Summary
Pyramidal neuron dendrites integrate synaptic input from multiple partners. Different inputs converging on the same dendrite have distinct structural and functional features, but the molecular mechanisms organizing input-specific properties are poorly understood. We identify the orphan receptor GPR158 as a binding partner for the heparan sulfate proteoglycan (HSPG) glypican 4 (GPC4). GPC4 is enriched on hippocampal granule cell axons (mossy fibers), whereas postsynaptic GPR158 is restricted to the proximal segment of CA3 apical dendrites receiving mossy fiber input. GPR158-induced presynaptic differentiation in contacting axons requires cell-surface GPC4 and the co-receptor LAR. Loss of GPR158 increases mossy fiber synapse density, but disrupts bouton morphology, impairs ultrastructural organization of active zone and postsynaptic density, and reduces synaptic strength of this connection, while adjacent inputs on the same dendrite are unaffected. Our work identifies an input-specific HSPG-GPR158 interaction that selectively organizes synaptic architecture and function of developing mossy fiber-CA3 synapses in the hippocampus.
Introduction
A key organizational feature of neural circuit connectivity is the diversification of pyramidal neuron inputs. The structural and functional properties, as well as the dendritic location, of a synaptic input are important factors in determining whether a pyramidal neuron generates an action potential (Spruston, 2008). The large dendritic trees of pyramidal neurons in hippocampus or cortex extend through multiple laminae and receive input from different presynaptic partners in each layer. These inputs can vary considerably in structure and function (O’Rourke et al., 2012), and are often found in close proximity on the same dendrite, suggesting that selective cell-surface interactions between pre- and postsynaptic cells specify synaptic properties (de Wit and Ghosh, 2016). While the molecular mechanisms regulating connectivity are emerging (Kolodkin and Tessier-Lavigne, 2011; Sanes and Yamagata, 2009; Shen and Scheiffele, 2010), it remains poorly understood how neurons organize the specific properties of their different inputs.
The hippocampal CA3 circuit plays an important role in the encoding of memory (Rebola et al., 2017). The proximal portion of the CA3 pyramidal neuron apical dendrite in stratum lucidum (SL) receives input from dentate gyrus (DG) granule cell (GC) axons, the mossy fibers (MFs) (Figure S1A). MF-CA3 synapses are large, morphologically complex, and strongly facilitating (Rollenhagen et al., 2007; Salin et al., 1996; Wilke et al., 2013). In comparison, associational/commissural (A/C) inputs from other CA3 neurons on the medial portion of the CA3 dendrite in stratum radiatum (SR) are smaller, less complex, and only mildly facilitating. The distal dendrite receives input from the entorhinal cortex. This precise organization of connectivity suggests that distinct cell-surface interactions between presynaptic inputs and postsynaptic target cell control the specific properties of CA3 inputs.
Heparan sulfate proteoglycan (HSPG) cell-surface molecules are emerging as regulators of synaptic connectivity across species (Condomitti and de Wit, 2018; Kamimura and Maeda, 2017; Saied-Santiago and Bülow, 2018). Transmembrane and secreted HSPGs comprise a protein core with covalently attached heparan sulfate (HS) polysaccharide chains, which are enzymatically modified and interact with diverse protein ligands (Xu and Esko, 2014). Glypican 4 (GPC4), a glycosylphosphatidylinositol (GPI)-anchored HSPG (Watanabe et al., 1995), is an evolutionary conserved synaptic organizing protein. The Drosophila GPC4 ortholog Dally-like controls active zone (AZ) morphogenesis at the neuromuscular synapse by binding to the presynaptic receptor protein tyrosine phosphatase (RPTP) leukocyte common antigen-related (LAR) (Johnson et al., 2006). In vertebrates, astrocyte-derived soluble GPC4 binds presynaptic RPTPs to induce release of the secreted glycoprotein neuronal pentraxin 1, which clusters the GluA1 AMPA receptor subunit on retinal ganglion cells (Allen et al., 2012; Farhy-Tselnicker et al., 2017). Neuron-derived, presynaptic GPC4 forms a trans-synaptic complex with the postsynaptic adhesion molecule LRRTM4 in hippocampal GCs to regulate excitatory synapse formation (Siddiqui et al., 2013; de Wit et al., 2013). GPC4 requires presynaptic RPTPσ to mediate LRRTM4’s synaptogenic activity (Ko et al., 2015). Finally, mutations in GPC4 have been linked to autism spectrum disorders and intellectual disability (Doan et al., 2016). In the hippocampal CA3 circuit, GPC4 is highly enriched on MFs, but LRRTM4 is not detected in CA3 neurons (Siddiqui et al., 2013; de Wit et al., 2013), suggesting the presence of another, yet unknown, GPC4 binding partner to organize MF-CA3 synaptic connectivity.
Here, we identify the orphan receptor GPR158 as a binding partner for GPC4. Gpr158 expression is highly enriched in CA3 pyramidal neurons during synaptogenesis, and GPR158 localization is restricted to the proximal segment of CA3 dendrites that receives MF input. Postsynaptic GPR158 induces presynaptic differentiation in a cell-surface GPC4-dependent manner. GPC4 requires RPTPs to mediate GPR158’s synaptogenic activity, in a manner distinct from LRRTM4 or astrocyte-derived GPC4. Loss of GPR158 increases MF-CA3 synapse density, but MF boutons are smaller, and AZ and postsynaptic density (PSD) ultrastructure are disrupted. Synaptic transmission at MF-CA3 inputs is impaired in the absence of GPR158, while neighboring A/C inputs are unaffected. Together, these results demonstrate that an input-specific HSPG-GPR158 interaction organizes developing MF-CA3 synapses.
Results
The orphan receptor GPR158 is a GPC4 binding partner
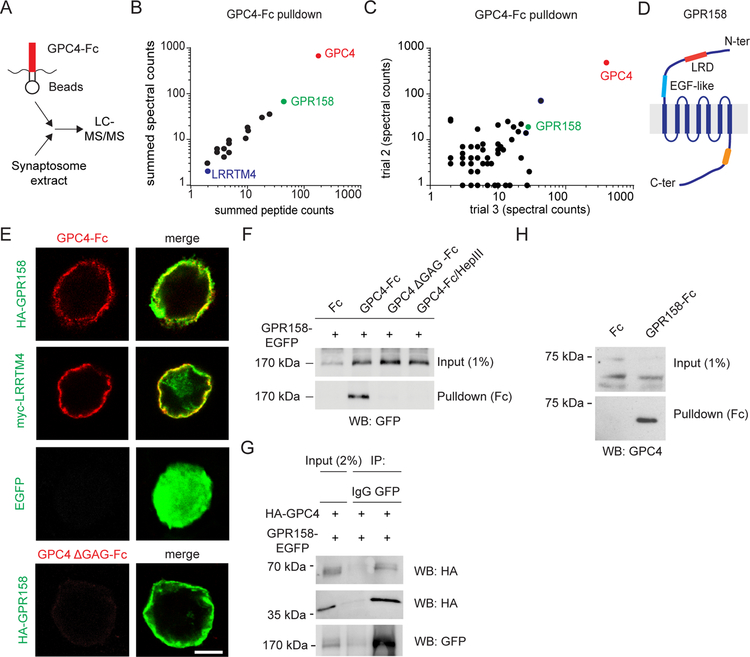
To identify novel GPC4 binding partners, we used recombinant GPC4-Fc protein as bait in pulldown experiments on detergent-solubilized synaptosomes. Whole rat brain was used to obtain sufficient prey material (Savas et al., 2014). Proteins bound to GPC4-Fc were analyzed by tandem mass spectrometry (Figure 1A). The major cell-surface protein identified in three independent experiments was GPR158 (Figure 1B, C and S1B), a G protein-coupled receptor (GPCR)-like orphan receptor with a large extracellular domain and homology to class C GPCRs (Bjarnadóttir et al., 2005; Orlandi et al., 2012) (Figure 1D). The GPC4 interactor LRRTM4 was also identified (Figure 1B). GPR158 and LRRTM4 were not detected in pulldown experiments with recombinant GPC4-Fc lacking HS glycosaminoglycan (GAG) chains (GPC4 ΔGAG-Fc) (Figure S1C), suggesting that GPR158 is an HS-dependent interactor.
Figure 1. GPR158 is a heparan sulfate-dependent GPC4 binding partner.
(A) Proteomic workflow for the identification of GPC4-interacting proteins.
(B) Identification of GPR158 as a GPC4 interactor by tandem mass spectrometry. GPC4-Fc protein was used as bait and synaptosome extract from P21 rat brains as prey. Graph shows summed peptide and spectral counts for all surface proteins after Fc background subtraction (n = 3 independent experiments).
(C) Freqency of detection of peptides (spectral count) for all proteins identified in two independent GPC4-Fc affinity purification experiments after Fc background subtraction.
(D) GPR158 domain organization. LRD, Leucine-Rich Domain; EGF-like, EGF-like Ca2+ binding motif. Yellow region, RGS7-binding site.
(E) Cell-surface binding assays in HEK293T cells. GPC4-Fc (red), but not GPC4 ΔGAG-Fc, binds the GPR158 ectodomain (green) expressed on the cell membrane. LRRTM4 and EGFP serve as positive and negative controls, respectively.
(F) Pulldown assay in HEK293T cells. GPC4-Fc, but not Fc alone, binds GPR158. HS removal by mutagenesis (GPC4 ΔGAG-Fc) or Heparinase III treatment (GPC4-Fc/HepIII) abolishes the interaction.
(G) GPC4 and GPR158 interact in trans. GPC4 co-immunoprecipitates with GPR158 following expression in separately transfected and co-cultured HEK293T cells. 65 kDa band represents full-length GPC4; 40 kDa band the N-terminal proteolytic fragment. IgG serves as negative control.
(H) Pulldown assay in hippocampal lysate. GPR158-Fc, but not Fc alone, binds endogenous GPC4.
Scale bar in (E) 10 µm.
See also Figure S1.
To validate the mass spectrometry results, we performed cell-surface binding assays using the pDisplay vector to express the hemagglutinin (HA)-tagged GPR158 ectodomain on the HEK293T cell membrane. GPC4-Fc bound to GPR158-expressing cells (Figure 1E). In control experiments, GPC4-Fc bound to LRRTM4-expressing cells, but not to cells expressing EGFP (Figure 1E). In addition, GPC4-Fc showed no detectable binding to cells expressing a panel of synaptic adhesion molecules (Figure S1D), indicating that the interaction with GPR158 is specific.
To further characterize the GPC4-GPR158 interaction, we performed pulldown assays in HEK293T cells expressing EGFP-tagged GPR158. GPC4-Fc, but not Fc alone, pulled down GPR158-EGFP from cell lysate (Figure 1F), confirming the interaction. Removal of HS, by mutagenesis (GPC4 ΔGAG-Fc) or by enzymatic treatment with Heparinase III (GPC4-Fc/HepIII), abolished the interaction between GPC4-Fc and GPR158-EGFP (Figure 1F). GPC4 ΔGAG-Fc showed no detectable binding to GPR158-expressing cells (Figure 1E), indicating that HS is a key determinant for the interaction with GPR158. To test whether GPC4 and GPR158 can interact in trans, we co-cultured HEK293T cells that were separately transfected with HA-GPC4 and GPR158-EGFP, and immunoprecipitated GPR158 from cell lysate using a GFP antibody. HA-GPC4 co-precipitated with GPR158, but not with control IgG, supporting a trans interaction (Figure 1G). Finally, we used recombinant GPR158-ecto-Fc protein as bait in pulldown assays in hippocampal lysate. GPR158-Fc, but not Fc protein alone, pulled down GPC4 (Figure 1H), confirming GPC4 as a GPR158 interactor. Together, these results identify GPR158 as a novel, HS-dependent GPC4 binding partner.
GPR158 is expressed in CA3 pyramidal neurons and enriched in stratum lucidum
To characterize GPR158 expression and localization in developing hippocampus, we used Gpr158 knockout (KO) mice, which contain a LacZ-neomycin reporter cassette in the Gpr158 locus replacing its initial coding sequence (Figure 2A). X-gal staining of hippocampal sections from postnatal day P7, P14 and P28 heterozygous Gpr158 mice showed strong Gpr158 expression in the CA3 region (Figure 2B). Gpr158 was not detected in the DG GC layer, but weaker expression appeared in CA1 at P28 (Figure 2B). To characterize Gpr158 cell type-specific expression patterns, we analyzed ß-galactosidase expression in GC, CA1 and CA3 P14 neurons using immunofluorescence. CA3 pyramidal neurons were the major Gpr158-expressing population (Figure 2C, D and S2A), in agreement with RNA sequencing analysis of hippocampal principal cell types (Cembrowski et al., 2016). These results indicate that Gpr158 expression is highly enriched in CA3 pyramidal neurons during MF-CA3 synapse development.
Figure 2. GPR158 is expressed in CA3 pyramidal neurons and enriched in stratum lucidum.
(A) Targeting strategy used to generate Gpr158 KO mouse. Boxes indicate coding exons.
(B) Gpr158 expression analysis using X-gal staining (blue) in Gpr158 heterozygous hippocampus.
(C) Representative insets of β-galactosidase immunofluorescence (green) in GC, CA3 and CA1 regions. Upper panels (greyscale) are inverted for visualization. Hoechst (blue), nuclear marker.
(D) Quantification of ß-gal expression in P14 principal hippocampal neurons (GC 0.065 ± 0.005 (N=3 mice, n=4051 cells) vs CA3 3.55 ± 0.06 (N=3, n=1864) vs CA1 0.98 ± 0.03 (N=3, n=1827)); ****p<0.0001, one-way ANOVA with multiple comparisons.
(E) Lamina-specific GPR158 immunoreactivity (green) in CA3 SL (arrowheads) in P28 WT hippocampus. No signal is detected in Gpr158 KO hippocampus. PSD95, postsynaptic marker (red); Bassoon, presynaptic marker (Bsn; blue). SP, stratum pyramidale; SL, stratum lucidum; SR, stratum radiatum.
(F) Higher magnification of GPR158 immunoreactivity in CA3 SL in WT and Gpr158 KO.
(G) Western blot analysis of rat brain fractionation probed for GPR158, PSD95 and presynaptic marker synaptophysin (SYP). GPR158-specific band is at 150 kDa, asterisk indicates aspecific band.
(H) HA-GPR158 (red) localization in EGFP-positive (green) P21 hippocampal CA3 pyramidal neuron in utero electroporated with EGFP--T2A--HA-GPR158. HA-GPR158 localizes to the proximal portion of the CA3 apical dendrite in SL.
(I) HA-GPR158 puncta localize to dendritic spine heads (arrowheads).
(J) Quantification of HA-GPR158 distribution in CA3 dendrites (SL 1.28 ± 0.17 (N=3 mice, n=13 dendrites/ 13 cells) vs SR 0.01 ± 0.0046 (N=3, n=8/8)); ****p<0.0001, Welch ‘s t-test.
Bar graphs show mean ± SEM.
Scale bar represents 300 µm in (B); 150 µm in (E); 50 µm in (F); 10 µm in (C), (H); and 5 µm in (I).
See also Figure S2.
We next analyzed GPR158 protein expression in mouse hippocampal membrane extracts by western blot, using a polyclonal antibody against the GPR158 N-terminal region. GPR158 was detectable by P14 and remained expressed into adulthood (Figure S2B). Immunohistochemistry of mouse hippocampal sections revealed layer-specific GPR158 labeling in CA3 SL (Figure 2E, F and S2C), which contains MF inputs. The lamina-specific distribution of GPR158 in CA3 SL was absent in Gpr158 KO hippocampus (Figure 2E, F), and was confirmed with a second antibody directed against the GPR158 intracellular region (Figure S2D, E), demonstrating specificity of GPR158 labeling. GPC4 immunoreactivity was strongly enriched on MFs in CA3 SL, confirming previous observations (de Wit et al., 2013) (Figure S2F).
To determine whether GPR158 localizes to synapses, we used subcellular fractionation. GPR158 was strongly enriched in the Triton-insoluble fraction containing the excitatory PSD protein PSD95 (Figure 2G), suggesting a postsynaptic localization. The antibodies used to detect endogenous GPR158 in hippocampus did not permit imaging of SL synapses at high-resolution, owing to the tissue conditions required for these antibodies to work. We therefore resorted to in utero electroporation of embryonic day E15.5 mouse embryos to express a GFP reporter and HA-tagged GPR158 (GFP--T2A--HA-GPR158) in CA3 pyramidal neurons, and imaged sparsely transduced cells using structured illumination microscopy (SIM) at P21. HA-GPR158 displayed a strikingly specific, punctate distribution that was restricted to the proximal portion of GFP-labeled CA3 apical dendrites (Figure 2H). Quantification of HA puncta density confirmed that HA-GPR158 immunofluorescence was restricted to SL and was absent in SR (Figure 2J). HA-GPR158 puncta localized to the heads of thorny excrescences (Figure 2I). Interestingly, not all spines displayed detectable HA immunofluorescence. Taken together, these results show that Gpr158 expression is enriched in CA3 pyramidal neurons during MF-CA3 synaptogenesis; that GPR158 displays a lamina-specific distribution in CA3 SL containing MF inputs; and that GPR158 is enriched in the excitatory PSD.
GPR158 induces presynaptic differentiation via cell-surface GPC4
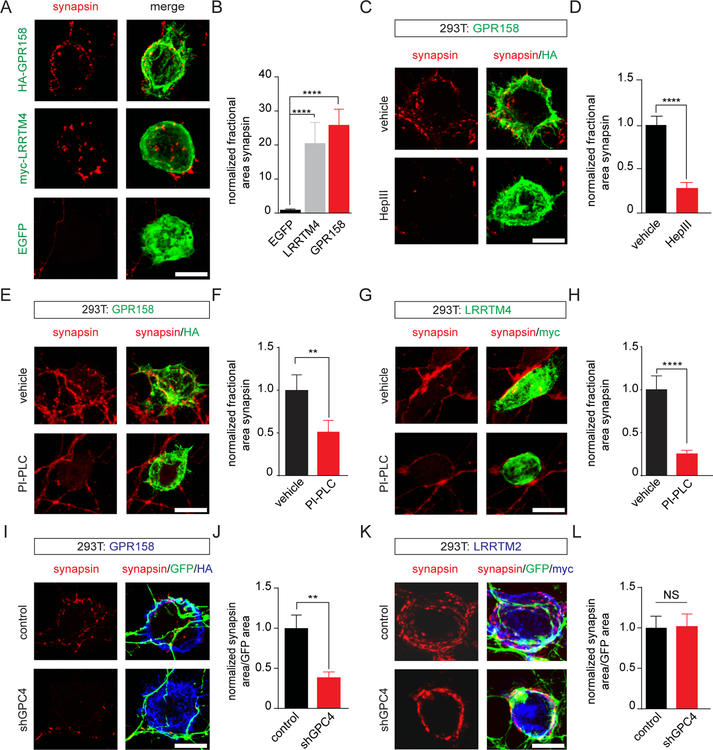
Our results indicate that GPR158 is a postsynaptic receptor interacting with GPC4, a presynaptic organizer. To test whether GPR158 displays synaptogenic activity similar to the postsynaptic adhesion molecule LRRTM4, which also interacts with GPC4 (Siddiqui et al., 2013; de Wit et al., 2013), we co-cultured HEK293T cells expressing the HA-tagged GPR158 ectodomain, myc-LRRTM4, or EGFP with hippocampal neurons and assessed clustering of the presynaptic marker synapsin in contacting axons. HA-GPR158 induced strong synapsin clustering on the HEK293T cell surface, similar to myc-LRRTM4, whereas GFP-expressing cells did not (Figure 3A, B). These results indicate that the extracellular domain of GPR158 induces presynaptic differentiation in contacting axons. GPR158’s synaptogenic activity did not require the EGF-like or leucine-rich domains (Figure S3A, B).
Figure 3. GPR158 induces presynaptic differentiation via cell-surface GPC4.
(A) The HA-GPR158 ectodomain (green) expressed on the surface of HEK293T cells co-cultured with 7DIV (days in vitro) hippocampal neurons induces clustering of the presynaptic marker synapsin (red) in axons. LRRTM4, positive control; EGFP, negative control.
(B) Quantification of (A). Fractional synapsin area: synapsin area per HA/myc-labeled surface area normalized to EGFP-expressing control cells; EGFP 1.00 ± 0.24 (n=17 cells) vs LRRTM4 20.6 ± 6.1 (n=21) and vs GPR158 25.9 ± 4.6 (n=34). ****p<0.0001, Kruskal-Wallis test, Dunn’s multiple comparisons post hoc test.
(C) Heparinase III (HepIII) treatment reduces GPR158-mediated synapsin clustering.
(D) Quantification of (C); vehicle 1.00 ± 0.16 (n=26 cells) vs HepIII 0.32 ± 0.06 (n=31); ****p<0.0001, Mann-Whitney test.
(E) PI-PLC treatment reduces GPR158-mediated synapsin clustering.
(F) Quantification of (E); vehicle 1.00 ± 0.17 (n=26 cells) vs PI-PLC 0.54 ± 0.12 (n=26); **p<0.001, Mann-Whitney test.
(G) PI-PLC treatment impairs LRRTM4-mediated presynaptic differentiation.
(H) Quantification of (G); vehicle 1.00 ± 0.1 (n=22 cells) vs PI-PLC 0.26 ± 0.04 (n=26); ****p<0.0001, Mann-Whitney test.
(I) Neuronal GPC4 knockdown (KD) decreases synapsin clustering in GFP-positive axons contacting GPR158-expressing cells (blue). The KD vector contains an EGFP reporter to visualize neuronal processes.
(J) Quantification of (I); control 1.00 ± 0.16 (n=23 cells) vs shGPC4 0.39 ± 0.07 (n=22); **p<0.001, Mann-Whitney test.
(K) Neuronal GPC4 KD does not affect LRRTM2-mediated presynaptic differentiation.
(L) Quantification of (K); control 1.00 ± 0.15 (n=21 cells) vs shGPC4 1.03 ± 0.15 (n=25); NS, p=0.93, Mann-Whitney test.
Bar graphs show mean ± SEM.
Scale bar represents 10 µm in (A), (C), (E), (G), (I) and (K).
See also Figure S3 and S4.
GPR158’s interaction with GPC4 is HS-dependent (Figure 1E, F). To determine whether GPR158- induced presynaptic differentiation requires HS, we treated neurons with HepIII to remove HS from the neuronal surface and co-cultured them with HEK293T cells expressing HA-GPR158, myc-LRRTM4, or myc-LRRTM2. The efficiency of HepIII treatment in removing HS was assessed by staining with the 3G10 HS stub antibody (data not shown). HS removal strongly reduced GPR158’s synaptogenic activity (Figure 3C, D). In control experiments, HepIII treatment also strongly impaired LRRTM4-induced synapsin clustering (Figure S3C, D), but did not affect LRRTM2’s synaptogenic activity (Figure S3E, F), which does not require HS (de Wit et al., 2013). These results show that GPR158 requires HS to induce presynaptic differentiation.
GPC4 is a GPI-anchored protein that can be released from the cell surface (Watanabe et al., 1995). To test whether GPC4 is specifically required on the neuronal membrane to mediate GPR158-induced presynaptic differentiation, we treated neurons with phosphatidylinositol-specific phospholipase C (PI-PLC) to remove GPI-anchored proteins, and co-cultured them with HEK293T cells expressing GPR158, LRRTM4, or LRRTM2. Surface labeling with an antibody against the GPI-anchored protein LSAMP confirmed efficient removal of LSAMP from the cell surface following PI-PLC treatment (Figure S3G). Enzymatic removal of GPI- anchored proteins strongly impaired GPR158’s ability to induce presynaptic differentiation (Figure 3E, F) and largely abolished LRRTM4-mediated presynaptic differentiation (Figure 3G, H). However, PI-PLC treatment did not affect the synaptogenic activity of LRRTM2 (Figure S3H, I), which acts via presynaptic neurexin, a transmembrane adhesion molecule (Ko et al., 2009; Siddiqui et al., 2010; de Wit et al., 2009). These results indicate that GPR158 requires GPC4 (and possibly other, unidentified GPI-anchored interactors) to be anchored on the neuronal membrane to mediate presynaptic differentiation.
To determine whether GPR158-induced presynaptic differentiation requires GPC4 in axons, we electroporated hippocampal neurons with a vector expressing an shRNA against GPC4 (de Wit et al., 2013) or a control plasmid, and co-cultured HEK293T cells expressing GPR158 or LRRTM2. GPC4 knockdown (KD) in neurons strongly decreased synapsin clustering on the surface of GPR158-expressing cells (Figure 3I, J), but did not affect LRRTM2-induced presynaptic differentiation (Figure 3K, L). Thus, axonal GPC4 is required for GPR158-mediated presynaptic differentiation.
Gpc4 is expressed in neurons, but also in astrocytes (Allen et al., 2012; Farhy-Tselnicker et al., 2017). To determine whether glial-derived GPC4 might contribute to presynaptic differentiation mediated by GPR158, we infected glial cultures with a lentiviral vector harboring an shRNA against GPC4 (De Wit 2013) or a control virus. We then performed HEK293T-neuron coculture assays in the presence of conditioned medium from control- or shGPC4-infected astrocytes. Conditioned medium from shGPC4-infected astrocytes did not affect the synaptogenic activity of GPR158, LRRTM4, or LRRTM2 (Figure S4A–F), although we observed a non-significant increase in GPR158-mediated presynaptic differentiation. These results indicate that glial-derived GPC4 is not required for GPR158-mediated presynaptic differentiation. In addition, we performed single molecule fluorescent in situ hybridization for Gpc4, the DG GC marker Prox1, and the astrocytic marker Aldh1l1 on P7 and P21 hippocampal sections to determine the source of GPC4 in the CA3 circuit (Figure S4G). At P7, Gpc4 was expressed in Prox1-positive GCs and in Aldh1l1-positive astrocytes in DG and CA3 (Figure S4G, H). Gpc4-positive astrocytes were prominently present in CA3 SL at this age. At P21, we observed robust expression of Gpc4 in GCs, but expression in astrocytes in DG and CA3 had strongly decreased (Figure S4G, H). These observations are in agreement with a previous study reporting a switch from glial to neuronal Gpc4 expression between P6 and P24 (Allen et al., 2012), and indicate that during the time that GPR158 protein levels are detectable (Figure S2B), GPC4 is predominantly present on MFs.
GPR158-mediated synaptic differentiation requires the presynaptic receptor LAR
LAR family RPTPs have previously been shown to bind glypicans and act as functional co-receptors on the presynaptic membrane (Coles et al., 2011; Johnson et al., 2006). The three RPTP family members LAR, RPTPδ and RPTPσ are all expressed in developing DG (Kwon et al., 2010). LAR, RPTPδ and RPTPσ were also detected in our GPC4-Fc pulldowns on detergent-solubilized synaptosomes (Figure S5A). To determine whether GPR158-mediated presynaptic differentiation requires RPTPs as co-receptors of GPC4, we infected neurons with lentiviral vectors harboring shRNAs against LAR, RPTPδ or RPTPσ (Yim et al., 2013) and cocultured HEK293T cells expressing GPR158, LRRTM4, or LRRTM2.
We first assessed the role of LAR, which was previously shown not to be required for LRRTM4- mediated presynaptic differentiation (Ko et al., 2015). In agreement with these results, we observed that LAR KD did not affect the synaptogenic activity of LRRTM4 (Figure 4A, B). As expected, LAR KD also did not affect LRRTM2-mediated presynaptic differentiation (Figure S5B, C). In contrast, the ability of GPR158 to cluster synapsin on the HEK293T surface was decreased following LAR KD (Figure 4C, D), indicating that LAR acts as a co-receptor for GPC4 on the presynaptic membrane in mediating GPR158-induced presynaptic differentiation.
Figure 4. GPR158-mediated synaptic differentiation requires the presynaptic receptor LAR.
(A) Neuronal LAR KD does not impair synapsin (red) clustering in axons contacting LRRTM4-expressing HEK293T cells (green).
(B) Quantification of (A); control 1.00 ± 0.15 (n=25 cells) vs shLAR 1.26 ± 0.13 (n=26); NS, p=0.11, Mann-Whitney test.
(C) Neuronal LAR KD reduces synapsin clustering in axons contacting co-cultured GPR158-expressing HEK293T cells.
(D) Quantification of (C); control 1.00 ± 0.17 (n=25 cells) vs shLAR 0.66 ± 0.17 (n=25); *p<0.01, Mann-Whitney test.
(E) Neuronal PTPσ KD, but not PTPδ KD, impairs LRRTM4-mediated presynaptic differentiation.
(F) Quantification of (E); control 1.00 ± 0.11 (n=29 cells) vs shPTPδ 0.99 ± 0.12 (n=29) vs shPTPσ 0.61 ± 0.12 (n=27); *p<0.01,one-way ANOVA with multiple comparisons.
(G) Neuronal PTPσ KD, but not PTPδ KD, increases GPR158-mediated presynaptic differentiation.
(H) Quantification of (G); control 1.00 ± 0.17 (n=26 cells) vs shPTPδ 1.27 ± 0.18 (n=26) vs shPTPσ 2.02 ± 0.29 (n=21); *p<0.01,one-way ANOVA with multiple comparisons.
Bar graphs show mean ± SEM.
Scale bar represents 10 µm in (A), (C), (E) and (G).
See also Figure S5.
We next tested the role of RPTPδ and RPTPσ. RPTPσ is a co-receptor for LRRTM4-mediated presynaptic differentiation via GPC4 (Ko et al., 2015). RPTPδ is specifically required for the synaptogenic effect of glial-derived GPC4 (Farhy-Tselnicker et al., 2017). Consistent with previous results (Ko et al., 2015), LRRTM4-mediated presynaptic differentiation was impaired by RPTPσ KD, but unaffected by RPTPδ KD (Figure 4E, F). Unexpectedly, we observed a robust increase in GPR158-mediated presynaptic differentiation following RPTPσ KD, whereas RPTPδ KD had no effect (Figure 4G, H). Neither RPTP KD affected LRRTM2’s synaptogenic activity, which required neurexin (Figure S5D, E). Thus, downregulation of RPTP family members has opposing effects on GPR158-mediated presynaptic differentiation: LAR KD decreases, whereas RPTPσ KD increases, GPR158’s synaptogenic activity. These results indicate that the mechanism of GPR158-mediated presynaptic differentiation via GPC4 is distinct from LRRTM4-mediated presynaptic differentiation, and from the synaptogenic effect of glial-derived GPC4.
GPR158 regulates MF-CA3 synapse density and size
GPR158’s ability to induce presynaptic differentiation, expression in CA3 pyramidal neurons during MF-CA3 synaptogenesis, and restricted localization to the SL portion of apical CA3 dendrites suggest a role as an organizer of MF-CA3 synapses. We therefore next examined MF-CA3 synapse development in Gpr158 KO mice. Gross MF pathway organization and hippocampal architecture was normal in KO mice compared to wild-type (WT) littermates (Figure S6A). GPC4 enrichement in CA3 SL and presynaptic localization was preserved in the absence of GPR158 (Figure S6B, C).
To visualize CA3 dendrites and spines, we filled CA3 pyramidal neurons in P21 acute hippocampal slices of WT and Gpr158 KO littermates with Alexa568 fluorescent dye using a patch pipette. The SL portion of Gpr158 KO CA3 apical dendrites displayed a 37% increase in spine density compared to WT dendrites (Figure 5A, B). Spine density on CA3 dendrites in SR from the same neurons was unaffected by loss of GPR158 (Figure 5C, D), indicating input-specificity of the effect.
Figure 5. GPR158 regulates MF-CA3 synapse density and size.
(A) Representative images of Alexa568-filled CA3 apical dendrites in stratum lucidum (SL) of P21 WT and Gpr158 KO littermates.
(B) Quantification of spine density in CA3 SL; WT 1.38 ± 0.11 spine/µm (N=3 mice, n=22 dendrites/18 cells) vs Gpr158 KO 1.89 ± 0.08 spine/µm (N=3, n=20/15); ***p<0.0001, Welch’s t test.
(C) Representative images of CA3 dendrites in stratum radiatum (SR) of P21 WT and Gpr158 KO littermates.
(D) Quantification of spine density in CA3 SR; WT 1.67 ± 0.07 spine/µm (N=3 mice, n=19 dendrites/18 cells) vs Gpr158 KO 1.55 ± 0.07 spine/µm (N=3, n=17/15); NS, p=0.25; Welch’s t test.
(E) Representative images of GFP-electroporated CA3 apical dendrites in SL of P21 WT and Gpr158 KO mice.
(F) Quantification of spine density in CA3 SL; WT 1.14 ± 0.09 spine/µm (N=3 mice, n=19 dendrites/17 cells) vs Gpr158 KO 1.94 ± 0.17 spine/µm (N=5, n=19/15); ****p<0.0001, Mann-Whitney test.
(G) Representative images of CA3 dendrites in stratum radiatum (SR) of P21 WT and Gpr158 KO mice.
(H) Quantification of spine density in CA3 SR; WT 1.51 ± 0.18 spine/µm (N=3 mice, n=14 dendrites/12 cells) vs Gpr158 KO 1.46 ± 0.08 spine/µm (N=5, n=14/13); NS, p=0.734; Mann-Whitney test.
(I) Electron micrographs of P14 WT and Gpr158 KO MF-CA3 synapses. Orange, presynaptic element. Lower panel, masked boutons (white).
(J, K) Quantification of MF-CA3 synapse density (WT 0.018 ± 0.001 (N=3 mice, n=50 synapses) vs Gpr158 KO 0.037 ± 0.002 (N=3, n=41)) [J], and presynaptic area (WT 6.84 ± 0.40 µm2 (N=3, n=87) vs Gpr158 KO 3.074 ± 0.17 µm2 (N=3, n=138)) [K]. ****p<0.00001; Mann-Whitney test.
(L) SBF-SEM 3D reconstructions of P14 WT and Gpr158 KO MF boutons.
(M, N) Quantification of MF bouton surface area (WT 62.9 ± 8.20 µm2 (N=2 mice, n=13 boutons) vs Gpr158 KO 34.8 ± 4.79 µm2 (N=2, n=12)) [M], and volume (WT 13.6 ± 1.68 µm3 (N=2 mice, n=13 boutons) vs Gpr158 KO 6.27 ± 0.94 µm2 (N=2, n=12)) [N]. **p<0.001; Mann-Whitney test.
Bar graphs show mean ± SEM.
Scale bar represents 10 µm in (A) (C), (E) and (G), 2 µm in (I) and (L).
See also Figure S6.
Dye filling in acute slices has the disadvantage that spine density and morphology could be affected by the whole-cell recording configuration and slicing procedure. As a complementary approach, we therefore used in utero electroporation of E15.5 embryos with a GFP expression plasmid and imaged transduced neurons using SIM in P21 WT and Gpr158 KO. The proximal portion of GFP-positive CA3 apical dendrites displayed a marked overgrowth of dendritic spines in Gpr158 KO mice compared to WT (Figure 5E, S6D). Spine morphology appeared more immature in Gpr158 KO mice, with filopodia protruding from spines (Figure 5E, S6D). Quantification revealed a 70% increase in the density of thorny excrescences in CA3 SL in Gpr158 KO mice (Figure 5F). This effect was again input-specific, as spine density on the same CA3 dendrites in SR was unaffected by loss of GPR158 (Figure 5G, H). Thus, loss of GPR158 results in an input-specific increase in spine density in CA3 apical dendrites.
To determine whether GPR158 loss results in a corresponding increase in presynaptic density, we used transmission electron microscopy (TEM) to analyze MF boutons in CA3 SL. MF boutons were identified by the presence of a large and elaborate presynaptic compartment with high synaptic vesicle content. Quantification of the number of MF presynaptic terminals per area revealed a strong increase in bouton density in Gpr158 KO mice compared to WT littermates (Figure 5I, J). Gpr158 KO MF terminals appeared noticeably smaller, and quantification confirmed a strong decrease in bouton area following loss of GPR158 (Figure 5K).
MF boutons are morphologically complex, potentially complicating area measurements in thin sections. We therefore performed 3D reconstructions of individual boutons in CA3 SL of P14 WT and Gpr158 KO littermates using serial block-face scanning electron microscopy (SBF-SEM) (Figure 5L and S6E). Analysis of reconstructed MF boutons revealed a ~50% decrease in bouton surface and volume in Gpr158 KO mice (Figure 5M, N). Taken together, these results show that loss of GPR158 increases MF-CA3 pre- and postsynaptic density, but reduces bouton size.
GPR158 organizes active zone and postsynaptic density ultrastructure
We next examined the ultrastructure of WT and Gpr158 KO MF-CA3 synapses, using TEM (Figure 6A). We observed an approximately two-fold increase in the number of PSDs per MF bouton and a strong reduction in PSD area in Gpr158 KO synapses compared to WT (Figure 6B, C). Thus, loss of GPR158 results in an increase in the number and a decrease in the area of PSDs at MF-CA3 synapses. The increase in PSD number per bouton is consistent with the increase in spine density in Gpr158 KO CA3 dendrites in SL.
Figure 6. GPR158 organizes MF-CA3 synaptic architecture.
(A) Electron micrographs of P14 WT and Gpr158 KO MF-CA3 synapses. Orange, presynaptic element. Lower panel, masked bouton outline (red) and PSDs (white).
(B, C) Quantification of the number of PSDs per MF-CA3 presynaptic profile (WT 11.7 ± 0.76 (N=3 mice, n=50 synapses) vs KO 25.2 ± 1.54 (N=3, n=41)) [B], and PSD area (WT 0.034 ± 0.0008 µm2 (N=3 mice, n=581 PSDs) vs KO 0.015 ± 0.0003 µm2 (N=3, n=1033)) [C]. ****p<0.0001, Mann-Whitney test.
(D) Electron micrographs of non-MF synapses in P14 WT and Gpr158 KO CA3 SR. Orange, presynaptic; green, postsynaptic element.
(E) Quantification of the number of PSDs per non-MF synapse in CA3 SR (WT 1.23 ± 0.05 (N=3 mice, n=47 synapses) vs KO 1.16 ± 0.06 (N=3, n=39)). NS, p=0.10, Mann-Whitney test.
(F) Electron micrographs of PTA-stained synapses in P14 WT and Gpr158 KO CA3 SL. Enlarged images show boxed regions.
(G, H) Quantification of PSD length (WT 0.29 ± 0.01 µm (N=3 mice, n=121 PSDs) vs KO 0.22 ± 0.005 µm (N=3, n=221)) [G], and AZ length (WT 0.27 ± 0.01 µm (N=3 mice, n=121 AZs) vs KO 0.20 ± 0.005 µm (N=3, n=221)) [H]. ****p<0.0001, Mann-Whitney test.
(I, J) Quantification of DP number per synapse (WT 3.31 ± 0.11 (N=3 mice, n=54 DPs) vs KO 2.1 ± 0.11 (N=3, n=49)) [I], and DP area (WT 1961 ± 46.5 nm2 (N=3 mice, n=212 DPs) vs KO 1588 ± 47.9 nm2 (N=3, n=115)) [J]. ****p<0.0001, Mann-Whitney test.
Bar graphs show mean ± SEM.
Scale bar in (A) 1 µm; (D) 0.2 µm; (F) 0.1 µm.
See also Figure S6.
We then asked whether loss of GPR158 specifically impairs MF-CA3 synapse structure, or whether adjacent A/C inputs on CA3 dendrites are also affected. We analyzed asymmetric synapses at the CA3 SL/SR border, where A/C-CA3 synapses are located (Figure 6D). Because the identity of these asymmetric synapses cannot be determined with certainty, we designated them as ‘non-MF’ synapses. In contrast to MF-CA3 synapses in CA3 SL (Figure 6B), the number of PSDs per synapse was not changed in Gpr158 KO non- MF synapses (Figure 6E). These results indicate that GPR158 exerts input-specific effects on synaptic ultrastructure in CA3 neurons.
To gain a more detailed insight in the organization of MF-CA3 synaptic structure in the absence of GPR158, we performed EM analysis on CA3 SL synapses in P14 WT and Gpr158 KO hippocampal sections subjected to negative staining with phosphotungstic acid (PTA), which labels the macromolecular complexes of the PSD and AZ (Bloom and Aghajanian, 1966) (Figure 6F). We found that PSD length in Gpr158 KO synapses was decreased by 27% (Figure 6G). PSD thickness was also decreased (Figure S6F). AZ length was reduced by 26% in Gpr158 KO synapses (Figure 6H). This reduction is similar to the decrease in PSD length, indicating that AZ and PSD alignment is not affected in Gpr158 KO MF-CA3 synapses. Synaptic cleft width was unaffected (Figure S6G).
PTA staining reveals specific AZ structures called dense projections (DPs), macromolecular complexes opposing the PSD that become apparent after chemical fixation. DPs are thought to link synaptic vesicles with the release machinery (Südhof, 2012). The number of DPs juxtaposed to labeled PSDs, as a readout of the number of DPs per synapse, was decreased by 36% (Figure 6I) and DP area was reduced by 20% (Figure 6J) in Gpr158 KO synapses compared to WT. DP perimeter, width, and height were also decreased in Gpr158 KO synapses (Figure S6H–J). Together, these results indicate that GPR158 acts in an input-specific manner to organize MF-CA3 synaptic ultrastructure.
GPR158 regulates MF-CA3 synaptic transmission
To examine whether GPR158 contributes to MF-CA3 synaptic function, we performed whole-cell voltage-clamp recordings in CA3 pyramidal neurons in acute hippocampal slices from P21–23 WT and Gpr158 KO littermates. Frequency and amplitude of spontaneous excitatory postsynaptic currents (sEPSCs) were reduced in Gpr158 KO compared to WT CA3 neurons (Figure 7A–E), whereas sEPSC decay time was not affected (Figure S7A). A comparison of sEPSC amplitudes revealed a preferential decrease in larger (>40 pA) amplitude events in Gpr158 KO CA3 neurons (Figure S7B, C), suggesting an impairment of MF-CA3 inputs, which give rise to larger amplitude events (Henze et al., 1997). To test this, we recorded sEPSCs while blocking MF inputs using the group II mGluR agonist DCG-IV, which strongly suppresses evoked MF-CA3 synaptic responses (Figure S7D). In the presence of DCG-IV, sEPSC frequency, amplitude, and decay time were similar between WT and Gpr158 KO CA3 neurons (Figure S7E–J), suggesting that MF-CA3 synapses are selectively impaired in the absence of GPR158.
Figure 7. GPR158 regulates MF-CA3 synaptic transmission.
(A) Representative sEPSC traces from CA3 pyramidal neurons in P21 WT (grey) and Gpr158 KO (red) acute hippocampal slices.
(B) Cumulative distribution of sEPSC inter-event intervals in WT and Gpr158 KO CA3 neurons.
(C) Quantification of sEPSC frequency (WT 4.61±0.75 Hz (N=3 mice, n=18 cells) vs KO 3.10±0.62 Hz (N=3, n=19)). *p<0.05, Mann-Whitney test.
(D) Cumulative distribution of sEPSC amplitudes in WT and Gpr158 KO CA3 neurons.
(E) Quantification of sEPSC amplitude (WT 56.7±3.7 pA (N=3 mice, n=18 cells) vs KO 44.3±3.1 pA (N=3, n=19)). **p<0.01, Mann Whitney test.
(F) Representative traces and quantification of AMPAR-mediated EPSC amplitude at MF-CA3 synapses (WT 303.4±45.8 pA (N=3 mice, n=18 cells) vs KO 154.6±15.9 pA (N=3, n=19)). *p<0.05, Mann Whitney test.
(G) Representative traces and quantification of NMDAR-mediated EPSC amplitude at MF-CA3 synapses (WT 78.5±11.8 pA (N=3 mice, n=9 cells) vs KO 47.8±5.3 pA (N=3, n=9)). *p<0.05, Mann Whitney test.
(H) AMPAR/NMDAR ratio (WT 4.11±0.49 (N=3 mice, n=9 cells) vs KO 3.41±0.43 (N=3, n=9)). NS, p=0.302. Two-tailed Student’s t-test.
(I) Representative traces and quantification of paired pulse ratio of MF-CA3 synapses (N=3 mice, n=18 cells). ***p<0.001, **p<0.01, *p<0.05, Mann Whitney test.
(J) Representative traces and quantification of paired pulse ratio of A/C-CA3 synapses (N=3, n=12 cells). NS, p>0.05. Two tailed Student’s t-test.
Bar graphs show mean ± SEM.
See also Figure S7.
To determine whether the altered structure of Gpr158 KO MF-CA3 synapses is reflected in a change in synaptic strength, we stimulated MFs with an electrode placed in the dentate hilar region, while performing whole-cell voltage-clamp recordings from CA3 neurons. The amplitude of AMPAR-mediated EPSCs was strongly reduced in Gpr158 KO CA3 neurons compared to WT (Figure 7F). NMDAR-mediated EPSCs, measured 50 ms after the stimulus at a +40 mV holding potential, were proportionally reduced in Gpr158 KO MF-CA3 synapses (Figure 7G, H). These results show that loss of GPR158 strongly impairs MF-CA3 synaptic transmission.
To assess presynaptic function of Gpr158 KO MF-CA3 synapses, we analyzed paired pulse facilitation (PPF), a form of presynaptic short-term plasticity dependent on residual Ca2+ in the terminal that increases the probability of release. MF-CA3 synapses are highly facilitating, and accordingly, the amplitude of the second EPSC was strongly increased in WT MF-CA3 synapses upon a paired stimulus (Figure 7I). In Gpr158 KO MF-CA3 synapses however, the increase in paired pulse ratio (PPR) was strongly reduced at shorter interstimulus intervals compared to WT synapses (Figure 7I), indicating impaired presynaptic short-term plasticity of MF-CA3 synapses in the absence of GPR158.
Finally, we tested whether the impaired facilitation in Gpr158 KO mice was specific to MF-CA3 synapses. We stimulated A/C inputs in CA3 SR in the presence of DCG-IV and performed whole-cell voltage-clamp recordings from CA3 neurons to analyze the PPR of A/C-CA3 synapses. The A/C-CA3 synapse PPR was not affected over a range of interstimulus intervals in the Gpr158 KO (Figure 7J), indicating that short-term plasticity of A/C-CA3 synapses is not impaired in these animals. In conclusion, these results show that GPR158 is selectively required for the synaptic function of MF, but not A/C, inputs onto CA3 pyramidal neurons.
Discussion
The molecular mechanisms by which pyramidal neurons organize the specific properties of their synaptic inputs are poorly understood. We identify an input-specific GPR158-HSPG interaction that selectively organizes synaptic architecture and function of MF inputs onto CA3 pyramidal neurons (Figure S7K).
A synapse-organizing GPR158-HSPG complex
We identify GPC4 as a binding partner for the orphan receptor GPR158. To our knowledge, no synaptic interactions of HSPGs with GPCRs have previously been reported. The interaction with GPC4 is required for GPR158-mediated presynaptic differentiation in contacting axons. GPR158 thus bears resemblance to adhesion-GPCRs, such as BAI3 and LPHNs (Anderson et al., 2017; Kakegawa et al., 2015; O’Sullivan et al., 2012; Sigoillot et al., 2015), but lacks defining features such as a GAIN domain (Langenhan et al., 2013). As the GPC4-GPR158 interaction is HS-dependent, additional HS carriers at the MF-CA3 synapse, such as syndecan-2 (SDC2) and SDC3 (Hsueh and Sheng, 1999), may also interact with GPR158. We were unable to demonstrate a SDC3-GPR158 interaction using pulldown assays in HEK293T cells (data not shown), which may reflect a lower affinity compared to the GPC4-GPR158 interaction. However, multiple classes of HSPGs were identified in affinity purification experiments with the GPR158 ectodomain (Orlandi et al., submitted), and enzymatic removal of GPI-anchored proteins including glypicans did not completely abolish GPR158’s synaptogenic activity (Figure 3F), suggesting that GPR158 may also interact with transmembrane HSPGs to induce presynaptic differentiation. The GPR158 ectodomain also contains protein-protein interaction motifs. Although these were not required for GPR158’s synaptogenic effect, the presence of these domains suggests that additional, non-HSPG, binding partners may exist. Thus, GPR158 likely integrates multiple extracellular signals at the synaptic cleft.
Our results show that GPC4 is required on the neuronal membrane to mediate GPR158-induced presynaptic differentiation. Glial-derived soluble GPC4 is not required for GPR158-mediated presynaptic differentiation. In the hippocampus, Gpc4 was detected in both GCs and astrocytes at P7, but was predominantly expressed in GCs at P21, consistent with previous observations indicating a shift to neuronal expression of Gpc4 occuring between P6 and P24 (Allen et al., 2012). MF boutons undergo a dramatic increase in complexity in the second postnatal week, as well as a major increase in density and volume of thorny excrescences (Wilke et al., 2013). GPR158 protein levels strongly increase during the same period. Thus, neurons appear to be the predominant source of GPC4 during rapid MF-CA3 synaptic differentiation in the second postnatal week. However, the prominent expression of Gpc4 in astrocytes in CA3 SL at P7 suggests a contribution from glial-derived GPC4 at early stages of MF-CA3 synapse development.
Although GPR158 and LRRTM4 both induce presynaptic differentiation via GPC4 in hippocampal neurons, the presynaptic co-receptors required for this effect are surprisingly distinct. GPR158’s synaptogenic effect depends in part on presynaptic LAR, which is not required for LRRTM4-induced presynaptic differentiation (Ko et al., 2015). LRRTM4’s synaptogenic activity requires RPTPσ (Ko et al., 2015) (this study), which unexpectedly inhibits GPR158’s synaptogenic activity. Thus, GPR158 and LRRTM4 both act via presynaptic HSPGs, but depend on different co-receptors. Glial-derived soluble GPC4 induces synaptic differentiation in retinal ganglion cells via both RPTPδ and RPTPσ (Farhy-Tselnicker et al., 2017), but RPTPδ KD did not affect GPR158- or LRRTM4- mediated presynaptic differentiation, further underscoring mechanistic differences between these proteins.
The underlying molecular mechanism for the difference in co-receptor requirement for GPR158- and LRRTM4-mediated presynaptic differentiation is not yet clear. LAR and RPTPσ have opposing roles in mediating GPR158-induced presynaptic differentiation. Possibly, GPR158 interacts with additional binding partners on the presynaptic membrane, which in turn could influence the activity of RPTPs. Cell type-specific modifications of HS might also play a role. GPC4 expressed in neurons or astrocytes likely differs in HS composition, depending on the complement of HS-modifying enzymes expressed in these cells (Condomitti and de Wit, 2018), which might influence interactions with co-receptors. Regardless of the exact mechanism involved, the opposing effects of LAR KD and RPTPσ KD on GPR158-mediated presynaptic differentiation suggest that GPR158 may promote or limit synaptic differentiation depending on the type of RPTP present. Whether RPTP protein levels at MF-C3 synapses vary during development is not known, but Rptp mRNA levels in DG change during postnatal development (Kwon et al., 2010). An interesting possibility would be that our observations of increased MF-CA3 synapse density but decreased differentiation of AZ and PSD in the absence of GPR158 relate to dynamic RPTP developmental expression profiles.
Cell-surface expression of the GPR158 ectodomain was sufficient to induce presynaptic differentiation in contacting axons. An outstanding question is whether GPC4 binding to GPR158 activates downstream signaling. GPR158 acts as a membrane anchor recruiting the intracellular G protein regulator RGS7 (Orlandi et al., 2012), suggesting that an interaction with presynaptic GPC4 might nucleate an assembly of a larger signaling complex around GPR158. However, whether GPR158 has a G protein-mediated signaling role, and whether GPC4 binding regulates RGS7-mediated GPR158 signaling or activates other downstream signaling pathways, is currently unknown. The cytoplasmic domain of GPR158 has been reported to translocate to the nucleus (Patel et al., 2013), suggesting possible additional signaling mechanisms.
Input-specific control of synaptic density and architecture by GPR158
GPR158 acts in an input-specific manner to control density and architecture of MF-CA3 synapses. Loss of GPR158 caused a strong increase in MF-CA3 bouton and spine density. An increase in spine density has also been observed in Gpr158 KO prefrontal cortex layer 2/3 neurons (Sutton et al., 2018). In CA3 dendrites, the increase in spine density following loss of GPR158 is input-specific, as the neighboring dendritic segment receiving A/C-CA3 inputs was unaffected. Consistent with its input-specific effects in CA3 neurons, GPR158 is highly restricted to dendritic spines in SL. The underlying mechanism for this input-specific distribution of GPR158 is not known. Trans-synaptic interactions (Nishimura-Akiyoshi et al., 2007) or intracellular scaffolding mechanisms (Ango et al., 2004) might play a role.
In line with the increase in synapse density, Gpr158 KO MF-CA3 synapses contained an increased number of PSDs per bouton. However, AZs and PSDs were shorter in KO synapses. Mutants for the Drosophila GPC4 homolog Dally-like show decreased AZ size and increased AZ numbers per bouton at the neuromuscular junction (Johnson et al., 2006). These findings, which are strikingly reminiscent of our observations at the Gpr158 KO MF-CA3 synapse, support the notion that GPR158 regulates presynaptic differentiation via GPC4. The GPC4-associated co-receptors LAR and PTPσ (Johnson et al., 2006; Ko et al., 2015) recruit the presynaptic scaffolding protein liprin-α. Knockdown of liprin-α2, the predominant liprin family member at MF-CA3 boutons (Spangler et al., 2011; Zürner et al., 2011), in cultured hippocampal neurons reduces AZ length (Spangler et al., 2013), similar to our findings at Gpr158 KO synapses. In addition to changes in AZ and PSD length, we also observed a decrease in number and area of DPs, pyramid-shaped macromolecular complexes in the AZ that form a hexagonal grid with intercalated synaptic vesicles (Pfenninger et al., 1972) and may link synaptic vesicles with Ca2+ channels and the vesicle release machinery (Südhof, 2012). C. elegans liprin-α/SYD-2 controls DP size and neurotransmission (Kittelmann et al., 2013; Stigloher et al., 2011). The decrease in DP number and area following loss of GPR158 is remarkable, as the only genetic deletion thus far reported to alter these presynaptic structures in hippocampal synapses is quadruple KO of the AZ proteins RIMs and RBPs (Acuna et al., 2016). Together, these observations suggest a mechanism whereby a trans-synaptic interaction of GPR158 with presynaptic GPC4 recruits co-receptors, scaffolds and AZ proteins to organize MF-CA3 presynaptic architecture and function.
Orphan receptor-HSPG interactions shape synapse identity
Despite severely impaired structure and function, MF-CA3 synapses are still established in the absence of GPR158, suggesting that other synaptic organizing protein complexes (Jang et al., 2017) operate in parallel to control MF-CA3 synapse development. Cadherin 9, a classic type II cadherin, is expressed in DG and CA3 and regulates MF-CA3 bouton size and dendritic spine morphology (Williams et al., 2011). C1ql2 and C1ql3 proteins are secreted from MF boutons and bridge presynaptic splice site 5-containing neurexin 3β and postsynaptic kainate receptors to regulate MF-CA3 synapse function (Matsuda et al., 2016). The synaptic adhesion protein SynCAM1 contributes to dendritic spine size (Park et al., 2016), and EphB2 contributes to synaptic plasticity (Contractor et al., 2002) at MF-CA3 synapses. This suggests that both synapse type-specific and broadly expressed proteins cooperate to regulate the specific structure, function, and molecular composition of MF-CA3 synapses. Thus, combinatorial codes of cell-surface molecules acting in particular cell types (Földy et al., 2016) and at specific inputs pattern the structural and functional properties of the synaptic network.
GPR158 is expressed in other brain regions, such as cortex and striatum, and has been implicated in cognition and depression (Khrimian et al., 2017; Sutton et al., 2018). It will be of interest to further investigate GPR158’s role in these circuits to determine to what extent GPR158 function is similar to that at MF-CA3 synapses. The molecular context in which GPR158 operates, such as the types of presynaptic HSPGs, co-receptors, and scaffolding proteins expressed at these synapses, seems an important factor in determining GPR158’s effects on synaptic development. On the other hand, the finding that closely related GPR179 interacts with the secreted extracellular matrix protein Pikachurin to regulate synaptic connectivity in the retina (Orlandi et al., submitted) suggests that, despite varying molecular contexts, GPR158/179 extracellular interactions organize specific synaptic connectivity across neural circuits.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joris de Wit (joris.dewit@kuleuven.vib.be).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal experiments were conducted according to the KU Leuven ethical guidelines and approved by the KU Leuven Ethical Committee (approved protocol numbers P015/2013 and P026/2013). Mice were maintained in a specific pathogen-free facility under standard housing conditions with continuous access to food and water. The health and welfare of the animals was supervised by a designated veterinarian. The KU Leuven animal facilities comply with all appropriate standards (cages, space per animal, temperature, light, humidity, food, water), and all cages are enriched with materials that allow the animals to exert their natural behavior. Mice used in the study were 2–4 weeks old and were maintained on a diurnal 12-hour light/dark cycle. For euthanasia, newborn pups were immediately decapitated and adult animals were injected with an irreversible dose of ketamine-xylazine. Both males and females were used for all experiments. To the best of our knowledge, we are not aware of an influence of sex on the parameters analyzed in this study.
The following mouse line was used in this study:
The Gpr158 knock out mouse line (Gpr158tm1(KOMP)Vlcg) was generated by Regeneron Pharmaceuticals Inc. (Valenzuela et al., 2003) and made into live mice by the KOMP Repository and the Mouse Biology Program at the University of California Davis.
Neuronal Cultures
Hippocampal neurons were cultured from E18 or P0 Long-Evans rats (Charles River) and plated on poly-D-lysine (Millipore) and laminin (Invitrogen)-coated glass coverslips (Glaswarenfabrik Karl Hecht). Neurons were maintained in Neurobasal or Neurobasal-A medium (Invitrogen) supplemented with B27, glucose, glutamax, penicillin/streptomycin (Invitrogen), and 25 µm β-mercaptoethanol (Sigma-Aldrich). For knockdown experiments, neurons were electroporated with DNA just before plating using a Bio-Rad Gene Pulser Xcell. Electroporation efficiency was typically around 40%.
Cell Lines
HEK293T-17 human embryonic kidney cells (available source material information: fetus) were obtained from American Type Culture Collection (ATCC) cat# CRL-11268. HEK293T-17 cells were grown in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin (Invitrogen).
METHODS DETAILS
Plasmids
Full-length cDNAs encoding mouse GPR158 (BC141527) and GPC4 (BC006622) were purchased from Open Biosystems (GE Healthcare). Mouse GPR158 was cloned into the pEGFP-N1 vector (Clontech) placing a C- terminal EGFP tag on GPR158. The mouse GPR158 extracellular domain, lacking the signal peptide, was cloned into a pDisplay vector (Invitrogen), placing an N-terminal HA (YPYDVPDYA) epitope tag on GPR158. GPC4-Fc was generated by PCR-amplifying mouse GPC4 lacking the signal peptide and GPI anchor attachment site (aa 23–528) and subcloning the PCR fragment between and in frame with the Caspr2 signal peptide and human Fc in the p3CPro vector (a gift from Dr. Davide Comoletti, Child Health Institute of New Jersey and Department of Neuroscience and Cell Biology, UMDNJ\Robert Wood Johnson Medical School, New Brunswick, NJ, USA). The GPC4-Fc glycosylation mutant (GPC4 ΔGAG-Fc) was generated by PCR from the GPC4-Fc construct using the following primer: cagtgatgagagtGCtggagagggaGCtggaGCcggatgtgaatatc. The capital GC bases are the introduced mutations, changing the AGT, AGT and AGC serine codons to GCT, GCT and GCC alanine codons. The GPR158 deletion mutants, GPR158 ΔEGF-like (aa 314–359 deleted from GPR158 extracellular domain) and GPR158 ΔLRD domain (aa108–136 deleted from GPR158 extracellular domain), were generated by PCR using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Myc-tagged LRRTM4, myc- LRRTM2, myc-FLRT2, FLAG-Neuroligin 1, HA-Neurexin 1β (-SS4) and FUGW-shGPC4 were all described previously (O’Sullivan et al., 2012; de Wit et al., 2009, 2013).
Histology/Immunocytochemistry
Immunohistochemistry
To generate hippocampal sections, WT and Gpr158 KO mouse brains were quickly dissected, embedded in OCT (Thermo Fisher) and frozen in 2-Methylbutane (Sigma-Aldrich). Frozen brains were cut on a cryostat (NX70, Thermo Fisher) and 16 µm coronal sections containing hippocampus were collected on super frost glass slides (Thermo Fisher). Sections were postfixed for 5 minutes at −20°C with acetone-methanol 1:1, washed in PBS and permeabilized for 20 minutes in 0.5% Triton X-100 in PBS. Sections were then blocked for 2 hours at 4°C in PBS containing 10 % FBS, 0.5% Triton X-100 and 30 µg/ml donkey anti-mouse IgG (Jackson ImmunoResearch) and incubated with primary antibodies overnight at 4°C. Primary antibodies were the following: rabbit anti-GPR158 N-terminal (1:500, Sigma-Aldrich), rabbit anti-GPR158 C-terminal (1:1000, Sigma-Aldrich), mouse anti-PSD95 7E3–1B8 (1:250, Thermo Fisher), guinea pig anti-bassoon (1:1000, Synaptic Systems), chicken anti-MAP2 (1:5000, Abcam), rabbit anti-synaptoporin (1:5000, Synaptic Systems), guinea pig anti-VGlut1 (1:5000, Millipore). Sections were subsequently washed in PBS and incubated for 2 hours with the secondary antibodies at room temperature. Fluorophore-conjugated secondary antibodies were from Jackson ImmunoResearch or Invitrogen. Sections were imaged using a Leica SP5 or Leica SP8 confocal microscope (Leica Microsystems) with 20X and 40X objectives.
For fluorescent β-galactosidase detection and HA-GPR158 localization experiments, WT and Gpr158 heterozygous mice were transcardially perfused for 7 minutes with ice-cold 4% PFA in PBS. The brains were dissected, post-fixed one hour at 4°C with 4% PFA and then embedded in 3% agarose (Sigma-Aldrich). Sixty µm coronal sections containing hippocampus were cut on a vibratome (VTS1000S, Leica Biosystems). Free- floating sections were permeabilized for 20 minutes in 0.5% Triton X-100 in PBS and blocked overnight at 4°C in PBS containing 10 % Normal Horse Serum, 0.5% Triton X-100, 0.2M glycine (Sigma) and 30 µg/ml donkey anti mouse IgG (Jackson ImmunoResearch). Sections were then briefly washed in 0.5% Triton X-100 in PBS and incubated for 48 hours with primary antibodies. Primary antibodies were the following: chicken anti-GFP (1:1000, Aves Labs), mouse anti-HA (1:500, Covance), mouse anti-β-galactosidase (1:1000, Millipore), mouse anti-NeuN (1:1000, Millipore), rabbit anti-GPC4 aa88–101 (1:1000, Immundiagnostik). Sections were subsequently washed in 0.5% Triton X-100 in PBS and incubated for 24 hours with the secondary antibodies at room temperature. Fluorophore-conjugated secondary antibodies were from Jackson ImmunoResearch or Invitrogen. Coverslips were mounted using ProLong Gold Antifade Mountant (Thermo Fisher). Sections were imaged using Leica SP8 confocal microscope (Leica Microsystems) with 40X and 63X objectives. For HA-GPR158 localization experiments, images were acquired from sections obtained from three independent mice. Elactroporated CA3 pyramidal neurons were imaged on a Zeiss ELYRA S.1 structured illumination microscope with 63X objective. For the analysis of HA-GPR158 localization, images were thresholded using ImageJ and the total number of HA-GPR18 puncta was measured and normalized to the total GFP-positive dendrite length. Analysis of β-galactosidase fluorescence was conducted using sections obtained from three independent Gpr158+/− mice. The analysis was performed using a custom-made protocol in NIS Microscope Imaging Software (Nikon).
X-galactosidase staining
Sixteen µm coronal hippocampal sections were generated from P7-P28 Gpr158 heterozygous mice as described above. Sections were fixed in PBS containing 0.2% glutaraldehyde (Sigma-Aldrich), briefly washed in PBS and permeabilized for 10 minutes at room temperature using a Detergent Rinse Solution (0.02% Igepal, 0.01% sodium deoxycholate, 2 mM MgCl2 in 0.1M Phosphate Buffer (PB) pH 7.3). Sections were then incubated with 1 mg/ml X-Gal (Millipore) in X-Gal Staining solution (0.02% Igepal, 0.01% sodium deoxycholate, 2 mM MgCl2, 5 mM Potassium Ferricyanide, 5mM Potassium Ferrocyanide in 0.1M PB buffer pH 7.3) for 2 hrs at 37°C in the dark. Subsequently the sections were post-fixed in 4% PFA in PBS, washed in PBS and in distilled water and finally dehydrated using increasing concentrations of ethanol and xylene. Coverslips were mounted using Micromount mounting medium (Leica Biosystems). Images were acquired using a Leica DM2500 widefield microscope (Leica Microsystems) with a 5X objective.
Single Molecule Fluorescent In Situ Hybridization
Sixteen micrometer coronal hippocampal sections were obtained from flash frozen P7 and P21 C57BL/6 mouse brains. RNAscope hybridization was performed using the RNAscope 2.5 HD detection kit (Advanced Cell Diagnostics). Heating steps were performed using the HybEZTM oven (Advanced Cell Diagnostics). The sections were treated with pretreatment solution and then incubated with custom-synthesized RNAscope probes (Gpc4, NM_008150.2, Cat #442821; Prox1, NM_008937.2, Cat# 488591-C2; Aldh1l1, NM_027406.2, Cat # 320269-C3), each targeting all predicted transcript variants, followed by amplifying hybridization processes. DAPI was used as nuclear stain. Coverslips were mounted using Prolong Gold Antifade (ThermoScientific). Images were acquired using a Leica SP8 confocal microscope (Leica Microsystems) with 40X and 63X objectives.
Immunocytochemistry
HEK293T cells and cultured hippocampal neurons were fixed in 4% paraformaldehyde, 4% sucrose in PBS, washed in PBS and blocked for one hour at room temperature in 3% BSA, 0.2% Triton X-100 in PBS. Primary antibodies were the following: chicken anti-GFP (1:1000, Aves Labs), mouse anti-HA clone 16B12 (1:500, Covance), mouse anti-myc clone 9E10 (1:1000, Santa Cruz Biotechnology), rabbit anti-synapsin (1:500, Millipore), mouse anti-FLAG M2 (1:1000, Sigma-Aldrich), mouse anti-LSAMP (DHSB). Fluorophore- conjugated secondary antibodies were from Jackson ImmunoResearch or Invitrogen.
Mixed-cultured assay
Mixed culture assays were performed as previously described (de Wit et al., 2013). Briefly, HEK293T cells were transfected with myc-tagged LRRTM2, myc-LRRTM4, EGFP, HA-GPR158, HA-GPR158 ∆EGF-like and HA-GPR158 ∆LRD using Fugene6 (Promega), mechanically dissociated and cocultured with 7 DIV hippocampal neurons for 8 or 24 hours, depending on the experiment. Following coculture, the cells were immunostained for synapsin, GFP, myc and HA. For the analysis of heparinase III treatment, hippocampal neurons (7 DIV) were treated with 1 U/ml heparinase III (Sigma-Aldrich) or vehicle (20 mM Tris-HCl [pH 7.5], 0.1 mg/ml BSA, 4mM CaCl2) for 2 hours at 37°C. Cells were then washed twice with hippocampal feeding medium and subsequently cocultured with transfected HEK293T cells for an additional 8 hours. To evaluate the effect of PI-PLC treatment, hippocampal neurons (7 DIV) were treated with 2 U/ml PI-PLC (Sigma-Aldrich) or vehicle (10 mM Tris-HCl [pH 7.4], 144mM NaCl, 0.05% BSA) for 2 hours at 37°C. Cells were then processed as described above. Images were then acquired using a Leica SP5 and Leica SP8 confocal microscope (Leica Microsystems) using a 63X objective. To test the effect of neuronal GPC4 knockdown, hippocampal neurons were electroporated with a previously validated shRNA against GPC4 (de Wit et al., 2013) just before plating. At 7–8 DIV hippocampal neurons were cocultured for 24 hours with HEK293T cells expressing myc-tagged LRRTM2 or HA-GPR158. To analyze the contribution of astrocyte-secreted GPC4,rat glial cells were infected with high-titer virus expressing a previously validated shRNA against GPC4 (de Wit et al., 2013) two days after plating. One week later, conditioned medium was transfered to 8 DIV hippocampal neurons and coculture was performed as described above.
To test the effect of neuronal LAR, PTPδ, PTPσ and neurexin knockdown, 1 DIV hippocampal neurons were transduced with low-titer viruses expressing previously validated shRNAs against LAR, PTPδ, PTPσ (Yim et al., 2013; Zhang et al., 2010). At 8 DIV, neurons were cocultured for 24 hours with HEK293T cells expressing myc-tagged LRRTM2, myc-tagged LRRTM4 or HA-GPR158.
For quantification of mixed-culture assays, images were thresholded using ImageJ and the total area of synapsin puncta was measured and normalized to the total GFP-positive area per cell. Individual puncta could not be measured in this assay because thresholding resulted in the fusion of individual puncta due to their high density. Measurements were performed in a minimum of three independent preparations and in each experiment for any given condition a minimum of eight cells were acquired. Imaging and analysis were conducted blind to the condition.
Binding Assays/Fractionation
Cell Surface Binding Assay
HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% FBS (Invitrogen), penicillin/streptomycin and transfected with EGFP, myc-LRRTM4 and HA-GPR158 plasmids using Fugene6 (Promega). Twenty-four hours after transfection, the cells were incubated with Fc or GPC4-Fc proteins (40 µg/ml) in DMEM supplemented with 20 mM Hepes (pH 7.4) for 1 hour at room temperature. Following two brief washes in DMEM/20 mM Hepes pH 7.4, cells were fixed in 4% PFA - 4% sucrose and blocked in 3% BSA for 1 hour at room temperature. The cells were then incubated for 1 hour at room temperature with donkey anti-human IgG Cy3-conjugated (1:1000, Jackson ImmunoResearch). Following three brief washes with PBS, the cells were permeabilized with 0.2% Triton in 3% BSA and then immunostained for GFP, myc and HA using the primary antibodies mentioned above). Fluorophore- conjugated secondary antibodies were from Jackson ImmunoResearch or Invitrogen. The cells were imaged with a Leica SP5 confocal microscope (Leica Microsystems) using a 63X objective.
Fc Pulldown Assays
For pulldown assays on HEK293T cells, cells were grown in 10 cm dishes in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin, and transfected with GPR158-EGFP expression construct using Fugene6 (Promega). Twenty-four hours after transfection, the media was changed to OptiMEM (Invitrogen) for 2 hours. Cells were then lysed in 1 ml ice-cold RIPA buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100 and protease inhibitors (Roche)) for 1 hour at 4°C on a rocking platform. Lysates were spun at 13,000 rpm for 30 minutes at 4°C. Three µg of human Fc, GPC4-Fc and GPC4ΔGAG-Fc was added to 1 ml of supernatant and rotated overnight at 4°C. Protein-A agarose beads (50 µl slurry) were added and rotated for 1 hour at 4°C. Beads were washed 3 times in cold RIPA buffer and once in PBS, boiled in 50 µl 2X sample buffer and analysed by Western blotting.
Co-Immunoprecipitation
HEK293T cells were grown as described, and separate dishes were transfected with GPR158-EGFP and HA-GPC4 expression constructs using Fugene6 (Promega). To test whether GPC4 and GPR158 can interact in trans, cells were enzymatically dissociated 24 hours after transfection, mixed at a 1:1 ratio, and co-cultured together for an additional 48 hours. Cells were then lysed in 1ml ice-cold IP buffer containing 1% CHAPSO (Sigma-Aldrich) and protease inhibitors (Roche) for 1 hour at 4°C on a rocking platform. Lysates were spun at 14,800 rpm for 20 minutes at 4°C, then incubated with 3 µg of rabbit IgG or anti-GFP antibody (Aves Labs) and rotated end-over-end overnight at 4°C. Protein-G agarose beads (50 µl slurry) were added and rotated for 1 hour at 4°C. Beads were washed 3 times in cold IP buffer and once in PBS, heated at 42°C in 50 µl 2X sample buffer and analysed by Western blotting.
Subcellular Fractionation
Synaptic fractionation was based on a previously described method (Carlin et al., 1980). In brief, 10 P21 rat brains were homogenized in 12 ml per brain with Homogenization Buffer (0.32 M sucrose, 4 mM Hepes pH 7.4, 1 mM MgCl2 and protease inhibitors) (homogenate), centrifuged at 1,500 x g for 15 minutes, and the supernatant was collected (post nuclear supernatant). The supernatant was then centrifuged at 18,000 x g for 20 minutes, and the resulting supernatant (cytosol) and pellet (crude membrane) collected. The pellet was re-suspended in Homogenization Buffer and loaded onto 0.85 M/1.0 M/1.2 M discontinuous sucrose gradients and centrifuged at 78,000 x g for 120 minutes. The material at the 1.0 M/ 1.2 M interface was collected (synaptosome). Triton X-100 was added to 0.5% and extracted at 4°C by end-over-end agitation for 20 minutes. The extract was centrifuged at 32,000 x g for 20 minutes, the supernatant collected (soluble synaptosome/Triton-soluble fraction) and the pellet was re-suspended in Homogenization Buffer, loaded onto a 1.0 M/1.5 M/2.0 M sucrose gradient, and centrifuged at 170,000 x g for 2 hours. Material was collected at the 1.5M/2.0M interface (PSD). 0.5% Triton X-100 was added and detergent-soluble material extracted at 4°C by end-over-end agitation for 10 minutes. Lastly, the extract was centrifuged at 100,000 x g for 20 minutes and the pellet re-suspended in Homogenization Buffer (purified PSD/Triton-insoluble fraction).
Fc-Protein Purification
GPC4-Fc, GPC4ΔGAG-Fc and GPR158-Fc proteins were produced by transient transfection of HEK293T cells using PEI (Polysciences). Six hours after transfection, media was changed to OptiMEM (Invitrogen) and harvested 5 days later. Conditioned media was centrifuged, sterile-filtered and run over a fast-flow Protein-A sepharose (GE Healthcare) column. In the case of GPC4-Fc and GPC4 ΔGAG-Fc, after extensive washing with wash buffer (50 mM Hepes pH 7.4, 300 mM NaCl and protease inhibitors), the column was eluted with Pierce elution buffer. Eluted fractions containing proteins were pooled and dialyzed with PBS using a Slide- A-Lyzer (Pierce) and concentrated using Amicon Ultra centrifugal units (Millipore). The integrity and purity of the purified ecto-Fc proteins was confirmed with SDS-PAGE and Coomassie staining, and concentration was determined using Bradford protein assay.
Affinity Chromatography
Affinity chromatography experiments were performed as previously described (Savas et al., 2014). For the preparation of crude synaptosome extracts, ten P21–22 rat brains were homogenized in homogenization buffer (4 mM Hepes pH 7.4, 0.32 M sucrose and protease inhibitors) using a Dounce homogenizer. Homogenate was spun at 1,000 x g for 10 minutes at 4°C. Supernatant was spun at 14,000 x g for 20 minutes at 4°C. The pellet (P2) was resuspended in Extraction Buffer (50 mm Hepes pH 7.4, 0.1 M NaCl, 2 mM CaCl2 , 2.5 mM MgCl2 and protease inhibitors), extracted with 1% Triton X-100 for 2 hours and centrifuged at 100,000 x g for 1 hour at 4°C to pellet insoluble material. Fast-flow Protein-A sepharose beads (GE Healthcare) (250 µl slurry) pre-bound in Extraction Buffer to 100 µg human Fc, GPC4-Fc or GPC4 ΔGAG-Fc were added to the supernatant and rotated overnight at 4°C.
Beads were packed into Poly-prep chromatography columns (BioRad) and washed with 50 ml of high- salt wash buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 0.1 mM CaCl2, 5% glycerol and protease inhibitors), followed by a wash with 10 ml low-salt wash buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 0.1 mM CaCl2, 5% glycerol and protease inhibitors). Bound proteins were eluted from the beads by incubation with Pierce elution buffer and TCA-precipitated overnight. The precipitate was resuspended in 8 M Urea with ProteaseMax (Promega) per the manufacturer’s instruction. The samples were subsequently reduced by 20- minute incubation with 5mM TCEP0 (tris(2carboxyethyl)phosphine) at room temperature and alkylated in the dark by treatment with 10 mM Iodoacetamide for 20 additional minutes. The proteins were digested overnight at 37°C with Sequencing Grade Modified Trypsin (Promega) and the reaction was stopped by acidification.
Mass Spectrometry (MS)
For LTQ MS analysis, the protein digest was pressure-load onto a 250 µm i.d. capillary packed with 2.5 cm of 10 µm Jupiter C18 resin (Phenomenex) followed by an additional 2.5 cm and 5 µm Partisphere strong cation exchanger (Whatman). The column was washed with buffer containing 95% water, 5% acetonitrile and 0.1% formic acid. After washing, a 100 µm i.d. capillary with a 5 µm pulled tip packed with 15 cm of 4 µm Jupiter C18 resin (Phenomenex) was attached to the filter union and the entire split-column (desalting column- filter union-analytical column) was placed in-line with an Agilent 1100 quaternary HPLC and analysed using a modified 5-step separation described previously (Washburn et al., 2001). The buffer solutions used were 5% acetonitrile /0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). Step 1 consisted of a 75-minute gradient from 0–100% buffer B. Steps 2–5 had a similar profile except 3 minutes of 100% buffer A, 5 minutes of X% buffer C, a 10 minutes gradient from 0–15% buffer B, and a 102-minutes gradient from 15–45% buffer B. The 5 minutes buffer C percentages (X) were 10, 40, 60, and 100% respectively for the 5-step analysis. As peptides eluted from the microcapillary column, they were electrosprayed directly into a LTQ mass spectrometer (ThermoScientific) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan mass spectrum (400–1400 m/z) followed by 3-data dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcaliber datasystem.
For Orbitrap Fusion Tribrid MS analysis, the tryptic peptides were purified with Pierce C18 spin columns (Thermo Scientific). Three micrograms of peptide was auto-sampler loaded with a Thermo EASY nLC 1200 UPLC pump onto a vented Acclaim Pepmap 100, 75 μm × 2 cm, nanoViper trap column coupled to a nanoViper analytical column (Thermo, 3 μm, 100 Å, C18, 0.075 mm, 500 mm) with stainless steel emitter tip assembled on the Nanospray Flex Ion Source with a spray voltage of 2.0 kV. Buffer A contained 94.785% H2O with 5% acetonitrile and 0.125% formic acid, and buffer B contained 99.875% acetonitrile with 0.125% formic acid. The chromatographic run was for 4h in total with the following profile: 0–7% over 7 min, ramp to 10% over 6 min, ramp to 25% over 160 min, ramp to 33% over 40 min, ramp to 50% over 7 min, ramp to 95% for 5 min and stay at 95% for 15 additional minutes. Additional MS parameters include: ion transfer tube temperature to 300°C, Easy-IC internal mass calibration, the default charge state was set to 2 and cycle time was set to 3s. Detector type set to Orbitrap, with 60,000 resolution, with wide quad isolation, mass range was set to normal, scan range was set to 300–1,500 (m/z), max injection time was set to 50ms, AGC target was set to 200,000, microscans was set to 1, S-lens RF level was set to 60, without source fragmentation, and data type was set to positive and centroid. Monoisotopic precursor selection was set as on, included charge states equal to 2–6 (and reject unassigned). Dynamic exclusion enabled and set to 1 for 30s and 45s exclusion duration at 10 p.p.m. for high and low. Precursor selection decision was set to most intense, top 20, isolation window was set to 1.6, scan range was set to auto normal, first mass was set to 110, collision energy was set to 30%. For CID, we used the ion trap detector, ion trap resolution was set to 30K, ion trap scan rate was set to rapid, maximum injection time was set to 75ms, AGC target was set to 10,000, and Q was set to 0.25, finally we injected ions for all available parallelizable time.
Spectrum raw files were extracted into ms1 and ms2 files using the in-house program RawXtractor or RawConverter (http://fields.scripps.edu/downloads), and the tandem mass spectra were searched against UniProt rat database (downloaded on 04–01-2013) and matched to sequences using the ProLuCID database search program (ProLuCIDver. 3.1). ProLuCID searches were done on an Intel Xeon cluster running under the Linux operating system. The search space included all fully and half-tryptic peptide candidates that fell within the mass tolerance window with no miscleavage constraint. Carbamidomethylation (+57.02146 Da) of cysteine was considered as a static modification. The validity of peptide/spectrum matches (PSMs) was assessed in DTASelect2 (using two SEQUEST-defined parameters, the cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, +3, and greater than +3) and tryptic status (fully tryptic, half-tryptic, and nontryptic), resulting in 12 distinct subgroups. In each of these subgroups, the distribution of Xcorr, DeltaCN, and DeltaMass values for (1) direct and (2) decoy database PSMs was obtained; then the direct and decoy subsets were separated by discriminant analysis. Full separation of the direct and decoy PSM subsets is not generally possible; therefore, peptide match probabilities were calculated on the basis of a nonparametric fit of the direct and decoy score distributions. A peptide confidence of 0.95 was set as the minimum threshold. The false discovery rate (FDR) was calculated as the percentage of reverse decoy PSMs among all the PSMs that passed the confidence threshold. Each protein identified was required to have a minimum of one half-tryptic peptide; however, this peptide had to be an excellent match with an FDR less than 0.001 which represents at least one excellent peptide match. For high resolution data we also required a delta mass ≤10ppm. After this last filtering step, protein FDRs were below 1% for each sample analysis based on decoy hits.
In Utero Electroporation
Hippocampi of E15.5-day-old embryos (E15.5) of timed-pregnant C57BL/6 or WT and Gpr158 KO mice were unilaterally electroporated with pFUGW-EGFP or pFUGW-EGFP—T2A—HA-GPR158 constructs. Briefly, the dam was anesthesized with isoflurane and the uterus exposed. A solution of 1 µg/µl DNA and 0.01% fast green dye was injected into the embryonic lateral ventricle with a bevelled glass capillary. The embryo’s head was then placed between the paddles of pair of platinum tweezer-type electrodes (Napagene) with the catode lateral to the filled ventricle and seven 50 ms, 40 V pulses were delivered at at 1 Hz by a ECM830 electroporator (Harvard Apparatus). After electroporation, the uterus was replaced, the incision sutured closed and the dam allowed to give birth normally.
Lentivirus production
Second generation VSV.G pseudotype lentiviruses were produced by transfecting HEK293T cells with control or shRNA containing pFUGW vector plasmids (Dittgen et al., 2004) and helper plasmids PAX2 and VSVG using polyethylenimine (Polysciences). Supernatant was collected 48 hours after transfection, spun at 2000 rpm to remove debris filtered trough a 0.45 µm filter (Corning). In order to maximize the purity of viral particles pellets, a small amount of sucrose solution (20% sucrose, 100 mM NaCl, 20 mM EDTA, at pH 7.4, filtered 0.22 µm filter) was placed at the bottom of the centrifuge tubes before adding filtered media. Viral particles were pelleted by centrifugation at 25000 rpm for two hours. The final pellet was re-suspended in 200 µl of PBS and stored at −80°C in 10 µl aliquots.
For low-titer virus production, the supernatant was collected 48 hours after transfection and filtered to remove debris through a 0.22 µm filter (Millipore). 250 µl aliquots were then prepared and stored at −80°C.
Spine Imaging and Analysis
For dye filling experiments, P21–22 WT and Gpc158 KO littermates were used. Briefly, after decapitation, the brain was quickly removed and transferred into ice-cold cutting solution (83 mM NaCl, 2.5 mM KCl, 1 mM NaH2PO4, 22 mM glucose, 26.2 mM NaHCO3, 0.5 mM CaCl2, 3.3 mM MgSO4, 72 mM sucrose, pH 7.4 with 5% CO2/ 95% O2). After brain dissection, whole brain coronal slices (300 µm) were cut with a vibratome (Leica VT1200). Afterwards slices were transferred to 33°C cutting solution for 1 hour to recover and finally maintained at room temperature until used for the experiment. Glass borosilicate recording pipettes (resistance 3–4 MΩ) were filled with 10 mM Alexa568 (Life Technologies) dissolved in internal solution (15 mM CsMSF, 20 mM CsCl, 10 mM HEPES, 2.5 mM MgCl2, 4 mM ATP, 0.4 mM GTP, 10 mM Creatine Phosphate and 0.6 mM EGTA). The tip of the pipette was moved through the tissue toward the cell of interest. Whole-cell configuration was used to contact and penetrate the targeted CA3 neurons. CA3 pyramidal cells were filled for fifteen minutes. Filled neurons were then fixed for twelve hours at 4°C using 4% PFA adjusted to mOsm with sucrose and pH 7.4. Sections were then washed three times with 0.1 M PB solution and coverslips were mounted using Prolong Gold Antifade (ThermoScientific).
For in utero electroporation experiments, P21 electroporated WT and Gpr158 KO mice were transcardially perfused with ice-cold 4% PFA adjusted to mOsm with sucrose and pH 7.4. Brains were then dissected and postfixed for one hour at 4°C. Brains were then embedded in 3% agarose (Sigma-Aldrich) and 60 µm coronal sections were cut on a vibratome (VTS1000S, Leica Biosystems). Sections in which CA3 pyramidal neurons visibly expressed GFP were immunostained with a chicken polyclonal anti-GFP primary antibody (1:1000, Aves Labs) and an Alexa 488-conjugated goat-anti chicken secondary antibodiy (1:1000, Invitrogen). Coverslips were then mounted using Prolong Gold Antifade (ThermoScientific).
Dendrites were imaged from sections obtained from a minimum of three independent mice. Primary CA3 apical dendrites were imaged on a Zeiss ELYRA S.1 structured illumination microscope. Dendritic protrusions were counted in Z-stack and the length of the dendritic segment was measured in Fiji using with the Simple Neurite Tracer plugin.
Transmission Electron Microscopy
P14 WT and Gpr158 KO littermates were prepared for Transmission Electron Microscopy (TEM) by transcardial perfusion with 4% paraformaldehyde, 2.5% glutaraldehyde, 0.2% picric acid in 0.1M PB. Brains were removed, embedded in 3% low gelling temperature agarose (Sigma-Aldrich) and 100 µm sections were cut on a vibratome (VTS1000S, Leica Biosystems). Selected sections were then post-fixed in a solution of 1% OsO4 containing 1.5% potassium ferrocyanide for 60 minutes at room temperature, stained with 0.2% tannic acid for 30 minutes, fixed with 1% OsO4 for 30 minutes, stained with 1% thiocarbohydrazide and incubated again with 1% OsO4. The sections were then contrasted with 0.5% uranyl acetate in 25% methanol overnight at 4°C. The samples were subsequently dehydrated in increasing concentrations of ethanol and finally infiltrated and embedded in resin (epon 812). Ultrathin sections (70 nm) were cut using a UC7 Ultratome (Leica Biosystems) and collected on copper grids. Sections were analysed and imaged using a JEOL JEM1400 TEM equipped with an Olympus SIS Quemesa camera operated at 80 kV.
For visualization of postsynaptic densities and dense projections, dehydrated sections were treated for 1 hour at room temperature in 1% ethanolic phosphotungstic acid (PTA, MP Biomedicals) and then processed as described above but without OsO4 postfixation.
Mossy Fiber (MF)-CA3 synapses were identified by their morphology, presence of multiple postsynaptic densities and high synaptic vesicle content. Image segmentation/analysis was performed initially by using Microscope Image Browser (MIB, University of Helsinki) in order to segment individual MF-CA3 synapses, postsynaptic densities, active zones and dense projections. Images were then thresholded using ImageJ software and the analysis of MF-CA3 surface area, active zone, postsynaptic density area, length and perimeter was performed by using ‘analyse particles’ function. For the analysis of dense projection number, area, perimeter and length a custom-made ImageJ/Fiji software macro was used. Measurements were performed in a minimum of three independent preparations.
Serial Block-Face Scanning Electron Microscopy
For 3D serial block-face scanning electron microscopy (SBF-SEM 3D), the tissue was prepared as described above. Portions of an embedded section were then mounted on a pin, pre-trimmed in a microtome and placed in a SEM (Zeiss VP Sigma) equipped with an internal microtome (Gatan, 3View). The serial sectioning was performed at 200 nm steps. Initially, serial electron images of the block face were acquired at 1.80 kV with 3K zoom. Simultaneously, serial electron images of specific region of interests were acquired at 1.80 kV with an 9.78 K zoom. Images were then aligned using Digital Micrograph Software (Gatan) and individual MF-CA3 synapses were segmented and reconstructed using Imaris software (Bitplane). Analysis of reconstructed MF- CA3 volume and surface was performed using Imaris software (Bitplane). Measurements were performed in a minimum of five reconstructed MF-CA3 synapses from each individual condition. The analysis was conducted on two independent preparations.
Acute Slice Electrophysiology
Sagittal slices were prepared from postnatal day P21–23 WT and Gpr158 KO littermates. Briefly, after decapitation, the brain was quickly removed and transferred into ice-cold cutting solution (83 mM NaCl, 2.5 mM KCl, 1 mM NaH2PO4, 22 mM glucose, 26.2 mM NaHCO3, 0.5 mM CaCl2, 3.3 mM MgSO4, 72 mM sucrose, pH 7.4 with 5% CO2/ 95% O2). After brain dissection, whole brain sagittal slices (300 µm) were cut with a vibratome (Leica VT1200). Afterwards slices were transferred to 33°C cutting solution for 1 hour to recover and finally maintained at room temperature until used for recordings.
Brain slices were continuously perfused in a submerged chamber (Warner Instruments) at a rate of 3– 4 ml/min with 119 mM NaCl, 2.5 mM KCl, 1mM NaH2PO4, 26 mM NaHCO3, 4 mM MgCl2, 4 mM CaCl2, 11 mM glucose at pH 7.4 with 5% CO2/ 95% O2 . For paired-pulse and spontaneous release recordings, 20 µM bicuculline, 100 µM AP-5 and 150 nM CNQX was added to the ACSF. For AMPA/NMDA ratio recordings we added 20 nM CNQX and 20 µM bicuculline. Whole-cell patch-clamp recordings were done using borosilicate glass recording pipettes (resistance 3–4 MΩ) filled with the following internal solution: 115 mM CsMSF, 20 mM CsCl, 10 mM HEPES, 2.5 mM MgCl2, 4 mM ATP, 0.4 mM GTP, 10 mM Creatine Phosphate and 0.6 mM EGTA. Spontaneous input to CA3 pyramidal neurons was recorded by whole-cell voltage-clamp recordings (Vm=−70 mV) from visually identifiable CA3 pyramidal neurons, using a Multiclamp 700B amplifier (Axon Instruments) and analysed using Mini Analysis program (Synaptosoft). An extracellular stimulation electrode (CE2C55, FHC) was placed in the hilar region of the dentate gyrus and used for initiating evoked AMPAR and NMDAR-mediated currents. AMPAR-mediated evoked EPSCs were measured in whole-cell voltage- clamp at a holding potential of −60 mV, while the NMDAR-mediated component was measured 50 ms after initiation of the combined AMPAR-and NMDAR-mediated EPSCs recorded at +40 mV. For paired-pulse ratio analysis, paired extracellular stimulations (interstimulus interval (ISI): 25, 50, 100, 200, 400, 1000 and 2000 ms) were delivered every 20 seconds (each ISI was repeated 4 times) and calculated as the EPSC2/EPSC1 ratio. MF-CA3 synaptic responses were identified by a marked PPF (PPF > 3 at 25 ms interstimulus interval) and by their sensitivity to the metabotropic glutamate receptor II agonist 2-(2,3-dicarboxy-cyclopropyl)glycine (DCG-IV, 1 µM), which was applied at the end of experiments. Paired-pulse ratio analysis of CA3-CA3 synaptic connections was performed as described above but in this case the stimulation electrode was placed in the stratum radiatum layer of CA3 and 1 µM DCG-IV was added to the recording solution. Measurements were performed in a minimum of three independent preparations.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are graphed as mean ± SEM. Data analysis was performed in GraphPad Prism 7 (GraphPad software), Clampfit 10.6 (Molecular Devices), MiniAnalysis (Synaptosoft) and ImageJ (NIH). In order to compare differences between two or more groups, ANOVA tests (one-way, with multiple comparisons) were performed. For all experiments, datasets were tested for normality using D’Agostino and Pearson test. If the datasets passed the test, the data were analyzed using Student’s unpaired t test (Welch’s correction). Otherwise, the datasets were analyzed using nonparametric unpaired t tests (Mann-Whitney). In Figure legends, for mixed cultured assays the designation ‘n’ indicates the total number of cells analyzed in three independent experiments for any given condition. In case of electrophysiology, immunohistochemistry and in utero electroporation analysis, the designation ‘N’ indicates the number of animals used, while ‘n’ indicates the number of cells analyzed. For the analysis of electron microscopy experiments, the designation ‘N’ indicates the number of animals used, while ‘n’ indicates the number of synapses or synaptic elements analyzed. Data from all subjects were included with no exclusions. The analysis of the coculture assay experiments was conducted blind to the conditions.
Supplementary Material
Acknowledgements
We thank Patrik Verstreken and Dietmar Schmucker for critical reading of the manuscript, and de Wit lab members for discussion and comments. We thank Heather Rice, Jeroen Vanderlinden, and Julie Nys for experimental help. We thank Katlijn Vints and Benjamin Pavie (VIB BioImaging Core, KU Leuven) for electron microscopy reagents and ImageJ scripts, respectively. We thank Jaewon Ko for generously providing LAR RPTP shRNA constructs. We thank Fahri Küçükali and Kristel Sleegers (VIB-UAntwerp Center for Molecular Neurology) for analysis of GPR158 variants in neurodevelopmental disorders. Zeiss Elyra S.1 was acquired through a CLME grant from Minister Lieten to the VIB BioImaging Core. Leica SP8x confocal microscope provided by InfraMouse (KU Leuven-VIB) through a Hercules type 3 project (ZW09–03). This work was supported by an FWO PhD fellowship to A.S. (11Z3715N/17N); an FWO postdoctoral fellowship to S.E.R. (12N3515N); NIH grants MH105482 and DA026405 (K.A.M); NIH grant R00DC013805 (J.N.S.); and ERC Starting Grant (#311083), FWO Odysseus Grant, FWO Project grants G094016N and G0C4518N, Methusalem grant of KU Leuven/Flemish Government, and ERA-NET NEURON SynPathy 2015 to J.d.W.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Acuna C, Liu X, and Südhof TC (2016). How to Make an Active Zone: Unexpected Universal Functional Redundancy between RIMs and RIM-BPs. Neuron 91, 792–807. [DOI] [PubMed] [Google Scholar]
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, and Barres BA (2012). Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 486, 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Maxeiner S, Sando R, Tsetsenis T, Malenka RC, and Südhof TC (2017). Postsynaptic adhesion GPCR latrophilin-2 mediates target recognition in entorhinal-hippocampal synapse assembly. J Cell Biol 216, 3831–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, di Cristo G, Higashiyama H, Bennett V, Wu P, and Huang ZJ (2004). Ankyrin- Based Subcellular Gradient of Neurofascin, an Immunoglobulin Family Protein, Directs GABAergic Innervation at Purkinje Axon Initial Segment. Cell 119, 257–272. [DOI] [PubMed] [Google Scholar]
- Bjarnadóttir TK, Fredriksson R, and Schiöth HB (2005). The gene repertoire and the common evolutionary history of glutamate, pheromone (V2R), taste(1) and other related G protein-coupled receptors. Gene 362, 70–84. [DOI] [PubMed] [Google Scholar]
- Bloom FE, and Aghajanian GK (1966). Cytochemistry of synapses: selective staining for electron microscopy. Science 154, 1575–1577. [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, and Siekevitz P (1980). Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol 86, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Sugino K, Shields BC, and Spruston N (2016). Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. ELife 5, e14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles CH, Shen Y, Tenney AP, Siebold C, Sutton GC, Lu W, Gallagher JT, Jones EY, Flanagan JG, and Aricescu AR (2011). Proteoglycan-Specific Molecular Switch for RPTPσ Clustering and Neuronal Extension. Science 332, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condomitti G, and de Wit J (2018). Heparan Sulfate Proteoglycans as Emerging Players in Synaptic Specificity. Front. Mol. Neurosci 11, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Rogers C, Maron C, Henkemeyer M, Swanson GT, and Heinemann SF (2002). Trans-synaptic Eph receptor-ephrin signaling in hippocampal mossy fiber LTP. Science 296, 1864–1869. [DOI] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, and Osten P (2004). Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl. Acad. Sci 101, 18206–18211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan RN, Bae B-I, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. (2016). Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167, 341–354.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy-Tselnicker I, van Casteren ACM, Lee A, Chang VT, Aricescu AR, and Allen NJ (2017). Astrocyte-Secreted Glypican 4 Regulates Release of Neuronal Pentraxin 1 from Axons to Induce Functional Synapse Formation. Neuron 96, 428–445.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földy C, Darmanis S, Aoto J, Malenka RC, Quake SR, and Südhof TC (2016). Single- cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. U. S. A 113, E5222–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze DA, Card JP, Barrionuevo G, and Ben-Ari Y (1997). Large amplitude miniature excitatory postsynaptic currents in hippocampal CA3 pyramidal neurons are of mossy fiber origin. J. Neurophysiol 77, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, and Sheng M (1999). Regulated expression and subcellular localization of syndecan heparan sulfate proteoglycans and the syndecan-binding protein CASK/LIN-2 during rat brain development. J. Neurosci. Off. J. Soc. Neurosci 19, 7415–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Lee H, and Kim E (2017). Synaptic adhesion molecules and excitatory synaptic transmission. Curr. Opin. Neurobiol 45, 45–50. [DOI] [PubMed] [Google Scholar]
- Johnson KG, Tenney AP, Ghose A, Duckworth AM, Higashi ME, Parfitt K, Marcu O, Heslip TR, Marsh JL, Schwarz TL, et al. (2006). The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 49, 517–531. [DOI] [PubMed] [Google Scholar]
- Kakegawa W, Mitakidis N, Miura E, Abe M, Matsuda K, Takeo YH, Kohda K, Motohashi J, Takahashi A, Nagao S, et al. (2015). Anterograde C1ql1 Signaling Is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum. Neuron 85, 316–329. [DOI] [PubMed] [Google Scholar]
- Kamimura K, and Maeda N (2017). Heparan sulfate proteoglycans in Drosophila neuromuscular development. Biochim. Biophys. Acta BBA - Gen. Subj 1861, 2442–2446. [DOI] [PubMed] [Google Scholar]
- Khrimian L, Obri A, Ramos-Brossier M, Rousseaud A, Moriceau S, Nicot A-S, Mera P, Kosmidis S, Karnavas T, Saudou F, et al. (2017). Gpr158 mediates osteocalcin’s regulation of cognition. J. Exp. Med 214, 2859–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann M, Hegermann J, Goncharov A, Taru H, Ellisman MH, Richmond JE, Jin Y, and Eimer S (2013). Liprin-α/SYD-2 determines the size of dense projections in presynaptic active zones in C. elegans. J. Cell Biol 203, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, and Südhof TC (2009). LRRTM2 Functions as a Neurexin Ligand in Promoting Excitatory Synapse Formation. Neuron 64, 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JS, Pramanik G, Um JW, Shim JS, Lee D, Kim KH, Chung G-Y, Condomitti G, Kim HM, Kim H, et al. (2015). PTPσ functions as a presynaptic receptor for the glypican- 4/LRRTM4 complex and is essential for excitatory synaptic transmission. Proc. Natl. Acad. Sci. U. S. A 112, 1874–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, and Tessier-Lavigne M (2011). Mechanisms and Molecules of Neuronal Wiring: A Primer. Cold Spring Harb. Perspect. Biol 3, a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S-K, Woo J, Kim S-Y, Kim H, and Kim E (2010). Trans-synaptic Adhesions between Netrin-G Ligand-3 (NGL-3) and Receptor Tyrosine Phosphatases LAR, Protein-tyrosine Phosphatase δ (PTPδ), and PTPσ via Specific Domains Regulate Excitatory Synapse Formation. J. Biol. Chem 285, 13966–13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T, Aust G, and Hamann J (2013). Sticky signaling--adhesion class G protein- coupled receptors take the stage. Sci. Signal 6, re3. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Budisantoso T, Mitakidis N, Sugaya Y, Miura E, Kakegawa W, Yamasaki M, Konno K, Uchigashima M, Abe M, et al. (2016). Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron 90, 752–767. [DOI] [PubMed] [Google Scholar]
- Nishimura-Akiyoshi S, Niimi K, Nakashiba T, and Itohara S (2007). Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc. Natl. Acad. Sci 104, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Omori Y, Wang Y, Roux MJ, Condomitti G, de Wit J, Furukawa T, and Martemyanov KA (submitted). Trans-synaptic interaction of orphan receptor GPR179 with dystroglycan-pikachurin complex is essential for the synaptic organization of photoreceptors [DOI] [PMC free article] [PubMed]
- Orlandi C, Posokhova E, Masuho I, Ray TA, Hasan N, Gregg RG, and Martemyanov KA (2012). GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol 197, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Xie K, Masuho I, Fajardo-Serrano A, Lujan R, and Martemyanov K. a (2015). Orphan Receptor GPR158 is an Allosteric Modulator of Regulator of G Protein Signaling 7 (RGS7) Catalytic Activity with Essential Role in Dictating its Expression and Localization in the Brain. J. Biol. Chem 290, 13622–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke NA, Weiler NC, Micheva KD, and Smith SJ (2012). Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat Rev Neurosci 13, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, and Ghosh A (2012). FLRT Proteins Are Endogenous Latrophilin Ligands and Regulate Excitatory Synapse Development. Neuron 73, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KA, Ribic A, Laage Gaupp FM, Coman D, Huang Y, Dulla CG, Hyder F, and Biederer T (2016). Excitatory Synaptic Drive and Feedforward Inhibition in the Hippocampal CA3 Circuit Are Regulated by SynCAM 1. J. Neurosci. Off. J. Soc. Neurosci 36, 7464–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Itakura T, Jr, J.M.G., Schwartz SG, and Fini ME (2013). GPR158, an Orphan Member of G Protein-Coupled Receptor Family C: Glucocorticoid-Stimulated Expression and Novel Nuclear Role. PLOS ONE 8, e57843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger K, Akert K, Moor H, and Sandri C (1972). The fine structure of freeze-fractured presynaptic membranes. J. Neurocytol 1, 129–149. [DOI] [PubMed] [Google Scholar]
- Rebola N, Carta M, and Mulle C (2017). Operation and plasticity of hippocampal CA3 circuits: implications for memory encoding. Nat. Rev. Neurosci 18, 208–220. [DOI] [PubMed] [Google Scholar]
- Rollenhagen A, Sätzler K, Rodríguez EP, Jonas P, Frotscher M, and Lübke JHR (2007). Structural Determinants of Transmission at Large Hippocampal Mossy Fiber Synapses. J. Neurosci 27, 10434–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saied-Santiago K, and Bülow HE (2018). Diverse roles for glycosaminoglycans in neural patterning. Dev. Dyn 247, 54–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salin PA, Scanziani M, Malenka RC, and Nicoll RA (1996). Distinct short-term plasticity at two excitatory synapses in the hippocampus. Proc. Natl. Acad. Sci 93, 13304–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Yamagata M (2009). Many Paths to Synaptic Specificity. Annu. Rev. Cell Dev. Biol 25, 161–195. [DOI] [PubMed] [Google Scholar]
- Savas JN, De Wit J, Comoletti D, Zemla R, Ghosh A, and Yates JR 3rd (2014). Ecto-Fc MS identifies ligand-receptor interactions through extracellular domain Fc fusion protein baits and shotgun proteomic analysis. Nat Protoc 9, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Scheiffele P (2010). Genetics and Cell Biology of Building Specific Synaptic Connectivity. Annu. Rev. Neurosci 33, 473–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, and Craig AM (2010). LRRTMs and Neuroligins Bind Neurexins with a Differential Code to Cooperate in Glutamate Synapse Development. J. Neurosci 30, 7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Tari PK, Connor SA, Zhang P, Dobie FA, She K, Kawabe H, Wang YT, Brose N, and Craig AM (2013). An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron 79, 680–695. [DOI] [PubMed] [Google Scholar]
- Sigoillot SM, Iyer K, Binda F, González-Calvo I, Talleur M, Vodjdani G, Isope P, and Selimi F (2015). The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells. Cell Rep 10, 820–832. [DOI] [PubMed] [Google Scholar]
- Spangler SA, Jaarsma D, De Graaff E, Wulf PS, Akhmanova A, and Hoogenraad CC (2011). Differential expression of liprin-α family proteins in the brain suggests functional diversification. J. Comp. Neurol 519, 3040–3060. [DOI] [PubMed] [Google Scholar]
- Spangler SA, Schmitz SK, Kevenaar JT, de Graaff E, de Wit H, Demmers J, Toonen RF, and Hoogenraad CC (2013). Liprin-α2 promotes the presynaptic recruitment and turnover of RIM1/CASK to facilitate synaptic transmission. J. Cell Biol 201, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N (2008). Pyramidal neurons: dendritic structure and synaptic integration. Nat. Rev. Neurosci 9, 206–221. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Zhan H, Zhen M, Richmond J, and Bessereau J-L (2011). The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J. Neurosci. Off. J. Soc. Neurosci 31, 4388–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC (2012). The presynaptic active zone. Neuron 75, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton LP, Orlandi C, Song C, Oh WC, Muntean BS, Xie K, Filippini A, Xie X, Satterfield R, Yaeger JDW, et al. (2018). Orphan receptor GPR158 controls stress-induced depression. ELife 7, e33273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol 21, 652–659. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, and Yates JR 3rd (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Yamada H, and Yamaguchi Y (1995). K-glypican: a novel GPI-anchored heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J Cell Biol 130, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke SA, Antonios JK, Bushong EA, Badkoobehi A, Malek E, Hwang M, Terada M, Ellisman MH, and Ghosh A (2013). Deconstructing Complexity: Serial Block-Face Electron Microscopic Analysis of the Hippocampal Mossy Fiber Synapse. J. Neurosci 33, 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, et al. (2011). Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 71, 640–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2016). Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci 17, 4. [DOI] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, Comoletti D, Taylor P, and Ghosh A (2009). LRRTM2 Interacts with Neurexin1 and Regulates Excitatory Synapse Formation. Neuron 64, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, O’Sullivan ML, Savas JN, Condomitti G, Caccese MC, Vennekens KM, Yates JR 3rd, and Ghosh A (2013). Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron 79, 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, and Esko JD (2014). Demystifying Heparan Sulfate–Protein Interactions. Annu. Rev. Biochem 83, 129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim YS, Kwon Y, Nam J, Yoon HI, Lee K, Kim DG, Kim E, Kim CH, and Ko J (2013). Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc. Natl. Acad. Sci 110, 4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Araç D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, and Südhof TC (2010). Neurexins Physically and Functionally Interact with GABAA Receptors. Neuron 66, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürner M, Mittelstaedt T, tom Dieck S, Becker A, and Schoch S (2011). Analyses of the spatiotemporal expression and subcellular localization of liprin-α proteins. J. Comp. Neurol 519, 3019–3039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.