Tan et al. demonstrate that an Alzheimer’s disease polygenic hazard score predicts regional amyloid PET deposition and longitudinal cortical atrophy, and is also associated with regional postmortem amyloid-beta and tangles, plus Lewy body and cerebrovascular pathology. Polygenic information may help identify individuals at high risk of multi-aetiology dementia.
Keywords: polygenic hazard score, amyloid, neurodegeneration, Lewy body, cerebrovascular disease
Abstract
Mounting evidence indicates that the polygenic basis of late-onset Alzheimer’s disease can be harnessed to identify individuals at greatest risk for cognitive decline. We have previously developed and validated a polygenic hazard score comprising of 31 single nucleotide polymorphisms for predicting Alzheimer’s disease dementia age of onset. In this study, we examined whether polygenic hazard scores are associated with: (i) regional tracer uptake using amyloid PET; (ii) regional volume loss using longitudinal MRI; (iii) post-mortem regional amyloid-β protein and tau associated neurofibrillary tangles; and (iv) four common non-Alzheimer’s pathologies. Even after accounting for APOE, we found a strong association between polygenic hazard scores and amyloid PET standard uptake volume ratio with the largest effects within frontal cortical regions in 980 older individuals across the disease spectrum, and longitudinal MRI volume loss within the entorhinal cortex in 607 older individuals across the disease spectrum. We also found that higher polygenic hazard scores were associated with greater rates of cognitive and clinical decline in 632 non-demented older individuals, even after controlling for APOE status, frontal amyloid PET and entorhinal cortex volume. In addition, the combined model that included polygenic hazard scores, frontal amyloid PET and entorhinal cortex volume resulted in a better fit compared to a model with only imaging markers. Neuropathologically, we found that polygenic hazard scores were associated with regional post-mortem amyloid load and neuronal neurofibrillary tangles, even after accounting for APOE, validating our imaging findings. Lastly, polygenic hazard scores were associated with Lewy body and cerebrovascular pathology. Beyond APOE, we show that in living subjects, polygenic hazard scores were associated with amyloid deposition and neurodegeneration in susceptible brain regions. Polygenic hazard scores may also be useful for the identification of individuals at the highest risk for developing multi-aetiological dementia.
Introduction
Beyond the ɛ4 allele of apolipoprotein E (APOE), there is increasing awareness that the genetic basis or architecture of late-onset Alzheimer’s disease is polygenic (Escott-Price and Jones, 2017). Alzheimer’s disease associated single nucleotide polymorphisms (SNPs) from large genome-wide association studies (GWAS) have been aggregated into polygenic scores for disease prediction (Sabuncu et al., 2012; Escott-Price et al., 2015, 2017; Chouraki et al., 2016; Mormino et al., 2016). Given the recent recommendations of the National Institute of Aging-Alzheimer’s Association for a research framework focusing on the diagnosis of Alzheimer’s disease with biomarkers in living people (Jack et al., 2018), it is increasingly important to elucidate the relationship of Alzheimer’s disease polygenic scores with markers of amyloid deposition, pathologic tau and neurodegeneration.
Although polygenic scores have been found to be associated with neurodegeneration in select brain regions (Sabuncu et al., 2012; Harrison et al., 2016; Desikan et al., 2017; Kauppi et al., 2018) and whole brain amyloid deposition (Mormino et al., 2016; Voyle et al., 2017; Ge et al., 2018; Tan et al., 2018), few, if any, studies have evaluated the relationship between Alzheimer’s disease polygenic scores, longitudinal volume loss and amyloid deposition in regions of interest across the entire cerebrum. Integrating common genetic variants into an age-dependent survival analysis framework, we have shown that a polygenic hazard score (PHS) can quantify Alzheimer’s disease dementia age of onset, even among APOE ɛ3/3 individuals, who constitute the majority of all individuals with Alzheimer’s disease dementia (Desikan et al., 2017). We have previously shown that a high PHS was associated with (i) faster longitudinal cognitive decline; and (ii) post-mortem and CSF/PET amyloid and tangle pathology, even in cognitively normal older individuals who are APOE ɛ4 non-carriers (Tan et al., 2017, 2018). However, the observation of individuals with a high PHS but low levels of in vivo and post-mortem Alzheimer’s disease pathology, and vice versa (Tan and Desikan, 2018) suggests the need to further understand the associations of polygenic risk with Alzheimer’s and non-Alzheimer’s pathobiology.
Using multimodal data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), Religious Orders Study and Rush Memory and Aging Project (ROSMAP), and the National Alzheimer’s Coordinating Center (NACC), we systematically investigated the relationship between Alzheimer’s disease polygenic risk and regional in vivo and post-mortem markers of Alzheimer’s associated pathology and neurodegeneration. Beyond APOE, we examined whether PHS is associated with: (i) regional tracer uptake using amyloid PET; (ii) regional volume loss using longitudinal MRI; (iii) post-mortem regional amyloid-β protein and tau-associated neurofibrillary tangles; and (iv) four common non-Alzheimer’s disease pathologies.
Materials and methods
Participants and clinical characterization
Alzheimer’s Disease Neuroimaging Initiative
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). Launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of the ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease. We first assessed 980 individuals (323 cognitively normal, 481 MCI, 176 Alzheimer’s dementia) with genetic, clinical, and florbetapir data, from ADNIGO and ADNI2. Next, we evaluated a group of 607 individuals (177 cognitively normal, 297 MCI, 133 Alzheimer’s dementia) with genetic, clinical and at least one longitudinal MRI scan (range = 0.43–3.46 years, mean = 2.03 years) from ADNI1. Lastly, we evaluated a subset of the ADNIGO and ADNI2 non-demented individuals [Clinical Dementia Rating (CDR) < 1; 229 cognitively normal, 403 MCI] with concurrent florbetapir and MRI scans at baseline. Cohort demographics are summarized in Table 1.
Table 1.
ADNI demographics of participants contributing amyloid PET data and those contributing longitudinal MRI data
| Florbetapir (n = 980) | Longitudinal MRI (n = 607) | |
|---|---|---|
| Age at baseline ± SD | 73.92 (7.52) | 75.48 (6.76) |
| Education in years ± SD | 16·26 (2.71) | 15.68 (2.93) |
| Sex (% female) | 540 (55.10) | 248 (41.36) |
| APOE ɛ4 carriers (%) | 431 (43.98) | 309 (51.41) |
| Diagnosis (CN/MCI/AD dementia) | 323/481/176 | 117/297/133 |
| PHS ± SD | 0.31 (0.76) | 0.50 (0.86) |
AD = Alzheimer’s disease; CN = cognitively normal; MCI = mild cognitive impairment.
ROSMAP
We evaluated 485 deceased individuals (age at death range = 71.3–108.3 years) with genetic, clinical, and neuropathology data from ROSMAP (Bennett et al., 2012a, b). We recalculated PHS as previously described (Desikan et al., 2017), after excluding all ROSMAP individuals. Cohort demographics from ROSMAP are summarized in Table 2.
Table 2.
ROSMAP demographics
| ROSMAP (n = 485) | |
|---|---|
| Age at death ± SD | 89.42 (6.34) |
| Sex (% female) | 331 (68.25) |
| APOE ɛ4 carriers (%) | 136 (28.04) |
| Diagnosis (CN/MCI/AD dementia) | 194/23/268 |
| PHS ± SD | −0.06 (0.45) |
AD = Alzheimer’s disease; CN = cognitively normal; MCI = mild cognitive impairment.
National Alzheimer’s Coordinating Center
We evaluated up to a total of 603 individuals (age at death range = 64.9–105.8 years) with genetic and post-mortem pathology data from the NACC. Cohort demographics from the NACC are summarized in Table 3. See Supplementary Fig. 1 for a distribution of PHS values for all cohorts and Supplementary Table 1 for demographics by diagnostic groups for all cohorts.
Table 3.
NACC demographics
| NACC (n = 603) | |
|---|---|
| Age at death ± SD | 86.03 (7.15) |
| Sex (% female) | 304 (50.08) |
| APOE ɛ4 carriers (%) | 302 (49.75) |
| Diagnosis (CN/MCI/AD dementia) | 129/63/411 |
| PHS ± SD | 0.62 (0.89) |
On average, individuals from the ROSMAP cohort have lower PHS as it is a community sample, unlike ADNI and NACC, which are clinical trial and memory clinic cohorts, respectively. AD = Alzheimer’s disease; CN = cognitively normal; MCI = mild cognitive impairment.
Polygenic hazard score
For all participants in this study, we computed their individual PHS, as described previously (Desikan et al., 2017). Briefly, we first delineated 1854 Alzheimer’s-associated SNPs at P < 10−5 using genotyped data from 17 008 Alzheimer’s disease dementia cases and 37 154 controls from stage 1 of the International Genomics of Alzheimer’s Project. Next, using genotyped data from 6409 Alzheimer’s disease dementia patients and 9386 older controls from phase 1 of the Alzheimer’s Disease Genetics Consortium dataset and corrected for baseline allele frequencies from the European genotypes from 1000 Genomes Project, we integrated these SNPs into a stepwise Cox proportional hazards model. The survival model involves the stepwise selection of one SNP that most minimized the Martingale regression residuals at every step, and stops if no SNPs can further minimize the residuals. The top five genetic principal components (to control for population substructure), sex, and APOE ɛ2 and ɛ4 variants were controlled for during the stepwise procedure to generate the parameter estimates (i.e. log hazard ratios) for each of the final 31 SNPs identified. A 1000× bootstrapping procedure (i.e. sampling with replacement) was used for model averaging and deriving the parameter estimates for each of the 31 SNPs to minimize any model overfitting in the training phase. The predictive value of PHS on Alzheimer’s dementia age of onset was then replicated in an independent cohort of 6984 Alzheimer’s dementia cases and 10 972 controls from the Alzheimer’s Disease Genetics Consortium phase 2 dataset. In this study, the PHS calculated for all participants represents the vector product of an individual’s genotype for the 31 SNPs and the corresponding parameter estimates from the Alzheimer’s Disease Genetics Consortium phase 1 Cox proportional hazard model, in addition to the APOE effects. A full description of the development and validation of PHS, including the 31 SNPs and their corresponding parameter estimates, are described in Desikan et al. (2017).
Statistical analysis
Alzheimer’s Disease Neuroimaging Initiative
First, we used linear regression to evaluate the relationship of PHS with cross-sectional regional florbetapir standard uptake volume ratio (SUVR) in 980 individuals across the disease spectrum from ADNIGO and ADNI2 (Table 1). In these analyses, we controlled for age at baseline, sex, education, and APOE status (binarized as having at least one copy of the ɛ4 allele versus none) to assess the effects of PHS beyond APOE (Ge et al., 2018). Florbetapir SUVR data in ADNI were determined by mean uptake in the cortical grey matter normalized by the whole cerebellum, based on Freesurfer 5.3 segmentation and parcellation of the individuals’ MRI that has been coregistered with the corresponding PET scan (Jagust et al., 2009). Left and right hemisphere’s florbetapir SUVR (normalized by the whole cerebellum) in 34 regions of interest from the Desikan-Killiany atlas in Freesurfer (Desikan et al., 2006) were averaged and we controlled for multiple comparisons at a false discovery rate (FDR) of <0.05. As follow-up, we conducted the same analysis in a subset of 323 cognitively normal individuals, co-varying for age, sex, and education.
Next, we used linear mixed-effects models to evaluate the relationship of PHS with longitudinal volume change in 33 regions of interest from the Desikan-Killiany atlas in Freesurfer (Desikan et al., 2006) in 607 individuals across the disease spectrum from ADNI1 (Table 1). Longitudinal volume change was quantified using quantitative anatomical regional change (QUARC; Holland et al., 2009; Holland and Dale, 2011). Left and right hemisphere’s volume change were averaged, and we controlled for multiple comparisons at an FDR of <0.05. In these analyses, we controlled for age at baseline, sex, education, and APOE status. We then examined the simple effects by comparing slopes of volume loss over time for individuals at high and low levels of PHS. We defined high PHS by 1 standard deviation (SD) above the mean (at ∼84 percentile) and low PHS by 1 SD below the mean (at ∼16 percentile) (Aiken and West, 1991; Tan et al., 2017).
Next, we used linear-mixed effects models to examine whether PHS was independently associated with longitudinal cognitive decline and clinical progression beyond frontal florbetapir SUVR, cross-sectional entorhinal cortex volume, and APOE. We restricted analyses to 632 non-demented individuals classified at baseline as cognitively normal (n = 229) or MCI (n = 403), and with both amyloid PET and MRI scans at baseline. To minimize multiple comparisons, we focused on frontal florbetapir SUVR given the known frontal cortex vulnerability for amyloid accumulation (Grimmer et al., 2010; Guo et al., 2017). To minimize multiple comparisons, we focused on the entorhinal cortex as it is one of the earliest regions susceptible to Alzheimer’s neurodegeneration (Van Hoesen et al., 1991; Gómez-Isla et al., 1996; Desikan et al., 2009; Thaker et al., 2017). We defined cognitive decline using change scores in two domains, namely executive function (Gibbons et al., 2012) and memory (Crane et al., 2012) based on composite standardized scores developed using the ADNI neuropsychological battery and validated using confirmatory factor analysis. We defined clinical progression using change scores in Cognitive Dementia Rating - Sum of Boxes (CDR-SB). We examined whether the higher PHS was associated with greater cognitive decline and clinical progression rate, controlling for frontal PET SUVR, entorhinal cortex volume, baseline age, sex, education, APOE status, and their interactions with time, using the following linear-mixed effects model:
| (1) |
Here, Δc = cognitive decline (executive function or memory) or clinical progression (CDR-SB) rate, Δt = change in time from baseline visit (years), and (1|subject) specifies the random intercept. We were specifically interested in PHS × Δt, whereby a significant interaction indicates differences in rates of decline, as a function of differences in PHS, after accounting for effects of frontal amyloid deposition, cross-sectional entorhinal cortex volume, APOE, and other covariates. If the interaction was significant, we then examined the simple effects by comparing slopes of cognitive decline and clinical progression over time for individuals who were high (at∼84 percentile) or low PHS (at ∼16 percentile). As follow-up, we conducted the same analysis in the subset of 229 cognitively normal individuals. In all analyses, continuous variables were centred and scaled prior to analysis to generate standardized effect estimates.
ROSMAP
We used linear regression to investigate the relationships of PHS (centred and scaled) with regional neuropathology (specifically, amyloid-β protein and tau associated neuronal neurofibrillary tangles identified through molecularly specific immunohistochemistry and quantified by image analysis). We investigated regional amyloid and tangles in eight regions of interest, namely hippocampus, entorhinal cortex, midfrontal cortex, inferior temporal cortex, angular gyrus, calcarine cortex, anterior cingulate cortex, and superior frontal cortex. Amyloid and tangle measures are the square-root of the percentage area occupied by amyloid or tangles in each of these regions. In all analyses, we co-varied for sex and age at death. We also assessed this relationship with the inclusion of APOE ɛ4 status to determine whether PHS is associated with regional neuropathology beyond APOE.
National Alzheimer’s Coordinating Center
We used linear and logistic regressions to investigate if PHS (centred and scaled) was associated with neuropathology other than the previously established associations with Consortium to Establish a Registry for Alzheimer’s disease (CERAD) score and neurofibrillary tangle scores assessed with Braak stages (Desikan et al., 2017), controlling for sex, and age at death. Specifically, we investigated whether PHS was associated with cerebrovascular disease pathology, Lewy body pathology, medial temporal lobe sclerosis, and frontotemporal lobar degeneration (FTLD) pathology (Pick’s, corticobasal degeneration, progressive supranuclear palsy, FTLD with TDP-43, and other taupathies). All statistical analyses were carried out with R version 3.5.1. See Fig. 1 for a flowchart delineating the hypotheses tested in the ADNI, ROSMAP, and NACC cohorts.
Figure 1.
Flowchart delineating the hypotheses tested in the study. Hypotheses were tested using data from the Alzheimer’s disease neuroimaging initiative (ADNI), Religious Orders Study and Rush Memory and Aging Project (ROSMAP), and the National Alzheimer’s Coordinating Center (NACC). AD = Alzheimer’s disease; CN = cognitively normal; MCI = mild cognitive impairment.
Data availability
The data that support the findings of this study are openly available for request in (i) LONI at https://ida.loni.usc.edu/; (ii) RADC Research Resource Sharing Hub at https://www.radc.rush.edu/; and (iii) NACC at https://www.alz.washington.edu/.
Results
PHS is associated with cross-sectional regional amyloid PET SUVRs
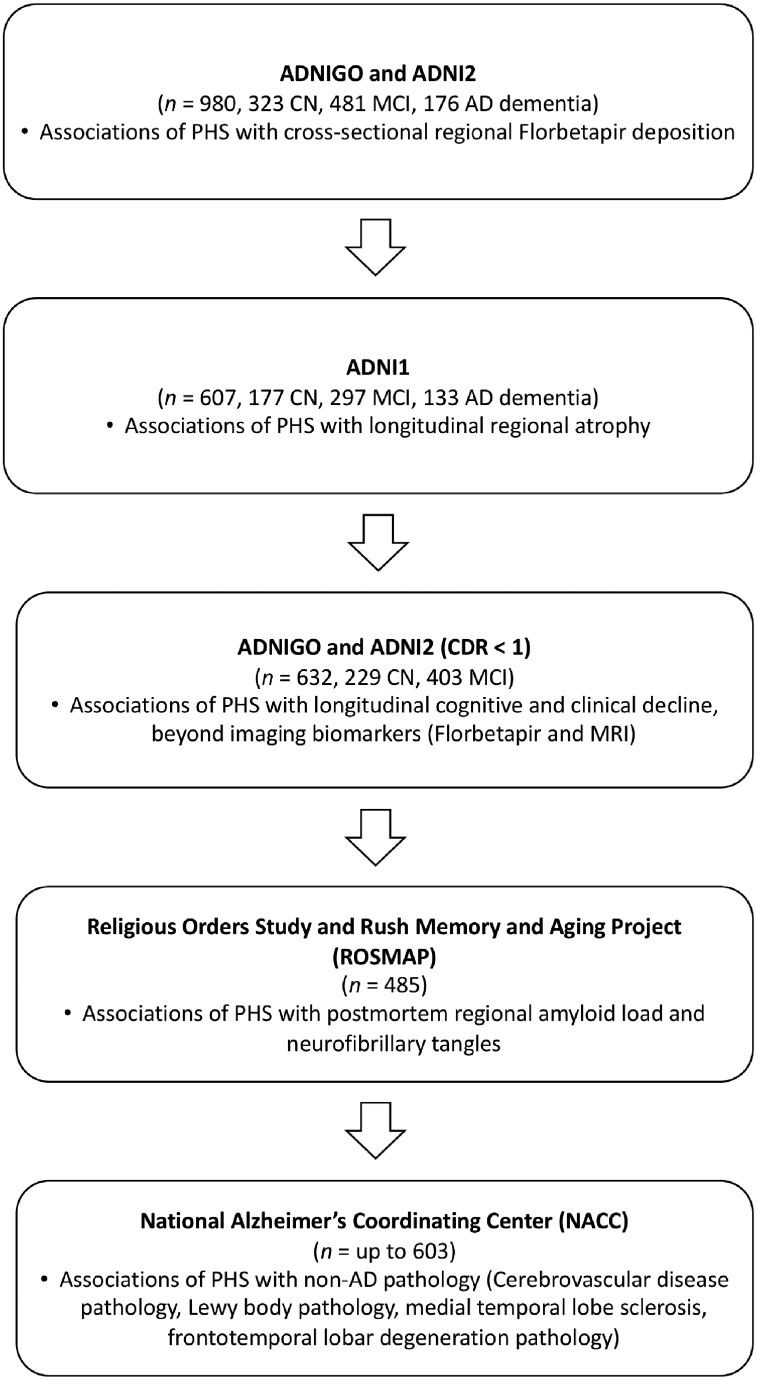
Beyond APOE, we found that higher PHS was associated with higher amyloid PET SUVR across all 34 regions of interest, with the largest effects in frontal regions such as the rostral anterior cingulate and rostral middle frontal gyrus (Fig. 2A and Supplementary Table 2) among all individuals from across the disease spectrum. In all regions of interest, these effects remained significant at an FDR of <0.05. In cognitively normal individuals, PHS was similarly associated with regional amyloid PET SUVR across all regions of interest, even at FDR < 0.05 (Supplementary Table 3).
Figure 2.
PHS is associated with regional amyloid accumulation and cortical atrophy. Beta estimates of (A) cross-sectional associations of PHS with regional florbetapir SUVRs. (B) Longitudinal change in regional cortical volumes in individuals with high (1 SD above mean, ∼ 84 percentile) PHS.
PHS is associated with longitudinal regional change in cortical volumes
In linear-mixed effects analysis, beyond APOE, PHS was significantly associated with change in 11 cortical volumes (Supplementary Table 4) in individuals across the disease spectrum, eight of which remained statistically significant at an FDR of <0.05, with high PHS individuals showing greater atrophy (Fig. 2B) compared to low PHS individuals. These effects were strongest for volume loss in regions such as the entorhinal cortex, inferior parietal cortex, inferior and middle temporal cortex.
PHS is associated with cognitive and clinical decline beyond amyloid PET and entorhinal volume
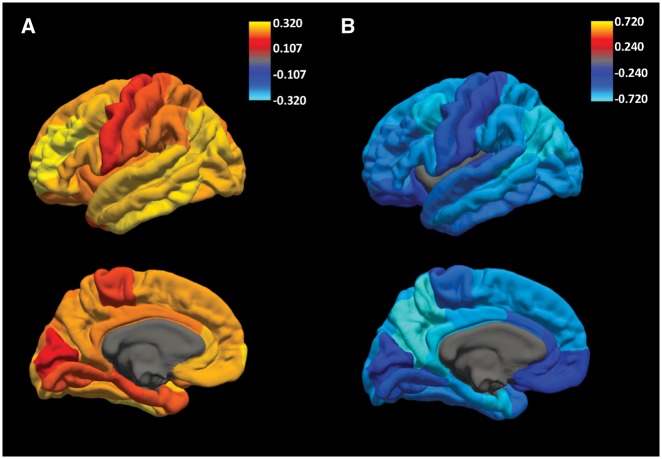
In linear-mixed effects analyses involving only non-demented individuals, PHS was associated with rates of decline over time in executive function [β = −0.03, 95% confidence interval (CI) = −0.06 to −0.003, standard error (SE) = 0.02, P = 2.94 × 10−2], memory (β = −0.04, 95% CI = −0.07 to −0.01, SE = 0.01, P = 3.65 × 10−3), and CDR-SB (β = 0.27, 95% CI = 0.19–0.35, SE = 0.04, P = 2.44 × 10−10), even when including frontal amyloid PET SUVR, entorhinal cortex volume, and other covariates (age, education, sex, and APOE status) in the model. Simple slope analyses showed that high PHS individuals experience greater rates of decline in executive function (β = −0.09, 95% CI = −0.16 to −0.01, SE = 0.04, P = 3.39 × 10−2), memory (β = −0.14, 95% CI = −0.21 to −0.07, SE = 0.03, P = 3.15 × 10−5), and CDR-SB (β = 0.49, 95% CI = 0.29–0.70, SE = 0.10, P = 2.66 × 10−6) compared to individuals with low PHS (Fig. 3). In addition, using likelihood ratio tests, we found that the full linear-mixed effects model including PHS resulted in a better fit than a reduced non-PHS linear-mixed effects model for executive function [χ2(2) = 6.86, P = 3.23 × 10−2], memory [χ2(2) = 15.55, P = 4.20 × 10−4], and CDR-SB [χ2(2) = 44.90, P = 1.78 × 10−10]. In cognitively normal individuals, PHS was only associated with rates of decline over time in CDR-SB (β = 0.08, 95% CI = 0.01–0.15, SE = 0.04, P = 3.25 × 10−2), with high PHS individuals experiencing greater rates of clinical decline (β = 0.28, 95% CI = 0.09–0.46, SE = 0.09, P = 3.45 × 10−3)
Figure 3.
PHS is associated with cognitive and clinical decline beyond imaging markers. Differences in rate of decline in (A) executive function, (B) memory, (C) clinical dementia sum-of-boxes (CDR-SB) over time for low (1 SD below mean, at ∼16 percentile; blue lines) and high (1 SD above mean, at ∼84 percentile; red lines) PHS non-demented individuals, after accounting for the effects of frontal florbetapir deposition, entorhinal cortex volume, APOE ɛ4 carrier status, age, sex, and education. Dashed lines are estimated standard error.
PHS is associated with post-mortem regional amyloid and tangles
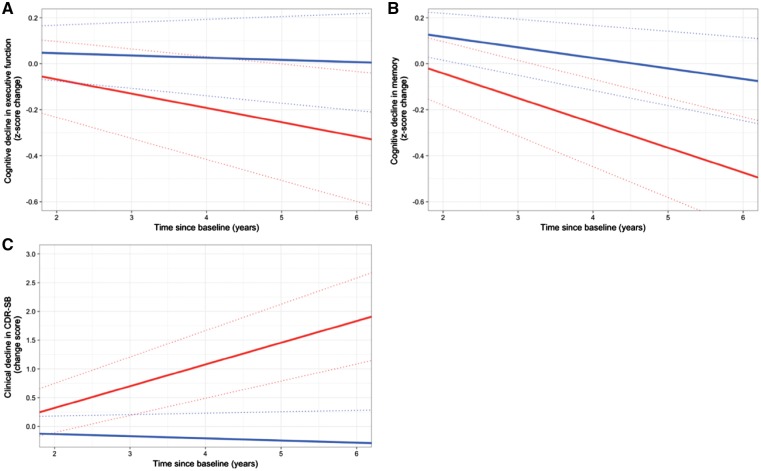
In the ROSMAP cohort, we found that higher PHS was associated with higher post-mortem amyloid-β across all eight regions of interest (Fig. 4A): angular gyrus (β = 0.37, 95% CI = 0.26–0.49, SE = 0.06, P = 7.29 × 10−10), calcarine cortex (β = 0.37, 95% CI = 0.28–0.45, SE = 0.04, P = 8.79 × 10−16), anterior cingulate cortex (β = 0.43, 95% CI = 0.29–0.56, SE = 0.07, P = 8.79 × 10−16), entorhinal cortex (β = 0.33, 95% CI = 0.22–0.44, SE = 0.06, P = 1.01 × 10−8), hippocampus (β = 0.26, 95% CI = 0.18–0.34, SE = 0.04, P = 1.97 × 10−10), inferior temporal cortex (β = 0.39, 95% CI = 0.28–0.49, SE = 0.05, P = 1.33 × 10−12), midfrontal cortex (β = 0.37, 95% CI = 0.26–0.49, SE = 0.06, P = 5.36 × 10−10), and superior frontal cortex (β = 0.38, 95% CI = 0.26–0.50, SE = 0.06, P = 1.59 × 10−9). Higher PHS was also associated with more post-mortem tangles across all eight regions of interest (Fig. 4B): angular gyrus (β = 0.43, 95% CI = 0.30–0.57, SE = 0.07, P = 4.57 × 10−10), calcarine cortex (β = 0.18, 95% CI = 0.12–0.23, SE = 0.03, P = 4.52 × 10−9), anterior cingulate cortex (β = 0.47, 95% CI = 0.34–0.60, SE = 0.07, P = 6.51 × 10−12), entorhinal cortex (β = 0.39, 95% CI = 0.24–0.55, SE = 0.08, P = 5.10 × 10−7), hippocampus (β = 0.50, 95% CI 0.31–0.68 = SE = 0.09, P = 2.28 × 10−7), inferior temporal cortex (β = 0.73, 95% CI = 0.55–0.92, SE = 0.09, P = 5.14 × 10−14), midfrontal cortex (β = 0.44, 95% CI = 0.33–0.56, SE = 0.06, P = 5.68 × 10−13), and superior frontal cortex (β = 0.43, 95% CI = 0.30–0.57, SE = 0.07, P = 1.11 × 10−9) These results were statistically significant when controlling for APOE status, with the exception of tangles in the hippocampus (Supplementary Table 5).
Figure 4.
PHS is associated with regional post-mortem neuropathology. Beta estimates of the associations between PHS and regional post-mortem (A) amyloid load and (B) neurofibrillary tangles.
PHS is associated with other pathologies
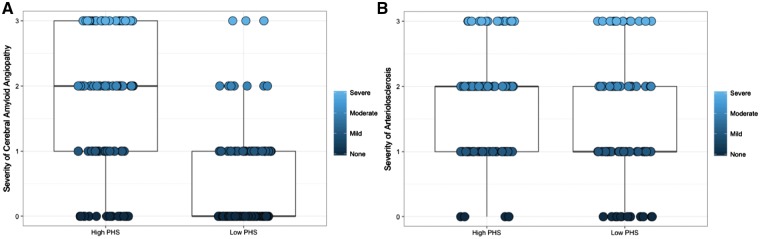
High PHS was found to be associated with three types of cerebrovascular pathology and presence of Lewy body pathology [odds ratio (OR) = 1.21, 95% CI = 1.01–1.45, P = 3.80 × 10−2]. Specifically, for cerebrovascular pathology, PHS was associated with cerebral amyloid angiopathy (β = 0.37, 95% CI = 0.29–0.45, SE = 0.04, P = 2.79 × 10−18, Fig. 5A) arteriolosclerosis (β = 0.11, 95% CI = 0.03–0.19, SE = 0.04, P = 4.84 × 10−3, Fig. 5B), and presence of haemorrhages and microbleeds (OR = 1.63, 95% CI = 1.17–2.29, P = 4.35 × 10−3). PHS was not associated with presence of large arterial infarcts (OR = 1.26, 95% CI = 0.89–1.76, P > 0.05), one or more lacunas (OR = 1.25, 95% CI = 0.97–1.62, P > 0.05), presence of multiple microinfarcts (OR = 1.13, 95% CI = 0.89–1.44, P > 0.05), laminar necrosis (OR = 1.27, 95% CI = 0.51–2.99, P > 0.05), or presence of subcortical arteriosclerotic leukoencephalopathy (OR = 1.27, 95% CI = 0.98–1.66, P > 0.05). PHS was also not associated with FTLD with tau pathology or other tauopathy pathologies (OR = 1.32, 95% CI = 0.85–2.05, P > 0.05), progressive supranuclear palsy (OR = 1.43, 95% CI = 0.74–2.75, P > 0.05), FTLD with TDP-43 pathology (OR = 0.45, 95% CI = 0.18–1.00, P > 0.05), other taupathies (OR = 0.60, 95% CI = 0.32–1.06, P > 0.05), and presence of medial temporal lobe sclerosis (OR = 0.83, 95% CI = 0.60–1.11, P > 0.05). There were too few individuals with Pick’s and corticobasal degeneration to generate estimates for these two FTLD pathologies.
Figure 5.
PHS is associated with post-mortem cerebrovascular disease pathology. Box plots of severity of (A) cerebral amyloid angiopathy and (B) arteriolosclerosis for low (1 SD below mean, ≤16 percentile, n = 92 and n = 86, respectively) and high (1 SD above mean, ≥84 percentile, n = 98 and n = 90, respectively) PHS individuals.
Discussion
Beyond APOE, we show that PHS is associated with cross-sectional regional amyloid PET SUVR and longitudinal regional brain atrophy in individuals across the disease spectrum. The strongest effects for amyloid deposition were found in frontal brain regions such as the rostral middle frontal cortex and frontal pole while the strongest effects for brain atrophy were found in the entorhinal cortex, inferior parietal cortex, middle and inferior temporal cortex. Together, these results illustrate that in living persons PHS was associated with amyloid deposition and neurodegeneration in susceptible brain regions.
Building on prior work (Kauppi et al., 2018; Tan et al., 2018), we found that PHS was associated with cognitive and clinical decline in non-demented older individuals, even after controlling for frontal amyloid PET and entorhinal cortex volume. Although strongly associated with PET and MRI biomarkers, PHS may provide additional information for identifying older individuals at risk of Alzheimer’s disease associated decline beyond imaging biomarkers. In addition, the combined model that included PHS, frontal amyloid PET and entorhinal cortex volume resulted in a better fit for quantifying cognitive and clinical decline than a model with only imaging markers.
The current study adds to the literature that PHS may have utility in disease screening and risk stratification (Kauppi et al., 2018; Tan et al., 2018). This approach implies that the most relevant use of polygenic scores may be for disease prognosis through risk stratification, as opposed to diagnosis (Tan and Desikan, 2018; Torkamani et al., 2018). While genetics by themselves do not quantify pathological change (Jack et al., 2018), our combined results illustrate that polygenic information may be useful as an ‘enrichment’ marker for in vivo amyloid deposition and neurodegeneration and improves quantification of imaging biomarkers for detecting individuals most likely to develop Alzheimer’s disease associated changes along the amyloid-β deposition, pathological tau, and neurodegeneration [AT(N)] biological construct. Although there is currently no disease-modifying therapeutic for Alzheimer’s disease, our results suggest that clinical risk (quantified by PET and MRI biomarkers) and polygenic risk may help determine the action threshold for therapeutic intervention by evaluation of combined risk (Torkamani et al., 2018).
In the ROSMAP community-based cohort, we found that PHS was associated with regional post-mortem amyloid load and neuronal neurofibrillary tangles, validating our imaging findings. While regional differences were not evident for amyloid load, PHS was most strongly associated with tangles in the inferior temporal cortex. Crucially, we found these relationships even when accounting for APOE status, suggesting that polygenic information beyond APOE is associated with regional amyloid load and tangles. The absence of a statistically significant association of PHS with hippocampal tangles after controlling for APOE may suggest that genetic relationships with hippocampal tangles are driven relatively more by APOE compared to polygenic influence, or greater variation as a function of polygenic influence. The strength of the associations of PHS with amyloid load appears to be relatively homogenous across regions, while showing the strongest associations with tangles in the inferior temporal cortex, and weakest in the pericalcarine cortex. These relationships are consistent with the pathological staging of amyloid and tau propagation (Godert, 2015), suggesting that PHS may be useful for identifying individuals who will eventually develop amyloid and tau pathology corresponding to the most severe disease post-mortem pathological stages.
In the NACC cohort, we found that besides Alzheimer’s pathology, PHS was also associated with Lewy body and cerebrovascular disease pathology. These findings are consistent with cerebrovascular pathology being extremely common (up to 50%) as a mixed pathology in individuals with Alzheimer’s dementia, especially in individuals aged 85 or older (Alzheimer’s Association, 2017), suggesting a tight link between vascular and neurodegeneration in Alzheimer’s disease. Further, given that the typical clinical diagnosis of Alzheimer’s disease dementia is usually due to heterogenous pathologic aetiology (Corriveau et al., 2017), PHS may be useful in identifying individuals with the highest risk of developing dementia due to a combination of vascular and Alzheimer’s aetiologies. The association of PHS with Lewy body pathology is also consistent with existing findings showing that dementia with Lewy bodies (DLB) shares genetic risk with Alzheimer’s disease (Meeus et al., 2012; Guerreiro et al., 2016) and that individuals diagnosed with DLB had higher levels of Alzheimer’s pathology (Irwin et al., 2017).
Why is an Alzheimer’s associated polygenic score associated with cardiovascular and DLB pathology? We consider several possibilities. First, these findings may reflect underlying genetic pleiotropy/overlap between Alzheimer’s disease, cardiovascular traits and DLB (Desikan et al., 2015; Guerreiro et al., 2016; Broce et al., 2018). Second, co-occurrence of these pathologies may make it more likely that individuals with Alzheimer’s disease also have cardiovascular and DLB pathology. Third, compatible with recent findings (Bennett et al., 2018), Alzheimer’s pathology may lead to cardiovascular and possibly DLB pathologies. Fourth, a combination of these such that shared genetic risk may make it more likely that Alzheimer’s, cardiovascular and DLB pathologies co-occur.
In conclusion, we show that PHS shows regional associations with amyloid PET and neurodegeneration across the disease spectrum, and may be useful for tracking disease progression even after accounting for amyloid deposition in the frontal cortex and entorhinal cortex volume. PHS also showed a degree of spatial specificity for post-mortem tangles in the inferior cortex beyond APOE ɛ4, and may therefore be useful for tracking tau deposition across the disease spectrum. In addition, PHS was also associated with cerebrovascular and Lewy body pathology. Taken together, these findings further suggest a nuanced relationship between polygenic risk and markers of amyloid deposition, pathological tau and neurodegeneration. Our results also suggest that PHS may be useful for the identification of individuals at the highest risk for developing multi-aetiological dementia.
Supplementary Material
Acknowledgements
We thank UCSF Memory and Aging Center, UCSF Laboratory for Precision Neuroimaging, and the Shiley-Marcos Alzheimer’s Disease Research Center at UCSD for continued support.
Glossary
Abbreviations
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- CDR-SB
Clinical Dementia Rating-Sum of Boxes
- DLB
dementia with Lewy bodies
- NACC
National Alzheimer’s Coordinating Center
- PHS
polygenic hazard score
- ROSMAP
Religious Orders Study and Rush Memory and Aging Project
- SNP
single nucleotide polymorphism
- SUVR
standard uptake volume ratio
Funding
This work was supported by the Radiological Society of North America’s Resident/Fellow Award and RMS1741, American Society of Neuroradiology Foundation Alzheimer’s Disease Imaging Award, National Alzheimer’s Coordinating Center Junior Investigator award, National Institutes of Health Grants (NIH-AG046374, K01AG049152, P20AG10161, P30AG10161, R01AG15819, R01AG17917), the Research Council of Norway (#213837, #225989, #223273, #237250/European Union Joint Programme–NeurodegenerativeDisease Research), the South East Norway Health Authority (2013–123), Norwegian Health Association, the Kristian Gerhard Jebsen Foundation. Please see Supplementary material for Alzheimer’s Disease Neuroimaging Initiative, National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site and Alzheimer’s Disease Genetics Consortium funding sources.
Competing interests
A.M.D. is a founder of and holds equity in CorTechs Labs, Inc., and serves on its Scientific Advisory Board. He is also a member of the Scientific Advisory Board of Human Longevity, Inc. (HLI), and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References
- Aiken LS, West SG. Multiple regression: testing and interpreting interactions. Thousand Oaks, CA, US: Sage Publications, Inc.; 1991. [Google Scholar]
- Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimers Demen 2017; 12: 459–509. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the Religious Orders Study. Curr Alzheimer Res 2012a; 9: 628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the rush memory and aging project. Curr Alzheimer Res 2012b; 9: 646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimers Dis 2018; 64: S161–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broce IJ, Tan CH, Fan CC, Jansen I, Savage JE, Witoelar A et al. Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer's disease. Acta Neuropathol 2018; 1–18. doi: 10.1007/s00401-018-1928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L et al. Evaluation of a genetic risk score to improve risk prediction for Alzheimer’s disease. J Alzheimers Dis 2016; 53: 921–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corriveau RA, Koroshetz WJ, Gladman JT, Jeon S, Babcock D, Bennett DA et al. Alzheimer’s Disease-Related Dementias Summit 2016: national research priorities. Neurology 2017; 89: 2381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain Imaging Behav 2012; 6: 502–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Cabral HJ, Fischl B, Guttmann CR, Blacker D, Hyman BT et al. Temporoparietal MR imaging measures of atrophy in subjects with mild cognitive impairment that predict subsequent diagnosis of Alzheimer disease. AJNR Am J Neuroradiol 2009; 30: 532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples A et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med 2017; 14: e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Schork AJ, Wang Y, Thompson WK, Dehghan A, Ridker PM et al. Polygenic overlap between C-reactive protein, plasma lipids, and Alzheimer disease. Circulation 2015; 131: 2061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006; 31: 968–80. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Jones L. Genomic profiling and diagnostic biomarkers in Alzheimer’s disease. Lancet Neurol 2017; 16: 582–3. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015; 138: 3673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol 2017; 82: 311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Sabuncu MR, Smoller JW, Sperling RA, Mormino EC. Dissociable influences of APOE e4 and polygenic risk of AD dementia on amyloid and cognition. Neurology 2018; 90: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P et al. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav 2012; 6: 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godert M. Alzheimer’s and Parkinson’s diseases: the prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 2015; 349: 1255555. [DOI] [PubMed] [Google Scholar]
- Gómez-Isla T, Price JL, McKeel DW Jr, Morris JC, Growdon JH, Hyman BT. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci 1996; 16: 4491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer T, Tholen S, Yousefi BH, Alexopoulos P, Förschler A, Förstl H et al. Progression of cerebral amyloid load is associated with the apolipoprotein E ɛ4 genotype in Alzheimer’s disease. Biol Psychiatry 2010; 68: 879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Escott-Price V, Darwent L, Parkkinen L, Ansorge O, Hernandez DG et al. Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson’s and Alzheimer’s diseases. Neurobiol Aging 2016; 38: 214.e7–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Brendel M, Grimmer T, Rominger A, Yakushev I. Predicting regional pattern of longitudinal β-amyloid accumulation by baseline PET. J Nul Med 2017; 58: 639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison TM, Mahmood Z, Lau EP, Karacozoff AM, Burggren AC, Small GW et al. An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro 2016; 3 pii: ENEURO.0098–16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D, Brewer JB, Hagler DJ, Fenema-Notestine C, Dale AM. Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc Natl Acad Sci 2009; 106: 20954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D, Dale AM. Nonlinear registration of longitudinal images and measurement of change in regions of interest. Medical Image Analysis 2011; 15: 489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DJ, Grossman M, Weintraub D, Hurtig HI, Duda JE, Xie SX et al. Neuropathological and genetic correlates of survival and dementia onset in synnucleinopathies: a retrospective analysis. Lancet Neurol 2017; 16: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB et al. NIA-AA research-framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018; 14: 535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM et al. Relationships between biomarkers in aging and dementia. Neurology 2009; 73: 1192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi K, Fan CC, McEvoy LK, Holland D, Tan CH, Chen CH et al. Combining polygenic hazard score with volumetric MRI and cognitive measures improves prediction of progression from mild cognitive impairment to Alzheimer’s disease. Front Neurosci 2018; 12: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeus B, Theuns J, VanBroeckhoven C. The genetics of dementia with Lewy bodies: what are we missing? Arch Neurol 2012; 69: 1113–8. [DOI] [PubMed] [Google Scholar]
- Mormino EC, Sperling RA, Holmes AJ, Buckner RL, De Jager PL, Smoller JW et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 2016; 5: 481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012; 22: 2653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, Desikan RS. Interpreting Alzheimer’s disease polygenic scores. Ann Neurol. 2018; 83: 443–5. [DOI] [PubMed] [Google Scholar]
- Tan CH, Fan CC, Mormino EC, Sugrue LP, Broce IJ, Hess CP et al. Polygenic hazard score: an enrichment marker for Alzheimer’s associated amyloid and tau deposition. Acta Neuropathol 2018; 135: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, Hyman BT, Tan JJX, Hess CP, Dillon WP, Schellenberg GD et al. Polygenic hazard scores in preclinical Alzheimer disease. Ann Neurol 2017; 82: 484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker AA, Weinberg BD, Dillon WP, Hess CP, Cabral HJ, Fleischman DA et al. Entorhinal cortex: antemortem cortical thickness and postmortem neurofibrillary tangles and amyloid pathology. AJNR Am J Neuroradiol 2017; 38: 961–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nav Rev Genet 2018; 19: 581–90. [DOI] [PubMed] [Google Scholar]
- Van Hoesen GW, Hyman BT, Damasio AR. Entorhinal cortex pathology in Alzheimer’s disease. Hippocampus 1991; 1: 1–8. [DOI] [PubMed] [Google Scholar]
- Voyle N, Patel H, Folarin A, Newhouse S, Johnstone C, Visser PJ et al. Genetic risk as a marker of amyloid-b and tau burden in cerebrospinal fluid. J Alzheimers Dis 2017; 55: 1417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available for request in (i) LONI at https://ida.loni.usc.edu/; (ii) RADC Research Resource Sharing Hub at https://www.radc.rush.edu/; and (iii) NACC at https://www.alz.washington.edu/.