Abstract
Myocardial infarction occurs every 36 s or nearly 1 million times in the United States. The treatment of acute myocardial infarction (AMI) has been revolutionized with coronary reperfusion ensuring over 96% in-hospital survival. There has, however, been a paucity in technological advancement in the field of acute coronary syndrome, with nearly 30% of individuals progressing toward heart failure after AMI. This has engendered a pandemic of ischemic heart failure worldwide, mandating the development of off-the-shelf regenerative interventions, including gene-encoded therapies, capable to acutely target the injured myocardium. However, the main challenge in realizing gene-encoded therapy for AMI has been the inadequate induction of gene expression following intracoronary delivery. To address this challenge, we, in this study, report the use of synthetic modified messenger RNA, engineered to reduce lag time. Termed M3RNA (microencapsulated modified messenger RNA), this platform achieved expeditious induction of protein expression in cell lines (HEK293, human dermal and cardiac fibroblasts) and primary cardiomyocytes. Expression was documented as early as 2–4 h and lasted up to 7 days without impact on electromechanical coupling, as tracked by patch clamp electrophysiology and calcium imaging in transfected cardiomyocytes. In vivo, firefly luciferase (FLuc) and mCherry M3RNA myocardial injections in mice using ∼100 nm nanoparticles yielded targeted and temporally restricted expression of FLuc protein within 2 h, and sustained for 72 h as assessed by Xenogen and mCherry expression using immunofluorescence. In a porcine model of myocardial infarction, protein expression targeted to the area of injury was demonstrated following intracoronary delivery of alginate carrying M3RNA encoding mCherry. M3RNA thus enables rapid protein expression in primary cardiomyocytes and targeted expression in mouse and porcine hearts. This novel technology, capable of inducing rapid simultaneous protein expression, offers a platform to achieve targeted gene-based therapies in the setting of AMI.
Impact Statement
The M3RNA (microencapsulated modified messenger RNA) platform is an approach to deliver messenger RNA (mRNA) in vivo, achieving a nonintegrating and viral-free approach to gene therapy. This technology was, in this study, tested for its utility in the myocardium, providing a unique avenue for targeted gene delivery into the freshly infarcted myocardial tissue. This study provides the evidentiary basis for the use of M3RNA in the heart through depiction of its performance in cultured cells, healthy rodent myocardium, and acutely injured porcine hearts. By testing the technology in large animal models of infarction, compatibility of M3RNA with current coronary intervention procedures was verified.
Keywords: M3RNA, myocardial infarction, gene therapy, targeted nucleotide delivery
Introduction
Cardiovascular diseases, exemplified by acute myocardial infarction (AMI), remain a leading cause of morbidity and mortality.1–5 Biologics-based therapies have been proposed as a means to expand therapies available to secure cardiac protection and ensure regeneration. A focal point of investigation has been viral vectors or oligonucleotide-mediated gene therapy.6–9 Viral gene delivery has been challenged due to serious safety concerns, including risk of mutation and triggering of the innate immune response.10 Lentiviral vectors have the capability to infect nondividing cells within the myocardium leading to long-term expression, but are limited due to integration into the host genome and poor transduction efficiency.11 Conversely, plasmid DNA, considered safer, is hampered by lower transfection efficiency due to inefficient nuclear translocation and risk of inflammation at the injection site. Moreover, modification of plasmid DNA using a strong constitutive promotor can increase expression, but harbors risk of integration inducing unexpected modifications.10,12
Small interfering RNA (siRNA) due to their specificity in downregulating proteins have been used as therapeutic targets in cardiovascular diseases, for example, PCKS9 inhibition to ameliorate hypercholesterolemia13 and silencing CCR2, a chemokine receptor-improved MI recovery.14 MicroRNAs (miRNAs), just as siRNAs, are endogenously produced by noncoding short nucleotides that inhibit protein expression and play a critical role in maintaining cardiac function, for example, cardiomyocyte-specific deletion of dgcr8 caused dilated cardiomyopathy and heart failure in mice heart.15 Although signals of benefit have been reported at preclinical stage, gene delivery technology has not been broadly realized in the setting of AMI.
The main limitation centers on a lack of ready-to-use technologies that can be delivered acutely at the time of percutaneous coronary intervention (PCI), with the capacity to target the site of injury and achieve rapid induction of desired proteins.16,17 To this end, the use of mRNA to generate therapeutic proteins has recently emerged potentially, offering advantages over viral or DNA-based therapeutics.18–20 Implementation of an mRNA-based approach would create an effective tool to induce protein expression in the setting of AMI.
RNA administration offers several advantages over DNA, protein, or viral technologies as the protein expression using mRNA is rapid, as its translation into proteins does not require nuclear transfer.18,20–25 In recent years, mRNA constructs have become more prevalent in therapeutic applications as they are easier to design and their modifications such as 5′ mRNA anti-reverse cap analogues (ARCA) and poly (A) tails during their synthesis significantly promote and prolong efficient translation of foreign mRNA within cells with minimal immune response.26 However, a major hurdle in the use of therapeutic mRNA is delivery efficiency and intact functionality needed to induce protein expression within cells and tissues of interest.
In this study, we test M3RNA to deliver and achieve protein expression in vitro and in vivo. We, in this study, provide data establishing this off-the-shelf gene delivery platform to rapidly induce protein expression for a defined time horizon within synchronously beating nondividing cardiomyocytes, in intact heart tissue and ultimately within acutely infarcted tissue.
Materials and Methods
Ethical consideration
This study followed the NIH guidelines and received IACUC and Biosafety Committee approval.
Cell lines and primary cell cultures
Cell lines used were human dermal fibroblasts (HDF; passage number P25–P29), human cardiac fibroblasts (HCF; Passage number P10–P14), and HEK293 cells (Passage number P38–P42) purchased from ATCC. HDF were isolated from skin biopsy of a 46-year-old male. Other than Figure 1, the cells utilized are from primary cardiomyocyte cultures, which were freshly isolated and were never passaged. Cells were maintained and passaged in DMEM (with Glucose), 10% fetal bovine serum, 1% Pen/Strep, and 1% glutamine. Initial plating density of cell lines was 200,000 human embryonic kidney (HEK) cells and 350,000 HDF and HCF cells/well in six-well plates. All cell lines were checked periodically for mycoplasma contamination. Time-pregnant rats were purchased from Charles River and rat cardiomyocytes were obtained from 19-day-old embryos, and cardiomyocytes were isolated according to manufacturer's instructions using Pierce neonatal primary cardiomyocyte isolation kit (ThermoFisher Scientific, Waltham, MA).
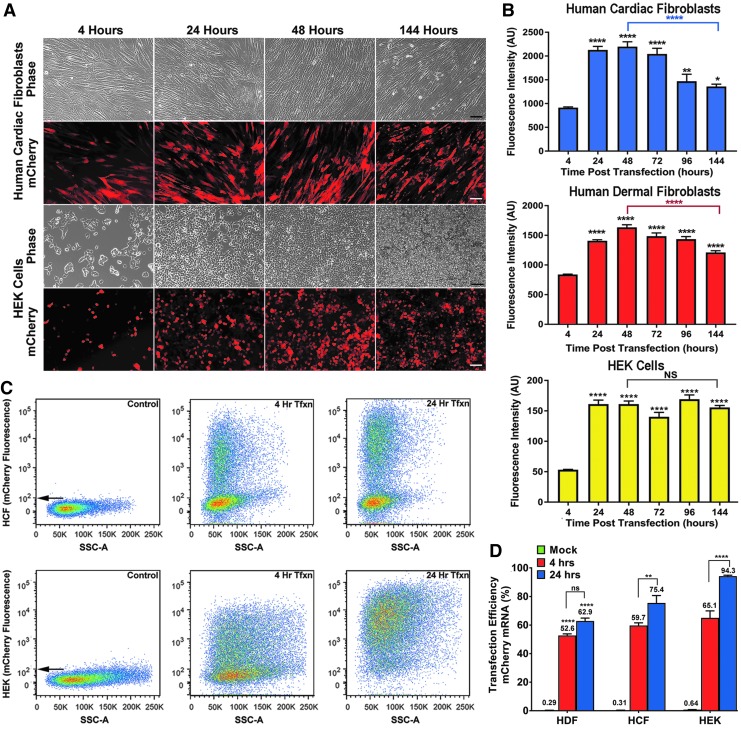
FIG. 1.
M3RNA-mCherry expression in multiple cell lines. (A) Representative fluorescence and phase contrast images of HCF and HEK cells upon mCherry mRNA transfection at indicated time periods. Scale bar = 100 μm. (B) Fluorescence of mCherry protein expression was quantified in HDF, HCF, and HEK cells. Plots indicate mean ± SEM of average fluorescence intensity (arbitrary units) at indicated time points (n = 3). ANOVA using GraphPad Prism with Tukey's multiple comparison test revealed significant differences between 48 and 144 h in HCF and HDF (****p < 0.0001), and significant expression at all time points in all three cell lines when compared to 4 h (****p < 0.0001, **p < 0.01, and *p < 0.05). (C) Representative flow cytometry plots of HCF and HEK cells upon M3RNA-mcherry transfection. (D) Percent transfection efficiency of sorted HDF, HCF, and HEK cells was using mock-transfected cells as control and transfected cells at 4 and 24 h. Results are plotted as mean ± SEM (n = 3) of three different cell lines. ANOVA using Dunnett's multiple comparison test revealed significant differences between the 4- and 24-h transfection with M3RNA-mCherry in HCF and HEK cells (**p < 0.01, ****p < 0.0001), and significant expression levels at 4 and 24 h transfection in all three cell lines when compared to mock transfected cells (****p < 0.0001). ANOVA, analysis of variance; HCF, human cardiac fibroblasts; HDF, human dermal fibroblasts; HEK, human embryonic kidney cells; M3RNA, microencapsulated modified messenger RNA; NS, not significant.
Antibodies, mRNAs, and transfections
Antibodies used are anti-mCherry (Rat IgG2a Monoclonal, 1:1000; ThermoFisher Scientific), anti-Cardiac Troponin T (Mouse IgG1 Monoclonal, 1:200; ThermoFisher Scientific), and anti-FLuc (Goat Polyclonal; 1:250; Novus Biologicals, Littleton, CO). Enhanced green fluorescent protein (EGFP), mCherry, and firefly luciferase (FLuc) mRNAs (Trilink Biotechnologies; San Diego, CA) featured modifications such as ARCA cap, polyadenylated tail, and modified nucleotides (5-methyl cytidine and pseudouridine) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tea). In vitro transfection studies for all the cell lines were carried out at ∼60–65% confluent cells using Lipofectamine MessengerMAX transfection reagent (ThermoFisher Scientific). We used 2.5 μg each of indicated mRNAs/well in six-well dishes for single transfection or co-transfections. For mice studies, we used 12 μg each of indicated mRNA for intracardiac injections in mice and 250 μg mCherry mRNA/pig for porcine studies.
Flow cytometry
Transfection efficiency was determined using FACS CantoX. Briefly, cells were mock or mCherry-mRNA transfected, trypsinized, and collected at 4 and 24 h (1 × 106 cells/mL) in 4% formaldehyde, in clear polystyrene tubes fitted with a cell filter. Tubes were then introduced into the FACS CantoX for analysis.
Calcium imaging
Calcium transients in cardiomyocytes were visualized using Cal520AM.27 Briefly, cardiomyocytes were transfected with mCherry mRNA overnight and were assessed if the cardiomyocytes were beating posttransfection under the microscope. On the following day, cells were loaded with Cal520AM (5 mM) 1:1 with powerload (Invitrogen, Carlsbad, CA) at the final concentration of 10 μM in Tyrode buffer (in mM) 1.33 CaCl2, 1 MgCl2, 5.4 KCl, 135 NaCl, 0.33 NaH2PO4, 5 glucose, and 5 HEPES. Cells were incubated for 30 min in an incubator, washed, and further incubated for 15 min to allow complete de-esterification of Cal520AM. Complete medium was added to cells and imaging was performed on Zeiss upright LSM5 live confocal microscope using 20 × objective (NA 0.8) in a 37°C humidified chamber with 5% CO2. Transfected and nontransfected cells in the same area were identified in the mCherry 543 nm excitation. Ca2+ transients in rat primary cardiomyocytes were collected at 488 nm excitation. Two hundred fifty single image frames were collected at 10 fps and the data were analyzed by measuring the emitted fluorescence from regions of interest (ROI) over single cardiomyocytes using Zen software and exported to excel, and the graphs were created to show Ca2+ transients.
Patch-clamp recording in primary cardiomyocytes
Patch clamp recording was performed with the modification of protocol.28,29 Neonatal rat primary cardiomyocytes were transfected with mCherry-modified mRNA using the whole-cell configuration of the patch-clamp technique in the voltage-clamp mode. Patch electrodes, with 5–7 MΩ resistance, were filled with (in mM) 120 KCl, 1 MgCl2, 5 EGTA, and 10 HEPES with 5 mM of ATP 9 (pH 7.3), and cells were superfused with (in mM) 136.5 NaCl, 5.4 KCl, 1 MgCl2, 1.8 CaCl2, and 5.5 HEPES plus glucose 1 g/L (pH 7.3). Membrane currents were measured using an Axopatch 200B amplifier (Molecular Devices). Cellular membrane resistance and cell capacitance were defined online based on analysis of capacitive transient currents. Series resistance (15–20 MΩ), was compensated by 50–60%, and along with uncompensated cell capacitances were continuously monitored throughout experiments. Current density was obtained by normalizing measured currents to cell capacitance. Protocol of stimulation, determination of cell parameters, and data acquisition were performed using the custom BioQuest software.28–30 Experiments were performed at 33°C ± 1.8°C.
Image analysis
Imaging of cell lines was performed using either upright Zeiss Axioplan epifluorescence widefield microscope (10 × objective, NA 0.3) or LSM780 confocal microscope (40 × water objective, NA 1.2). Data for quantitation of fluorescence intensity were then analyzed by importing the figures into Tiff format and analyzed using Image J. Average fluorescence intensity for the whole image was quantified and plotted.31,32
In vivo delivery of FLuc, EGFP, and mCherry-modified mRNA
In vivo delivery of modified mRNAs were carried out in FVB/NJ mice (18–22 g, 6–8 weeks of age) Jackson laboratory using modified protocol.33 Under anesthesia, the heart was exposed and indicated M3RNA at 12.5 μg/mRNA/mice (as indicated) was injected in the myocardium of left ventricle. Animals were then imaged or processed for immunohistochemistry at indicated times. We used total of 20 animals for M3RNA injections and 10 animals for controls in these experiments.
Injectable alginate M3RNA preparation
Calcium crosslinked alginate solution was prepared by mixing 1 mL of 2% alginate (FMC Corporation, Philadelphia, PA) with 0.5 mL of 0.6% Ca gluconate (Sigma), and 0.5 mL of water was mixed to yield 2 mL of alginate solution. Five hundred microliters of encapsulated mCherry mRNA (250 μg/pig) was prepared using nanoparticle in vivo transfection reagent (Altogen Biosystems, Las Vegas, NV) according to the manufacturer's instructions. Solutions were mixed together and injected intracoronary in porcine heart as described below.
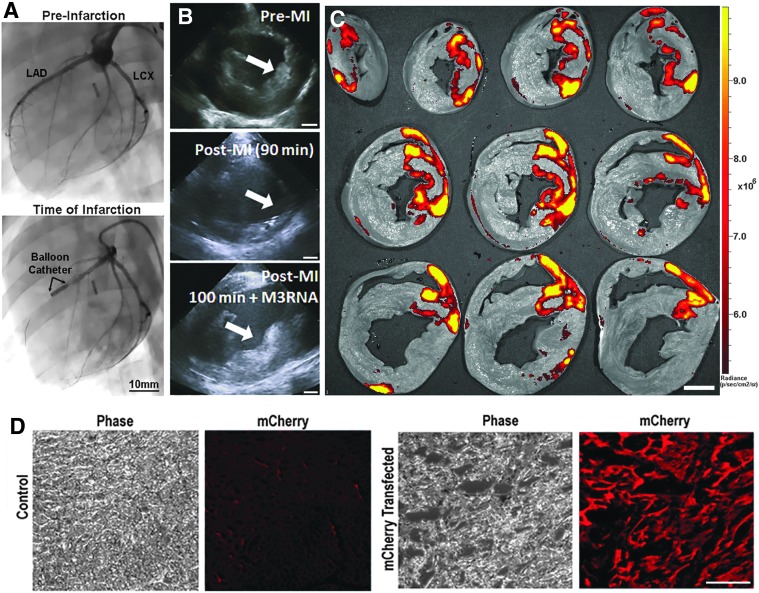
M3RNA expression in porcine myocardial infarction
Four Yorkshire pigs underwent myocardial infarction using a 90-min balloon occlusion of left anterior descending coronary artery. An intracardiac echocardiography (ICE) probe was placed in the right atrium for real-time LV monitoring. Using an AR-2 style coronary catheter, the left main artery of the pig was accessed and visualized by fluoroscopy with instilment of Omnipaque. A 0.014” balanced middleweight coronary wire was advanced into the distal left anterior descending coronary artery (LAD). Utilizing stored guiding angiographic imaging, a 2.5–3 mm balloon was advanced to be positioned across the second diagonal vessel of the LAD. The balloon was inflated to occlude the LAD for 90 min followed by reperfusion. Ischemic damage was monitored by ICE as well as continuous electrocardiography telemetry. Following reperfusion, a perfusion catheter was placed at the location of the balloon. Encapsulated mRNA combined with an alginate solution was introduced into the LAD over a 5-min period and infarct zone-targeted gene delivery was documented at day 3.
Statistics
Data are expressed as mean ± SEM. Statistical significance was determined by GraphPad Prism 7 using one-way or two-way analysis of variance with multiple comparisons. p-Values less than 0.05 were taken as a statistically significant difference. The “n” values refer to the number of times experiments are repeated or the number of animals.
Results
M3RNA transfection in multiple cell lines
M3RNA was transfected (as indicated) in HDF, HCF, and HEK293 cells and fluorescent protein expression was imaged live in a 37°C humidified chamber with 5% CO2. mCherry protein expression was detected at 2 h, but reliable and quantifiable expression was observed at 4 h. Fluorescent images of HCF and HEK293 cells (Fig. 1A) demonstrated rapid mCherry protein expression that sustained for 6 days. Simultaneous delivery of M3RNAs encoding mCherry and EGFP resulted in co-expression (Supplementary Fig. S2). Quantification of fluorescence intensity within three different cell lines (>10 fields of cells/time period/cell line) noted an increase in intensity over the initial 24–48 h, followed by a steady decline in two of three cell lines (Fig. 1B). Since protein expression peaked at 24 h, we then evaluated transfection efficiency at 4 and 24 h using flow cytometry upon mCherry M3RNA transfection. Scatter plots of fluorescence intensity on the x-axis and sideward scattering signal on the y-axis revealed a consistent bimodal population following transfection (Fig. 1C), with the transition revealing the number of transfected cells seen at 4 and 24 h. Transfection efficiency was quantified and compared to mock-transfected cells (Fig. 1D). Note the high transfection efficiency at the 24-h time point, especially in the HEK population.
M3RNA transfection in rat neonatal primary cardiomyocytes
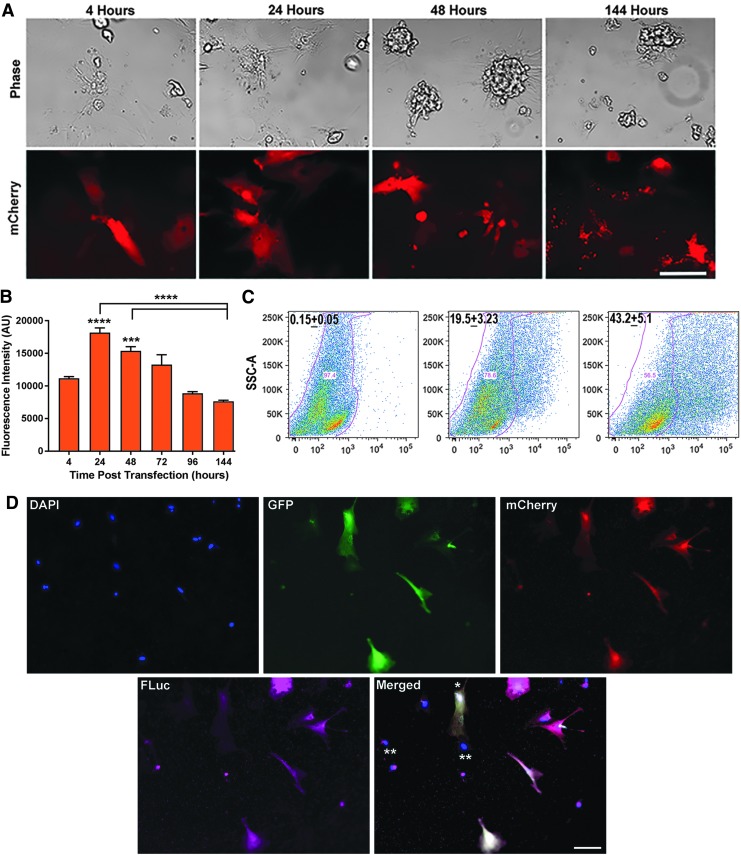
We then carried out transfection in hard-to-transfect primary cardiomyocytes. Cardiomyocyte-enriched cultures, following documentation of a synchronous beating pattern, were transfected with mCherry M3RNA. Fluorescence images at 4 h up to 6 days were acquired. Representative images showed rapid and sustained protein expression within primary cardiomyocytes (Fig. 2A). Quantification of the fluorescence intensity revealed maximum expression at 24 h, declining in linear manner for 6 days (Fig. 2B). Significant transfection efficiency was seen at 4 h (∼20%) and 24 h (43%) using flow cytometry from two independent experiments (Fig. 2C). Multigene transfection showed simultaneous expression of three proteins (EGFP, mCherry, and FLuc) within the same cardiomyocytes (Fig. 2D).
FIG. 2.
M3RNA-mCherry expression in neonatal rat primary cardiomyocytes. (A) Representative phase contrast and fluorescence images exhibiting rapid and declining mCherry expression at indicated time points within the rat primary neonatal cardiomyocytes. Scale bar = 100 μm. (B) Quantitation of mCherry protein expression in cardiomyocytes upon transfection (n = 3 with >10 images/time point) plotted as mean ± SEM average fluorescence intensity. ANOVA using GraphPad Prism with Tukey's multiple comparison test revealed significant differences between 24, 48, and 144 h (****p < 0.0001), and significant expression levels at 24 and 48 h when compared to 4-h time point (***p < 0.001, ****p < 0.001). (C) Representative flow cytometry plots of cardiomyocytes at the indicated time points posttransfection with mCherry mRNA using mock-transfected cells as control. Transfection efficiency was calculated from sets of three different experiments (n = 3). (D) Primary cardiomyocytes were co-transfected with mCherry, GFP, and FLuc mRNA and the representative image exhibiting triple transfection in the same cells is shown. Scale bar = 100 μm. * indicates the cardiac fibroblast transfection with three genes and ** represents the nontransfected cells in the same field. FLuc, firefly luciferase; GFP, green fluorescent protein.
Transfection does not alter cardiomyocyte structure and function
To test that transfection of primary cardiomyocytes does not alter their structural integrity, we transfected cardiomyocytes with mCherry M3RNA and stained with cardiac-specific troponin antibody and SiR-Actin staining. Actin staining was used to differentiate cardiomyocytes from fibroblasts (#). No significant differences between the cardiomyocyte-specific troponin staining were identified in the transfected versus nontransfected cells (##) (Fig. 3), indicating intact structural integrity of cardiomyocytes.
FIG. 3.
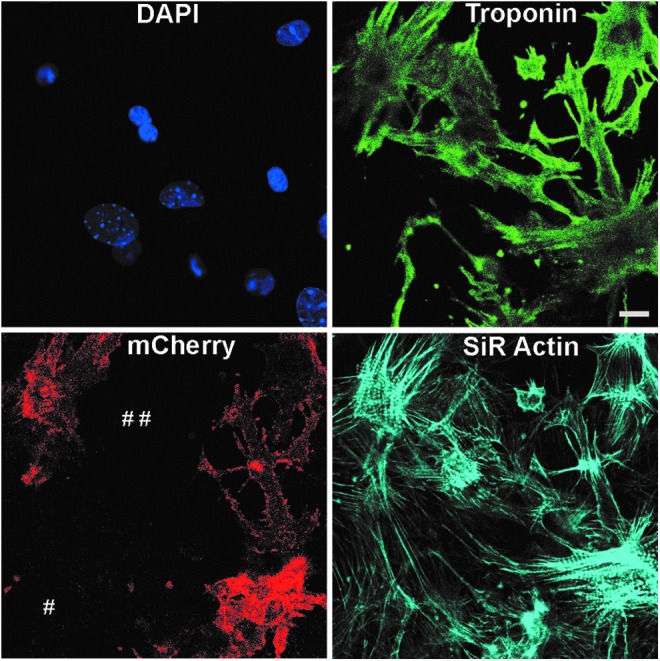
Transfection of neonatal primary cardiomyocytes with M3RNA-mCherry does not alter the structural integrity of cardiomyocytes. Structural integrity of transfected cardiomyocytes was observed using SiR Actin and cardiac troponin staining of transfected cells. Note the similar troponin staining in transfected and nontransfected cells (indicated by # #) and specificity of troponin staining with cells positive for SiR Actin, but negative for troponin staining (#). Scale bar = 10 μm.
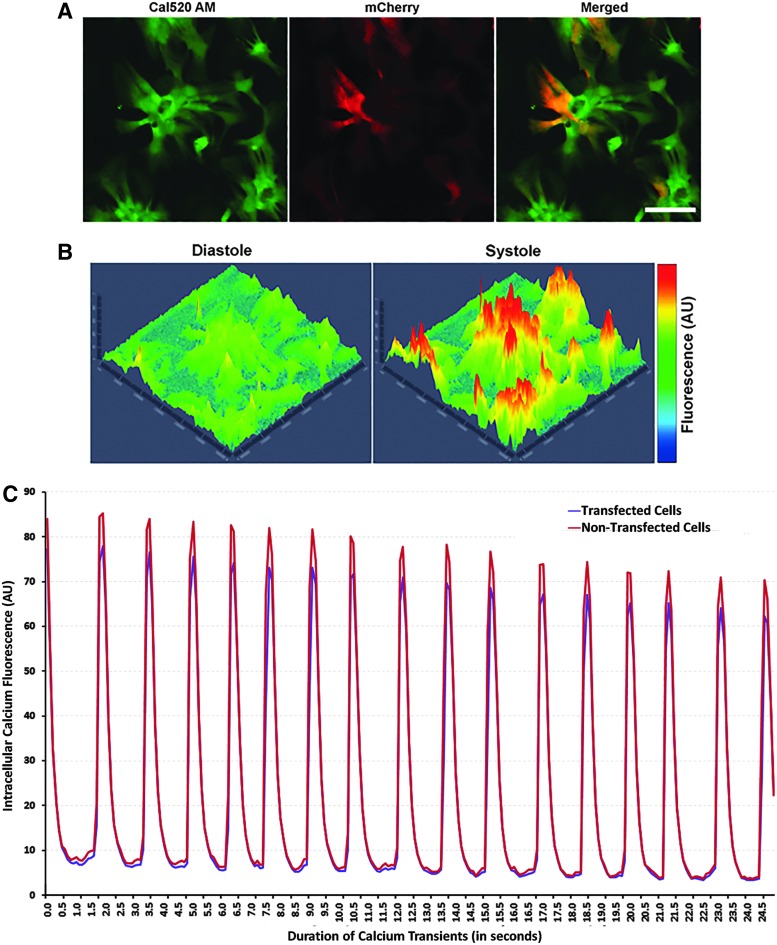
To determine if the transfection alters the central electrical properties of transfected cells, two intrinsic functional parameters of primary cardiomyocytes were compared: (1) calcium channel transients and (2) voltage-current relationships. [Ca2+]i (Intracellular calcium) transients from primary cardiomyocytes were recorded using the free intracellular Ca2+ binding dye Cal520AM. Primary cardiomyocytes meeting the beating pattern criterion were transfected with mCherry M3RNA. To image Ca2+ transients using Cal520AM, fields featuring both transfected and nontransfected cells were selected using the mCherry filter (Fig. 4A). Robust [Ca2+]i transients were observed in the primary cardiomyocyte cultures (Supplementary Video S1) having mCherry expression (Supplementary Video S2). Representative annotation of fluorescence intensity created at systole and diastole revealed the rhythmic and coordinated (in both transfected and nontransfected cells) [Ca2+]i transients with synchronous rapid [Ca2+]i bursts during systole with its absence during diastole (Fig. 4B). ROI were created from transfected and nontransfected cells (Fig. 4A) and intracellular fluorescence intensity (Y-axis) versus duration of Ca2+ transients (X-axis) was plotted (Fig. 4C). Note the similar [Ca2+]i transients in transfected and nontransfected cells.
FIG. 4.
Transfection of neonatal primary cardiomyocytes with M3RNA-mCherry expression does not alter the functionality of cardiomyocytes as revealed by identical calcium channel currents within transfected cells. (A) Representative image of Ca2+ transients using Cal520AM dye (left) and mCherry (middle) exhibiting transfected cells and the overlay (right). Scale bar = 100 μm (B) Cal520AM-labeled primary cardiomyocytes demonstrating 3D representation of systole and diastole state (pseudo color). (C) Representative annotation of fluorescence intensity (arbitrary units) versus time for Ca2+ transients from the average fluorescence intensity of 28 transfected and 19 nontransfected cardiomyocytes showing similar rhythmic and coordinated Ca2+ transients in transfected and nontransfected cardiomyocytes within the same area.
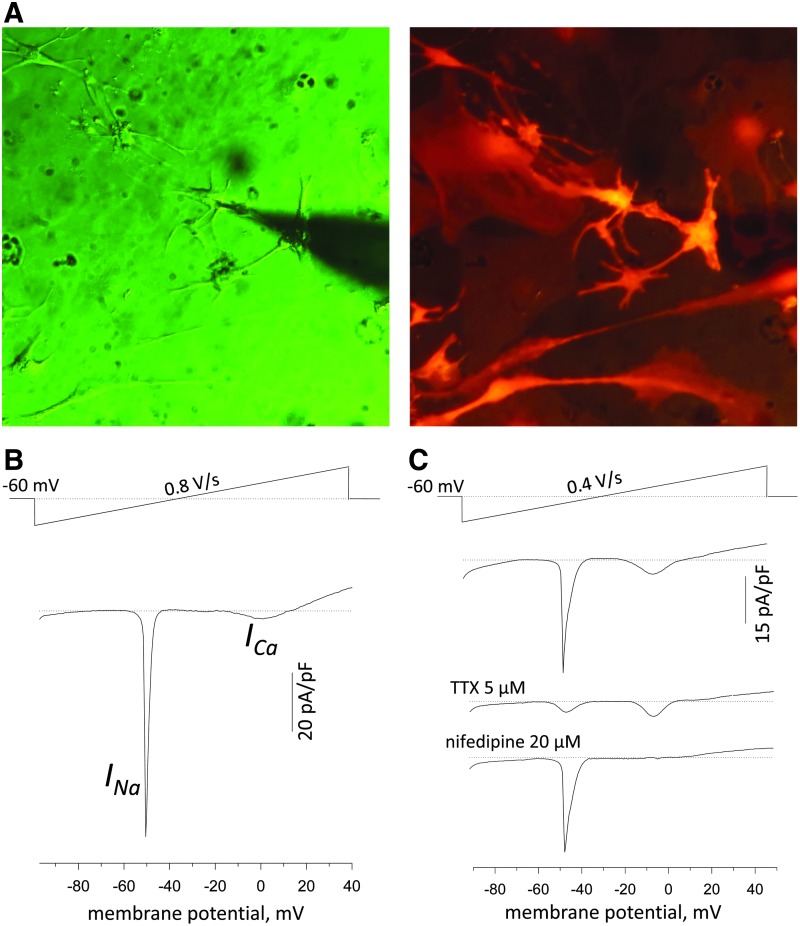
To test the intactness of electrical excitability of transfected primary cardiomyocytes, we tested intactness of the cardiomyocyte excitability and contraction. The beating cells were transfected overnight using mCherry mRNA, and transfected cells were identified using the fluorescence microscope (Fig. 5A). To discriminate inward currents components responsible for the cell excitation, the transfected neonatal cardiomyocytes were exposed to the ramp stimulation protocol in the whole-cell patch-clamp mode of recordings. Under such conditions, a ramp pulse from −90 to +40 mV induced two typical inward current components that were different in voltage-gating properties (Fig. 5B). The first component with the peak value at ∼50 mV was typically sensitive to tetrodotoxin (TTX, 5 μM), a selective inhibitor of voltage-gated Na+ channels (Fig. 5C). The second component at peak value ∼0 mV membrane potential was sensitive to nifedipine, a voltage-dependent L-type Ca2+ channels inhibitor (ICa). Thus, the obtained voltage-current relationships revealed intact profile of INa and ICa current components under mCherry transfection.
FIG. 5.
Transfection of neonatal primary cardiomyocytes with M3RNA-mCherry does not alter basic electrophysiological features of cardiomyocytes as revealed by voltage-current relationships in response to voltage-ramp stimulation in the whole cell mode of patch-clamp technique. (A) Image on the left shows the transmitted light image of the patched transfected neonatal cardiomyocytes with the corresponding image of mCherry fluorescence at 615 nm emission on the right. (B) Representative voltage-current relationships in mCherry-transfected neonatal cardiomyocytes obtained using the ramp protocol (shown on the top) revealed typical INa and ICa current components. (C) Voltage-sensitive inward currents were sensitive to tetrodotoxin (TTX, 5 μM), a selective inhibitor of voltage-gated Na+ channels (INa), and second current component, at peak value ∼0 mV membrane potential, was blocked by nifedipine, an inhibitor of voltage-dependent L-type Ca2+ channels (ICa).
M3RNA-FLuc myocardial injection induces prompt protein expression
To confirm if the rapid expression in primary cardiomyocytes holds under in vivo conditions, we performed in vivo M3RNA-FLuc transfection using direct myocardial injections into the left ventricle of FVB mice, using nanoparticle-based encapsulation of M3RNA-FLuc. For in vivo studies, we used nanoparticles (∼100 nm) that act as carriers of mRNA coated with positively charged biological polymers, which form complexes with negatively charged mRNA molecules due to ionic interactions (Fig. 6). Upon in vivo administration of M3RNA, nanoparticles enter the cells by endocytosis and release mRNA molecules for translation. Nanoparticles composed of iron moieties get degraded and the released iron enters the normal iron metabolic pathway.34 FLuc is used to determine protein expression kinetics in live animals. Bioluminescence imaging documented cardiac-targeted expression within the heart in as early as 2 h postinjection, increasing nearly 3.5 times in 24 h and fading to nearly background levels by 72 h (Fig. 7A, B). No off-target transfection was observed as the entire signal was primarily detected in the heart area (Fig. 7A). Furthermore, serial sections upon 24-h M3RNA-mCherry intracardiac injection revealed significant mCherry protein expression (middle panel) in heart tissue injected with mCherry mRNA (bottom panels) compared to vehicle control (top panels), with mCherry expression confirmed by anti-mCherry antibody in the green channel (left panels) (Fig. 7C). Troponin antibody revealed mCherry expression in the cardiomyocytes and note (#) the identical expression of mCherry in noncardiomyocytes areas as well. Finally, multiple gene expression with a single epicardial injection was performed using GFP, mCherry, and M3RNA-FLuc versus vehicle only in rat hearts. FLuc imaging can be performed on live animals; therefore, we first confirmed FLuc expression within mouse heart at 24 h using Xenogen (Supplementary Fig. S3) and the animal was then sacrificed, and heart tissues were processed for immunofluorescence (IF) analysis. IF revealed that GFP, mCherry, and FLuc protein (using anti-FLuc antibody) expression overlapped in M3RNA-injected rats (lower panels) versus no expression in sham (upper panels) (Fig. 7D).
FIG. 6.
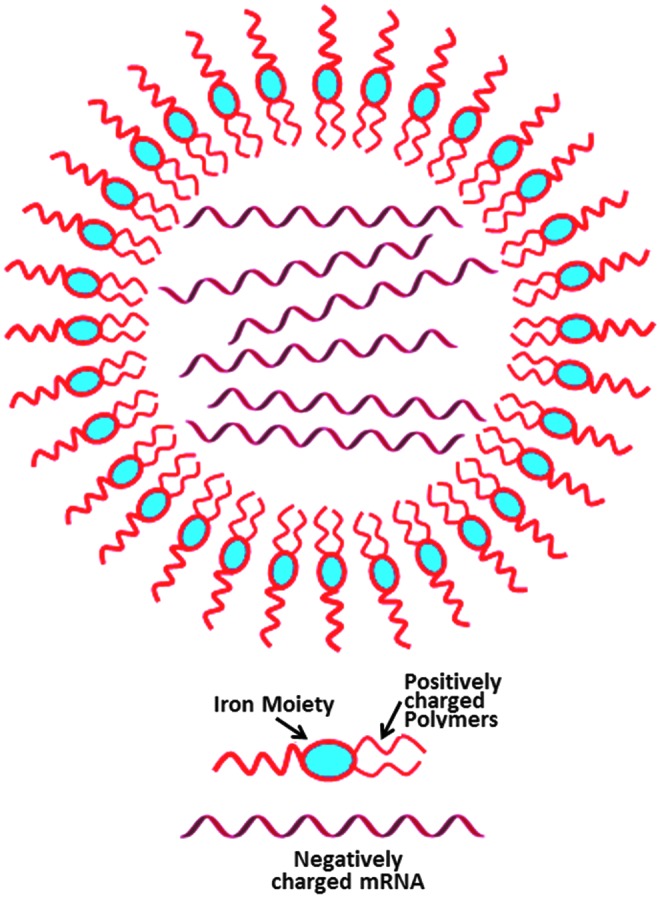
Graphical representation of nanoparticle. Nanoparticles (∼100 nm) act as carriers of mRNA coated with positively charged biological polymers that form complexes with negatively charged mRNA molecules due to ionic interactions.
FIG. 7.
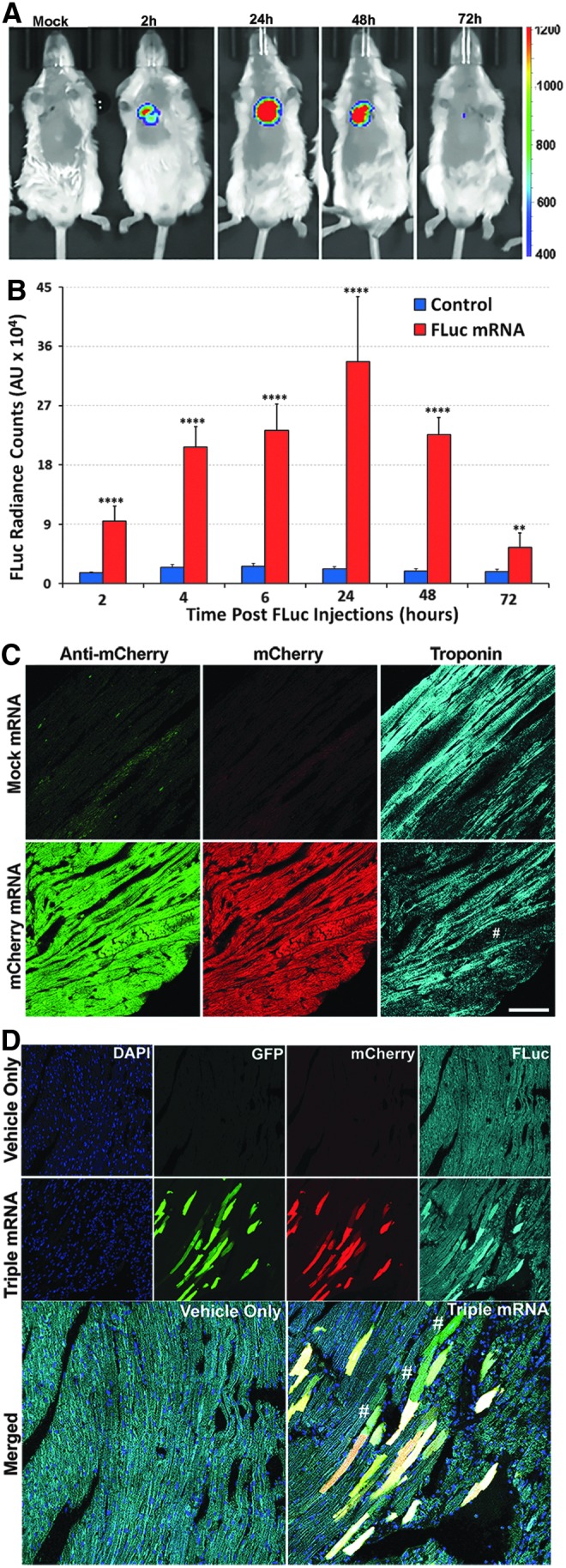
Gene expression within heart upon direct myocardial injection of M3RNAs. (A) Representative bioluminescence images showing the time course of luciferase expression following direct injections of M3-FLuc mRNA into the anterior left ventricle wall. Mice were intubated and intracardiac injections were performed upon open chest surgery (see Materials and Methods). In vivo bioluminescence images of mice in the supine position are shown at 2, 24, 48, and 72 h postsurgery. (B) Quantitative bioluminescence levels as average radiance (photons/cm2) from three different experiments (five animals each; three FLuc M3RNA and two Vehicle only) were obtained from the regions of interest and were plotted against time. ****p < 0.0001, **p < 0.01. (C) IF analysis showing the mCherry expression following direct injections into the anterior left ventricle wall. mCherry-M3RNA was injected directly into the left ventricle and heart tissues were obtained 24 h postsurgery. Hearts were fixed, sectioned, and stained with anti-mCherry and troponin antibody. Upper panel shows the staining in mock-transfected hearts, while the bottom middle panel shows the mCherry expression in heart tissue (red channel), while the left panel shows the anti-mCherry antibody and the right panel is revealing the sarcomeric structures using troponin antibody. Note the structures positive for mCherry and anti mCherry, but negative for sarcomeric structures (indicated by #). (D) IF analysis of hearts (as in C above) showing GFP, mCherry, and FLuc expression upon direct injections of M3RNAs into the left ventricle and heart tissues was processed as in C above. Images were acquired using confocal microscope LSM780 with 3 × 3 tiles using the tiling tool to acquire bigger area with sharp images using a 40 × , 1.2 W lens. Upper panel shows the GFP, mCherry, and FLuc in green, red, and far-red channel, respectively, in mock-transfected hearts, while the bottom panel shows indicated gene expressions in heart tissue. In merged image, # represents areas where transfection of a single gene was observed. Scale bar = 50 μm. IF, immunofluorescence.
Targeted expression of mCherry M3RNA in a porcine model of acute MI
M3RNA-mCherry was encapsulated within a calcium alginate solution. Using an acute porcine model of myocardial infarction (Fig. 8A), an intracoronary bolus of ∼250 μg M3RNA-mCherry was infused into the LAD using the distal opening of the infracting over-the-wire balloon. Following intracoronary delivery, alginate was visualized to preferentially gel in the site of acute injury as monitored by ICE (Fig. 8B). The heart was harvested at 72 h, flushed with chilled normal saline and sliced using the ProCUT sampling tool. Imaging of sliced heart sections on Xenogen using mCherry filter showed significant mCherry protein expression localized to the area of infarction (Fig. 8C and Supplementary Fig. S4). IHC on 1 cm slices from areas of infarction versus noninfarcted regions featured significantly higher mCherry staining (Fig. 8D), confirming targeted induction of protein expression within the injured portion of the heart.
FIG. 8.
Targeted mCherry mRNA within infarcted porcine heart using alginate gel (A) Angiography of the pig heart showing the induction of acute myocardial infarction. Left panel shows the distribution of arterial blood flow along the anterior portion of the left ventricle to determine proper balloon placement. Right panel shows the inflated balloon blocking completely the distal blood flow. Scale bar = 10 mm. (B) Serial intracardiac echocardiographic images document transition of the anterior wall of the left ventricle (arrow) from normal (top) to injured after 90 min of LAD occlusion (middle) to loaded with alginate gel having M3RNA (lower panel) over 10 min immediately after reperfusion (total time 100 min). Scale bar = 10 mm. (C) Pig heart removed and flushed with chilled normal saline, and sectioned was imaged to document mCherry expression surrounding the anterior wall infarcted area in most of the areas starting from the apex to the mid-papillary portion of the left ventricle. Scale bar = 10 mm. (D) Heart areas from the anterior wall infarcted region (mCherry transfected) versus remote regions (control) demonstrate significant mCherry expression on IF probing. Total number of pig studies n = 10 for alginate optimization and n = 4 for documentation of infarct targeting. Scale bar = 100 μm. LAD, left anterior descending coronary artery; LCx, left circumflex artery.
Discussion
Gene therapy is a promising strategy for the treatment and regeneration in cardiovascular diseases. Some clinical scenarios require gene expression or gene editing to reverse the course of disease such as clotting disorders, enzymatic deficiency, or gene mutation.35–37 However, within the healthy population, an adverse inflammatory response to acute events may result in tissue nonhealing or chronic injury.38–41 DNA and viral vectors are great tools for treating diseases where long-term expression of an encoded protein is required.
mRNA vector may be more appropriate where transient expression may be preferable such as in attenuating acute inflammation and CRISPR-mediated genome editing, where off-target events are a problem.21,42 RNA vectors have advantages, including a reduction in the risk of genome integration or invoking an immune response, and can initiate rapid and transient protein expression.43,44 We, in this study, present a novel M3RNA-based approach to induce rapid expression within multiple cell lines, primary cardiomyocytes, and murine heart, and in acutely injured porcine myocardium. This platform showed controlled expression kinetics in multiple cell lines and primary cells, with transfection having little or no impact on the structural properties and functional characteristics, including calcium transients and electrical excitability of primary cardiomyocytes. Myocardial injection of M3RNA encoding FLuc, mCherry, and GFP, reproducibly induced rapid and consistent protein expression within the murine heart. Furthermore, this approach was found flexible enough for simultaneous delivery of multiple genes within the murine heart and could be targeted into acutely injured tissue in porcine models of myocardial infarction. Thus, proof of concept of utilizing reporter genes to allow expedited and time-limited expression of multiple genes within cardiomyocytes and murine heart with a single delivery platform is provided in this study.
During AMI, a rapid sequence of molecular events occurs during injury and following reperfusion, it ultimately culminates in damage to tissue. Injury can be fully aborted with rapid PCI if the patient presents within a very short period of time (<90 min).45,46 However, in those who present (>90 min to <12 h), PCI is still indicated, but the scope of damage to myocardium becomes increasingly worse due to ischemia and hypoxia.47,48 In most individuals, restoration of blood flow even after the initial 90-min results in recovery of myocardial function and restoration or organ performance to near normal. However in about 30% of the population, this is not the case with fulminant loss of myocardium, ensuing despite reperfusion.49,50 Efforts to mitigate this phenomenon have been focused on antiplatelet agents and neurohormonal antagonism. However, a compendium of recent evidence suggests that a deregulation of the inflammatory response to injury may be at the root cause of catastrophic myocardial damage.51–53
Beyond revascularization, many regenerative platforms have been utilized to try and attenuate myocardial injury post-AMI. Initial interventions targeting cardioprotection focused on activation of the potassium ATP channel in an effort to augment native cardioprotective mechanisms.54 Beyond cardioprotection, cell therapeutic efforts to improve outcomes at the time of acute myocardial infarction have been pursued, delivering bone marrow mononuclear cells, mesenchymal stem cells, and lineage-specified cells to the myocardium.55–60 Gene-encoded therapies have been increasingly considered in both heart failure and myocardial infarction. The most advanced effort in gene delivery was the modulation of SERCA2A in the setting of heart failure61 through the use of an adeno-associated viral gene delivery platform. Although this platform did not meet efficacy endpoints in phase 2b testing,62,56 it did establish a novel paradigm of broad-based gene delivery to the heart. RNA and DNA platforms have been utilized to deliver vascular endothelial growth factor (VEGF) into the myocardium through direct epicardial injection.63,64 The results of these platforms were, however, not favorable, primarily due to a paucity in gene regulation and likely due to the fact that only a single gene approach was utilized.
Furthermore, siRNA and nonencoding miRNA have also been increasingly suggested as potential therapeutic platforms to alter the myocardial microenvironment post-AMI.65,66 Although preclinical efforts have successfully demonstrated biopotency, the translatability of such platforms have to date remained quite limited, likely due to the systemic nature of delivery and potential for off-target impact. Thus, an increasing need for improving myocardial delivery of biologics has been called for. Unfortunately, the technological approaches utilized to date have been based on cell-based delivery and have not been tailored to the newer biologics, which do not have the size or survival restrictions imposed by cytotherapeutics.
The work presented herein attempts to address these issues by taking steps toward a novel approach that is complementary with the current interventional practice, introducing modified mRNA for increased stability and expression and reduced immunogenicity in vivo.67,68 M3RNA complexes were created by microencapsulating modified mRNA in 100 nm iron-based lipid nanoparticles (Fig. 6). Given the complex nature of the postinjury myocardial microenvironment, single gene expression within the heart is likely insufficient to achieve any mitigating impact on cardiovascular morbidity, as was seen with the VEGF experience. We designed the M3RNA system to be compatible with simultaneous gene delivery and indeed show that injection of three M3RNAs within murine heart can yield expression of three distinct proteins. Furthermore, M3RNA biopotentiation of alginate to target the infarcted bed provides a unique opportunity to achieve rapid gene expression in the setting of AMI. One can envisage delivery of complementary genes serving the angiogenic, cytoprotective, and immunomodulatory needs of the myocardium postinfarction. A gene-based delivery approach that can target cell survival and impede inflammatory pathways would need to come on rapidly after restoration of blood flow if it is to have a beneficial effect. However, given that these pathways change within a 48–72 h period, long-term expression may not be of significant benefit and may pose a risk of harm.69 Thus, this initial system was designed to offer acute gene therapy intervention as a step toward development of a more comprehensive biologics-based approach.
The spontaneous crosslinking of alginate in the presence of Ca2+ 342 at the infarcted site provides localized in situ alginate matrix70–72 for encapsulating therapeutic RNA for treatment of infraction. We, in this study, have shown a novel approach for the use of alginate gel to achieve targeted gene delivery and expression in acutely infarcted porcine heart to achieve targeted and significant protein expression in 3 days. This approach could be beneficial for patients suffering from heart attack to achieve rapid, transient, and targeted protein expression within the heart. In addition, although alginate gel provides a means to achieve targeting to the infarcted bed, it rapidly biodegrades and is essentially absent 3 days postcardiac administration.
To date, the use of biologics-based therapy has eluded acute care, due to the fact that the logistics of cell processing is incompatible with emergency procedures. Truly, off-the-shelf platforms provide a unique and novel opportunity to pursue the use of regenerative approaches in the acute setting. In this way, gene-encoded therapeutics can now be envisaged as a next-generation paradigm in the treatment of AMI, complementing the current standard of care. In this article, we highlight the M3RNA platform that has been developed as a novel technology, fully compatible with current procedural standards utilized in PCI. Given the homing capability afforded by this approach, one can now envision interventional delivery of genes immediately after PCI with a time horizon tailored for acute events. Beyond the heart, as this technology can induce gene expression in any cell phenotype, its use in other acute events such as musculoskeletal injury, stroke, and sepsis serves as avenues of future exploration.
Supplementary Material
Acknowledgments
We thank Dr. Vanda Lennon's Neuroimmunology Research laboratory for sharing their pregnant rats for the isolation of neonatal rat primary cardiomyocytes. Funding sources include the following: National Institutes of Health HL134664, VanCleve Cardiac Regeneration Medicine Program, Marriott Foundation, NIH Mayo-MSTP Training Grant T32GM065841, NIH-MPET Training Grant T32GM072474, and NIH Mayo-IMSD R25GM055252.
Disclosure Statement
M3RNA technology has been patented by Mayo Clinic and Licensed to Rion LLC. Mayo Clinic, A.B. and A.T. have interests in Rion LLC.
References
- 1. Terzic A., and Behfar A. Stem cell therapy for heart failure: ensuring regenerative proficiency. Trends Cardiovasc Med 26, 395, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kochegarov A., and Lemanski L.F. New trends in heart regeneration: a review. J Stem Cells Regen Med 12, 61, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hastings C.L., Roche E.T., Ruiz-Hernandez E., Schenke-Layland K., Walsh C.J., and Duffy G.P. Drug and cell delivery for cardiac regeneration. Adv Drug Deliv Rev 84, 85, 2015 [DOI] [PubMed] [Google Scholar]
- 4. Smit F.E., and Dohmen P.M. Cardiovascular tissue engineering: where we come from and where are we now? Med Sci Monit Basic Res 21, 1, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behfar A., Faustino R.S., Arrell D.K., Dzeja P.P., Perez-Terzic C., and Terzic A. Guided stem cell cardiopoiesis: discovery and translation. J Mol Cell Cardiol 45, 523, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hajjar R.J., and Ishikawa K. Introducing genes to the heart: all about delivery. Circ Res 120, 33, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McErlean E.M., McCrudden C.M., and McCarthy H.O. Delivery of nucleic acids for cancer gene therapy: overcoming extra- and intra-cellular barriers. Ther Deliv 7, 619, 2016 [DOI] [PubMed] [Google Scholar]
- 8. Barata P., Sood A.K., and Hong D.S. RNA-targeted therapeutics in cancer clinical trials: current status and future directions. Cancer Treat Rev 50, 35, 2016 [DOI] [PubMed] [Google Scholar]
- 9. Thomas C.E., Ehrhardt A., and Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 4, 346, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Sheridan C. Gene therapy finds its niche. Nat Biotechnol 29, 121, 2011 [DOI] [PubMed] [Google Scholar]
- 11. Mátrai J., Chuah M.K., and VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol Ther 18, 477, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das S.K., Menezes M.E., Bhatia S., et al. Gene therapies for cancer: strategies, challenges and successes. J Cell Physiol 230, 259, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frank-Kamenetsky M., Grefhorst A., Anderson N.N., et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci U S A 105, 11915, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Majmudar M.D., Keliher E.J., Heidt T., et al. Monocyte-directed RNAi targeting CCR2 improves infarct healing in atherosclerosis-prone mice. Circulation 127, 2038, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rao P.K., Toyama Y., Chiang H.R., et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105, 585, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madonna R., Van Laake L.W., Davidson S.M., et al. Position paper of the european society of cardiology working group cellular biology of the heart: cell-based therapies for myocardial repair and regeneration in ischemic heart disease and heart failure. Eur Heart J 37, 1789, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matkar P.N., Leong-Poi H., and Singh K.K. Cardiac gene therapy: are we there yet? Gene Ther 23, 635, 2016 [DOI] [PubMed] [Google Scholar]
- 18. Turnbull I.C., Eltoukhy A.A., Fish K.M., et al. Myocardial delivery of lipidoid nanoparticle carrying modRNA induces rapid and transient expression. Mol Ther 24, 66, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schott J.W., Morgan M., Galla M., and Schambach A. Viral and synthetic RNA vector technologies and applications. Mol Ther 24, 2016, 1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pardi N., Secreto A.J., Shan X., et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun 8, 14630, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sahin U., Karikó K., and Türeci Ö. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov 13, 759, 2014 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Su H.H., Yang Y., et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther 21, 358, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sultana N., Magadum A., Hadas Y., et al. Optimizing cardiac delivery of modified mRNA. Mol Ther 25, 1306, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McIvor R.S. Therapeutic delivery of mRNA: the medium is the message. Mol Ther 19, 822, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zangi L., Lui K.O., von Gise A., et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 31, 898, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitehead K.A., Langer R., and Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 8, 129, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh R.D., Gibbons S.J., Saravanaperumal S.A., et al. Ano1, a Ca2+-activated Cl− channel, coordinates contractility in mouse intestine by Ca2+ transient coordination between interstitial cells of Cajal. J Physiol 592, 4051, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alekseev A.E., Gomez L.A., Aleksandrova L.A., Brady P.A., and Terzic A. Opening of cardiac sarcolemmal KATP channels by dinitrophenol separate from metabolic inhibition. J Membr Biol 157, 203, 1997 [DOI] [PubMed] [Google Scholar]
- 29. Pitari G.M., Zingman L.V., Hodgson D.M., et al. Bacterial enterotoxins are associated with resistance to colon cancer. Proc Natl Acad Sci U S A 100, 2695, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakipova O.V., Averin A.S., Evdokimovskii E.V., et al. Store-operated Ca2+ entry supports contractile function in hearts of hibernators. PLoS One 12, e0177469, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burgess A., Vigneron S., Brioudes E., Labbé J.C., Lorca T., and Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A 107, 12564, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh R.D., Holicky E.L., Cheng Z.J., et al. Inhibition of caveolar uptake, SV40 infection, and beta1-integrin signaling by a nonnatural glycosphingolipid stereoisomer. J Cell Biol 176, 895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamada S., Arrell D.K., Martinez-Fernandez A., et al. Regenerative therapy prevents heart failure progression in dyssynchronous nonischemic narrow QRS cardiomyopathy. J Am Heart Assoc 4, pii: , 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oh N., and Park J.H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomater 9, 51, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subramanian S., Shahaf G., Ozeri E., et al. Sustained expression of circulating human alpha-1 antitrypsin reduces inflammation, increases CD4+FoxP3+ Treg cell population and prevents signs of experimental autoimmune encephalomyelitis in mice. Metab Brain Dis 26, 107, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Rafi M.A., Zhi Rao H., Passini M.A., et al. AAV-mediated expression of galactocerebrosidase in brain results in attenuated symptoms and extended life span in murine models of globoid cell leukodystrophy. Mol Ther 11, 734, 2005 [DOI] [PubMed] [Google Scholar]
- 37. Saura C.A., Parra-Damas A., and Enriquez-Barreto L. Gene expression parallels synaptic excitability and plasticity changes in Alzheimer's disease. Front Cell Neurosci 9, 318, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koh T.J., and DiPietro L.A. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med 13, e23, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White E.S., and Mantovani A.R. Inflammation, wound repair, and fibrosis: reassessing the spectrum of tissue injury and resolution. J Pathol 229, 141, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eming S.A., Krieg T., and Davidson J.M. Gene therapy and wound healing. Clin Dermatol 25, 79, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eming S.A., Krieg T., and Davidson J.M. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol 127, 514, 2007 [DOI] [PubMed] [Google Scholar]
- 42. Finn J.D., Smith A.R., Patel M.C., et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep 22, 2227, 2018 [DOI] [PubMed] [Google Scholar]
- 43. Nallagatla S.R., Toroney R., and Bevilacqua P.C. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol 5, 140, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karikó K., Buckstein M., Ni H., and Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Chen F.C., Lin Y.R., Kung C.T., Cheng C.I., and Li C.J. The association between door-to-balloon time of less than 60 minutes and prognosis of patients developing ST segment elevation myocardial infarction and undergoing primary percutaneous coronary intervention. Biomed Res Int 2017, 1910934, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blankenship J.C., Scott T.D., Skelding K.A., et al. Door-to-balloon times under 90 min can be routinely achieved for patients transferred for ST-segment elevation myocardial infarction percutaneous coronary intervention in a rural setting. J Am Coll Cardiol 57, 272, 2011 [DOI] [PubMed] [Google Scholar]
- 47. Siontis K.C., Barsness G.W., Lennon R.J., et al. Pharmacoinvasive and primary percutaneous coronary intervention strategies in ST-elevation myocardial infarction (from the Mayo Clinic STEMI Network). Am J Cardiol 117, 1904, 2016 [DOI] [PubMed] [Google Scholar]
- 48. Hoffman S.J., Routledge H.C., Lennon R.J., et al. Procedural factors associated with percutaneous coronary intervention-related ischemic stroke. JACC Cardiovasc Interv 5, 200, 2012 [DOI] [PubMed] [Google Scholar]
- 49. Spoon D.B., Lennon R.J., Psaltis P.J., et al. Prediction of cardiac and noncardiac mortality after percutaneous coronary intervention. Circ Cardiovasc Interv 8, e002121, 2015 [DOI] [PubMed] [Google Scholar]
- 50. Wu C., Camacho F.T., King S.B., et al. Risk stratification for long-term mortality after percutaneous coronary intervention. Circ Cardiovasc Interv 7, 80, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dick S.A., and Epelman S. Chronic heart failure and inflammation: what do we really know? Circ Res 119, 159, 2016 [DOI] [PubMed] [Google Scholar]
- 52. Pohl J., Hendgen-Cotta U.B., Stock P., et al. Myocardial expression of macrophage migration inhibitory factor in patients with heart failure. J Clin Med 6, pii: , 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruparelia N., Godec J., Lee R., et al. Acute myocardial infarction activates distinct inflammation and proliferation pathways in circulating monocytes, prior to recruitment, and identified through conserved transcriptional responses in mice and humans. Eur Heart J 36, 1923, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zingman L.V., Hodgson D.M., Bast P.H., et al. Kir6.2 is required for adaptation to stress. Proc Natl Acad Sci U S A 99, 13278, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schächinger V., Erbs S., Elsässer A., et al. Intracoronary bone marrow-derived progenitor cells in acute myocardial infarction. N Engl J Med 355, 1210, 2006 [DOI] [PubMed] [Google Scholar]
- 56. Hare J.M., Fishman J.E., Gerstenblith G., et al. Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. JAMA 308, 2369, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Makkar R.R., Smith R.R., Cheng K., et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet 379, 895, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Behfar A., Yamada S., Crespo-Diaz R., et al. Guided cardiopoiesis enhances therapeutic benefit of bone marrow human mesenchymal stem cells in chronic myocardial infarction. J Am Coll Cardiol 56, 721, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bartunek J., Behfar A., Dolatabadi D., et al. Cardiopoietic stem cell therapy in heart failure: the C-CURE (Cardiopoietic stem Cell therapy in heart failURE) multicenter randomized trial with lineage-specified biologics. J Am Coll Cardiol 61, 2329, 2013 [DOI] [PubMed] [Google Scholar]
- 60. Bartunek J., Davison B., Sherman W., et al. Congestive heart failure cardiopoietic regenerative therapy (CHART‐1) trial design. Eur J Heart Fail 18, 160, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jaski B.E., Jessup M.L., Mancini D.M., et al. Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID Trial), a first-in-human phase 1/2 clinical trial. J Card Fail 15, 171, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Greenberg B., Butler J., Felker G.M., et al. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): a randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet 387, 1178, 2016 [DOI] [PubMed] [Google Scholar]
- 63. Kormann M.S., Hasenpusch G., Aneja M.K., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29, 154, 2011 [DOI] [PubMed] [Google Scholar]
- 64. Fujii H., Li S.H., Wu J., et al. Repeated and targeted transfer of angiogenic plasmids into the infarcted rat heart via ultrasound targeted microbubble destruction enhances cardiac repair. Eur Heart J 32, 2075, 2011 [DOI] [PubMed] [Google Scholar]
- 65. Pratt A.J., and MacRae I.J. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284, 17897, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Quiat D., and Olson E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest 123, 11, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kauffman K.J., Mir F.F., Jhunjhunwala S., et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Karikó K., Muramatsu H., Welsh F.A., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16, 1833, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Springer M.L., Chen A.S., Kraft P.E., Bednarski M., and Blau H.M. VEGF gene delivery to muscle: potential role for vasculogenesis in adults. Mol Cell 2, 549, 1998 [DOI] [PubMed] [Google Scholar]
- 70. Liberski A., Latif N., Raynaud C., Bollensdorff C., and Yacoub M. Alginate for cardiac regeneration: from seaweed to clinical trials. Glob Cardiol Sci Practice 2016, e201604, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leor J., Tuvia S., Guetta V., et al. Intracoronary injection of in situ forming alginate hydrogel reverses left ventricular remodeling after myocardial infarction in Swine. J Am Coll Cardiol 54, 1014, 2009 [DOI] [PubMed] [Google Scholar]
- 72. Landa N., Miller L., Feinberg M.S., et al. Effect of injectable alginate implant on cardiac remodeling and function after recent and old infarcts in rat. Circulation 117, 1388, 2008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.