Abstract
Changes to the cellular microenvironment are an integral characteristic of numerous pathologies, including cancer, fibrosis, and autoimmune disease. Current in vitro methodologies available to study three-dimensional tissue remodeling are ill-suited for high-throughput studies as they are not scalable for large-scale experiments. Combining droplet microfluidics and patterned low-adhesion culture surfaces, we have engineered a workflow to incorporate cell–extracellular matrix (ECM) interactions in a versatile and high-throughput platform that is compatible with existing high-throughput liquid handling systems, enables long-term experiments (>1 month), and is well suited for traditional and novel biological measurements. With our platform, we demonstrate the feasibility of high-throughput ECM remodeling studies with collagen microtissues as one application of a tissue-level function. In this study, we use our workflow to examine ECM remodeling at the tissue, cell, and subcellular levels, leveraging assays ranging from immunohistochemistry and live cell imaging, to proliferation and contraction assays. With our unique culture system, we can track individual constructs over time and evaluate remodeling on several scales for large populations. Finally, we demonstrate the ability to cryopreserve our microtissues while retaining high viability and cell function, an invaluable method that could allow for dissemination and freezing of microtissues after mass production. Using these methods, our ECM-based system becomes a viable platform for modeling diseases characterized by tissue reorganization as well as a scalable method to conduct in vitro cell-based assays for drug screening and high-throughput biological discovery.
Impact Statement
The described microtissue–microwell workflow is uniquely suited for high-throughput study of extracellular matrix (ECM) remodeling at the molecular, cellular, and tissue levels and demonstrates possibilities of studying progressive, heterogeneous diseases in a way that is meaningful for drug discovery and development. We outline several assays that can be utilized in studying tissue-level diseases and functions that involve cell–ECM interactions and ECM remodeling (e.g., cancer, fibrosis, wound healing) in pursuit of an improved three-dimensional cell culturing system. Finally, we demonstrate the ability to cryopreserve cells encapsulated in microtissue constructs while remaining highly viable, proliferative, and retaining cell functions that are involved in ECM remodeling.
Keywords: 3D cell culture, microtissues, droplets, microfluidics, high-throughput screening, ECM remodeling
Introduction
Alteration to extracellular matrix (ECM) composition and organization is a key hallmark of several diseases, including cancer,1–4 fibrosis,2,5,6 and autoimmune disease.6–8 The ECM not only provides structural support and enables cell–ECM signaling, but acts as a reservoir for endogenous growth factors,9–12 behaves as a barrier to therapy,13–15 and participates in generation of bioactive protein fragments involved in diseased state cell signaling.16,17 Furthermore, these pathologies progress and evolve over time, and tissue disorganization requires prolonged periods to fully develop.
Mechanisms that involve cell–ECM interactions and remodeling over time are prominent in cancer, fibrosis, and autoimmune research and are favored targets for drug development; however, there remains an unmet need for model systems that could be used in the initial stages of drug development at a tissue level.18,19 Furthermore, ECM remodeling is difficult to visualize and quantify especially in the case of degenerative disease as these timescales can be difficult to recapitulate in vitro, revealing a need for platform that is compatible with high-throughput screening and retains complex cell–cell, cell–ECM, and architectural cues while enabling quantification of cell behavior over time.
Current in vitro methodologies available to perform high-throughput and long-term culture studies include both standard two-dimensional (2D) culture and spheroids,20–23 but they are ill-suited for probing the mechanisms of matrix remodeling as they lack an ECM component.24,25 Platforms that are more appropriate for addressing these questions exist, however fabrication is rate limiting and handling is time consuming, ultimately reducing sample size and preventing use for high-throughput applications. The most relevant examples are large hydrogels and microfluidic model systems. Hydrogels allow for incorporation of ECM proteins and are compatible with automated liquid handling systems, but they require substantial volumes of matrix and cells, elevating cost and reducing throughput with finite cell sources. Furthermore, these gels are often so large they are: (1) limited by diffusion, reducing nutrient transport and constraining staining choices,26,27 and (2) restricted in compatible imaging modalities.28–30
Microfluidic devices meanwhile provide opportunities to incorporate spatial organization, can integrate multiple matrix proteins, and require smaller reagent volumes than other methods.31–33 However, these platforms depend on consistent tissue geometry to maintain complex flow patterns, development of chemical gradients, and the ability to track movement of cells over time, making compaction to physiologically relevant ECM densities difficult to study. In addition, chip scalability is limited even with advanced device design, indicating a persistent need for high-throughput ECM remodeling platforms.
Leveraging droplet microfluidics, we have previously demonstrated use of protein-based microtissues for high-throughput, short-term studies of cellular phenotypes and interactions with ECM.34,35 To expand the capabilities of our microtissue platform, we engineered a workflow that couples microtissue fabrication with culture in patterned agarose microwells in a manner that is compatible with existing liquid handling techniques, enables long-term experiments (>1 month), and is well-suited for several traditional and novel biological measurements.
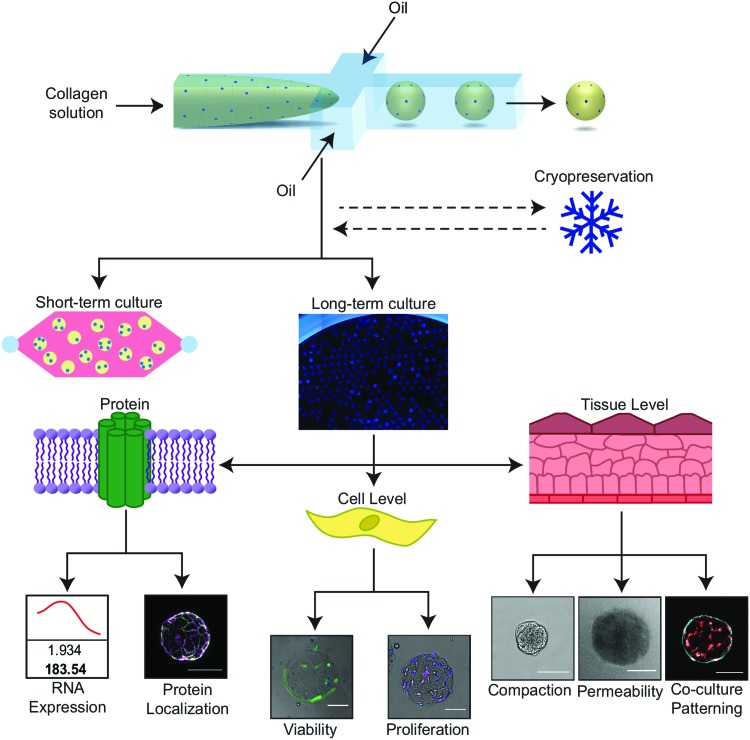
In this study, we demonstrate compatibility of our long-term ECM-based culture platform with several molecular, cellular, and tissue-level measurements to assess cell phenotype and function (Fig. 1). In this study, we focus on ECM remodeling as a tissue-level function that is quantifiable in our platform and is highly relevant to developing therapeutics for cancer, fibrosis, and autoimmune disease. We demonstrate that we can quantify local and global compaction, reversible and irreversible remodeling, and heterogeneity in compaction rates of individual microtissues. Additionally, we demonstrate that microtissues incorporating cells can be cryopreserved for distribution or use in later experiments. Using these methods, our ECM-based system becomes a viable platform for modeling diseases characterized by tissue reorganization as well as a scalable method to conduct in vitro assays for drug screening and high-throughput biological discovery.
FIG. 1.
The microtissue–microwell workflow facilitates functional and mechanistic analysis at multiple scales. Our workflow facilitates studies of tissue remodeling and disease progression on tissue, cellular, and subcellular scales in short- and long-term studies, while visualizing global and local behaviors. This system is compatible with a wide range of cell types, ECM proteins, and biological assays, making this a practical solution for high-throughput fabrication and handling of 3D tissue engineering construct. Scale bars are 100 μm. 3D, three-dimensional; ECM, extracellular matrix. Color images available online at www.liebertpub.com/tec
Materials and Methods
Cell culture
Normal human lung fibroblasts (NHLFs) were cultured in FGM-2 (Lonza) supplemented with 1 × antibiotic–antimycotic and used between passage 3 and 7. National Institute of Health 3T3 (NIH 3T3) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 1 × antibiotic–antimycotic and were used between passage 15 and 20. MDA-MB-231 cells were cultured in the same DMEM-based media and used at passage 9. Lastly, human umbilical vein endothelial cells (HUVECs) were cultured on gelatin-coated flasks in EGM-2 (Lonza) and used at passage 2. Cells were released from flasks with 0.05% trypsin-EDTA and trypsin was neutralized with respective growth media before counting cells.
Microtissue fabrication
Collagen microtissues ∼300 μm in diameter were fabricated using previously established protocols.34,35 Briefly, high-concentration rat tail collagen I (Corning) was buffered with 10 × Dulbecco's phosphate-buffered saline (DPBS), neutralized to pH 7.4, and diluted to a concentration of 6 mg/mL. For cell-laden microtissues, cells were resuspended in the collagen solution to a final concentration of 2 million cells/mL. At 4°C, the collagen solution was partitioned into droplets using a flow-focusing polydimethylsiloxane (PDMS; Dow Corning) microfluidic device. The continuous phase from the droplet generation (FC-40 with 2% 008-FluoroSurfactant; Ran Biotechnologies), was collected with the collagen microtissues in a low-retention Eppendorf tube and polymerized for 20 min at room temperature. The oil phase was removed and the collagen microtissues were resuspended in 1 × DPBS with a manual micropipette.
Microwell fabrication
Polystyrene multiwell plates were coated with a thin layer of 2% agarose and dehydrated in a sterile laminar flow hood overnight. PDMS stamps featuring a radial pattern of 300 μm diameter posts were plasma treated for 2 min to produce a hydrophilic surface and sterilized with boiling water. For a 24-well plate, 175 μL of molten 2% agarose solution was pipetted into each well and the hydrophilic PDMS stamp placed immediately onto the agarose. After cooling for 5 min, stamps were removed gently from the polymerized agarose and hydrated with DPBS. Wells were washed with appropriate culturing media before adding microtissues with a manual micropipette. The stamped plates are referred to as “microwell plates.”
Microtissue coating protocol
Fifty thousand cells were added to each well of a microwell plate and allowed to settle into the microwells for 5 min. Acellular microtissues were then added and allowed to settle into microwells. Finally, an additional 50,000 cells were added, sandwiching the tissues between two layers of cells.
Cryopreservation
Microtissues were resuspended in freezing solution (90% FBS, 10% dimethyl sulfoxide [DMSO]) and frozen in 500 μL aliquots. Tissues were cooled gently using a Mr. Frosty (Thermo Scientific) overnight and then transferred to liquid nitrogen for 1 week. Control tissues from the same experiment were pipetted into plates containing microwells immediately after fabrication to compare the effects of freezing.
Viability and proliferation assays
Microtissues containing encapsulated cells were fabricated and cultured for up to 1 month. At various time points, tissues were gently pipetted and removed from microwells and collected in an Eppendorf tube. For viability staining, constructs were washed thoroughly with DPBS and then incubated with a staining solution of 5 μM DRAQ5 (Invitrogen) and 5 μM Calcein AM at 37°C for 20 minutes. A Zeiss Axio Observer was used to image z-positions throughout each microtissue with a step size of 10 μm, which is sufficiently small to account for each cell. Collected z-stacks were analyzed and the total number of cells (from the nuclear stain) and the number of live cells (from the Calcein) were counted for at least 24 microtissues to calculate the percentage of live cells.
To visualize proliferation, microtissues were incubated for 4 h with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) before fixing with 3.7% formalin and the Click-iT EdU Alexa Fluor 647 Kit (Invitrogen) was used following the manufacturer's instructions. A Zeiss Axio Observer was used to image 20 z-positions throughout each microtissue with a step size of 10 μm. At least 28 microtissues were examined for each condition.
Compaction assay
Projected areas and shape descriptors were measured by thresholding images of encapsulated fluorescent, 1 μm diameter Fluoro-Max Microspheres (Thermo Scientific). Outliers, determined as areas three standard deviations above or below the average, were removed from the data sets.
Second-harmonic generation imaging and analysis
NHLF-coated microtissues were incubated in 0.1%, 2%, and 10% FBS and collected and fixed with 3.7% formalin after 1 and 3 days of culture. Nuclei were stained with 80 nM Hoechst before the constructs were immobilized in 1% agarose for imaging. Second-harmonic generation (SHG) signal from the collagen fibers was collected with a custom multiphoton laser-scanning microscope (Prairie Technologies/Bruker) with a Mai Tai Ti:Sapphire laser (Spectra Physics). Fibers were excited at 880 nm. SHG signal intensity and construct circularity were measured using ImageJ (n = 4–6 microtissues per condition).
EDTA release
Microtissues were cultured for up to 5 days, with collections at 2 h and 1, 2, and 5 days. At each time point, half of the tissues were fixed with 3.7% formalin and the remaining microtissues were washed with DPBS and incubated in 5 mM EDTA solution at 37°C overnight before fixation with formalin.
Immunofluorescent staining
Microtissues were collected in low-retention Eppendorf tubes and fixed with 3.7% formalin overnight at 4°C. Formalin was quenched with 0.1 M glycine and constructs were blocked and permeabilized with 10% FBS and 0.1% Triton X-100 for 1 h at room temperature. The tissues were then incubated with primary antibody (rabbit anti-fibronectin [1:200; Abcam], mouse anti-collagen IV [1:200;Abcam]) at 4°C overnight. The microtissues were washed thoroughly, and incubated with secondary antibodies (1:200; Jackson) and/or phalloidin (1:100; Santa Cruz Biotech) at 4°C overnight. Nuclei were visualized with 20 μM Hoechst. A Zeiss Axio Observer with Apotome was used to obtain optical sections.
Tracking and multiplexing
Collagen microtissues containing 1 μm Fluoro-Max Microspheres and encapsulated NHLFs were cultured in 0.1%, 2%, and 10% FBS for a period of 6 days and imaged at several time points. Cultures were maintained with half-volume media changes every other day. On the final day of the experiment, tissues were fixed in microwells with 3.7% formalin and then stained with phalloidin (1:100; Santa Cruz Biotech). Microtissues were then imaged on widefield in microwells using a Zeiss Axio Observer. A semiautomated custom macro was used to calculate the projected tissue area for all time points and, for the final time point, actin fluorescence was also quantified.
Statistical analysis
All experiments reported in this article were completed at least three times with similar results. Statistical significance of EDTA release experiments was performed with paired t-tests and significance was determined using Bonferroni's correction. Significance of tracking and cryopreservation experiments was calculated with a student's t-test. Lastly, analysis of variance with Scheffé's method, where p-values <0.05 were considered significant, was used to determine significance in SHG experiments.
Results
The combination of droplet technology and agarose microwells enables a high-throughput workflow and long-term culture of protein-based microtissues
A major hurdle that must be overcome in any high-throughput tissue engineering application is scalability of construct fabrication as well as the cost and availability of required reagents. To address these concerns, our protocol leverages a microfluidic flow-focusing device to produce tens of thousands of nanoliter-volume microtissues per hour (Fig. 1). We fabricate more than 20,000 microtissues using the same reagent volume as is used to cast 6 traditional hydrogels in a 96-well plate. Additionally, all described experiments utilized a maximum of 20 cells per construct, meaning these assays could be scaled for large compound-screens with a limited number of cells.
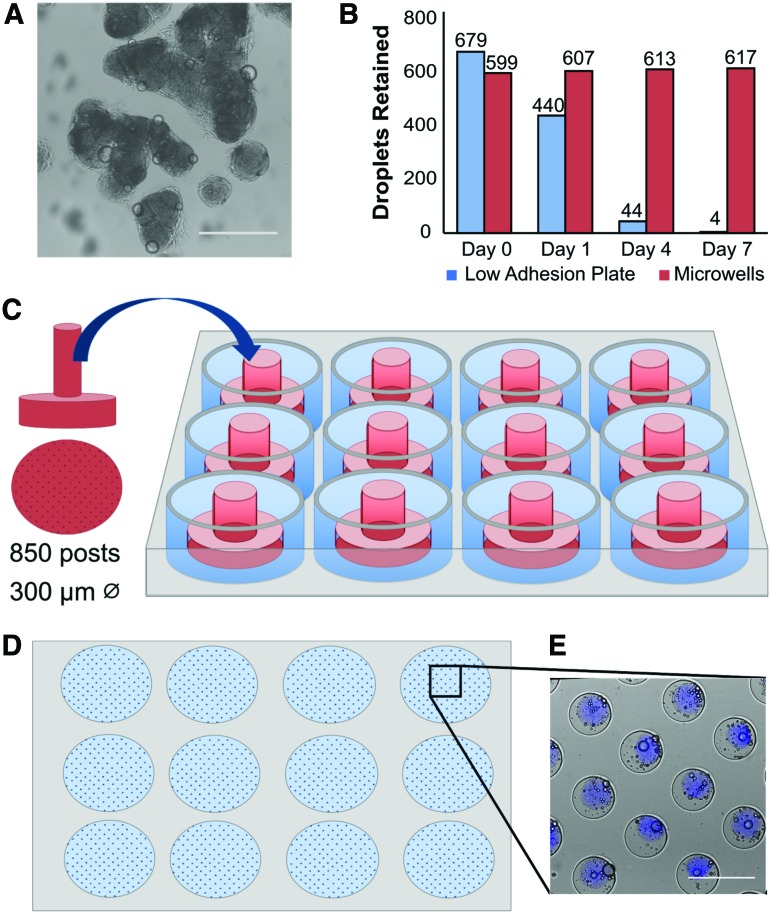
Although we have previously demonstrated that these protein-based microtissues are useful for observing short-term tissue remodeling, it has been widely documented that most three-dimensional (3D) tissue culture systems, including our microtissues, typically aggregate in static culture (Fig. 2A), limiting their use for long-term culture and high-content readouts.36–38 To address this challenge, we fabricated patterned agarose microwells using PDMS molds (Fig. 2C, D). We chose agarose as it is affordable, but also biocompatible, nonadhesive for cells, amenable to patterning, and has precedence in the field for this purpose.39–42 These microwells physically separated microtissues and prevented aggregation (Fig. 2E).
FIG. 2.
Microwells enabled long-term culture of large microtissue populations. (A) Brightfield imaging reveals that microtissues aggregate when cultured for 4 days in standard low adhesion plates. (B) Quantification of individual microtissues after 1 week of culture in standard low adhesion plates (blue bars) reveals that the population is reduced by over 100-fold. By contrast, microtissues cultured in patterned agarose microwells (red bars), remained separated over the same 1-week period. (C) To fabricate microwells, plasma-treated PDMS stamps with 300-μm diameter posts were placed into 2% agarose solution and (D) removed once polymerized. (E) Brightfield combined with blue fluorescence imaging shows microtissues contained in agarose microwells in a 12-well plate. Microtissues are labeled blue by encapsulating blue fluorescent beads within the polymerized collagen matrix. Scale bars 500 μm. PDMS, polydimethylsiloxane. Color images available online at www.liebertpub.com/tec
When microtissues were cultured in commercially available low-adhesion plates, we observed a reduction in individual microtissues over a period of 1 week as they coalesced at the center of the well and formed aggregates (Fig. 2A), resulting in a 600-fold reduction in individual microtissues (Fig. 2B). By contrast, microtissues cultured in microwells demonstrate no loss in the population over 1 week (Fig. 2B). With the microwell culturing method, we cultured microtissues as independent entities for up to 1 month (>90% viability of encapsulated NHLFs after 30 days). Below, we demonstrate the utility of this platform and workflow for quantifying tissue remodeling.
Long-term compaction of microtissues reveals reversible and irreversible ECM remodeling by fibroblasts and endothelial cells
Wound healing is an ECM remodeling process involving fibroblast-driven wound contraction followed by ECM deposition and crosslinking to return mechanical integrity to a tissue. In vivo, this process occurs over the span of days to weeks, making it difficult to study with short-term in vitro model systems lacking an ECM component. Using our microtissue–microwell platform and leveraging its long-term culturing abilities, we modeled this dynamic wound-healing process with NHLFs in collagen constructs. For the purposes of this study, we used short-term culture to refer to time points that occur before 48 h of culture, a conservative estimate for protein synthesis to occur [Yan, Suzuki], and use the phrase long term to denote culture >2 days.
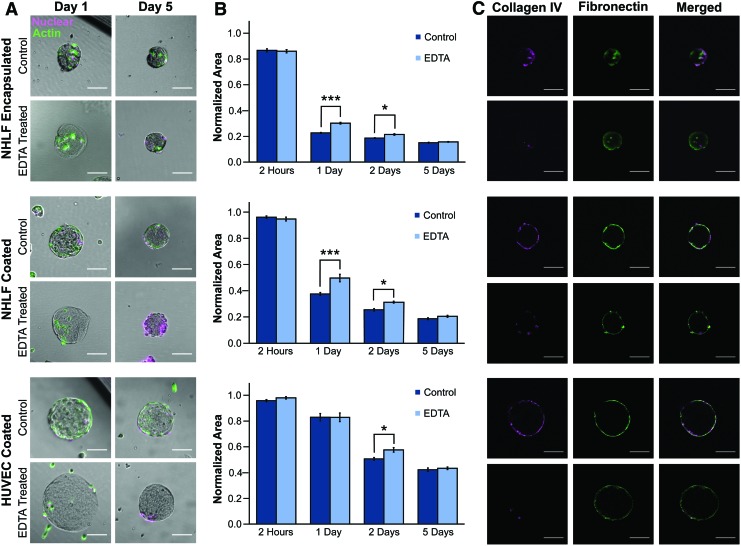
To identify reversible and irreversible changes to ECM over short and long timescales, we collected tissues at various stages of remodeling and examined ECM compaction and deposition before and after cell release. There are several enzymatic methods to remove cells from ECM, but many of these treatments are detrimental to the structural integrity of the ECM itself. Thus, we elected to use EDTA as it destabilizes calcium- and magnesium-dependent integrin linkages and is a gentle and minimally disruptive agent for both the cells and the matrix. From widefield imaging, we found that the fibroblasts compacted the collagen microtissues (Fig. 3A), but permanence of this remodeling was not immediately evident.
FIG. 3.
EDTA release allows for studying microtissue remodeling on short and long timescales. (A) Brightfield imaging of microtissues after 1 and 5 days of culture with encapsulated NHLFs or coated with NHLFs or HUVECs. The actin cytoskeleton was visualized with phalloidin (green) and nuclei with Draq5 (magenta). After 1 day, we observed a reduction in construct size, which was partially reversed after releasing cells with EDTA. After 5 days, this reversal was not observed. To quantify these effects with construct size, we measured (B) microtissue projected area and confirmed that on a population scale at early time points, compaction was partially reversible, while after 5 days of culture remodeling was not reversible. We calculated significance with a paired t-test with a Bonferroni correction for multiple comparisons (***p < 0.00025, *p < 0.013, standard error shown). Projected area is normalized to the average area of each microtissue batch after fabrication. (C) Fluorescence imaging with optical sectioning shows that fibronectin (green) and collagen IV (magenta) were deposited on the surface of the microtissues; however, the collagen IV was disrupted by EDTA treatment. All scale bars 100 μm and sample sizes are an average of 38 microtissues per condition. HUVECs, human umbilical vein endothelial cells; NHLFs, normal human lung fibroblasts. Color images available online at www.liebertpub.com/tec
Cell–ECM interactions likely stabilize microtissue compaction, and thus releasing cells would result in an increase in projected area for the tissue constructs. For both encapsulated and surface-coated NHLF conditions, we treated constructs with EDTA after 24 h of culture and we found significant expansion of 33.3% and 24.5%, respectively (p < 0.00025). The reversibility of ECM compaction decreased over time, with the cell release resulting in no significant expansion for encapsulated and surface-coated conditions (Fig. 3B).
After observing irreversible matrix expansion after 5 days, we hypothesized that ECM deposition may contribute to remodeling as microtissue compaction did not increase greatly from days 1 and 2 to 5. Using immunofluorescence, we found that after 5 days of culture, cells deposit collagen IV and fibronectin when encapsulated in or coated on collagen I microtissues. Furthermore, in addition to removing the cells, EDTA disrupted collagen IV, a finding observed previously.43,44 Incubating in EDTA solution, however, did not affect synthesized fibronectin, and deposition of this and other matrix proteins not vulnerable to chelating agents may have prevented microtissue expansion (Fig. 3C).
In addition to fibroblasts, endothelial cells also participate in ECM remodeling, and to probe this endothelial function, we measured microtissue compaction of HUVEC-coated microtissues. The endothelialized constructs were only minimally compacted after 24 h (Fig. 3B) and thus did not show significant expansion after cell release, a distinct difference to the nearly total amount of compaction performed by NHLFs in the first 24 h. After 2 days, however, we observed a significant expansion of 12.2% (p < 0.012) with EDTA treatment (Fig. 3B) as the HUVECs were able to contract and compact the matrix beneath them, but again calculated no significant difference (2.6%) after 5 days of culture. This inability to detect reversible changes in construct size at late time points mimicked the behavior of both coated and encapsulated NHLFs.
Although our results show that HUVECs compact microtissues more slowly than NHLFs, endothelial cells are also able to make irreversible changes to the ECM (Fig. 3B). Staining of ECM proteins confirmed that matrix was deposited at later time points in endothelial cell conditions as well (Fig. 3C). These results show that ECM remodeling by different cell types follow unique time courses, but it may also be possible that there is heterogeneity within the remodeling time course even within the same cell type. As microwells enable long-term culture and retention of individual microtissues in a consistent spatial arrangement, we can gather additional time scale-related data through tracking compaction time courses of discrete microtissues over the entire duration of an experiment.
Long-term tracking of individual microtissues reveals heterogeneity in cell–ECM interactions
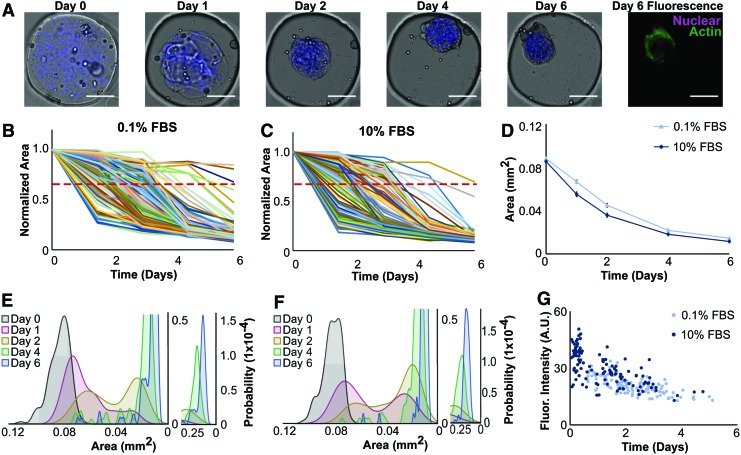
We have previously shown that ECM-based microtissues can be used for high-throughput, short-term compaction assays,34 and we demonstrated in this study how microwells can extend the experiment duration (Fig. 2A). In addition, our ability to retain microtissues as discrete samples in the same spatial arrangement allows for identifying and tracking constructs as unique entities to enable characterization of subpopulations. Over 1 week, we tracked compaction for over 125 fibroblast-laden microtissues per condition (Fig. 4A) and observed a distribution of compaction rates, with most major changes in size occurring on days 1, 2, and 4 (Fig. 4B, C).
FIG. 4.
Microwells facilitate tracking of discrete microtissues and coupling live imaging data with endpoint staining. (A) Brightfield and fluorescence imaging of collagen microtissues containing NHLFs and encapsulated fluorescent beads (blue) to mark construct borders were cultured in 0.1% (or 10%) serum for 1 week. All scale bars 100 μm. (B, C) Areas of 121 individual microtissues cultured in 0.1% FBS (B) and 137 constructs cultured in 10% FBS (C) were measured and plotted. A red dashed line indicates a threshold compaction level of 30% compaction. Projected area is normalized to day 0 for each microtissue compaction trajectory. (D) Population averages show similar compaction trajectories between 0.1% and 10% serum conditions. Shown with standard error. (E, F) Probability density functions of microtissue area at each time point reveal heterogeneity in the populations, indicated by bimodal distributions on day 1 (red) and day 2 (yellow) populations. (G) We observed a strong negative correlation between actin intensity and the time at which a 30% reduction in projected area occurred, as indicated by correlation coefficients of −0.68 and −0.65 for 0.1% and 10% FBS conditions, respectively. This suggests that a faster compaction rate is correlated to increased actin expression. We also found that higher serum conditions corresponded to elevated actin intensity and increased compaction rates. FBS, fetal bovine serum. Color images available online at www.liebertpub.com/tec
Despite different compaction rates we observed that nearly all microtissues compacted to similar sizes (final area of 13,000 μm2), with the greatest difference in projected area observed on days 1 and 2 (Fig. 4D). Plotting the probability density function for each serum condition revealed a bimodal population on days 1 and 2 that returned to a unimodal distribution by day 4 (Fig. 4E, F) indicating that a heterogeneous population of microtissues existed in each condition and that our system enables us to quantify these divergences within an experimental condition. Characterizing construct-level heterogeneity in our microtissue populations is beneficial as histology of pathologies, such as cancer and fibrosis, are variable throughout the diseased tissue.
To better understand the molecular origin of this heterogeneity, we visualized the cytoskeleton of cells within microtissues and found that tissues cultured in high serum expressed more actin than those cultured in low serum conditions (p < 0.0001) (Fig. 4G). As expected, we found that microtissues in 10% serum were not only on average brighter but compacted faster, decreasing to 30% of their original projected area more quickly than microtissues in the 0.1% condition (p = 0.002).
Additionally, we also found variation in the actin intensity within each condition. As the staining protocol did not disturb microtissue positions, we compared the compaction profile and the actin signal on an individual microtissue basis. For each microtissue, we calculated the time point at which the projected area had decreased by 30% and compared this value with the final phalloidin intensity. We found microtissues that compacted faster demonstrated increased actin intensity: we calculated a Pearson's correlation coefficient of −0.68 and −0.65 for 0.1% and 10% FBS conditions, respectively, between these variables, indicating a strong negative linear relationship for both conditions (Fig. 4G). By combining the contraction assay with immunofluorescence, we observed that actin intensity was related to but not directly correlated with final microtissue size.
Cells remodel microtissues on a local and global scale
Increased density of collagen and other ECM components is proportional to the stiffness of a tissue, an important factor in cancer and fibrosis.45–47 Multiphoton microscopy is a well-established tool used to investigate changes to global and local collagen densities through visualization of SHG signal. Increased signal correlates to a higher density of fibers and it is known that cells will organize and bundle fibers both in vivo and in vitro.48–50 Using SHG, we visualized changes in local collagen density and observed changes in both global and local ECM remodeling. First, we confirmed the fibrillar nature of acellular microtissues. We observed collagen fibers throughout the constructs and found they projected out of the microtissue free boundary surface, demonstrating their entangled nature and complex topography (Fig. 5A).
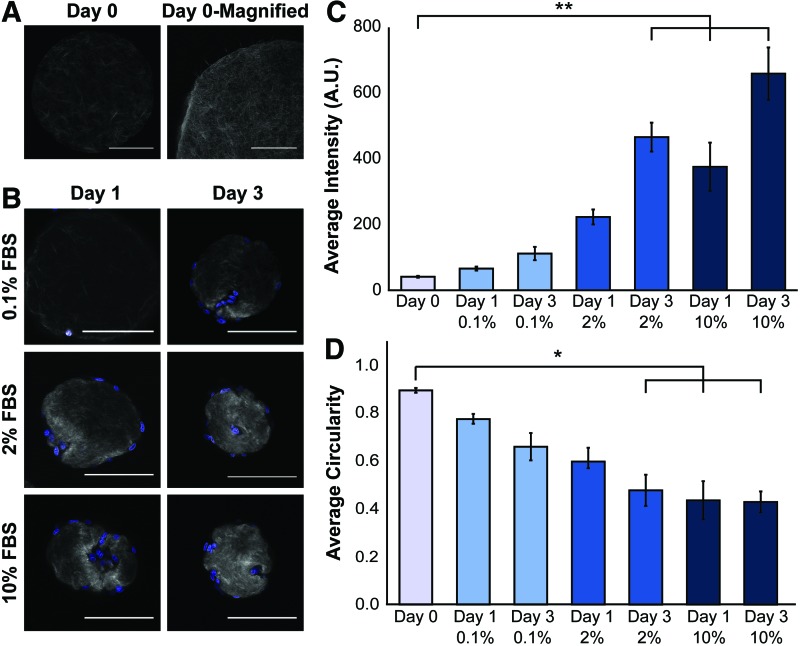
FIG. 5.
SHG microscopy revealed local ECM remodeling of collagen constructs. (A) Acellular collagen microtissues were coated with NHLFs and cultured for 3 days in varying serum concentrations. Collagen compaction and ECM remodeling were observed using SHG to visualize collagen fibers (gray) and Hoechst to stain the nuclei (blue). A section of an acellular slice is magnified (scale bar 50 μm) and the brightness adjusted to demonstrate the complex topography and fibrillar nature of the constructs. (B) Increased SHG signal intensity correlates to increased collagen fiber density and we qualitatively observed that with increasing serum and incubation time, cells interact with and increasingly modify their microenvironment. All other scale bars are 100 μm. (C) Average signal intensity from the collagen fibers was determined for each optical slice for each condition (n = 4–6 microtissues per condition). We observed that as serum concentrations increase, collagen fiber density increases due to compaction of fibroblasts. (D) Circularity of each construct is reported and with increasing serum concentrations and time, we found that microtissues became less circular (*p < 0.05, **p < 0.01, shown with standard error). SHG, second-harmonic generation. Color images available online at www.liebertpub.com/tec
Then, to assess ECM remodeling through collagen fiber organization and bundling, we examined coated collagen microtissues. As found in analogous large gel studies, we observed high serum conditions resulted in significant ECM remodeling,51,52 with a statistically significant increase in SHG signal with 10% serum (Fig. 5B). At early time points for all conditions, we observed increased collagen fiber intensity adjacent to nuclei, indicating that local remodeling and compaction of fibers is predictably occurring nearest to cells. After 3 days in culture, the global ECM remodeling was more evident as this localized intensity change was less apparent and the entire construct exhibited elevated SHG signal (Fig. 5A). We also observed that higher serum concentrations resulted in more significant changes to overall construct shape, changing the circularity from 0.88 to 0.47 after 3 days of culture in 10% FBS (Fig. 5C).
These differences in collagen density may help explain the discrepancy in actin intensity staining observed in NHLFs cultured in low (0.1%) and high (10%) serum culturing conditions (Fig. 4G), despite similar construct size as was quantified from widefield imaging. Furthermore, the transition from local changes in collagen density at early time points to more uniform, global increases in collagen density may explain the irreversible remodeling that occurs at later time points (Fig. 3). Lastly, we observed that cells had migrated through the constructs as well as caused local collagen degradation. These migration tracks throughout the constructs are especially apparent in the 10% FBS conditions (Fig. 5A). This is another form of ECM remodeling that fundamentally changes the constructs and it is observable in our microtissue system.
Cells retain their ability to remodel ECM and to proliferate after cryopreservation in microtissues
Cryopreservation is important for preserving precious cell populations and, as an experimental tool, maximizes the number of experiments that can be completed out of a single cell source. Freezing large tissues is challenging because diffusion limitations prevent cryoprotectants from fully penetrating the tissue and results in damage during freezing. The small size scale of our microtissues, however, allows for sufficient transport of cryoprotectant necessary for freezing while retaining the ability to study tissue-level functions.
To assess the compatibility of our microtissues with cryopreservation protocols, we fabricated constructs with various cell types encapsulated in 6 mg/mL collagen. Immediately after microtissue fabrication, we cultured half of our microtissues following standard protocols and cryopreserved the remainder using distributor-recommended freezing solutions. Cryopreserved NIH 3T3, MDA-MB-231, and NHLF cells encapsulated in 6 mg/mL collagen microtissues demonstrated high viability 1 day after thawing (84%, 87%, and 83%, respectively), which was similar to their controls (86%, 94%, and 90%). The percentage of live cells improved after a week of culture (92%, 98%, and 88%) (Fig. 6A), which again was similar to the corresponding controls (90%, 97%, and 89%), with viability at the 2-week time point remaining similarly high (data not shown).
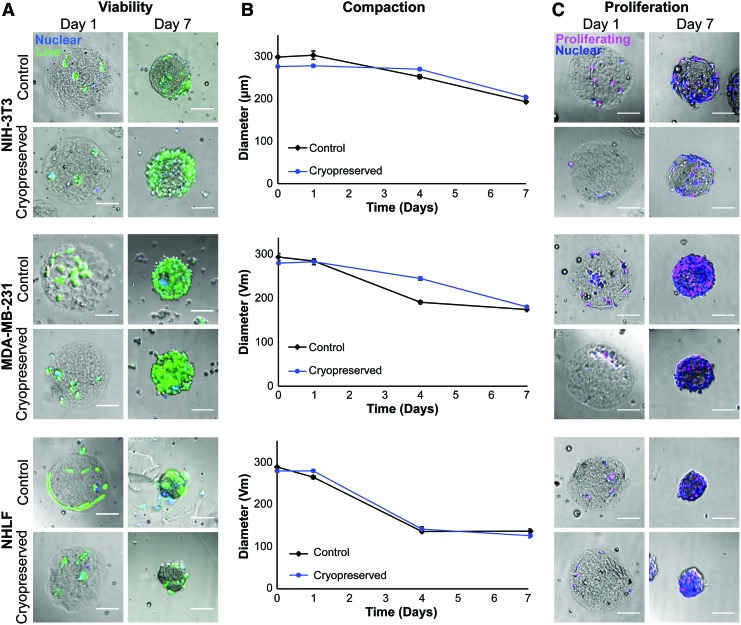
FIG. 6.
Microtissues remain highly viable and functional after cryopreservation. (A) Brightfield and widefield fluorescence imaging of microtissues with encapsulated NIH 3T3, MDA-MB-231, and NHLF cells (nuclei shown in blue). Calcein AM (green) shows that all cell types had high viability (>83%) after 1 week of freezing and were over 90% viable after 1 week of culture in standard conditions, which was comparable to tissues that never underwent freezing. (B) Compaction of encapsulated cells indicated that before and after cryopreservation, cells were similarly contractile, compacting microtissues at similar rates and with the same trajectories. Shown with standard error and an average of 82 microtissues per condition. (C) Brightfield and widefield fluorescence imaging of cell proliferation after cryopreservation. Proliferative capacity, as determined with a Click-It EdU assay (nuclei shown in blue, proliferating cells in magenta). All microtissues in all conditions had cells that proliferated, indicating that cells retained their proliferative capacity after cryopreservation in the microtissues. All scale bars 100 μm. NIH 3T3, National Institute of Health 3T3. EdU, 5-ethynyl-2′-deoxyuridine. Color images available online at www.liebertpub.com/tec
We further assessed cell function after cryopreservation through a compaction assay. To determine the difference in compaction between control and frozen tissues, we compared final microtissue diameters for each condition and found them to be insignificantly different (p > 0.05) (Fig. 6B). We also observed that at intermediate time points each cell type demonstrated similar compaction between cryopreserved and control microtissues. Lastly, in addition to retaining contractile function, all three cell types retained proliferative ability after freezing, as all microtissues in every condition tested had cells that entered the S phase (Fig. 6C).
Discussion
In the context of high-throughput screening, maintaining large populations of discrete microtissues is critical for feasibly completing large-scale experiments, and our agarose microwells enable such a workflow. When used in conjunction with agarose microwells, we cultured upward of 500 microtissues per well in a standard 24-well plate, which equates to large number of conditions and hundreds of replicates per condition in any given experiment. Additionally, the familiar well-plate format of our microtissue–microwell platform is compatible with automated liquid handling techniques and automated imaging systems, making this culture method a practical system for large-scale screening. We conducted our studies in 24-well plates as a proof of concept to show the potential of this platform for use in larger-scale studies. However, this microwell system could be adapted to 96- or 384-well plates as the agarose patterning method is independent of well size.
In this study, the application of this platform allowed us to track individual constructs over time, measuring dynamic changes in overall construct geometry as well as performing endpoint readouts with a consistent spatial arrangement. Coupling compaction with other nondestructive live cell measurements (e.g., reporter lines or cell migration), we envision that tracking microtissues over time could provide time-lapse information on the development and progression of disease states in vitro. This would allow for testing the efficacy of therapeutics at several stages of disease as well as the long-term effects of drugs on specific populations. This could be particularly important for identifying drug-resistant populations in diseases such as cancer. If used in conjunction with primary cells from patient biopsies, these methods offer additional metrics to investigate drug efficacy on diseased cells' ability to remodel and interact with their microenvironment.
By tracking individual microtissues over time, we were able to observe that ECM remodeling by different cell types follow unique time courses and that there is heterogeneity within the remodeling time course even within the same cell type. One likely driver of heterogeneity in the microtissue compaction rates is the variance in the initial cell seeding density. in large collagen hydrogels, the rate of collagen compaction is dependent on initial cell density, and in droplet systems, it is well characterized that cell encapsulation methods follow a Poisson distribution.53 Notably, the microtissues eventually converge to a narrower distribution, which is consistent with previous literature suggesting that as gels approach a common smaller size, cell density reaches a critical threshold and becomes a less significant factor.54 The workflow described in this study facilitates the simultaneous study of a large number of replicate microtissues and thus enables the possibility to study heterogeneity that may derive from factors, such as cell number, variability in underlying ECM, and heterogeneity in the underlying cell population.
We also found that compaction in our system was reversible at early time points, and over time the remodeling became more permanent. We examined the effects of encapsulating versus coating cells on the surface and found the surface-coated version to produce irreversible remodeling more quickly. Differences in cell numbers between these two conditions may also explain this observed effect. Because we encapsulated relatively low numbers of cells (<20 per microtissue), we may have had more cells adhered to the surface of the microtissues then were encapsulated. An alternative hypothesis is that the surface-coated NHLFs create a more stable ECM modification because the deposited ECM is supporting the remodeled geometry consistently from the outer edge of the microtissue.
In comparison, the encapsulated fibroblasts deposit matrix throughout the microtissues, which provides less organized modification near the surface of the microtissues on average, as alluded to by the ECM staining results (Fig. 3C). Thus, the surface deposition may provide more resistance to tissue expansion and thus would result in a diminished increase in size after releasing cells with EDTA.
In combination with measuring tissue compaction as a functional assay, we have demonstrated the ability to measure molecular readouts that provide insight into the mechanistic drivers of the function. Notably, our microtissue platform permits simultaneous measurement and correlation of cell function (compaction) and molecular mechanism (actin) in the same cells and in the same microenvironment. Previously, correlation of actin polymerization and its effect on gel contraction have been reported by separately measuring actin in a 2D culture system while separately measuring compaction in a 3D bulk gel.55,56 This separation of experimental readouts was required because the large size of the hydrogels hindered imaging.28–30 Thus, the scale of our system enables a more direct connection between mechanism and function.
We also present proof of concept that microtissues can be easily cryopreserved with methods typically used for single cell suspensions. The use of standard tissue culture cryopreservation protocols for a whole-construct application is unusual as diffusion limitations typically prevent the use of DMSO, as large tissues are difficult to saturate efficiently. PEG is an extremely popular material for cell encapsulation, but transport limitations in these systems are severe, with pore sizes of the hydrogels ranging from ∼40 to 200 Å.57 This is a stark contrast to collagen gels that have pore sizes >1 μm.58 Because our tissue constructs are small and highly porous, we can use a range of cryoprotectants not usually available for whole tissue constructs, as we can quickly saturate the tissues with cryoprotectant agents and avoid deleterious effects.59
Combined with the ability to cryopreserve the constructs, we can preserve precious cell populations to maximize the timescale and number of experiments that can be completed with a single cell source. As our assays and protocols are modular and can be combined and used to assess tissue-level behaviors for a variety of applications, this platform becomes flexible and amenable to study wide classes of diseases.
Conclusion
We have developed an in vitro platform that enables long-term tissue remodeling on a cellular scale for large populations. Using agarose microwells to culture collagen microtissues, we have cultured microtissues as independent entities for up to 1 month and have tracked and measured them as discrete tissues for 1 week. We have also demonstrated our ability to cryopreserve our microtissues while retaining high viability and cell function, an invaluable method that could allow for freezing and dissemination after mass production. Additionally, we have quantified both local and global as well as short- and long-term remodeling that occurs in our culturing system. Establishing high-throughput metrics of remodeling is crucial for the development of drug-screening models for fibrotic and cancerous disease, although the merit of the platform extends beyond these pathologies to include any disease in which ECM interactions are considered important.
Acknowledgments
Portions of this work were conducted in the Minnesota Nano Center, which is supported by the National Science Foundation through the National Nanotechnology Coordinated Infrastructure (NNCI) Network under Award Number ECCS-1542202. The authors thank the American Heart Association (13SDG6450000), the National Heart, Lung, and Blood Institute (R21 HL132256), the National Institute of Environmental Health Sciences (R21 ES027622), and the National Science Foundation (CBET 1704332) for financial support. A.L.C. acknowledges the National Science Foundation Graduate Research Fellowship Program (00039202) for support. They also thank Dr. Paolo Provenzano for the use of his multiphoton microscope and donating MDA-MB-231 cells, Dr. Daniel Tschumperlin for donating NHLFs, Dr. Wei Shen for donating NIH 3T3 cells, and Julia Nguyen and Dr. Gregory Vercellotti for isolating and generously donating HUVECs.
Disclosure Statement
No competing financial interests exist.
References
- 1. Lu P., Weaver V.M., and Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196, 395, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cox T.R., and Erler J.T. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Models Mech 4, 165, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venning F.A., Wullkopf L., and Erler J.T. Targeting ECM disrupts cancer progression. Front Oncol 5, 224, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gialeli C., Theocharis A.D., and Karamanos N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278, 16, 2011 [DOI] [PubMed] [Google Scholar]
- 5. Klingberg F., Hinz B., and White E.S. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol 229, 298, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Desai O., Winkler J., Minasyan M., and Herzog E.L. The role of immune and inflammatory cells in idiopathic pulmonary fibrosis. Front Med (Lausanne) 5, 43, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei L. Immunological aspect of cardiac remodeling: T lymphocyte subsets in inflammation-mediated cardiac fibrosis. Exp Mol Pathol 90, 74, 2011 [DOI] [PubMed] [Google Scholar]
- 8. Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol 10, 712, 2010 [DOI] [PubMed] [Google Scholar]
- 9. Kanematsu A., Marui A., Yamamoto S., et al. . Type I collagen can function as a reservoir of basic fibroblast growth factor. J Control Release 99, 281, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Bashkin P., Doctrow S., Klagsbrun M., Svahn C.M., Folkman J., and Vlodavsky I. Basic fibroblast growth factor binds to subendothelial extracellular matrix and is released by heparitinase and heparin-like molecules. Biochemistry 28, 1737, 1989 [DOI] [PubMed] [Google Scholar]
- 11. Gospodarowicz D., Neufeld G., and Schweigerer L. Fibroblast growth factor: structural and biological properties. J Cell Physiol Suppl (Suppl 5), 15, 1987 [DOI] [PubMed] [Google Scholar]
- 12. Jones J.I., Gockerman A., Busby W.H., Camacho-Hubner C., and Clemmons D.R. Extracellular matrix contains insulin-like growth factor binding protein-5: potentiation of the effects of IGF-I. J Cell Biol 121, 679, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keane T.J., Dziki J., Sobieski E., et al. . Restoring mucosal barrier function and modifying macrophage phenotype with an extracellular matrix hydrogel: potential therapy for ulcerative colitis. J Crohns Colitis 11, 360, 2017 [DOI] [PubMed] [Google Scholar]
- 14. Tannock I.F., Lee C.M., Tunggal J.K., Cowan D.S.M., and Egorin M.J. Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res 8, 878, 2002 [PubMed] [Google Scholar]
- 15. Choi I.K., Lee Y.S., Yoo J.Y., et al. . Effect of decorin on overcoming the extracellular matrix barrier for oncolytic virotherapy. Gene Ther 17, 190, 2010 [DOI] [PubMed] [Google Scholar]
- 16. Demidova-Rice T.N., Geevarghese A., and Herman I.M. Bioactive peptides derived from vascular endothelial cell extracellular matrices promote microvascular morphogenesis and wound healing in vitro. Wound Repair Regen 19, 59, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martino M.M., Tortelli F., Mochizuki M., et al. . Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci Transl Med 3, 100ra89, 2011 [DOI] [PubMed] [Google Scholar]
- 18. Pampaloni F., Stelzer E.H.K., and Masotti A. Three-dimensional tissue models for drug discovery and toxicology. Recent Pat Biotechnol 3, 103, 2009 [DOI] [PubMed] [Google Scholar]
- 19. Weigelt B., Ghajar C.M., and Bissell M.J. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev 69–70, 42, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sant S., and Johnston P.A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 23, 27, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klingelhutz A.J., Gourronc F.A., Chaly A., et al. . Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci Rep 8, 523, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hagemann J., Jacobi C., Hahn M., et al. . Spheroid-based 3D cell cultures enable personalized therapy testing and drug discovery in head and neck cancer. Anticancer Res 37, 2201, 2017 [DOI] [PubMed] [Google Scholar]
- 23. Edmondson R., Broglie J.J., Adcock A.F., and Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 12, 207, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mehta G., Hsiao A.Y., Ingram M., Luker G.D., and Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J Control Release 164, 192, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Breslin S., and O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 18, 240, 2013 [DOI] [PubMed] [Google Scholar]
- 26. Corstorphine L., and Sefton M.V. Effectiveness factor and diffusion limitations in collagen gel modules containing HepG2 cells. J Tissue Eng Regen Med 5, 119, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McMurtrey R.J. Analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in three-dimensional tissue constructs with applications and insights in cerebral organoids. Tissue Eng Part C Methods 22, 221, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith L.E., Smallwood R., and Macneil S. A comparison of imaging methodologies for 3D tissue engineering. Microsc Res Tech 73, 1123, 2010 [DOI] [PubMed] [Google Scholar]
- 29. Appel A.A., Anastasio M.A., Larson J.C., and Brey E.M. Imaging challenges in biomaterials and tissue engineering. Biomaterials 34, 6615, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langhans S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol 9, 6, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X.J., Valadez A.V., Zuo P., and Nie Z. Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioanalysis 4, 1509, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halldorsson S., Lucumi E., Gómez-Sjöberg R., and Fleming R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron 63, 218, 2015 [DOI] [PubMed] [Google Scholar]
- 33. Chiu D.T., deMello A.J., Di Carlo D., et al. . Small but perfectly formed? Successes, challenges, and opportunities for microfluidics in the chemical and biological sciences. Chem 2, 201, 2017 [Google Scholar]
- 34. Brett M.-E., Crampton A.L., and Wood D.K. Rapid generation of collagen-based microtissues to study cell–matrix interactions. Technology 4, 80, 2016 [Google Scholar]
- 35. Crampton A.L., Cummins K.A., and Wood D.K. A high-throughput microtissue platform to probe endothelial function in vitro. Integr Biol (Camb) 10, 555, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kinney M.A., Sargent C.Y., and McDevitt T.C. The multiparametric effects of hydrodynamic environments on stem cell culture. Tissue Eng Part B Rev 17, 249, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lock L.T., Farance I., Baraniak P., Tsai A.-C., Ma T., and Rowley J.A. Rapid and economic generation of consistent hMSC spheroids for macroscopic tissue biofabrication and therapeutic applications. Cytotherapy 17, S77, 2015 [Google Scholar]
- 38. McGuigan A.P., and Sefton M.V. Vascularized organoid engineered by modular assembly enables blood perfusion. Proc Natl Acad Sci U S A 103, 11461, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wood D.K., Weingeist D.M., Bhatia S.N., and Engelward B.P. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci U S A 107, 10008, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong X., Lin C., Cheng J., et al. . Generation of multicellular tumor spheroids with microwell-based agarose scaffolds for drug testing. PLoS One 10, e0130348, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dahlmann J., Kensah G., Kempf H., et al. . The use of agarose microwells for scalable embryoid body formation and cardiac differentiation of human and murine pluripotent stem cells. Biomaterials 34, 2463, 2013 [DOI] [PubMed] [Google Scholar]
- 42. Choi Y.Y., Chung B.G., Lee D.H., Khademhosseini A., Kim J.H., and Lee S.H. Controlled-size embryoid body formation in concave microwell arrays. Biomaterials 31, 4296, 2010 [DOI] [PubMed] [Google Scholar]
- 43. Blomlöf J., Blomlöf L., and Lindskog S. Effect of different concentrations of EDTA on smear removal and collagen exposure in periodontitis-affected root surfaces. J Clin Periodontol 24, 534, 1997 [DOI] [PubMed] [Google Scholar]
- 44. Simpson J.P., Penkman K.E.H., Demarchi B., et al. . The effects of demineralisation and sampling point variability on the measurement of glutamine deamidation in type I collagen extracted from bone. J Archaeol Sci 69, 29, 2016 [Google Scholar]
- 45. Mohammadi H., Arora P.D., Simmons C.A., Janmey P.A., and McCulloch C.A. Inelastic behaviour of collagen networks in cell-matrix interactions and mechanosensation. J R Soc Interface 12, 20141074, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barcus C.E., Keely P.J., Eliceiri K.W., and Schuler L.A. Stiff Collagen Matrices Increase Tumorigenic Prolactin Signaling in Breast Cancer Cells. J Biol Chem 288, 12722, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Riegler J., Labyed Y., Rosenzweig S., et al. . Tumor elastography and its association with collagen and the tumor microenvironment. Clin Cancer Res 24, 4455, 2018 [DOI] [PubMed] [Google Scholar]
- 48. Provenzano P.P., Inman D.R., Eliceiri K.W., et al. . Collagen density promotes mammary tumor initiation and progression. BMC Med 6, 11, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carey S.P., Martin K.E., and Reinhart-King C.A. Three-dimensional collagen matrix induces a mechanosensitive invasive epithelial phenotype. Sci Rep 7, 42088, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lakshman N., and Petroll W.M. Growth factor regulation of corneal keratocyte mechanical phenotypes in 3-D collagen matrices. Invest Ophthalmol Vis Sci 53, 1077, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goulet S., Bihl M.P., Gambazzi F., Tamm M., and Roth M. Opposite effect of corticosteroids and long-acting β2-agonists on serum- and TGF-β1-induced extracellular matrix deposition by primary human lung fibroblasts. J Cell Physiol 210, 167, 2007 [DOI] [PubMed] [Google Scholar]
- 52. Kobayashi T., Liu X., Kim H.J., et al. . TGF-β1 and serum both stimulate contraction but differentially affect apoptosis in 3D collagen gels. Respir Res 6, 141, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Collins D.J., Neild A., deMello A., Liu A.Q., and Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip 15, 3439, 2015 [DOI] [PubMed] [Google Scholar]
- 54. Bell E., Ivarsson B., and Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci U S A 76, 1274, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Janson L.W., Kolega J., and Taylor D.L. Modulation of contraction by gelation/solation in a reconstituted motile model. J Cell Biol 114, 1005, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sohail M.A., Hashmi A.Z., Hakim W., et al. . Adenosine induces loss of actin stress fibers and inhibits contraction in hepatic stellate cells via Rho inhibition. Hepatology 49, 185, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nicodemus G.D., and Bryant S.J. Cell encapsulation in biodegradable hydrogels for tissue engineering applications. Tissue Eng Part B Rev 14, 149, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harjanto D., Maffei J.S., and Zaman M.H. Quantitative analysis of the effect of cancer invasiveness and collagen concentration on 3D matrix remodeling. PLoS One 6, e24891, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Best B.P. Cryoprotectant toxicity: facts, issues, and questions. Rejuvenation Res 18, 422, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]