Abstract
We propose to use segment graph convolutional and recurrent neural networks (Seg-GCRNs), which use only word embedding and sentence syntactic dependencies, to classify relations from clinical notes without manual feature engineering. In this study, the relations between 2 medical concepts are classified by simultaneously learning representations of text segments in the context of sentence syntactic dependency: preceding, concept1, middle, concept2, and succeeding segments. Seg-GCRN was systematically evaluated on the i2b2/VA relation classification challenge datasets. Experiments show that Seg-GCRN attains state-of-the-art micro-averaged F-measure for all 3 relation categories: 0.692 for classifying medical treatment–problem relations, 0.827 for medical test–problem relations, and 0.741 for medical problem–medical problem relations. Comparison with the previous state-of-the-art segment convolutional neural network (Seg-CNN) suggests that adding syntactic dependency information helps refine medical word embedding and improves concept relation classification without manual feature engineering. Seg-GCRN can be trained efficiently for the i2b2/VA dataset on a GPU platform.
Keywords: graph convolutional networks, bidirectional long short-term memory networks, medical relation classification, natural language processing
INTRODUCTION AND RELATED WORK
Relation extraction in biomedical literature and clinical narratives is an important step for downstream tasks, including computational phenotyping, clinical decision making, and trial screening, which has drawn extensive research efforts in recent years.1–5 Participants in the 2010 i2b2/VA challenge and follow-up publications have showcased part of recent progress of classifying relations among medical concepts.6–16 One of the major tasks of the challenge workshop focuses on classifying relations of concept pairs, such as medical treatments–problems, medical tests–problems, and medical problems–problems. The classifiers developed by challenge participants are all equipped with many engineered features, either from customarily constructed regular expressions, from knowledge-based annotations produced by natural language processing (NLP) pipelines including MetaMap17 and cTakes,18 or from annotated and unannotated external data to improve the classification performance.
One significant drawback of the abovementioned participating systems is that they all rely on extensive feature engineering, which does not generalize well to different datasets.19 To reduce the burden of feature engineering and improve the system generalizability, recent studies tackled clinical text modeling using convolutional or recurrent neural networks.15,20–27 These models can navigate the large parameter space to automatically learn feature representations.28 For medical relation classification, Sahu et al.15 applied the convolutional neural networks (CNNs) on the i2b2/VA dataset to learn a sentence-level representation, but their method did not outperform the top challenge participants. Luo et al. proposed segment long short-term memory (Seg-LSTM)21 and Segment CNN (Seg-CNN)26 by observing the need to distinguish the segments that form the relations14,29 (ie, preceding, concept1, middle, concept2, and succeeding), as they play different roles in determining the relation class. Both systems used only word embedding for medical relation classification and modeled segments’ sequences of words using the same order as they appeared in the original sentence. Moreover, Seg-CNN outperformed all i2b2/VA challenge participants, and was comparable to the follow-up study by Zhu et al.30 in overall micro F-measure. However, both Seg-LSTMs and Seg-CNNs are syntax-agnostic, which was suggested as a potential deficiency in NLP tasks such as semantic role labeling.31 Thus, we are motivated to design neural network models that integrate both natural order word sequence and syntactic dependency information, and test whether such models can improve relation classification. Our proposed system achieved state-of-the-art micro F-measure in relation classification on the i2b2/VA dataset.
METHODS AND MATERIALS
Dataset
The 2010 i2b2/VA relation dataset contains clinical corpus and concept relations and is available at https://www.i2b2.org/NLP/Relations/.14 There are 3 relation categories: medical treatment–problem (TrP) category, medical test–problem (TeP) category, and medical problem–problem (PP) category. There are various possible relations in each category. For example, the PP relation category includes: 2 medical problems are related to each other (PIP) and 2 medical problems have no relation. Furthermore, the named entities for i2b2/VA relation classification are given, so the named entity recognition is not necessary. Segments were also provided by the i2b2/VA dataset, so no detection is necessary. Detailed relation classes, their descriptions, and class distributions in the i2b2/VA datasets are included in the Supplementary Material. We perform preprocessing steps on the data, including tokenization, syntactic dependency parsing, and segment detection on the sentences. More detailed descriptions on preprocessing steps are included in the Supplementary Materials as well.
Fine-tune word embedding using syntactic dependency
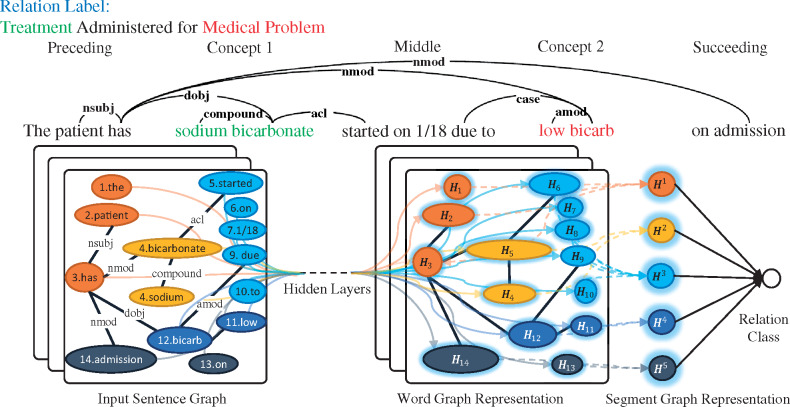
We use word2vec to train word embedding using Medical Information Mart for Intensive Care (MIMIC)-III clinical notes corpus26 as input to our model. However, the embedding trained on external corpora may not generalize well to the i2b2/VA challenge dataset. Kim32 introduced a fine-tuning method to his CNN classifier to learn task-specific word embedding. However, CNN is syntax-agnostic and ignores the important information in sentences’ syntactic dependencies. We instead use syntax-aware graph convolutional networks (GCNs)33 to fine-tune word embedding based on syntactic dependencies, as shown in Figure 1.
Figure 1.
An example of the schematic of a multi-layer GCN. The syntactic dependencies are extracted for a sentence from the i2b2/VA challenge dataset, with 5 segments in different colors. The 2 concepts with the relation to be classified are shown in green and red on the top. GCN allows for bridging words that are far away in the original sentence but connected through syntactic dependency [eg, “sodium bicarbonate” and “low bicarb” are bridged by 2 dependency links (nmod and dobj) with only 1 word in between as opposed to 5 words in the sentence].
Concept relation classification using Seg-GCRN
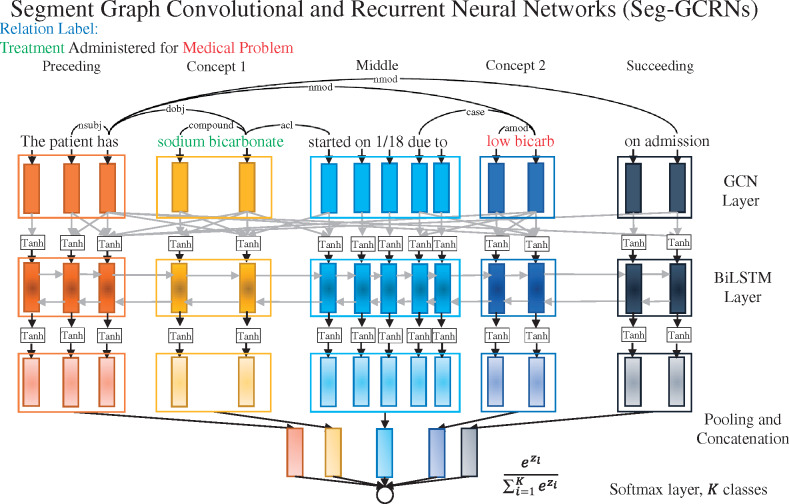
We propose segment graph convolutional and recurrent neural networks (Seg-GCRNs) to make the representation learning both syntax-aware and sequence-aware. Seg-GCRNs use GCN layers to integrate syntactic dependency information and recurrent neural network layers to integrate word sequence information. Figure 2 shows the schematic of the Seg-GCRNs.
Figure 2.
Segment graph convolutional and recurrent neural networks (Seg-GCRNs). Context texts are divided into 5 segments: before the first concept (preceding), the first concept (concept1), between the 2 concepts (middle), the second concept (concept2), and after the second concept (succeeding). Tanh stands for hyperbolic tangent activation function. The dependencies are generated from the McCCJ parser trained using the GENIA treebank and PubMed abstracts.34
The GCN layers of Seg-GCRNs focus on fine-tuning word embedding using syntactic dependencies.33 Compared with CNN, GCN allows for bridging words that are far away in the original sentence but connected through syntactic dependencies31 (see Figure 1). We denote the syntactic dependency parse of the relation-containing sentence as an undirected graph . Let the embedding matrix of the sentence be with each row containing node (word) ’s -dimensional embedding. Sentences shorter than words are padded with zeros. Denote as the graph ’s adjacency matrix, as the matrix of eigenvectors from the normalized graph Laplacian of , and as a Fourier domain filter matrix parametrized with a scalar as its diagonal elements. We followed the simplified parametrization for computational feasibility.35 The graph convolution for the -dimensional embedding (for words) is
| (1) |
where is -dimensional convolved signal, and is essentially the fine-tuned word embedding. In this formulation, the original word embedding is first transformed into the Fourier domain as , then multiplied with the filter matrix to become , and transformed back into the original domain as . Following Kipf and Welling,33 we use Chebyshev polynomial approximation to simplify the graph convolution and improve the computational efficiency, and extend the embedding and convolved signal to -dimensional and respectively. Let be the hyperbolic tangent activation function, then
| (2) |
where , is an identity matrix, and is the tunable weight factor to introduce self-connections in the graph, and is the graph convolution matrix. A single-layer GCN with 1st-order Chebyshev polynomial approximation performs convolution only among nodes that are 1 hop away from each other.33 Stacking layers of GCN can encode up to j-hop neighborhoods.36 To generate the syntactic dependencies and define the node adjacencies, we adopted the McCCJ parser trained using a biomedical domain treebank.34 To zoom in on the concept pairs of the candidate relation, we use the Dijkstra’s algorithm37 to find the shortest path between 2 concepts in the graph. Then, only edges having at least 1 end (node) on the shortest path are preserved in for graph convolution.
For recurrent neural network layers, Seg-GCRN integrates the long short-term memory (LSTM) network, as Luo et al.21,26 showed that sequence-aware modeling also recovers important information in clinical texts for relation classification. Marcheggiani et al.31 also validated the complementarity of GCNs (syntax-aware) and bi-directional LSTMs (BiLSTMs) (sequence-aware) in semantic role labeling. Therefore, we adopt BiLSTMs and define the network unit for each word as consisting of 2 LSTMs as
| (3) |
where , , and are the forward and backward passing LSTMs, each with a hidden state of dimension . Compared with Seg-LSTM,21 which learns 5 individual LSTM models for 5 sentence segments, Seg-GCRN learns 1 BiLSTM model through concatenating all 5 sentence segments (as shown in Figure 2) but pools separately to respect segment boundaries. This greatly saves the computational cost on the LSTM layer (as shown in Table 2) and enables sequence-aware information encoding across segments. The encoded feature vectors, collectively as , are then split into 5 segments representing preceding, concept1, middle, concept2, and succeeding segments. Each segment of word features is pooled using max or min pooling (tuned for different datasets using validation data) individually into , which are concatenated into a vector . We then pass to a fully connected layer, which contains weight and bias , to produce a size- vector , where is the number of relation classes. A softmax layer is finally used to compute the probability for the th class as
| (4) |
and gives the relation class for the 2 concepts.
Table 2.
Running time of the Seg-GCRN on 3 datasets
| System | Medical treatment–problem relations | Medical test–problem relations | Medical problem–problem relations |
|---|---|---|---|
| Seg-GCRN | 55s | 108s | 220s |
| Seg-GCN | 31s | 67s | 100s |
| Seg-CNN | 120s | 217s | 413s |
| Seg-LSTM | 1901s | 2175s | 1550s |
The time is measured by number of seconds.
EXPERIMENTS AND RESULTS
To make a fair comparison, we adopted the same training and testing partition as the i2b2/VA challenge. We randomly used 10% of the training dataset as the validation dataset to guide the tuning of model hyperparameters. The word embedding is trained using MIMIC-III corpus with embedding dimension 300. The McCCJ parsers are trained using (1) GENIA treebank and PubMed abstracts (biomedical domain) and (2) Wall Street Journal (WSJ) corpus (general domain) to test the sensitivity of the Seg-GCRN under different parsers. The 2-class PP dataset is particularly imbalanced, which has approximately 8 times more None labels than PIP labels. Following the approach adopted by de Bruijn et al.13 and Luo et al.,26 we randomly down-sampled the PIP/None ratio to 1:4 in the training dataset.
To regularize the Seg-GCRN and prevent it from overfitting, we applied regularization on the parameter matrix of the GCN layer and dropout on the output of the BiLSTM layer. The relative weight of regularization33 and dropout probability was tuned using the validation dataset. For model performance evaluation, we adopted the same micro-averaged precision, recall, and F-measure from the i2b2/VA challenge (see Table 1). Without manual feature engineering, Seg-GCRN with the biomedical domain McCCJ parser achieved state-of-the-art performance for all 3 datasets on micro-averaged F-measure compared with the past systems, most of which required extensive feature engineering. Seg-GCRN with the general domain McCCJ parser achieved state-of-the-art performance on micro-averaged F-measure only for TrP and PP datasets, suggesting the importance of using domain-specific parsers for the best modeling performance. A more detailed comparison of the results using confusion matrices is also included in the Supplementary Material, where the class-by-class error analysis is performed. The analysis shows that Seg-GCN effectively improved the precision and recall rates on several classes across 3 categories, hence improving the overall classification performance. Furthermore, compared with Zhu et al.30 and Seg-CNN,26 which achieved the previously state-of-the-art overall evaluation on precision, recall, and F-measure as [0.755, 0.726, 0.742] (in30) and [0.748, 0.736, 0.742] (in26), Seg-GCRN improved the result to [0.772, 0.743, 0.758]. Additionally, as the generalized convolution, GCN can perform sequential convolution as CNN does, by setting the off-diagonal elements of the adjacency matrix as non-zeros.31 We report the testing result of the revised Seg-GCN, which can also perform the sequence-aware (SA) encoding, as “Seg-SAGCN” in Table 1. Compared with Seg-GCRN, however, the performance is slightly worse on TrP and PP datasets, and almost the same on the TeP dataset. This reveals that performing sequence-aware and syntax-aware encoding all through convolution might not be as effective as separately performing them through BiLSTMs and GCNs. Similar observations have also been made by Bastings et al.39 In summary, we can see that Seg-GCRN achieved state-of-the-art performance for the i2b2/VA challenge datasets with only word embedding features and automatically generated syntactic dependencies. We also include an additional performance report based on 10-fold cross-validation (CV) using the training dataset in order to quantify the modeling performance variation (see the Supplementary Material). Specifically, the standard deviation of the micro-averaged F-measures for TeP, TrP, and PP datasets over 10-fold CV are 0.005, 0.004, and 0.008, respectively, which are all smaller than the improvement made by Seg-GCRN over the previous state-of-the-art (0.006, 0.007, and 0.038 for TeP, TrP, and PP datasets). Therefore, we believe that the improvement made by Seg-GCRN is notable. Additionally, we include some examples showing how Seg-GCRN can help to correct misclassified samples by the previous state-of-the-art Seg-CNN,26 based on syntactic dependency information (see the Supplementary Material). In the examples, we can see that syntactic dependencies combined with 1-layer GCN can quickly bridge the medical concepts within 2 hops and enable efficient fine-tuning of word embedding.
Table 1.
Performance of the Seg-GCRN model with word embedding trained on the MIMIC-III corpus and sentences parsed with McCCJ parser trained using (1) GENIA treebank and PubMed corpora, and (2) WSJ corpus.34 Please refer to the Supplementary Table S1 for detailed relation classes and their definitions
| Medical treatment–problem relations |
Medical test–problem relations |
Medical problem–problem relations |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| System | P | R | F | P | R | F | P | R | F |
| Seg-GCRN (GENIA+PubMed) | 0.703 | 0.682 | 0.692 | 0.833 | 0.821 | 0.827 | 0.762 | 0.722 | 0.741 |
| Seg-GCRN (WSJ) | 0.684 | 0.683 | 0.683 | 0.842 | 0.802 | 0.821 | 0.787 | 0.702 | 0.742 |
| Seg-GCN | 0.673 | 0.684 | 0.679 | 0.818 | 0.795 | 0.807 | 0.641 | 0.717 | 0.677 |
| Seg-SAGCN | 0.663 | 0.676 | 0.670 | 0.845 | 0.777 | 0.809 | 0.635 | 0.702 | 0.667 |
| Seg-CNN26 | 0.687 | 0.685 | 0.686 | 0.836 | 0.804 | 0.820 | 0.700 | 0.704 | 0.702 |
| Seg-LSTM21 | 0.641 | 0.683 | 0.661 | 0.766 | 0.838 | 0.800 | 0.728 | 0.681 | 0.704 |
| Rink et al.38 | 0.672 | 0.686 | 0.679 | 0.798 | 0.833 | 0.815 | 0.664 | 0.726 | 0.694 |
| de Bruijn et al.13 | 0.750 | 0.583 | 0.656 | 0.843 | 0.789 | 0.815 | 0.691 | 0.712 | 0.701 |
| Grouin et al.12 | 0.647 | 0.646 | 0.647 | 0.792 | 0.801 | 0.797 | 0.670 | 0.645 | 0.657 |
| Patrick et al.11 | 0.671 | 0.599 | 0.633 | 0.813 | 0.774 | 0.793 | 0.677 | 0.627 | 0.651 |
| Jonnalagadda et al.10 | 0.581 | 0.679 | 0.626 | 0.765 | 0.828 | 0.795 | 0.586 | 0.730 | 0.650 |
| Divita et al.9 | 0.704 | 0.582 | 0.637 | 0.794 | 0.782 | 0.788 | 0.710 | 0.534 | 0.610 |
| Solt et al.16 | 0.621 | 0.629 | 0.625 | 0.801 | 0.779 | 0.790 | 0.469 | 0.711 | 0.565 |
| Demner-Fushman et al.8 | 0.642 | 0.612 | 0.626 | 0.835 | 0.677 | 0.748 | 0.662 | 0.533 | 0.591 |
| Anick et al.7 | 0.596 | 0.619 | 0.608 | 0.744 | 0.787 | 0.765 | 0.631 | 0.502 | 0.559 |
| Cohen et al.6 | 0.606 | 0.578 | 0.591 | 0.750 | 0.781 | 0.765 | 0.627 | 0.492 | 0.552 |
Testing performance of all i2b2/VA challenge participating systems and some recent studies are shown for comparison as gray. The Seg-GCRN’s best performance is attained with -layer GCN stacked with 1-layer BiLSTM and using 300 as embedding dimension. The best hyperparameter combinations are [2 as the -norm penalty coefficient, 0.5 as dropout possibility, and 30 as self-connection weight ] for TeP relations with min-pooling for each segment, [0.2, 0.5, 30] for TrP relations with min-pooling, and [1, 0.1, 30] for PP relations with max-pooling. The Seg-GCN’s best performance is attained with 1-layer GCN and using 300 as embedding dimension. The best hyperparameter combinations are [0.3 as the -norm coefficient of the GCN layer, 0.1 as the -norm coefficient of the fully connected layer, and 20 as self-connection weight ] for TeP relations with max-pooling, [0.2, 0.1, 30] for TrP relations with max-pooling, and [0.1, 0.1, 15] for PP relations with max-pooling. No dropout is used for the best Seg-GCN performance. The Seg-SAGCN’s best performance is attained with 1-layer GCN and using 300 as embedding dimension. The best window size is selected as 3, and the same hyperparameter combinations from Seg-GCN on -norm coefficients and self-connection weight adopted as the tuning paths for the 2 models are basically identical. No dropout is used for the best Seg-SAGCN performance. The best micro-averaged F-measures across different systems are displayed in bold.
Seg-GCRN is implemented in Tensorflow,40 and NVidia K40 GPU is adopted for model training and testing. We have released our code at https://github.com/yuanluo/seg_gcn. Table 2 shows that Seg-GCRN trains on all 3 datasets for under 4 minutes and is efficient for practical usage.
DISCUSSION
To evaluate the effectiveness of combining sequence-aware and syntax-aware models, we report the results of segment modeling with only LSTM layers21 and only GCN layers in Table 1 for comparison. We see significant performance increase using Seg-GCRN, which shows the complementarity of syntax-aware and sequence-aware models. Furthermore, Seg-GCRN achieved state-of-the-art performances across all 3 datasets, suggesting that Seg-GCRN is a robust tool for relation classification in clinical notes. The improvements achieved by Seg-GCRN are modest over the past studies for TeP and TrP datasets, but more significant for the PP dataset. This is not surprising, as the PP dataset’s training and testing sizes are approximately more than twice that of TeP’s and TrP’s. Such imbalanced improvements are more conspicuous than those in Seg-CNN and Seg-LSTM (see Table 1), suggesting that Seg-GCRN training can be more sensitive to sample size while training the graph convolution. Furthermore, this result shows that Seg-GCRN can effectively capture context variations in large clinical records better than sequence-aware, or traditional feature engineering-based approaches. The model tuning result shows that the 1-layer GCN performs the best for Seg-GCRN and Seg-GCN. This can indicate that most important information can be encoded among nodes’ immediate syntactic neighborhood. However, this may also suggest that multi-layer GCNs demand more training data for parameter estimation. We will systematically explore the impact of number of GCN layers in Seg-GCRN using larger training datasets in the future. Additionally, we focused mainly on using syntactic dependencies as the linguistic features in Seg-GCRN, but plan to investigate the usefulness of other linguistic and semantic features to further improve Seg-GCRN. Lastly, only the text information from a single sentence was used to classify the relation between 2 medical concepts, and such information can be limited for classification. In the future, we plan to integrate knowledge source in the learning process of Seg-GCRN. For example, knowledge source on possible adverse events of drugs can be useful in classifying the TrCP (treatment cause medical problem) relation. We will combine the knowledge-graph encoding41 with Seg-GCRN and aim at learning effective knowledge-guided features automatically, in order to further improve the relation classification accuracy to a higher level that is of more practical utility.
CONCLUSION
In this work, we addressed an unmet need of a medical relation classification system that requires no manual feature engineering and learns relation representation jointly from lexical and syntactic information. We built Seg-GCRN to learn relation representations using word sequence and dependency syntax of 5 segments within a sentence (preceding, concept1, middle, concept2, and succeeding) for classification. Seg-GCRN achieved state-of-the-art performance for the i2b2/VA challenge datasets with only word embedding features and syntactic dependencies. We demonstrate the advantage of using deep neural networks to integrate lexical and syntactic information, compared to using either information alone. Our results encourage further research on deep neural networks to better utilize syntactic structures and linguistic features for relation classification and other NLP tasks.
FUNDING
This work was supported in part by NIH Grant 1R21LM012618-01.
CONTRIBUTORS
YL formulated the original problem. YL and YFL designed Seg-GCRN. YFL implemented the system and performed experiments and analysis. YL and RJ guided the study and provided statistical expertise. All authors contributed to the writing of the paper.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank i2b2 National Center for Biomedical Computing, which is funded by U54LM008748, for creating the clinical records in the i2b2/VA relation classification challenge. Furthermore, the authors thank the NVidia GPU grant for providing the GPU, which is adopted in this research.
Conflict of interest statement. The authors have no competing interests to declare.
REFERENCES
- 1. Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB.. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc 2011; 18 (Suppl 1): 116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harpaz R, Vilar S, DuMouchel W, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc 2013; 20 (3): 413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y, Thompson WK, Herr TM, et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf 2017; 40 (11): 1075–89. [DOI] [PubMed] [Google Scholar]
- 4. Harpaz R, Callahan A, Tamang S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf 2014; 37 (10): 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo Y, Uzuner Ö, Szolovits P.. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Brief Bioinform 2017; 18 (1): 160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen AM, Ambert K, Yang J, et al Ohsu/portland vamc team participation in the 2010 i2b2/va challenge tasks. In: proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010; Boston, MA, USA: i2b2.
- 7.Anick P, Hong P, Xue N, Anick D. I2B2 2010 challenge: machine learning for information extraction from patient records. In: proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010; Boston, MA, USA: i2b2.
- 8.Demner-Fushman D, Apostolova E, Islamaj Dogan R. NLM’s system description for the fourth i2b2/VA challenge. In: proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010; Boston, MA, USA: i2b2.
- 9.Divita G, Treitler OZ, Kim YJ. Salt lake city VA’s challenge submissions. In: proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010; Boston, MA, USA: i2b2.
- 10. Jonnalagadda S, Cohen T, Wu S, Gonzalez G.. Enhancing clinical concept extraction with distributional semantics. J Biomed Inform 2012; 45 (1): 129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patrick JD, Nguyen DH, Wang Y, Li M.. A knowledge discovery and reuse pipeline for information extraction in clinical notes. J Am Med Inform Assoc 2011; 18 (5): 574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grouin C, Abacha AB, Bernhard D, et al. CARAMBA: concept, assertion, and relation annotation using machine-learning based approaches. i2b2 Medication Extraction Challenge Workshop; 2010; Villeurbanne, France: HAL online repository.
- 13. de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X.. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011; 18 (5): 557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uzuner Ö, South BR, Shen S, DuVall SL.. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sahu SK, Anand A, Oruganty K, Gattu M. Relation extraction from clinical texts using domain invariant convolutional neural network. arXiv preprint arXiv: 1606.09370 2016.
- 16.Solt I, Szidarovszky FP, Tikk D. Concept, assertion and relation extraction at the 2010 i2b2 relation extraction challenge using parsing information and dictionaries. In: proc. of i2b2/VA Shared-Task; 2010; Washington, DC.
- 17.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: proc. AMIA Symp.; 2001; Philadelphia, PA: American Medical Informatics Association. [PMC free article] [PubMed]
- 18. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Björne J, Salakoski T. Generalizing biomedical event extraction. In: proceedings of the BioNLP Shared Task 2011 Workshop; 2011; Stroudsburg, PA: Association for Computational Linguistics.
- 20.Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M. Medical image classification with convolutional neural network. Control Automation Robotics & Vision (ICARCV), 2014 13th International Conference on; 2014; Washington, DC: IEEE.
- 21. Luo Y. Recurrent neural networks for classifying relations in clinical notes. J Biomed Inform 2017; 72: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dernoncourt F, Lee JY, Uzuner O, Szolovits P.. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. In: proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting; 2016; Stroudsburg, PA: NIH Public Access. [DOI] [PMC free article] [PubMed]
- 24.Tamura A, Watanabe T, Sumita E. Recurrent neural networks for word alignment model. In: proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers); 2014; Stroudsburg PA: Association for Computational Linguistics.
- 25.Xu Y, Mou L, Li G, Chen Y, Peng H, Jin Z. Classifying relations via long short term memory networks along shortest dependency paths. In: proceedings of the 2015 conference on empirical methods in natural language processing; 2015; Stroudsburg PA: Association for Computational Linguistics.
- 26. Luo Y, Cheng Y, Uzuner Ö, Szolovits P, Starren J.. Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. J Am Med Inform Assoc 2018; 25 (1): 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JY, Dernoncourt F. Sequential short-text classification with recurrent and convolutional neural networks. arXiv preprint arXiv: 1603.03827 2016.
- 28. Miotto R, Wang F, Wang S, Jiang X, Dudley JT.. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 2017; 19:1236–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uzuner O, Mailoa J, Ryan R, Sibanda T.. Semantic relations for problem-oriented medical records. Artif Intell Med 2010; 50 (2): 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu X, Cherry C, Kiritchenko S, Martin J, De Bruijn B.. Detecting concept relations in clinical text: Insights from a state-of-the-art model. J Biomed Inform 2013; 46 (2): 275–85. [DOI] [PubMed] [Google Scholar]
- 31.Marcheggiani D, Titov I. Encoding sentences with graph convolutional networks for semantic role labeling. In: Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing; 2017; Stroudsburg PA: Association for Computational Linguistics.
- 32. Kim Y. Convolutional neural networks for sentence classification. arXiv preprint arXiv: 1408.5882 2014.
- 33. Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv: 1609.02907 2016.
- 34. McClosky D. Any Domain Parsing: Automatic Domain Adaptation for Natural Language Parsing [PhD thesis]. Brown University; 2010; Providence, RI, USA.
- 35.Defferrard M, Bresson X, Vandergheynst P. Convolutional neural networks on graphs with fast localized spectral filtering. Advances in Neural Information Processing Systems; 2016; Cambridge, MA: MIT Press.
- 36. LeCun Y, Bottou L, Bengio Y, Haffner P.. Gradient-based learning applied to document recognition. Proc IEEE 1998; 86 (11): 2278–324. [Google Scholar]
- 37. Johnson DB. A note on Dijkstra’s shortest path algorithm. JACM 1973; 20 (3): 385–8. [Google Scholar]
- 38. Rink B, Harabagiu S, Roberts K.. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc 2011; 18 (5): 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bastings J, Titov I, Aziz W, Marcheggiani D, Sima'an K. Graph convolutional encoders for syntax-aware neural machine translation. arXiv preprint arXiv: 1704.04675 2017.
- 40.Abadi M, Barham P, Chen J, et al Tensorflow: a system for large-scale machine learning. OSDI; 2016; PeerJ, San Diego, USA.
- 41. Sowa JF. Knowledge Representation: Logical, Philosophical, and Computational Foundations. Brooks/Cole; 1994; Boston, MA, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.