Mutations in neuronal sodium channels cause neurodevelopmental disorders and epilepsy. Bunton-Stasyshyn et al. develop a mouse model with conditional expression of the recurrent gain-of-function mutation SCN8A-R1872W. Activation by Cre recombinase in excitatory but not inhibitory neurons results in seizures and lethality, while activation in adult brain recapitulates the seizure disorder.
Keywords: sodium channel, Nav1.6, epileptic encephalopathy, conditional allele, mouse model
Abstract
De novo mutations of the sodium channel gene SCN8A result in an epileptic encephalopathy with refractory seizures, developmental delay, and elevated risk of sudden death. p.Arg1872Trp is a recurrent de novo SCN8A mutation reported in 14 unrelated individuals with epileptic encephalopathy that included seizure onset in the prenatal or infantile period and severe verbal and ambulatory comorbidities. The major biophysical effect of the mutation was previously shown to be impaired channel inactivation accompanied by increased current density. We have generated a conditional mouse mutation in which expression of this severe gain-of-function mutation is dependent upon Cre recombinase. Global activation of p.Arg1872Trp by EIIa-Cre resulted in convulsive seizures and lethality at 2 weeks of age. Neural activation of the p.Arg1872Trp mutation by Nestin-Cre also resulted in early onset seizures and death. Restriction of p.Arg1872Trp expression to excitatory neurons using Emx1-Cre recapitulated seizures and juvenile lethality between 1 and 2 months of age. In contrast, activation of p.Arg1872Trp in inhibitory neurons by Gad2-Cre or Dlx5/6-Cre did not induce seizures or overt neurological dysfunction. The sodium channel modulator GS967/Prax330 prolonged survival of mice with global expression of R1872W and also modulated the activity of the mutant channel in transfected cells. Activation of the p.Arg1872Trp mutation in adult mice was sufficient to generate seizures and death, indicating that successful therapy will require lifelong treatment. These findings provide insight into the pathogenic mechanism of this gain-of-function mutation of SCN8A and identify excitatory neurons as critical targets for therapeutic intervention.
Introduction
Epileptic encephalopathies are severe seizure disorders accompanied by cognitive, behavioural and movement disturbances (Berg et al., 2010). The first SCN8A mutation identified in epileptic encephalopathy was the de novo missense variant p.Asn1768Asp (N1768D) (Veeramah et al., 2012). More than 150 additional de novo missense mutations have now been reported (EIEE13, MIM#614558) (Larsen et al., 2015; Meisler et al., 2016). SCN8A encodes the voltage gated sodium channel Nav1.6, which is responsible for the initiation and propagation of neuronal action potentials. Nav1.6 is concentrated at the axon initial segment and nodes of Ranvier in neurons of the central and peripheral nervous system. Patient mutations of SCN8A are localized to the evolutionarily conserved transmembrane segments, intracellular inactivation gate loop, and C-terminal domain (Wagnon and Meisler, 2015). Most of the characterized patient mutations result in gain-of-function changes in biophysical properties resulting in elevated channel activity, either due to premature channel opening or impaired channel inactivation (Veeramah et al., 2012; de Kovel et al., 2014; Estacion et al., 2014; Blanchard et al., 2015; Barker et al., 2016; Patel et al., 2016; Wagnon et al., 2016). In contrast, loss-of-function mutations of SCN8A result in isolated intellectual disability, myoclonus, and movement disorders (Wagnon et al., 2017, 2018) with elevated sensitivity to anaesthesia in the mouse (Pal et al., 2015).
Clinical features of SCN8A encephalopathy include multiple seizure types with onset between prenatal and 18 months (average 4 months) (Larsen et al., 2015; Hammer et al., 2016). Seizures are refractory to therapy and often require multi-drug treatment. Impaired cognitive function and developmental arrest or regression after seizure onset are common. Intellectual disability ranges from mild to severe. Language is commonly impaired with many affected individuals having little or no speech. Approximately half of all diagnosed children are non-ambulatory. Those who do learn to sit or walk often have a movement disorder and progressive loss of ambulation. There is increased risk of death, with reported rates of 5 to 10% (Hammer et al., 2016; Johannesen et al., 2018).
The first mouse model of SCN8A encephalopathy was generated by knock-in of the patient mutation p.Asn1768Asp (Veeramah et al., 2012; Jones and Meisler, 2014; Wagnon et al., 2015). Heterozygous Scn8aN1768D/+ mice recapitulated spontaneous seizures and sudden death with 50% penetrance. Video-EEG recordings identified convulsive seizures concordant with ictal activity, as well as epileptiform discharges coincident with myoclonic jerks. Mild deficits in motor coordination and social discrimination were observed. Affected mice have onset at 2–4 months and rapid progression to death (Wagnon et al., 2015; Sprissler et al., 2017). Persistent sodium current and spontaneous firing are elevated in hippocampal neurons but not in cortical pyramidal neurons (Lopez-Santiago et al., 2017). Medial entorhinal cortex neurons, which provide the major input into the hippocampus, displayed impaired channel closing and increased persistent and resurgent currents (Ottolini et al., 2017). Both direct and indirect effects of mutant Nav1.6 on cardiac function were observed. Scn8aN1768D/+ mice are bradycardic but denervated hearts had normal heart rate, suggesting an indirect effect of parasympathetic innervation. Cell-autonomous abnormalities in dissociated cardiac myocytes from Scn8aN1768D/+ mice included hyperexcitability and arrhythmia (Frasier et al., 2016). These cardiac defects may contribute to the elevated risk of death in affected individuals.
To develop a conditional model of SCN8A encephalopathy that includes the more severe clinical features, we selected the recurrent mutation p.Arg1872Trp (R1872W). This mutation is located in a conserved region of the C-terminus that contributes to stability of the inactivated channel through ionic interaction of the positively charged arginine 1872 with negatively charged residues in the inactivation gate (Nguyen and Goldin, 2010). Replacement of arginine 1872 with uncharged amino acids, including tryptophan, results in impaired channel inactivation and elevated current density (Wagnon et al., 2016). More than 24 independent de novo substitutions at residue 1872 have been reported. Clinical data for four individuals with the R1872W mutation reveal seizure onset between 10 days and 4 months of age, status epilepticus in three individuals, and severe motor disabilities including hypertonia, hypotonia, tremor, spasticity and non-ambulation (Ohba et al., 2014; Larsen et al., 2015; Takahashi et al., 2015; Gardella et al., 2018). Sudden unexpected death in epilepsy (SUDEP) was reported for one child at 5 years of age.
We now describe the generation of a conditionally inducible mouse model of SCN8AR1872W and initial characterization of the underlying pathophysiology of seizures. Global activation of the p.Arg1872Trp mutation results in a more severe disease than in the original p.Asn1768Asp mouse, including earlier onset of seizures and shorter duration before lethality. We have been able to compare the effects of the mutation in excitatory and inhibitory neurons, and the effect of activation after development in the adult brain. Compared with the original model, the conditional mouse will permit more efficient evaluation of therapeutic interventions due to the earlier onset, complete penetrance and reproducible time course of the disorder.
Materials and methods
TALEN-mediated genome targeting
Genome targeting was carried out as previously described using two exon 26-directed TAL effector nuclease (TALEN) mRNAs (Jones and Meisler, 2014) and a new 9 kb targeting template (Supplementary Fig. 1A). The circular targeting construct DNA and two TALEN mRNAs were microinjected into the pronucleus of (C57BL/6J × SJL)F2 eggs in the University of Michigan Transgenic Animal Core (www.tamc.umich.edu). Potential founders were screened by PCR of genomic DNA from tail biopsies using two pairs of primers. Primer pair 1 (5′-CGAGTACCCAAGCCCAACAC-3′ and 5′-AGCCAGGTGTCCCCTGTAAG-3′) amplified a 276 bp product containing a BsrBI site that was ablated by the R1872W mutation. Digestion with BsrBI generated wild-type fragments of 56, 100 and 120 bp and mutant fragments of 56 and 220 bp. Intronic DNA upstream of exon 26a was amplified by primer pair 2 (5′-GCACGTGCTGAAAAAGTGG-3′ and 5′-CCTCCTCTTACCGTGCAGAC-3′) producing a product of 414 bp that is resistant to KpnI digestion from the wild-type allele and a 448 bp product from the conditional allele that is digested by Kpn1 to give fragments of 262 bp and 168 bp (Supplementary Fig. 2). The recombination junctions of the correctly targeted founder (#2176) were assayed by Southern blotting after digestion with HincII or KpnI, as described previously (Jones and Meisler, 2014). The HincII blot was probed with the previously described 1.8 kb downstream flanking fragment (Jones and Meisler, 2014) to detect a unique 3.5 kb restriction fragment resulting from correct recombination at the 3′ end of the targeted allele. The KpnI blot was probed with an 0.67-kb upstream fragment amplified from C57BL/6J genomic DNA with primer pair 3 (5′-TCGTGTGAGTGGGTGCTCATCTTG-3′ and 5′-GGTTTTGTCTGCAAGACTGGTTCC-3′) that hybridizes with a unique 5.4 kb fragment confirming the predicted recombination at the 5′ end of the targeted allele. The sequence of the correctly targeted conditional allele was confirmed by Sanger sequencing of two long-range junction PCR products amplified from genomic DNA using Invitrogen Platinum™ SuperFi™ PCR Master Mix with an annealing temperature of 63°C. A 7 kb fragment containing the 5′ junction was amplified using primer pair 4 (5′-CGACTGTGCCTTCTAGTTGC-3′ and 5′-GGTGGACAGATGGAAGGTGA-3′). A 4.5 kb fragment containing the 3′ junction was amplified with primer pair 5 (5′-AGATGCCCAGTACCAAAGCT-3′ and 5′-GCAACTAGAAGGCACAGTCG-3′). A comprehensive list of primers used in this study is given in Supplementary Table 1.
Genotyping assays
Mice carrying the conditional allele were routinely genotyped by PCR using primer pair 2 followed by digestion with KpnI to generate products of 414 bp from the wild-type allele, products of 262 and 168 bp from the non-activated conditional allele containing exons 26a and 26b, and products of 228, 168 and 40 bp from the activated conditional allele with deletion of exon 26a (Supplementary Fig. 2).
RT-PCR
Total RNA was isolated from whole brain by TRIzol® extraction. cDNA was synthesized using LunaScript® RT SuperMix Kit (NEB). RT-PCR was carried out using Promega GoTaq® reagents using standard PCR protocols. All of the Scn8a transcripts were amplified from exon 25 to 26 using primer pair 6 (5′-GGTCATTCTCTCCATTGTGG-3′ and 5′-CCTCCATTCTCCAGCTTGTT-3′). Transcripts containing exon 26a and the bGH 3′ untranslated region (UTR) were amplified in a 1.2 kb product using primer pair 7 (5′-GGTCATTCTCTCCATTGTGG-3′ and 5′-GCAACTAGAAGGCACAGTCG-3′). Transcripts containing exon 26b and the endogenous 3′ UTR were amplified in a 1.2 kb product using primer pair 8 (5′-GGTCATTCTCTCCATTGTGG-3′ and 5′-GCTCTGTGTCCGTGAGATTC-3′). Sanger sequencing of PCR products was carried out by the University of Michigan Sequencing Core.
Quantitative RT-PCR
TaqMan® gene expression assays were carried out in duplicate in a 20 µl reaction volume using the TaqMan® Gene Expression Master Mix. Reactions were run in quadruplicate on an ABI StepOnePlus™ cycler using StepOne™ Software v2.3 (Life Technologies). Total Scn8a transcript was assayed using FAM-labelled gene expression assay Mm00488110_m1 standardized to the VIC labelled Tbp endogenous control Mm01277042_m1. The adult-specific Scn8a transcript containing exon 18A (Plummer et al., 1998) was assayed using VIC labelled gene expression assay Mm00488119 and standardized to FAM-labelled total Scn8a transcript Mm00488110_m1. Relative transcript quantity was calculated using the ΔΔCT method (Livak and Schmittgen, 2001).
Animals
All mice were housed and cared for in accordance with NIH guidelines in a 12/12-h light/dark cycle with standard mouse chow and water available ad libitum. Experiments were approved by the Committee on the Use and Care of Animals at the University of Michigan and the University of Virginia. Scn8acond/cond mice homozygous for the conditional allele were crossed with the Cre lines EIIa-Cre (Jax 003724), Emx1-Cre (Jax 005628), Gad2-Cre (Jax 028867), Dlx5a-Cre (Jax 008199), Nes-Cre (Jax 003771), and ER-Cre (Jax 004682). Expression patterns and published references to the Cre lines are provided in Supplementary Table 2.
EEG recordings
Electrodes were implanted in pre-weaning Scn8acond/+,EIIa-Cre mice at postnatal Days 7–15 (P7–P15) and Scn8acond/+,Emx-1 mice at P21 as previously described (Zanelli et al., 2014). Mice were recorded for 2–4 h twice a day starting on P9–P14. Recordings were visually inspected for the presence of defined electrographic seizures (Zanelli et al., 2014). Adult Scn8acond/+,Gad2-Cre mice were implanted and recorded as previously described (Wagnon et al., 2015) with the following modifications: mice were anaesthetized with isoflurane, three burr holes were drilled to place left parietal, right parietal and cerebellar (reference) electrodes, and video/EEG recordings were obtained with a Natus Neuroworks system.
Tamoxifen treatment
Tamoxifen (Sigma T5648) was dissolved in a mixture of 90% corn oil and 10% ethanol at a concentration of 15 mg/ml. Eight-week-old mice were injected intraperitoneally on five successive days at 75 mg/kg body weight/day.
Effects of GS967/Prax330
For in vivo experiments, mouse chow containing GS967/Prax330 (8 mg/kg) was provided to mothers of treated mice beginning on P1 and continuing after weaning, as described previously (Baker et al., 2018). Control mice received unsupplemented chow.
For experiments in cultured cells, ND7/23 cells (Sigma Aldrich) were transfected with wild-type or mutant cDNA and biophysical properties were examined as previously described (Wagnon et al., 2016).
For experiments in brain slices, hippocampal CA1 neurons and layer V cortical neurons were recorded as previously described in P13–P17 mice (Ottolini et al., 2017).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
Design of the conditional R1872W allele of Scn8a
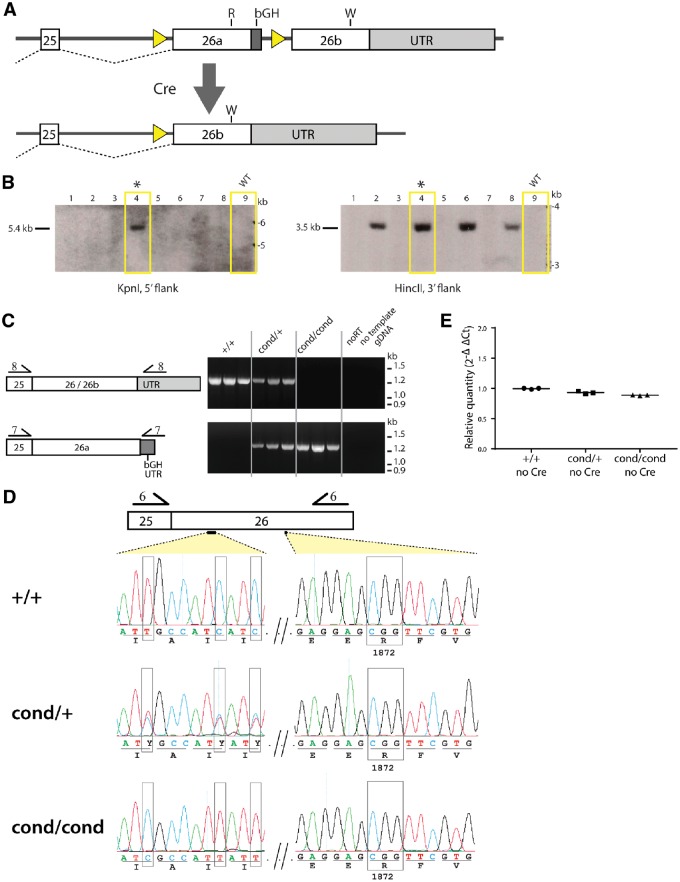
The conditional allele of Scn8a contains two copies of exon 26, the final exon of the gene (Fig. 1A). The upstream copy, designated exon 26a, is flanked by LoxP sites and encodes the wild-type arginine 1872 codon (CGG). Exon 26a also contains eight synonymous nucleotide substitutions for protection against TALEN digestion and terminates with the 282 bp 3′ UTR from the bovine growth hormone (bGH) gene (Goodwin and Rottman, 1992). Exon 26b contains the same eight silent substitutions but encodes the mutant tryptophan residue 1872 (TGG) and terminates with the endogenous 6 kb 3′ UTR (Drews et al., 2005). In the absence of Cre recombinase, the transcript of the conditional allele was designed to be spliced from exon 25 to exon 26a and to terminate in the bGH 3′ UTR. The exon 26a transcript will encode the wild-type arginine 1872 residue (Fig. 1A). After deletion of exon 26a by Cre recombinase, transcripts should be spliced from exon 25 to exon 26b, resulting in expression of the mutant tryptophan 1872 codon.
Figure 1.
Structure and basal expression of the conditional R1872W allele of Scn8a. (A) The conditional allele contains two copies of exon 26, the final exon of Scn8a. Both exons 26a and 26b were derived from a targeting vector with eight silent substitutions in exon 26 that were introduced to prevent digestion by the TALEN endonucleases used for targeting of the Scn8a locus (Jones and Meisler, 2014; see Supplementary Fig. 1 for details). The upstream exon 26a contains wild-type arginine codon 1872 (R) and the 3′ UTR from the bGH gene, and is flanked by loxP sites. The downstream exon 26b contains the same eight silent substitutions as well as the mutant tryptophan codon 1872 (W) and terminates in the endogenous 3′ UTR. Exposure to Cre recombinase will delete exon 26a and lead to expression of exon 26b encoding the mutant sodium channel. (B) Southern blot of genomic DNA from eight potential founder mice. Correct targeting of the endogenous Scn8a locus in DNA from founder 2176 (asterisk) is demonstrated by the 5.4 kb KpnI restriction fragment hybridizing with the 5′ probe 1 as well as the 3.5 kb HincII hybridizing with 3′ probe 2 (Supplementary Fig. 1B). The full gel is shown in Supplementary Fig. 1B. Related data in Supplementary Fig. 1C show the sequenced regions of long-range junction PCR products that confirmed the structure of the conditional allele. (C) RT-PCR products were amplified with forward primer in exon 25 and reverse primer either in the bGH 3′ UTR or the endogenous 3′ UTR. The wild-type allele expresses the endogenous 3′ UTR and the conditional allele expresses only the bGH 3′ UTR in the absence of Cre recombinase. (D) Sequences of the RT-PCR products amplified using primer pair 6 (forward primer in exon 25, reverse primer in exon 26) demonstrate expression of exon 26 from the wild-type allele and exon 26a from the conditional allele. Boxed nucleotides indicate the location of markers of the conditional allele: three of the synonymous nucleotide substitutions and tryptophan codon 1872. (E) Quantitative RT-PCR of whole brain RNA with the TaqManTM assay for Scn8a and comparison with the control transcript of the Tata-binding protein (Tbp) demonstrates comparable level of transcript from the wild-type allele and the conditional allele. Three animals of each genotype were assayed.
The targeting construct for the conditional allele (Supplementary Fig. 1A) was injected into fertilized mouse eggs together with two mRNAs encoding TALEN endonucleases directed to sequences in exon 26 near codon 1768 (Jones and Meisler, 2014). Sixty-one potential founder mice were obtained and genotyped to detect targeting into the Scn8a locus on mouse chromosome 15. Thirty-nine mice had changes in the endogenous Scn8a locus that were detectable by PCR. To identify correctly targeted alleles, we carried out Southern blotting with probes to the upstream and downstream homology arm regions of the targeting construct (Supplementary Fig. 1B). Genomic DNA from founder #2176 contained both the 5.4 kb KpnI fragment and the 3.5 kb HincII fragment that are diagnostic for the correctly targeted locus (Fig. 1B). The predicted structure of the conditional allele was confirmed by long range junction PCR using primers upstream and downstream of the targeting construct (Supplementary Fig. 1C) followed by Sanger sequencing of 4.8 kb from the PCR products (Supplementary Fig. 1C). The founder transmitted the conditional allele to 50% of offspring (24/49). Heterozygous Scn8acond/+ offspring of this founder were intercrossed to generate homozygous Scn8acond/cond mice. The homozygous carriers of the conditional allele were viable and fertile, with no visible abnormalities up to 1 year of age.
To determine which copy of exon 26 was expressed from the Scn8acond allele prior to exposure to Cre, we amplified the Scn8a transcript from Scn8a+/+, Scn8acond/+ and Scn8acond/cond mice using primer pair 6 located in exons 25 and 26. Sanger sequencing of the RT-PCR products detected the eight silent substitutions and the arginine codon from exons 26a in heterozygous state in Scn8acond/+ mice and in homozygous state in Scn8acond/cond mice, confirming the intended splicing from exon 25 to exon 26a (Fig. 1D). We investigated splicing of the conditional allele using RT-PCR to detect the bGH 3′ UTR located downstream of exon 26a (Fig. 1C). As predicted, transcripts containing the bGH 3′ UTR were present in Scn8acond/+ and Scn8acond/cond mice but not in Scn8a+/+ mice. Importantly, transcripts containing the endogenous 3′ UTR were present in Scn8a+/+ and Scn8acond/+ mice but were not present in Scn8acond/cond mice at levels detectable by 30 PCR cycles. The molecular analysis of the conditional allele in the absence of Cre confirmed the expression of exon 26a encoding wild-type channel and no expression of exon 26b encoding the R1872W mutant.
Quantitation of Scn8a transcripts in brain from mice of genotypes Scn8a+/+, Scn8acond/+ and Scnacond/cond demonstrated that the level of Scn8a transcripts is comparable in the three types of mice (Fig. 1E). Interestingly, replacement of the endogenous 3′ UTR by the bGH 3′ UTR does not alter the steady-state abundance of Scn8a transcript. We also found that the proportion of transcripts containing alternatively spliced exons 18A and 18N (Plummer et al., 1998) is comparable in mice of the three genotypes (Supplementary Fig. 3).
Heterozygous expression of the mutant transcript after global activation of the conditional allele
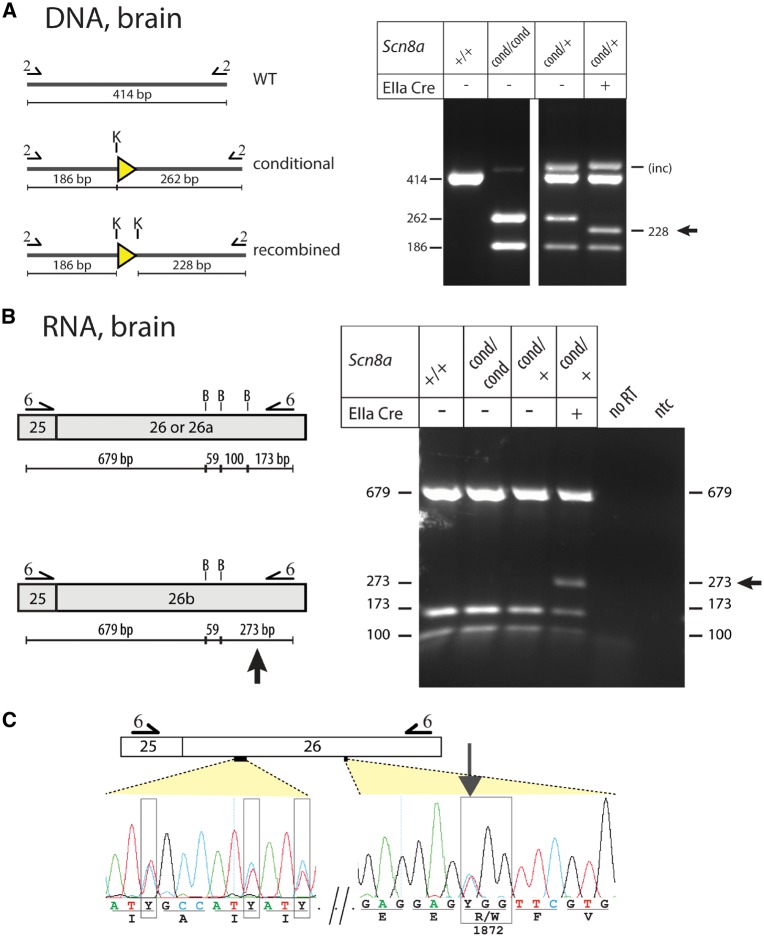
Male Scnacond/cond mice were crossed with female mice carrying the EIIa-Cre transgene, which is globally expressed in early pre-implantation embryos and also in female germ cells (Lakso et al., 1996; De Gasperi et al., 2008). We examined the deletion of exon 26a in offspring who inherited both the conditional allele and the Cre transgene (Scn8acond/+,EIIa-Cre mice). Genomic DNA was amplified with primer pair 2 flanking the LoxP site in intron 25, followed by digestion with KpnI (Supplementary Fig. 2). The recombined fragment with deletion of exon 26a was present in DNA from tail, forebrain and hindbrain of the Scn8acond/+,EIIa-Cre mice, but was not present in Scn8acond/+ mice lacking EIIa-cre (Fig. 2A). To detect the expression of exon 26b, we carried out RT-PCR of brain RNA using primer pair 6, and digested the amplified products with BsrB1 (Fig. 2B). The introduction of the tryptophan codon destroys a BsrBI site in this amplicon, producing a 273 bp BsrBI fragment from transcripts expressing exon 26b in place of the 100 bp and 173 bp fragments from the wild-type and exon 26a. The presence of 100, 173 and 273 bp BsrBI fragments in RNA from Scn8acond/+,EIIa-Cre tissues demonstrates heterozygous expression of exon 26b and wild-type exon 26 (Fig. 2B). Sanger sequencing of the RT-PCR product confirmed heterozygous expression of the R1872W codon from exon 26b as well as the eight silent substitutions (Fig. 2C). The molecular analysis of genomic DNA and brain RNA demonstrates successful activation by EIIa-Cre of exon 26b from the Scn8a conditional allele.
Figure 2.
Ubiquitous activation of the conditional Scn8a allele by EIIA-Cre. (A) Genotyping of genomic DNA samples from brain and tail of heterozygous Scn8acond/+,EIIa-Cre mice expressing one wild-type and one conditional allele demonstrates Cre-mediated deletion of exon 26a. Genomic DNA was amplified with primer pair 2 located in intron 25 (Supplementary Table 1 and Supplementary Fig. 2) and the PCR product was digested with KpnI. (B) RT-PCR of brain RNA using primer pair 6 in exons 25 and 26. There is expression of exon 26b from the conditional allele, due to the excision of exon 26a by Cre recombinase. In addition, exon 26 is expressed from the wild-type allele in the heterozygous mice. B = Bsrb1. (C) Sanger sequencing of the RT-PCR products demonstrates the heterozygous expression of wild-type exon 26 and exon 26b. Heterozygous synonymous mutations are boxed and the heterozygous R1872W mutation is marked with an arrow. The location of the sequenced nucleotides within the amplified exon is indicated by dotted lines.
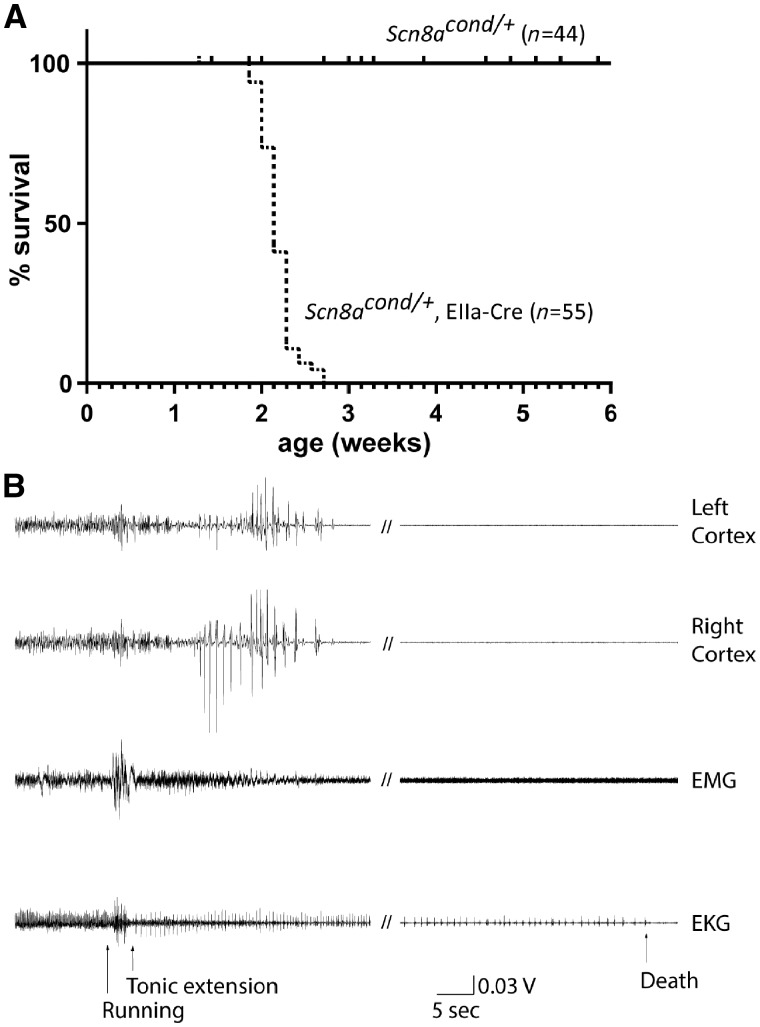
Early onset seizures and lethality in mice with global activation of the mutant channel
Scn8acond / +,EIIa-Cre mice with heterozygous expression of the R1872W mutant channel were indistinguishable from wild-type littermates for the first 2 weeks of postnatal life, with normal righting reflex and visible motor activity. At P14 to P16, severe spontaneous seizures were observed, with median survival to P15 (Fig. 3A). To monitor seizure onset, EEG electrodes were implanted in Scn8acond/+, EIIa-Cre mice at P7 to P15, and video-EEG monitoring was carried out for 2 h/day. The seizures began with abrupt onset of running that lasted for 1 to 3 s, immediately followed by the loss of upright posture and brief tonic extension (Supplementary Video 1). The movement artefact recorded during running was followed after 4.0 ± 0.6 s (n = 8) by an electrographic seizure or repetitive spike trains that lasted for 20.0 ± 3.0 s, followed by post-ictal suppression (Fig. 3B). The age at death for the implanted animals was P16 ± 0.4 days (n = 8), which did not differ from non-implanted mice. The time from running fit to death varied from 43 s to 8 min. In five of eight mice, death occurred immediately after the recording of a single seizure. ECG recording from three mice detected simultaneous tonic extension and slowing of heart rate (Fig. 3B). Reduction in heart rate from 756 beats per minute (bpm) to 156 bpm was followed by death at 3 min 20 s after the start of the running fit is shown for one of these mice.
Figure 3.
Phenotypic effects of activation of the conditional allele by ubiquitously expressed EIIa-Cre. (A) Scn8acond/+ heterozygotes expressing EIIa-Cre exhibited sudden lethality at the end of the second week of life, with median survival of 15 days. Scn8acond/+ heterozygotes that did not inherit EIIa-Cre exhibited normal life span. (B) EEG recording of a fatal seizure in an Scn8acond/+,EIIa-Cre mouse on P15. The recording was obtained from bipolar insulated stainless-steel electrodes in the left parietal cortex (top trace) and right parietal cortex (second trace). The EMG artefact in the third trace reflects the period of running evolving to tonic extension. Death occurred 3 min 20 s after the onset of running. The bottom trace was obtained from an electrode placed subcutaneously in the right thorax and demonstrates sudden slowing of the heart rate from 756 bpm prior to the running fit to 156 bpm immediately afterwards.
Three mice demonstrated multiple seizures with return to normal activity 2.0 ± 0.5 min after each seizure. Thorough visual review of EEG recordings from two age-matched wild-type mice between P9 and P14 did not detect epileptiform discharges or electrographic seizures.
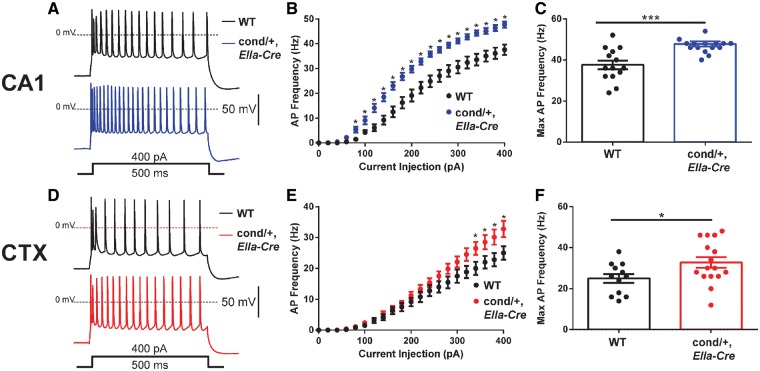
Neuronal hyperactivity in mice with global expression of R1872W
To investigate neuronal activity underlying the seizures in Scn8acond/+,EIIa-Cre mice, we carried out slice recordings from cerebral cortex and hippocampus. CA1 neurons from Scn8acond/+,EIIa-Cre mice were hyperexcitable over a range of current injection steps (Fig. 4A–C). At injection of 400 pA, the action potential frequency was increased in both mutant CA1 neurons (47.8 ± 1.2 Hz, n = 16 cells from five mice, P < 0.001) and mutant layer V cortical neurons (32.8 ± 2.6 Hz; n = 16 cells from 5 mice, P < 0.05) compared with wild-type CA1 (37.6 ± 2.2 Hz; n = 14 cells from four mice) and cortical neurons (25.0 ± 2.2 Hz; n = 12 cells from four mice) (Fig. 4D–F).
Figure 4.
Hyperactivity of cortical neurons expressing the Scn8aR1872W mutant sodium channel. (A and B) Representative slice recordings from control (14 cells from four mice) and Scn8acond/+,EIIa-Cre CA1 neurons (16 cells from five mice). (C) Action potential firing frequency at 400 pA current injection. (D and E) Representative slice recordings from control (12 cells from four mice) and Scn8acond/+,EIIa-Cre layer V cortical neurons (16 cells from five mice). (F) Action potential firing frequency at 400 pA current injection. Data shown represent means ± standard error of the mean (SEM) *P < 0.05, ***P < 0.001.
Analysis of membrane properties of CA1 neurons revealed significant increases in upstroke velocity, spike amplitude and input resistance and decreased rheobase in the mutant CA1 neurons (Table 1). The data demonstrate increased excitability of cortical and hippocampal neurons expressing Scn8aR1872W that is likely to increase overall network excitability and predispose to seizure initiation and propagation.
Table 1.
Membrane and action potential properties of CA1 pyramidal neurons
| Vm, mV | Threshold, mV | Rheobase, pA | Upstroke velocity, mV/ms | Downstroke velocity, mV/ms | Amplitude, mV | APD50, ms | Input resistance, MΩ | |
|---|---|---|---|---|---|---|---|---|
| Wild-type (n = 14, 4) | −67.0 ± 0.9 | −42.4 ± 1.0 | 84.33 ± 9.6 | 328 ± 35 | −57.8 ± 1.9 | 88.6 ± 2.5 | 1.39 ± 0.06 | 252 ± 28 |
| R1872W (n = 16, 5) | −67.1 ± 1.1 | −43.5 ± 1.1 | 51.3 ± 5.8* | 435 ± 25* | −55.8 ± 1.6 | 94.1 ± 0.9* | 1.47 ± 0.04 | 320 ± 29* |
Recordings were carried out on multiple cells from each animal (n = cells, animals). There was no current injection into the resting cells.
*Statistical significance at P < 0.05, Student’s t-test.
Neural expression of Scn8aR1872W is sufficient for seizures and lethality
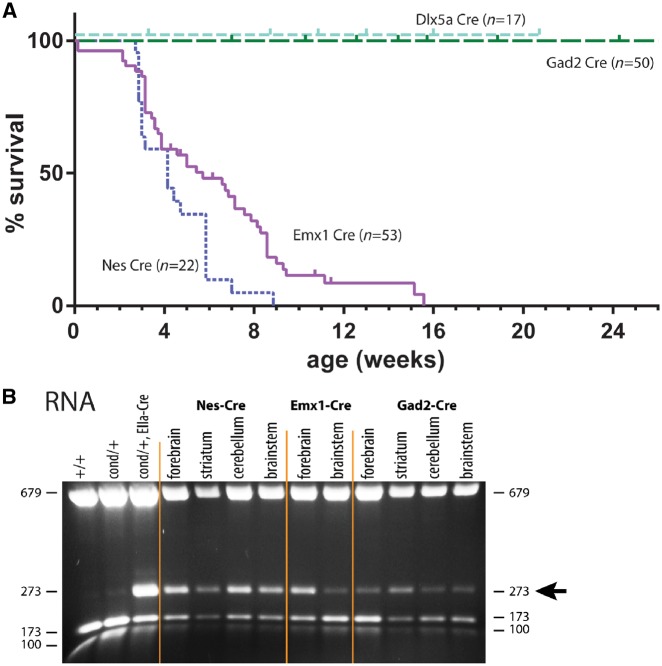
To obtain restricted neural expression of the R1872W mutant channel, we crossed Scn8acond/cond mice with mice expressing a Nestin-Cre transgene (Supplementary Table 2). Scn8acond/+,Nes-Cre mice developed severe spontaneous seizures at 3 to 4 weeks of age, leading rapidly to lethality (Fig. 5A). RT-PCR of RNA demonstrated inclusion of exon 26b encoding the R1872W codon in transcripts from forebrain, striatum, cerebellum and brainstem (273 bp fragment), at similar levels to transcripts containing the wild-type exon (173 bp), demonstrating efficient deletion of exon 26a throughout the brain (Fig. 5B). The median survival of mice expressing Nestin-Cre was 21 days postnatal. The rapid progression of the seizure disorder in Scn8acond/+, Nes-Cre mice indicates that neural expression of the mutant channel is sufficient to replicate the effects of global expression of R1872W. The 6-day delay of seizure onset and lethality in the Nes-Cre mice compared with the Scn8acond/+,EIIa-Cre mice [χ2(1) = 39.82; P < 0.0001] suggests that non-neural expression of Scn8a may contribute to the phenotype.
Figure 5.
Effects of expression of Scn8aR1872W in forebrain neurons. (A) Survival of conditional mice expressing cell-specific Cre constructs. Nestin-Cre is broadly expressed in neural cells; median survival 21 days. Emx1-Cre is expressed in excitatory neurons of the forebrain; median survival 46 days. Gad2-Cre is expressed in inhibitory neurons. (B) Brain transcripts encoding arginine 1872 (173 bp RT-PCR fragment) and tryptophan 1872 (273 bp RT-PCR fragment) in mice expressing various Cre recombinases.
Expression of Scn8aR1872W in forebrain excitatory neurons results in seizures and lethality
To dissect the contributions of neuronal subpopulations, we compared the effect of activating the R1872W mutation in excitatory versus inhibitory neurons. Scn8acond/cond mice were crossed with heterozygous mice carrying an Emx1-Cre from the endogenous Emx1 locus that is expressed in forebrain excitatory neurons (Supplementary Table 2). All of the Scn8acond/+,Emx1-Cre mice experienced lethal seizures, beginning at 1 month of age, with median survival to 6 weeks (Fig. 5A). Many mice exhibited multiple seizure types that included facial spasms, mouth clicking, opisthotonus (head rearing), circular running with tonic extension of the tail, retropulsion with clonic movements of the hind limbs, rearing, falling, clonic limb seizures and tonic-clonic seizures. In some mice, episodes of clustered seizures led to periods of weakness requiring provision of soft food. EEG recording of a seizure in an Scn8acond/+,Emx1-Cre mouse is shown in Supplementary Fig. 4A.
RT-PCR was carried out on brain RNA from the Scn8acond/+,Emx1-Cre mice and expression of transcripts from the mutant (273 bp fragment) and wild-type (173 bp fragment) alleles was compared. The transcripts were comparable in RNA from forebrain, demonstrating efficient deletion of exon 26a (Fig. 5B). The proportion of mutant transcripts was much lower in brainstem, as predicted (Fig. 5B).
The data demonstrate that expression of mutant Scn8a in excitatory neurons of the forebrain is sufficient to generate seizures, with 100% penetrance.
Expression of Scn8aR1872W in inhibitory neurons does not result in seizures
Scn8acond / cond homozygotes were crossed with mice expressing a Gad2-Cre that is active in a subset of inhibitory neurons (Supplementary Table 2). No behavioural seizures or abnormal phenotypes were observed in Scn8acond/+, Gad2-Cre mice up to 6 months of age (Fig. 5B). We also carried out 8 days of 24-h video-EEG monitoring of three Scn8acond/+,Gad2.Cre mice, without any evidence of seizures or epileptiform discharges (see sample recordings in Supplementary Fig. 4B). In the heterozygous Scn8acond/+, Gad2-Cre offspring, the level of mutant transcripts in RNA from striatum (273 bp) was comparable to wild-type (173 bp) (Fig. 5B), with a much lower proportion in other brain regions (Fig. 5B).
To confirm the ineffectiveness of expression of the mutant transcript in inhibitory neurons, we carried out a second cross with the Dlx5a-Cre line (Supplementary Table 2). The Scn8acond/+,Dlx5a-Cre offspring also lack an overt phenotype and all of these mice survived to 5 months (current age) (Fig. 5A). We did not observe impaired gait or other abnormal phenotype in the Scn8acond/+,Dlx5a-Cre mice. We thus conclude that expression of the R1872W mutation in inhibitory neurons does not result in seizures or overt neurological disease.
Adult onset expression of Scn8aR1872W recapitulates seizures and lethality
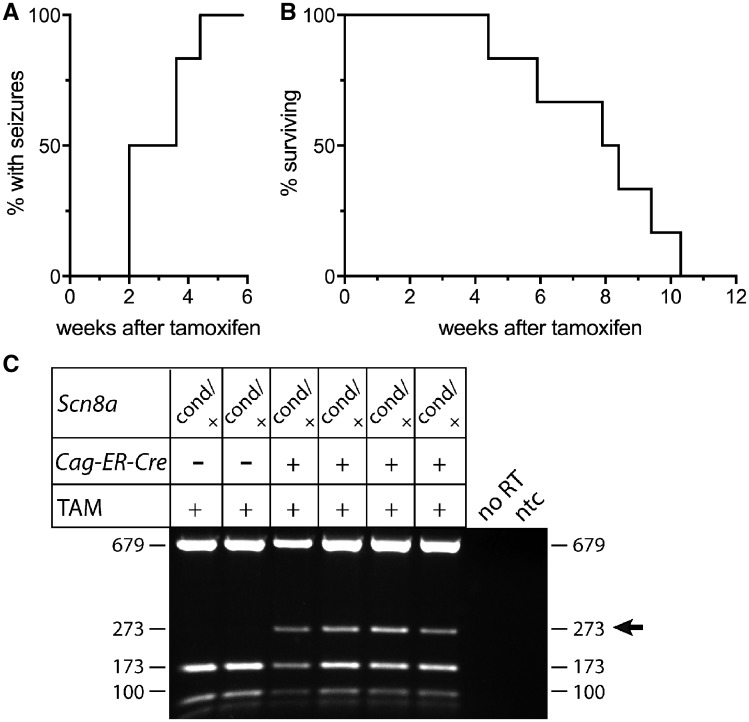
The mutant channel was activated by treatment of 8-week-old Scn8acond/+,CAG-Cre-ER mice with five daily intraperitoneal injections of tamoxifen. Mice were monitored visually for 8 h per day to detect behavioural seizures. The first seizures were observed 2 weeks after the final tamoxifen injection, with an average delay of 21 ± 7 days (n = 6) after treatment (Fig. 6A). Typical seizures included a running fit followed by tonic extension lasting ∼30 s, similar to Scn8acond/+,EIIa-Cre mice. Other phenotypes included freezing with staring, head bobbing, mouth twitching, and forelimb clonus resembling the focal seizures in Scn8acond/+,Emx1-Cre mice. Myoclonic jerks and periods of repetitive grooming were observed between seizures. Seizures were often provoked by handling or cage changing. The average survival after tamoxifen-treatment was 6 weeks (54 ± 15 days, n = 6) (Fig. 6B). Thus the mature brain, subsequent to completion of development, is not protected from the pathogenic consequences of expression of the Scn8aR1872W channel.
Figure 6.
Activation of Scn8aR1872W in adult mice leads to seizures and lethality. Scn8acond / +,CAG-Cre-ER mice (n = 6) were treated at 8 weeks of age with five daily injections of tamoxifen. Seizures were monitored visually for 8 h per day. (A) The first seizures were observed between 2 and 4 weeks after tamoxifen injection. (B) Death occurred between 1 and 3 months after seizure onset. No seizures or deaths were observed in Scn8acond/+ littermates lacking Cre (n = 3). (C) RT-PCR of brain RNA using primer pair 6 in exons 25 and 26. Brain transcripts encoding tryptophan 1872 (273 bp RT-PCR fragment) are present only in tamoxifen-treated Scn8acond/+,CAG-Cre-ER mice and not in littermate controls lacking CAG-Cre-ER.
Analysis of brain RNA demonstrated robust expression of the mutant transcript in Scn8acond/+,CAG-Cre-ER mice (Fig. 6C). Control Scn8acond/+littermates that did not inherit the CAG-Cre-ER transgene did not undergo activation of the mutant allele (Fig. 6C).
The sodium channel modulator GS967/Prax330 lengthens survival of Scn8aR1872W/+ mice
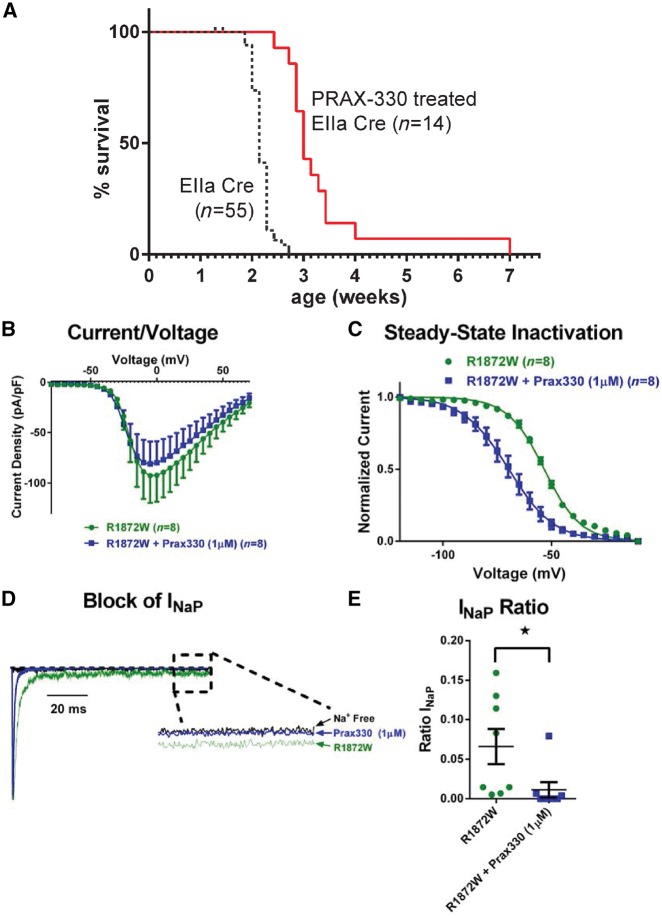
GS967/Prax330 is a polycyclic sodium channel modulator that reduces persistent current more than peak current (Anderson et al., 2014; Baker et al., 2018). Mice expressing the SCN8A patient mutation N1768D were protected from seizures by administration of GS967/Prax330 in the food (Baker et al., 2018). To evaluate the effectiveness of the drug against the more severe SCN8AR1872W allele, we provided mothers of Scn8acond/+,EIIa-Cre mice with food containing the drug beginning 1 day after delivery (P1 for the offspring). In Scn8acond/+,EIIa-Cre offspring whose mothers received the drug, seizure onset and lethality were postponed for an average of 6 days longer than those whose mothers received regular chow, a 25% increase in lifespan (Fig. 7A). Prior to seizure onset, the treated mice did not differ from untreated controls in behaviour or motor activity. This is comparable to the protective effect of the drug on homozygous Scn8aN1768D/N1768D mice, whose survival was increased from 2 weeks to 3 weeks by GS967/Prax330 (Baker et al., 2018).
Figure 7.
GS967/Prax330 extends the survival of Scn8acond/+,EIIa-Cre mice and reduces activity of the mutant channel in transfected cells. (A) GS967/Prax330 was added to the mouse chow at 8 mg/kg beginning on P1. Untreated mice received normal chow. Mean survival was increased from 15 days for untreated mice (n = 55) to 21 days for treated mice (n = 14). Kaplan-Meier log-rank (Mantel-Cox) test, χ2(1) = 32, P < 0.0001. (B–E) ND7/23 cells were transfected with Scn8aR1872W cDNA and incubated in the presence or absence of 1 µM GS967/Prax330. (B) Averaged current-voltage (I-V) relation for cells expressing mutant Nav1.6 ± GS967/Prax330. Peak currents were normalized to cell capacitance. (C) Voltage dependence of steady-state inactivation. Smooth lines correspond to the least squares fit when average data were fit to a single Boltzmann equation. (D) Representative normalized current traces recorded during a 100 ms depolarizing pulse from −120 mV to 0 mV illustrating the reduction of persistent sodium current for p.Arg1872Trp in the presence of GS967/Prax330. (E) Quantification of the ratio of peak to persistent sodium current. Data are means ± SEM statistical significance (*P < 0.05).
To examine the effect of the drug on the biophysical properties of the mutant channel, we transfected ND7/23 cells with the Scn8aR1872W cDNA and measured channel activity in the presence and absence of 1 µM GS967/Prax330. The presence of the drug did not reduce peak current (Fig. 7B). However, there was a 15 mV depolarizing shift in the voltage dependence of channel inactivation (Fig. 7C) and a reduction in persistent current from 6.6 ± 2.2% of peak current to 1.1 ± 1.0% (n = 8, P < 0.05, paired t-test) (Fig. 7D and E). Both of these effects counter the hyperactivity of the mutant channel. The data thus indicate that the effectiveness of Prax330 extends to more severe pathogenic SCN8A mutations, beyond the initially described amelioration of SCN8AN1768D.
Discussion
The conditional allele of Scn8a described here provides a new tool for molecular dissection of the disease process in SCN8A encephalopathy. Homozygous and heterozygous mice carrying the conditional allele (Scn8acond/cond and Scn8acond/+) are phenotypically normal in the absence of Cre recombinase and do not express the R1872W mutation, permitting maintenance of the line. In the absence of Cre, the conditional allele generates a transcript encoding the wild-type protein and terminating in the short 3′ UTR from the bGH gene in place of the endogenous 3′ UTR, which is 4–6 kb in length, depending on the polyadenylation site (Drews et al., 2005). The conserved microRNA binding sites in the endogenous 3′ UTR are evidently non-essential during normal development. Transport of mRNAs for local translation in dendrites and axons is thought to be mediated by non-coding sequences in the UTRs. The tolerance of the Scn8a mRNA to replacement of the 3′ UTR is consistent with the current view that voltage-gated sodium channels are translated in the cell body followed by transport of the protein to the axon initial segment, nodes of Ranvier, and other subcellular sites.
Neural expression of Scn8aR1872W is sufficient to cause seizures and sudden death
In the conditional mouse model, activation of the R1872W patient mutation by Cre recombinase permits spatial and temporal regulation of expression. Global activation of R1872W by EIIA-Cre results in seizures and early lethality between 2 and 3 weeks of age. Sudden death was frequently observed after a single seizure in these mice. Restricted activation of R1872W by Nestin-Cre also resulted in seizures and sudden death, demonstrating neural origin of the major phenotypes of SCN8A encephalopathy.
Major role of excitatory neurons
Activation of R1872W in forebrain excitatory neurons by Emx1-Cre is sufficient to cause seizures and sudden death in 100% of animals, demonstrating the important role of excitatory neurons in initiation of seizures in SCN8A encephalopathy. During disease progression, Scn8acond/+, Emx1-Cre mice experienced multiple seizure types, from focal to generalized seizures. In mice with globally activated Scn8aR1872W, electrophysiological measurements of cortical layer V and hippocampal CA1 neurons revealed increased neuronal excitability. This alteration is predicted to contribute directly to an increase in overall network excitability and seizure generation. It will be of interest in the future to evaluate the role of subcortical brain regions such as the thalamus and brainstem in the initiation and generalization of seizures and susceptibility to sudden death.
Inhibitory neurons
In striking contrast to the effect of expression in forebrain excitatory neurons, the activation of mutant channel in inhibitory neurons by Gad2-Cre or Dlx5a-Cre did not cause seizures or overt neurological impairment. These mice did not exhibit overt impaired movement, tremor or ataxia, which are common co-morbidities of SCN8A encephalopathy. It will be of interest to determine whether specific activation of Scn8aR1872W in Purkinje cells, cerebellar granule cells, or motor neurons would recapitulate these co-morbidities of SCN8A encephalopathy.
Adult expression of the gain-of-function mutation
To distinguish between the developmental consequences of expression of the gain-of-function R1872W mutation on neuronal circuits, and the cell-autonomous effects of neuronal hyperactivity, we initiated expression of the mutant channel in 8-week-old mice in which development of neuronal circuits is complete. The half-life of sodium channels appears to be shorter than 2 weeks in vivo (Makinson et al., 2014). We therefore anticipated that a substantial proportion of mutant channel would be incorporated into the axon initial segments within 2 weeks of the initiation of transcription using the tamoxifen-regulated CAG-Cre-ER transgene. In keeping with this prediction, seizures were observed within 2 weeks after the completion of tamoxifen treatment. This rapid response suggests that the cell-autonomous increase in neuronal excitability may be sufficient to explain the aetiology of seizures in mice expressing gain-of-function mutations of Scn8a. Because of the side effects of anti-epileptic drugs, withdrawal from treatment is often considered when a patient’s seizures are under control. The data on adult onset indicate that life-long treatment will be required to prevent seizure recurrence. This is supported by our earlier observation that termination of treatment of Scn8aN1768D/+ mice with the sodium channel modulator GS967 at 6 months of age results in recurrence of seizures (Baker et al., 2018).
Distinct mechanisms underlying Dravet syndrome and SCN8A encephalopathy
Previous work established fundamental differences between SCN8A encephalopathy and Dravet syndrome, a developmental and epileptic encephalopathy caused by loss-of-function mutations of the sodium channel gene SCN1A. SCN8A encephalopathy is clinically more severe and is caused by gain-of-function mutations that elevate channel activity, rather than loss-of-function (Meisler et al., 2016). As a consequence, sodium channel blockers are transiently effective in many patients with SCN8A encephalopathy, while this class of drugs is typically avoided for the sodium channel deficiency in Dravet syndrome. We have added another important distinction, the type of neuron directly responsible for the two disorders. In contrast to the observations reported here, inactivation of one allele of Scn1a in inhibitory neurons is sufficient to reproduce the major features of the disease in mice, while inactivation in excitatory neurons did not recapitulate the disorder (Yu et al., 2006; Ogiwara et al., 2007). Thus seizures result from two distinct mechanisms, reduced activity of inhibitory neurons (Dravet syndrome) versus elevated activity of excitatory neurons (SCN8A encephalopathy). This difference and its implications for treatment underscore the importance of early genetic testing to distinguish between these disorders (Oates et al., 2018).
SCN8AR1872W is more severe than SCN8AN1768D in human and mouse
Children with the de novo mutation R1872W in SCN8A are severely impaired, and do not develop the verbal and motor skills that were seen with the N1768D mutation (Veeramah et al., 2012). In the mouse, ubiquitous, global activation of Scn8aR1872W by EIIa-Cre causes seizure onset and death at 2–3 weeks of age, while heterozygous Scn8aN1768D/+ mice exhibit seizures after 8 weeks of age (Wagnon and Meisler, 2015). The penetrance of seizures is 100% for the R1872W heterozygotes but only 50% for N1768D heterozygotes. The seizures of R1872W mice are also more severe. A lethal, tonic seizure is often the first sign of disease in the R1872W mice, while N1768D mice often exhibit a brief tonic-clonic seizure followed by 30 s of immobility and rapid recovery.
The biophysical abnormalities of the R1872W channel are also more extreme than the N1768D channel. Expression of N1768D in primary neurons or transfected cells results in delayed channel inactivation and elevated persistent current (Veeramah et al., 2012; Lopez-Santiago et al., 2017). A greater delay in decay of macroscopic current is seen in transfected cells expressing R1872W, as well as elevation of peak current density, which may contribute to the more severe phenotype (Wagnon et al., 2016). The sodium channel modulator GS967/Prax330, which preferentially inhibits persistent sodium current, confers complete protection to heterozygous N1768D mice but only delays seizure onset in homozygous N1768D mice and heterozygous R1872W mice (Baker et al., 2018; this study). In addition to its direct effect on the mutant channel, the therapeutic effect of Prax330 may be mediated by reduction of persistent current generated by the wild-type allele in heterozygous patients.
The early onset, rapid progression, temporal regulation and fully penetrant lethal phenotype of the Scn8acond/+ mouse make it a useful test system for therapeutic intervention, and will accelerate the preclinical evaluation of new therapies for SCN8A encephalopathy. The ability to regulate expression of a deleterious mutation of Scn8a will facilitate future studies of pathogenic mechanisms underlying epilepsy.
Supplementary Material
Acknowledgements
We are grateful to Roman Giger and William Dauer for helpful discussions. We thank David Ginsburg for the EIIa-Cre mice. We acknowledge the important contributions of Thomas Saunders, Wanda Filipiak, and Anna LaForest in the Transgenic Model Animal Core at the University of Michigan.
Glossary
Abbreviation
- TALEN
TAL effector nuclease
Funding
R.K.B-S. was recipient of a Postdoctoral Fellowship from the Wishes for Elliott Foundation. This work was supported by National Institutes of Health R01 NS34509 (MHM) and NS103090 (MKP) and the Cute Syndrome Foundation.
Competing interests
The authors report no competing interests.
References
- Anderson LL, Thompson CH, Hawkins NA, Nath RD, Petersohn AA, Rajamani S et al. . Antiepileptic activity of preferential inhibitors of persistent sodium current. Epilepsia 2014; 55: 1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EM, Thompson CH, Hawkins NA, Wagnon JL, Wengert ER, Patel MK et al. . The novel sodium channel modulator GS-458967 (GS967) is an effective treatment in a mouse model of SCN8A encephalopathy. Epilepsia 2018; 59: 1166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker BS, Ottolini M, Wagnon JL, Hollander RM, Meisler MH, Patel MK. The SCN8A encephalopathy mutation p.Ile1327Val displays elevated sensitivity to the anticonvulsant phenytoin. Epilepsia 2016; 57: 1458–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, Van Emde Boas W et al. . Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 2010; 51: 676–85. [DOI] [PubMed] [Google Scholar]
- Blanchard MG, Willemsen MH, Walker JB, Dib-Hajj SD, Waxman SG, Jongmans MC et al. . De novo gain-of-function and loss-of-function mutations of SCN8A in patients with intellectual disabilities and epilepsy. J Med Genet 52; 2015: 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gasperi R, Rocher AB, Sosa MAG, Wearne SL, Perez GM, Friedrich VL et al. . The IRG mouse: a two-color fluorescent reporter for assessing Cre-mediated recombination and imaging complex cellular relationships in situ. Genesis 2008; 46: 308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CGF, Meisler MH, Brilstra EH, van Berkestijn FMC, van’t Slot R, van Lieshout S et al. . Characterization of a de novo SCN8A mutation in a patient with epileptic encephalopathy. Epilepsy Res 2014; 108: 1511–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews VL, Lieberman AP, Meisler MH. Multiple transcripts of sodium channel SCN8A (NaV1.6) with alternative 5′- and 3′-untranslated regions and initial characterization of the SCN8A promoter. Genomics 2005; 85: 245–57. [DOI] [PubMed] [Google Scholar]
- Estacion M, O’Brien JE, Conravey A, Hammer MF, Waxman SG, Dib-Hajj SD et al. . A novel de novo mutation of SCN8A (Nav1.6) with enhanced channel activation in a child with epileptic encephalopathy. Neurobiol Dis 2014; 69: 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasier CR, Wagnon JL, Bao YO, McVeigh LG, Lopez-Santiago LF, Meisler MH et al. . Cardiac arrhythmia in a mouse model of sodium channel SCN8A epileptic encephalopathy. Proc Natl Acad Sci USA 2016; 113: 12838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella E, Marini C, Trivisano M, Fitzgerald MP, Alber M, Howell KB et al. . The phenotype of SCN8A developmental and epileptic encephalopathy Neurology 2018; 91: e1112–124. [DOI] [PubMed] [Google Scholar]
- Goodwin EC, Rottman FM. The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J Biol Chem 1992; 267: 16330–334. [PubMed] [Google Scholar]
- Hammer MF, Wagnon JL, Mefford HC, Meisler MH. SCN8A-related epilepsy with encephalopathy. In: Adam MP, Ardinger HH, Pagon RA et al. editors. GeneReviews® [Internet]. Seattle, WA: University of Washington, Seattle; 2016, 1993–2018. Retrieved fromhttps://www-ncbi-nlm-nih-gov.proxy.lib.umich.edu/books/NBK379665/ [Google Scholar]
- Johannesen KM, Gardella E, Scheffer I, Howell K, Smith DM, Helbig I et al. . Early mortality in SCN8A-related epilepsies. Epilepsy Res 2018; 143: 79–81. [DOI] [PubMed] [Google Scholar]
- Jones JM, Meisler MH. Modeling human epilepsy by TALEN targeting of mouse sodium channel Scn8a. Genesis 2014; 52: 141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E et al. . Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 1996; 93: 5860–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Carvill GL, Gardella E, Kluger G, Schmiedel G, Barisic N et al. . The phenotypic spectrum of SCN8A encephalopathy. Neurology 2015; 84: 480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta delta CT method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- Lopez-Santiago LF, Yuan Y, Wagnon JL, Hull JM, Frasier CR, O’Malley HA et al. . Neuronal hyperexcitability in a mouse model of SCN8A epileptic encephalopathy. Proc Natl Acad Sci USA 2017; 114: 2383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinson CD, Tanaka BS, Lamar T, Goldin AL, Escayg A. Role of the hippocampus in Nav1.6 (Scn8a) mediated seizure resistance. Neurobiol Dis 2014; 68: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, Helman G, Hammer MF, Fureman BE, Gaillard WD, Goldin AL et al. . SCN8A encephalopathy: Research progress and prospects. Epilepsia 2016; 57: 1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HM, Goldin AL. Sodium channel carboxyl-terminal residue regulates fast inactivation. J Biol Chem 2010; 285: 9077–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates S, Tang S, Rosch R, Lear R, Hughes EF, Williams RE et al. . Incorporating epilepsy genetics into clinical practice: a 360° evaluation. NPJ Genom Med 2018; 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I et al. . Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci 2007; 27: 5903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Kato M, Takahashi S, Lerman-Sagie T, Lev D, Terashima H et al. . Early onset epileptic encephalopathy caused by de novo SCN8A mutations. Epilepsia 2014; 55: 994–1000. [DOI] [PubMed] [Google Scholar]
- Ottolini M, Barker BS, Gaykema RP, Meisler MH, Patel MK. Aberrant sodium channel currents and hyperexcitability of medial entorhinal cortex neurons in a mouse model of SCN8A encephalopathy. J Neurosci 2017; 37: 7643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Jones JM, Wisidagamage S, Meisler MH, Mashour GA. Reduced Nav1.6 sodium channel activity in mice increases in vivo sensitivity to volatile anesthetics. PLoS One 2015; 10: e0134960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RR, Barbosa C, Brustovetsky T, Brustovetsky N, Cummins TR. Aberrant epilepsy-associated mutant Nav1.6 sodium channel activity can be targeted with cannabidiol. Brain 2016; 139: 2164–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer NW, Galt J, Jones JM, Burgess DL, Sprunger LK, Kohrman DC et al. . Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics 1998; 54: 287–96. [DOI] [PubMed] [Google Scholar]
- Sprissler RS, Wagnon JL, Bunton-Stasyshyn RK, Meisler MH, Hammer MF. Altered gene expression profile in a mouse model of SCN8A encephalopathy. Exp Neurol 2017; 288: 134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Yamamoto S, Okayama A, Araki A, Saitsu H, Matsumoto N et al. . Electroclinical features of epileptic encephalopathy caused by SCN8A mutation. Pediatr Int 2015; 57: 758–62. [DOI] [PubMed] [Google Scholar]
- Veeramah KR, O’Brien JE, Meisler MH, Cheng X, Dib-Hajj SD, Waxman SG et al. . De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet 2012; 90: 502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Barker BS, Hounshell JA, Haaxma CA, Shealy A, Moss T et al. . Pathogenic mechanisms of recurrent mutations of sodium channel SCN8A in epileptic encephalopathy. Ann Clin Transl Neurol 2016; 3: 114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Barker BS, Ottolini M, Park Y, Volkheimer A, Valdez P et al. . Loss-of-function mutations of SCN8A in intellectual disability without seizures. Neurol Genet 2017; 3: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Korn MJ, Parent R, Tarpey TA, Jones JM, Hammer MF et al. . Convulsive seizures and SUDEP in a mouse model of SCN8A epileptic encephalopathy. Hum Mol Genet 2015; 24: 506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Meisler MH. Recurrent and non-recurrent mutations of SCN8A in epileptic encephalopathy. Front Neurol 2015; 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Mencacci NE, Barker BS, Wenger ER, Bhatia KP, Balint B et al. . Partial loss-of-function of sodium channel SCN8A in familial isolated myoclonus. Hum Mutat 2018; 39: 965–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 2006; 9: 1142–49. [DOI] [PubMed] [Google Scholar]
- Zanelli S, Goodkin HP, Kowalski S, Kapur J. Impact of transient acute hypoxia on the developing mouse EEG. Neurobiol Dis 2014; 68: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.