Abstract
Helicobacter pylori is a major human pathogen. Diagnosis of H. pylori infection and determination of its antibiotic susceptibility still mainly rely on culture and phenotypic drug susceptibility testing (DST) that is time-consuming and laborious. Whole genome sequencing (WGS) has recently emerged in medical microbiology as a diagnostic tool for reliable drug resistance prediction in bacterial pathogens. The aim of this study was to compare phenotypic DST results with the predictions based on the presence of genetic determinants identified in the H. pylori genome using WGS. Phenotypic resistance to clarithromycin, metronidazole, tetracycline, levofloxacin, and rifampicin was determined in 140 clinical H. pylori isolates by E-Test®, and the occurrence of certain single nucleotide polymorphisms (SNPs) in target genes was determined by WGS. Overall, there was a high congruence of >99% between phenotypic DST results for clarithromycin, levofloxacin, and rifampicin and SNPs identified in the 23S rRNA, gyrA, and rpoB gene. However, it was not possible to infer a resistance phenotype for metronidazole based on the occurrence of distinct SNPs in frxA and rdxA. All 140 H. pylori isolates analysed in this study were susceptible to tetracycline, which was in accordance with the absence of double or triple nucleotide substitutions in the 16S rRNA gene.
Keywords: Helicobacter pylori, whole genome sequencing, antibiotic resistance prediction, phenotypic drug susceptibility testing, laboratory automation
1. Introduction
Helicobacter pylori is a gram-negative bacterium that infects about half of the world’s human population. Unless treated, colonization in the human gastrointestinal tract persists lifelong. H. pylori infection represents a key factor in the aetiology of various gastroduodenal diseases, including chronic active gastritis, peptic or duodenal ulcers, gastric adenocarcinoma, and mucosa associated tissue lymphoma [1,2,3]. The current first-choice empiric regimen for H. pylori eradication in countries with clarithromycin resistance <15% consists of a clarithromycin-based triple therapy with a proton pump inhibitor (PPI) in combination with metronidazole or amoxicillin [4,5]. However, in the last years, the effectiveness of this empiric first-line therapy has been steadily reduced due to increasing clarithromycin and/or metronidazole resistance in H. pylori [6,7,8]. Alternatively, non-bismuth or bismuth containing quadruple therapy combined with tetracycline, levofloxacin, or rifabutin-based antibiotic regimens can be administered to patients [5,9]. However, the increase in quinolone resistant H. pylori strains [10,11], and the emergence of quadruple-resistant H. pylori clinical isolates in Western Europe [8,12] emphasize the need for more rapid and cost effective molecular methods that enable reliable prediction of antibiotic resistance phenotypes prior to the administration of antimicrobial therapy.
Diagnosis of H. pylori infection and determination of the pathogen’s antibiotic susceptibility mainly relies on bacterial culture and phenotypic drug susceptibility testing (DST), typically delivering results within two weeks. However, visual inspection of minimum inhibitory concentrations (MICs) on the E-Test® strips leaves great scope for personal interpretation and potentially leads to high inter-observer variability due to the fastidious growth of H. pylori [13].
Recently, whole-genome sequencing (WGS) from cultured bacterial isolates has emerged as an important tool for surveillance and antibiotic resistance control. This primarily owes to improvements in sequencing technologies, affordable instrument pricing, user friendly and simple workflows requiring little hands on time, availability of standardized protocols and reagents, and reasonable per sample costs (150 EUR per 5 MB genome in the case of the Illumina MiSeq), and are hence an attractive choice as a diagnostic tool to detect drug resistance in diagnostic microbiology laboratories. Within a clinically relevant timeframe (24 to 48 h), WGS provides a comprehensive view of the genotype of a bacterial isolate [14,15]. In principle, the genome sequence contains all the information required to determine the antibiotic resistance phenotype. The biggest challenge, however, lies in the identification of high-confidence single nucleotide polymorphisms (SNPs) associated with resistance against a certain drug. Genotype-based prediction of phenotypic resistance is less complex for H. pylori than for other bacteria as antibiotic resistance is primarily based on point mutations in the genome and resistance determinants in H. pylori seem not to be encoded on plasmids, transposons, or integrons [16].
In detail, clarithromycin resistance in H. pylori has been associated with point mutations in domain V of the 23S rRNA gene, namely at the nucleotide position, A2146 and A2147 (positions according to H. pylori reference strain 26695; correspond to nucleotides A2058 and A2059 in Escherichia coli) [17]. Some studies have reported point mutations outside these positions, but their association with clarithromycin resistance is still a matter of debate [18,19,20]. Amino acid (aa) exchanges in the quinolone resistance-determining region (QRDR) of the H. pylori gyrA gene at codons 87 and 91 (positions according to H. pylori reference strain 26695; corresponds to codons 83 and 87 in Escherichia coli) alone or in combination with mutations in gyrB have been reported to lead to resistance to fluoroquinolones [10,11,21]. Rifampicin resistance in H. pylori has been associated with aa exchanges in the rifampicin resistance-determining region (RRDR) of rpoB, mainly at codons 525 to 545, 547, and 586 (positions according to H. pylori reference strain 26695; corresponds to codons 512 to 573 in Escherichia coli) [22]. The mechanism of resistance to metronidazole is less clear. Frameshift mutations and truncations in genes that encode electron transfer proteins, such as the nicotinamide adenine dinucleotide phosphate hydrogen (NADPH)-flavin nitroreductase (frxA) and oxygen-insensitive NADPH nitroreductase (rdxA), have been implicated in elevated MIC values to metronidazole [23,24]. Thus far, it is unclear, whether elevated metronidazole MIC values lead to therapeutic failure [25]. For tetracycline, one resistance mechanism appears to be single, double, or triple base pair substitutions in the primary binding site of tetracycline (at nucleotide positions 926 to 928) in the 16S rRNA gene [26].
Here, we report on the first sequencing study that systematically applied WGS to H. pylori clinical isolates to detect specific point mutations in the 23S rRNA, gyrA, rpoB, frxA, rdxA, and 16S rRNA genes. We aimed at correlating the occurrence of SNPs in these target genes to phenotypic DST results for each drug investigated, thereby allowing the calculation of predictive sensitivities and specificities of each set of SNPs.
2. Materials and Methods
2.1. Clinical H. pylori Isolates and H. pylori Culture
This study was conducted using a set of 140 H. pylori strains from the bacterial strain collection of the Institute of Medical Microbiology (IMM), University of Zurich. The H. pylori strains were isolated between 2013 and 2017 from gastric biopsy specimens that were sent to the IMM for culture-based phenotypic DST. For this study, H. pylori strains were intentionally selected based on their antimicrobial resistance phenotype and are therefore not representative of H. pylori primary antibiotic resistance epidemiology in Switzerland. After thawing, the strains were incubated on in-house produced Brucella agar plates (with 5% horse blood) for 3 days at 37 °C under microaerobic conditions (90% N2, 5% CO2, 5% O2) using a gas generator (CampyGen, Thermo Scientific, Waltham, MA, USA). After 3 days, H. pylori isolates were subcultured on Brucella agar plates to obtain sufficient biomass for the E-Test® (bioMérieux, Marcy l’Etoile, France) and to perform DNA extraction for WGS.
2.2. Phenotypic DST by E-Test®
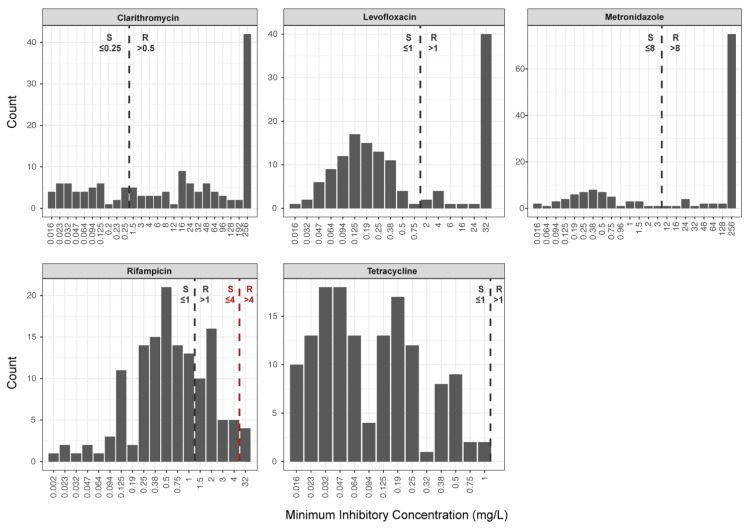
H. pylori cultures were adjusted to a McFarland standard of 3 [27]. Phenotypic DST was performed on Mueller Hinton agar plates containing 5% horse blood (bioMérieux), using the following E-Tests® (bioMérieux): Clarithromycin (0.016–256 mg/L), metronidazole (0.016–256 mg/L), levofloxacin (0.016–32 mg/L), rifampicin (0.016–32 mg/L), and tetracycline (0.016–32 mg/L). Agar plates were incubated under microaerobic conditions at 37 °C for 3 days. Subsequently, MICs were determined using a light microscope (Leica M80, Leica Microsystems, Heerbrugg, Switzerland). Susceptibility interpretation was performed according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [28], except for rifampicin, for which the susceptibility interpretation (MIC: S ≤ 4; R > 4) was done according to Hays et al. [22] and the Comité de l’Antibiogramme de la Société Française de Microbiologie (CASFM)/EUCAST recommendations for H. pylori [29] (Figure 1).
Figure 1.
Antimicrobial drug susceptibility determined by E-Test® in 140 clinical H. pylori isolates selected from the bacterial strain collection of the Institute of Medical Microbiology, University of Zurich, Switzerland. Clinical breakpoints issued by EUCAST [28] are displayed for all five drugs (black dashed line). For rifampicin, the clinical breakpoint of 4 mg/L [22,29] is displayed as a red dashed line; R denotes resistant and S denotes susceptible.
2.3. DNA Extraction, Library Preparation, and WGS of H. pylori Strains
DNA extraction from H. pylori cultures was performed with the DNeasy® UltraClean® Microbial kit (Qiagen, Hilden, Germany), following the producers’ recommendations. Library preparation was done using the Qiagen® QIAseq FX DNA kit (Qiagen, Hilden, Germany), according to the producers’ recommendations. Sequencing library quality and size distribution were analysed on a fragment analyser automated CE system (Advanced Analytical Technologies Inc., Heidelberg, Germany), according to the manufacturers’ instructions using the fragment analyser 474 HS next generation sequencing (NGS) kit. Sequencing libraries were pooled in equimolar concentrations and paired-end sequenced (2 × 150 bp) on an Illumina MiSeq platform (Illumina®, San Diego, CA, USA).
2.4. Bioinformatic Analysis
Raw sequencing reads (fastq) were filtered and trimmed using the FASTQ trimmer tool of the FASTX-Toolkit (Hannon Lab, Cold Spring Harbour Laboratories, Cold Spring Harbor, NY, USA) applying a threshold PHRED score of 25. To identify SNPs in genes conferring resistance, fastq files were analysed using the ARIBA pipeline [30], querying a custom-made database of gene sequences derived from the H. pylori reference strain 26695 (NCBI reference sequence: NC_000915.1). For the evaluation of metronidazole resistance, all SNPs in rdxA, frxA, mdaB, omp11, and rpsU that occurred more frequently in metronidazole resistant than susceptible H. pylori isolates were assessed.
In order to identify probable plasmid-encoded resistance genes, trimmed and filtered FASTQ sequencing files were analysed using the plasmidSPAdes pipeline (Version 3.10.1; [31]). Obtained contigs were used to perform a blastn search querying the NCBI Nucleotide database.
2.5. Discrepancy Analysis
H. pylori isolates showing discrepant phenotypic DST and WGS results were sent to the French National Reference Centre for Campylobacter and Helicobacter (Bordeaux, France) (www.cnrch.fr) for repetition of phenotypic DST. Briefly, H. pylori isolates were adjusted to a McFarland of 3 and grown on in-house produced Mueller Hinton agar plates (with 10% sheep blood and globular extract) for 48 h at 35 °C in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) in a special workstation (Baker, Ruskin, Concept, Bridget, UK). After 48 h, MICs were determined by E-Test® (bioMérieux), and antibiotic susceptibilities were assessed based on the CASFM/EUCAST recommendations for H. pylori [29].
In addition, all discrepant H. pylori strains were grown on in-house produced Brucella agar plates (with 5% horse blood), and DNA extraction for WGS was repeated at the Institute of Medical Microbiology, Zurich, Switzerland.
2.6. Statistics and Data Analysis
We used the 2 × 2 contingency table to calculate the agreement between WGS and phenotypic DST results [32,33]. In this context, specificity describes the likelihood that SNPs identified in a target gene lead to phenotypic drug resistance, while sensitivity describes the proportion of resistant cases that are identified through that set of SNPs. A major error occurred if a H. pylori strain that was identified as susceptible by phenotypic DST was determined resistant by WGS. In this case, an alternative antimicrobial regimen would have been prescribed to the patient, though first line treatment could be administered. A very major error occurred if an H. pylori strain that was identified as resistant by phenotypic DST was determined susceptible by WGS. In this case, the organism would be falsely assessed as susceptible, resulting in the administration of an ineffective first-line antibiotic therapy. A chi-square test (X2) of independence was performed to examine the relationship between the occurrence of certain SNPs and the antibiotic resistance phenotype. All statistics and data visualization was done in R [34].
2.7. Declarations
2.7.1. Ethics Approval and Consent to Participate
The study was conducted according to good laboratory practice and in accordance with the Declaration of Helsinki and national and institutional standards. The Swiss act on medical research involving human subjects does not apply to this study as solely bacterial strains and no human health-related data were used in this study.
2.7.2. Consent for Publication
No consent for publication is required. The Swiss act on medical research involving human subjects do not apply to this study as solely bacterial strains and no human health-related data were used.
3. Results
3.1. Occurrence of Plasmids Carrying Resistance Genes in H. pylori
In the 140 analysed H. pylori genomes, we did not detect a single plasmid encoding a resistance gene. This points towards the primary importance of SNPs in target genes in mediating phenotypic drug resistance in H. pylori and facilitates the prediction of resistance phenotypes.
3.2. Coherence in Prediction of Clarithromycin Resistance
Forty-three H. pylori strains showed a clarithromycin susceptible phenotype and, accordingly, displayed no mutations in domain V of the 23S rRNA gene at nucleotide position A2146 or A2147. Two of the 43 susceptible H. pylori isolates (with MICs of 0.016 and 0.023 mg/L) carried a G2224A mutation, which does not cause macrolide resistance [35].
One isolate carried a T2182C mutation and exhibited a low-level clarithromycin resistance with an MIC of 1.5 mg/L. As the relevance of the T2182C mutation in clarithromycin resistance is questionable [18], this strain was sent for retesting to the H. pylori reference centre in Bordeaux, where an MIC of 0.5 mg/L was determined (Table S1). Thus, initially, phenotypic DST falsely identified this H. pylori strain as clarithromycin resistant, representing a major error.
Two H. pylori strains initially showed an MIC of 0.19 and 0.5, but carried an A2146G and an A2147G mutation, respectively. Repetition of the E-Test® eventually revealed a mixed population of clarithromycin susceptible and resistant H. pylori strains (0.19 and 6; 0.5 and >256). These two samples were scored as clarithromycin resistant H. pylori strains for further statistical analysis (Table 1 and Table 2).
Table 1.
Point mutations identified by WGS in the 23S rRNA gene at nucleotide positions, A2146 and A2147, in 96 clarithromycin resistant H. pylori isolates and their corresponding MICs.
| Number of Clinical H. pylori Strains | MIC of Clarithromycin (mg/L) | 23S rRNA Mutation a |
|---|---|---|
| 1 | >256 | A2146C |
| 28 | 8 to 256 | A2146G |
| 67 | 1.5 to 256 | A2147G |
a Nucleotide exchange relative to H. pylori reference strain 26695 sequence. WGS: whole genome sequencing MICs: minimum inhibitory concentrations.
Table 2.
Agreement between phenotypic DST results and identified point mutations in the 23S rRNA gene at nucleotide positions, A2146 and A2147, conferring resistance to macrolides (N = 140).
| Phenotypic DST | |||
|---|---|---|---|
| Susceptible | Resistant | ||
| WGS | Susceptible a | 43 | 1 |
| Resistant b | 0 | 96 | |
a no mutation present at nucleotide positions, A2146 and A2147, in the 23S rRNA gene. b A mutation present at nucleotide positions, A2146 and A2147, in the 23S rRNA gene. DST: drug susceptibility testing.
Thus, in total, 96 H. pylori isolates were determined as clarithromycin resistant by phenotypic DST, and 28 A2146G (29%), 67 A2147G (70%), and one A2146C (1%) mutation were found (Table 1 and Table 2). Moreover, different point mutations resulted in different levels of clarithromycin resistance. While A2147G isolates showed MICs of 1.5 to >256, strains harbouring either the A2146G or the A2142C point mutation always displayed high clarithromycin resistance levels (MIC of 8 to ≥256 mg/L) (Table 1, Figure S1 Panel A).
Overall, the congruence between the phenotypic DST and WGS results was 99%.
Specificity, i.e., the likelihood that the presence of a mutation at nucleotide position A2146 and A2147 in the 23S rRNA gene resulted in a clarithromycin resistant phenotype, was 100%. Sensitivity, which describes the proportion of resistant isolates that were identified through the presence of a mutation at nucleotide positions, A2146 and A2147, was also 100%. Thus, the presence of point mutations, A2146C, A2146G, and A2147G, in the 23S rRNA gene of H. pylori could be significantly related to phenotypic clarithromycin resistance (X2 [1, N = 140] = 135.4, p < 0.001).
3.3. Coherence in Prediction of Metronidazole Resistance
Frameshift mutations and truncations in the genes, rdxA or frxA, are generally supposed to lead to metronidazole resistance in H. pylori [24]. Some frameshift mutations (occurring at codon positions, 105, 149, or 192, in frxA and 18, 38, and 112 in rdxA) were only detected in metronidazole resistant H. pylori isolates (Tables S2 and S3). Other frameshift mutations (occurring at codon positions 18 in frxA and 62, 96, and 162 in rdxA) occurred in both metronidazole susceptible and resistant H. pylori strains (Tables S2 and S3). SNPs resulting in the aa exchanges, A67V, A68E, K64N, P106S, and R16C, merely occurred in the rdxA gene of metronidazole resistant H. pylori strains (Table S4). All frameshift mutations and aa exchanges detected in rdxA and frxA of the H. pylori isolates and their corresponding metronidazole MIC are displayed in Tables S5 and S6. Interestingly, we did not detect any published SNPs associated with phenotypic metronidazole resistance within mdaB, omp11, or rpsU that were more prevalent in metronidazole resistant compared to susceptible H. pylori isolates.
3.4. Coherence in Prediction of Levofloxacin Resistance
In 90 of the 91 levofloxacin susceptible H. pylori isolates, the gyrA QRDR carried no SNP at codon 87 or 91. One H. pylori strain harboured an N87T aa exchange that has not been associated with levofloxacin resistance [11]. In 46 fluoroquinolone resistant H. pylori strains, WGS identified SNPs at codon 87 (N = 25; 54%), codon 91 (N = 19, 41%), and at codon 87 and 91 (N = 2; 5%) (Table 3 and Table 4). Single aa exchanges at codon 87 or 91 led to a relatively broad range of levofloxacin MICs (4 to 32 mg/L). High-level resistance (i.e., MIC > 32 mg/L) was exclusively found in the presence of aa exchanges at codons 87 and 91 (Table 3).
Table 3.
Amino acid exchanges identified by WGS in the QRDR of the gyrA gene at codon 87 and/or 91 in 46 levofloxacin resistant H. pylori isolates and their corresponding MICs.
| Number of Clinical H. pylori Strains | MIC of Levofloxacin (mg/L) | gyrA aa Exchange a |
|---|---|---|
| 1 | >32 | N87I |
| 22 | >32 | N87K |
| 2 | 4 to 32 | N87Y |
| 8 | 16 to 32 | D91G |
| 5 | 4 to 32 | D91N |
| 6 | 4 to 32 | D91Y |
| 2 | >32 | N87K D91N |
a Amino acid exchange relative to H. pylori reference strain 26,695 sequence. QRDR: Quinolone resistance determinig region.
Table 4.
Agreement between phenotypic DST results and identified aa exchanges in the gyrA gene at codon 87 and/or 91 conferring resistance to fluoroquinolones (N = 138).
| Phenotypic DST | |||
|---|---|---|---|
| Susceptible | Resistant | ||
| WGS | Susceptible a | 91 | 1 |
| Resistant b | 0 | 46 | |
a no aa exchange present in the QRDR of the gyrA gene at codon 87 and/or 91. b An aa exchange present in the QRDR of the gyrA gene at codon 87 and/or 91.
Three strains were phenotypically resistant to levofloxacin with MICs of 2 and 4 mg/L, respectively, but showed no SNPs in the gyrA QRDR (Figure S1, Panel B). Unfortunately, two of the three H. pylori strains showing discrepant results could not be regrown for discrepancy analysis. Thus, we were not able to verify the correctness of phenotypic DST or WGS and decided to subsequently exclude these two H. pylori strains from the study. The third H. pylori strain could be regrown and was sent for retesting to the H. pylori reference centre in Bordeaux. There, it showed an MIC of 1.5 mg/L (Table S1). The gyrB gene of this discrepant H. pylori isolate was analysed as well and showed multiple unpublished SNPs (compared to the gyrB sequence of the H. pylori reference strain 26695; Table S7).
The congruence between the phenotypic DST and WGS results was 99%. Specificity, i.e., the likelihood that aa exchanges in the QRDR of the gyrA gene at codon 87 and 91 confer levofloxacin resistance, was 100%. Sensitivity, i.e., the proportion of resistant cases that were identified through the presence of specific aa exchanges at codon 87 and 91 in the gyrA gene, was 98%. Overall, one major error occurred: One H. pylori isolate tested levofloxacin resistant by phenotypic DST was categorized as susceptible by WGS. Nevertheless, specific aa exchanges at codon 87 (N87I, N87K and N87Y) and 91 (D91N and D91Y) of the gyrA gene in H. pylori can be used to predict phenotypic levofloxacin resistance (X2 [1, N = 138] = 136.0, p < 0.001).
3.5. Coherence in Prediction of Rifampicin Resistance
All 136 rifampicin susceptible H. pylori isolates (MIC ≤ 4 mg/L) did not show an aa exchange in the RRDR of the rpoB gene. The four phenotypic resistant H. pylori isolates with MICs >32 mg/L had either an H540N (N = 2) or an L525P (N = 2) aa exchange (Table 5 and Table 6). Overall, an agreement of 100% was found between the WGS and phenotypic rifampicin susceptibility results.
Table 5.
Amino acid exchanges identified by WGS in the RRDR of the rpoB gene conferring resistance to rifamycins and their corresponding MICs.
| Number of Clinical H. pylori Strains | MIC of Levofloxacin (mg/L) | rpoB aa Exchange a |
|---|---|---|
| 2 | >32 | H540N |
| 2 | >32 | L525P |
a Amino acid exchange relative to H. pylori reference strain 26,695 sequence.
Table 6.
Agreement between phenotypic DST results and identified aa exchanges in the RRDR of the rpoB gene conferring resistance to rifamycins (N = 140).
| Phenotypic DST | |||
|---|---|---|---|
| Susceptible | Resistant | ||
| WGS | Susceptible a | 136 | 0 |
| Resistant b | 0 | 4 | |
a No aa exchange present in the RRDR of the rpoB gene. b An aa exchange present in the RRDR of the rpoB gene.
3.6. Coherence in Prediction of Tetracycline Resistance
All 140 H. pylori strains included in this study were tetracycline susceptible in phenotypic DST. In two isolates with MICs of 0.25 and 0.5 mg/L, an A926T mutation was detected in the 16S rRNA gene. One H. pylori isolate showed an A926G mutation with an MIC of 0.032 mg/L, and one carried an A928C mutation with an MIC of 0.125 mg/L (Table 7). It has been previously reported that an A926G or an A928C nucleotide exchange was associated with a broad range of MICs, ranging from susceptible (0.75 mg/L) to low level resistance (4 mg/L), whereas double mutants displayed MICs of >4 mg/L [36,37].
Table 7.
Point mutations identified by WGS in the primary binding site of tetracycline in the 16S rRNA gene at nucleotide positions, 926 to 928, and their corresponding MICs.
| Number of Clinical H. pylori Strains | MIC of Levofloxacin (mg/L) | 16S rRNA Mutation a |
|---|---|---|
| 2 | 0.25 to 0.5 | A926T |
| 1 | 0.032 | A926G |
| 1 | 0.125 | A928C |
a Nucleotide exchange relative to H. pylori reference strain 26,695 sequence.
4. Discussion
In the present study, we have applied massive parallel Illumina-based WGS on clinical H. pylori isolates to define the relationship between the occurrence of SNPs in selected target genes and phenotypic antimicrobial susceptibility. We could show a clear correlation between the occurrence of point mutations in the 23S rRNA, gyrA, and rpoB genes of H. pylori and macrolide, fluoroquinolone, and rifamycin resistance, respectively. To our knowledge, this is the first study showing that genetic determinants of antimicrobial resistance identified by WGS can be used for the prediction of drug resistance phenotypes in H. pylori. In contrast, there was no clear association between identified SNPs in frxA and rdxA and phenotypic metronidazole resistance.
During this study, we realized that assessment of antimicrobial susceptibility in H. pylori by culture based phenotypic DST can be challenging due to the pathogen’s fastidious growth requirements. Moreover, it was not always possible to determine an explicit MIC of a drug by E-Test® because of the small and transparent colonies that H. pylori forms. Variations in the redox potential of the test medium caused a broad range of MIC distributions, especially for metronidazole, when performing the E-Test®, thereby making standardized and reproducible interpretation difficult. Phenotypic DST may also be challenged when a mixed population of resistant and susceptible H. pylori strains is present in the same patient [38]. In two samples included in this study, phenotypic DST initially did not detect the resistant sub-population. This is problematic, as antimicrobial therapy is based on in vitro phenotypic susceptibly information. In contrast, the presence of susceptible and resistant H. pylori subpopulations can be predicted from the WGS sequencing data through the occurrence of hetero-resistance at specific nucleotide positions [38].
Molecular based methods, like polymerase chain reaction (PCR) and line probe assays, that enable specific detection of point mutations in the 23S rRNA (nucleotide positions, A2146 and A2147) or gyrA gene (codons 87 and 91) can be directly applied on clinical specimens [39,40]. Our data indicate that molecular assays targeting these point mutations would be sufficient to monitor clarithromycin and levofloxacin resistance in H. pylori. However, one advantage of WGS is that it delivers a more complete picture of resistance determinants present in a clinical isolate than targeted molecular approaches that can only examine a limited number of nucleotide positions. Thus, false negative results may occur, when new polymorphisms, potentially conferring drug resistance, arise that are not covered by the assay. In contrast, the relevance of these polymorphisms can easily be assessed by retrospective analysis of WGS data, whereas targeted molecular assays would need to be redesigned and samples retested.
In this study, we found clarithromycin resistance in H. pylori to be highly correlated with the presence of mutations A2146C, A2146G or A2147G in the domain V of the 23S rRNA gene. Recent studies reported additional point mutations outside the domain V and an active drug efflux mechanism to be involved in clarithromycin resistance [35,41]. Also, mutations in other target genes, like rpl22 (encodes a ribosomal protein that interacts with the 23S rRNA domains) and infB (encodes translation initiation factor, IF-2), were identified to induce low level clarithromycin resistance (MICs of 0.5 to 4 mg/L). Interestingly, these mutations led to a high level of clarithromycin resistance (MICs >256 mg/L) in combination with A2146 and A2147 mutations [42]. In a recent study, it was reported that mutations in multidrug efflux transporter genes may be involved in clarithromycin resistance [43]. In this study, however, we found that the occurrence of point mutations at nucleotide positions, A2146 and A2147, were sufficient for the reliable prediction of phenotypic clarithromycin resistance in H. pylori.
For metronidazole, there was no clear correlation between the isolates’ observed phenotype and their genotype. Firstly, frameshift mutations in rdxA and frxA that were reported to confer metronidazole resistance were also found in H. pylori strains assessed as metronidazole susceptible by phenotypic DST. Secondly, in contrast to recent reports suggesting that metronidazole resistance requires mutations in both genes, frxA and rdxA, one isolate was found to be resistant (MIC of 26 mg/L) though carrying only a mutation in frxA [44,45]. Some rdxA and frxA mutations were identified in metronidazole resistant H. pylori isolates; however, several metronidazole resistant strains harboured no mutations at all in rdxA and/or frxA. Recently, it has been reported that additional genes, such as mdaB, omp11, and rpsU, may be involved in metronidazole resistance in H. pylori [46,47]. However, consensual data are still lacking and a relationship between SNPs detected in one of these five genes (rdxA, frxA, mdaB, omp11, and rpsU) and phenotypic metronidazole resistance was not found in the H. pylori isolates investigated in this study. In sum, it remains difficult to predict metronidazole susceptibility based merely on genotypic data, as the correlation between detected SNPs and the phenotype to date is very poor.
On the basis of the presented data, we therefore suggest to reconsider the usefulness of phenotypic metronidazole susceptibility testing in H. pylori as it is stated in the CASFM/EUCAST recommendations for H. pylori [29]. Moreover, in vitro and in vivo resistance data are not congruent [48] as H. pylori strains tested resistant in vitro can still be eradicated with a combination therapy that contains an increased dosage of metronidazole [25].
Resistance to fluoroquinolones could be attributed to mutations in the QRDR of gyrA at codons 87 and 91 in 98% of the fluoroquinolone resistant H. pylori strains. One levofloxacin resistant H. pylori strain with an MIC of 1.5 mg/L showed no codon exchange in the QRDR of gyrA. The relevance of mutations in other target genes, like gyrB, in quinolone resistant H. pylori isolates without gyrA mutations is not consensual [49,50]. The aforementioned isolate harboured various SNPs in gyrB; however, none matched any mutations recently reported to be associated with quinolone resistance in H. pylori [49,51]. Notably, H. pylori lacks genes encoding topoisomerase IV (encoded by parC and parE genes), which is targeted by quinolones in other bacteria [52]. Moreover, active efflux pumps seem not to be involved in mediating fluoroquinolone resistance [53].
In our study, H. pylori strains with a rifampicin susceptible phenotype (i.e., MICs ≤ 4 mg/L) showed no mutation in the RRDR of rpoB. Amino acid substitutions, L525P and H540N, were associated with resistance and MICs of >32 mg/L. Noteworthy, application of the current EUCAST clinical breakpoint for rifampicin of 1 mg/L would split the wild-type population (Figure 1). This in turn introduces serious interpretation problems, since some H. pylori strains that do not carry point mutations in the RRDR of rpoB would be classified as rifampicin resistant, representing a major error. Given the rifampicin MIC distribution of H. pylori isolates analysed in this study, we recommend using the clinical breakpoint of 4 mg/L proposed by Hays et al. [22] and the CASFM/EUCAST guidelines [29].
Findings gained in this study can be useful for future research that may aim at performing culture-independent WGS directly from clinical specimens for the prediction of phenotypic drug susceptibility in H. pylori. Some studies have already successfully applied WGS directly on gastric biopsies for the detection of H. pylori [54,55]. Moreover, in-house developed and commercial protocols (e.g., [56,57]) are becoming available for the depletion of human DNA or the enrichment of bacterial DNA prior to performing WGS, thereby increasing the efficiency and cost-effectiveness of NGS due to less human DNA background in samples.
Our study has several limitations: It was designed as a single centre laboratory-based study using clinical H. pylori isolates. For some antibiotics, there were only a few (i.e., rifampicin) or no resistant H. pylori isolates (i.e., tetracycline) available. Thus, the analytical sensitivity of WGS in determining phenotypic rifampicin resistance based on mutations in the RRDR of rpoB might have been overestimated as rifampicin resistance has also been reported in H. pylori isolates lacking these mutations [8]. For tetracycline, calculation of the analytical sensitivity of WGS was not possible due to a lack of resistant H. pylori isolates. Moreover, genotype-based prediction of tetracycline resistance in H. pylori is additionally hampered since isolates without mutations at nucleotide positions, 926 to 928, in the 16S rRNA gene display a resistant phenotype [58]. Therefore, tetracycline resistance seems to be multifactorial, involving alterations in ribosomal binding, enzymatic degradation of antibiotics, a reduction of membrane permeability, and an active efflux [59,60,61].
In this study, we have not examined the possible role of active efflux mechanisms. Their involvement in intrinsic antibiotic resistance in H. pylori is still a matter of debate and would require transcriptional analyses of the genes encoding drug efflux systems (e.g., by using a meta-transcriptomics approach), which was beyond the scope of this study.
5. Conclusions
It is evident that increasing antimicrobial resistance represents a significant challenge in the successful management of H. pylori infections. This highlights the importance of rapid determination of drug susceptibility in tailoring treatments to increase H. pylori eradication success. In addition to culture based phenotypic DST, high-throughput sequencing is rapidly changing the landscape in medical microbiology. In this study, we could demonstrate that genetic determinants identified by WGS in the H. pylori genome are significantly correlated to phenotypic drug resistance. This allows the prediction of phenotypic clarithromycin, rifamycin, and levofloxacin resistance based on genotypic information in the 23S rRNA, gyrA, and rpoB genes with high specificity and good sensitivity in a clinically relevant timeframe.
Acknowledgments
We thank the technicians of the Institute of Medical Microbiology for expert help and assistance, especially Susanne Blasowitsch and Valeria Pereira Pires. We thank the Institute of Medical Microbiology, University of Zurich, for continuous support. We are grateful to Erik Christian Böttger and Patrice Courvalin for fruitful discussion.
Supplementary Materials
The following are available online at http://www.mdpi.com/2077-0383/8/1/53/s1. Table S1. H. pylori isolates showing discrepant phenotypic DST and WGS results were sent to the H. pylori reference centre in Bordeaux, France, for culture based phenotypic DST. Table S2. Comparison of frameshift mutations and SNPs detected in the frxA gene of metronidazole susceptible and resistant H. pylori isolates. Table S3. Comparison of frameshift mutations and truncations detected in the rdxA gene of metronidazole susceptible and resistant H. pylori isolates. Table S4. Comparison of SNPs detected in the rdxA gene of metronidazole susceptible and resistant H. pylori isolates. Table S5. List of all frameshift mutations and SNPs detected in the frxA gene of metronidazole susceptible and resistant H. pylori isolates. Table S6. List of all frameshift mutations and SNPs detected in the rdxA gene of metronidazole susceptible and resistant H. pylori isolates. Table S7. List of all SNPs detected in the gyrA and gyrB genes of the discrepant H. pylori isolate showing a levofloxacin resistant phenotype. Figure S1. Association between phenotypic drug resistance and mutations in the 23S rRNA gene at nucleotide positions A2146 and A2147 (panel A) and mutations in the gyrA QRDR (panel B). One discrepant H. pylori isolate showing phenotypic levofloxacin resistance without mutations in the gyrA QRDR is displayed in red. Availability of Data and Material: Whole genome sequences of the H. pylori strains analyzed in this study are available on NCBI under accession numbers.
Author Contributions
F.N.L., F.I., P.M.K. and K.W. designed the study. F.N.L., P.L., R.Z., A.B., L.B. and K.W. were responsible for the microbiological work-up of the samples. F.N.L., F.I., P.M.K., P.L. and K.W. did the data mining and analysed the data. F.N.L., F.I., P.M.K. and K.W. wrote the initial draft of the manuscript. All authors reviewed and edited the manuscript.
Funding
This study was funded by the Institute of Medical Microbiology, University of Zurich.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Thaker Y., Moon A., Afzali A. Helicobacter pylori: A review of epidemiology, treatment, and management. J. Clin. Gastroenterol. Treat. 2016;2:1–5. doi: 10.23937/2469-584X/1510019. [DOI] [Google Scholar]
- 2.Fischbach W., Goebeler-Kolve M., Dragosics B., Greiner A., Stolte M. Long term outcome of patients with gastric marginal zone B cell lymphoma of mucosa associated lymphoid tissue (MALT) following exclusive Helicobacter pylori eradication therapy: Experience from a large prospective series. Gut. 2004;53:34–37. doi: 10.1136/gut.53.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukase K., Kato M., Kikuchi S., Inoue K., Uemura N., Okamoto S., Terao S., Amagai K., Hayashi S., Asaka M., et al. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: An open-label, randomised controlled trial. Lancet. 2008;372:392–397. doi: 10.1016/S0140-6736(08)61159-9. [DOI] [PubMed] [Google Scholar]
- 4.Gisbert J., Gonzalez L., Calvet X., García N., López T., Roque M., Gabriel R., Pajares J. Proton pump inhibitor, clarithromycin and either amoxycillin or nitroimidazole: A meta-analysis of eradication of Helicobacter pylori. Aliment. Pharmacol. Ther. 2000;14:1319–1328. doi: 10.1046/j.1365-2036.2000.00844.x. [DOI] [PubMed] [Google Scholar]
- 5.Malfertheiner P., Megraud F., O’Morain C., Gisbert J., Kuipers E., Axon A., Bazzoli F., Gasbarrini A., Atherton J., Graham D., et al. Management of Helicobacter pylori infection—The Maastricht V/Florence consensus report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 6.Dore M.P., Leandro G., Realdi G., Sepulveda A.R., Graham D.Y. Effect of pretreatment antibiotic resistance to metronidazole and clarithromycin on outcome of Helicobacter pylori therapy. Dig. Dis. Sci. 2000;45:68–76. doi: 10.1023/A:1005457226341. [DOI] [PubMed] [Google Scholar]
- 7.Buzás G. First-line eradication of Helicobacter pylori: Are the standard triple therapies obsolete? A different perspective. World J. Gastroenterol. 2010;16:3865–3870. doi: 10.3748/wjg.v16.i31.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wueppenhorst N., Stueger H.P., Kist M., Glocker E. Identification and molecular characterization of triple-and quadruple-resistant Helicobacter pylori clinical isolates in Germany. J. Antimicrob. Chemother. 2009;63:648–653. doi: 10.1093/jac/dkp003. [DOI] [PubMed] [Google Scholar]
- 9.Gisbert J.P., Bermejo F., Castro-Fernández M., Pérez-Aisa A., Fernández-Bermejo M., Tomas A., Barrio J., Bory F., Almela P., Sánchez-Pobre P., et al. Second-line rescue therapy with levofloxacin after H. pylori treatment failure: A Spanish multicenter study of 300 patients. Am. J. Gastroenterol. 2008;103:71–76. doi: 10.1111/j.1572-0241.2007.01500.x. [DOI] [PubMed] [Google Scholar]
- 10.Bogaerts P., Berhin C., Nizet H., Glupczynski Y. Prevalence and mechanisms of resistance to fluoroquinolones in Helicobacter pylori strains from patients living in Belgium. Helicobacter. 2006;11:441–445. doi: 10.1111/j.1523-5378.2006.00436.x. [DOI] [PubMed] [Google Scholar]
- 11.Cattoir V., Nectoux J., Lascols C., Deforges L., Delchier J.C., Megraud F., Soussy C.J., Cambau E. Update on fluoroquinolone resistance in Helicobacter pylori: New mutations leading to resistance and first description of a gyrA polymorphism associated with hypersusceptibility. Int. J. Antimicrob. Agents. 2007;29:389–396. doi: 10.1016/j.ijantimicag.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Glocker E., Bogdan C., Kist M. Characterization of rifampicin-resistant clinical Helicobacter pylori isolates from Germany. J. Antimicrob. Chemother. 2007;59:874–879. doi: 10.1093/jac/dkm039. [DOI] [PubMed] [Google Scholar]
- 13.Hombach M., Zbinden R., Böttger E.C. Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witney A.A., Cosgrove C.A., Arnold A., Hinds J., Stoker N.G., Butcher P.D. Clinical use of whole genome sequencing for Mycobacterium tuberculosis. BMC Med. 2016;14 doi: 10.1186/s12916-016-0598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker T.M., Kohl T.A., Omar S.V., Hedge J., Elias C.D.O., Bradley P., Iqbal Z., Feuerriegel S., Niehaus K.E., Wilson D.J., et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: A retrospective cohort study. Lancet Infect. Dis. 2015;15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerrits M.M., van Vliet A.H., Kuipers E.J., Kusters J.G. Helicobacter pylori and antimicrobial resistance: Molecular mechanisms and clinical implications. Lancet Infect. Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 17.Versalovic J., Shortridge D., Kibler K., Griffy M.V., Beyer J., Flamm R.K., Tanaka S.K., Graham D.Y., Go M.F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 1996;40:477–480. doi: 10.1128/AAC.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burucoa C., Landron C., Garnier M., Fauchère J.L. T2182C mutation is not associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2005;49:868–870. doi: 10.1128/AAC.49.2.868-870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J.M., Kim J.S., Kim N., Kim Y.J., Kim I.Y., Chee Y.J., Lee C.H., Jung H.C. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J. Microbiol. Biotechnol. 2008;18:1584–1589. [PubMed] [Google Scholar]
- 20.Rimbara E., Noguchi N., Kawai T., Sasatsu M. Novel mutation in 23S rRNA that confers low-level resistance to clarithromycin in Helicobacter pylori. Antimicrob. Agents Chemother. 2008;52:3465–3466. doi: 10.1128/AAC.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L.H., Cheng H., Hu F.L., Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World J. Gastroenterol. 2010;16:2272–2277. doi: 10.3748/wjg.v16.i18.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hays C., Burucoa C., Lehours P., Tran C.T., Leleu A., Raymond J. Molecular characterization of Helicobacter pylori resistance to rifamycins. Helicobacter. 2018;23 doi: 10.1111/hel.12451. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.M., Kim N., Kwon Y.H., Nam R.H., Kim J.M., Park J.Y., Lee Y.S., Lee D.H. rdxa, frxa, and efflux pump in metronidazole-resistant Helicobacter pylori: Their relation to clinical outcomes. J. Gastroenterol Hepatol. 2018;33:681–688. doi: 10.1111/jgh.13906. [DOI] [PubMed] [Google Scholar]
- 24.Masaoka T., Suzuki H., Kurabayashi K., Nomoto Y., Nishizawa T., Mori M., Hibi T. Could frameshift mutations in the frxA and rdxA genes of Helicobacter pylori be a marker for metronidazole resistance? Aliment. Pharmacol. Ther. Symp. Ser. 2006;2:81–87. doi: 10.1111/j.1746-6342.2006.00029.x. [DOI] [Google Scholar]
- 25.Romano M., Cuomo A. Eradication of Helicobacter pylori: A clinical update. MedGenMed. 2004;6:19. [PMC free article] [PubMed] [Google Scholar]
- 26.Gerrits M.M., Berning M., van Vliet A.H., Kuipers E.J., Kusters J.G. Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2003;47:2984–2986. doi: 10.1128/AAC.47.9.2984-2986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFarland J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. JAMA. 1907;49:1176–1178. doi: 10.1001/jama.1907.25320140022001f. [DOI] [Google Scholar]
- 28.EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.0. [(accessed on 5 February 2018)];2018 Available online: http://www.eucast.org.
- 29.CASFM/EUCAST. [(accessed on 5 February 2018)];2018 Available online: http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFMV1_0_MARS_2018.pdf.
- 30.Hunt M., Mather A.E., Sánchez-Busó L., Page A.J., Parkhill J., Keane J.A., Harris S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microbial. Genom. 2017;3 doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antipov D., Hartwick N., Shen M., Raiko M., Lapidus A., Pevzner P. plasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 32.Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 33.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 34.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [(accessed on 6 January 2019)]. Available online: http://www.R-project.org. [Google Scholar]
- 35.Hao Q., Li Y., Zhang Z.J., Liu Y., Gao H. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J. Gastroenterol. 2004;10:1075–1077. doi: 10.3748/wjg.v10.i7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson A.J., Elviss N.C., Owen R.J. Real-time PCR detection and frequency of 16S rDNA mutations associated with resistance and reduced susceptibility to tetracycline in Helicobacter pylori from England and Wales. J. Antimicrob. Chemother. 2005;56:282–286. doi: 10.1093/jac/dki199. [DOI] [PubMed] [Google Scholar]
- 37.Seriki A.T., Smith S.I., Adeleye A.I., Fowora M.A. Molecular analysis of low-level tetracycline resistance in clinical isolates of Helicobacter pylori among dyspeptic patients in South-west Nigeria. J. Glob. Antimicrob. Resist. 2018;13:143–145. doi: 10.1016/j.jgar.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., Wang Y., Pang Y., Liu C. Comparison of different drug susceptibility test methods to detect the rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014;58:5632–5635. doi: 10.1128/AAC.02778-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cambau E., Allerheiligen V., Coulon C., Corbel C., Lascols C., Deforges L., Soussy C.J., Delchier J.C., Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J. Clin. Microbiol. 2009;47:3600–3607. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redondo J.J., Keller P.M., Zbinden R., Wagner K. A novel RT-PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn. Microbiol. Infect. Dis. 2018;90:1–6. doi: 10.1016/j.diagmicrobio.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Garrido L., Toledo H. Novel genotypes in Helicobacter pylori involving domain V of the 23S rRNA gene. Helicobacter. 2007;12:505–509. doi: 10.1111/j.1523-5378.2007.00506.x. [DOI] [PubMed] [Google Scholar]
- 42.Binh T.T., Shiota S., Suzuki R., Matsuda M., Trang T.T.H., Kwon D.H., Iwatani S., Yamaoka Y. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J. Antimicrob. Chemother. 2014;69:1796–1803. doi: 10.1093/jac/dku050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J., Ye L., Jin L., Xu X., Xu P., Wang X., Li H. Application of next-generation sequencing to characterize novel mutations in clarithromycin-susceptible Helicobacter pylori strains with A2143G of 23S rRNA gene. Ann. Clin. Microbiol. Antimicrob. 2018;17 doi: 10.1186/s12941-018-0259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Francesco V., Zullo A., Hassan C., Giorgio F., Rosania R., Ierardi E. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World J. Gastrointest. Pathophysiol. 2011;2:35–41. doi: 10.4291/wjgp.v2.i3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon D.H., El-Zaatari F.A., Kato M., Osato M.S., Reddy R., Yamaoka Y., Graham D.Y. Analysis of rdxA and involvement of additional genes encoding NAD (P) H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob. Agents Chemother. 2000;44:2133–2142. doi: 10.1128/AAC.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albert T.J., Dailidiene D., Dailide G., Norton J.E., Kalia A., Richmond T.A., Molla M., Singh J., Green R.D., Berg D.E. Mutation discovery in bacterial genomes: Metronidazole resistance in Helicobacter pylori. Nat. Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- 47.Binh T.T., Suzuki R., Trang T.T.H., Kwon D.H., Yamaoka Y. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob. Agents Chemother. 2015;59:2343–2348. doi: 10.1128/AAC.04852-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Courvalin P., Leclercq R., Rice L.B. Antibiogram. ESKA Pub.; Portland, OR, USA: 2010. 3rd State-of-the Art, International ed. [Google Scholar]
- 49.Miyachi H., Miki I., Aoyama N., Shirasaka D., Matsumoto Y., Toyoda M., Mitani T., Morita Y., Tamura T., Kinoshita S., et al. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter. 2006;11:243–249. doi: 10.1111/j.1523-5378.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 50.Tankovic J., Lascols C., Sculo Q., Petit J.C., Soussy C.J. Single and double mutations in gyrA but not in gyrB are associated with low-and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2003;47:3942–3944. doi: 10.1128/AAC.47.12.3942-3944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rimbara E., Noguchi N., Kawai T., Sasatsu M. Fluoroquinolone resistance in Helicobacter pylori: Role of mutations at position 87 and 91 of gyrA on the level of resistance and identification of a resistance conferring mutation in gyrB. Helicobacter. 2012;17:36–42. doi: 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- 52.Tomb J.F., White O., Kerlavage A.R., Clayton R.A., Sutton G.G., Fleischmann R.D., Ketchum K.A., Klenk H.P., Gill S., Dougherty B.A., et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 53.Bina J., Alm R., Uria-Nickelsen M., Thomas S., Hancock R. Helicobacter pylori uptake and efflux: Basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob. Agents Chemother. 2000;44:248–254. doi: 10.1128/AAC.44.2.248-254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Z., Andersson A.F., Ye W., Nyrén O., Normark S., Engstrand L. A method for metagenomics of Helicobacter pylori from archived formalin-fixed gastric biopsies permitting longitudinal studies of carcinogenic risk. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0026442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C., Cleveland K., Schnoll-Sussman F., McClure B., Bigg M., Thakkar P., Schultz N., Shah M.A., Betel D. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasan M.R., Rawat A., Tang P., Jithesh P.V., Thomas E., Tan R., Tilley P. Depletion of human DNA in spiked clinical specimens to improve the sensitivity of pathogen detection by next generation sequencing. J. Clin. Microbiol. 2016;54:919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thoendel M., Jeraldo P.R., Greenwood-Quaintance K.E., Yao J.Z., Chia N., Hanssen A.D., Abdel M.P., Patel R. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J. Microbiol. Methods. 2016;127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J.Y., Kim J.J., Reddy R., Wang W., Graham D.Y., Kwon D.H. Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob. Agents Chemother. 2005;49:578–583. doi: 10.1128/AAC.49.2.578-583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy S.B. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 1992;36:695–703. doi: 10.1128/AAC.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nikaido H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 61.Putman M., van Veen H.W., Konings W.N. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 2000;64:672–693. doi: 10.1128/MMBR.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.