Abstract
Multicellular tumor spheroid (MCTS) cultures represent more physiologically relevant in vitro cell tumor models that recapitulate the microenvironments and cell–cell or cell–extracellular matrix interactions which occur in solid tumors. We characterized the morphologies, viability, and growth behaviors of MCTSs produced by 11 different head and neck squamous cell carcinoma (HNSCC) cell lines seeded into and cultured in ultra-low attachment microtiter plates (ULA-plates) over extended periods of time. HNSCC MCTS cultures developed microenvironments, which resulted in differences in proliferation rates, metabolic activity, and mitochondrial functional activity between cells located in the outer layers of the MCTS and cells in the interior. HNSCC MCTS cultures exhibited drug penetration and distribution gradients and some developed necrotic cores. Perhaps the most profound effect of culturing HNSCC cell lines in MCTS cultures was their dramatically altered and varied growth phenotypes. Instead of the exponential growth that are characteristic of two-dimensional HNSCC growth inhibition assays, some MCTS cultures displayed linear growth rates, categorized as rapid, moderate, or slow, dormant MCTSs remained viable but did not grow, and some MCTSs exhibited death phenotypes that were either progressive and slow or rapid. The ability of MCTS cultures to develop microenvironments and to display a variety of different growth phenotypes provides in vitro models that are more closely aligned with solid tumors in vivo. We anticipate that the implementation MCTS models to screen for new cancer drugs for solid tumors like HNSCC will produce leads that will translate better in in vivo animal models and patients.
Keywords: multicellular tumor spheroids, head and neck squamous cell carcinoma, 3D microenvironments
Introduction
Head and neck cancers (HNCs) are the eighth leading cause of cancer worldwide with ∼600,000 new cases and ∼300,000 deaths occurring per annum.1–3 In 2018, it's estimated that 51,540 people in the United States will develop oral cavity or pharynx cancer and 10,030 will die of these cancers. Smoking, alcohol, genetics, and human papillomavirus (HPV) infection are major risk factors for the development of head and neck squamous cell carcinoma (HNSCC).1–3 HNC incidence is rising in developed countries despite reductions in cigarette smoking rates, with much of the increase attributable to HPV infection.1–4 Surgical resection and chemo-radiotherapy are the frontline therapies for localized HNC.5–9 Although surgical and radiation therapies have improved, cure rates have remained stationary at ∼50% for >30 years.1–3,7,9,10 Patients with advanced, recurrent, or metastatic HNC have a poor prognosis with median survival rates of 6–12 months.1,2,9 The Federal Drug Administration (FDA) has approved only seven drugs for HNC; methotrexate and 5-fluorouracil in the 1950s, bleomycin and cisplatin in the 1970s, docetaxel and cetuximab in 2006, and pembrolizumab in 2016. However, only 10%–25% of HNC patients respond to monotherapy with the five older chemotherapeutics and/or the epidermal growth factor (EGF)-receptor targeted antibody, and these agents have failed to significantly improve either 5-year survival or cure rates.1,2 The FDA approved the monoclonal antibody pembrolizumab (Keytruda®) in patients with recurrent or metastatic HNSCC that continued to progress despite standard-of-care chemotherapy. Pembrolizumab blocks the immune checkpoint anti-programmed death-ligand 1 (PD1) receptor interactions with its ligands, PD-L1 and PD-L2.11–13 The FDA accelerated Keytruda approval was based on early clinical trials where it was well tolerated and produced clinically relevant antitumor activity in recurrent or metastatic HNSCC.11–13 Although tumor responses lasted ≥6 months in 82% of responders, and some patients experienced lasting (>2 years) and/or complete responses, only 16% of HNSCC patients responded to Keytruda treatment.11–13 Despite Keytruda's success, its low response rate together with the overall lack of efficacy of the other approved drugs for HNSCC underscores the urgent unmet need to discover new and effective HNSCC therapies.
Despite significant investments in cancer research, drug discovery, and development, <5% of all new small molecule cancer drugs entering phase I clinical trials gain FDA approval.8,14–17 New cancer drug leads are usually discovered in growth inhibition (GI) assays conducted in tumor cell line panels maintained and assayed in two-dimensional (2D) culture conditions that are compatible with high throughput screening (HTS).8,18–20 2D GI HTS assays typically utilize low cell numbers to reduce the cell culture burden and to maximize drug sensitivity by promoting exponential growth during the compound exposure period.6,20–22 Cells in 2D GI assays experience uniform drug concentrations in a homogenous environment where cell interactions with the extracellular matrix (ECM) and cell–cell contacts are either nonexistent or substantially reduced.6,20–22 Genomic comparisons of established HNC cell lines to primary tumors revealed that although 51% of genetic alterations were shared at similar mutation frequencies, subsets of mutations were unique to patient tumors and cell lines, respectively.7 Mutations unique to cell lines favored immortalization and continuous maintenance in tissue culture.7 In addition, tumor cell lines adapted to growth in 2D proliferate faster than cells from primary tumors, display altered drug resistance patterns, and respond preferentially to antiproliferative agents while overlooking the self- and population-renewing tumor stem cells, which contribute to recurrence and metastasis.22–27 2D tumor GI assays fail to recapitulate the complex three-dimensional (3D) architecture, cell–cell and cell–ECM interactions, microenvironments, drug diffusion kinetics, and drug responses of solid tumors in vivo.15,22–24,27–35 To identify better leads that have the potential to improve the clinical development success rates of new cancer drugs for solid tumors like HNSCC, more physiologically relevant in vitro 3D tumor models that better represent the growth and tumor microenvironments observed in preclinical in vivo mouse models and patient tumors are being deployed for lead generation.22,24,26,27,29,30,32,34–42
A wide variety of anchorage-dependent and anchorage-independent in vitro 3D tumor cell culture models have been developed to investigate and understand how cellular and functional responses are altered by 3D culture conditions and to provide 3D assays for cancer drug discovery.30,34,40 Multicellular tumor spheroids (MCTSs) resemble avascular tumor nodules, micro-metastases, or the intervascular regions of large solid tumors with respect to morphological features and volume growth kinetics, and they develop microenvironments produced by gradients of nutrient distribution and oxygen concentration resulting in differential zones of proliferation and drug permeability.22,24,25,29,32,34,41,43,44 Several groups have utilized U-bottomed ultra-low attachment 96-, 384-, or 1536-well microtiter plates (ULA-plates) treated with a hydrophilic neutrally charged coating covalently bound to the polystyrene well surface to prevent cell adhesion to the plate surface and promote tumor cell line self-assembly into tight MCTSs or cell aggregates.22,24,29,34–36,38,41 We have shown that the production of HNSCC MCTSs in 384-well ULA-plates is both compatible with automation and scalable for HTS because MCTSs form within 1–3 days and require relatively few cells (≤2.5 K) per well, and both compound exposure and homogeneous assay detection can be performed in situ.29,34 We have used Cal33 and FaDu HNSCC MCTSs generated in 384-well ULA-plates to demonstrate that cancer drug accumulation increases as cell numbers and MCTS sizes increase and that drugs exhibit restricted penetration and distribution gradients, accumulating preferentially in cells in the outer layers of MCTSs relative to those in the inner cores.22 In addition, 2D Cal33 monolayers are 6-fold, 20-fold, 10-fold, and 16-fold more sensitive than Cal33 MCTSs to GI produced by ellipticine, idarubicin, daunorubicin, and doxorubicin, respectively.22 The present article extends these studies to characterize the distinct morphologies, viability, and growth phenotypes of MCTSs formed in 384-well ULA-plates by 11 HNSCC cell lines and an immortalized human “normal” esophageal epithelial cell line. For five HNSCC cell lines with distinct MCTS growth phenotypes, we show the development of microenvironments and uneven drug accumulation in different regions of MCTSs and demonstrate that these more physiologically relevant 3D tumor models can be readily adapted to measure drug-induced cytotoxicity.
Materials and Methods
Reagents
Thirty-seven percent formaldehyde was purchased from Sigma-Aldrich (St. Louis, MO). Hoechst 33342 was purchased from Life Technologies (Thermo Fisher Scientific, Waltham, MA). Dimethyl sulfoxide (DMSO) 99.9% high-performance liquid chromatography grade was obtained from Alfa Aesar (Ward Hill, MA). Dulbecco's Mg2+- and Ca2+-free phosphate-buffered saline (PBS) was purchased from Gibco (Grand Island, NY). Dulbecco's modified Eagle's medium (DMEM) and Dulbecco's modified Eagle's medium/Ham's F12 50/50 (DMEM/F12) were purchased from Corning (Manassas, VA). Fetal bovine serum (FBS), l-glutamine, and penicillin/streptomycin (P/S) were purchased from Thermo Fisher Scientific. CellTiter-Blue® (CTB) was purchased from Promega Corporation (Madison, WI). Calcein AM (CAM), Ethidium Homodimer (EHD), MitoTracker® Orange (MTO) a thiol-reactive chloromethyl derivative of tetra-methyl-rosamine, tetra-methyl-rhodamine methyl ester (TMRM), Click-iT® 5-ethynyl-2′-deoxyuridine (EdU), and 5-(and-6)-(((4-chloromethyl)benzoyl)amino)-tetra-methyl-rhodamine CellTracker® Orange (CTO) were all purchased from Life Technologies (Thermo Fisher Scientific). Doxorubicin was provided by the National Cancer Institute (NCI).
Cells and Tissue Culture
Eleven human HNSCC cell lines were provided by Dr. Jennifer Grandis of the HNC Spore at the University of Pittsburgh Medical Center Hillman Cancer Center and were maintained in a humidified incubator at 37°C, 5% CO2, and 95% humidity; Cal33, Cal27, FaDu, UM-22B, BICR56, OSC-19, PCI-13, PCI-52, Detroit-562, UM-SCC-1, and SCC-9. The human Het-1A (CRL-2692™) esophageal squamous epithelial cell line transfected with the SV40 large T antigen was purchased from the American Type Culture Collection (ATCC®, Manassas, VA). All cell lines except SCC-9 were cultured in DMEM supplemented with 10% FBS, 1% l-glutamine, and 1% P/S. The culture medium for the FaDu and OSC-19 cell lines was also supplemented with 1% nonessential amino acids, and the medium for the BICR56 and UM-SCC-1 cell lines was supplemented with 0.4 μg/mL hydrocortisone. The SCC-9 cell line was cultured in DMEM/F12 medium supplemented with 10% FBS, 1% l-glutamine, and 1% P/S. HNSCC cell lines were passaged or used to generate MCTS after isolated cell suspensions were prepared from tissue culture flasks by dissociating cells with trypsin and centrifugation at 270 × g for 5 min at room temperature and resuspension in growth media. The number of viable trypan blue excluding cells in the cell suspension was counted using a hemocytometer.
Generation of HNSCC MCTSs in Ultra-Low Attachment Microtiter Plates
We have previously described the generation of MCTSs after seeding several HNSCC cell lines into 384-well U-bottomed ultra-low attachment microtiter plates (ULA-plates; Cat. No. 4516; Corning, Tewksbury, MA).22,29,34 Briefly, 384-well ULA-plates were rehydrated by the addition of 50 μL of serum-free culture medium to each well and incubation in a humidified incubator for 15 min. Media was removed from the wells of the ULA-plates, and 45 μL of a single-cell suspension of the HNSCC cell lines at different seeding densities (625, 1,250, 2,500, 5,000, 10,000, or 20,000 cells/well) in the appropriate growth medium was transferred into each well using a Matrix automated multichannel pipette (Thermo Fisher Scientific); ULA-plates were centrifuged at 17 × g for 1 min and then placed in an incubator at 37°C, 5% CO2, and 95% humidity for the indicated time periods. In time course experiments where HNSCC MCTS cultures were maintained in the ULA-plates beyond 3 days, spent media was exchanged for fresh medium every 3 days using a Janus MDT Mini (PerkinElmer, Waltham, MA) automated liquid handler platform equipped with a 384-well transfer head. Each medium exchange cycle consisted of 2 × 20 μL aspiration and discard steps followed by 2 × 20 μL fresh media dispense steps. Three media exchange cycles were performed to achieve ∼85% exchange of fresh medium for spent medium and a uniform volume of 45 μL per well.
Investigation of HNSCC MCTS Morphology, Viability, and Growth in Ultra-Low Attachment Microtiter Plates by High Content Imaging
We used an ImageXpress Micro (IXM) automated wide field high content imaging platform integrated with MetaXpress Imaging and Analysis software (Molecular Devices, LLC, Sunnyvale, CA) to acquire and analyze images of HNSCC MCTSs. The IXM optical drive uses a 300 W Xenon lamp broad spectrum white light source and a 1.4-megapixel 2/3″ chip Cooled CCD Camera and optical train for standard fluorescence imaging and a transmitted light (TL) module with phase contrast. The IXM is equipped with Zero Pixel Shift (ZPS) filter sets; DAPI, FITC/ALEXA 488, CY3/TRITC, CY5, and Texas Red. A four-position objective turret can be loaded with various objectives; a 4 × Plan Apo 0.20 NA objective, a 10 × Plan Fluor 0.3 NA objective, a 20 × Ph1 Plan Fluor extra-long working distance (ELWD) dark medium objective, a 20 × S Plan Fluor ELWD 0.45 NA objective, and a 40 × S Plan Fluor ELWD 0.60 NA objective.
Single images of HNSCC MCTSs were sequentially acquired using a 4 × Plan Apo 0.20 NA objective in both the TL and fluorescent image acquisition modes; DAPI, FITC, and TRITC.22,29,34,45 To acquire best focus images of MCTSs we used the IXM automated image-based focus algorithm to acquire both a coarse focus (large μm steps) set of images of Hoechst stained objects in the DAPI channel for the first MCTS to be imaged, followed by a fine (small μm steps) set of images to select the best focus image. For all subsequent wells and channels to be imaged, only a fine focus set of images was acquired to select the best focus Z-plane.22,29,34,45 MCTS morphology and growth were assessed daily by the acquisition of 4 × TL images on the IXM, and we used the line-scan tool of the MetaXpress image analysis software to measure the diameters of the HNSCC MCTSs.45 The change in MCTS diameter over time in culture was used as an indicator of MCTS growth or death.
To label viable and/or dead cells within the HNSCC MCTS cultures, we incubated HNSCC MCTSs with a cocktail of the Hoechst (8 μg/mL) DNA stain, the CAM (2.5 μM) live reagent, and the EHD (5 μM) dead reagent for 1 h, and single images of HNSCC MCTSs were sequentially acquired on the IXM using a 4 × objective in both the TL and fluorescent image acquisition modes; DAPI, FITC, and Texas Red channels. We used the multiwavelength cell scoring (MWCS) image analysis module to analyze the HNSCC MCTS fluorescent images as described previously.22,29,34 To create a whole MCTS mask, we set the approximate minimum width of the Hoechst stained nuclei of the MCTS to be 150 μm with an approximate maximum width to be 550 μm and applied a threshold intensity above local background of 70. The total MCTS mask from the Hoechst channel was used to count the number of MCTSs per image, typically one. After applying user defined background average intensity thresholds, typically 50–70 in both the FITC and Texas Red channels, the MWCS module image segmentation then created total MCTS masks in all three fluorescent channels. The derived HNSCC total MCTS masks in channels 2 and 3 were then used to quantify the mean integrated fluorescence intensity (MIFI) of the CAM live cell signal in the FITC channel and the EHD dead cell signal in the Texas Red channel. The MIFI values represent the total pixel fluorescent intensities in channels 1, 2, or 3 within the total MCTS masks of positively stained MCTSs above the preset background thresholds.
Pseudo-color fluorescence intensity data visualizations were also used to illustrate fluorescent dye/drug uptake, accumulation, and distribution in HNSCC MCTS cultures.22 The relative fluorescent intensities of the pixels in the image were represented as distinct colors with the “hotter” and “brighter” colors (low to high, yellow, red, white) representing higher intensity signals and cooler colors (low to high, purple, cyan, green) representing lower intensity signals. Line-scan fluorescence intensity plots were also used to portray fluorescent dye/drug uptake, accumulation, and distribution in HNSCC MCTS cultures.22 Line-scan fluorescence intensity plots were created using the line scanning tool of the MetaXpress image analysis software to draw a line across the image and plot the fluorescent intensity values versus distance in μm across the image to provide an intensity profile graph.
Determination of HNSCC MCTS Proliferation
After 3 days of culture in ULA-plates to allow MCTSs to form, the cell culture medium was exchanged and the HNSCC MCTSs were exposed to EdU reagent (Life Technologies) according to the manufacturer's instructions for 12, 24, and 48 h before fixation in 3.7% formaldehyde and permeabilization with 0.1% Triton X-100. The modified nucleoside EdU, which becomes incorporated into DNA by proliferating cells undergoing DNA synthesis, is detected using an Alexa Fluor 488 picolyl azide fluorescent label, which is incorporated using a quick “click chemistry” reaction. Images of HNSCC MCTSs were sequentially acquired on the IXM using a 4 × objective in both the TL and FITC channels.
Characterization of HNSCC MCTS Mitochondrial Mass and Membrane Potential
After 3 days of culture in ULA-plates to allow MCTSs to form, the cell culture medium was exchanged and the HNSCC MCTSs were exposed to 500 nM MTO or 250 nM TMRM (Life Technologies) for 1 h at 37°C, 5% CO2, and 95% humidity. MTO labeled HNSCC MCTSs were fixed and stained with 3.7% formaldehyde containing 8 μg/mL Hoechst and then washed 3 × with PBS before images were acquired on the IXM using a 4 × objective in the TL, DAPI, and TRITC channels. Images of TMRM labeled live HNSCC MCTSs were acquired on the IXM using a 4 × objective in both the TL and TRITC channels. HNSCC MCTS images were analyzed using the MWCS image analysis module described above to quantify the MIFI of the MTO and TMRM fluorescent dyes, and both pseudo-color visualizations and line scan plots were used to illustrate their distribution throughout the HNSCC MCTSs.
Drug Penetration and Distribution in HNSCC MCTSs
Analysis of drug penetration was performed as previously described for the Cal33 and FaDu HNSCC cell lines.22 Briefly, HNSCC cell lines were cultured for 3 days to allow MCTSs to form before a media exchange was performed, and the MCTSs were then exposed to 10 μM doxorubicin and incubated at 37°C, 5% CO2, and 95% humidity for 0.5, 3, 6, 12, and 24 h before fixation in 3.7% formaldehyde for 30–45 min, 3 × washes with PBS, and acquisition of 4 × images on the IXM in the TL and TRITC channels. The MWCS image analysis module described above was used to quantify the fluorescent intensity of doxorubicin in HNSCC MCTS TRITC channel images using the doxorubicin stained nuclei in the TRITC channel to identify and define the whole MCTS mask. The MIFI parameter represents the total pixel fluorescent intensity of doxorubicin within the MCTS.22 Pseudo-color visualizations and line-scan plots were also used to illustrate doxorubicin accumulation and distribution throughout HNSCC MCTSs.
Analysis of HNSCC MCTS Viability and Growth using the CTB Reagent
The homogeneous CTB cell viability reagent provides a fluorescent method for monitoring cell viability and/or GI that is based on the ability of living cells to convert the redox dye resazurin into a fluorescent end product resorufin. HNSCC cell lines were seeded at the indicated cell densities into 384-well ULA-plates in 45 μL of growth medium incubated at 37°C, 5% CO2, and 95% humidity for the indicated time periods. After the prescribed time in culture, 10 μL of the CTB cell viability detection reagent was dispensed into the wells of HNSCC MCTS assay plates and incubated for 4 h at 37°C, 5% CO2, and 95% humidity before capturing the relative fluorescent unit (RFU) signals (Ex. 560 nm/EM. 590 nm) on a SpectraMax M5e (Molecular Devices, LLC) microtiter plate reader platform.
Data Processing, Analysis, and Curve Fitting
For HNSCC MCTS growth assays in 384-well ULA-plates, the measured diameters of the HNSCC MCTSs and their corresponding CTB RFU signals were fit to a linear regression model. The MIFI data from HNSCC MCTSs exposed to CAM, EHD, MTO, TMRM, or doxorubicin are presented as the mean ± standard deviation (n = 3). For HNSCC MCTS GI (50% growth inhibitory concentration [GI50]) assays, the DMSO control wells (Max controls, n = 14) and 200 μM doxorubicin control wells (Min controls, n = 32) were used to represent uninhibited growth and 100% cytotoxicity, respectively. The mean maximum and minimum plate control CTB RFUs were used to normalize the RFU data from the compound treated wells as percent inhibition of growth. The GI50 data were fit to a nonlinear sigmoidal log (inhibitor) versus normalized response variable slope model using the equation: Y = 100/(1 + 10^[{LogIC50 − X} × Hillslope]), where Y was the percent GI and X was the corresponding log10 of the compound concentration. The GI50 is the concentration of compound that gives a 50% response, half way between 0% and 100%. The Hillslope describes the steepness of the curve. All curve fitting, linear regression analysis, and graphs were created using the GraphPad Prism 6 software.
Results
Morphology and Viability of HNSCC MCTSs Formed in 384-Well U-Bottomed Ultra-Low Attachment Microtiter Plates
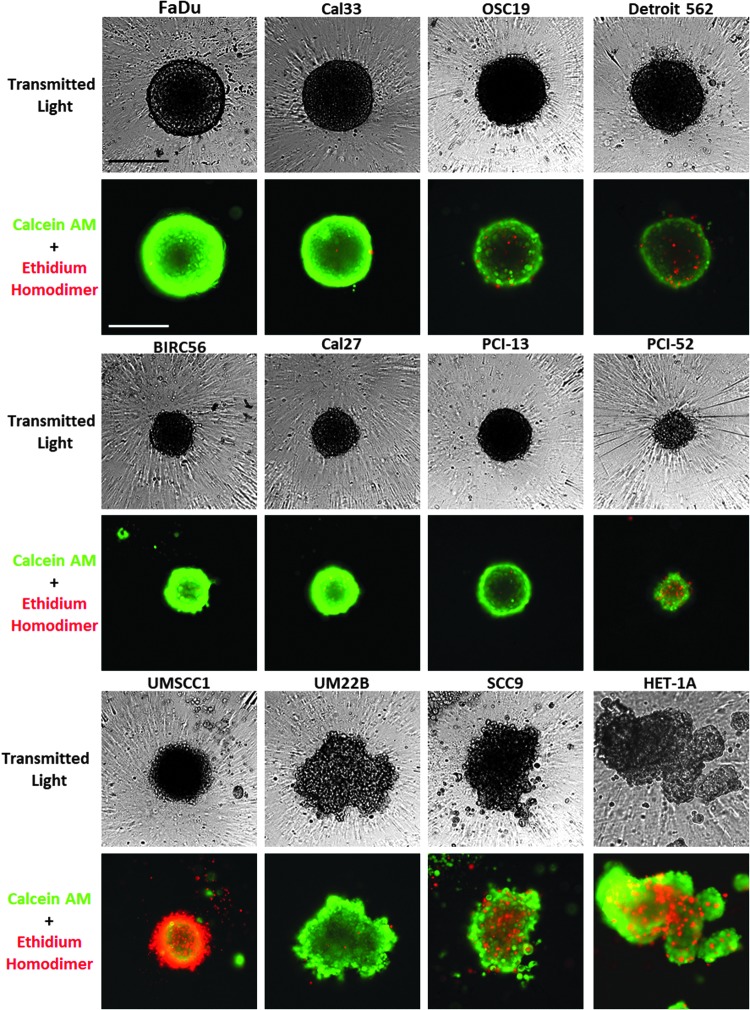
We have previously shown that six HNSCC tumor cell lines form MCTSs within 24 h of seeding into 384-well ULA-plates.22,29,34 To extend these studies, we selected five additional HNSCC cell lines that have previously been used for HNC drug discovery.6,7,21 The Het-1A SV40 T-antigen immortalized human esophageal epithelial cell line was included to represent a “normal” control cell line model, since these cells do not grow as xenograft tumors in immunodeficient mice.46–49 To characterize the morphology and viability of the HNSCC MCTSs formed, 11 HNSCC cell lines and Het-1A cells were seeded into 384-well ULA-plates and cultured for 3 days before they were stained with live cell CAM and dead cell EHD reagents, and 4 × images were acquired on the IXM in the TL, FITC, and Texas Red channels (Fig. 1). Although some HNSCC cell lines formed MCTSs within 24 h of seeding into 384-well ULA-plates, others required 2–3 days to self-assemble. The morphologies (shape and compactness) and sizes of the MCTSs varied with each HNSCC cell line (Fig. 1). After 72 h in culture, FaDu, Cal33, Cal27, and PCI-13 cell lines formed condensed MCTSs with a smooth and even periphery. OSC19, Detroit 562, BICR56, PC1–52, and UMSCC1 cell lines produced rounded MCTSs with uneven perimeters, and the Het-1A, UM22B, and SCC9 cell lines formed cell aggregates with irregular outer margins. FaDu, Cal33, OSC19, and Detroit 562 cells formed larger MCTSs approximately 350–400 μm in diameter, while BICR56, Cal27, PC13, PCI-52, and UMSCC1 formed smaller MCTSs with approximately 200–250 μm diameters. FaDu, Cal33, BICR56, and PC1–52 MCTSs and UM22B aggregates exhibited strong CAM staining with little or no EHD staining, indicating that all the cells were viable. OSC19, Detroit 562, and PCI-52 MCTSs also exhibited strong CAM staining, and although there was some EHD staining, most cells were viable. In contrast, UMSCC1 MCTSs exhibited strong EHD staining and much lower CAM staining, indicating that most cells were dead. The SCC9 and Het-1A irregular aggregates exhibited strong CAM staining together with some significant EHD staining.
Fig. 1.
Characterization of HNSCC MCTS morphology and viability in Ultra-low Attachment Plates. Eleven HNSCC cell lines and Het-1A cells were seeded at 2,500 cells per well in 384-well ULA-plates and cultured for 3 days. MCTSs were then stained with the live cell CAM and dead cell EHD reagents, and 4 × images were acquired on the IXM in the TL, FITC, and Texas Red channels. Grayscale TL images are presented along with color composite fluorescent images of live cell CAM and dead cell EHD staining depicted as green and red, respectively. Representative images from multiple independent experiments are presented. All scale bars represent 300 μm. CAM, Calcein AM; EHD, Ethidium Homodimer; HNSCC, head and neck squamous cell carcinoma; IXM, ImageXpress Micro; MCTS, multicellular tumor spheroid; TL, transmitted light; ULA-plates, ultra-low attachment microtiter plates.
HNSCC MCTS Growth Phenotypes in ULA-Plates
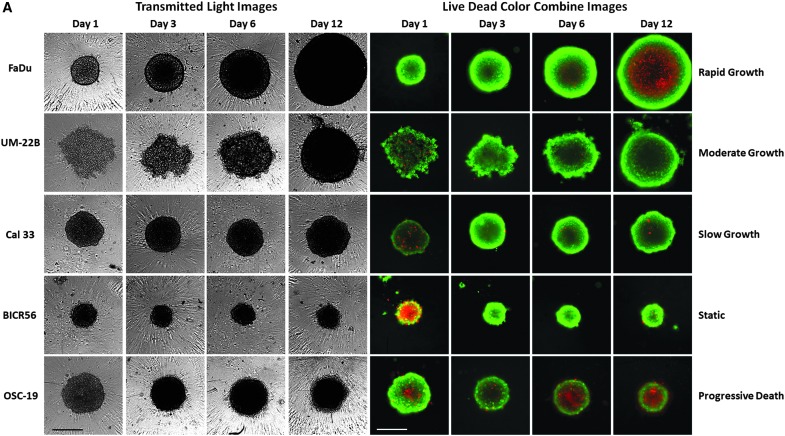
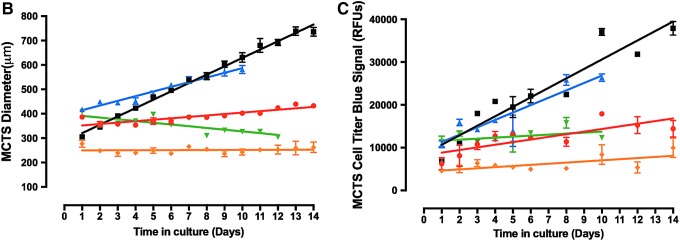
To further characterize the morphologies, viability, and growth behavior of HNSCC MCTSs cultured in 384-well ULA-plates over time, we conducted a series of cell seeding density and time course experiments. The 11 HNSCC cell lines were seeded into 384-well ULA-plates at densities ranging between 625 and 20,000 cells per well and were cultured for 12–14 days, with media exchanges every 3 days. MCTS growth and viability were evaluated by TL imaging, CAM/EHD staining, and CTB RFUs. The 11 HNSCC cell lines exhibited 6 distinct MCTS growth phenotypes; rapid growth (FaDu), moderate growth (UM22B), slow growth (Cal33), dormant (BICR56, Cal27, Detroit 562, and PCI-13), progressive slow death (OSC19, PCI-52, and SCC9), and rapid death (UMSCC1). Figure 2 shows time course data for 5 HNSCC cell lines seeded at 2,500 cells per well in 384-well ULA-plates that represent 5 of the MCTS growth phenotypes. Consistent with Figure 1, the TL images indicated that MCTS morphologies and sizes were different for each HNSCC cell line and, furthermore, that these changed over time in culture (Fig. 2A). In general, HNSCC MCTS TL images became progressively darker with longer time in culture, especially in the MCTS core regions, perhaps indicating that the cells were becoming more densely packed. The UM22B line exhibited the most dramatic change in morphology with increasing time in culture, coalescing from a loose aggregate of cells into a compact MCTS with a smooth and even periphery after several days (Fig. 2A). To assess the changes in HNSCC MCTS size over time, we used the line-scan tool of the image analysis software to measure the diameters of the HNSCC MCTSs in TL images acquired daily (Fig. 2B). The diameters of the FaDu, UM22B, and Cal33 MCTSs increased linearly with respect to time in culture throughout the 10- to 14-day culture period. In contrast, BICR56 MCTSs exhibited almost no change in diameter, and OSC19 MCTS diameters became progressively smaller with extended time in culture. Based on the observed changes in MCTS diameter over time, the rank order of HNSCC MCTS growth was FaDu > UM22B >> Cal33. BICR56 MCTS diameters remained constant over time, while OSC19 MCTS diameters declined gradually with time in culture.
Fig. 2.
Growth phenotypes of MCTSs produced by five representative HNSCC cell lines seeded and maintained in Ultra-low Attachment Plates. (A) Five HNSCC cell lines were seeded at 2,500 cells per well in 384-well ULA-plates and were cultured for 12 days with fresh media exchanges performed every 3 days. MCTSs were stained after 1, 3, 6, or 12 days in culture with the live cell CAM and dead cell EHD reagents, and 4 × images were acquired on the IXM in the TL, FITC, and Texas Red channels. Grayscale TL images are presented along with color composite fluorescent images of live cell CAM and dead cell EHD staining depicted as green and red, respectively. Representative images from multiple independent experiments are presented. All scale bars represent 300 μm. (B) The line-scan tool of the MetaXpress image analysis software was used to measure the diameters of the five HNSCC MCTSs in TL images that were acquired daily throughout the 10- to 14-day culture period. (C) CTB RFU signals of the five HNSCC MCTS cultures were measured at the indicated time points throughout the 10- to 14-day culture period. MCTS diameters in μm (B) and CTB RFUs (C) were measured in triplicate wells (n = 3) and are presented as the mean ± SD for the following HNSCC cell lines; UM-22B (blue triangle), OSC-19 (green triangle), FaDu (black square), BICR56 (yellow diamond), and Cal33 (red circle). A linear regression of the mean MCTS diameter and CTB RFU data for each of the HNSCC cell lines was performed using the GraphPad Prism 6 software. Representative data from three independent experiments are presented. CTB, CellTiter-Blue®; RFU, relative fluorescent unit; SD, standard deviation.
The homogeneous CellTiter Glo® (CTG) ATP detection reagent has frequently been used to measure growth and compound mediated cytotoxicity in MCTS cultures produced and maintained in 96- and 384-well ULA-plates.22,24,29,34,36,41 However, we switched to CTB when we found that the relative light units produced by CTG for the same number of cells in 2D and 3D cultures were ∼10-fold lower in 3D cultures, whereas we observed little or no difference in the CTB RFUs between 2D and 3D cultures composed of the same cell numbers (data not shown). To illustrate the correlation between CTB RFUs and the number of viable HNSCC cells in MCTS cultures, we seeded between 625 and 20,000 FaDu cells per well into 384-well ULA-plates and cultured them for 24 h to allow MCTSs to form. After 24 h we acquired 4 × TL images of the MCTSs on the IXM (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/adt), measured the diameters of the MCTSs using the line-scan tool, and measured the CTB RFUs 4 h after reagent addition (Supplementary Fig. S1B). In the 625–5,000 cells per well range, MCTS diameters and CTB RFUs increased linearly with respect to the number of viable FaDu cells seeded per well, and although MCTS diameters and CTB RFUs continued to increase as more cells were added to wells, the increase was no longer linear with respect to cell number (Supplementary Fig. S1B). The MCTS diameter and CTB RFU data were almost superimposable indicating that they are closely correlated (Supplementary Fig. S1B). Similar CTB data were observed for all HNSCC MCTS cultures (data not shown). Based on the observed changes in CTB signal over time the rank order of MCTS growth rates was the same as that indicated by the change in MCTS diameter (Fig. 2B, C).
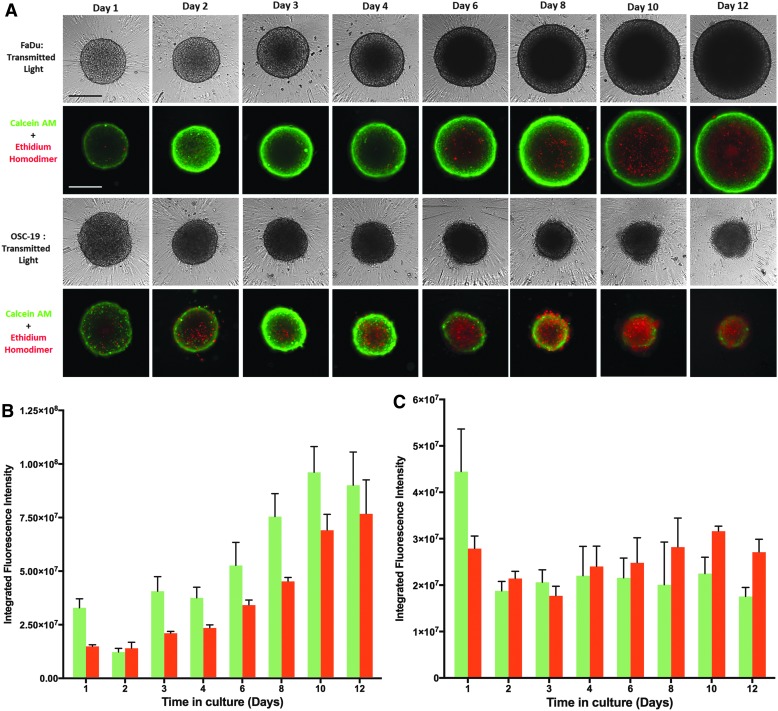
The corresponding CAM/EHD time course images suggest that at most time points a large majority of cells in the FaDu, UM22B, Cal33, and BICR56 MCTSs were viable, although FaDu MCTSs appeared to develop necrotic cores by day 6 through 12 as they became progressively larger, and OSC19 MCTSs appeared to have a necrotic core at all time points (Fig. 2A). To further explore the development of necrotic cores in HNSCC MCTSs, we seeded the FaDu and OSC19 cell lines into 384-well ULA-plates and cultured them for up to 12 days, with media exchanges every 3 days. MCTS growth and viability were evaluated by TL microscopy and CAM/EHD staining (Fig. 3A), and we used the MWCS image analysis module to quantify and compare the CAM and EHD MIFI signals in FaDu (Fig. 3B) and OSC-19 (Fig. 3C) MCTSs. Consistent with its rapid growth phenotype (Fig. 2), FaDu MCTS size increased over time in culture (Fig. 3A), and the corresponding CAM MIFI signals also increased (Fig. 3B). At most time points the FaDu MCTS CAM MIFIs were greater than the corresponding EHD MIFIs, but after day 4 the development of a necrotic core was indicated by both the CAM/EHD images and by an apparent larger increase in EHD MIFIs (Fig. 3A, B). The slow progressive death phenotype of OSC19 cell line was illustrated by the decrease in MCTS size observed in the TL and CAM/EHD images (Fig. 3A) and the lack of any change in CAM MIFIs between day 2 and 12, with a trend toward increased EHD MIFIs over the same period (Fig. 3C).
Fig. 3.
Development of a necrotic core in MCTSs formed by the FaDu and OSC19 HNSCC cell lines seeded and maintained in Ultra-low Attachment Plates. (A) The FaDu and OSC19 HNSCC cell lines were seeded at 2,500 cells per well in 384-well ULA-plates and were cultured for 12 days with fresh media exchanges performed every 3 days. MCTSs were stained after the indicated days in culture with the live cell CAM and dead cell EHD reagents, and 4 × images were acquired on the IXM in the TL, FITC, and Texas Red channels. Grayscale TL images are presented along with color composite fluorescent images of live cell CAM and dead cell EHD staining depicted as green and red, respectively. Representative images from multiple independent experiments are presented. All scale bars represent 300 μm. The MWCS image analysis module of the MetaXpress software was used to quantify the live cell CAM and dead cell EHD MIFI signals in (B) FaDu and (C) OSC-19 MCTSs over time in culture. The MWCS image module segmented Hoechst channel images to create a whole MCTS mask and then applied user defined background average intensity thresholds to create whole MCTS masks for the FITC and Texas Red channels. The whole MCTS masks were used to quantify the MIFIs of the CAM live cell signal (green) in the FITC channel and the EHD dead cell signal (red) in the Texas Red channel. MIFIs were measured in triplicate wells (n = 3) at the indicated time points and are presented as the mean ± SD. Representative data from three independent experiments are presented. MIFI, mean integrated fluorescence intensity; MWCS, multiwavelength cell scoring.
Images of HNSCC MCTSs stained with the live cell CAM reagent exhibited a donut-like staining pattern, where cells in the outer edges of the MCTSs exhibited higher intensities relative to cells in the inner cores, irrespective of MCTS size or HNSCC cell line (Figs. 1, 2A, and 3A). To show that the differential distribution of CAM observed in the outer layers of HNSCC MCTSs was not due to an imaging artifact caused by inefficient excitation light penetration and/or fluorescent emission light detection, we prelabeled HNSCC cell lines with CTO before seeding them into 384-well ULA-plates to form MCTSs (Supplementary Fig. S2). The TRITC images, pseudo-color visualizations, and line-scan plots showed that CTO fluorescence was distributed uniformly throughout the MCTSs or, if anything, the fluorescent intensities were slightly higher in the inner cores of MCTSs relative to the outer layers. Since the CAM reagent must be metabolized by live cells into the fluorescent end product, it is possible that the higher CAM fluorescent intensities observed in cells in the outer layers of HNSCC MCTSs reflect their higher metabolic activity relative to cells in the inner cores.
HNSCC MCTS Cell Proliferation
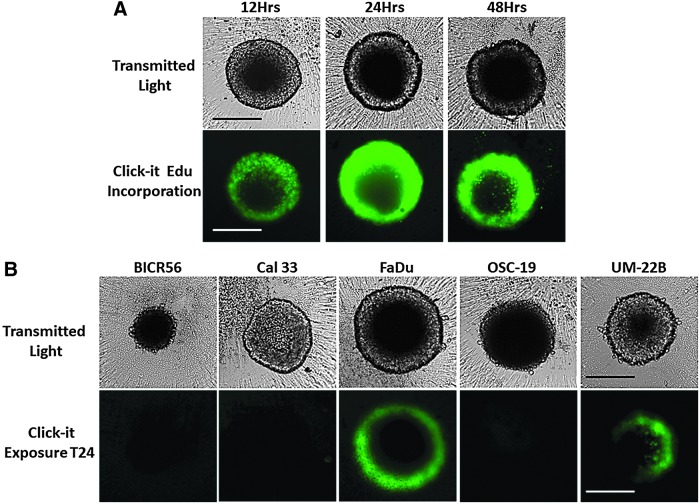
To investigate whether cells in HNSCC MCTSs formed and maintained in ULA-plates exhibited different rates of cell proliferation in distinct MCTS regions, we seeded 2,500 HNSCC cell lines into 384-well ULA-plates and allowed MCTSs to form for 3 days before the culture media was exchanged and during the next 3 days MCTSs were exposed to EdU for 12, 24, or 48 h (Fig. 4). Six-day FaDu MCTS cultures that were exposed to EdU for 12, 24, or 48 h exhibited a distinct gradient of EdU staining, with cells in the outer layers of the MCTSs exhibiting higher intensities relative to cells in the inner cores at all EdU exposure periods (Fig. 4A). In 6-day HNSCC MCTS cultures representing the five growth phenotypes, only the rapidly (FaDu) and moderately (UM22B) growing MCTSs displayed evidence of detectable EdU incorporation after 24 h of exposure (Fig. 4B). Similar results were observed after 48 h of EdU exposure (data not shown). These data indicate that in HNSCC MCTSs that exhibit rapid and moderate growth rates in ULA-plates, the more quiescent cells in the interior are surrounded by outer layers of proliferating cells.
Fig. 4.
Cell proliferation in HNSCC MCTSs produced and cultured in Ultra-low Attachment Plates. HNSCC cell lines were seeded at 2,500 cells per well into 384-well ULA-plates and allowed to form MCTSs for 3 days before the culture media was exchanged, and over the next 3 days MCTSs were exposed to EdU for the indicated time periods and developed with the Click-iT® reagent according to the manufacturer's recommendations. MCTSs were fixed in 3.7% formaldehyde, and 4 × images were acquired on the IXM in the TL and FITC channels to capture the Click-iT EdU (green) fluorescent signal. (A) Six-day FaDu MCTS cultures that were exposed to EdU for 12, 24, or 48 h before fixation. (B) Six-day MCTS cultures prepared from each of the five representative HNSCC cell lines (UM-22B, OSC-19, FaDu, BICR56, and Cal33) that were exposed to EdU for 24 h before fixation. All scale bars represent 300 μm. EdU, 5-ethynyl-2′-deoxyuridine.
HNSCC MCTS Mitochondrial Mass and Membrane Potential
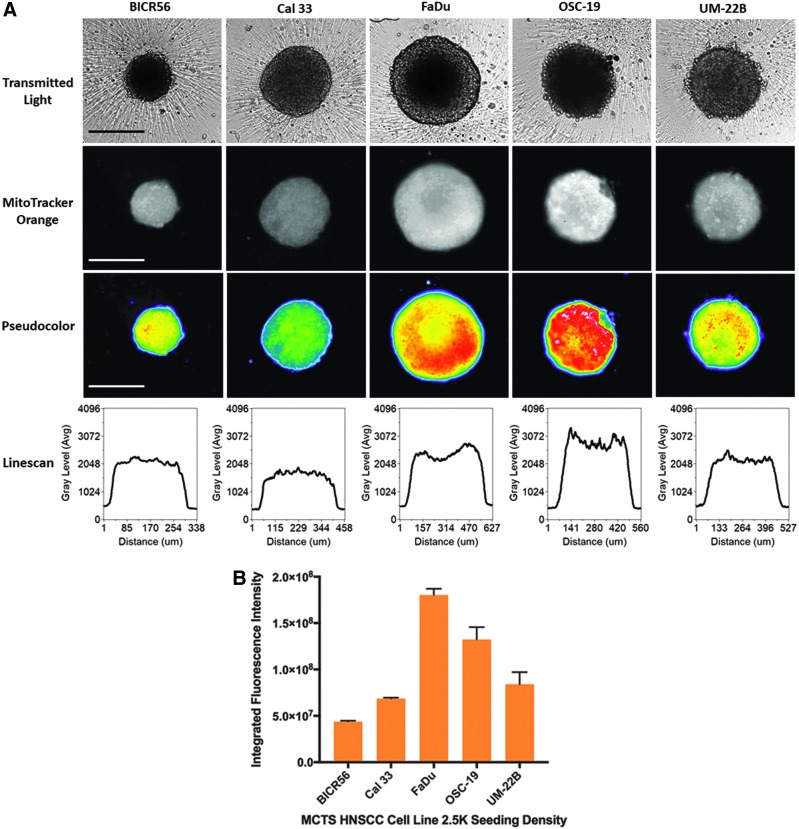
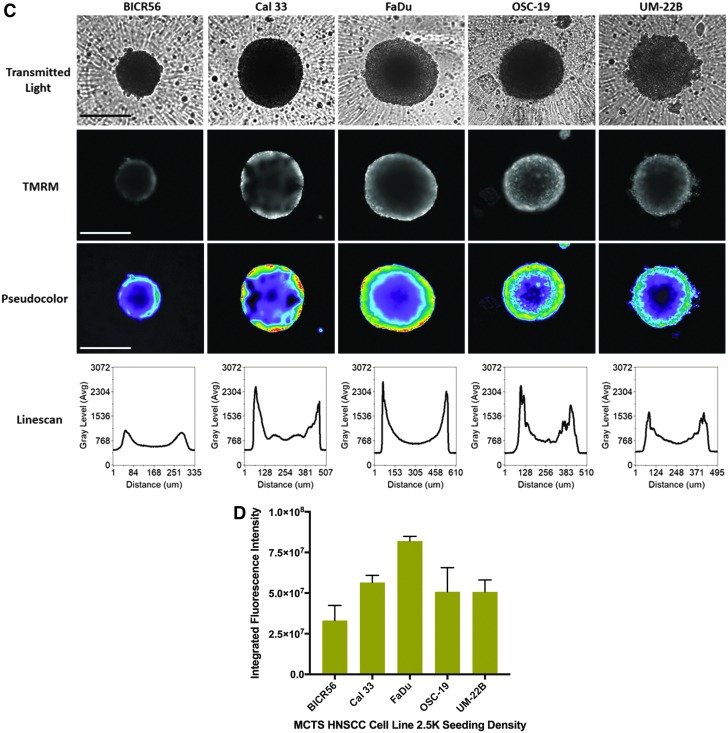
We used the MTO and TMRM dyes to determine the mass and membrane potential of active mitochondria in the cells of HNSCC MCTS cultures (Fig. 5).50,51 HNSCC cell lines were seeded into 384-well ULA-plates, and after 3 days in culture the culture media was exchanged and MCTS were exposed to MTO (Fig. 5A, B) or TMRM (Fig. 5C, D) for 1 h. The TRITC images, pseudo-color visualizations, and line-scan plots showed that the MTO fluorescence was distributed uniformly throughout formaldehyde fixed HNSCC MCTSs, although the fluorescent intensities were perhaps marginally higher in cells in the inner cores of MCTSs relative to the outer layers (Fig. 5A). These data indicate that cells in HNSCC MCTS cultures contain active mitochondria irrespective of their location within the MCTS. Using the MWCS image analysis module to quantify MCTS MTO MIFI showed that mitochondrial mass tracks with MCTS size; FaDu > OSC19 > UM22B > Cal33 > BICR56 (Fig. 5B). In contrast, TRITC images, pseudo-color visualizations, and line-scan plots of HNSCC MCTSs stained with the live cell TMRM mitochondrial potential dye exhibited a differential staining pattern, where cells in the outer layers of MCTSs exhibited higher intensities relative to cells in the inner cores, irrespective of MCTS size or HNSCC cell line (Fig. 5C). These data show that mitochondria in cells in the outer layers of the HNSCC MCTSs had higher membrane potentials than mitochondria in cells in the inner regions, indicating that mitochondria in cells in the outer layers were more functionally active. MCTS TMRM MIFI values among the five HNSCC cell lines did not however track with MCTS size; FaDu > OSC19 = UM22B = Cal33 > BICR56 (Fig. 5D).
Fig. 5.
Mitochondrial mass and membrane potential in HNSCC MCTSs produced in Ultra-low Attachment Plates. Two thousand and five hundred cells of the five representative HNSCC cell lines were seeded into 384-well ULA-plates, and after 3 days in culture the culture media was exchanged and MCTS were exposed to 500 nM MTO or 250 nM TMRM for 1 h. (A) HNSCC MCTSs that were stained with MTO were fixed in 3.7% formaldehyde, and 4 × images were acquired on the IXM in the TL and TRITC channels. Representative grayscale TL and MTO (TRITC) images are presented along with the corresponding pseudo-color pixel intensity visualizations and line-scan fluorescent intensity plots. In pseudo-color pixel intensity visualizations, the relative fluorescent intensities of the pixels in the images are indicated as distinct colors. The “hotter” and “brighter” colors (low to high, yellow, red, white) represent higher intensity signals, and cooler colors (low to high, purple, cyan, green) represent lower intensity signals. In line-scan fluorescence intensity plots the line scanning tool of the MetaXpress image analysis software to draw a line across the image and plot the fluorescent intensity values versus distance in μm across the image to provide an intensity profile graph. All scale bars represent 300 μm. (B) The MWCS image analysis module was used to create a whole MCTS mask and quantify MTO MIFI signals in the HNSCC MCTSs. HNSCC MCTS MTO MIFIs were measured in triplicate wells (n = 3) and are presented as the mean ± SD. Representative data from three independent experiments are presented. (C) 4 × images of HNSCC MCTSs that were stained with TMRM were acquired on the IXM in the TL and TRITC channels. Representative grayscale TL and TMRM (TRITC) images are presented along with the corresponding pseudo-color pixel intensity visualizations and line-scan fluorescent intensity plots. All scale bars represent 300 μm. (D) The MWCS image analysis module was used to create a whole MCTS mask and quantify TMRM MIFI signals in the HNSCC MCTSs. HNSCC MCTS TMRM MIFIs were measured in triplicate wells (n = 3) and are presented as the mean ± SD. Representative data from three independent experiments are presented. MTO, MitoTracker® Orange; TMRM, tetra-methyl-rhodamine methyl ester.
Doxorubicin Penetration and Cytotoxicity in HNSCC MCTS Cultures
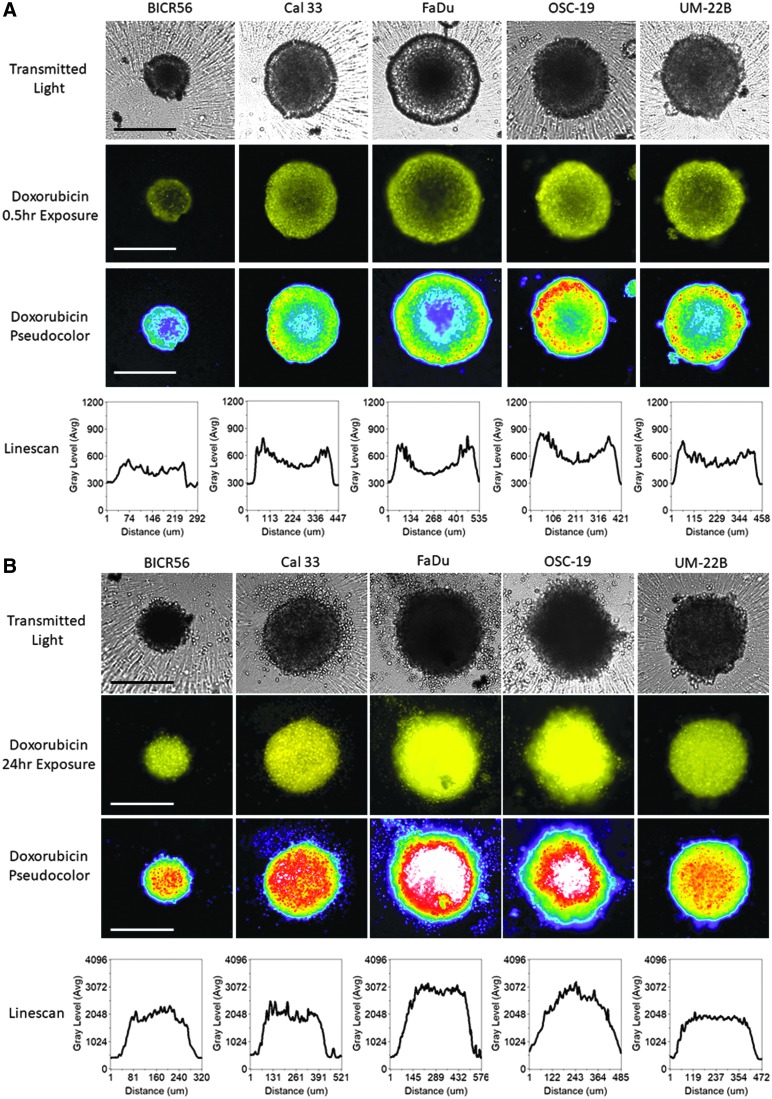
To investigate cancer drug uptake, penetration, and distribution in HNSCC MCTS, we seeded 2,500 cells of the HNSCC cell lines into 384-well ULA-plates and after 3 days the culture media was exchanged and MCTSs were exposed to 10 μM doxorubicin for 0.5 (Fig. 6A) and 24 h (Fig. 6B). As expected, the TRITC images, pseudo-color visualizations, and line-scan plots of all five HNSCC MCTSs exposed to doxorubicin for 0.5 h exhibited an apparent donut-like staining pattern, where cells in the outer edges of the MCTSs had higher doxorubicin staining intensities relative to cells in the inner cores (Fig. 6A). After 24 h of doxorubicin exposure, however, the TRITC images, pseudo-color visualizations, and line-scan plots showed that doxorubicin fluorescence was distributed throughout the HNSCC MCTSs and was perhaps slightly higher in cells in the inner cores of MCTSs relative to cells in the outer layers (Fig. 6A). A comparison of the TL images of HNSCC MCTS exposed to 10 μM doxorubicin for 0.5 (Fig. 6A) and 24 h (Fig. 6B) revealed that the longer drug exposure had a significant morphological impact producing less rounded MCTSs with uneven perimeters surrounded by a halo of single cells and generally darker inner cores.
Fig. 6.
Doxorubicin Drug Distribution in MCTSs formed in Ultra-low Attachment Plates. HNSCC cell lines were seeded at 2,500 cells per well into 384-well ULA-plates and after 3 days in culture the culture media was exchanged and MCTS were then exposed to 10 μM doxorubicin for either 0.5 h (A) or 24 h (B). HNSCC MCTSs that were exposed to 10 μM doxorubicin were then fixed in 3.7% formaldehyde, and 4 × images were acquired on the IXM in the TL and TRITC channels. Representative grayscale TL and doxorubicin (TRITC) images are presented along with the corresponding pseudo-color pixel intensity visualizations and line-scan fluorescent intensity plots. Representative data from three independent experiments are presented. All scale bars represent 300 μm.
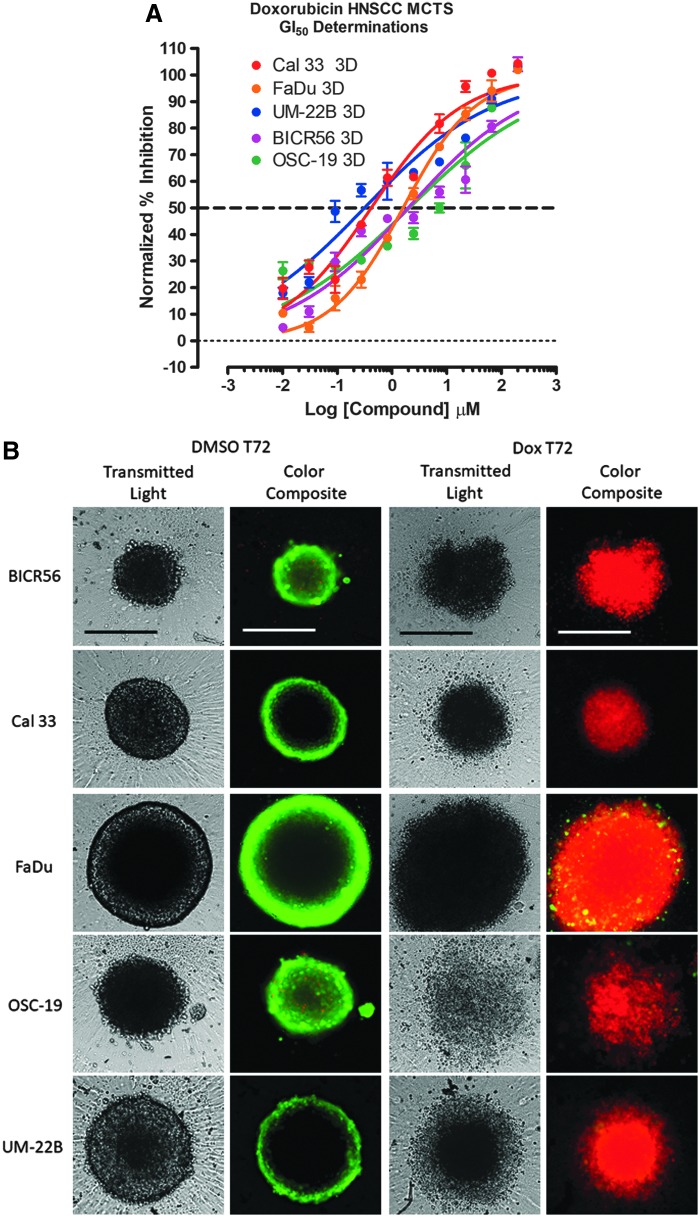
To investigate doxorubicin-induced cytotoxicity in HNSCC MCTS, we seeded 2,500 cells of the HNSCC cell lines into 384-well ULA-plates and after 3 days in culture the culture media was exchanged. The MCTSs were then exposed to 0.01–200 μM doxorubicin for 72 h before the addition of the CTB detection reagent and subsequent measurement of the RFU signals. To define the dynamic range of the 72 h compound exposure in HNSCC MCTS assays, we used 0.2% DMSO control wells to represent uninhibited growth (Max controls, n = 32) and 200 μM Doxorubicin +0.2% DMSO control wells to represent 100% GI (Min controls, n = 32), respectively. Doxorubicin inhibited the growth of all five HNSCC MCTS cultures in a concentration dependent manner with GI50s in the low- to sub-micromolar range; 0.31, 0.42, 1.59, 1.97, and 2.07 μM for the UM22B, Cal33, FaDu, BICR56, and OSC19 cell lines, respectively (Fig. 7A). To determine the effects of 72 h doxorubicin exposure on HNSCC MCTS morphology and viability, we acquired TL and CAM/EHD fluorescent images of doxorubicin treated HNSCC MCTSs (Fig. 7B). All five HNSCC MCTSs were totally disrupted after 72-h exposure to the top three concentrations of doxorubicin tested (200, 66.7, and 22.2 μM), and the images provided no additional information (not shown). After 72-h exposure to 7.4 μM doxorubicin, however, the TL and CAM/EHD images reveal profound differences compared to images acquired from DMSO control wells. In TL images of DMSO control wells, FaDu, Cal33, and UM22B cell lines formed condensed MCTSs with a smooth and even periphery, while OSC19 and BICR56 formed rounded MCTSs with uneven perimeters. Exposure to 7.4 μM doxorubicin for 72 h dramatically altered HNSCC MCTS morphologies: OSC19 MCTSs became irregular loose cell aggregates; BICR56 MCTSs became dark irregular cell aggregates with uneven outlines surrounded by single cells; Cal33 and UM22B MCTS became smaller and less rounded with dark inner cores and uneven perimeters surrounded by a halo of single cells; and FaDu MCTSs became bigger and darker with a rounded uneven perimeter. However, the CAM/EHD images revealed the most dramatic differences between doxorubicin treated and DMSO control wells. The strong CAM and almost nonexistent EHD fluorescence in the CAM/EHD composite images of the five HNSCC MCTSs from DMSO control wells indicated that most of the cells in the MCTSs were viable. In sharp contrast, the very weak CAM and strong EHD fluorescence in the CAM/EHD composite images of the five HNSCC MCTSs from wells exposed to 7.4 μM doxorubicin indicated that most of the cells in the MCTSs were dead. These data demonstrate that although different HNSCC cell lines seeded into ULA-plates produced MCTSs with distinct morphologies and growth phenotypes, these more physiologically relevant 3D tumor models can be readily adapted to measure compound mediated cytotoxicity.
Fig. 7.
Doxorubicin-induced cytotoxicity in MCTSs formed in Ultra-low Attachment Plates. HNSCC cell lines were seeded at 2,500 cells per well in 384-well ULA-plates, and after 3 days in culture the culture media was exchanged and then MCTSs were exposed to the indicated concentrations of doxorubicin for 72 h before the addition of the CTB detection reagent and subsequent measurement of the RFU signals. (A) The mean maximum (0.5% DMSO) and minimum (200 μM doxorubicin +0.5% DMSO) plate control CTB RFUs were used to normalize the RFU data from the compound treated wells as percent inhibition of growth, and the GI50 data were fit to a nonlinear sigmoidal log inhibitor concentration versus the normalized response variable slope model using the GraphPad Prism 6 software. The normalized mean ± SD (n = 3) growth inhibition data from triplicate wells for each compound concentration are presented. Representative experimental data from one of four independent experiments are shown. (B) To examine the effects of 72 h doxorubicin exposure on HNSCC MCTS morphology and viability, we acquired 4 × TL and CAM/EHD fluorescent images of HNSCC MCTSs exposed to 7.4 μM doxorubicin for 72 h. Grayscale TL images are presented along with color composite fluorescent images of live cell CAM and dead cell EHD staining depicted as green and red, respectively. Representative images from four independent experiments are presented. All scale bars represent 300 μm. DMSO, dimethyl sulfoxide; GI50, 50% growth inhibitory concentration.
Discussion
MCTS cultures represent more physiologically relevant in vitro cell tumor models that recapitulate the microenvironments and cell–cell or cell–ECM interactions which occur in solid tumors.32,34,40,41,52 We set out to characterize the morphologies, viability, and growth behaviors of MCTSs produced by 11 different HNSCC cell lines seeded into and cultured in ULA-plates over extended periods of time (Figs. 1 and 2). The 11 HNSCC cell lines form xenografts in immunodeficient mice, have similar genetic profiles to patient tumors, and have previously been used for HNSCC drug discovery.6,7,21 The Het-1A SV40 T-antigen immortalized human esophageal epithelial cell line does not grow as xenograft tumor in immunodeficient mice.46–49 We characterized the development of distinct microenvironments and the penetration and distribution of drugs in different regions of the MCTSs (Figs. 3–6) and investigated drug-induced cytotoxicity (Fig. 7). Our goal was to obtain a better understanding of how HNSCC MCTS cultures behaved so that we could apply these models in HTS campaigns designed to identify cancer drug leads that will be more translatable into efficacy in in vivo animal models and patients.
Nine of the 11 HNSCC cell lines seeded into 384-well ULA-plates formed condensed MCTSs with either a smooth or uneven periphery after 3 days. The UM22B and SCC9 HNSCC cell lines and Het-1A esophageal epithelial cell line formed cell aggregates with irregular outer margins. After several days in culture, however, the UM22B cell line coalesced into a compact MCTS with an even periphery. The HNSCC MCTS morphologies are consistent with our previous observations,22,29,34 and similar HNSCC MCTS morphologies were described in studies conducted in 96-well ULA-plates.41,53 In a study of six HNSCC cell lines, which included the Cal27 and Detroit 562 cell lines of the present study, four formed tight spheroids and two formed compact cell aggregates.41 In a study of 10 HNSCC cell lines, which included the Cal27 and FaDu cell lines of the present study, 6 formed tight compact spheroids, 3 formed intermediate spheroids, and 1 produced loose irregular cell aggregates.53 It has been reported that H-RAS-transformed fibroblast cell lines form MCTSs that grow when seeded into 96-well ULA-plates, whereas nontransformed fibroblasts do not.54 It was suggested that the ability to form MCTS in ULA-plates was similar to soft agar colony formation clonogenicity assays, where only transformed cells can form colonies. In our study, however, both the SCC9 HNSCC cell line and the Het-1A SV40 T-antigen immortalized human esophageal epithelial cell line formed irregularly shaped cell aggregates in ULA-plates that did not self-assemble into MCTSs. Some HNSCC MCTSs, most noticeably those formed by the FaDu, Cal33, Cal27, and PCI-13 cell lines, exhibited two distinct spheroid regions, a darker or denser inner core surrounded by lighter outer layers that were two to several layers of cells thick depending upon the cell line. The two distinct spheroid regions have been observed previously in MCTSs formed by the FaDu and Cal33 cell lines.22,53 In addition to morphology differences, we observed that HNSCC cell lines produced MCTSs of two different size ranges when they were seeded into ULA-plates at the same cell number, either smaller more compact MCTSs approximately 200–250 μm in diameter or larger MCTSs approximately 350–400 μm in diameter. HNSCC develops at multiple anatomic sites in different cell types within the head and neck region; paranasal sinuses, nasal cavity, oral cavity, pharynx, larynx, salivary gland, and thyroid.3,55–57 Histology, molecular characteristics, and clinical outcomes vary widely across different sites.3,55–57 In the present study, UMSCC1, SCC9, PCI-13, Cal33, Cal27, and BICR56 HNSCC cell lines were established from oral cavity tumors, the OSC-19 tumor originated in the tongue but the cell line was established from a metastatic site in the cervical lymph node, the FaDu and UM22B cell lines were established from hypopharynx tumors, the PCI-52 cell line was established from a larynx tumor, and the Detroit 562 tumor originated in the larynx but the cell line was established from a pleural effusion metastasis.7 HNSCC patient tumors have on average 130 coding mutations and 141 gene copy number alterations (gains/deletions) per tumor3,56,57 and exhibit 4 gene expression signature subtypes; basal, mesenchymal, atypical, and classical.58 It is likely that the complexity and heterogeneity of HNSCC contribute to the variety in MCTS sizes and morphologies that we observed.
We measured the changes in MCTS diameters and CTB RFUs over time in culture to assess the growth of HNSCC MCTS cultures in ULA-plates (Fig. 2B, C). MCTS diameters and CTB RFUs both increased linearly over time in culture throughout the 10- to 14-day culture period for FaDu, UM22B, and Cal33 MCTSs. In contrast, BICR56 MCTSs were static, and OSC19 MCTSs exhibited gradual declines for both indicators over time. Time course CAM/EHD images demonstrated that most of the cells in UM22B, Cal33, and BICR56 MCTSs were viable at all time points (Figs. 2A and 3A). FaDu MCTSs appeared to develop necrotic cores as they became progressively larger, and OSC19 MCTSs appeared to have a necrotic core at all time points. The presence of necrotic cores in OSC19 and FaDu MCTSs was confirmed by both the CAM/EHD images and the quantitative MIFI data presented in Figure 3. The necrotic core in FaDu MCTSs developed between day 4 and 6 in culture after the MCTSs reached ∼500 μm in diameter (Fig. 2B), and we speculate that diffusion was no longer able to support efficient nutrient and/or oxygen uptake and distribution to interior cells and/or to remove waste products. Several methods have been used to label actively proliferating cells in MCTS cultures, including Ki67 staining and the incorporation of either H3-thymidine or H3-bromo-deoxyuridine incorporation into DNA.34,39–41,59 Previous MCTS proliferation studies have shown that MCTS cultures exhibit differential zones of proliferation characterized by outer layers of proliferating cells surrounding inner layers of quiescent cells and inner cores that may become necrotic.34,39–41,59 Consistent with these observations, cells in the outer layers of the FaDu and UM22B MCTSs exhibited EdU incorporation (Fig. 4). However, none of the cells in Cal33, BICR56, and OSC19 MCTSs exhibited detectable levels of EdU incorporation, even with 48 h EdU exposure. Only HNSCC MCTSs that showed substantial increases in MCTS diameters and CTB RFUs with time in culture contained actively proliferating cells detectable by EdU incorporation. Despite the apparent viability of the cells in Cal33 and BICR56 MCTSs demonstrated by the CAM/EHD (live/dead) stains, their rates of proliferation were below the detection limits of EdU incorporation. OSC19 MCTSs exhibit a slow progressive death phenotype, and none of the cells in OSC19 MCTSs was positive for EdU incorporation. Based on Ki67 staining, the rates of proliferation of 8 of 9 HNSCC cell lines that formed MCTS cultures in 96-well ULA-plates decreased substantially compared to the corresponding 2D cultures, especially for those that formed tight spheroids.53 Only the FaDu cell line exhibited comparable Ki67 staining in 2D and 3D cultures.53
The rates of proliferation and growth of HNSCC cell lines in ULA-plate MCTS cultures are dramatically different from that in 2D cultures.6,21 When HNSCC cell lines are seeded at 1,000 cells per well into standard tissue culture treated 384-well microtiter plates, they proliferate exponentially throughout a 96-h culture period and typically undergo >2 but <3 doublings.6,21 Tumor cell lines adapted to growth in 2D proliferate faster than cells from primary tumors, exhibit altered drug response profiles, and are very sensitive to antiproliferative agents while under-representing the self- and population-renewing tumor stem cells that contribute to recurrence and metastasis.22–27 Dormant or quiescent tumor cells which have stopped replicating, or that proliferate slowly due to reduced nutrient and/or oxygen microenvironments, are resistant to molecules targeting cell proliferation mechanisms.60–62 The reduced proliferation and growth of HNSCC cell lines that we observed in MCTS culture conditions may more accurately align with tumor growth in vivo, which may be critical to screening for new solid tumor cancer drug leads that will translate better in in vivo animal models and in patients.
Images of CAM stained HNSCC MCTS cultures indicated that cells in the outer layers of MCTSs exhibited higher intensities relative to cells in the interior, irrespective of MCTS size or HNSCC cell line (Figs. 1, 2A, and 3A). The uniform distribution of fluorescence observed in HNSCC MCTSs prelabeled with CTO before MCTS formation (Supplementary Fig. S2) suggests that the gradient of CAM fluorescence was not due to an imaging artifact caused by inefficient excitation light penetration into MCTS cores and/or fluorescent emission light detection from these regions. Since the CAM reagent is metabolized by live cells into a fluorescent end product, the higher CAM signals in cells in the outer layers of HNSCC MCTSs may be due to their higher metabolic activity relative to cells in the inner cores. We cannot however exclude the possibility that poor CAM reagent permeability might have produced uneven reagent penetration and distribution within the MCTSs and that the apparent gradient of CAM staining was due to its preferential uptake, accumulation, and metabolism by cells in the outer layers of HNSCC MCTSs. Mitochondria are critical to cellular energy production, and we used the MTO and TMRM dyes to evaluate the mass and membrane potential of active mitochondria in the cells of HNSCC MCTS cultures (Fig. 5).50,51 The MTO data indicated that all of the cells in HNSCC MCTS cultures contain active mitochondria irrespective of their location within the MCTS (Fig. 5A) and that MCTS mitochondrial mass appears to track with MCTS size (Fig. 5B). The TMRM data indicated that the mitochondria in cells in the outer layers of HNSCC MCTSs had higher membrane potentials than mitochondria in cells in the inner regions, indicating that cells in the outer layers of MCTS have more functionally active mitochondria. Both the CAM and TMRM data indicated that cells in the outer layers of HNSCC MCTSs were more metabolically active than cells in the interior regions.
When MCTSs are initially exposed to doxorubicin, cells in the outer layers of HNSCC MCTSs exhibit higher doxorubicin fluorescent intensities than cells in the inner cores (Fig. 6A). After 24 h, however, doxorubicin alters the integrity and morphologies of MCTSs and becomes uniformly distributed throughout MCTSs (Fig. 6B). These data are consistent with our previous observations that Cal33 and FaDu MCTSs exhibited permeability barriers that resulted in uneven drug distribution and exposure gradients which coincided with enhanced resistance relative to monolayer cultures.22 Adhesion junctions between adjacent cells, cell–ECM contacts, and high tumor cell packing densities constitute drug permeability barriers that limit cancer drug penetration, distribution, and efficacy.60–65 Drug physiochemical properties also affect their distribution in tissues, and cells in solid tumors which are distal to blood vessels experience lower drug concentrations due to reduced drug access.62–64 Exposure to doxorubicin for 72 h inhibited the growth and viability of all five HNSCC MCTS cultures in a concentration dependent manner with GI50s in the low- to sub-micromolar range; 0.31, 0.42, 1.59, 1.97, and 2.07 μM for the UM22B, Cal33, FaDu, BICR56, and OSC19 MCTSs, respectively (Fig. 7A). The top three concentrations of doxorubicin totally disrupted the integrity and viability of all five HNSCC MCTSs. At 7.4 μM doxorubicin, HNSCC MCTS integrity and morphologies were profoundly altered, and the CAM/EHD images indicated that most of the cells were dead (Fig. 7B).
HNSCC MCTS cultures develop microenvironments, which result in differences in proliferation rates, metabolic activity, and mitochondrial functional activity between cells located in the outer layers of the MCTS and cells in the interior. Some HNSCC MCTS cultures have necrotic cores that gradually increase as MCTS size and viability decline, while others develop necrotic cores surrounded by viable layers of cells as MCTSs achieve a critical size threshold. HNSCC MCTS cultures also exhibit drug penetration and distribution gradients that coincide with enhanced resistance. Perhaps the most profound effect of culturing HNSCC cell lines in MCTS cultures was their dramatically altered and varied growth phenotypes. Instead of the exponential growth that are characteristic of 2D HNSCC GI assays, some MCTS cultures displayed linear growth rates, categorized as rapid, moderate, or slow; dormant MCTSs remained viable but did not grow; and some MCTSs exhibited death phenotypes that were either progressive and slow or rapid. The ability of MCTS cultures to develop microenvironments and to display a variety of different growth phenotypes provides in vitro models that are more closely aligned with solid tumors in vivo. We anticipate that the implementation of MCTS models to screen for new cancer drugs for solid tumors like HNSCC will produce leads that will translate better in in vivo animal models and patients.
Supplementary Material
Acknowledgments
The studies were supported, in part, by a Development Research Project award (P.A.J., Principal Investigator) from the HNC Spore P50 (Ferris and Grandis, CA097190) of the University of Pittsburgh Medical Center Hillman Cancer Center.
Abbreviations Used
- 2D
two-dimensional
- 3D
three-dimensional
- CAM
Calcein AM
- CTB
CellTiter-Blue®
- CTG
CellTiter Glo®
- CTO
CellTracker® Orange
- DMEM
Dulbecco's modified Eagle's medium
- DMEM/F12
Dulbecco's modified Eagle's medium/Ham's F12 50/50
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- EdU
5-ethynyl-2′-deoxyuridine
- EHD
Ethidium Homodimer
- ELWD
extra-long working distance dark medium
- FBS
fetal bovine serum
- FDA
The Federal Drug Administration
- GI
growth inhibition
- GI50
50% growth inhibitory concentration
- HNC
head and neck cancer
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- HTS
high throughput screening
- IXM
ImageXpress Micro
- MCTSs
multicellular tumor spheroids
- MIFI
mean integrated fluorescence intensity
- MTO
MitoTracker® Orange
- MWCS
Multiwavelength Cell Scoring
- P/S
penicillin/streptomycin
- PBS
phosphate-buffered saline
- RFUs
relative fluorescent units
- TL
transmitted light
- TMRM
tetra-methyl-rhodamine methyl ester
- ULA-plates
Ultra-Low Attachment microtiter plates
Disclosure Statement
No competing financial interests exist.
References
- 1. Brockstein B: Management of recurrent head and neck cancer: recent progress and future directions. Drugs 2011;71:1551–1559 [DOI] [PubMed] [Google Scholar]
- 2. Goerner M, Seiwert TY, Sudhoff H: Molecular targeted therapies in head and neck cancer—an update of recent developments. Head Neck Oncol 2010;2:8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stransky N, Egloff AM, Tward AD, et al. : The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramqvist T, Dalianis T: Oropharyngeal cancer epidemic and human papillomavirus. Emerg Infect Dis 2010;16:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cattley R, Radinsky RR: Cancer therapeutics: understanding the mechanism of action. Toxicol Pathol 2004;32:116–121 [DOI] [PubMed] [Google Scholar]
- 6. Johnston P, Sen M, Hua Y, et al. : High-content pSTAT3/1 imaging assays to screen for selective inhibitors of STAT3 pathway activation in head and neck cancer cell lines. Assay Drug Dev Technol 2014;12:55–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H, Wawrose JS, Gooding WE, et al. : Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res 2014;12:571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ocana A, Pandiella A, Siu LL, Tannock IF: Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol 2011;8:200–209 [DOI] [PubMed] [Google Scholar]
- 9. Denaro N, Russi EG, Adamo V, Colantonio I, Merlano MC: Postoperative therapy in head and neck cancer: state of the art, risk subset, prognosis and unsolved questions. Oncology 2011;81:21–29 [DOI] [PubMed] [Google Scholar]
- 10. Perez-Ordonez B, Beauchemin M, Jordan RCK: Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol 2006;59:445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Addeo R, Caraglia M, Iuliano G: Pembrolizumab: the value of PDL1 biomarker in head and neck cancer. Expert Opin Biol Ther 2016;16:1075–1078 [DOI] [PubMed] [Google Scholar]
- 12. Chow L, Haddad R, Gupta S, et al. : Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol 2016;34:3838–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seiwert T, Burtness B, Mehra R, et al. : Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–965 [DOI] [PubMed] [Google Scholar]
- 14. Toniatti C, Jones P, Graham H, Pagliara B, Draetta G: Oncology drug discovery: planning a turnaround. Cancer Discov 2014;4:397–404 [DOI] [PubMed] [Google Scholar]
- 15. Hait W: Anticancer Drug Development: the grand challenges. Nat Rev Drug Discov 2010;9:253–254 [DOI] [PubMed] [Google Scholar]
- 16. Hutchinson L, Kirk R: High drug attrition rates—where are we going wrong? Nat Rev Clin Oncol 2011;8:189–190 [DOI] [PubMed] [Google Scholar]
- 17. Kamb A, Wee S, Lengauer C: Why is cancer drug discovery so difficult? Nat Rev Drug Discov 2007;6:115–120 [DOI] [PubMed] [Google Scholar]
- 18. Barretina J, Caponigro G, Stransky N, et al. : The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim N, He N, Yoon S: Cell line modeling for systems medicine in cancers. Int J Oncol 2014;44:371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shoemaker R: The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–823 [DOI] [PubMed] [Google Scholar]
- 21. Johnston P, Sen M, Hua Y, et al. : HCS campaign to identify selective inhibitors of IL-6-induced STAT3 pathway activation in head and neck cancer cell lines. Assay Drug Dev Technol 2015;13:356–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shan F, Close DA, Camarco DP, Johnston PA: High-content screening comparison of cancer drug accumulation and distribution in two-dimensional and three-dimensional culture models of head and neck cancer. Assay Drug Dev Technol 2018;16:27–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baker B, Chen CS: Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 2012;125:3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ekert J, Johnson K, Strake B, et al. : Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro—implication for drug development. PLoS One 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA: Spheroid-based drug screen: considerations and practical approach. Nat Protoc 2009;4:309–324 [DOI] [PubMed] [Google Scholar]
- 26. Härmä V, Virtanen J, Mäkelä R, et al. : A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hongisto V, Jernström S, Fey V, et al. : High-throughput 3D screening reveals differences in drug sensitivities between culture models of JIMT1 breast cancer cells. PLoS One 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abbot A: Biology's new dimension. Nature 2003;424:870–872 [DOI] [PubMed] [Google Scholar]
- 29. Close DA, Camarco DP, Shan F, Kochanek SJ, Johnston PA: The generation of three-dimensional head and neck cancer models for drug discovery in 384-well ultra-low attachment microplates. Methods Mol Biol 2018;1683:355–369 [DOI] [PubMed] [Google Scholar]
- 30. Fang Y, Eglen RM: Three-dimensional cell cultures in drug discovery and development. SLAS Discov 2017;22:456–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA: Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol 2010;148:3–15 [DOI] [PubMed] [Google Scholar]
- 32. Lovitt C, Shelper TB, Avery VM: Advanced cell culture techniques for cancer drug discovery. Biology 2014;3:345–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pampaloni F, Reynaud EG, Stelzer EH: The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007;8:839–845 [DOI] [PubMed] [Google Scholar]
- 34. Sant S, Johnston PA: The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 2017;23:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selby M, Delosh R, Laudeman J, et al. : 3D models of the NCI60 cell lines for screening oncology compounds. SLAS Discov 2017;22:473–483 [DOI] [PubMed] [Google Scholar]
- 36. Howes A, Richardson RD, Finlay D, Vuori K: 3-Dimensional culture systems for anti-cancer compound profiling and high-throughput screening reveal increases in EGFR inhibitor-mediated cytotoxicity compared to monolayer culture systems. PLoS One 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lovitt C, Shelper TB, Avery VM: Miniaturized three-dimensional cancer model for drug evaluation. Assay Drug Dev Technol 2013;11:435–448 [DOI] [PubMed] [Google Scholar]
- 38. Madoux F, Tanner A, Vessels M, et al. : A 1536-well 3D viability assay to assess the cytotoxic effect of drugs on spheroids. SLAS Discov 2017;22:516–524 [DOI] [PubMed] [Google Scholar]
- 39. Onozato Y, Kaida A, Harada H, Miura M: Radiosensitivity of quiescent and proliferating cells grown as multicellular tumor spheroids. Cancer Sci 2017;108:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryan S, Baird AM, Vaz G, et al. : Drug discovery approaches utilizing three-dimensional cell culture. Assay Drug Dev Technol 2016;14 19–28 [DOI] [PubMed] [Google Scholar]
- 41. Vinci M, Gowan S, Boxall F, et al. : Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 2012;10:29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wenzel C, Riefke B, Gründemann S, et al. : 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res 2014;323:131–143 [DOI] [PubMed] [Google Scholar]
- 43. Foty R: A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp 2011;51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C, Tang Z, Zhao Y, Yao R, Li L, Sun W: Three-dimensional in vitro cancer models: a short review. Biofabrication 2014;6. [DOI] [PubMed] [Google Scholar]
- 45. Singh M, Close DA, Mukundan S, Johnston PA, Sant S: Production of uniform 3D microtumors in hydrogel microwell arrays for measurement of viability, morphology, and signaling pathway activation. Assay Drug Dev Technol 2015;13:570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ahrens T, Timme S, Hoeppner J, et al. : Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and azacytidine. Epigenetics 2015;10:431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahrens T, Timme S, Ostendorp J, et al. : Response of esophageal cancer cells to epigenetic inhibitors is mediated via altered thioredoxin activity. Lab Invest 2015;96:307–316 [DOI] [PubMed] [Google Scholar]
- 48. Fichter C, Gudernatsch V, Przypadlo CM, et al. : ErbB targeting inhibitors repress cell migration of esophageal squamous cell carcinoma and adenocarcinoma cells by distinct signaling pathways. J Mol Med (Berl) 2014;92:1209–1223 [DOI] [PubMed] [Google Scholar]
- 49. Stoner G, Kaighn ME, Reddel RR, et al. : Establishment and characterization of SV40 T-antigen immortalized human esophageal epithelial cells. Cancer Res 1991;51:365–371 [PubMed] [Google Scholar]
- 50. Kholmukhamedov A, Schwartz JM, Lemasters JJ: MitoTracker probes and mitochondrial membrane potential. Shock 2013;39:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scaduto RJ, Grotyohann LW: Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J 1999;76:469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Das V, Bruzzese F, Konecny P, Iannelli F, Budillon A, Hajduch M: Pathophysiologically relevant in vitro tumor models for drug screening. Drug Discov Today 2015;20:848–855 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt M, Scholz CJ, Polednik C, Roller J: Spheroid-based 3-dimensional culture models: gene expression and functionality in head and neck cancer. Oncol Rep 2016;35:2431–2440 [DOI] [PubMed] [Google Scholar]
- 54. Rotem A, Janzer A, Izar B, et al. : Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc Natl Acad Sci U S A 2015;112:5708–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kang H, Kiess A, Chung CH: Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol 2015;12:11–26 [DOI] [PubMed] [Google Scholar]
- 56. Cancer Genome Atlas Network: Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun W, Califano JA: Sequencing the head and neck cancer genome: implications for therapy. Ann N Y Acad Sci 2014;1333:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walter V, Yin X, Wilkerson MD, et al. : Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PloS One 2013;8:e56823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hagemann J, Jacobi C, Hahn M, et al. : Spheroid-based 3D cell cultures enable personalized therapy testing and drug discovery in head and neck cancer. Anticancer Res 2017;37:2201–2210 [DOI] [PubMed] [Google Scholar]
- 60. Kerr D, Kaye SB: Aspects of cytotoxic drug penetration, with particular reference to anthracyclines. Cancer Chemother Pharmacol 1987;19:1–5 [DOI] [PubMed] [Google Scholar]
- 61. Minchinton A, Tannock IF: Drug penetration in solid tumours. Nat Rev Cancer 2006;6:583–592 [DOI] [PubMed] [Google Scholar]
- 62. Tannock I, Lee CM, Tunggal JK, Cowan DS, Egorin MJ: Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res 2002;8:878–884 [PubMed] [Google Scholar]
- 63. Grantab R, Sivananthan S, Tannock IF: The penetration of anticancer drugs through tumor tissue as a function of cellular adhesion and packing density of tumor cells. Cancer Res 2006;66:1033–1039 [DOI] [PubMed] [Google Scholar]
- 64. Grantab R, Tannock IF: Penetration of anticancer drugs through tumour tissue as a function of cellular packing density and interstitial fluid pressure and its modification by bortezomib. BMC Cancer 2012;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tredan O, Galmarini CM, Patel K, Tannock IF: Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst 2007;99:1441–1454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.