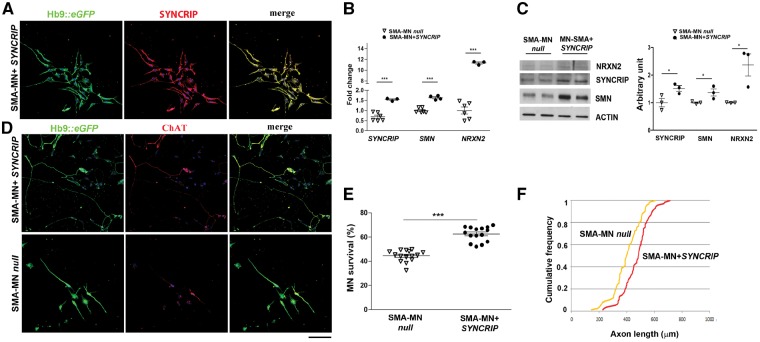
Figure 5.
SYNCRIP protects human SMA motor neurons from degeneration by increasing NRXN2 and SMN levels. (A) Representative images of SMA motor neurons after SYNCRIP transfection (Hb9::eGFP, green; SYNCRIP, red). Nuclei are labelled with DAPI (blue). (B) q-PCR analysis showed a significant increase in the expression of SYNCRIP, SMN, and NRXN2 transcripts in SMA-motor neurons compared to null SMA-motor neurons. ***P < 0.001, student t-test. Data are presented as mean ± SEM from three independent experiments. (C) Western blot representative image of SYNCRIP, SMN, and NRXN2 in SMA motor neurons after SYNCRIP overexpression compared to SMA-null cells. Western blot analysis revealed an increased level of SYNCRIP (*P < 0.05), SMN (*P < 0.05), and NRXN2 (*P < 0.05) in SMA motor neurons after SYNCRIP overexpression compared to SMA-null cells. Data are presented as mean ± SEM of three independent experiments. (D) Representative images of SMA motor neurons (Hb9::eGFP, green; ChAT, red) with and without SYNCRIP transfection. Nuclei are labelled with DAPI (blue). (E) The number of SYNCRIP SMA-motor neurons in long-term culture was significantly increased compared to SMA motor neurons null. SYNCRIP overexpression was protective to motor neurons. ***P < 0.0001, Student t-test. Data are presented as mean ± SEM of five independent experiments. (F) At 8 weeks, SMA motor neurons overexpressing SYNCRIP showed longer axons than SMA motor neurons null. P < 0.001, Kolmogorov-Smirnov test. Scale bar = 75 µm.
Figure 4.
NRXN2 protects human SMA-motor neurons from degeneration. (A) Significant motif 7 peaks along the NRXN2 transcript (RefSeq ID NM_015080). Only significant peaks are reported (Bonferroni corrected P < 0.01). Red and blue shades represent exon boundaries, thinner lines the UTRs. (B) Quantitative PCR analysis showed a significant decrease in the expression of NRXN2 transcript in SMA-motor neurons compared to wild-type. *P < 0.05, Student’s t-test. Data are presented as mean ± SEM from three independent experiments. (C) Representative image of NRXN2 western blot in wild-type motor neurons, SMA motor neurons null and SMA-NRXN2. NRXN2 overexpression in SMA motor neurons was confirmed with respect to SMA null (***P < 0.01, Student’s t-test). Data are presented as mean ± SEM from three independent experiments. (D) Representative images of SMA-motor neurons after NRXN2 transfection (Hb9::eGFP, green; NRXN2, red). Nuclei are labelled with DAPI (blue). (E) Representative images of SMA-motor neurons (ChAT, red; Hb9::eGFP, green) with and without NRXN2 transfection. Nuclei are labelled with DAPI (blue). (F) The number of NRXN2-treated SMA motor neurons in long-term culture was significantly increased compared to SMA motor neurons null. NRXN2 overexpression was protective to motor neurons. ***P < 0.0001, Student t-test. Data are presented as mean ± SEM of five independent experiments. (G) At 8 weeks, SMA-motor neurons overexpressing NRXN2 had longer axons than SMA motor neurons null. P < 0.001, Kolmogorov-Smirnov test. Scale bar = 75 µm.
To confirm the role of NRXN2 in SMA pathogenesis, we investigated whether the upregulation of NRXN2 can halt the degeneration of human patient-specific SMA iPSC-derived motor neurons. SMA motor neurons (monitored by Hb9::eGFP) were transfected with a vector encoding the human cDNA for NRXN2, confirming its upregulation after infection (P < 0.01 Fig 4C and D). SMA motor neurons present apparent cell autonomous degeneration in vitro after 8 weeks of culturing (Corti et al., 2012; Allodi et al., 2016), and in the present study overexpression of NRXN2 in SMA-motor neurons significantly improved their survival (P < 0.0001, Fig. 4E and F) and increased neurite length compared to null-treated SMA motor neurons (P < 0.001, Fig. 4G). These data suggest that NRXN2 can rescue the disease phenotype in a human SMA model.
SYNCRIP protects vulnerable SMA motor neurons, increasing SMN and NRXN2 expression
We investigated whether the upregulation of SYNCRIP could rescue the expression levels of NRXN2, which harbours motif 7, halting the degeneration of human patient-specific SMA iPSC-derived motor neurons (Fig. 5).
SMA motor neurons (monitored by Hb9::eGFP) were transfected with a vector expressing the human cDNA for SYNCRIP (Fig. 5A and B). A subsequent increase in NRXN2 was detected by both qPCR (P < 0.0001, Fig. 5B) and western blot (P < 0.05, Fig. 5C), confirming our hypothesis of SYNCRIP having a role in its expression. SYNCRIP has also been described to modulate the splicing of SMN2, promoting the inclusion of exon 7 (Chen et al., 2008). In line with this finding, we demonstrated increased expression of SMN in SYNCRIP-treated SMA-motor neurons by q-PCR (P < 0.001, Fig. 5B) and western blot (P < 0.05, Fig. 5C). Overexpression of SYNCRIP in SMA-motor neurons significantly improved their survival (P < 0.0001, Fig. 5D and E) and increased neurite length compared to null-treated SMA motor neurons (P < 0.001, Fig. 5F). Therefore, the missing interaction between SMN and SYNCRIP could be the cause of NRXN2 reduction, contributing to the SMA phenotype. Our hypothesis is that the complex SYNCRIP/SMN can bind NRXN2 3′UTR through motif 7 regulating its mRNA and protein expression.
To validate the hypothesis of a direct interaction between SMN, SYNCRIP, and NRXN2, we downregulated SYNCRIP with siRNA in the human neural SH-SY5Y cell line. SYNCRIP silencing induced a significant reduction in SMN levels, as expected (mRNA P < 0.001, protein P < 0.05), and a marked decrease in NRXN2 levels in q-PCR and western blot analysis (P < 0.001, Supplementary Fig. 5A–C), confirming the presence of a positive loop among the three proteins (Supplementary Fig. 5D). Intriguingly, we found that even SYT13 and STMN2 expression, deregulated in SMA, directly correlate with SYNCRIP expression (Supplementary Fig. 4, P < 0, 0001), suggesting that the interaction between SMN and SYNCRIP may account for the observed dysregulation of other key motor neuron genes in SMA.
Syncrip overexpression ameliorates pathological phenotype in SMA in vivo models
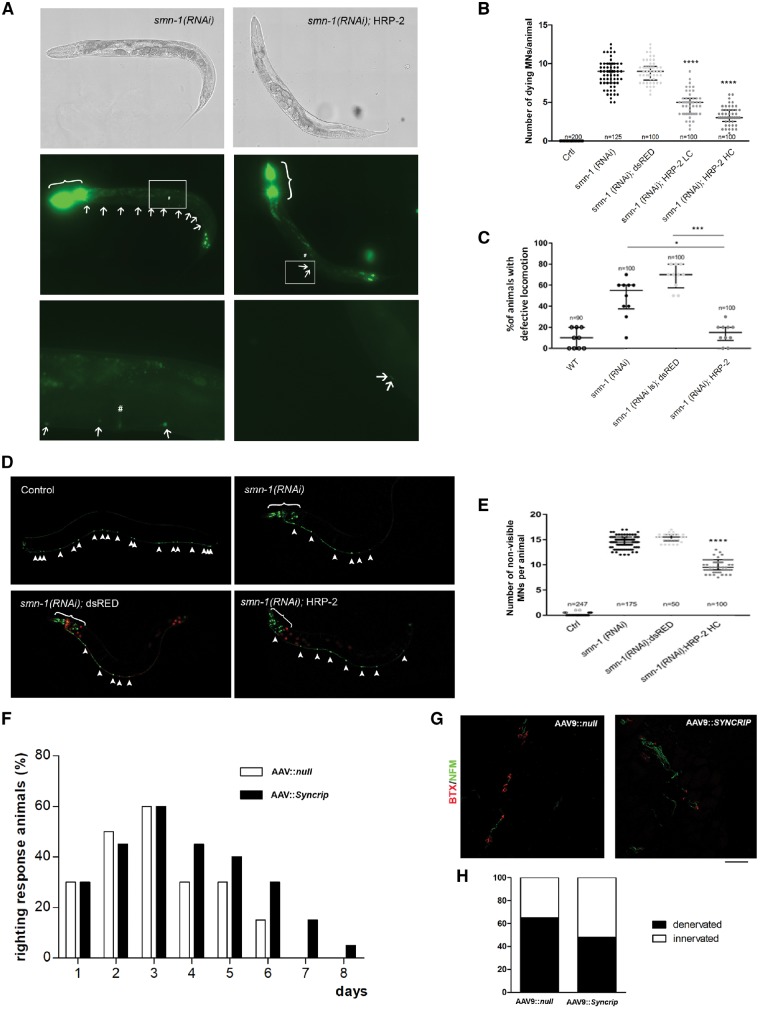
We next investigated if overexpression of Syncrip in SMA animal models could improve the disease phenotypes. First, we used C. elegans depleted of smn-1 in D-type motor neurons (Gallotta et al., 2016). The SYNCRIP homolog in C. elegans is hrp-2 (NM_060648.3), a gene expressed in all tissues, including D-type motor neuron (Blazie et al., 2017), during embryogenesis, larval development, and in adult animals (Kinnaird et al., 2004). The protein is localized in the nuclei and has nucleic acid binding activity, and its depletion causes defects in embryonic and larval development, fertility (Kinnaird et al., 2004), lifespan (Heintz et al., 2017), and splicing (Kabat et al., 2009; Heintz et al., 2017). To test whether hrp-2 genetically interacts with smn-1 (Smn1 homologue), we expressed hrp-2 under the control of a pan-neuronal promoter (punc-119::hrp-2, referred to as HRP-2), in smn-1(RNAi) knocked-down C. elegans. The smn-1(RNAi) animals used in this work are transgenic for D-type motor neurons with specific silencing of smn-1 at a very high dose (Gallotta et al., 2016). These animals are viable and fertile but with motor neuron degeneration, followed by their death and strong impairment in backward locomotion (Gallotta et al., 2016). Overexpression of HRP-2 at two different concentrations (low and high) halved the number of dying motor neurons per animal, from 8.6 in smn-1(RNAi) to 4.7 in smn-1(RNAi);HRP-2 LC and 3.3 in smn-1(RNAi);HRP-2 HC (P < 0.0001 in both cases, Fig. 6A and B). The rescue obtained is dose-dependent and gene-specific because the expression of an unrelated protein under the same conditions, such as dsRED fluorescent protein, did not rescue this phenotype (nine dying motor neurons/animal). Therefore, we used the most effective concentration (high concentration) to determine whether HRP-2 was able to rescue another earlier phenotype caused by smn-1(RNAi), neuronal degeneration. HRP-2 partially rescued the disappearance of viable motor neurons expressing GFP; 51.7% of the 19 expected neurons degenerated in smn-1(RNAi);HRP-2 HC compared to 76.5% in smn-1(RNAi) animals and 80.7% in smn-1(RNAi);dsRED (P < 0.0001 in both cases, Fig. 6D and E). Finally, we tested the ability of HRP-2 to rescue the lack of smn-1 at the functional level. Notably, the locomotion defect with smn-1(RNAi) was fully rescued by HRP-2 expression, reducing the number of animals with an abnormal locomotion behaviour from 48% in smn-1(RNAi) and 68% in smn-1(RNAi);dsRED to 14% in smn-1(RNAi);HRP-2 HC (P = 0.0303 and P = 0.0001, respectively), which is similar to that observed in wild-type animals (9%, P > 0.05, Fig. 6C). Our results demonstrate that HRP-2 plays a conserved role in the SMN pathway in neurons, and its overexpression can rescue neuron degeneration and death, and the locomotion defects caused by smn-1 silencing.
Figure 6.
SYNCRIP ameliorated the disease phenotype in in vivo models of SMA. (A–E) C. elegans; (F–H) mouse model. (A) Transgenic animals expressing HRP-2 in all neurons and silenced for smn-1 (right) present fewer dying neurons (arrows) than smn-1(RNAi) animals (left). In all panels, anterior is left and ventral is down. The animals were observed using brightfield (top) or epifluorescence (middle and bottom). The top and middle panels were reconstructed using the Tile Scan function of the Leica LAS X program. Middle: The co-injection marker was expressed in the pharynx and the nuclei of the most posterior intestinal cells (brackets). Bottom: Enlargements of the areas outlined in the middle panels around the vulva (#). (B) Quantification of dying motor neurons in non-silenced control (Ctrl), smn-1(RNAi), smn-1(RNAi) expressing dsRED (negative control), and HRP-2 at low (LC) and high (HC) concentrations. ****P < 0.0001 between smn-1(RNAi);HRP-2 LC and HC versus smn-1(RNAi) and smn-1(RNAi); dsRED (one-way ANOVA with Kruskal-Wallis test). Each dot represents the number of apoptotic fluorescently-labeled dying motor neurons in a single animal. The median with interquartile range is shown. n is the total number of animals observed from two independent transgenic lines. (C) Knock-down of smn-1 in D-type motor neurons leads to a locomotion defect not observed in wild-type (WT). This defect is rescued by HRP-2 but not by dsRED expression. *P = 0.0303, smn-1(RNAi);HRP-2 versus smn-1(RNAi) and ***P = 0.0001 smn-1(RNAi);HRP-2 versus smn-1(RNAi);dsRED (one-way ANOVA with Kruskal-Wallis test). Each dot represents the percentage of animals with a defective backward, among 10 animals tested. The median with interquartile range is shown. n is the total number of animals tested from at least two independent transgenic lines. (D) Transgenic animals expressing HRP-2 in all neurons and silenced for smn-1 (bottom right) present more visible/viable neurons labelled with GFP (arrowheads) than smn-1(RNAi) (top right) or smn-1(RNAi);dsRED animals (bottom left). All 19 motor neurons are visible in the ventral cord in control animals (top left). Motor neurons are visible because of the expression of GFP in the ventral cord, transgene oxIs12[punc-47::GFP]. Additional cells express GFP in the head because of the expression of the pchs-2::GFP co-injection marker (brackets, top right and bottom). Red fluorescence is visible in all neurons in dsRED animals (bottom left) or in the intestinal nuclei because of pelt-2::RFP co-injection marker (bottom right). (E) Quantification of the percentage of degenerating neurons in wild-type (Ctrl), smn-1(RNAi), and smn-1(RNAi) expressing dsRed or hrp-2. The smn-1 knock-down induced the degeneration of motor neurons, and this defect was partially rescued by HRP-2 expression. ****P < 0.0001 smn-1(RNAi);HRP-2 versus smn-1(RNAi) and smn-1(RNAi); dsRED (one-way ANOVA with Kruskal-Wallis test). Each dot represents the number of degenerated motor neurons in a single animal, among 19 that are visible in the wild-type. The median with interquartile range is shown. n is the total number of animals observed from two independent transgenic lines. (F) The righting reflex was significantly improved in SMAΔ mice injected ICV at P1 with AAV9::Syncrip compared to AAV9::null SMA mice beginning on Day 4. Notably, after P6, no AAV9::null SMA mice were able to right, whereas a percentage of SMA mice overexpressing Syncrip maintained the righting reflex function. **P < 0.01, contingency chi-squared test (χ2 = 23.76), n = 5 mice/group. (G and H) Quantification of presynaptic NF-M (green) and postsynaptic α-bungarotoxin (red) signal in intercostal muscles showed a significant increase in NMJ innervation in AAV9::Syncrip SMA mice compared to AAV9::null SMA mice. *P < 0.05, contingency chi-squared test (χ2 = 5.879), n = 100 NMJs analysed for each animal, three mice/group.
We also evaluated the effect of Syncrip overexpression in a more complex in vivo SMA model, SMAΔ7, which presents a severe SMA phenotype type 1, characterized by loss of motor neurons and neuromuscular function, NMJ denervation, and dramatically reduced survival (Le et al., 2005). We intracerebroventricularly injected SMA mice at P1 with AAV9, a vector with a particular tropism for the CNS (Foust et al., 2009), carrying Syncrip. Syncrip overexpression was able to improve righting performance in SMA mice (Fig. 6F, P < 0.01, n = 5, contingency and χ2 = 23.76) and increase neuromuscular junction innervation (Fig. 6G, P < 0.05, n = 3, contingency and χ2 = 5.879). However, the treatment did not show significant results in terms of survival elongation (data not shown).
As demonstrated by western blot (Supplementary Fig. 6), we detected significantly higher levels of SYNCRIP (P < 0.05), as expected, but also SMN and NRXN2 (both P < 0.05) in the brains of treated mice, confirming in vivo the hypothesis of a positive loop among these three genes.
Discussion
SMA is a severe, disabling, inherited neurological disease characterized by motor neuron degeneration (Pellizzoni et al., 1998) and SMN protein depletion. Nusinersen, the first approved therapeutic compound, became available at the end of 2016 (Finkel et al., 2016) (www.curema.org), even if it is unclear whether this approach can be resolutive for all types of patients. The implementation of successful therapy requires better understanding of the molecular mechanisms underlying the disease, particularly regarding the selectivity of motor neuron loss. Patient-specific iPSCs represent a novel tool for in vitro disease modelling and therapeutic discovery in neurodegenerative diseases that involve a specific neuronal population (Frattini et al., 2015). Here, we differentiated SMA patient iPSCs into motor neurons, confirming their reduced survival and axonal length in culture. The role of SMN in mRNA processing is well established (Pellizzoni et al., 1998), even if the mechanisms of this function are still not completely elucidated.
To understand mRNA-related dysregulation linked to selective motor neuron vulnerability, we performed whole RNA-Seq on SMA and wild-type motor neurons, targeting 40 M clusters per sample. The gene expression profiles clearly distinguished the two groups, specifically 1084 downregulated and 808 upregulated genes. Among the most deregulated gene sets, we observed a significant reduction in transcripts linked to specific neural assets in SMA motor neurons, including axon-related proteins (STMN2 and PLP1), ion channels, particularly potassium channel, and synapses (i.e. SYT13 and NRXN1, NRXN2, NRXN3). We validated the relevant genes by quantitative PCR and/or western blot analysis, including genes critical for motor neuron function, such as GRIA1, GRIA12, GRIA14 and IGF1/IGF12. Remarkably, we confirmed the downregulation of some key transcripts already found to be altered in other SMA models (Pellizzoni et al., 1998), such as CHODL1, NRN1, and CRABP1.
Based on the known role of SMN in the biogenesis of spliceosomal snRNPs (Pellizzoni et al., 2002), an effect of SMN deficiency on splicing is expected. Therefore, we analysed the exon alternatively spliced in SMA motor neurons compared to wild-type motor neurons. We found 12 144 deregulated cassette exons belonging to genes involved in motor neuron diseases and function, such as MARK2, CAMK2D, AGRN, and MPHOSPH9.
We determined whether differentially spliced genes share some common gene sequence motifs, which could predict either silenced or enhanced exons in the whole set of cassette exons analysed. We found 15 significant motifs and compared them to the most complete set of known RBP motifs (Ray et al., 2013) to detect significant binding probabilities. Interestingly, motifs identified in differentially spliced exons, particularly motif 7, match those of known RBPs involved in mRNA splicing, transcription, and post-transcriptional regulation. As these processes could have an effect in determining the level of gene expression, we evaluated the enrichment of these motifs in differentially expressed genes. Strikingly, some genes with high motif 7 enrichment in the 3′ UTR exhibit axonal and synaptic functions, including NRXN1, NRXN2, and SYT13.
We focused our attention on NRXN2, which is associated with axonogenesis, synaptogenesis, and synaptic function and is relevant for motor neuron survival and function. NRXN2, a neuron-specific gene encoding a presynaptic cell adhesion protein that associates with postsynaptic proteins, such as neuroligin, is involved in Ca2+-evoked neurotransmitter release (Ushkaryov et al., 1992; Missler et al., 2003). Moreover, reduced expression and decreased inclusion of exon 12 has been shown in SMN-deficient zebrafish embryos. Nrxn2a knock-down in zebrafish embryos led to motor axon branching defects, as well as reduced Ca2+ influx into presynaptic terminals, resembling SMA disease (See et al., 2014). Reduced expression and altered alternative splicing of Nrxn2a mRNA have also been identified in both the spinal cord and motor neurons of mammals, particularly in severe transgenic SMA mice (See et al., 2014).
Here, we detected for the first time reduced expression of NRXN2 and decreased inclusion of exon 12 in human SMA motor neurons. Furthermore, our in vitro experiments showed that NRXN2 upregulation was protective for SMA motor neurons, improving survival and increasing neurite length. Based on these data, we hypothesize that NRXN2 alteration can account for the axon and pre-synaptic defects at neuromuscular endplates in SMA pathophysiology, representing a potential non-SMN therapeutic target.
The RBPs with a higher probability of binding motif 7, the polyA binding protein, include SYNCRIP (hnRNPQ), which was described previously as directly interacting with RG-rich domains of full-length SMN protein (Mourelatos et al., 2001). Interestingly, SYNCRIP interacts only with full-length SMN, and not with truncated or mutant SMN forms identified in SMA; this may account for the inefficient pre-mRNA expression and splicing in SMA patients (Mourelatos et al., 2001). On the other hand, SYNCRIP upregulation has been described in pauci-symptomatic siblings of SMA patients, supporting its role as a protective gene (Pellizzoni et al., 1998). Finally, SYNCRIP is a splicing modulator of SMN, promoting the inclusion of exon 7 in SMN2, probably by activating the use of its upstream 3′ splice site (Chen et al., 2008). Intriguingly, SYNCRIP emerged as one of the top RPBs with high probability of interacting with motif 7, which is also present in NRXN2 mRNA. We demonstrated that SYNCRIP upregulation rescued NRXN2 levels, suggesting that it can stabilize the NRXN2 mRNA and increase its expression by interacting with motif 7. Furthermore, increased expression of SMN protein was found in SYNCRIP-treated SMA-motor neurons, confirming the ability of SYNCRIP to enhance SMN levels (Chen et al., 2008). We demonstrated that SYNCRIP overexpression increased in vitro SMA motor neuron survival and axonal outgrowth, likely due to the upregulation of SMN and downstream target NRXN2. Finally, we demonstrated that SYNCRIP overexpression rescued neuron survival and function in vivo in a C. elegans SMA model and ameliorated the pathological phenotype in SMNΔ7 mice. These data highlighted its conserved role in the SMN pathway in neurons, making SYNCRIP a promising therapeutic target and modifier in SMA. We hypothesized the presence of a positive loop among SYNCRIP, SMN, and NRXN2, which was further demonstrated in our experiments on SH-SY5Y cells in which SYNCRIP was silenced and in the brain of mice overexpressed for Syncrip with AAV9. Because motif 7 is present in other motor neuron altered genes, such as SYT13, we propose that the interaction between SMN and SYNCRIP is crucial in SMA, regulating the expression of several motor neuron genes. Furthermore, this hypothesis is supported by the correlation detected between SYNCRIP expression and the expression of two other key motor neuron genes deregulated in SMA SYT13 and STMN2.
Overall, we performed the first detailed analysis of the correlation between SMN defects, specific gene expression, and splicing alterations in human SMA motor neurons, identifying common motif sequences in altered genes. This study led us to identify non-SMN therapeutic targets, such as NRXN2 and especially SYNCRIP. Moreover, we demonstrated that the SMN/SYNCRIP interaction is likely responsible for the altered mRNA processing of several key motor neuron genes in SMA, contributing to motor neuron degeneration, and that its modulation can be crucial to modifying the SMA phenotype. The therapeutic approach presented in this study can also be combined with the already approved SMN-targeted therapy (nusinersen), to complement the beneficial effect and in particular when it is not completely successful, for example in patients who are already symptomatic.
Supplementary Material
Acknowledgements
The authors wish to thank Associazione Amici del Centro Dino Ferrari for support, Giuseppina Zampi and Federica Cieri for technical support, Prof. J.D. McGhee (University of Calgary, Canada) for plasmids and the Caenorhabditis Genetics Center (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for strains.
Glossary
Abbreviations
- AAV9
adenovirus-associated vector serotype 9
- DAVID
database for annotation, visualization, and integrated discovery
- GO
gene ontology
- hnRNP
heterogeneous nuclear ribonucleoprotein
- iPSC
induced pluripotent stem cell
- NMJ
neuromuscular junction
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- RBP
RNA binding protein
- RNA-Seq
RNA sequencing
Funding
This study was supported by the Cariplo Foundation (to S.C. and U.P., 2012–0513), the Joint Programme Neurodegenerative Disease (JPND) Research Grant DAMNDPATHS (2014) and Italian Telethon Foundation (to M.N., GGP14025 and to E.D.S., GGP16203) and the Italian Telethon Foundation. Animal experiments partially funded by Italian fiscal contribution ‘5x1000’ 2014 – MIUR – devolved to Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico.
Competing interests
The authors report no competing interests.
References
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D et al. . Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA 2011; 108: 10337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allodi I, Comley L, Nichterwitz S, Nizzardo M, Simone C, Benitez JA et al. . Differential neuronal vulnerability identifies IGF-2 as a protective factor in ALS. Sci Rep 2016; 6: 25960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM et al. . Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 2009; 5: e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie SM, Geissel HC, Wilky H, Joshi R, Newbern J, Mangone M. Alternative polyadenylation directs tissue-specific miRNA targeting in caenorhabditis elegans somatic tissues. Genetics 2017; 206: 757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereda M, Sironi M, Cavalleri M, Pozzoli U. GeCo++: a C++ library for genomic features computation and annotation in the presence of variants. Bioinformatics 2011; 27: 1313–5. [DOI] [PubMed] [Google Scholar]
- Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol 2008; 28: 6929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, Ronchi D et al. . Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med 2012; 4: 165ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosi N, Rossi S, Gerbino V, Cozzolino M. Rac1 at the crossroad of actin dynamics and neuroinflammation in Amyotrophic Lateral Sclerosis. Front Cell Neurosci 2014; 8: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA et al. . Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009; 457: 277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli I, Nizzardo M, Comi GP, Corti S. Spinal muscular atrophy–recent therapeutic advances for an old challenge. Nat Rev Neurol 2015; 11: 351–9. [DOI] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC et al. . Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 2016; 388: 3017–26. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S et al. . Ensembl 2014. Nucl Acids Res 2014; 42: D749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 2009; 27: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM et al. . Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol 2010; 28: 271–4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Frattini E, Ruggieri M, Salani S, Faravelli I, Zanetta C, Nizzardo M et al. . Pluripotent stem cell-based models of spinal muscular atrophy. Mol Cell Neurosci 2015; 64: 44–50. [DOI] [PubMed] [Google Scholar]
- Gallotta I, Mazzarella N, Donato A, Esposito A, Chaplin JC, Castro S et al. . Neuron-specific knock-down of SMN1 causes neuron degeneration and death through an apoptotic mechanism. Hum Mol Genet 2016; 25: 2564–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilina TO, McGovern VL, Workman E, Crawford TO, Gogliotti RG, DiDonato CJ et al. . Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum Mol Genet 2008; 17: 1063–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet 2016; 135: 851–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582: 1977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C, Doktor TK, Lanjuin A, Escoubas C, Zhang Y, Weir HJ et al. . Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature 2017; 541: 102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 2002; 32: 728–30. [DOI] [PubMed] [Google Scholar]
- Honda D, Ishigaki S, Iguchi Y, Fujioka Y, Udagawa T, Masuda A et al. . The ALS/FTLD-related RNA-binding proteins TDP-43 and FUS have common downstream RNA targets in cortical neurons. FEBS Open Bio 2013; 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, Zahler AM. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J Biol Chem 2009; 284: 28490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird JH, Maitland K, Walker GA, Wheatley I, Thompson FJ, Devaney E. HRP-2, a heterogeneous nuclear ribonucleoprotein, is essential for embryogenesis and oogenesis in Caenorhabditis elegans. Exp Cell Res 2004; 298: 418–30. [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD et al. . SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 2005; 14: 845–57. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L et al. . Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–65. [DOI] [PubMed] [Google Scholar]
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK et al. . An SMN-dependent U12 splicing event essential for motor circuit function. Cell 2012; 151: 440–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Harris AW, Kingham BF, Lumpkin CJ, Opdenaker LM, McCahan SM et al. . Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PLoS One 2014; 9: e106818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 2008; 3: 649–57. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J et al. . ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol 2009; 327: 551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature 1993; 364: 337–41. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature 1997; 389: 870–6. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 1991; 10: 3959–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K et al. . Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 2003; 423: 939–48. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J 2001; 20: 5443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkis G, Ofir R, Manor E, Landau D, Elbedour K, Birk OS. Lethal congenital contractural syndrome type 2 (LCCS2) is caused by a mutation in ERBB3 (Her3), a modulator of the phosphatidylinositol-3-kinase/Akt pathway. Am J Hum Genet 2007; 81: 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 2008; 582: 1997–2003. [DOI] [PubMed] [Google Scholar]
- Ng SY, Soh BS, Rodriguez-Muela N, Hendrickson DG, Price F, Rinn JL et al. . Genome-wide RNA-Seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell 2015; 17: 569–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzardo M, Simone C, Dametti S, Salani S, Ulzi G, Pagliarani S et al. . Spinal muscular atrophy phenotype is ameliorated in human motor neurons by SMN increase via different novel RNA therapeutic approaches. Sci Rep 2015; 5: 11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzardo M, Simone C, Rizzo F, Ruggieri M, Salani S, Riboldi G et al. . Minimally invasive transplantation of iPSC-derived ALDHhiSSCloVLA4+ neural stem cells effectively improves the phenotype of an amyotrophic lateral sclerosis model. Hum Mol Genet 2014a; 23: 342–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzardo M, Simone C, Salani S, Ruepp MD, Rizzo F, Ruggieri M et al. . Effect of combined systemic and local morpholino treatment on the spinal muscular atrophy Delta7 mouse model phenotype. Clin Ther 2014b; 36: 340–56.e5. [DOI] [PubMed] [Google Scholar]
- Pagliarini V, Pelosi L, Bustamante MB, Nobili A, Berardinelli MG, D’Amelio M et al. . SAM68 is a physiological regulator of SMN2 splicing in spinal muscular atrophy. J Cell Biol 2015; 211: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep 2007; 8: 340–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 1998; 95: 615–24. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002; 298: 1775–9. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X et al. . A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013; 499: 172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Ramirez A, Compagnucci C, Salani S, Melzi V, Bordoni A et al. . Genome-wide RNA-seq of iPSC-derived motor neurons indicates selective cytoskeletal perturbation in Brown-Vialetto disease that is partially rescued by riboflavin. Sci Rep 2017; 7: 46271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Ronchi D, Salani S, Nizzardo M, Fortunato F, Bordoni A et al. . Selective mitochondrial depletion, apoptosis resistance, and increased mitophagy in human Charcot-Marie-Tooth 2A motor neurons. Hum Mol Genet 2016; 25: 4266–81. [DOI] [PubMed] [Google Scholar]
- Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet 2002; 11: 93–105. [DOI] [PubMed] [Google Scholar]
- See K, Yadav P, Giegerich M, Cheong PS, Graf M, Vyas H et al. . SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum Mol Genet 2014; 23: 1754–70. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN et al. . rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA 2014; 111: E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C, Nizzardo M, Rizzo F, Ruggieri M, Riboldi G, Salani S et al. . iPSC-Derived neural stem cells act via kinase inhibition to exert neuroprotective effects in spinal muscular atrophy with respiratory distress type 1. Stem Cell Rep 2014; 3: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013; 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science 1992; 257: 50–6. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M et al. . SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008; 133: 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pinto AM, Wan L, Wang W, Berg MG, Oliva I et al. . Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci USA 2013; 110: 19348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in GEO at https://www.ncbi.nlm.nih.gov/geo/ reference number GSE108094.