Summary
N-linked glycosylation affects the potency, safety, immunogenicity, and pharmacokinetic clearance of several therapeutic proteins including monoclonal antibodies. A robust control strategy is needed to dial in appropriate glycosylation profile during the course of cell culture processes accurately. However, N-glycosylation dynamics remains insufficiently understood owing to the lack of integrative analyses of factors that influence the dynamics, including sugar nucleotide donors, glycosyltransferases, and glycosidases. Here, an integrative approach involving multi-dimensional omics analyses was employed to dissect the temporal dynamics of glycoforms produced during fed-batch cultures of CHO cells. Several pathways including glycolysis, tricarboxylic citric acid cycle, and nucleotide biosynthesis exhibited temporal dynamics over the cell culture period. The steps involving galactose and sialic acid addition were determined as temporal bottlenecks. Our results show that galactose, and not manganese, is able to mitigate the temporal bottleneck, despite both being known effectors of galactosylation. Furthermore, sialylation is limited by the galactosylated precursors and autoregulation of cytidine monophosphate-sialic acid biosynthesis.
Subject Areas: Process Biotechnology, Industrial Biotechnology, Metabolomics, Transcriptomics
Graphical Abstract
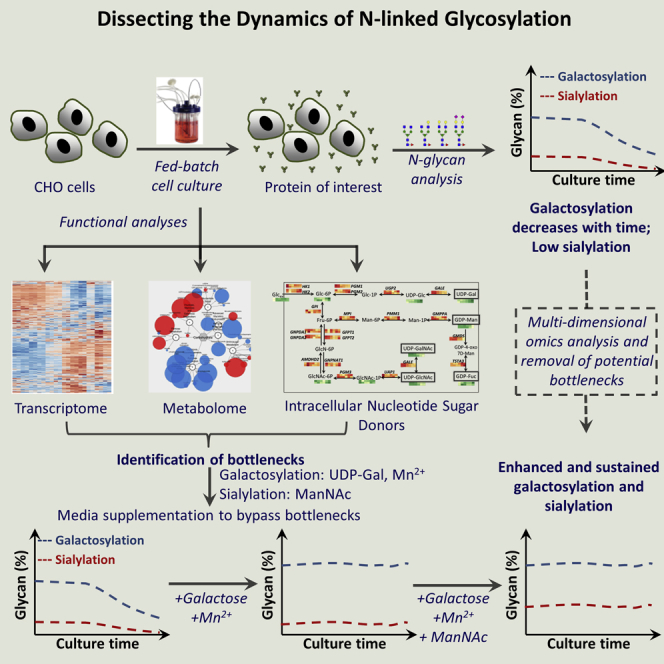
Highlights
-
•
Major glycosylated species exhibit temporal dynamics during fed-batch processes
-
•
Key metabolic pathways linked to N-glycosylation exhibit significant temporal dynamics
-
•
Dynamics in nucleotide sugar donors (NSDs) directly influences glycoform heterogeneity
-
•
Glycoform heterogeneity can be mitigated by supplementing NSD biosynthetic precursors
Process Biotechnology; Industrial Biotechnology; Metabolomics; Transcriptomics
Introduction
Over the past decade, monoclonal antibody (mAb) therapeutics has shown significant success in clinical studies and constitutes the majority of biologically derived drugs (Niwa and Satoh, 2015, Sha et al., 2016). More than 30% of the new drugs licensed currently are based on mAbs and biological therapeutics (Niwa and Satoh, 2015, Sathish et al., 2013). N-linked glycosylation (N-glycosylation) is known to play a key role in governing the potency, safety, immunogenicity, and pharmacokinetic clearance of these proteins. It can therefore be a critical quality attribute of such biologically derived therapeutic proteins (Berger et al., 2011, Eon-Duval et al., 2012, Maverakis et al., 2015). N-glycosylation is a complex network of reactions resulting in a number of possible glycoform structures (Hossler et al., 2006, Krambeck and Betenbaugh, 2005). As a result, despite consistent protein backbones, a number of different glycosylation isoforms (glycoforms) are observed on recombinant proteins purified from the harvest of a Chinese hamster ovary (CHO) cells fed-batch process. Therefore, a robust control strategy is needed during the course of a culture to dial in a desired harvest glycosylation profile accurately.
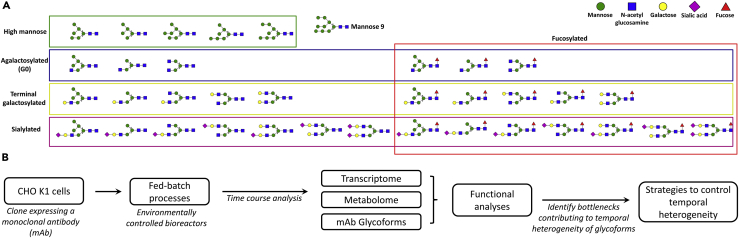
Glycoform produced at any given time point in a cell culture process is heterogeneous. For example, there could be 5% mAbs with high mannose species, another 25% mAbs with galactosylated species, etc. (Figure 1A). However, this heterogeneity in glycoform also changes with time, or exhibits temporal dynamics. Both of these contribute toward the heterogeneity in glycoform observed at the harvest, also called cumulative glycoform. We hypothesize that the temporal variation in glycoform distribution is likely due to the kinetics of the metabolic components in the glycosylation network during CHO fed-batch cultures. Such temporal dynamics in glycosylation network activity can be due to changes in factors including sugar nucleotide levels, glycosylation enzyme levels, and availability of cofactors such as metal ions. These factors are, in turn, influenced by changes in cell culture process parameters including, but not limited to, culture nutrient levels (Fan et al., 2015), pH (Lin et al., 2015, Villiger et al., 2016a), levels of by-products such as lactate and ammonia (Fan et al., 2015, Gawlitzek et al., 2000), and temperature (Ivarsson et al., 2014, Sou et al., 2015). Several groups have developed mathematical models to probe the heterogeneity of glycoforms in a cell culture process (Hossler et al., 2007, Jedrzejewski et al., 2014, Jimenez del Val et al., 2011, Jiménez del Val et al., 2013, Kaveh et al., 2013, Krambeck et al., 2009, Villiger et al., 2016b). Only recently, with the availability CHO genome data (Lewis et al., 2013), several groups have implemented time course omics analysis of CHO fed-batch cultures to probe the system as a whole (Hsu et al., 2017, Mulukutla et al., 2012, Opdam et al., 2017, Sha et al., 2018). However, our understanding of the dynamics in the intracellular factors influencing N-glycosylation, including levels of nucleotide sugar donors (NSDs), glycosyltransferases, and glycosidases, remains insufficient owing to the lack of an integrative and quantitative analyses of the same. In this study, a systems approach that employs multi-prong omics analyses was used to probe the glycosylation physiology of CHO cells in fed-batch cultures with the specific goal of dissecting the temporal dynamics in NSD synthesis, glycosylation-related gene expression, and levels of cofactors that play a key role in the glycosylation reaction network.
Figure 1.
Schematic of the Study
(A) Glycoform heterogeneity in the monoclonal antibodies observed in the harvest of a fed-batch culture.
(B) Clonally propagated CHO-K1 cells with random integration of a monoclonal antibody gene were grown under two different processes, and time course analysis was performed on the transcriptome, metabolome, and mAb glycoform data to identify key bottlenecks.
Briefly, in this study, a CHO-K1-derived cell line expressing a model mAb was cultivated in environmentally controlled bioreactors using two distinct fed-batch processes (Figure 1B). Time course transcriptomics (RNA sequencing [RNA-seq] analysis), metabolomics, and glycan analyses were performed on the two processes. Statistical and functional analyses, including principal-component analysis (PCA), gene set enrichment analysis (GSEA) (Subramanian et al., 2005), time course gene set analyses (TCGSA) (Hejblum et al., 2015), and time course differential gene and metabolite expression analyses (maSigPro) (Conesa et al., 2006), were performed on the omics data to identify pathways that were differentially regulated between the two processes or, within a process as a function of time. This helped identify potential factors that could be responsible for temporal dynamics in glycosylation reaction kinetics. Additional experiments were performed to validate and confirm the role of these factors in temporal dynamics of glycosylation.
Results
Major Glycosylated Species Show Similar Temporal Dynamics across the Two Fed-Batch Processes Employed but Vary in Their Absolute Values
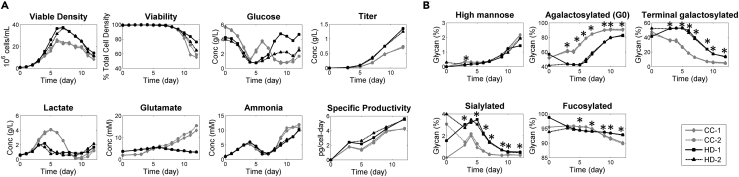
A CHO-K1 host-cell-derived clone producing a model mAb was cultivated in two distinct 12-day fed-batch cell culture processes, namely, platform (CC) and HiPDOG (HD), carried out in 1-L working volume bioreactors (see the Transparent Methods for details on the process differences between CC and HD). Higher peak cell densities and titers were observed in the HD process than those observed in the CC process (Figure 2A). Peak lactate levels were significantly lower in the HD (∼2 g/L) than in the CC process (∼4 g/L). However, cells in CC culture underwent a strong metabolic shift to lactate consumption on day 5 of the process. The higher viable cell density observed in the HD process could therefore be due to the relatively lower peak lactate levels when compared with the CC process. Specific productivity or the amount of protein produced per cell per day (qP) was similar across both processes and increased over time until day 9 (Figure 2A). The dynamics of other process-related parameters and extracellular metabolites is shown in Figure S1A.
Figure 2.
Glycan Species from the mAbs Exhibit Similar Time Dynamics Despite Process Variation
(A) The two processes resulted in widely varying lactate profiles as well as cell growth and titer.
(B) Agalactosylated (G0) and high-mannose species increase over time, whereas terminal galactosylated and sialylated species decrease over time. Slight decrease in fucosylated species is also observed over time. A detailed classification of the glycan structures is provided in Data S2. Asterisks (*) represent p < 0.05 for two-tailed unpaired t tests comparing CC and HD samples from each day.
Hydrophilic interaction chromatography-based glycan analysis was performed on the mAb molecules present in the spent media samples collected from different days of the two processes across the exponential “growth phase” (days 0, 3, 5) and the stationary “production phase” (days 7, 9, and 12) (Data S1). These glycan structures were then classified into major glycosylated species (Data S2). Each major glycosylated species had similar temporal dynamic patterns in both processes, but differed in the absolute level of the species measured (Figures 2B and S1B). For example, the levels of high mannose species, which is primarily the glycan core with five or more mannose moieties, and the agalactosylated species (G0) increased over time in both processes. However, terminally galactosylated species (term-gal) that have either one (G1) or two (G2) galactoses terminating the glycan chain decreased significantly over time across both processes. Furthermore, a slight decrease in the sialylated and fucosylated species was also observed over time in both processes. To understand the glycan profile of the mAbs at specific time points during the culture, instantaneous glycan profiles were calculated by taking the time derivative of the cumulative glycans with respect to the titer produced (Figure S1C, see Transparent Methods). These results suggested that there are two observations that needed explanation, namely, the absolute differences in the levels of major species (specifically galactosylation/agalactosylation) between the two fed-batch processes and the temporal changes in the major glycan species (which are relatively conserved across the two processes).
To explain the absolute differences in the major species between the two processes, differences in the spent culture media for the two processes were probed. As the two processes were implemented in distinct basal and feed media, a detailed metal ion analysis was performed to understand the differences between the processes (Figure S1D). Manganese, a known cofactor (Witsell et al., 1990) for the enzyme beta-1,4-galactosyltransferase-1 (B4GALT1), which catalyzes the addition of galactose to the extending glycan chain, was observed to be significantly lower in the CC samples. Although other metal ions such as Mg2+, Ca2+, and Cu2+ also showed differences between the two fed-batch processes and they could be potential covariates in contributing to the absolute differences, the goal was to identify the major influencing factor. As Mn2+ is directly linked to the galactosylation step, we hypothesized that manganese level could be the main reason for the differences in the absolute levels of term-gal species between the two processes. To test this, a fed-batch experiment was conducted in shake flasks with pH adjustment, by inoculating cells in the two basal media but with the manganese levels swapped between them (Figure S1E). Cells were inoculated in original or modified versions of CC and HD media. Modified versions of each medium had Mn2+ levels adjusted to match the starting levels in the original formulation of the other medium. This experiment was part of a bigger experiment described later in the last subsection of the Results section. The absolute levels of term-gal produced in the two media were found significantly different and corresponded to the swapped manganese concentrations. This established that the absolute difference in the galactosylated (and agalactosylated) species was mainly due to the difference in the culture manganese levels between the two processes.
From the instantaneous glycan profiles, it was apparent that throughout the fed-batch process, most of the glycans produced have the mannose sugars trimmed and N-acetylglucosamine (GlcNAc) added to them (Figure S1C). However, the amount of agalactosylated species increased with time, whereas those of the galactosylated and sialylated species decreased over time. This suggested that the conversion of high-mannose species into low-mannose species and the subsequent addition of GlcNAc in the Golgi might not have bottlenecks, but the steps involving galactosyl and sialyl transfer might be rate-limiting steps in the glycosylation reaction network. The reaction steps involving glycan chain extension in the N-glycosylation process are highly ordered and sequential. Under pseudo-steady-state assumption, the turnover rate (which includes factors such as precursor, substrate, enzyme concentrations and enzyme activity influenced by enzymatic rate constants, as well as steric hindrance) of reactants to products governs the fraction of substrate converted into product. For successive reaction steps, under no rate limitation, one can plot the substrate and product levels on x and y axes, respectively. For example, if all the glycans attached to mAbs exist as the terminal species, i.e., completely sialylated species, on a scatterplot of nth species as y axis and (n-1)th species as x axis, the data should fall on the y axis, i.e., (n-1)th species ∼0% and nth species ∼100%. For the rest of the intermediate species, because their levels are near zero, on a scatterplot of ith (y axis) and (i-1)th species (x axis), the data should fall at the intersection of the x and y axes, i.e., (i-1)th species ∼0% and ith species ∼0%. Alternatively, under rate limitation scenario, the scatterplot would be shifted to non-zero values due to significant buildup of the intermediate species, as seen in case of several glycosylated species (Figure S2). This alternative approach using quasi-steady-state analysis of instantaneous glycan profiles supports the same conclusion that the steps involving galactosyl and sialyl transfer might be rate-limiting steps in the glycosylation reaction network (Figure S2). Temporal variation observed in the glycoforms produced could be an outcome of these bottlenecks in the galactosylation and sialylation steps. To investigate the other potential reasons for the dynamic nature of the glycosylation profile, and to identify the bottlenecks in the galactosylation and sialylation steps, systems analysis approach integrating transcriptomic and metabolomic data was undertaken.
Systems Analysis Suggests that CHO Cells Undergo a Shift in Their Transcriptional, Metabolic, and Glycome Program along the Course of a Fed-Batch Culture
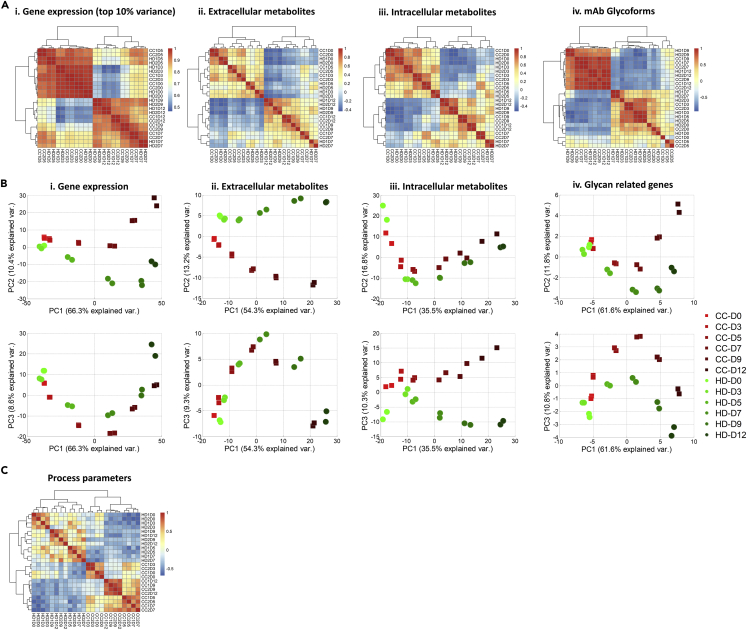
Time course analyses of global transcript levels (RNA-seq) as well as intra- and extracellular metabolite levels were performed for the samples collected from both CC and HD processes (see the Transparent Methods and Data S3, S4, and S5). Pairwise clustering, using the Spearman correlation was used to cluster samples based on the transcriptional profile of the genes constituting the top 10% variance (Figure 3A [i]). Irrespective of the process employed, the samples within each phase, i.e., growth or production phase, clustered together. Next, pairwise clustering was performed on the time course extracellular and intracellular metabolomic data (Figure 3A [ii, iii]). Similar to the transcriptomic data, the metabolomics data also showed clustering of samples based on growth or production phase, irrespective of the processes. Alternatively, PCA performed on the transcriptome data showed that the trajectory of change in the variance over the course of the culture, as explained by principal component 1 (PC1) and PC2 or PC3, appears to be primarily dictated by the growth or production phase of the culture, together explaining 75% of the total variance (Figure 3B [i]). PCA of the metabolomics data revealed similar shift in the metabolic state of the cells during the growth and production phases, together explaining 63% and 52% of the variance for the extracellular and intracellular metabolites, respectively (Figure 3B [ii, iii]). These analyses showed that the metabolic and transcriptional footprints of samples correlated well within growth or production phase, despite different processes being employed.
Figure 3.
CHO Cells Undergo a Shift in Transcriptional and Metabolic Profile during Cell Culture
(A) Time course data for (i) genes that constitute top 10% variance across the samples, (ii) the intracellular metabolites, (iii) extracellular metabolites, and (iv) mAb glycoforms were correlated with all other samples, and the samples were clustered based on their pairwise Spearman correlation coefficients.
(B) Principal-component analysis of the transcriptomic, metabolomic, and glycan data. The first and second (top panel) or third (bottom panel) principal components together account for ∼60%–90% of the variance in the omics data and show temporal shift in the CHO transcriptional and metabolic profile from growth phase (day 0 to day 7) to production phase (day 7 to day 12).
(C) Time course data for process parameters were correlated with all other samples, and the samples were clustered based on their pairwise Spearman correlation coefficients showing that the parameters cluster separately for the two process conditions (HD and CD), regardless of the growth phase and production phase. Process variables included in the analysis are glucose, lactate, glutamate, pH, media feed, base feed, glucose feed, and ICP metals (fed differently in the two processes). As the units of each of the variables differ, the data were z-normalized for each variable before calculating the Spearman correlations.
Similar clustering analysis of the standardized (Z scored) glycan data for the time course samples suggested that the glycan profiles also appeared to be dependent on the stage of the culture (Figure 3A [iv]). Interestingly, HD1D7 and HD2D7 samples from HD process clustered with growth phase (days 0, 3, 5). Glycan addition to the mAbs is downstream of all the steps, including transcription, translation, and metabolism (nucleotide synthesis). Therefore, a time delay (or lag) is possible, explaining why HD1D7 and HD2D7 glycoforms cluster with growth phase rather than the production phase. In addition, PCA analysis was performed on a list of glycosylation-related genes curated from the literature (Nairn et al., 2008). Only those genes that were expressed at least for one time point for both the processes were considered in the analysis (Data S6). Similar to the clustering analysis, variance in the glycan-related genes was a function of the state of cells and appeared to be independent of the process (Figure 3B [iv]). Next, correlation analysis was performed on the process parameter data from different days of the two processes, spanning growth and production phases (see Figure 3 legends). Interestingly, unlike the transcriptome and metabolome, the process parameters clustered together based on the process employed (CC or HD) (Figure 3C). Together, the results suggested that although the cells are subjected to relatively different process conditions, the transcriptome, metabolome, and to an extent the N-glycan signature seemed to be dictated primarily by the culture stages (growth or production phase). These results motivated a detailed functional analysis of the time course omics data to gain insights into the complex regulation of N-glycosylation dynamics.
Functional Analysis of the Omics Data Identified Significant Time Dynamics in Key Metabolic Pathways Linked to N-Glycosylation
An integrative approach, employing three orthogonal methods, was taken to analyze the multi-dimensional omics data. These methods include GSEA, TCGSA, time course metabolite set analysis (TCMSA), and maSigPro. GSEA was employed to identify functional classes that are enriched in either the growth phase or the production phase of the culture. TCGSA or TCMSA were used to capture the functional classes (gene sets or metabolic sets) that may or may not have dynamics in one phase or the other, but might show dynamics over the course of the complete culture. maSigPro was used to identify key genes or metabolites that exhibit significant temporal dynamics (i.e., the genes and metabolites that were significantly perturbed over time) using a regression-based analysis (see Transparent Methods). The analyses were performed on data from both processes separately, and unless otherwise stated, the results/trends from these analyses hold true for both processes.
For GSEA and TCGSA functional class analyses, a composite curated gene set list was used comprising gene sets from KEGG, BioCarta, Reactome, and additional gene sets related to glycosylation from Gene Ontology (Data S7, see also Transparent Methods). Employing GSEA, several pathways were found to be enriched in the growth phase including central energy metabolism, cell cycle, apoptosis, and protein synthesis (Data S8A and S8B, Figure S3). In contrast, several other pathways including those involved in glycosylation, aminoglycan metabolism, glycerolipid metabolism, sphingolipid metabolism, and cell adhesion were enriched in the production phase. TCGSA analysis identifies the gene sets that exhibit time dynamics over the complete duration of the culture irrespective of the changes in the growth or production phases of the culture (see methods section in Transparent Methods for more details). Employing TCGSA and using the same gene set list that was used for GSEA, gene sets such as those related to NSD biosynthetic pathways, pentose phosphate pathway, and oxidative phosphorylation, among others, were identified to have time dynamics over the complete time span of the culture. Interestingly, these gene sets were not enriched in GSEA (Data S9).
Apart from transcription level control of enzyme/metabolic pathway activity, it is well known that changes in metabolic pathway flux can also be influenced significantly by intermediate metabolite levels (Hackett et al., 2016, Mulukutla et al., 2016). To investigate temporal changes in metabolite levels across the fed-batch processes, TCMSA was performed on the intracellular metabolomic dataset. A curated list of metabolites based on their functional classes was employed for the analysis (Data S10). The analysis suggested that several pathways that exhibit significant temporal dynamics at the transcriptional level also vary significantly at the metabolite level. These include sphingolipid metabolism, fatty acid metabolism, purine and pyrimidine metabolism, nucleotide sugar metabolism, amino acid metabolism, and glycolytic pathway (Data S11). In addition, a statistical test (maSigPro) was employed to identify key transcripts and metabolites that show significant time dynamics. Top transcripts and intracellular metabolites that vary significantly over time were ranked based on their q-values for the maSigPro test, and a ranked list is shown for overall data as well as for various functional groups (Data S12) and metabolic pathways (Data S13).
Combining the results from all the functional set and statistical analyses, a list of key pathways that exhibit significant time dynamics in CHO cells during fed-batch culture time course was curated (Table 1). For each functional class listed in the table, gene/metabolites that had significant time dynamics are also included. Several pathways from this list were identified as potential regulators of N-glycosylation dynamics because of their direct or indirect linkage with the N-glycosylation process (Figure 4). Next, temporal changes in these specific pathways were explored.
Table 1.
Key Functional Groups and Pathways that Exhibit Significant Temporal Dynamics over the Cell Culture Period during Fed-Batch Processes
| Pathway/Functional Sets (GSEA Enrichment Phase) | Significant Gene Sets (GSEA and TCGSA) | Significant Metabolic Sets (TCMSA) | Top Significant Genes (maSigPro) | Top Significant Metabolites (maSigPro) |
|---|---|---|---|---|
| Sphingolipid metabolism (production phase) | REACTOME_SPHINGOLIPID_METABOLISM REACTOME_GLYCOSPHINGOLIPID_METABOLISM KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES KEGG_SPHINGOLIPID_METABOLISM REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS |
Sphingolipid metabolism | ASAH1 SMPD1 PSAP GM2A |
Glycosyl-N-nervonoyl-sphingadienine N-nervonoyl-sphingadiene N-erucoyl-sphingosine sphingomyelin |
| Cell adhesion &extracellular matrix (production phase) | KEGG_CELL_ADHESION_MOLECULES_CAMS KEGG_ECM_RECEPTOR_INTERACTION REACTOME_COLLAGEN_FORMATION REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX |
NA | NEO ICAM1 SDC1 PTPRM |
NA |
| Amino acid metabolism (growth phase) | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVESa REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERSa |
Lysine and histidine metabolism Urea cycle, arginine, and proline metabolism Gammaglutamyl amino acid Tyrosine and phenylalanine metabolism Tryptophan metabolism |
SMS AHCY SLC25A15 CBS GOT1 |
Taurine Pipecolate Argininate N-acxetylhistidinemethionine sulfoxide |
| Phospholipid metabolism (production phase) | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS REACTOME_PHOSPHOLIPID_METABOLISM KEGG_GLYCEROPHOSPHOLIPID_METABOLISM |
Phosphatidylglycerol/phosphatidylinositol Phosphatidylcholine Lysophospholipid |
PNPLA2 PNPLA8 PCYT1A DGAT2 PLD3 |
CDP-ethanolamine 1,2-dioleoyl-GPG 1-palmitoleoyl-2-oleoyl-glycerophosphoglycerol 1-palmitoyl-2-stearoyl-glycerophosphocholine 1,2-dipalmitoleoyl-glycerophosphocholine |
| Amino sugar and NSD metabolism (NA) | KEGG_AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISMa GO_NUCLEOTIDE_SUGAR_BIOSYNTHETIC_PROCESSa |
Amino sugar and nucleotide sugar | GALE UGDH FPGT PGM2 GMDS |
Glucuronate UDP-galactose UDP-glucose UDP-GlcNAc/UDP-GalNAc CMP-N-acetylneuraminic acid |
| Pyrimidine metabolism (growth phase) | GO_PYRIMIDINE_NUCLEOTIDE_BIOSYNTHETIC_PROCESS GO_PYRIMIDINE_NUCLEOTIDE_METABOLIC_PROCESS KEGG_PYRIMIDINE_METABOLISM REACTOME_PYRIMIDINE_METABOLISM |
Pyrimidine metabolism Pyrimidine metabolism uracil containing |
PRPS1 TYMS UNG DUT TDG |
Thymidine 2′-O-methylcytidine 5-methylcytidine 5-methyl-2′-deoxycytidine beta-alanine |
| Purine metabolism (growth phase) | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE KEGG_PURINE_METABOLISM REACTOME_PURINE_METABOLISM |
Purine metabolism | RRM2 IMPDH2 RRM1 |
N1-methylinosine N2,N2-dimethylguanosine 7-methylguanine |
| Cell cycle, mitosis, and apoptosis (growth phase) | KEGG_CELL_CYCLE REACTOME_CELL_CYCLE REACTOME_CELL_CYCLE_CHECKPOINTS REACTOME_CELL_CYCLE_MITOTIC REACTOME_REGULATION_OF_APOPTOSIS |
NA | LMNB1 ZWILCH POLE CDC6 |
NA |
| Glycosylation (production phase) | GO_GLYCOSYLATION GO_N_GLYCAN_PROCESSING GO_PROTEIN_N_LINKED_GLYCOSYLATION REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER |
NA | MAN1B1 DDOST ALG13 B4GALT1 |
NA |
| Amino glycans (production phase) | KEGG_GLYCOSAMINOGLYCAN_DEGRADATION REACTOME_GLYCOSAMINOGLYCAN_METABOLISM KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_ |
NA | GNS CHPF HMMR |
NA |
| TCA cycle (growth phase) | MOOTHA_TCA KEGG_CITRATE_CYCLE_TCA_CYCLE REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE REACTOME_METABOLISM_OF_CARBOHYDRATES |
TCA cycle | FH ACLY ME1 SDHC SDHB |
Malate Aconitate Citrate Succinate 2-methylcitrate/homocitrate |
| Glycolysis and gluconeogenesis (growth phase) | REACTOME_GLUCOSE_TRANSPORT REACTOME_REGULATION_OF_GLUCOKINASE REACTOME_GLUCONEOGENESIS BIOCARTA_GLYCOLYSIS_PATHWAY REACTOME_METABOLISM_OF_CARBOHYDRATES |
Glycolysis and gluconeogenesis | ADPGK PFKFB4 TPI1 HK2 ALDOC |
Lactate Dihydroxyacetone phosphate Hexose diphosphates Glucose 6-phosphate Pyruvate |
| Glutamate/glutathione metabolism (growth phase) | KEGG_GLUTATHIONE_METABOLISM | Glutamate and glutathione metabolism | RRM2 SMS GSTM2 |
5-oxoproline glutamine N-acetylglutamine |
| Pentose phosphate pathway (NA) | KEGG_PENTOSE_PHOSPHATE_PATHWAYa KEGG_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONSa |
Pentose metabolism and pentose pathway | H6PD TKT RPIA |
Arabitol/xylitol Ribitol Arabonate/xylonate |
| Oxidative phosphorylation (NA) | KEGG_OXIDATIVE_PHOSPHORYLATION | NA | NDUFA7 COX6B1 ETFDH |
NA |
| Glycosidases and deglycosylation (production phase) | GO_PROTEIN_DEGLYCOSYLATION | NA | FUCA1 HEXA NEU1 |
NA |
| Regulators of N-glycosylation (NA) | GO_REGULATION_OF_PROTEIN_GLYCOSYLATIONa | NA | TMEM59 IL15 |
NA |
NSD, nucleotide sugar donors; GPG, glycerophosphoglycerol ; GPC, glycerophosphocholine. Three orthogonal analyses (GSEA, TCGSA, and maSigPro) on the transcriptome and metabolome data were performed to identify key pathways that change over time. Top transcripts and metabolites based on maSigPro analysis are also shown (see also Data S15).
Found significant in time course analysis, but not in the GSEA analysis.
Figure 4.
Transcriptional and Metabolic Factors that Can Potentially Affect N-Glycosylation Dynamics
Glycosylation dynamics can potentially be regulated via multiple ways as suggested by the time course and gene set enrichment analyses. Dynamics of top few transcripts (green blocks)/metabolites (red blocks) from each group that vary significantly with time are shown. A p value based on the time course analysis (maSigPro) is indicated by asterisks as follows: ***p <0.005, **0.005 > p > 0.01, *0.01 > p > 0.05.
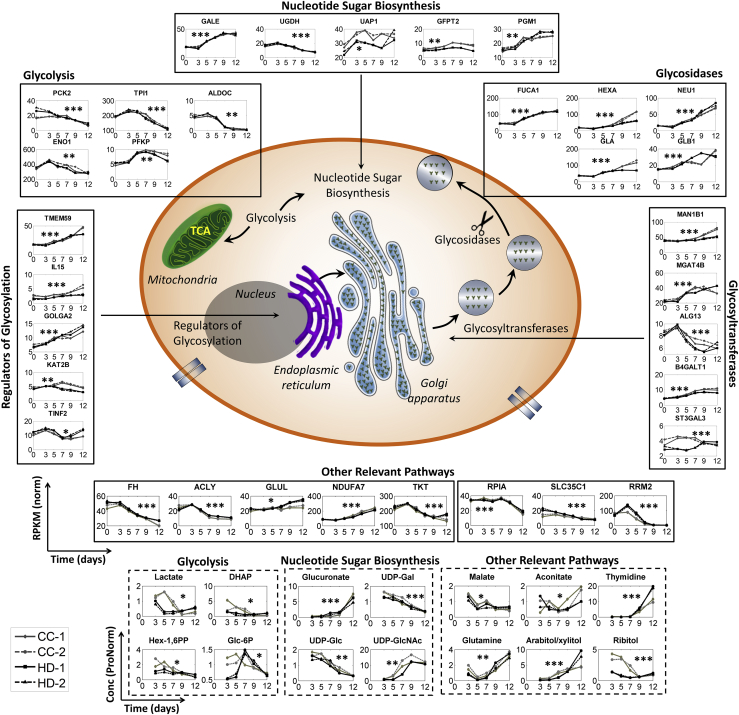
Temporal Dynamics in Levels of Enzyme Transcripts and Intermediate Metabolites of Metabolic Pathways Influencing N-Glycosylation Dynamics
Glycolysis
The glycolytic pathway was found to be enriched in the growth phase (GSEA). TCGSA suggested that glycolytic gene sets exhibit significant temporal perturbation or dynamics (Table 1). Glycolytic metabolites such as glucose-6-phosphate (G6P) and fructose-6-phosphate (F6P) serve as precursors to the NSD biosynthetic pathways. Therefore, changes in the transcript levels of glycolytic enzymes and in the corresponding metabolites can potentially influence the flux directed toward NSD biosynthesis. Glucose consumption rates (qGlc) and lactate production rates (qLac) have been used as indicators of the dynamics in central energy metabolism. For both processes, the metabolic state of the cells in the bioreactors shifted from lactate-producing (qLac/qGlc>0) to lactate-consuming state (qLac/qGlc<0) and again to a marginally lactate-producing state toward the end of the culture (Figure S4A). Such metabolic shifts are known to be outcomes of changes in the glycolytic gene expression and intermediate metabolite levels (Hartley et al., 2018, Mulukutla et al., 2012). Several glycolytic enzymes, including ADP-dependent glucokinase (ADPGK), 6-phosphofructo-2-kinase (PFKFB4), triosephosphate isomerase (TPI), hexokinase-2 (HK2), and fructose bisphosphate aldolase C (ALDOC), changed significantly over time (Data S12). Similarly, metabolites such as lactate, dihydroxyacetone phosphate, hexose diphosphates, and pyruvate were also found varying significantly with time (Data S13, Figures 4 and S4B).
Purine and Pyrimidine Metabolism
Purine and pyrimidine metabolism provides essential nucleotides during NSD biosynthesis such as cytidine triphosphate, guanosine triphosphate, and uridine triphosphate. Both these pathways are enriched in the growth phase (GSEA, Data S8B) and also show significant time dynamics in the time course analyses (Data S9 and S11). Several key enzymes involved in these pathways also vary significantly with time, such as ribose phosphate pyrophosphokinase 1 (PRPS1), thymidylsynthatase (TYMS), uracil-DNA glycosylase (UNG), ribonucleoside-diphosphate reductase M2 (RRM2), and inosine-5′-monophosphate dehydrogenase 2 (IMPDH2). Several key metabolites also exhibit significant temporal dynamics, including thymidine, 2′-O-methylcytidine, and N1-methylinosine (Table 1).
TCA Cycle
The tricarboxylic citric acid (TCA) cycle is directly linked to glycolysis, glutamine, and purine and pyrimidine metabolic pathways (Figure S4B). Several gene sets related to the TCA cycle were enriched in the growth phase (GSEA, Data S8B). These gene sets also exhibited significant time dynamics (TCGSA, Data S9). The levels of several genes including those encoding for fumarate hydratase (FH), ATP citrate synthase (ACLY), NADP-dependent malic enzyme (ME1), and succinate dehydrogenase (SDHC) varied over time (maSigPro, Data S12). Interestingly, although the TCA cycle did not show up in TCMSA analysis, a few of its metabolites including aconitate, citrate, and malate were either up- or downregulated over time (Data S13).
Other Central Energy Metabolism Pathways
Other pathways linked to glycolysis and TCA cycle such as glutamine/glutamate metabolism (which serves as a precursor to N-acetylated NSD intermediates), pentose phosphate pathway (PPP), and oxidative phosphorylation showed significant time dynamics in their corresponding gene sets, transcripts, and metabolites (Table 1). For example, transketolase (TKT) and ribose-5-phosphate isomerase (RPIA) enzymes in the PPP decreased with time, whereas NADH dehydrogenase 1 alpha subcomplex 7 (NDUFA7) from oxidative phosphorylation increased with time (Figure 4, Data S12).
Glycosyltransferases
The concentration of glycosyltransferases can directly affect the reaction rates for each of the glycosylation steps. GSEA analysis suggested that gene sets containing glycosyltransferases were primarily enriched in the production phase (Data S8B). These gene sets also showed up significant in TCGSA analysis (Data S9). Several genes encoding for mannosidases and glycosyltransferases varied over time (Data S12, Figure 4). For example, 1,2-alpha mannosidases (MAN1B1, MAN2A2, MAN2B1, MAN2B2, and MAN1A1), N-acetylglucosamineyltransferases (MGAT4B and MGAT1), and beta-1,4-galactosyltransferase 1 (B4GALT1) increased over time (Figure S5). Other mannosidases (MAN1C1, MAN2A1, MAN2C1), fucosyltransferase (FUT8), and sialyltrasferases (ST3GALT3 and ST3GAL4) showed marginal or no dynamics.
Glycosidases
Glycosidases catalyze cleavage of specific sugar moieties from the glycan structures on therapeutic proteins (and other host cell proteins). TCGSA strongly suggested that gene sets linked to glycan degradation vary significantly over time (Data S9), and GSEA analysis suggested that these gene sets are enriched in the production phase (Data S8B). Most of the glycosidase enzymes showed up in the maSigPro analysis including, fucosidase (FUCA1), galactosidases (GLA, GLB1), sialidases (NEU1, NEU2, NEU3, NEU4), and hexosidases (HEXA, HEXB) (Figure 4).
Other Regulators of Glycosylation
Apart from enzymes or gene sets that are directly linked to glycosylation, a gene set comprising regulators of glycosylation also showed up significant in GSEA and TCGSA analyses. The expression levels of most of the genes in this gene set increased over time (Figure 4, Data S12). These include transmembrane protein 59 (TMEM59), interleukin-15 (IL15) and golgin subfamily A member 2 (GOLGA2). For example, TMEM59 has been reported to be an inhibitor of galactosylation and sialylation (Ullrich et al., 2010). Temporal dynamics in transcript levels of TMEM59 levels can affect the glycoforms produced.
In summary, functional analyses of time course omics data identified a number of enriched pathways and functional classes that can influence either the levels of substrates for NSD biosynthesis or the enzymes linked with the N-glycosylation process. These include genes and metabolites involved in glycolysis, TCA cycle, purine and pyrimidine biosynthesis, and amino sugar and NSD biosynthesis. Key genes and metabolites involved in these pathways that could be potential limiting factors for N-glycosylation are listed in Table 1 and Data S15. In the next section, a detailed analysis of the NSD biosynthetic pathway dynamics is performed.
Analysis of Temporal Dynamics in Nucleotide Sugar Donor Biosynthetic Pathway Suggests Galactosylation and Sialylation Enzymatic Steps Might Be Substrate Limited
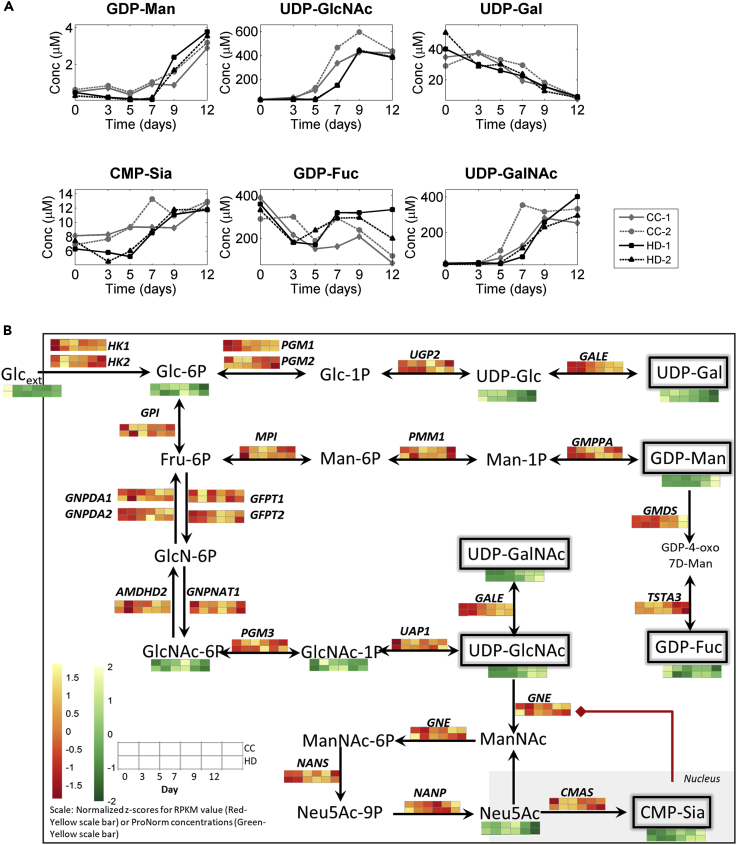
NSDs serve as substrates for the N-glycosylation process. Intracellular levels of NSDs were quantified from the cell pellet samples taken at multiple time points from the bioreactors for both fed-batch processes. The NSD concentrations varied over the course of fed-batch culture, and the dynamics for each NSD is conserved between the two fed-batch processes employed (Figure 5A). The dynamics in NSDs could be a result of temporal changes in the precursors from glycolysis and/or the enzymes and intermediate metabolites involved in the NSD biosynthetic pathways (Figure 5B). Time course transcriptome and metabolome data were used to probe biosynthetic pathway changes for each NSD, to identify the cause for temporal variation observed in the corresponding NSDs.
Figure 5.
Time Dynamics of Nucleotide Sugar Donors and NSD Intermediates Partially Explains N-Glycosylation Dynamics Observed Experimentally
(A) Intracellular concentration of nucleotide sugar donors varies significantly over time for both processes.
(B) Overview of the temporal dynamics for the nucleotide sugar intermediates and transcripts that encode for enzymes involved in the nucleotide sugar biosynthetic pathways. A p value based on the time course analysis (maSigPro) for the RNA-seq and metabolomic data are available in Supplemental Information.
GDP-Mannose
Glucose is converted to G6P by hexokinase (HK1 and HK2). G6P enters the NSD biosynthetic pathway and serves as a precursor to all the NSDs (Figure 5B). G6P exhibited significant time dynamics over the culture period (Data S13, Figure 5B). It is converted to F6P, which serves as the precursor for guanosine triphospho-mannose (GDP-Man) synthesis (Figure 5B). The intracellular concentration of GDP-Man increased during the course of the culture.
GDP-Fucose
Levels of guanosine triphospho-L-fucose (GDP-Fuc), which serves as the substrate for fucose addition to the glycan chain, for most part were relatively constant over the course of the culture for both the processes. GDP-Fuc is synthesized from GDP-Man in a two-step enzymatic process catalyzed by GDP-mannose 4,6 dehydratase (GMDS) and GDP-L-fucose synthase (encoded by the gene tissue-specific transplantation antigen P35B [TSTA3]) (Figure 5B). Marginal dynamics were observed in the transcript levels of the two enzymes, with GMDS going up with time and TSTA3 decreasing over time.
UDP-GlcNAc
Uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) serves as the substrate for GlcNAc addition to the glycan chain. It is synthesized from G6P via intermediates including glucosamine-6-phosphate (GlcNAc6P) and glucosamine-1-phosphate (GlcNAc1P) (Figure 5B). UDP-GlcNAc increases significantly with time (∼15-fold in CC; ∼12-fold in HD), whereas no significant dynamics is observed for GlcNAc6P and GlcNAc1P (Data S13). The expression levels of the genes that encode the corresponding enzymes involved in the synthesis of GlcNAc1P also did not exhibit any significant time dynamics (Data S12). UDP-GlcNAc is also converted to Uridine diphospho-N-acetylgalactosamine (UDP-GalNAc) by UDP-glucose 4-epimerase, which is encoded by the gene GALE. UDP-GalNAc shows similar trends as UDP-GlcNAc despite significant increase in the expression level of GALE (Figure 5, Data S12).
UDP-Galactose
Uridine diphospho-galactose (UDP-Gal) serves as the substrate for galactose addition to the glycan chain. It is generally synthesized from G6P via a multi-step process (Figure 5B). G6P is initially converted to UDP-glucose (UDP-Glc), which is then converted to UDP-Gal by GALE. Transcript levels of enzymes involved in the UDP-Glc biosynthesis did not change significantly with time (Data S12), whereas the transcript levels of GALE were observed to increase with time. However, both UDP-Gal and UDP-Glc levels decreased significantly during the production phase (Figure 5A). As G6P levels decreased over time, the drop in UPD-Gal could be a direct result of decrease in G6P levels.
CMP-Sialic Acid
Cytidine monophospho-sialic acid (CMP-Sia) serves as the substrate for sialic acid addition to the glycan chain. Formation of CMP-Sia from UDP-GlcNAc is a multi-step process (Figure 5B). UDP-GlcNAc is converted to N-acetyl mannosamine (ManNAc) by the enzyme glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (GNE). ManNAc is converted to 5-acetyl neuraminic acid (Neu5Ac) by a series of enzymatic steps, which is then converted to CMP-Sia. CMP-Sia is known to strongly inhibit (with Hill coefficient = 4.1) the epimerase function of GNE, thus maintaining an autoregulation of CMP-Sia levels (Yardeni et al., 2011). The levels of CMP-Sia were in the lower concentration range, although a slight increase was observed over the course of the fed-batch processes (Figure 5A). Inhibition of the epimerase function of GNE by slightly higher concentrations of CMP-Sia might result in inhibition of UDP-GlcNAc conversion to downstream intermediates including Neu5Ac. This is observed to be the case as the Neu5Ac levels decreased over time, even though the levels of UDP-GlcNAc increased over time (Figure 5B).
Galactosylation and sialylation were observed to decrease over time (Figure 2B). Decrease in galactosylated species can now be explained by the decrease in the UDP-Gal levels over the course of the fed-batch culture. The decrease in sialylation can be explained by the decrease in galactosylated species over time. Relatively low availability of CMP-Sia does not help the drop in sialylation incurred due to the decrease in galactosylated species. With the help of the above-described understanding for the cause of the drop in the UDP-Gal levels and low levels of CMP-Sia, we next designed and performed experiments to overcome these bottlenecks in an attempt to increase and maintain a sustained level of galactosylation and sialylation in fed-batch cultures.
Temporal Rate Limitations in Galactosylation and Sialylation Can Be Mitigated by Supplementation of Corresponding NSD Biosynthetic Pathway Intermediates
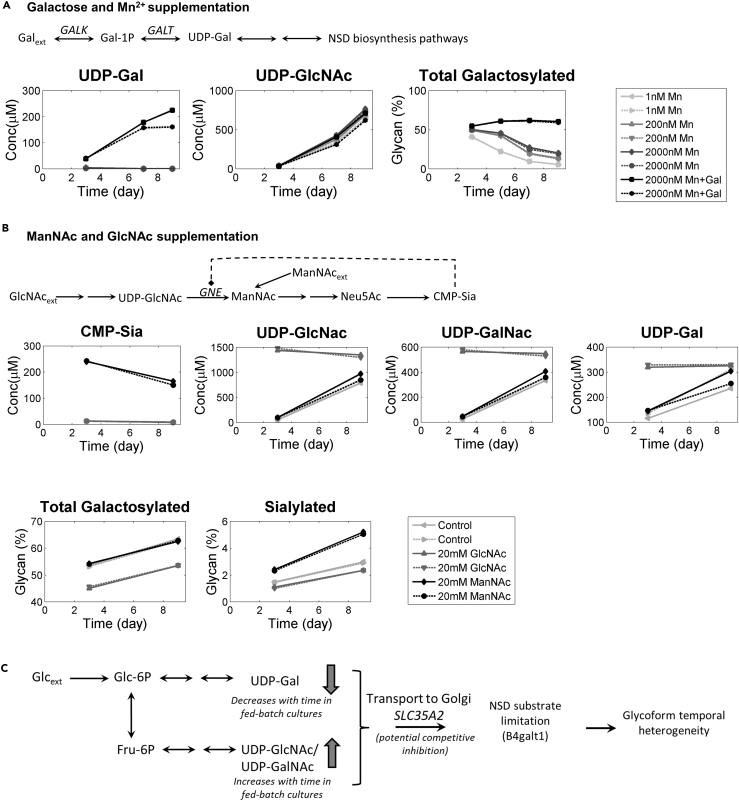
To explore if the drop in the substrate (UDP-Gal) and/or cofactor (Mn2+) levels of the enzyme that catalyzes galactosylation (B4galt1) is the root cause for the drop in the galactosylation of mAb glycoform produced, a pH-controlled fed-batch experiment was conducted in shake flasks with additional supplementation of galactose and/or Mn2+. Galactose can be converted intracellularly to UDP-Gal by the action of enzymes encoded by GALK and GALT, the expression of which was observed to be relatively constant over the course of the fed-batch culture (Figure S6A). The shake flask experiment was performed using media used in the CC and HD bioreactor runs, with varying levels of Mn2+ (0.9 nM, 200 nM and 2,000 nM) or with combined supplementation of 5 g/L galactose and 2,000 nM Mn2+. The additives were added to the culture on day 0. Manganese supplementation resulted in a decrease in cell growth in CC medium, whereas no such effect was observed in HD medium (Figure S6B). Time course glycan analysis suggested that although Mn2+ enhanced galactosylation, Mn2+ alone was insufficient to achieve high and sustained levels of galactosylation across the course of the culture (Figures 6A and S6). Interestingly, additional supplementation of galactose resulted in high and sustained levels of galactosylated species, which could be explained by the significantly higher UDP-Gal levels (Figure 6A). This established that maintaining higher levels of UDP-Gal and Mn2+ can help mitigate the drop in the galactosylation level over the course of a fed-batch culture.
Figure 6.
Media Supplementation Can Potentially Bypass the NSD Biosynthetic Pathway to Partially Mitigate Galactosylation and Sialylation Bottlenecks
(A) Supplementation of manganese to enhance B4galt1 activity alone is insufficient to reduce the bottleneck at galactosylation level, whereas galactose supplementation (5 g/L) leads to 25- to 50-fold increase in intracellular pool of UDP-Gal (left), resulting in sustained levels of galactosylation throughout the cell culture (right).
(B) Supplementation of 20 mM ManNAc bypasses the self-regulation on CMP-Sia biosynthesis (shown by dotted arrow) and leads to ∼30-fold increase in intracellular concentrations of CMP-sialic acid (left), resulting in around 2-fold increase in sialylated species.
(C) Temporal variations in the intracellular concentrations of the NSD substrates and/or potential substrate competition for intra-Golgi transport due to shared transport protein might result in the temporal heterogeneity in glycoforms observed in fed-batch cultures.
Next, the effect of enhancing the substrate level for sialyltransferases, namely, CMP-Sia, on total sialylation was probed. As inferred in the previous section, it was hypothesized that low levels of CMP-Sia in fed-batch cultures could be due to the presence of active inhibitory feedback regulation of epimerase activity of GNE by CMP-Sia, limiting its own formation (autoregulation). The epimerase encoded by GNE converts UDP-GlcNAc to ManNAc. Supplementation of downstream metabolite of GNE, namely, ManNAc, can bypass this autoregulation by CMP-Sia resulting in higher levels of CMP-Sia, which has been previously demonstrated (Gu and Wang, 1998). However, supplementation of an upstream metabolite, namely, GlcNAc, should not result in increase in CMP-Sia. To test this, a fed-batch experiment with either no supplementation (control) or with supplementation of 20 mM ManNAc or GlcNAc was performed in pH-controlled shake flasks. For this experiment, HD medium supplemented with 2,000 nM Mn2+ and 5 g/L of galactose was used to ensure maximum possible levels of galactosylation. Day 3 and day 9 intracellular NSD and mAb glycan levels were measured for all conditions (Figures 6B and S7A). A 2-fold increase in sialylation was observed in case of ManNAc-supplemented media, but no such increase in sialylation was observed for GlcNAc-supplemented or control conditions (Figures 6B and S7B). In fact, a small drop in the sialylation was observed in the GlcNAc-supplemented condition when compared with the control condition, which could be attributed to a proportional drop in galactosylation observed (when compared with control) (Figures 6B and S7). The increase in the mAb sialylation in case of ManNAc supplementation could be explained by an ∼24-fold increase in intracellular CMP-Sia levels compared to the control condition (Figure 6B). No significant change in CMP-Sia levels was observed in the GlcNAc-supplemented condition when compared to the control condition.
Discussion
A fundamental challenge with therapeutic protein production using CHO fed-batch cultures is the heterogeneous glycoforms observed on the recombinant proteins at harvest. Glycoform heterogeneity at harvest is a cumulative effect of transient glycoform distribution integrated over the cell culture period. The transient glycoform distribution varies from one time point in culture to another. This heterogeneity in the transient glycoform distribution appears to be contributed by the changing glycosylation state of the cells with the progress of a fed-batch culture. In other words, the glycoforms produced at any given time point seem to change with time in a fed-batch culture (Figure S1C). Such temporal changes in glycosylation state could be a result of variation in multiple intrinsic factors including glycosyltransferase enzymes (Könitzer et al., 2015), NSDs (Hutter et al., 2017, Sha et al., 2016), by-products from central energy metabolism like ammonia (Gawlitzek et al., 2000), cofactors for glycosylation enzymes (Witsell et al., 1990), and host cell protein production (Park et al., 2017). To understand the underlying cause for dynamics of protein N-glycosylation across fed-batch cultures and identify key factors contributing to it, we employed a systems approach combining time course transcriptomics, metabolomics, and glycan analysis for a model CHO cell line producing a recombinant antibody in two variants of fed-batch culture (Figures 1 and 2).
Functional analysis of the omics data showed that a significant part of the time course variance in transcriptome, metabolome, and glycoform, at a global level, appeared to be a function of culture time and that a smaller fraction of the variance depended on the type of medium/process employed (Figure 3). Analysis of the glycoforms produced showed that one of the bottlenecks in the glycosylation pathway could be at the reaction step, which adds galactose to the extending glycan moiety (Figure S2). Employing statistical analysis approaches including GSEA, TCGSA, TCMSA, and maSigPro on transcriptome and metabolome data, it was observed that a number of glycosylation-affecting factors became more dynamically perturbed over the course of the culture, including metabolic pathways such as glycolysis, TCA cycle, pyrimidine/purine metabolism, nucleotide sugar synthesis, and other functional classes such as glycosidases (Table 1). The changes observed across the various metabolic pathways were due to changes at transcript or metabolite intermediate levels or in some cases due to both occurring simultaneously.
Glycosyltransferases are the primary factors that affect glycoform heterogeneity as they catalyze addition of sugar moieties to the extending glycan chain on a secretory protein (McDonald et al., 2016). The transcript levels of a number of glycosyltransferases were observed to change significantly over the culture time (Figure S5). However, these changes did not correlate with the glycoforms produced. For example, levels of galactosyltransferase (B4GALT1) increased with time, whereas galactosylation decreased with time. Similarly, 2-beta-N-acetylglucosaminyltransferase (MGAT2) decreased with time, whereas agalactosylated species increased with time. No significant dynamics was observed for alpha-(1,6)-fucosyltransferase (FUT8), whereas a marginal decrease in fucosylated species was observed over time. Sialylated species decreased over time in both processes, which could not be explained by the constant or marginal increased transcript levels of sialyltransferases (ST3GALT3 and ST3GAL4). This suggested that the temporal variation of galactosylation and sialylation could be at the biochemical levels, i.e., enzyme cofactors and substrates (NSDs) (Hutter et al., 2017, Sha et al., 2016).
Glycolytic intermediates G6P and F6P are key carbon sources for all the NSD biosynthesis. Dynamics in the levels of glycolytic pathway activity can directly affect the NSD levels. Glycolytic pathway was observed to be enriched in growth phase with a number of glycolytic enzymes and glycolytic intermediates demonstrating time dynamics (Table 1, Data S12 and S13). Levels of G6P (and other upper glycolysis intermediates) decreased over time in the production phase of the cultures. Interestingly, the dynamics of intermediates in lower glycolytic pathway intermediates, namely 3-Phosphoglycerate (3PG) and Phophoenolpyruvate (PEP), increased over time. Glycolytic intermediates are known to have feedforward and feedback allosteric regulation on the upstream and downstream glycolysis enzymes imparting non-linear behavior to the flux and intermediate concentrations (Mulukutla et al., 2014, Mulukutla et al., 2015). The dynamics observed in the levels of G6P and lower glycolysis intermediates could be an outcome of such non-linear allosteric regulations at play in addition to enzyme level changes.
Decrease in the levels of G6P in the production phase should result in a corresponding fall in the intracellular levels of the UDP-Gal, UDP-Man, and UDP-GlcNAc, which are NSDs produced directly from G6P through a chain of reactions. However, only UDP-Gal levels decreased with time, whereas UDP-GlcNAc and GDP-Man increased over time in the production phase of the cultures. This suggests that there are additional regulations within the UPD-GlcNAc and GDP-Man biosynthetic pathway segments. For example, increase in the UDP-GlcNAc levels could be due to increase in the levels of UDP-GlcNAc pyrophosphorylase encoded by the gene UAP1, an enzyme that catalyzes the conversion of GlcNAc-1P to UDP-GlcNAc (Figure 5B, Data S12). The increase in UDP-GlcNAc happens even though the levels of its direct precursor, GlcNAc-1P, decrease with time, which further strengthens the argument that increase in the levels of UAP1 could be at play here.
Levels of GDP-Fuc and CMP-Sia could be influenced by the levels of their precursors, GDP-Man and UDP-GlcNAc, respectively. Interestingly, the intracellular levels of GDP-Fuc remain relatively constant, although GDP-Man increases over the last 2 days of the culture. One explanation for this could be the decrease in the levels of fucose synthase enzyme (TSTA3) in the last 2 days of the culture. Similarly, even though UDP-GlcNAc levels increase significantly, only marginal increase in the levels of CMP-Sia were observed. This could be explained by the negative feedback regulation of an irreversible enzyme, GNE, in CMP-Sia biosynthetic pathway by itself. Supplementation of the intermediates downstream of the GNE, ManNAc, led to a significant increase in the CMP-Sia, further supporting the argument that auto feedback regulation could be at play. This regulation was also previously reported to be at play in CHO cells (Gu and Wang, 1998).
UDP-Gal, the NSD for the step of galactosylation, decreases with time for both the conditions (CC and HD) (Figure 5A). Assessing if the temporal changes in the NSD levels affect the glycoforms being produced, a clear correlation was observed between temporal decrease in protein galactosylation and intracellular levels of UDP-Gal. Another well-known factor that affects the level of galactosylation is manganese (Mn2+), a cofactor that enhances B4galt1 activity (Witsell et al., 1990). Mn2+ concentration decreases with time even for the condition with higher Mn2+ (HD-1 and HD-2) (Figure S1D). Although manganese enhances galactosylation in a dose-dependent manner, supplementation of manganese alone fails to sustain high levels of galactosylation throughout the course of the culture (Figure 6A). Interestingly, in the presence of Mn2+, supplementation of galactose to fed-batch cultures, which increased the intracellular UDP-Gal levels by 25- to 100-fold results in sustained levels of galactosylation for the entire cell culture period (Figure 6A). Galactose supplementation bypasses the glycolytic pathway for UDP-Gal biosynthesis, and galactose is converted to UPD-Gal by the Leloir pathway. The transcript levels of enzymes in the Leloir pathway, namely, GALK1, GALK2, GALT, and GALE, either increase or stay relatively constant across the fed-batch cultures (Figure S6A).
Furthermore, temporal reduction in galactosylation could also be partly due to the inhibition of UDP-Gal transport into trans-Golgi cisternae by increased levels of UDP-GalNAc and UDP-GlcNAc, observed in the late stages of the culture (Figure 5A). The NSD transport protein encoded by SLC35A2 not only serves as a transporter for UDP-Gal, but is also reported to transport UDP-GalNAc (Song, 2013) and UDP-GlcNAc (Hadley et al., 2014). Therefore, each one of them can competitively inhibit transport of the other substrate. When cells were cultivated in GlcNAc-supplemented media, a significant increase in intracellular concentrations of UDP-Gal, UDP-GlcNAc, and UDP-GalNAc were observed (Figures 6B and S7A). This resulted in reduction in galactosylation when compared with control (Figures 6B and S7B). Interestingly, the ratio of UDP-Gal to UDP-GalNAc or UDP-GlcNAc was lower in the GlcNAc-treated condition when compared with control. These data further substantiate the hypothesis that increase in UDP-GalNAc and/or UDP-GlcNAc might negatively influence galactosylation (Figure 6C).
We also notice that UDP-Gal showed an increasing trend over time in a fed-batch culture upon galactose supplementation. Based on our hypothesis UDP-HexNAc (UDP-GlcNAc and UDP-GalNAc) competes with UDP-Gal for transport to Golgi. UDP-HexNAc was observed to increase over time intracellularly (Figure 6A). In the presence of such higher levels of UDP-HexNAc, and under sufficient availability of galactose, it is possible that to maintain the same flux of UDP-Gal through Golgi, the cytosolic UDP-Gal concentrations could increase.
Our study also shows that sialylation levels are very low and decrease with time. Galactose supplementation did not increase the peak levels of sialylation observed across the culture but helped prevent the drop in the sialylation observed during the production phase of the culture (Figure S6). This suggested that one reason behind temporal heterogeneity of sialylation is the heterogeneity in galactosylation levels. Another reason for low levels of sialylation could be the relatively lower levels of the corresponding NSD substrate (CMP-Sia). Evidently, supplementation of ManNAc increased the intracellular levels of CMP-Sia, which in turn enhanced the levels of sialylation. Interestingly, although ManNAc supplementation resulted in ∼30-fold increase in the intracellular pool of CMP-Sia, the overall increase in sialylation was significantly low (from ∼3% to ∼5.5%). This suggests that, apart from the substrate limitations (availability of galactosylated species and CMP-Sia), other factors might be at play. These include, but are not limited to, low expression levels of sialyltransferases, reduced residence time for sialylation in trans-Golgi networks, and steric hindrance for sialyltransferases for glycan chain extension in case of mAbs (Dekkers et al., 2016).
Another potential factor that can influence temporal heterogeneity in glycan profile is the glycosidase activity. Glycosidases are prominently present in the extracellular matrix of the CHO fed-batch cultures for mAb production (Park et al., 2017). Addition of neuraminidases (glycosidases for sialic acid) results in reduced levels of sialylation in fed-batch cultures (Gramer et al., 1995). Functional analyses of the transcriptome data suggested that these glycosidases are significantly expressed and are enriched during production phase. We tested the effect of glycosidases present in the extracellular matrix by incubating the day 9 cell-free supernatants from cell cultures for 3 days at 36.5°C. When these samples were analyzed for changes in the glycan profile over the 3 days, only a slight reduction in the sialylation levels were observed (from ∼2.9% to ∼2.5%), whereas no significant differences were observed for other glycosylated species (data not shown). A possible reason why the effect was minimal is because the fed-batch cultures were maintained in the strict pH range of 6.9–7.2, whereas the optimum pH for most of these glycosidase activity is within the pH range of 4–6 (Gramer and Goochee, 1993, Miyagi and Yamaguchi, 2012). Intracellularly, these glycosidases are known to localize primarily in lysosomes (Aronson and Kuranda, 1989, Miyagi and Yamaguchi, 2012) where pH is lower and suitable for their activity, such as participating in salvage pathways for misfolded proteins. Thus, despite exhibiting significant time dynamics, barring sialidases, other glycosidases might not directly influence the N-glycosylation dynamics in fed-batch cultures.
Functional analysis of the omics data also suggested genes and functional classes that might act as potential competitors for the common substrates (NSDs) and might affect the glycosylation dynamics. These include enzymes/genes that are involved in O-linked glycosylation, glycosaminoglycan, and glycosphingolipid synthesis. Although not completely understood, given the co-localization of some of these enzymes with those involved in N-glycosylation, these enzymes might compete for NSDs and even cofactors, thus influencing the dynamics. Similarly, the level and type of host cell glycoproteins secreted over the course of the culture might also affect the temporal changes in glycosylation of the mAb being produced. In addition, the specific productivity can potentially affect the residence time of the mAb within Golgi cisternae. Temporal changes in specific productivity can, therefore, result in the changes in the glycoforms produced with time. Additional work is needed to assess the role of these factors on glycosylation.
In conclusion, N-glycosylation is a complex dynamic process influenced by several pathways including NSD biosynthesis, glycolysis, and other processes that compete for enzymes or substrates. Systematic analysis of time course omics data identified the primary cause for glycosylation dynamics in fed-batch cultures as changes in the substrate availability. The holistic understanding of glycosylation network dynamics helped contrive strategies to overcome the bottlenecks toward reduction in the heterogeneity observed in fed-batch culture. Going forward, this understanding can be used to implement additional strategies to perform both process- and genetic-engineering-related modifications toward further reducing the glycoform heterogeneity observed in fed-batch cultures.
Limitations of the Study
In this study, in addition to identifying the cause for dynamics in N-glycosylation pathway activity and the glycoform produced, we made a number of observations that are intriguing but need to be probed further experimentally. First, we showed using one cell line with two distinct cell culture processes that glycoform dynamics are dominantly time dependent and less process dependent. Further experimentation with additional producing CHO cell lines across different lineages employing different fed-batch processes will be needed to be analyzed before fully establishing the generality of the above argument. Second, we identified other glycosylation-related pathways, including sphingolipid, glycerolipid, and aminoglycan biosynthesis, to be modulated in growth or production phases of the fed-batch processes. More experimentation is needed to probe the effect of competition for resources shared (nucleotide sugars, cofactors, and glycosylation enzymes) by these pathways along with protein glycosylation on the recombinant protein glycoform. Third, we observed that an increase in UPD-GlcNAc levels results in lower galactosylation. Experimentation is needed for identifying the mechanism at play.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Authors thankfully acknowledge Gregory Hiller and Dana L. DiNino for their comments and suggestions during manuscript preparation.
Author Contributions
M.S. and B.C.M. conceived and designed the experiments. M.S. performed the experiments. M.S. and S.D. analyzed the data. M.S., S.D., and B.C.M. wrote the manuscript. A.-H.A.C. and J.J.S. generated the experimental cell clone. A.-H.A.C., K.C., and J.K.M. performed glycan and nucleotide sugar analyses. A.-H.A.C., J.J.S.,A.A.C., J.K.M., R.W., D.A.L., B.F., and B.C.M. consulted on data analysis and edited the manuscript.
Declaration of Interests
The authors declare no conflict of interest.
Published: February 22, 2019
Footnotes
Supplemental Information includes Transparent Methods, 7 figures, and 15 data files and can be found with this article online at https://doi.org/10.1016/j.isci.2019.01.006.
Supplemental Information
References
- Aronson N.N., Kuranda M.J. Lysosomal degradation of Asn-linked glycoproteins. FASEB J. 1989;3:2615–2622. doi: 10.1096/fasebj.3.14.2531691. [DOI] [PubMed] [Google Scholar]
- Berger M., Kaup M., Blanchard V. Genomics and Systems Biology of Mammalian Cell Culture. Springer; 2011. Protein glycosylation and its impact on biotechnology; pp. 165–185. [DOI] [PubMed] [Google Scholar]
- Conesa A., Nueda M.J., Ferrer A., Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- Dekkers G., Plomp R., Koeleman C.A.M., Visser R., Von Horsten H.H., Sandig V., Rispens T., Wuhrer M., Vidarsson G. Multi-level glyco-engineering techniques to generate IgG with defined Fc-glycans. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep36964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eon-Duval A., Broly H., Gleixner R. Quality attributes of recombinant therapeutic proteins: an assessment of impact on safety and efficacy as part of a quality by design development approach. Biotechnol. Prog. 2012;28:608–622. doi: 10.1002/btpr.1548. [DOI] [PubMed] [Google Scholar]
- Fan Y., Jimenez Del Val I., Müller C., Wagtberg Sen J., Rasmussen S.K., Kontoravdi C., Weilguny D., Andersen M.R. Amino acid and glucose metabolism in fed-batch CHO cell culture affects antibody production and glycosylation. Biotechnol. Bioeng. 2015;112:521–535. doi: 10.1002/bit.25450. [DOI] [PubMed] [Google Scholar]
- Gawlitzek M., Ryll T., Lofgren J., Sliwkowski M.B. Ammonium alters N-glycan structures of recombinant TNFR-IgG: degradative versus biosynthetic mechanisms. Biotechnol. Bioeng. 2000;68:637–646. doi: 10.1002/(sici)1097-0290(20000620)68:6<637::aid-bit6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Gramer M.J., Goochee C.F. Glycosidase activities in Chinese hamster ovary cell lysate and cell culture supernatant. Biotechnol. Prog. 1993;9:366–373. doi: 10.1021/bp00022a003. [DOI] [PubMed] [Google Scholar]
- Gramer M.J., Goochee C.F., Chock V.Y., Brousseau D.T., Sliwkowski M.B. Removal of sialic acid from a glycoprotein in CHO cell culture supernatant by action of an extracellular cho cell sialidase. Biotechnology (N. Y.) 1995;13:692–698. doi: 10.1038/nbt0795-692. [DOI] [PubMed] [Google Scholar]
- Gu X., Wang D.I. Improvement of interferon-gamma sialylation in Chinese hamster ovary cell culture by feeding of N-acetylmannosamine. Biotechnol. Bioeng. 1998;58:642–648. [PubMed] [Google Scholar]
- Hackett S.R., Zanotelli V.R.T., Xu W., Goya J., Park J.O., Perlman D.H., Gibney P.A., Botstein D., Storey J.D., Rabinowitz J.D. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science. 2016;354:aaf2786. doi: 10.1126/science.aaf2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley B., Maggioni A., Ashikov A., Day C.J., Haselhorst T., Tiralongo J. Structure and function of nucleotide sugar transporters: current progress. Comput. Struct. Biotechnol. J. 2014;10:23–32. doi: 10.1016/j.csbj.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley F., Walker T., Chung V., Morten K. Mechanisms driving the lactate switch in Chinese hamster ovary cells. Biotechnol. Bioeng. 2018;115:1890–1903. doi: 10.1002/bit.26603. [DOI] [PubMed] [Google Scholar]
- Hejblum B.P., Skinner J., Thiébaut R. Time-course gene set analysis for longitudinal gene expression data. PLoS Comput. Biol. 2015;11:1–21. doi: 10.1371/journal.pcbi.1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossler P., Goh L.-T., Lee M.M., Hu W.-S. GlycoVis: Visualizing glycan distribution in the proteinN-glycosylation pathway in mammalian cells. Biotechnol. Bioeng. 2006;95:946–960. doi: 10.1002/bit.21062. [DOI] [PubMed] [Google Scholar]
- Hossler P., Mulukutla B.C., Hu W.-S.S. Systems analysis of N-glycan processing in mammalian cells. PLoS One. 2007;2:e713. doi: 10.1371/journal.pone.0000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-H., Araki M., Mochizuki M., Hori Y., Murata M., Kahar P., Yoshida T., Hasunuma T., Kondo A. A systematic approach to time-series metabolite profiling and RNA-seq analysis of Chinese hamster ovary cell culture. Sci. Rep. 2017;7:43518. doi: 10.1038/srep43518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S., Villiger T.K., Brühlmann D., Stettler M., Broly H., Soos M., Gunawan R. Glycosylation flux analysis reveals dynamic changes of intracellular glycosylation flux distribution in Chinese hamster ovary fed-batch cultures. Metab. Eng. 2017;43:9–20. doi: 10.1016/j.ymben.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Ivarsson M., Villiger T.K., Morbidelli M., Soos M. Evaluating the impact of cell culture process parameters on monoclonal antibody N-glycosylation. J. Biotechnol. 2014;188:88–96. doi: 10.1016/j.jbiotec.2014.08.026. [DOI] [PubMed] [Google Scholar]
- Jedrzejewski P.M., del Val I.J., Constantinou A., Dell A., Haslam S.M., Polizzi K.M., Kontoravdi C. Towards controlling the glycoform: a model framework linking extracellular metabolites to antibody glycosylation. Int. J. Mol. Sci. 2014;15:4492–4522. doi: 10.3390/ijms15034492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez del Val I., Nagy J.M., Kontoravdi C. A dynamic mathematical model for monoclonal antibody N-linked glycosylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnol. Prog. 2011;27:1730–1743. doi: 10.1002/btpr.688. [DOI] [PubMed] [Google Scholar]
- Jiménez del Val I., Constantinou A., Dell A., Haslam S., Polizzi K.M., Kontoravdi C. A quantitative and mechanistic model for monoclonal antibody glycosylation as a function of nutrient availability during cell culture. BMC Proc. 2013;7:O10. [Google Scholar]
- Kaveh O., Hengameh A., Johannes G., Murray M.-Y., Legge R.L., Jeno S., Budman H.M. Novel dynamic model to predict the glycosylation pattern of monoclonal antibodies from extracellular cell culture conditions. IFAC. 2013;46(31):30–35. [Google Scholar]
- Könitzer J.D., Müller M.M., Leparc G., Pauers M., Bechmann J., Schulz P., Schaub J., Enenkel B., Hildebrandt T., Hampel M. A global RNA-seq-driven analysis of CHO host and production cell lines reveals distinct differential expression patterns of genes contributing to recombinant antibody glycosylation. Biotechnol. J. 2015;10:1412–1423. doi: 10.1002/biot.201400652. [DOI] [PubMed] [Google Scholar]
- Krambeck F.J., Betenbaugh M.J. A mathematical model of N-linked glycosylation. Biotechnol. Bioeng. 2005;92:711–728. doi: 10.1002/bit.20645. [DOI] [PubMed] [Google Scholar]
- Krambeck F.J., Bennun S.V., Narang S., Choi S., Yarema K.J., Betenbaugh M.J. A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology. 2009;19:1163–1175. doi: 10.1093/glycob/cwp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N.E., Liu X., Li Y., Nagarajan H., Yerganian G., O’Brien E., Bordbar A., Roth A.M., Rosenbloom J., Bian C. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat. Biotechnol. 2013;31:759–765. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- Lin N., Mascarenhas J., Sealover N.R., George H.J., Brooks J., Kayser K.J., Gau B., Yasa I., Azadi P., Archer-Hartmann S. Chinese hamster ovary (CHO) host cell engineering to increase sialylation of recombinant therapeutic proteins by modulating sialyltransferase expression. Biotechnol. Prog. 2015;31:334–346. doi: 10.1002/btpr.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maverakis E., Kim K., Shimoda M., Gershwin M.E., Patel F., Wilken R., Raychaudhuri S., Ruhaak L.R., Lebrilla C.B. Glycans in the immune system and the altered glycan theory of autoimmunity: a critical review. J. Autoimmun. 2015;57:1–13. doi: 10.1016/j.jaut.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald A.G., Hayes J.M., Davey G.P. Metabolic flux control in glycosylation. Curr. Opin. Struct. Biol. 2016;40:97–103. doi: 10.1016/j.sbi.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Miyagi T., Yamaguchi K. Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- Mulukutla B.C., Gramer M., Hu W.S. On metabolic shift to lactate consumption in fed-batch culture of mammalian cells. Metab. Eng. 2012;14:138–149. doi: 10.1016/j.ymben.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Mulukutla B.C., Yongky A., Daoutidis P., Hu W.S. Bistability in glycolysis pathway as a physiological switch in energy metabolism. PLoS One. 2014;9:e98756. doi: 10.1371/journal.pone.0098756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulukutla B.C., Yongky A., Grimm S., Daoutidis P., Hu W.S. Multiplicity of steady states in glycolysis and shift of metabolic state in cultured mammalian cells. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0121561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulukutla B.C., Yongky A., Le T., Mashek D.G., Hu W.S. Regulation of glucose metabolism – a perspective from cell bioprocessing. Trends Biotechnol. 2016;34:638–651. doi: 10.1016/j.tibtech.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Nairn A.V., York W.S., Harris K., Hall E.M., Pierce J.M., Moremen K.W. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 2008;283:17298–17313. doi: 10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R., Satoh M. The current status and prospects of antibody engineering for therapeutic use: focus on glycoengineering technology. J. Pharm. Sci. 2015;104:930–941. doi: 10.1002/jps.24316. [DOI] [PubMed] [Google Scholar]
- Opdam S., Richelle A., Kellman B., Li S., Zielinski D.C., Lewis N.E. A Systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst. 2017;4:318–329.e6. doi: 10.1016/j.cels.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Jin J.H., Lim M.S., An H.J., Kim J.W., Lee G.M. Proteomic analysis of host cell protein dynamics in the culture supernatants of antibody-producing CHO cells. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep44246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish J.G., Sethu S., Bielsky M.-C., de Haan L., French N.S., Govindappa K., Green J., Griffiths C.E.M., Holgate S., Jones D. Challenges and approaches for the development of safer immunomodulatory biologics. Nat. Rev. Drug Discov. 2013;12:306–324. doi: 10.1038/nrd3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha S., Agarabi C., Brorson K., Lee D.-Y., Yoon S. N-glycosylation design and control of therapeutic monoclonal antibodies. Trends Biotechnol. 2016;34:835–846. doi: 10.1016/j.tibtech.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Sha S., Bhatia H., Yoon S. An RNA-seq based transcriptomic investigation into the productivity and growth variants with Chinese hamster ovary cells. J. Biotechnol. 2018;271:37–46. doi: 10.1016/j.jbiotec.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Song Z. Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol. Aspects Med. 2013;34:590–600. doi: 10.1016/j.mam.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Sou S.N., Sellick C., Lee K., Mason A., Kyriakopoulos S., Polizzi K.M., Kontoravdi C. How does mild hypothermia affect monoclonal antibody glycosylation? Biotechnol. Bioeng. 2015;112:1165–1176. doi: 10.1002/bit.25524. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Münch A., Neumann S., Kremmer E., Tatzelt J., Lichtenthaler S.F. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J. Biol. Chem. 2010;285:20664–20674. doi: 10.1074/jbc.M109.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiger T.K., Roulet A., Perilleux A., Stettler M., Broly H., Morbidelli M., Soos M. Controlling the time evolution of mAb N-linked glycosylation, Part I: Microbioreactor experiments. Biotechnol. Prog. 2016;32:1123–1134. doi: 10.1002/btpr.2305. [DOI] [PubMed] [Google Scholar]
- Villiger T.K., Scibona E., Stettler M., Broly H., Morbidelli M., Soos M. Controlling the time evolution of mAb N-linked glycosylation - Part II: model-based predictions. Biotechnol. Prog. 2016;32:1135–1148. doi: 10.1002/btpr.2315. [DOI] [PubMed] [Google Scholar]
- Witsell D.L., Casey C.E., Neville C. Divalent cation of galactosyltransferase in native activation mammary golgi vesicles. J. Biol. Chem. 1990;265:15731–15737. [PubMed] [Google Scholar]
- Yardeni T., Choekyi T., Jacobs K., Ciccone C., Patzel K., Anikster Y., Gahl W.A., Kurochkina N., Huizing M. Identification, tissue distribution, and molecular modeling of novel human isoforms of the key enzyme in sialic acid synthesis, UDP-GlcNAc 2-epimerase/ManNAc kinase. Biochemistry. 2011;50:8914–8925. doi: 10.1021/bi201050u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.