Abstract
Cytotoxic T lymphocyte-associated protein 4 (CTLA-4) is a coinhibitory checkpoint protein expressed on the surface of T cells. A recent study by our working group revealed that the rs231775 single nucleotide polymorphism (SNP) in the CTLA-4 gene was associated with the survival of patients with sepsis and served as an independent prognostic variable. To further investigate the impact of CTLA-4 genetic variants on sepsis survival, we examined the effect of two functional SNPs, CTLA-4 rs733618 and CTLA-4 rs3087243, and inferred haplotypes, on the survival of 644 prospectively enrolled septic patients. Kaplan–Meier survival analysis revealed significantly lower 90-day mortality for rs3087243 G allele carriers (n = 502) than for AA-homozygous (n = 142) patients (27.3% vs. 40.8%, p = 0.0024). Likewise, lower 90-day mortality was observed for TAA haplotype-negative patients (n = 197; compound rs733618 T/rs231775 A/rs3087243 A) than for patients carrying the TAA haplotype (n = 447; 24.4% vs. 32.9%, p = 0.0265). Carrying the rs3087243 G allele hazard ratio (HR): 0.667; 95% confidence interval (CI): 0.489–0.909; p = 0.0103) or not carrying the TAA haplotype (HR: 0.685; 95% CI: 0.491–0.956; p = 0.0262) remained significant covariates for 90-day survival in the multivariate Cox regression analysis and thus served as independent prognostic variables. In conclusion, our findings underscore the significance of CTLA-4 genetic variants as predictors of survival of patients with sepsis.
Keywords: sepsis, CTLA-4, single nucleotide polymorphisms, haplotypes, survival, predictors
1. Introduction
Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infection [1] and remains a leading cause of death and critical illness worldwide [2]. The reported incidence of sepsis is increasing in aging populations with more comorbidities [1,3,4] and is accompanied by rising health care costs. The multifaceted pathophysiology of sepsis and sepsis-related organ dysfunction is highly diverse and therefore has been intensively investigated in the recent past [5]. Host genetic characteristics, such as gene variants encoding innate immune effectors or inflammatory mediators, seem to contribute considerably to the disease course and sepsis outcome [6,7,8,9]. The genetic component of host susceptibility to sepsis is clearly polygenetic [10], and identification of responsible variants will help elucidate the pathophysiology and biology of the disease. Identifying sepsis-associated genetic variants and evaluating their impacts on gene expression, protein function and host physiology will potentially reveal future targets for sepsis treatment [11].
CTLA-4 has gained recognition since James P. Allison and Tasuku Honja were awarded the Nobel Prize for Physiology or Medicine in 2018 for their discovery of cancer therapy by inhibition of negative immune regulation [12]. CTLA-4 is a coinhibitory cell surface protein expressed on T cells that competes with CD28 for binding to CD80 and CD86 on antigen-presenting cells (APCs) [13,14] and thereby reduces T cell activation and proliferation [15]. Thus, an increased CTLA-4 expression has an inhibitory effect on the host immune reaction [16].
Coinhibitory immune checkpoint proteins, such as PD-1 and CTLA-4, have been reported to play key roles in the course of sepsis [13,17,18,19], and our working group has revealed genetic associations of their genes with the survival of septic patients [9,20]. Our recent study revealed that the CTLA-4 rs231775 GG genotype, which was reported to have an attenuated inhibitory effect on T cell reactions [21] and was associated with greater T cell activation and proliferation and autoimmune diseases [22], was also associated with favorable 90- and 28-day survival in septic patients compared to the survival rates of A allele carriers at this position [20].
The CTLA-4 gene is located on chromosome 2q33 and contains two further functional polymorphisms, the CTLA-4 rs733618 and CTLA-4 rs3087243, which are associated with enhanced T cell reactions and higher susceptibility to autoimmune diseases, such as myasthenia gravis [23], systemic lupus erythematosus [24,25], Hashimoto thyroiditis [26], latent autoimmune diabetes [27], multiple sclerosis [28] and acute liver transplant rejection [29]. This study aimed to investigate the impact of these two functional single nucleotide polymorphisms (SNPs) and inferred haplotypes compound of rs733618, rs231775, and rs3087243 on the 90-day mortality of septic patients in order to further reveal the role of CTLA-4 genetic variants in sepsis.
2. Methods
2.1. Patients
The present study included 644 prospectively enrolled adult Caucasian patients with sepsis. Patients were recruited from three surgical intensive care units (ICUs) through the GENOSEP database of the Department of Anesthesiology at the University Medical Center Goettingen [20]. Enrolled patients were identified by daily screening of the three ICUs according to the actual sepsis definitions and guidelines [1,30]. The exclusion criteria were defined in previous studies [20,31] and were an age below 18 years, pregnancy, therapy with immunosuppressive drugs or chemotherapy, myocardial infarction within six weeks before enrollment, human immunodeficiency virus (HIV) infection, chronic heart failure classified as New York Heart Association (NYHA) stage IV, end-stage incurable disease, a persistent vegetative state (apallic syndrome), and a “Do Not Treat” (DNT) or “Do Not Resuscitate” (DNR) order. Upon enrollment, blood was drawn within 72 h, and the patients were followed up for 90 days unless previously dismissed or deceased.
This investigation and the experimental protocols were approved under ethical project identification code 15/1/12 by the institutional ethics committee of the University of Goettingen in Goettingen, Germany. The study was performed in accordance with the provisions of the Declaration of Helsinki and relevant guidelines and regulations. The methods were carried out in accordance with approved guidelines. Written informed consent was obtained from either the patient or their legal representative.
2.2. Data Collection
All patient data were recorded through the GENOSEP database. At enrollment, relevant baseline characteristics were obtained, such as comorbidities, preexisting medication, and the initial Sequential Organ Failure Assessment (SOFA) and Acute Physiology and Chronic Health Evaluation (APACHE II) scores. Patients were followed up for 90 days, and 90-day mortality was recorded as the primary outcome parameter. Within the first 28 days after sepsis onset, relevant clinical data were generated through a standardized clinical report form (CRF) on a daily basis. The data were collected from the electronic patient record system (Intellispace Critical Care and Anesthesia (ICCA), Phillips Healthcare, Andover, MA, USA). Parameters included SOFA score-relevant organ dysfunction variables; information about organ support requirements; and inflammatory, kidney and liver values.
2.3. CTLA-4 Genotyping and Haplotyping
DNA extraction, CTLA-4 rs733618 and rs3087243 genotyping, and haplotyping were performed entirely in the laboratories of the Department of Clinical Pharmacology of the University Medical Center Goettingen. The DNA extraction and genotyping methods were performed as previously described [20]. DNA was automatically extracted from either 200 µL of ethylene diamine tetraacetic acid (EDTA) blood using the QIAamp® DNA Blood Kit in the QIAcube®, 350 µL of EDTA blood using the EZ1® DNA Blood Kit in the BioRobot EZ1®, or peripheral blood mononuclear cells (PBMCs) using the AllPrep DNA Mini Kit according to the manufacturer’s instructions (all from Qiagen, Hilden, Germany). The quantity and quality of the DNA were determined by spectrophotometric measurement. Genotyping of the extracted DNA was performed via TaqMan polymerase chain reaction (PCR) using the predesigned TaqMan® SNP Genotyping Assays C_2415791_10 and C_3296043_10 and the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions (Life Technologies, Darmstadt, Germany). The genotyping outcomes were generated using the 7900HT Fast Real-Time PCR System Software (SDS v2.4.1 for Windows 7, Applied Biosystems, Foster City, CA, USA). Twenty percent of the samples were genotyped in duplicate for reliability assessment and yielded concordant results.
The CTLA-4 haplotypes were inferred using the PHASE software (Matthew Stephens, Washington, DC, USA, version 2.1), which is a Bayesian statistical method that is used to reconstruct haplotypes from population genotype data [32,33]. CTLA-4 haplotype reconstruction was performed in 100 iterations to yield the most likely haplotype combination (best pair) for each individual, with a minimum probability for the most likely combination of 99.9%.
2.4. Statistical Analyses
The data analysis was performed using the STATISTICA 13 software (version 13.0, StatSoft, Tulsa, OK, USA). The patient baseline characteristics and disease severity data were evaluated in univariate analyses with Pearson’s Chi-square test, a two-sided Fisher’s exact test, the Mann–Whitney U test or the Kruskal-Wallis test, if applicable. The Kaplan–Meier time-to-event data were compared with the log-rank test. Multivariate Cox regression analyses were used to adjust for possible effect confounders. The appropriateness of the genotype distributions and allele frequencies of the study population was evaluated with a Chi-square test of the Hardy–Weinberg equilibrium. The CTLA-4 rs733618 CC and CT genotypes, as well as the rs3087243 GG and AG genotypes, were pooled to compare the clinical course of the functionally relevant rs733618 homozygous TT (n = 550) with C allele carriers (n = 94) and the rs3087243 G allele carriers (n = 502) with homozygous AA (n = 142) patients.
A p-value <0.05 was considered statistically significant. Data are presented as absolute numbers or percentages for categorical variables and the mean ± standard deviation for continuous variables.
2.5. Data Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
3. Results
3.1. Baseline Characteristics
Our cohort of 644 prospectively enrolled adult Caucasian patients with sepsis, which was previously described [20], was examined in the present study. The average age was 63 ± 15 years; 66% were of male gender; and the average SOFA and APACHE II scores were 9.4 ± 3.9 and 22 ± 7, respectively, upon enrollment. A total of 93% of the patients were mechanically ventilated, 79% received vasopressor therapy, 21% received renal replacement therapy, and 51% were in septic shock during the observation period (Table 1 and Table 2).
Table 1.
Patient baseline characteristics with regard to the cytotoxic T lymphocyte-associated protein 4 (CTLA-4) rs3087243 genotypes.
| Parameter | All (n = 644) | rs3087243 | p-Value | |
|---|---|---|---|---|
| AA (n = 142) | GG/AG (n = 502) | |||
| Age (years) | 63 ± 15 | 67 ± 14 | 62 ± 15 | 0.0033 |
| Male (%) | 66 | 64 | 67 | 0.5560 |
| Body mass index | 28 ± 6 | 27 ± 6 | 28 ± 6 | 0.2881 |
| Sepsis severity | ||||
| Septic shock (%) | 51 | 52 | 51 | 0.8467 |
| SOFA score day 1 | 9.4 ± 3.9 | 9.4 ± 3.9 | 9.4 ± 3.9 | 0.8047 |
| APACHE II score day 1 | 22 ± 7 | 22 ± 7 | 21 ± 7 | 0.2844 |
| Comorbidities, n (%) | ||||
| Hypertension | 54 | 53 | 55 | 0.7094 |
| History of myocardial infarction | 5 | 6 | 5 | 0.9057 |
| Chronic obstructive pulmonary disease | 15 | 12 | 16 | 0.2436 |
| Renal dysfunction | 10 | 8 | 11 | 0.3879 |
| Non-insulin-dependent diabetes mellitus | 9 | 8 | 9 | 0.8492 |
| Insulin-dependent diabetes mellitus | 11 | 12 | 10 | 0.5349 |
| Chronic liver disease | 6 | 6 | 6 | 0.9875 |
| History of cancer | 16 | 15 | 16 | 0.7830 |
| History of stroke | 6 | 9 | 5 | 0.0623 |
| Recent surgical history, n (%) | ||||
| Elective surgery | 29 | 37 | 27 | |
| Emergency surgery | 53 | 46 | 55 | |
| No history of surgery | 18 | 17 | 18 | |
| Site of infection, n (%) | ||||
| Lung | 62 | 64 | 61 | |
| Abdomen | 20 | 20 | 20 | |
| Bone or soft tissue | 4 | 3 | 4 | |
| Surgical wound | 2 | 2 | 1 | |
| Urogenital | 2 | 1 | 3 | |
| Primary bacteremia | 7 | 6 | 7 | |
| Other | 3 | 4 | 4 | |
| Organ support (%) | ||||
| Used during observation period | ||||
| Mechanical ventilation | 93 | 96 | 93 | 0.1850 |
| Use of vasopressor | 79 | 82 | 78 | 0.4063 |
| Renal replacement therapy | 21 | 20 | 21 | 0.7173 |
| Used on sepsis onset | ||||
| Mechanical ventilation | 86 | 86 | 86 | 0.9381 |
| Use of vasopressor | 67 | 66 | 67 | 0.9048 |
| Renal replacement therapy | 9 | 9 | 9 | 0.8851 |
SOFA, sequential organ failure assessment; APACHE, acute physiology and chronic health evaluation.
Table 2.
Patient baseline characteristics with regard to the CTLA-4 haplotype H1: TAA.
| Parameter | All (n = 644) | H1: TAA | p-Value | |
|---|---|---|---|---|
| Positive (n = 447) | Negative (n = 197) | |||
| Age (years) | 63 ± 15 | 64 ± 15 | 62 ± 14 | 0.1014 |
| Male (%) | 66 | 67 | 64 | 0.4356 |
| Body mass index | 28 ± 6 | 28 ± 6 | 28 ± 7 | 0.2388 |
| Sepsis severity | ||||
| Septic shock (%) | 51 | 51 | 53 | 0.6384 |
| SOFA score day 1 | 9.4 ± 3.9 | 9.3 ± 3.9 | 9.7 ± 3.8 | 0.1802 |
| APACHE II score day 1 | 22 ± 7 | 21 ± 7 | 22 ± 7 | 0.1474 |
| Comorbidities, n (%) | ||||
| Hypertension | 54 | 54 | 54 | 0.9671 |
| History of myocardial infarction | 5 | 6 | 3 | 0.0758 |
| Chronic obstructive pulmonary disease | 15 | 15 | 16 | 0.5779 |
| Renal dysfunction | 10 | 10 | 11 | 0.6734 |
| Non-insulin-dependent diabetes mellitus | 9 | 9 | 8 | 0.6654 |
| Insulin-dependent diabetes mellitus | 11 | 9 | 13 | 0.1480 |
| Chronic liver disease | 6 | 6 | 8 | 0.3893 |
| History of cancer | 16 | 12 | 17 | 0.0731 |
| History of stroke | 6 | 6 | 6 | 0.8209 |
| Recent surgical history, n (%) | ||||
| Elective surgery | 29 | 31 | 26 | |
| Emergency surgery | 53 | 52 | 54 | |
| No history of surgery | 18 | 17 | 20 | |
| Site of infection, n (%) | ||||
| Lung | 62 | 64 | 58 | |
| Abdomen | 20 | 19 | 21 | |
| Bone or soft tissue | 4 | 3 | 6 | |
| Surgical wound | 2 | 2 | 2 | |
| Urogenital | 2 | 2 | 3 | |
| Primary bacteremia | 7 | 7 | 6 | |
| Other | 3 | 3 | 4 | |
| Organ support (%) | ||||
| Used during observation period | ||||
| Mechanical ventilation | 93 | 93 | 93 | 0.9580 |
| Use of vasopressor | 79 | 79 | 80 | 0.6749 |
| Renal replacement therapy | 21 | 18 | 26 | 0.0204 |
| Used on sepsis onset | ||||
| Mechanical ventilation | 86 | 86 | 85 | 0.8341 |
| Use of vasopressor | 67 | 66 | 69 | 0.4944 |
| Renal replacement therapy | 9 | 6 | 15 | 0.0005 |
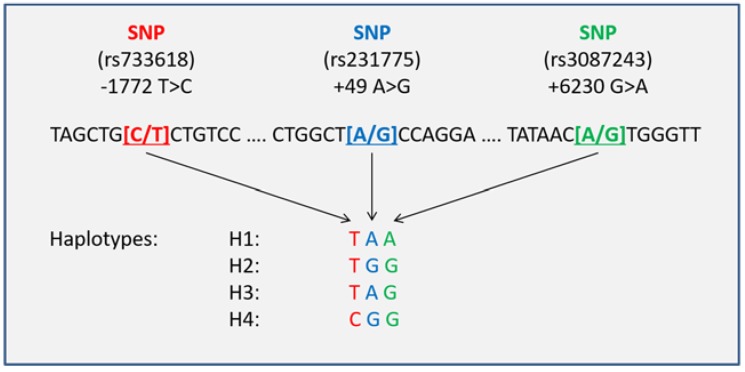
All patients were successfully genotyped, and the relevant haplotypes were inferred. The CTLA-4 rs733618 and rs3087243 genotype distributions were 4:90:550 (CC:CT:TT) and 142:305:197 (AA:AG:GG), with observed minor allele frequencies (MAFs) of 0.08 and 0.47, respectively. The observed frequencies were consistent with the Hardy–Weinberg equilibrium (χ2 test p = 0.8789 and p = 0.2448) and almost equaled the reported MAFs from reference populations, such as HapMap CEPH (CEU) with MAFs of 0.06 and 0.46 respectively (dbSNP database, accession numbers ss79540 [34] and ss44315036 [35]). The CTLA-4 haplotype analysis compound of the rs733618, rs231775, and rs3087243 SNPs revealed four common haplotypes (H1: TAA, H2: TGG, H3: TAG, and H4: CGG) (Figure 1).
Figure 1.
Compound cytotoxic T lymphocyte-associated protein 4 (CTLA-4) haplotypes. SNP, single nucleotide polymorphism.
The corresponding haplotype distributions can be obtained from Table 3. Linkage disequilibrium (LD) analysis of the three SNPs revealed a squared correlation coefficient (r2) of 0.39 for rs231775 and rs3087243, 0.23 for rs733618 and rs231775, and 0.09 for rs733618 and rs3087243.
Table 3.
CTLA-4 haplotype distributions.
| Haplotype CTLA-4-1772 T>C/+49 A>G/+6230 G>A | Number of Haplotypes | n (%) |
|---|---|---|
| H1: TAA | 0 | 197 (30.59) |
| 1 | 305 (47.36) | |
| 2 | 142 (22.05) | |
| H2: TGG | 0 | 316 (49.07) |
| 1 | 263 (40.84) | |
| 2 | 65 (10.09) | |
| H3: TAG | 0 | 451 (70.03) |
| 1 | 178 (27.64) | |
| 2 | 15 (2.33) | |
| H4: CGG | 0 | 550 (85.40) |
| 1 | 90 (13.98) | |
| 2 | 4 (0.62) |
Analysis of the baseline characteristics with regard to the genotype distributions revealed a significant difference in age between rs3087243 G allele carriers (n = 502) and AA homozygous patients (n = 142) at sepsis onset (62 ± 15 vs. 67 ± 14 years; p = 0.0033; Table 1). In addition, noncarriers of the inferred haplotype H1: TAA (TAA-negative, n = 197) received significantly more renal replacement therapy upon sepsis onset (15% vs. 6%, p = 0.0005; Table 2) as well as over the observation period (26% vs. 18%; p = 0.0204; Table 2) than carriers of the H1: TAA haplotype (TAA-positive, n = 447). These significant differences in patient baseline characteristics were adjusted by their inclusion as potential confounders in the Cox regression analysis.
3.2. Outcomes
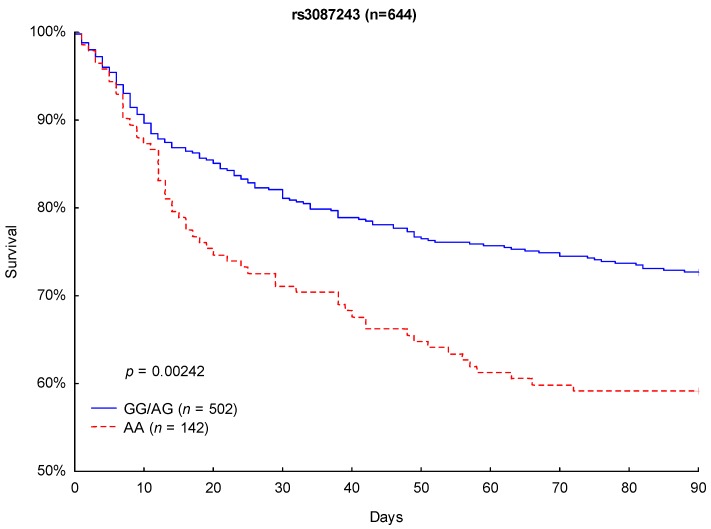
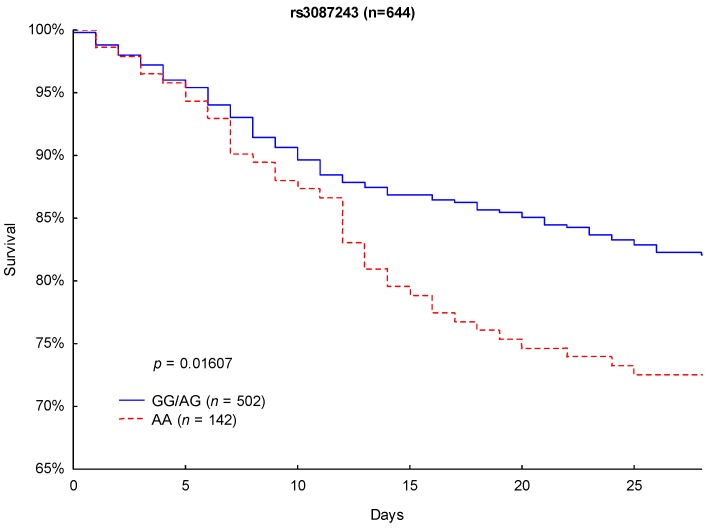
Although Kaplan–Meier analysis of 90- and 28-day survival revealed no significant differences between the CTLA-4 rs733618 TT homozygous and C allele carriers (p = 0.6864 and p = 0.4693), significant differences in survival were observed between the CTLA-4 rs3087243 G allele carriers and the AA-homozygous patients. CTLA-4 rs3087243 G allele carriers showed significantly lower 90-day mortality (27.3% vs. 40.8%, respectively; p = 0.0024; Figure 2) and 28-day mortality (17.9% vs. 27.5%; p = 0.0161; Figure 3) than AA homozygous patients.
Figure 2.
Kaplan–Meier survival analysis (90 days) with regard to the CTLA-4 rs3087243 genotypes.
Figure 3.
Kaplan–Meier survival analysis (28 days) with regard to the CTLA-4 rs3087243 genotypes.
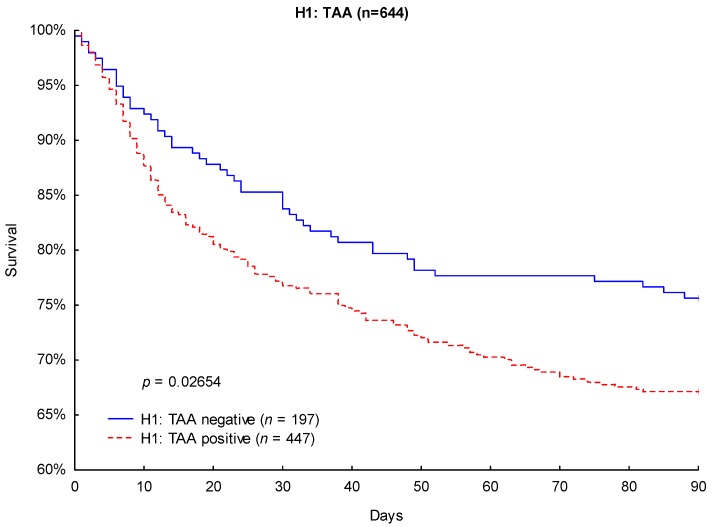
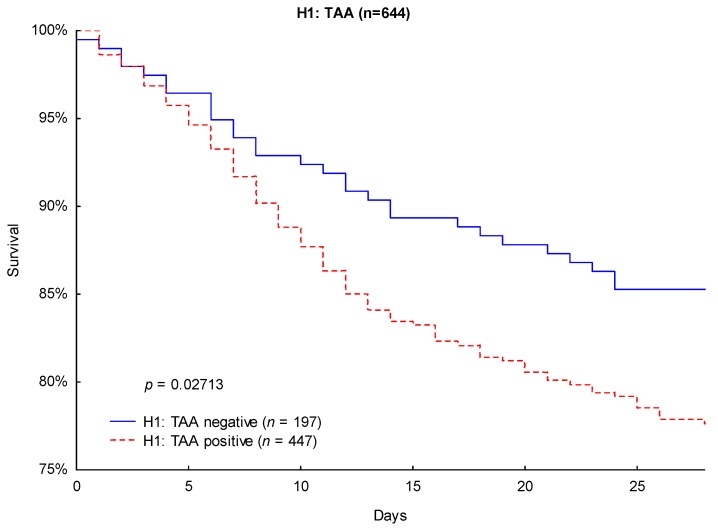
Likewise, Kaplan–Meier survival analysis revealed significantly lower 90-day mortality (24.4% vs. 32.9%; p = 0.0265; Figure 4) and 28-day mortality (14.7% vs. 22.4%; p = 0.0271; Figure 5) in TAA-negative patients compared to that of patients carrying the H1: TAA haplotype.
Figure 4.
Kaplan–Meier survival analysis (90 days) with regard to the CTLA-4 haplotype H1: TAA.
Figure 5.
Kaplan–Meier survival analysis (28 days) with regard to the CTLA-4 haplotype H1: TAA.
No other significant differences in mortality were observed for the remaining inferred haplotypes (H2: TGG, H3: TAG, and H4: CGG).
3.3. Cox Regression Analysis
Univariate and multivariate Cox regression analyses with regard to the CTLA-4 rs3087243 genotypes and the H1: TAA haplotype were performed to adjust for possible effects of baseline characteristics and other potential confounders on 90- and 28-day mortality. In addition to the baseline variables age, male gender, body mass index (BMI), and SOFA and APACHE II scores, statin therapy was included in accordance with findings from our previous study [36]. Application of renal replacement therapy upon sepsis onset and during the observation period was included in the Cox regression analysis for the H1: TAA haplotype based on our findings in the patient baseline characteristics. Multivariate Cox regression analyses revealed that carrying the CTLA-4 rs3087243 G allele was a significant positive predictor for 90-day survival (hazard ratio: 0.667; 95% confidence interval (CI): 0.489–0.909; p = 0.0103; Table 4) and 28-day survival (hazard ratio: 0.676; 95% CI: 0.463–0.987; p = 0.0423; Table 4), as was not carrying the H1:TAA haplotype (for 90-day survival, hazard ratio: 0.685; 95% CI: 0.491–0.956; p = 0.0262; for 28-day survival, hazard ratio: 0.621; 95% CI: 0.407–0.947; p = 0.0270; Table 5).
Table 4.
Cox regression analysis with regard to the CTLA-4 rs3087243 genotypes.
| Variable | 90 Days | |||||
| Univariate Analysis | Multivariate Analysis | |||||
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Age | 1.032 | 1.020–1.043 | 0.000000 | 1.025 | 1.013–1.038 | 0.000047 |
| Male gender | 1.093 | 0.810–1.474 | 0.558922 | 1.076 | 0.796–1.454 | 0.633436 |
| Body mass index | 0.985 | 0.961–1.009 | 0.218173 | 0.979 | 0.954–1.005 | 0.109574 |
| SOFA score day 1 | 1.104 | 1.065–1.143 | 0.000000 | 1.072 | 1.025–1.122 | 0.002432 |
| APACHE II score day 1 | 1.069 | 1.046–1.092 | 0.000000 | 1.033 | 1.004–.1062 | 0.026207 |
| Statin therapy | 1.114 | 0.807–1.536 | 0.511644 | 0.979 | 0.702–1.366 | 0.901706 |
| CTLA-4 rs3087243 G allele | 0.612 | 0.450–0.833 | 0.001760 | 0.667 | 0.489–0.909 | 0.010307 |
| 28 Days | ||||||
| Univariate Analysis | Multivariate Analysis | |||||
| Age | 1.029 | 1.015–1.043 | 0.000035 | 1.023 | 1.009–1.039 | 0.001824 |
| Male gender | 1.278 | 0.875–1.866 | 0.204154 | 1.270 | 0.868–1.860 | 0.218759 |
| Body mass index | 0.982 | 0.953–1.013 | 0.252686 | 0.976 | 0.945–1.008 | 0.135751 |
| SOFA score day 1 | 1.119 | 1.071–1.169 | 0.000000 | 1.085 | 1.026–1.146 | 0.003854 |
| APACHE II score day 1 | 1.073 | 1.045–1.102 | 0.000000 | 1.033 | 0.998–1.070 | 0.063787 |
| Statin therapy | 0.964 | 0.641–1.451 | 0.861906 | 0.841 | 0.552–1.281 | 0.420505 |
| CTLA-4 rs3087243 G allele | 0.624 | 0.428–0.908 | 0.013778 | 0.676 | 0.463–0.987 | 0.042323 |
Table 5.
Cox regression analysis with regard to the CTLA-4 haplotype H1: TAA.
| Variable | 90 Days | |||||
| Univariate Analysis | Multivariate Analysis | |||||
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Age | 1.032 | 1.020–1.043 | 0.000000 | 1.025 | 1.013–1.038 | 0.000049 |
| Male gender | 1.093 | 0.810–1.474 | 0.558922 | 1.079 | 0.798–1.459 | 0.620112 |
| BMI | 0.985 | 0.961–1.009 | 0.218173 | 0.975 | 0.950–1.001 | 0.061158 |
| SOFA score day 1 | 1.104 | 1.065–1.143 | 0.000000 | 1.038 | 0.988–1.091 | 0.139057 |
| APACHE II score day 1 | 1.069 | 1.046–1.092 | 0.000000 | 1.027 | 0.997–1.057 | 0.078734 |
| Statin therapy | 1.114 | 0.807–1.536 | 0.511644 | 1.086 | 0.777–1.519 | 0.629022 |
| Renal replacement therapy during observation period | 2.854 | 2.134–3.817 | 0.000000 | 2.724 | 1.867–3.974 | 0.000000 |
| Renal replacement therapy upon sepsis onset | 1.565 | 1.021–2.399 | 0.040064 | 1.973 | 1.192–3.268 | 0.008241 |
| H1: TAA negative | 0.698 | 0.504–0.967 | 0.030812 | 0.685 | 0.491–0.956 | 0.026202 |
| 28 Days | ||||||
| Univariate Analysis | Multivariate Analysis | |||||
| Age | 1.029 | 1.015–1.043 | 0.000035 | 1.023 | 1.009–1.039 | 0.001984 |
| Male gender | 1.278 | 0.875–1.866 | 0.204154 | 1.269 | 0.866–1.859 | 0.221286 |
| BMI | 0.982 | 0.953–1.013 | 0.252686 | 0.972 | 0.940–1.004 | 0.089311 |
| SOFA score day 1 | 1.119 | 1.071–1.169 | 0.000000 | 1.050 | 0.988–1.116 | 0.118403 |
| APACHE II score day 1 | 1.073 | 1.045–1.102 | 0.000000 | 1.026 | 0.990–1.064 | 0.155294 |
| Statin therapy | 0.964 | 0.641–1.451 | 0.861906 | 0.790 | 0.517–1.209 | 0.278234 |
| Renal replacement therapy during observation period | 3.028 | 2.131–4.301 | 0.000000 | 2.772 | 1.769–4.343 | 0.000009 |
| Renal replacement therapy upon sepsis onset | 1.616 | 0.970–2.691 | 0.065366 | 1.933 | 1.060–3.524 | 0.031464 |
| H1: TAA negative | 0.632 | 0.418–0.955 | 0.029340 | 0.621 | 0.407–0.947 | 0.027008 |
3.4. Disease Severity
The average SOFA and organ-specific SOFA subscores, as well as inflammatory, kidney, and liver parameters were included in the disease severity analysis (Table 6 and Table 7). Assessment of disease severity revealed that CTLA-4 rs3087243 G allele carriers had significantly lower average SOFA-Central Nervous System (SOFA-CNS) scores (1.9 ± 1.1 vs. 2.2 ± 1.0; p = 0.0094) over the observation period and a lower percentage of ventilated days as a fraction of observation days (65 ± 32% vs. 71 ± 32%; p = 0.0280) than AA homozygous patients.
Table 6.
Disease severity with regard to the CTLA-4 rs3087243 genotypes.
| Variable | All (n = 644) | rs3087243 | p-Value | |
|---|---|---|---|---|
| AA (n = 142) | GG/AG (n = 502) | |||
| SOFA | 7.0 ± 3.5 | 7.4 ± 3.8 | 6.9 ± 3.5 | 0.3005 |
| SOFA-Respiratory score | 2.0 ± 0.8 | 2.0 ± 0.8 | 2.0 ± 0.8 | 0.6043 |
| SOFA-Cardiovascular score | 1.5 ± 1.0 | 1.7 ± 1.0 | 1.5 ± 1.0 | 0.1284 |
| SOFA-Central Nervous System score | 2.0 ± 1.1 | 2.2 ± 1.0 | 1.9 ± 1.1 | 0.0094 |
| SOFA-Renal score | 0.8 ± 1.2 | 0.7 ± 1.1 | 0.8 ± 1.2 | 0.6669 |
| SOFA-Coagulation score | 0.4 ± 0.6 | 0.4 ± 0.7 | 0.3 ± 0.6 | 0.5521 |
| SOFA-Hepatic score | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.9780 |
| Organ support | ||||
| Ventilation days/observation days (%) | 66 ± 32 | 71 ± 32 | 65 ± 32 | 0.0280 |
| Vasopressor days/observation days (%) | 34 ± 30 | 37 ± 33 | 33 ± 29 | 0.2919 |
| Dialysis days/observation days (%) | 9 ± 23 | 9 ± 23 | 10 ± 22 | 0.8287 |
| Inflammatory values | ||||
| Leucocytes (1000/µL) | 13 ± 5 | 13 ± 5 | 13 ± 5 | 0.1026 |
| CRP (mg/L) | 152 ± 87 | 137 ± 81 | 156 ± 88 | 0.1356 |
| Procalcitonin (ng/dL) | 4.0 ± 9.3 | 3.5 ± 10.1 | 4.1 ± 9.1 | 0.0655 |
| Kidney values | ||||
| Urine output (mL/day) | 2978 ± 1337 | 2973 ± 1388 | 2979 ± 1324 | 0.9387 |
| Urine output (mL/kg/day) | 1.6 ± 0.8 | 1.6 ± 0.8 | 1.6 ± 0.78 | 0.9051 |
| Creatinine (mg/dL) | 1.2 ± 0.9 | 1.2 ± 0.9 | 1.3 ± 1.0 | 0.4218 |
| Liver values | ||||
| AST (GOT) (IU/L) | 169 ± 598 | 127 ± 238 | 182 ± 672 | 0.7476 |
| ALT (GPT) (IU/L) | 94 ± 188 | 101 ± 193 | 92 ± 187 | 0.2135 |
| Bilirubin (mg/dL) | 1.2 ± 2.1 | 1.2 ± 1.8 | 1.2 ± 2.1 | 0.5972 |
CRP, C-reactive protein; AST, aspartate transaminase; GOT, glutamic oxaloacetic transaminase; ALT, alanine transaminase; GPT, glutamate-pyruvate transaminase.
Table 7.
Disease severity with regard to the CTLA-4 haplotype H1: TAA.
| Variable | All (n = 644) | H1: TAA | p-Value | |
|---|---|---|---|---|
| Positive (n = 447) | Negative (n = 197) | |||
| SOFA | 7.0 ± 3.5 | 7.0 ± 3.6 | 7.1 ± 3.5 | 0.5088 |
| SOFA-Respiratory score | 2.0 ± 0.8 | 2.0 ± 0.8 | 1.9 ± 0.8 | 0.9796 |
| SOFA-Cardiovascular score | 1.5 ± 1.0 | 1.5 ± 1.0 | 1.5 ± 1.0 | 0.7476 |
| SOFA-Central Nervous System score | 2.0 ± 1.1 | 2.0 ± 1.1 | 1.9 ± 1.1 | 0.2731 |
| SOFA-Renal score | 0.8 ± 1.2 | 0.7 ± 1.1 | 0.9 ± 1.3 | 0.1230 |
| SOFA-Coagulation score | 0.4 ± 0.6 | 0.4 ± 0.6 | 0.3 ± 0.6 | 0.6741 |
| SOFA-Hepatic score | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.4 ± 0.8 | 0.4264 |
| Organ support | ||||
| Ventilation days/observation days (%) | 66 ± 32 | 67 ± 32 | 65 ± 32 | 0.3607 |
| Vasopressor days/observation days (%) | 34 ± 30 | 33 ± 30 | 35 ± 30 | 0.6474 |
| Dialysis days/observation days (%) | 9 ± 23 | 9 ± 22 | 11 ± 23 | 0.1246 |
| Inflammatory values | ||||
| Leucocytes (1000/µL) | 13 ± 5 | 13 ± 5 | 13 ± 4 | 0.4087 |
| CRP (mg/L) | 152 ± 87 | 147 ± 81 | 162 ± 99 | 0.4562 |
| Procalcitonin (ng/dL) | 4.0 ± 9.3 | 3.5 ± 9.1 | 5.1 ± 9.7 | 0.0002 |
| Kidney values | ||||
| Urine output (mL/day) | 2978 ± 1337 | 3060 ± 1370 | 2792 ± 1242 | 0.0229 |
| Urine output (mL/kg/day) | 1.6 ± 0.8 | 1.6 ± 0.8 | 1.5 ± 0.8 | 0.0529 |
| Creatinine (mg/dL) | 1.2 ± 0.9 | 1.2 ± 0.9 | 1.4 ± 1.1 | 0.0902 |
| Liver values | ||||
| AST (GOT) (IU/L) | 169 ± 598 | 184 ± 687 | 131 ± 272 | 0.9910 |
| ALT (GPT) (IU/L) | 94 ± 188 | 99 ± 206 | 81 ± 142 | 0.1307 |
| Bilirubin (mg/dL) | 1.2 ± 2.1 | 1.2 ± 1.8 | 1.3 ± 2.5 | 0.9584 |
The H1: TAA noncarriers showed highly significant higher average procalcitonin levels (5.1 ± 9.7 ng/dL vs. 3.5 ± 9.1 ng/dL; p = 0.0002) as well as significantly lower urine outputs (2792 ± 1242 mL/day vs. 3060 ± 1370 mL/day; p = 0.0229) over the observation period than the H1: TAA-positive patients.
4. Discussion
Identification of genetic variants associated with the outcome of sepsis is important for understanding the pathophysiology of the disease and identifying patients at a higher risk of death, and may even reveal future targets for personalized medicine. The present study aimed to identify associations between functional SNPs and haplotypes of the CTLA-4 gene with the outcomes of septic patients. The main finding was that in addition to our previous identification of the CTLA-4 rs231775 SNP as a prognostic variable for the outcome of septic patients, the functional CTLA-4 rs3087243 SNP and the inferred CTLA-4 haplotype H1: TAA were significantly associated with sepsis survival. Both CTLA-4 rs3087243 G allele carriers and CTLA-4 H1: TAA haplotype-negative patients had significantly better 90- and 28-day survival. To the best of our knowledge, this study is the first to report an association of these genetic variants with sepsis survival. The identified associations remained significant predictors for 90- and 28-day mortality in the multivariate Cox regression analysis, indicating that despite potential confounders, they were independent prognostic variables for the survival of septic patients.
The findings at the CTLA-4 rs3087243 position could be explained by observations that revealed lower CTLA-4 protein expression levels as well as higher susceptibility to autoimmune diseases in rs3087243 G allele carriers [24,26,27,28]. We can assume that lower expression levels of the coinhibitory regulatory CTLA-4 protein in rs3087243 G allele carriers result in an enhanced T lymphocyte host immune response [37], which may be beneficial in the immunosuppressive phase of sepsis. Our hypothesis that genetic variants associated with lower CTLA-4 expression levels and autoimmune disease susceptibility may be favorable during the course of sepsis is further supported by our findings for the H1: TAA haplotype. This most common CTLA-4 haplotype consists of the rs733618 T, rs231775 A, and rs3087243 A alleles, of which the second two are associated with unfavorable sepsis outcomes. Not carrying this haplotype was significantly associated with better survival during the course of sepsis.
Furthermore, our study revealed that CTLA-4 rs3087243 G allele carriers showed significantly better sepsis disease severity in terms of a lower average SOFA-CNS score and a reduced need for organ support, as represented by a reduced number of ventilation days as a fraction of observation days (Table 6). Significantly lower average urine outputs (mL/day) and higher procalcitonin levels were observed for the H1: TAA-negative patients (Table 7). Although the lower urine output levels can be explained by the significantly higher administration of renal replacement therapy to the H1: TAA-negative patients in the patient baseline characteristics, we do not have a satisfactory explanation for the higher average procalcitonin levels.
The present study has some limitations. This study is a single-center study focusing on a relatively large and prospectively enrolled cohort of severely ill patients from surgical ICUs; no patients from medical ICUs were included in the study. Therefore, our observations may not be completely applicable to other ICU cohorts. In addition, our findings should be validated in additional patient cohorts, ideally including patients of other ethnicities and from other ICUs (i.e., neurological or medical ICUs).
5. Conclusions
Although a sepsis-specific biomarker is nonexistent at present, potential for the use of genetic biomarkers for prognostic purposes is high. In this context, our study provides further evidence implicating CTLA-4 polymorphisms and haplotypes in determining sepsis severity and mortality, and thus they may serve as prognostic variables. However, the molecular mechanisms by which CTLA-4 polymorphisms and haplotypes influence the sepsis disease severity and mortality risk remain to be identified. Our findings raise hope that identification of patients at risk and development of tailored therapeutic approaches may be enabled by the effective use of modern molecular diagnostics, including evaluation of genetic variants, such as the SNPs and haplotypes identified in our present study.
Acknowledgments
The authors thank the staff of the ICUs of the Department of Anesthesiology and Department of General and Visceral Surgery, and all of those who were involved in patient care and monitoring. The authors thank Benjamin Liese, Simon Wilmers, Yvonne Klee, Sebastian Gerber, Chang Ho Hong, Fabian Kriesel, and Carolin Böhnke for their help with data acquisition. The authors thank Karolin Jobst of the Department of Clinical Pharmacology for her support and supervision on the DNA extraction and genotyping.
Author Contributions
Conceptualization, C.M., J.H., A.-F.P., M.G., T.B., M.T., M.Q., S.S.-O., I.B., and A.M.; Data curation, C.M., B.B., and J.R.; Formal analysis, C.M., J.H., T.B., and O.J.; Funding acquisition, A.M.; Investigation, C.M., B.B., A.A., A.-F.P., M.G., T.B., M.T., J.R., and A.M.; Methodology, C.M., A.A., T.B., S.S.-O., and A.M.; Project administration, A.A., A.-F.P., M.G., O.J., J.R., and I.B.; Resources, M.G., M.T., and M.Q.; Software, J.H.; Supervision, J.H., M.Q., and A.M.; Validation, T.B. and A.M.; Visualization, C.M.; Writing—original draft, C.M., J.H., and A.M.; Writing—review & editing, B.B., A.A., A.-F.P., M.G., T.B., M.T., O.J., J.R., M.Q., S.S.-O., and I.B.
Funding
This research was funded by the VolkswagenStiftung, grant ZN3168.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent J.-L., Marshall J.C., Ñamendys-Silva S.A., François B., Martin-Loeches I., Lipman J., Reinhart K., Antonelli M., Pickkers P., Njimi H., et al. Assessment of the worldwide burden of critical illness: The Intensive Care Over Nations (ICON) audit. Lancet Respir. Med. 2014;2:380–386. doi: 10.1016/S2213-2600(14)70061-X. [DOI] [PubMed] [Google Scholar]
- 3.Gaieski D.F., Edwards J.M., Kallan M.J., Carr B.G. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit. Care Med. 2013;41:1167–1174. doi: 10.1097/CCM.0b013e31827c09f8. [DOI] [PubMed] [Google Scholar]
- 4.Mansur A., Mulwande E., Steinau M., Bergmann I., Frederik Popov A., Ghadimi M., Beissbarth T., Bauer M., Hinz J. Chronic kidney disease is associated with a higher 90-day mortality than other chronic medical conditions in patients with sepsis. Sci. Rep. 2015;5:10539. doi: 10.1038/srep10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angus D.C., van der Poll T. Severe Sepsis and Septic Shock. [(accessed on 14 October 2018)]; Available online: https://www.nejm.org/doi/10.1056/NEJMra1208623?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov.
- 6.Namath A., Patterson A.J. Genetic polymorphisms in sepsis. Crit. Care Nurs. Clin. N. Am. 2011;23:181–202. doi: 10.1016/j.ccell.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Mansur A., Liese B., Steinau M., Ghadimi M., Bergmann I., Tzvetkov M., Popov A.F., Beissbarth T., Bauer M., Hinz J. The CD14 rs2569190 TT genotype is associated with an improved 30-day survival in patients with sepsis: A prospective observational cohort study. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0127761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansur A., von Gruben L., Popov A.F., Steinau M., Bergmann I., Ross D., Ghadimi M., Beissbarth T., Bauer M., Hinz J. The regulatory toll-like receptor 4 genetic polymorphism rs11536889 is associated with renal, coagulation and hepatic organ failure in sepsis patients. J. Transl. Med. 2014;12:177. doi: 10.1186/1479-5876-12-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansur A., Hinz J., Hillebrecht B., Bergmann I., Popov A.F., Ghadimi M., Bauer M., Beissbarth T., Mihm S. Ninety-day survival rate of patients with sepsis relates to programmed cell death 1 genetic polymorphism rs11568821. J. Investig. Med. 2014;62:638–643. doi: 10.2310/JIM.0000000000000059. [DOI] [PubMed] [Google Scholar]
- 10.Arcaroli J., Fessler M.B., Abraham E. Genetic polymorphisms and sepsis. Shock. 2005;24:300–312. doi: 10.1097/01.shk.0000180621.52058.e1. [DOI] [PubMed] [Google Scholar]
- 11.Man M., Close S.L., Shaw A.D., Bernard G.R., Douglas I.S., Kaner R.J., Payen D., Vincent J.-L., Fossceco S., Janes J.M., et al. Beyond single-marker analyses: Mining whole genome scans for insights into treatment responses in severe sepsis. Pharmacogenom. J. 2013;13:218–226. doi: 10.1038/tpj.2012.1. [DOI] [PubMed] [Google Scholar]
- 12.Nobel Media AB 2018 All Nobel Prizes. [(accessed on 15 October 2018)]; Available online: https://www.nobelprize.org/prizes/lists/all-nobel-prizes/
- 13.Fallon E.A., Biron-Girard B.M., Chung C.-S., Lomas-Neira J., Heffernan D.S., Monaghan S.F., Ayala A. A novel role for coinhibitory receptors/checkpoint proteins in the immunopathology of sepsis. J. Leukoc. Biol. 2018 doi: 10.1002/JLB.2MIR0917-377R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walunas T.L., Lenschow D.J., Bakker C.Y., Linsley P.S., Freeman G.J., Green J.M., Thompson C.B., Bluestone J.A. Pillars article: CTLA-4 can function as a negative regulator of T cell activation. immunity. 1994. 1: 405–413. J. Immunol. 2011;187:3466–3474. [PubMed] [Google Scholar]
- 15.Gardner D., Jeffery L.E., Sansom D.M. Understanding the CD28/CTLA-4 (CD152) pathway and its implications for costimulatory blockade. Am. J. Transpl. 2014;14:1985–1991. doi: 10.1111/ajt.12834. [DOI] [PubMed] [Google Scholar]
- 16.McCoy K.D., Le Gros G. The role of CTLA-4 in the regulation of T cell immune responses. Immunol. Cell Biol. 1999;77:1–10. doi: 10.1046/j.1440-1711.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- 17.Buchbinder E.I., Desai A. CTLA-4 and PD-1 pathways: Similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stearns-Kurosawa D.J., Osuchowski M.F., Valentine C., Kurosawa S., Remick D.G. The pathogenesis of sepsis. Annu. Rev. Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Li J., Lou J., Zhou Y., Bo L., Zhu J., Zhu K., Wan X., Cai Z., Deng X. Upregulation of programmed death-1 on T cells and programmed death ligand-1 on monocytes in septic shock patients. Crit. Care. 2011;15 doi: 10.1186/cc10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mewes C., Büttner B., Hinz J., Alpert A., Popov A.F., Ghadimi M., Beissbarth T., Tzvetkov M., Shen-Orr S., Bergmann I., et al. The CTLA-4 rs231775 GG genotype is associated with favorable 90-day survival in Caucasian patients with sepsis. Sci. Rep. 2018;8:15140. doi: 10.1038/s41598-018-33246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soskic B., Qureshi O.S., Hou T., Sansom D.M. Chapter Four—A Transendocytosis Perspective on the CD28/CTLA-4 Pathway. In: Alt F.W., editor. Advances in Immunology. Volume 124. Academic Press; Cambridge, MA, USA: 2014. pp. 95–136. [DOI] [PubMed] [Google Scholar]
- 22.Benhatchi K., Jochmanová I., Habalová V., Wagnerová H., Lazúrová I. CTLA4 exon1 A49G polymorphism in Slovak patients with rheumatoid arthritis and Hashimoto thyroiditis—Results and the review of the literature. Clin. Rheumatol. 2011;30:1319–1324. doi: 10.1007/s10067-011-1752-z. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Pirskanen R., Giscombe R., Lefvert A.K. Two SNPs in the promoter region of the CTLA-4 gene affect binding of transcription factors and are associated with human myasthenia gravis. J. Int. Med. 2008;263:61–69. doi: 10.1111/j.1365-2796.2007.01879.x. [DOI] [PubMed] [Google Scholar]
- 24.Torres B., Aguilar F., Franco E., Sánchez E., Sánchez-Román J., Alonso J.J., Núñez-Roldán A., Martín J., González-Escribano M.F. Association of the CT60 marker of the CTLA4 gene with systemic lupus erythematosus. Arthritis Rheum. 2004;50:2211–2215. doi: 10.1002/art.20347. [DOI] [PubMed] [Google Scholar]
- 25.Hudson L., Rocca K., Song Y., Pandey J. CTLA-4 gene polymorphisms in systemic lupus erythematosus: A highly significant association with a determinant in the promoter region. Hum. Genet. 2002;111:452–455. doi: 10.1007/s00439-002-0807-2. [DOI] [PubMed] [Google Scholar]
- 26.Zaletel K., Krhin B., Gaberšček S., Biček A., Pajič T., Hojker S. Association of CT60 cytotoxic T lymphocyte antigen-4 gene polymorphism with thyroid autoantibody production in patients with Hashimoto’s and postpartum thyroiditis. Clin. Exp. Immunol. 2010;161:41–47. doi: 10.1111/j.1365-2249.2010.04113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douroudis K., Prans E., Kisand K., Nemvalts V., Uibo R. Cytotoxic T-lymphocyte antigen 4 gene polymorphisms are associated with latent autoimmune diabetes in adults. Clin. Chim. Acta. 2009;403:226–228. doi: 10.1016/j.cca.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Karabon L., Kosmaczewska A., Bilinska M., Pawlak E., Ciszak L., Jedynak A., Jonkisz A., Noga L., Pokryszko-Dragan A., Koszewicz M., et al. The CTLA-4 gene polymorphisms are associated with CTLA-4 protein expression levels in multiple sclerosis patients and with susceptibility to disease. Immunology. 2009;128:787–796. doi: 10.1111/j.1365-2567.2009.03083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muro M., Rojas G., Botella C., Miras M., Campillo J.A., Minguela A., Sánchez-Bueno F., Bermejo J., Ramírez P., Álvarez-López M.R. CT60 A/G marker of the 3′-UTR of the CTLA4 gene and liver transplant. Transpl. Immunol. 2008;18:246–249. doi: 10.1016/j.trim.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Bone R.C., Balk R.A., Cerra F.B., Dellinger R.P., Fein A.M., Knaus W.A., Schein R.M.H., Sibbald W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 31.Hinz J., Büttner B., Kriesel F., Steinau M., Frederik Popov A., Ghadimi M., Beissbarth T., Tzvetkov M., Bergmann I., Mansur A. The FER rs4957796 TT genotype is associated with unfavorable 90-day survival in Caucasian patients with severe ARDS due to pneumonia. Sci. Rep. 2017;7:9887. doi: 10.1038/s41598-017-08540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens M., Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Submitted SNP(ss) Details: ss79540. [(accessed on 21 November 2018)]; Available online: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss79540.
- 35.Submitted SNP(ss) Details: ss44315036. [(accessed on 21 November 2018)]; Available online: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss44315036.
- 36.Mansur A., Steinau M., Popov A.F., Ghadimi M., Beissbarth T., Bauer M., Hinz J. Impact of statin therapy on mortality in patients with sepsis-associated acute respiratory distress syndrome (ARDS) depends on ARDS severity: A prospective observational cohort study. BMC Med. 2015;13:128. doi: 10.1186/s12916-015-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda H., Howson J.M.M., Esposito L., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M.D., Smith A.N., Di Genova G., et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]