Abstract
Recent evidence has demonstrated that remote responses in the brain, as well as local responses in the injured spinal cord, can be induced after spinal cord injury (SCI). Intravenous infusion of mesenchymal stem cells (MSCs) has been shown to provide functional improvements in SCI through local therapeutic mechanisms that provide neuroprotection, stabilization of the blood–spinal cord barrier, remyelination, and axonal sprouting. In the present study, we examined the brain response that might be associated with the functional improvements induced by the infused MSCs after SCI. Genome-wide RNA profiling was performed in the motor cortex of SCI rats at 3 days post-MSC or vehicle infusion. Then, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) data revealed that the “behaviorally-associated differentially expressed genes (DEGs)” were identified by the Pearson's correlation analysis with the behavioral function, suggesting that the “behaviorally-associated DEGs” may be related to the functional recovery after systemic infusion of MSCs in SCI. These results suggested that the infused MSCs alter the gene expression signature in the brain and that these expression changes may contribute to the improved function in SCI.
Keywords: MSC, SCI, transcriptome
Introduction
Recent studies in spinal cord injury (SCI) have shown that remote regenerative responses in the brain could be induced after SCI,1–8 as could local responses in the injured spinal cord.9–11 Brain network reorganization is critical for functional recovery after SCI in both nonhuman primate SCI models12,13 and clinical patients with SCI.14 Therefore, to develop a new therapy for SCI, strategic insights regarding the whole central nervous system; that is, not only the injured spinal cord but also the brain, should be considered.
Intravenous infusion of mesenchymal stem cells (MSCs) is one of the most promising therapies in improving functional outcomes after SCI.15–17 Cellular therapy with MSCs has previously been suggested to have multi-modal therapeutic effects in the injured spinal cord, including enhanced neuroprotection,15,18,19 stabilization of the blood–spinal cord barrier,17,20 remyelination,17 and axonal sprouting.17,21 In addition, the brain response against SCI following intravenous infusion of MSCs might also contribute to providing functional recovery.
Gene expression profiling in the brain post-SCI has shown that acute adaptive regenerative responses in the brain were induced after SCI,1 and that the SCI-induced molecular pathways in the brain may play an important role in recovery from the injury.1–3 However, as gene expression profiling in the brain after intravenous infusion of MSCs post-SCI has not yet been assessed, in this study, genome-wide RNA profiling in the motor cortex was performed following intravenous infusion of MSCs post-SCI induction in a rat model of contused SCI. Moreover, the gene expression signature in the brain that might be associated with the functional improvements induced by the infused MSCs after SCI was explored.
Methods
Animals
All experiments were conducted in accordance with the institutional guidelines of Sapporo Medical University. The use of animals in this study was approved by the Animal Care and Use Committee of Sapporo Medical University.
Preparation of MSCs from rat bone marrow
Rat MSCs were prepared following a protocol used in our previous studies.16,17 In brief, bone marrow was obtained from the femoral bones of adult rats. The marrow was then diluted to 20 mL with Dulbecco's modified Eagle's medium (DMEM) (Sigma, St. Louis, MO) and supplemented with 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific Inc., Waltham, MA), 2 mM l-glutamine (Sigma), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Thermo Fisher Scientific Inc.), and incubated for 3 days (5% CO2, 37°C). When cultures had almost reached confluence, the adherent cells were detached with trypsin-EDTA solution (Sigma) and subcultured at 1 × 104 cells/mL. Phenotypic analysis of the MSC surface antigen was performed using anti-CD45, CD73, CD90, and CD106 antibodies.22 Following three passages, the MSCs were collected for infusion.
SCI model
Contusive SCI was made as described previously.17,20 Briefly, adult male Sprague–Dawley rats at 8–9 weeks (250–300 g) were anesthetized with an intraperitoneal (IP) injection of ketamine (90 mg/kg) and xylazine (4 mg/kg). Following incision, the T9 vertebra was stabilized by clamping the rostral T8 and caudal T10 vertebral bodies with forceps, and a dorsal laminectomy was performed at the T9 to 10 level. A 130 kDyn contusion was then delivered using the Infinite Horizons impactor (Precision Systems and Instrumentation, LLC, Lexington, KY). After the injury, actual injured forces were recorded and compared with predicted severities. There was ∼ < 0.05% variation between these values, which is consistent with a previous report.23 Appropriate postoperative care, including manual bladder expression twice a day, was provided for all animals. Rats were housed at a temperature of 24 ± 2°C and 50% humidity.
Transplantation procedure
Only rats with Basso, Beattie, and Bresnahan (BBB) scores24 displaying 0 points at 1 day after SCI induction were included in this study. The SCI rats with BBB scores of 0 points were randomized and received a single intravenous infusion of MSCs at 1.0 × 106 cells in 1.0 mL of fresh DMEM or vehicle (1.0 mL fresh DMEM alone) via the femoral vein 1 day after SCI induction. All rats were injected daily with cyclosporine A (10 mg/kg, IP).16,17,25 In addition, intact rats were used as sham.
Behavioral testing
Open field locomotor function was assessed using the BBB scale by an evaluator blinded to treatment condition. Rats were scored at 1 day after SCI induction (immediately prior to infusion) and 3 days post-infusion (n = 7/group, time point).
RNA purification
Animals (MSC group, n = 7/time point; vehicle group, n = 7/time points; sham group, n = 5) were euthanized using deep anesthesia with ketamine (75 mg/kg) and xylazine (10 mg/kg, IP) and the brains were dissected. The cerebral cortex (motor cortical region) was extracted and stored at −80°C. After homogenization, total RNA was purified using the RNeasy Plus mini kit (QIAGEN, Venlo, The Netherlands). RNA quality was assessed using the Bioanalyzer RNA 6000 nano kit (Agilent Technologies, Santa Clara, CA). Samples showing RNA integrity number (RIN) >7.0 were selected for subsequent use in this study (Table S1) (see online supplementary material at http://www.liebertpub.com).
Microarray hybridization and analysis
The Clariom™ D/Gene Chip® Rat Transcriptome Array (RTA 1.0., Affymetrix, Santa Clara, CA) was used with 100 ng input. The images of hybridized microarrays were processed using Expression Console software (Affymetrix). Raw expression values obtained directly from CEL files were preprocessed using the Signal Space Transformation-Robust Multichip Analysis18 pipeline as previously described.26 A single microarray analysis was performed in this study. The differentially expressed genes (DEGs) were screened using one way ANOVA with a cutoff p value <0.05 and a fold change (FC) >1.5 or <−1.5. A volcano plot and heat map were generated using the statistical language R ver.3.4.2 (Fig. S1) (see online supplementary material at http://www.liebertpub.com).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
The Super Script® VILO™ cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) was used for reverse transcription. A 100 ng total mRNA input was used for qRT-PCR. TaqMan® Universal Master Mix II with Uracil-N glycosylase (UNG), and TaqMan® Gene Expression assays were purchased from Thermo Fisher Scientific Inc. (Gapdh, Rn01775763_g1; Kcnip2, Rn01411450_g1; lipid phosphate phosphatase-related protein type 4 [Lppr4], Rn01522267_m1; Pde10a, Rn00673152_m1; calcium binding protein 7 [Cabp7], Rn01443564_m1; plakophilins-2 [Pkp2], Rn01404502_m1; Scn3b, Rn01422019_m1; nuclear receptor binding protein 2 [Nrbp2], Rn01505756_m1; Sik1, Rn00572495_m1; Fos, Rn02396759_m1; Dusp1, Rn00678341_g1; Btg2, Rn00568504_m1; Ier2, Rn02531674_s1; Cyr61, Rn00580055_m1; Apold1, Rn02131262_s1; Id2, Rn01495280_m1). qRT-PCR was performed in triplicate using PRISM7500 with 7500 software v2.3 (Thermo Fisher Scientific Inc.). Thermal cycling was performed at 50°C for 2 min and at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and of 60°C for 1 min. The ΔCT was calculated against the endogenous control (Gapdh), and the ΔΔCT was calculated against the ΔCT of the sham. FC was also calculated using the comparative Ct method.27
Prediction of transcription factor binding domains
Transcriptional binding domains in the gene loci of interest were determined using the University of California at Santa Cruz (UCSC) genome browser (https://genome-asia.ucsc.edu/cgi-bin/hgGateway?redirect=manual&source=genome.ucsc.edu) with the JASPAR custom track (http://jaspar.genereg.net/). The information of functional genomic domains and cross-species conservation was downloaded from the VISTA Genome Browser (http://pipeline.lbl.gov/cgi-bin/gateway2).
Gene set enrichment analysis (GSEA)
GSEA28 was performed as detailed in the online supplementary materials (see online supplementary material at http://www.liebertpub.com).
Data deposition
The data reported in this article (Tables S2 and S3) have been deposited in the Gene Expression Omnibus (GEO) database: www.ncbi.nlm.nih.gov/geo (accession no. GSE108168) (see online supplementary material at http://www.liebertpub.com).
Statistical analysis
Data were subjected to statistical analyses using TAC version 3.1.0.5 (Affymetrix®) and JMP® version 12.2.0 (SAS Institute, Cary, NC). Comparison of the BBB behavioral scores between MSC and vehicle-treated animals was performed using the Mann–Whitney U test. Microarray statistical analyses were performed using one way ANOVA. Correlation between mRNA expression and BBB behavioral score was evaluated using Pearson's correlation coefficients (PCC). Continuous data were assessed for normality using the Shapiro–Wilk test. The continuous data with normality were analyzed by one way ANOVA, and the Tukey–Kramer test was used for post-hoc comparisons. Results were expressed as the means ± SEM.
Results
Infused MSCs provide improved locomotor function
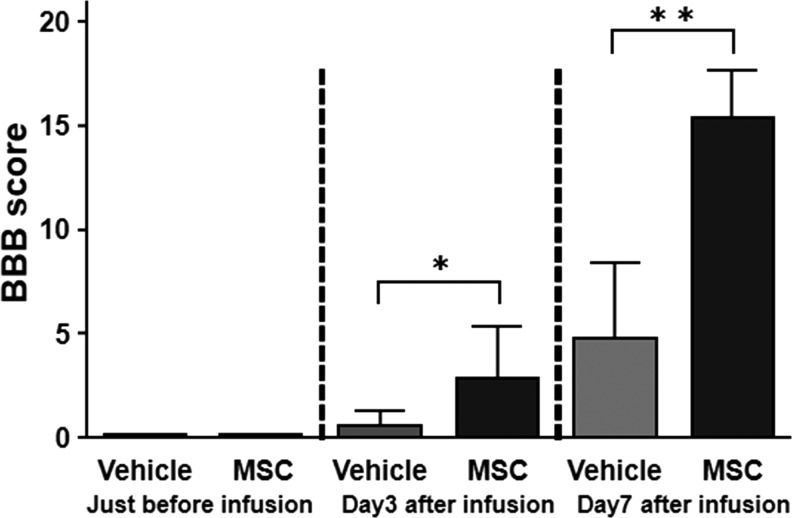
Open field locomotor function was assessed using the BBB behavioral score immediately prior to and 3 days following MSC or vehicle infusion for confirmation of injury equivalency across SCI animals, as well as to determine whether infused MSCs had an effect on overall locomotor ability. All included animals demonstrated complete hindlimb paraplegia prior to infusion (1 day after SCI). Scores on the BBB in the MSC-infused group were significantly higher than those of the vehicle-treated group at 3 and 7 days after MSC infusion (2.86 ± 2.5 vs. 0.57 ± 0.73; p < 0.05, Mann–Whitney U test: n = 7/group). The results are shown in Figure 1. These results indicated that the infused MSCs provided acute therapeutic effects in the rat contused SCI model system.
FIG. 1.
Locomotor function assessment using the Basso, Beattie, and Bresnahan (BBB) behavioral score. Both groups displayed complete hindlimb paraplegia immediately prior to infusion (1 day after SCI induction) (left). The BBB scores at 3 and 7 days after mesenchymal stem cells (MSC) or vehicle infusion (4 and 8 days after SCI induction) are shown (right). **p < 0.01.
Microarray analysis
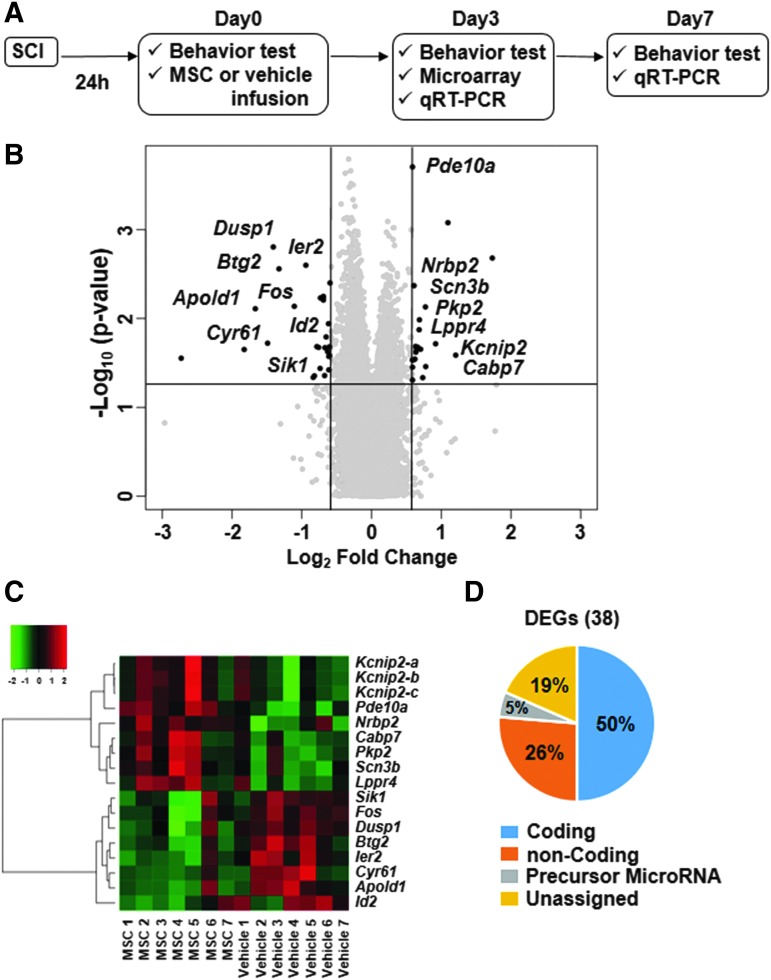
Microarray was conducted to screen for DEGs. A volcano plot showing 38 DEGs defined with the cutoff of FC at 1.5 or −1.5, and p < 0.05 by one way ANOVA was generated (Fig. 2B). A heat map of 15 coding DEGs was also used to illustrate the expression levels in both MSC and vehicle groups (Fig. 2C). Figure 2D shows the percentage of the 38 DEGs that belongs to each type of RNA including coding, non-coding, precursor microRNA, and unassigned DEGs. These results identified 38 DEGs including 15 coding genes between vehicle and control groups as screened by microarray analysis.
FIG. 2.
Experimental procedures for differentially expressed gene (DEG) analysis (A) Schematic drawing of the experimental procedure. (B) Volcano plot showing DEGs defined with the cutoff of fold change at 1.5 or −1.5, and p < 0.05 by one way ANOVA. The black dots represent the 38 screened DEGs. The 15 coding DEGs are displayed with the gene name. (C) Heat map of 15 coding DEGs showing the expression levels in each SCI+MSC and SCI+vehicle sample. The dendrogram was generated based on the Euclidean distance metric with Ward's method. (D) Charts showing the percentage of the 38 DEGs that belong to each type of RNA including coding, non-coding, precursor microRNA, and unassigned DEGs.
Identification of “behaviorally-associated DEGs”
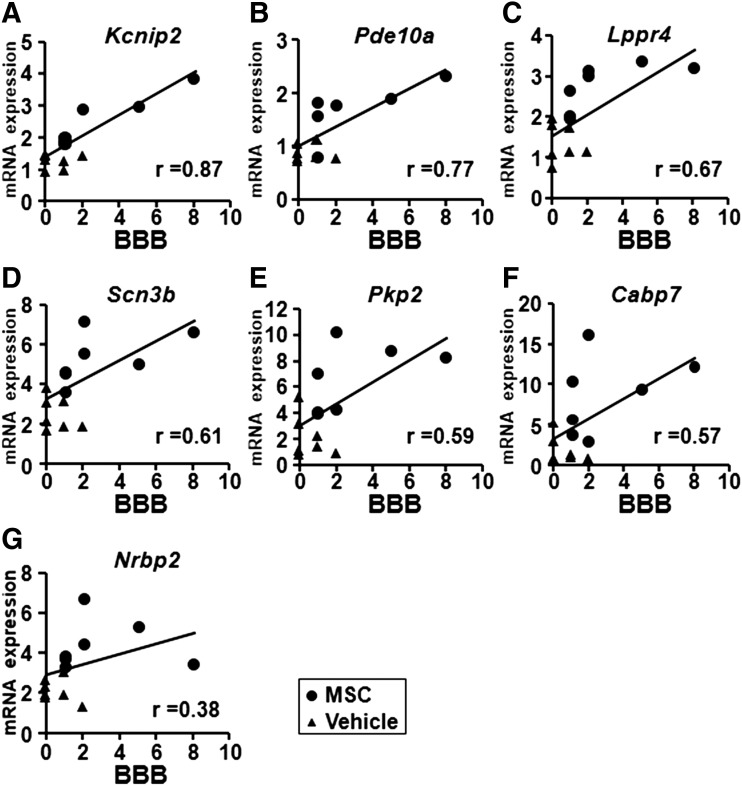
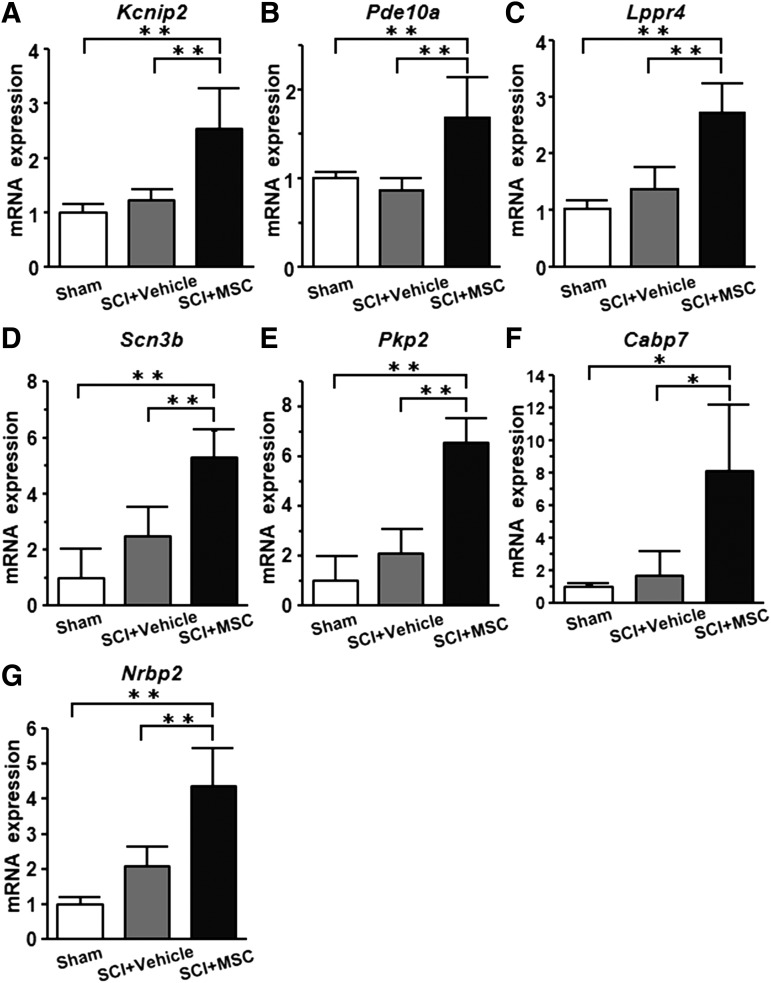
qRT-PCR data of the 15 coding DEGS that were screened by microarray analysis were used to identify the “behaviorally-associated DEGs,” Kcnip2 (r = 0.87), Pde10a (r = 0.77), Lppr4 (r = 0.67), Scn3b (r = 0.61), Pkp2 (r = 0.59), Cabp7 (r = 0.57), and Nrbp2 (r = 0.38), which were defined by Pearson's correlation analysis as exhibiting positive correlation with the BBB behavioral scores (Fig. 3A–G). Although qRT-PCR showed that the expression levels of “behaviorally-associated DEGs” in the SCI+vehicle group (n = 7) tended to be higher than those in the sham group (n = 5), the expression levels in the SCI+MSC group (n = 7) were highly elevated compared with those in the SCI+vehicle group (Fig. 4). These results suggest that seven “behaviorally-associated DEGs” were identified in this study.
FIG. 3.
Correlation between mRNA expression and behavioral function. Pearson's correlation analysis of the Basso, Beattie, and Bresnahan (BBB) scores and mRNA expression (2-ΔΔCT) of the indicated genes as detected by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Circles and triangles indicate SCI+MSC and SCI+vehicle samples, respectively. The X-axis represents the BBB score and the Y-axis shows the 2-ΔΔCT of mRNA gene expression.
FIG. 4.
mRNA expression of “behaviorally-associated differentially expressed genes (DEGs).” Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) data of the “behaviorally-associated DEGs” are shown. Sham, SCI+vehicle, and SCI+MSC groups were used. The Y-axis shows the 2-ΔΔCT of mRNA gene expression. Asterisks indicate *p < 0.05, **p < 0.01 by the Tukey–Kramer test.
Promoter analysis based on transcription factor binding sites
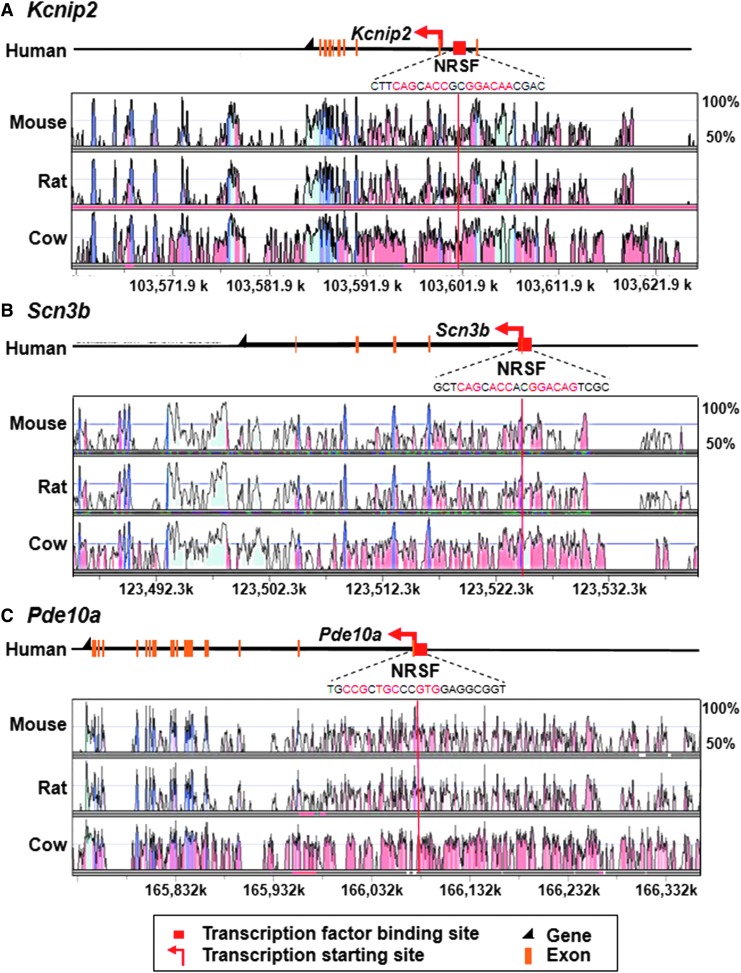
To obtain further mechanistic insights regarding the DEGs, we performed promoter analysis. In some “behaviorally-associated DEGs” including Kcnip2, Scn3b, and Pde10a, the predicted promoter regions possessed neuron-restrictive silencer factor (NRSF) binding domains upstream or in the introns in the 5′ region of these genes. These NRSF promoter regions are conserved across mammalian species including human, mouse, rat, and cow (Fig. 5). These results indicate that some “behaviorally-associated DEGs” might be involved in previously known cellular mechanisms of recovery.
FIG. 5.
The neuron-restrictive silencer factor (NRSF) binding domains in some “behaviorally-associated differentially expressed genes (DEGs).” Genomic loci of NRSF binding domains within 20 kb from the transcription start of the genes including (A) Kcnip2 (Base genome: human chromosome: chr10 103,561,373–103,626,222), (B) Scn3b (Base genome: human chromosome: chr11 123,481,778–123,540,867), and (C) Pde10a (Base genome: human chromosome: chr6 165,727,008–166,409,519) (red arrows) are shown. The binding motifs of NRSF are indicated with red characters.
Correlation between the overall gene expression profiles in the brain and behavioral function
Correlation analysis between the overall gene expression landscape and behavior function was performed by utilizing GSEA (Fig. S1). The higher enrichment score of the experimental dataset (Fig. S1A: ES = 0.22, q < 0.001) than that of the simulated data set (Fig. S1B: ES = −0.07, q = 0.86) suggested that the overall gene expression profiles may be associated with the functional recovery.
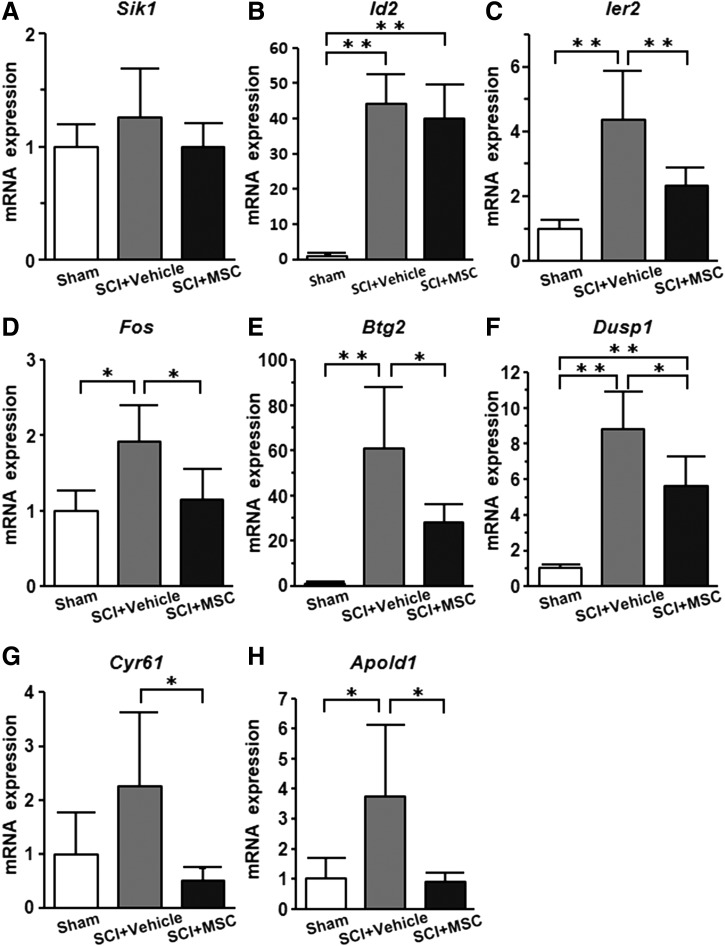
The non-“behaviorally-associated DEGs” might be associated with beneficial effects
qRT-PCR of the non-“behaviorally-associated DEGs”, including Sik1, Id2, Ier2, Fos, Btg2, Dusp1, Cyr61, and Apold1, screened using microarray analysis showed that whereas the expression levels of non-“behaviorally-associated DEGs” in the SCI+vehicle group (n = 7) were elevated compared with those in the sham group (n = 5), the expression levels in the SCI+MSC group (n = 7) were suppressed compared with those in the vehicle group (Fig. 6). These results suggest that eight non-“behaviorally-associated DEGs” were identified in this study.
FIG. 6.
mRNA expression of non-“behaviorally-associated differentially expressed genes (DEGs).” Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) data of the non-“behaviorally-associated DEGs” are shown. The Y-axis represents the 2-ΔΔCT of mRNA gene expression. Asterisks indicate *p < 0.05, **p < 0.01 by the Tukey–Kramer test.
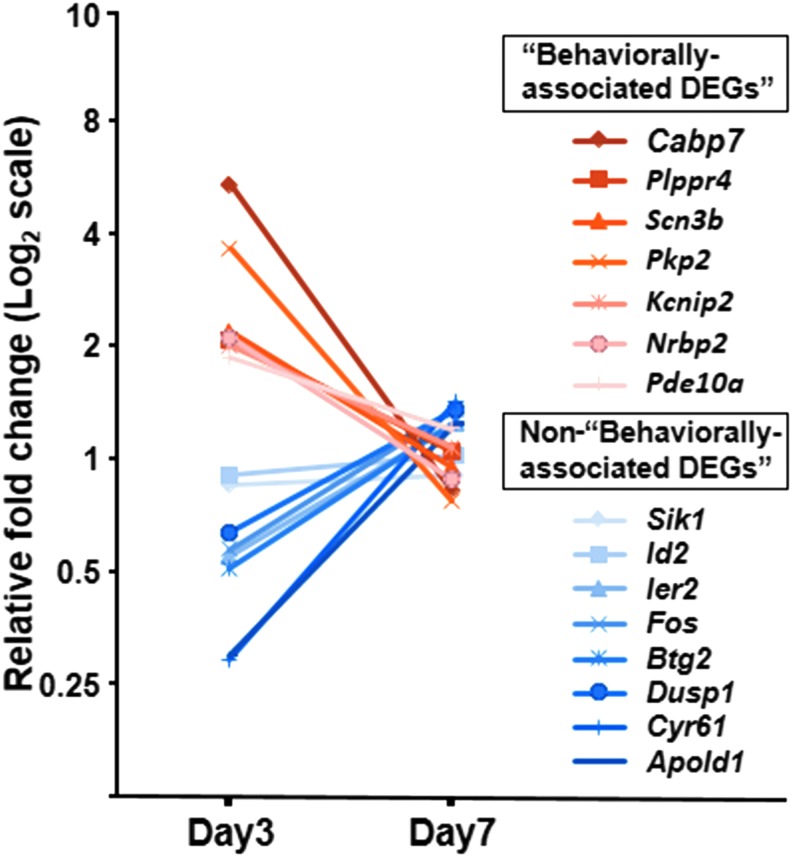
Temporal expression of DEGs in the acute phase
We also examined the time-course expression of coding DEGs by qRT-PCR. None of the 15 coding DEGs at 3 days post-infusion exhibited significant differential expression at 7 days (n = 7/group) (Fig. 7).
FIG. 7.
Differential gene expression is transient in the acute phase. Relative fold change of mRNA expression in all coding differentially expressed genes (DEGs) at 3 days post-infusion is shown as red (“behaviorally-associated DEGs”) and blue (non-“behaviorally-associated DEGs”). No significant differences were observed in the comparisons between SCI+MSC and SCI+vehicle groups at day 7 post-infusion.
Discussion
In this study, acute therapeutic efficacy was observed following intravenous infusion of MSCs in a rat model of contused SCI. We then explored the therapeutic molecular mechanisms in the brain underlying the functional improvements facilitated by the MSC. Microarray analysis identified 15 coding DEGs in the motor cortices between the MSC- and vehicle-infused rats after SCI. Among these, Pearson's correlation analysis indicated positive correlation of seven “behaviorally-associated DEGs” with behavioral function.
Further, qRT-PCR showed that the expression levels of “behaviorally-associated DEGs” in the SCI+MSC group were higher than those of the sham and the SCI+vehicle groups. The increased expression levels of “behaviorally-associated DEGs” in the SCI+MSC group in the brain might be associated with promotion of the mechanisms to facilitate functional recovery. Although the expression levels of “behaviorally-associated DEGs” in the SCI+vehicle group tended to be higher than those in the sham group, this increase was not statistically significant and appeared insufficient for inducing endogenous therapeutic functional improvement. This implies the possibility that the infused MSCs may upregulate the “behaviorally-associated DEGs” in the brain to provoke positive functions with regard to restoring the neuronal functions, likely through enhanced neuronal adaptation in the cortex after SCI,12,13,29 which would be consistent with the known functions of these “behaviorally-associated DEGs”. For example, these include the promotion of axonal outgrowth during development and regenerative sprouting after injury (Lppr4),30 cellular homeostasis (Pkp2),31 synaptic connectivity (Cabp7),32 and neural stem/progenitor cell differentiation and survival (Nrbp2).33 We also found several “behaviorally-associated DEGs” including Kcnip2, Pde10a, and Scn3b that might be involved in previously known cellular mechanisms of recovery, including NRSF-mediated regulation of gene expression. As NRSF constitutes the critical transcriptional repressor of neuronal fate,34–38 overall neurogenesis and neuronal maturation may be augmented by the infusion of MSCs. In addition, we performed GSEA to examine the correlation between the overall gene expression landscape in the brain and behavioral function, which suggested that the overall gene signature may also be associated with the improved behavioral outcome following MSC infusion in SCI.
The remainder of the DEGs (Sik1, Id2, Ier2, Fos, Btg2, Dusp1, Cyr61, and Apold1), which did not comprise the “behaviorally-associated DEGs,” were associated with immediate early responses.39–45 Immediate early response genes are stimulated in response to both cell-extrinsic and cell-intrinsic signals including cellular stress.46 qRT-PCR analysis showed that the expression levels of these genes in the SCI+vehicle group were highly elevated compared with those in the sham group. Infused MSCs reduced the increased expression of these genes mediated by the injury, which may be indicative of a reduced excitotoxicity response.47 This also suggests the possibility that the infused MSCs may mitigate the unfavorable conditions in the post-SCI brain.
Notably, the acute gene expression changes after intravenous infusion of MSCs in the brain defined at 3 days post-infusion were not detectable 7 days post-infusion. These phenomena may, therefore, contribute to the early manifestation of therapeutic effects by the intravenous infusion of MSCs. Morita and coworkers demonstrated a rapid increase of behavioral function following infused MSCs even in a chronic SCI model.17 These early behavioral improvements might thus be associated with the transient gene expression change in the brain following systemic administration of MSCs, as well as with the local effects mediated by the MSCs.48 It is also possible that these initial changes of gene expression, identified as DEGs detected at day 3 in this study, might trigger additional downstream gene expression to improve the functional outcome following intravenous infusion of MSCs in SCI.
A previous report that profiled injured spinal cord tissue after local transplantation of MSCs into the injury loci in SCI showed a marked increase in the expression of genes associated with foreign body response and adaptive immune response in the injured spinal cord tissue.18 In comparison, we examined the gene expression in the brain, which was not directly injured; accordingly, the gene signature in our study may be associated with enhanced neuronal adaptation in the cortex facilitating functional recovery.
Conclusion
In summary, we have shown that intravenous infusion of MSCs alters acute gene expression signature in the brain. We stress that the brain also reacts to facilitate functional recovery following systemic administration of MSCs in SCI, and that intravenous infusion represents a more appropriate route to act on the brain. Thus, profiling gene signature change in the brain following infused MSCs in SCI may offer an opportunity to elucidate the potential mechanisms providing functional improvement and may also shed some light on the unknown molecular bases related to the neuroprotection and regeneration effected by MSCs, which have yet to be determined.
Supplementary Material
Acknowledgments
We thank Dr. Alexander Son for critical reading of the manuscript. This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI grant numbers 16K10830 (to T.O.), 17K10902 (to T.M.), 16K10794 (to M.S.), the AMED Translational Research Network Program JP16lm0103003 (to M.S.), National Institute of Health Grants R21AA024882 (to K.H.-T.), R01AA025215 (to K.H.-T.), and the Scott-Gentle Foundation (to K.H.-T.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Thompson K., DiBona V., Dubey A., Crockett D., and Rasin M.-R. (2010). Acute adaptive responses of central sensorimotor neurons after spinal cord injury, in: Translational Neuroscience, De Gruyter, Warsaw, Poland: pps. 268 [Google Scholar]
- 2. Baek A., Cho S.R., and Kim S.H. (2017). Elucidation of gene expression patterns in the brain after spinal cord injury. Cell Transplant. 26, 1286–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jaerve A., Kruse F., Malik K., Hartung H.P., and Muller H.W. (2012). Age-dependent modulation of cortical transcriptomes in spinal cord injury and repair. PLoS One 7, e49812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Endo T., Spenger C., Tominaga T., Brene S., and Olson L. (2007). Cortical sensory map rearrangement after spinal cord injury: fMRI responses linked to Nogo signalling. Brain 130, 2951–2961 [DOI] [PubMed] [Google Scholar]
- 5. Aguilar J., Humanes-Valera D., Alonso-Calvino E., Yague J.G., Moxon K.A., Oliviero A., and Foffani G. (2010). Spinal cord injury immediately changes the state of the brain. J. Neurosci. 30, 7528–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim B.G., Dai H.N., McAtee M., Vicini S., and Bregman B.S. (2006). Remodeling of synaptic structures in the motor cortex following spinal cord injury. Exp. Neurol. 198, 401–415 [DOI] [PubMed] [Google Scholar]
- 7. Brock J.H., Rosenzweig E.S., Blesch A., Moseanko R., Havton L.A., Edgerton V.R., and Tuszynski M.H. (2010). Local and remote growth factor effects after primate spinal cord injury. J. Neurosci. 30, 9728–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falnikar A., Stratton J., Lin R., Andrews C.E., Tyburski A., Trovillion V.A., Gottschalk C., Ghosh B., Iacovitti L., Elliott M.B., and Lepore A.C. (2018). Differential response in novel stem cell niches of the brain following cervical spinal cord injury and traumatic brain injury. J. Neurotrauma [Epub ahead of print; DOI: 10.1089/neu.2017.5487] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu B., and Ren X. (2009). Promoting axonal myelination for improving neurological recovery in spinal cord injury. J. Neurotrauma 26, 1847–1856 [DOI] [PubMed] [Google Scholar]
- 10. Fawcett J.W. (2006). Overcoming inhibition in the damaged spinal cord. J. Neurotrauma 23, 371–383 [DOI] [PubMed] [Google Scholar]
- 11. Siddiqui A.M., Khazaei M., and Fehlings M.G. (2015). Translating mechanisms of neuroprotection, regeneration, and repair to treatment of spinal cord injury. Prog. Brain Res. 218, 15–54 [DOI] [PubMed] [Google Scholar]
- 12. Isa T. (2017). The brain is needed to cure spinal cord injury. Trends Neurosci. 40, 625–636 [DOI] [PubMed] [Google Scholar]
- 13. Nishimura Y., Onoe H., Morichika Y., Perfiliev S., Tsukada H., and Isa T. (2007). Time-dependent central compensatory mechanisms of finger dexterity after spinal cord injury. Science 318, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 14. Hawasli A.H., Rutlin J., Roland J.L., Murphy R.K.J., Song S.-K., Leuthardt E.C., Shimony J.S., and Ray W.Z. (2017). Spinal cord injury disrupts resting-state networks in the human brain. J. Neurotrauma 35, 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quertainmont R., Cantinieaux D., Botman O., Sid S., Schoenen J., and Franzen R. (2012). Mesenchymal stem cell graft improves recovery after spinal cord injury in adult rats through neurotrophic and pro-angiogenic actions. PLoS One 7, e39500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osaka M., Honmou O., Murakami T., Nonaka T., Houkin K., Hamada H., and Kocsis J. (2010). Intravenous administration of mesenchymal stem cells derived from bone marrow after contusive spinal cord injury improves functional outcome. Brain Res. 1343, 226–235 [DOI] [PubMed] [Google Scholar]
- 17. Morita T., Sasaki M., Kataoka-Sasaki Y., Nakazaki M., Nagahama H., Oka S., Oshigiri T., Takebayashi T., Yamashita T., Kocsis J.D., and Honmou O. (2016). Intravenous infusion of mesenchymal stem cells promotes functional recovery in a model of chronic spinal cord injury. Neuroscience 335, 221–231 [DOI] [PubMed] [Google Scholar]
- 18. Torres-Espin A., Hernandez J., and Navarro X. (2013). Gene expression changes in the injured spinal cord following transplantation of mesenchymal stem cells or olfactory ensheathing cells. PLoS One 8, e76141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakajima H., Uchida K., Guerrero A.R., Watanabe S., Sugita D., Takeura N., Yoshida A., Long G., Wright K.T., Johnson W.E., and Baba H. (2012). Transplantation of mesenchymal stem cells promotes an alternative pathway of macrophage activation and functional recovery after spinal cord injury. J. Neurotrauma 29, 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsushita T., Lankford K.L., Arroyo E.J., Sasaki M., Neyazi M., Radtke C., and Kocsis J.D. (2015). Diffuse and persistent blood–spinal cord barrier disruption after contusive spinal cord injury rapidly recovers following intravenous infusion of bone marrow mesenchymal stem cells. Exp. Neurol. 267, 152–164 [DOI] [PubMed] [Google Scholar]
- 21. Sasaki M., Radtke C., Tan A., Zhao P., Hamada H., Houkin K., Honmou O., and Kocsis J. (2009). BDNF-hypersecreting human mesenchymal stem cells promote functional recovery, axonal sprouting, and protection of corticospinal neurons after spinal cord injury. J. Neurosci. 29, 14,932–14,941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S., Honmou O., Kato K., Nonaka T., Houkin K., Hamada H., and Kocsis J. (2006). Neural differentiation potential of peripheral blood- and bone-marrow-derived precursor cells. Brain Res. 1123, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao Q., Zhang Y.P., Iannotti C., DeVries W.H., Xu X.M., Shields C.B. and Whittemore S.R. (2005). Functional and electrophysiological changes after graded traumatic spinal cord injury in adult rat. Exp. Neurol. 191, Suppl. 1, S3–S16 [DOI] [PubMed] [Google Scholar]
- 24. Basso D.M., Beattie M.S., and Bresnahan J.C. (1995). A sensitive and reliable locomotor rating scale for open field testing in rats. J. Neurotrauma 12, 1–21 [DOI] [PubMed] [Google Scholar]
- 25. Davies S.J., Shih C.H., Noble M., Mayer-Proschel M., Davies J.E., and Proschel C. (2011). Transplantation of specific human astrocytes promotes functional recovery after spinal cord injury. PLoS One 6, e17328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashimoto-Torii K., Kawasawa Y.I., Kuhn A., and Rakic P. (2011). Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol. Proc. Natl. Acad. Sci. U. S. A. 108, 4212–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmittgen T.D., and Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 28. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15,545–15,550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishimura Y., and Isa T. (2009). Compensatory changes at the cerebral cortical level after spinal cord injury. Neuroscientist 15, 436–444 [DOI] [PubMed] [Google Scholar]
- 30. Brauer A.U., Savaskan N.E., Kuhn H., Prehn S., Ninnemann O., and Nitsch R. (2003). A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat. Neurosci. 6, 572–578 [DOI] [PubMed] [Google Scholar]
- 31. Bass-Zubek A.E., Hobbs R.P., Amargo E.V., Garcia N.J., Hsieh S.N., Chen X., Wahl J.K., 3rd, Denning M.F., and Green K.J. (2008). Plakophilin 2: a critical scaffold for PKC alpha that regulates intercellular junction assembly. J. Cell Biol. 181, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haynes L.P., McCue H.V., and Burgoyne R.D. (2012). Evolution and functional diversity of the calcium binding proteins (CaBPs). Front. Mol. Neurosci. 5, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsson J., Forsberg M., Brannvall K., Zhang X.Q., Enarsson M., Hedborg F., and Forsberg-Nilsson K. (2008). Nuclear receptor binding protein 2 is induced during neural progenitor differentiation and affects cell survival. Mol. Cell Neurosci. 39, 32–39 [DOI] [PubMed] [Google Scholar]
- 34. Yang Y., Li Y., Lv Y., Zhang S., Chen L., Bai C., Nan X., Yue W., and Pei X. (2008). NRSF silencing induces neuronal differentiation of human mesenchymal stem cells. Exp. Cell Res. 314, 2257–2265 [DOI] [PubMed] [Google Scholar]
- 35. Li H.T., Jiang F.X., Shi P., Zhang T., Liu X.Y., Lin X.W., and Pang X.N. (2012). In vitro reprogramming of rat bone marrow-derived mesenchymal stem cells into insulin-producing cells by genetically manipulating negative and positive regulators. Biochem. Biophys. Res. Commun. 420, 793–798 [DOI] [PubMed] [Google Scholar]
- 36. Ishii T., Hashimoto E., Ukai W., Tateno M., Yoshinaga T., Saito S., Sohma H., and Saito T. (2008). Lithium-induced suppression of transcription repressor NRSF/REST: effects on the dysfunction of neuronal differentiation by ethanol. Eur. J. Pharmacol. 593, 36–43 [DOI] [PubMed] [Google Scholar]
- 37. Zhao Y., Zhu M., Yu Y., Qiu L., Zhang Y., He L., and Zhang J. (2017). Brain REST/NRSF Is not only a silent repressor but also an active protector. Mol. Neurobiol. 54, 541–550 [DOI] [PubMed] [Google Scholar]
- 38. Ballas N., Grunseich C., Lu D.D., Speh J.C., and Mandel G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 [DOI] [PubMed] [Google Scholar]
- 39. Aitken S., Magi S., Alhendi A.M., Itoh M., Kawaji H., Lassmann T., Daub C.O., Arner E., Carninci P., Forrest A.R., Hayashizaki Y., Khachigian L.M., Okada-Hatakeyama M., and Semple C.A. (2015). Transcriptional dynamics reveal critical roles for non-coding RNAs in the immediate-early response. PLoS Comput. Biol. 11, e1004217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carmel J.B., Kakinohana O., Mestril R., Young W., Marsala M., and Hart R.P. (2004). Mediators of ischemic preconditioning identified by microarray analysis of rat spinal cord. Exp. Neurol. 185, 81–96 [DOI] [PubMed] [Google Scholar]
- 41. Lapointe N.P., Ung R.V., and Guertin P.A. (2007). Plasticity in sublesionally located neurons following spinal cord injury. J. Neurophysiol. 98, 2497–2500 [DOI] [PubMed] [Google Scholar]
- 42. Kalita K., Kharebava G., Zheng J.J., and Hetman M. (2006). Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J. Neurosci. 26, 10,020–10,032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horita H., Wada K., Rivas M.V., Hara E., and Jarvis E.D. (2010). The dusp1 immediate early gene is regulated by natural stimuli predominantly in sensory input neurons. J. Comp. Neurol. 518, 2873–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang K.R., Nemoto T., and Yokota Y. (2007). RFX1 mediates the serum-induced immediate early response of Id2 gene expression. J. Biol. Chem. 282, 26167–26177 [DOI] [PubMed] [Google Scholar]
- 45. Regard J.B., Scheek S., Borbiev T., Lanahan A.A., Schneider A., Demetriades A.M., Hiemisch H., Barnes C.A., Verin A.D., and Worley P.F. (2004). Verge: a novel vascular early response gene. J. Neurosci. 24, 4092–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bahrami S. and Drablos F. (2016). Gene regulation in the immediate-early response process. Adv. Biol. Regul. 62, 37–49 [DOI] [PubMed] [Google Scholar]
- 47. Voulgari-Kokota A., Fairless R., Karamita M., Kyrargyri V., Tseveleki V., Evangelidou M., Delorme B., Charbord P., Diem R., and Probert L. (2012). Mesenchymal stem cells protect CNS neurons against glutamate excitotoxicity by inhibiting glutamate receptor expression and function. Exp. Neurol. 236, 161–170 [DOI] [PubMed] [Google Scholar]
- 48. Honmou O., Onodera R., Sasaki M., Waxman S.G., and Kocsis J.D. (2012). Mesenchymal stem cells: therapeutic outlook for stroke. Trends Mol. Med. 18, 292–297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.