Abstract
A high-throughput screening assay for modulators of Trp53/NF1 mutant astrocytoma cell growth was adapted for use with natural product extracts and applied to a novel collection of prefractionated/partially purified extracts. Screening 68 427 samples identified active fractions from 95 unique extracts, including the terrestrial plant Millettia ichthyotona. Only three of these extracts showed activity in the crude extract form, thus demonstrating the utility of a partial purification approach for natural product screening. The NF1 screening assay was used to guide purification of active compounds from the M. ichthyotona extract, which yielded the two rotenones deguelin (1) and dehydrodeguelin (2). The deguelins have been reported to affect growth of a number of cancer cell lines. They potently inhibited growth of only one of a panel of NF1/Trp53 mutant murine astrocytoma cell lines, possibly related to epigenetic factors, but had no effect on the growth of normal astrocytes. These results suggest the potential utility of deguelins as tools for further investigating NF1 astrocytoma cell growth. These bioprobes were identified only as a result of screening partially purified natural product extracts.
Graphical Abstract
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disorder in which patients carry a loss-of-function mutation on the NF1 gene (Nf1 in mice), resulting in increased risk for a variety of malignancies.1–3 Neurofibromin, the product of the NF1 gene, is a ras GTPase activator protein (RasGAP) that acts as a tumor suppressor protein. Loss or mutational inactivation of neurofibromin contributes to tumorigenesis via uncontrolled ras signaling, leading to increased cellular proliferation and survival. In order to better understand NF1-driven oncogenesis and to develop potential therapeutics, a mouse model for astrocytoma/glioblastoma was developed by incorporating mutations of two tumor suppressor genes, Nf1 and Trp53, encoding the p53 protein, on the same copy of Chr 11 (in cis).4 Astrocytoma cell lines have been derived from these Nf1−/+;Trp53−/+cis mice for characterization of astrocytoma/glioblastoma development4,5 and configuration of a high-throughput screening (HTS) assay for modulators of astrocytoma growth and survival.6
Anaplastic astrocytoma (WHO grade III) is a malignant brain tumor that is currently incurable. It makes up 6.1% of all gliomas7 and can progress to glioblastoma multiforme (GBM). The five-year survival rate for anaplastic astrocytoma is 27.3%, highlighting the need to develop new therapies. The NF1 gene is mutated in 18% of anaplastic astrocytomas and 11% of GBM. In addition, the tumor suppressor TP53 (homologous to the Trp53 in mice) is mutated in 56% of anaplastic astrocytoma (TCGA Research Network http://cancergenome.nih.gov).8 Since few cell lines exist to study anaplastic astrocytoma, the use of astrocytoma cell lines from the Nf1−/+;Trp53−/+cis mouse model facilitates screening for candidate therapeutics for this disease.
This assay has now been optimized for application to natural product extracts. Natural products have been and continue to be rich sources for biological probes and drugs. For example, the majority of current anticancer drugs are natural products or natural product derivatives.9 The starting points for initial identification of biologically active natural products are crude extracts from a wide variety of organisms. However, screening with natural product extracts is fraught with difficulties, including but not limited to the presence of nonspecific or cytotoxic compounds.10 In order to maximize the probability of finding specific antiastrocytoma activities in natural product extracts, two approaches were taken. The first was the application of a well-validated assay able to identify growth inhibitory activity via a dual-luciferase reporter system6 that may be less susceptible to interference by nonspecific effects. Second, a library of natural product extracts was screened that had been partially purified by “prefractionation” (i.e., chromatographic separation of crude extracts into five fractions prior to assay) in order to concentrate minor constituents into a chromatographic fraction, which resulted in trace metabolites being screened at higher effective concentrations in the assay. Prefractionation can also serve to remove or sequester interfering or nonspecifically cytotoxic components of an extract. The combination of a dual readout assay and the screening of prefractionated samples led to the identification of an active extract derived from the plant Millettia ichthyotona Drake (Fabaceae) that yielded deguelin (1) and dehydrodeguelin (2).11–13 These compounds subsequently showed significant, potent growth inhibitory activity against Nf1- and Trp53-null astrocytoma cells derived from the Nf1−/+;Trp53−/+cis mouse model4,5 without affecting growth of primary astrocytes.
RESULTS AND DISCUSSION
It is well recognized that extract prefractionation can greatly reduce many of the problematic issues associated with screening intact extracts, and the use of prefractionated libraries to facilitate screening, dereplication, and compound identification has been reported by both academic14–19 and industrial20–22 natural products programs. The most common approach is to employ a high-resolution HPLC separation with UV, ELSD, and MS detection and automated collection of the resulting fractions in a plate format that is compatible with bioassay requirements. This results in fractions that contain only a small number of constituents, but the mass of each resulting fraction and therefore the concentration used for screening are unknown. Potential limitations to this approach are the increased screening requirements for each extract that is processed and the time and resources necessary to perform the HPLC separations. An alternative strategy involves adsorption of an extract onto a solid-phase extraction (SPE) cartridge and elution based on polarity. This is a high-throughput, low-cost method that can be applied to a large number of samples, but the resulting fractions are still relatively complex mixtures. An SPE-based prefractionation protocol was developed and applied to a large taxonomically diverse collection of crude natural product extracts sourced from the NCI Natural Products Repository. Organic solvent extracts were separated into five fractions of increasing polarity on a diol stationary phase column: fraction A, eluted with hexane/CH2Cl2 (9:1); fraction B, eluted with CH2Cl2/EtOAc (20:1); fraction C, eluted with 100% EtOAc; fraction D, eluted with EtOAc/MeOH (5:1); and fraction E, eluted with 100% MeOH. Aqueous extracts were chromatographed on a fully water-wettable macroporous poly(divinylbenzene-co-N-vinylpyrrolidone) polymeric reversed-phase sorbent (Oasis HLB) to give five fractions of increasing lipophilicity: fractions A and B, eluted with 100% H2O; fraction C, eluted with H2O/MeOH (3:1); fraction D, eluted with H2O/MeOH (1:3); and fraction E, eluted with MeOH/CH2Cl2 (9:1). The SPE fractions and an aliquot of unprocessed crude extract (six samples total for each extract) comprise the “prefractionated natural product library” (PNPL) used for screening. To date 29 183 extracts have been processed to generate 172 961 samples in the PNPL.23
The dual-luciferase screening assay, described in detail in the Experimental Section, consists of a cell proliferation readout (green luminescence) and a cell number/cell health readout (red luminescence). In order to assess applicability of the assay to natural products, a selection of 1408 prefractionated samples was chosen to be representative of the types of extracts present in the library. These were then run in the screening assay twice each at 1 and 10 μg/mL to assess reproducibility of the assay with natural product samples and to establish the most appropriate concentration to use in subsequent screening, as well as to define hit criteria. Figure 1 shows the results for 10 μg/mL and Table 1 summarizes results for both concentrations.
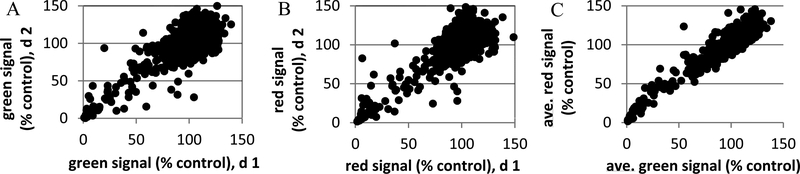
Figure 1.
Applicability and stability of dual luminescence assay for natural product samples run on two different days. Cells were treated with 1408 representative prefractionated samples as described in the text. Signals were normalized to untreated controls on the same plate and expressed as % of control value: (A) green (cell proliferation) signal, (B) red (cell health) signal, (C) comparison of the two signals (average values, n = 2 days for each signal). Values representative of reproducibility are presented in Table 1.
Table 1.
Assay Repeatability with 1408 Samples over Two Days
| 1 μg/mL |
10 μg/mL |
|||
|---|---|---|---|---|
| green | red | green | red | |
| overall average | 103.2 | 101.9 | 99.2 | 99.6 |
| SD | 18.2 | 18.6 | 24.7 | 24.3 |
| slope (day 1 vs day 2) | 0.69 | 0.58 | 0.84 | 0.90 |
| corr | 0.64 | 0.45 | 0.85 | 0.79 |
| average difference (day 1 vs day 2) | 12.3 | 13.9 | 10.8 | 10.4 |
As would be expected across a moderately large number of samples, the “average result” is no effect in the assay. Table 1 shows that, overall, signals from sample-treated wells were on average the same as those of untreated controls. Consistency of the signal from 1 day to the next was quite good for the samples at 10 μg/mL (slope and correlation coefficients near 1), but less so for 1 μg/mL. Repeat reads averaged within 10–11% of each other for the samples at 10 μg/mL, but were less consistent at 1 μg/mL. On the basis of these results, the subsequent screening procedure utilized samples at 10 μg/mL. Given that the first goal of the assay was to identify samples able to inhibit growth (i.e., reduce the green signal), a hit cutoff for screening of <20% of the control green signal was chosen. Definition of a hit cutoff for the red signal was more problematic in that it essentially measures overall transcriptional activity, thus reflecting cell numbers and general cell health, and the pilot assay statistics did not provide an obvious hit cutoff. However, assuming requirements for recovery of cells upon plating, lag times for depletion of cell cycle machinery upon initiation of growth inhibition (reflected by the green signal, reduction in E2F transcription), and cell growth kinetics, along with intrinsic variation in the signals (Figure 1, Table 1), a red signal of >50% of untreated control was chosen as a second cutoff. Figure 1C shows the comparison of green and red signals for the pilot experiment. None of the samples from this pilot experiment met these hit criteria, although several had <20% green signal and >40% red (i.e., borderline). The previously described screening assay for modulators of Nf1/Trp53-null astrocytoma growth thus proved to be adaptable, with minor modifications, for use with natural product samples. The derived hit definition resulted in identification of samples that significantly inhibit proliferation (i.e., low green luminescence signal) with minimal effect on cell health/viability (i.e., high red luminescence signal).
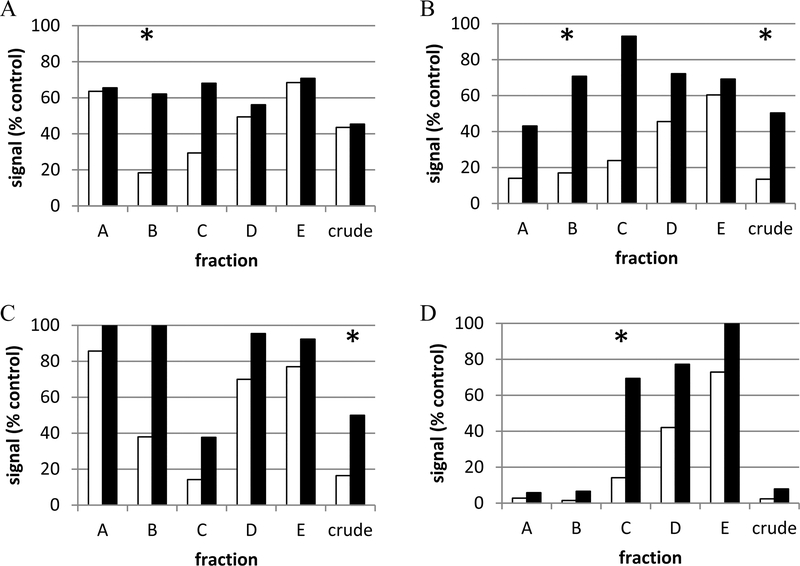
From a total of 68 427 natural product samples tested,24 101 fractions derived from 95 individual extracts (representing 86 different genera) were confirmed active after primary screening and reassay. Of these 95, there were 57 corresponding crude (unfractionated) extracts available for more detailed chemical study.25 Figure 2 shows examples of four types of activity profiles for fractions and the parental crude extracts. Figure 2A is an example of an extract for which the crude was inactive, but in this particular example, fraction B was active. Figure 2B shows an example of an extract for which the crude was active and there was also an active fraction (fraction B). Figure 2D shows a cytotoxic extract and prefractionation of this extract separated cytotoxic material (fractions A and B) from the desired activity (fraction C). There were also several apparently cytotoxic extracts from which active fraction(s) were obtained, but no cytotoxic material was recovered (not shown). Table 2 summarizes these results.
Figure 2.
Representative examples of the types of extracts yielding active samples. (A) Active fraction and inactive crude extract; (B) fraction and crude extract are active; (C) only crude extract is active; (D) toxic crude extract and active fraction. Open bars represent a green signal (i.e., proliferative index); black bars represent a red signal (i.e., cell health index) (* indicates active sample by the hit criteria discussed in the Results and Discussion section).
Table 2.
Categories of Extracts that Yielded Active Samples in the Screen
| category | crude extract | fraction(s) | % of total hits |
|---|---|---|---|
| A | inactive | active | 73.7 |
| B | active | active | 8.8 |
| C | active | inactive | 3.5 |
| D | toxic | active | 14.0 |
For the majority of active samples (86%), the crude extract was not a hit in the screening assay, but one or more of the fractions were. Thus, in this study, prefractionation resulted in the identification of at least 50 active samples that would not have been identified without partial purification prior to screening. Conversely, there were two examples for which the crude extract was active, but activity was not present in the fractions (category C in Table 2; example C in Figure 2). In order to maximize the probability of success in screening natural products, both crude extracts and partially purified samples should be assessed whenever possible.
The organic solvent extract of a Vietnamese collection of M. ichthyotona was part of the PNPL that was tested in the NF1 screen. It provided a prefractionation sample B, eluted from diol SPE with CH2Cl2/EtOAc (20:1), that had confirmed activity in the NF1 assay. A 2 g portion of the extract was separated by liquid–liquid extraction, which concentrated the NF1 activity into the methyl tert-butyl ether (MTBE)-soluble fraction. Further purification by gel permeation on Sephadex LH-20 (CH2Cl2/MeOH, 1:1) followed by C18 HPLC provided deguelin (1) and dehydrodeguelin (2) as the principal NF1active constituents. Compounds 1 and 2 were identified by comparison of their spectroscopic data with corresponding literature values.26–29
On the basis of the configuration of the screening assay and the activities of deguelin (1) and dehydrodeguelin (2) in this assay, these compounds would be expected to have cytostatic activity against astrocytoma cell lines. They were therefore tested in a growth assay against three mouse grade III astrocytoma lines as well as primary mouse astrocytes. The results are summarized in Table 3. Both compounds clearly had a cytostatic effect on the K5001 astrocytoma cell line (i.e., maximal inhibition of growth: 70–80% suggesting survival of nonproliferating cells based on cellular growth characteristics) and some effect on KR158 without significantly affecting growth of normal primary astrocytes or the K1492 astrocytoma line. Neither compound affected the growth of primary astrocytes derived from Nf1−/+;Trp53−/+cis mice (IC50 for growth inhibition, >80 μM). Half-maximal activity for both deguelin (1) and dehydrodeguelin (2) occurred at 1.2 ± 0.1 μM for K5001 growth inhibition and at >80 μM for all of the other cells tested, demonstrating a very strong bias toward this cell line. The astrocytoma cell lines tested in these experiments were derived from Nf1−/+;Trp53−/+cis mice, and the mice from which they were derived have been previously characterized.5,30 The three grade III lines, K5001, KR158, and K1492, were derived from female mice, and the parental source of the mutant chromosome was the father for K5001, but the mother for KR158 and K1492. This may be relevant in that it has been previously demonstrated that parentally derived epigenetic factors contribute to development of astrocytomas in the progeny in the mice from which the cell lines were developed.30
Table 3.
Effects of Deguelin (1) and Dehydrodeguelin (2) on Astrocytoma Cell Lines and Primary Astrocytes
| IC50 for growth inhibition (av ± SD) (μM) |
||||
|---|---|---|---|---|
| compound | K1492 | KR158 | K5001 | astrocytes |
| deguelin (1) | >80 | 12.6 ± 8.1 | 1.2 ± 0.11 | >80 |
| dehydrodeguelin (2) | >80 | >80a | 1.2 ± 0.07 | >80 |
40% growth inhibition at 80.
Interestingly, most of the mechanisms of action described in the literature for deguelin derivatives are related to apoptosis rather than inhibition of proliferation.13,31–33 However, inhibition of proliferation rather than apoptosis induction by deguelins in these astrocytoma cells was corroborated by an absence of detection of DNA fragmentation (an apoptosis end point) after up to 48 h treatment of the cell lines with deguelin (1) or dehydrodeguelin (2) (Figure S8, Supporting Information). Similarly, although inhibition of AKT was reported to lead to inhibition of growth of these cell lines5 and deguelin has been reported to be an AKT inhibitor,33,34 the deguelins were ineffective in inhibiting AKT activation as monitored by AKT phosphorylation (Figure S9, Supporting Information). It is possible that some effects of 1 may be mediated by generation of reactive oxygen species (ROS).35,36 When assessed using a fluorogenic probe, treatment of K5001 cells for 6 h with 10 μM deguelin was found to increase cellular ROS to (3.5 ± 0.25) × control. However, ROS increase was similar for the other two, nonsusceptible, cell lines, KR158 and K1492 ((3.9 ± 1.99) and (5.6 ± 0.34) × control, respectively). Thus, deguelin (1) induces ROS generation, but this does not account for its differential effects on these three astrocytoma cell lines. These differences in susceptibility to growth inhibition by the deguelins, suggestion of involvement of epigenetic factors or secondary mutations in the cell lines, and potential differential resistance to ROS may provide a novel means for further investigating factors relevant to astrocytoma development and progression. Finally, other as yet untested possible deguelin targets may give additional insights into these factors.
In conclusion, it is clear that deguelin derivatives may prove to be extremely valuable probes for increasing the understanding of astrocytoma development. These probes were identified as a result of a focused program for development of partially purified natural product extracts as a valuable high-throughput screening resource. Although the deguelins are well-known compounds, they have potential as novel reagents for studying astrocytoma. Screening of additional deguelin analogues, as well as other structurally related rotenones and pterocarpan derivatives, for NF1 related activity would also be warranted. In addition, the application of partially purified natural product extracts with carefully developed screening assays holds promise for identification of additional compounds that can serve as biological probes.
EXPERIMENTAL SECTION
Chemicals and Reagents.
General cell culture components were from Invitrogen (Carlsbad, CA, USA). Growth medium for primary astrocytes was obtained from ScienCell Research Laboratories (Carlsbad, CA, USA). Sterile polylysine-coated 384-well, white-wall, white bottom plates and sterile polylysine-coated 384-well, clear-wall, clear-bottom plates were from PerkinElmer (Waltham, MA, USA). 2,3-Bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide (XTT) was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (Frederick, MD, USA). Natural product extracts were obtained from the Natural Products Support Group of the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (Frederick, MD, USA). Chroma-Glo cell lysis and dualluciferase substrate solutions were obtained from Promega (Madison, WI, USA).
Prefractionation of Natural Product Extracts.
Organic solvent and aqueous extracts from the NCI Natural Products Repository were prepared as previously described.37 Organic solvent extracts (100 mg) were dissolved in 1.8 mL of CH2Cl2/MeOH (1:1) and adsorbed onto Diol SPE cartridges (2 g, 6 mL, Applied Separations, Allentown, PA, USA). After evaporation of the solvent they were eluted with a step gradient of 6 mL of hexane/CH2Cl2 (9:1), CH2Cl2/EtOAc (20:1), 100% EtOAc, EtOAc/MeOH (5:1), and 100% MeOH. Aqueous extracts (200 mg) were dissolved in 2 mL of H2O, applied onto Oasis HLB reversed-phase cartridges (500 mg, 6 mL, Waters, Milford, MA, USA), and eluted with 6 mL of H2O (×2), H2O/MeOH (3:1), H2O/ MeOH (1:3), and MeOH/CH2Cl2 (9:1). Groups of 22 extracts were fractionated with SPE cartridges mounted on a vacuum manifold with the elution solvents collected in 24-well plates. Solvents were removed by centrifugal evaporation, and the fractions were then resolubilized in 1 mL of the elution solvent and transferred to tared 2D barcoded vials (1.4 mL). After solvent removal from the vials, they were reweighed to establish the mass of each prefractionation sample. In addition to the five fractions that were generated, an aliquot of each crude extract was also included in the prefractionation library.
Plant Material.
Leaves of the tropical evergreen tree Millettia ichthyotona were collected in Bac Can Providence, Vietnam, on March 21, 1988 (longitude 135 east, latitude 10 north). Collections were coordinated by Djaja Djendoel Soejarto, University of Illinois at Chicago, and taxonomic identification was provided by T. N. Ninh. A voucher specimen is maintained at the Smithsonian Institution, Washington, DC (collection number 0GHA339).
Extraction and Isolation.
The dried plant material (892 g) was ground and processed using the standard NCI method for terrestrial plant samples.37 It was sequentially extracted with CH2Cl2/MeOH (1:1) and 100% MeOH to give a combined organic extract of 22.6 g, after removal of the solvents under reduced pressure (NSC #N125389). A 2 g portion of the extract was utilized for NF-1based bioassay-guided isolation of the cytotoxic compounds. The extract was solubilized in 100 mL of 90% aqueous MeOH and partitioned with 3 × 100 mL hexanes (fraction A, 1.09 g). An additional 100 mL of H2O was added to the aqueous MeOH layer, which was then extracted with 2 × 100 mL of MTBE (fraction B, 316 mg). The MeOH was removed by evaporation under reduced pressure, and the aqueous layer extracted with 3 × 100 mL of EtOAc (fraction C, 30 mg). The H2O-soluble material was designated fraction D (248 mg). The NF1 activity concentrated in fraction B, which was separated on a 90 × 2.0 cm i.d. column of Sephadex LH-20 eluted with CH2Cl2/MeOH (1:1) into five main fractions. The NF1active fraction (116 mg) was the third one collected off of LH-20, and a 75 mg portion of this material was purified by C18 HPLC eluted with a gradient of MeOH/H2O (1:1) to 100% MeOH over 20 min to provide deguelin (1, 3.0 mg; 0.23% yield) and dehydrodeguelin (2, 2.7 mg; 0.21% yield). The identities of deguelin and dehydrodeguelin were established by comparing their spectroscopic data with appropriate literature values.26–29
Dual-Luciferase Screening Assay.
The G/R-luc cell culture and maintenance and basic assay conditions were as described6 with some modifications. For use in 384-well plates, optimal cell number for the assay was 1000 cells/well. Compounds or extracts were diluted to 10× final concentration in medium and added to assay plates 4–6 h after plating cells. After 2 days of incubation, cells were lysed in the presence of Chromo-Glo reagent, and green and red luminescence was read on a PheraStar plate reader (BMG, Durham, NC, USA). Readouts for treated cells were normalized to those in untreated (i.e., actively growing cells) wells. Due to the difficulty in replacing medium in 384-well plates as described in the original paper, growth-inhibited controls were plated directly in starving medium rather than replacing medium after cell attachment. As a result, in order to ensure adherence, polylysine-coated plates were utilized. However, this control was inadequate for toxicity calculations6 since it did not reproduce initiation of growth inhibition upon addition of samples, so serum-starved cells were used only for monitoring assay quality. As derived from the original assay development,6 inhibition of the green signal represents inhibition of cell growth, whereas inhibition of the red luminescence signal is an index of cell number and health. Hit criteria are defined in the results section, and hit confirmation was based on a repeat assay in a dose–response format starting at a high concentration of 10 μg/mL (i.e., screening concentration) and a fivepoint serial 1:2 dilution series.
Growth Inhibition Studies.
Grade III murine Nf1- and Trp53-null astrocytoma cell lines were maintained as described.5 Primary mouse astrocytes were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) or isolated from Nf1−/+;Trp53−/+cis mice and maintained as described.5,6 For assessment of growth inhibition, cells were plated at 1250 cells per well in polylysine-coated 384-well plates. After overnight recovery, compounds were added and incubated with cells for 2 days followed by estimation of cell numbers using the XTT assay.38 The XTT signal was normalized to untreated control values.
Reactive Oxygen Species Detection.
Generation of reactive oxygen species was quantitated using DCFDA, a compound that fluoresces upon oxidation by ROS. A 10 mM stock solution of carboxymethyl H2DCFDA (Invitrogen) was prepared in DMSO. Cells were plated at 1000 cells/well in black-wall, clear-bottom, polylysine-coated 384-well plates (Corning, Inc., Corning, NY, USA) and treated with DMSO (control), deguelin, or dehydrodeguelin (10 μM). DCFDA was added to a final concentration of 25 μM for the last 1 h of incubation. After washing with PBS (2 × 50 μL), cell-associated fluorescence was measured on a Tecan fluorometer at Ex 495 nm, Em 529 nm (bottom read mode). Resultant fluorescence was normalized to that of untreated controls and reported as a T/C ratio. Increasing fluorescence is indicative of the presence of ROS in the cells.
Calculations.
Data from each assay plate were handled separately. Apparent IC50 values were calculated using SigmaPlot (SPSS, Inc., Chicago, IL, USA) four-parameter logistic nonlinear regression analysis. Unless otherwise noted, all data are presented as means ± SD.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Natural Products Support Group at NCI-Frederick for extract preparation and S. Tarasov and M. Dyba (Biophysics Resource Core, Structural Biophysics Laboratory, CCR) and H. Bokesch (MTL) for assistance with high-resolution mass spectrometry. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project was also funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.5b00753.
1H NMR, 13C NMR, MS, DNA fragmentation, and AKT phosphorylation data for compounds 1 and 2 (PDF)
REFERENCES
- (1).Lin AL; Gutmann DH Nat. Rev. Clin. Oncol 2013, 10, 616–624. [DOI] [PubMed] [Google Scholar]
- (2).Patil S; Chamberlain RS Oncologist 2012, 17, 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chen YH; Gutmann DH Oncogene 2014, 33, 2019–2026. [DOI] [PubMed] [Google Scholar]
- (4).Reilly KM; Loisel DA; Bronson RT; McLaughlin ME; Jacks T Nat. Genet 2000, 26, 109–113. [DOI] [PubMed] [Google Scholar]
- (5).Gursel DB; Connell-Albert YS; Tuskan RG; Anastassiadis T; Walrath JC; Hawes JJ; Amlin-Van Schaick JC; Reilly KM Neuro Oncol. 2011, 13, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hawes JJ; Nerva JD; Reilly KM J. Biomol. Screening 2008, 13, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ostrom QT; Gittleman H; Liao P; Rouse C; Chen Y; Dowling J; Wolinsky Y; Kruchko C; Barnholtz-Sloan J Neuro Oncol. 2014, 16, 1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cerami E; Gao J; Dogrusoz U; Gross BE; Sumer SO; Aksoy BA; Jacobsen A; Byrne CJ; Heuer ML; Larsson E; Antipin Y; Reva B; Goldberg AP; Sander C; Schultz N Cancer Discovery 2012, 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cragg GM; Newman DJ Biochim. Biophys. Acta, Gen. Subj 2013, 1830, 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Henrich CJ; Beutler JA Nat. Prod. Rep 2013, 30, 1284–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Clark EP Science 1931, 73, 17–18. [DOI] [PubMed] [Google Scholar]
- (12).Takatsuki A; Nakatani N; Morimoto M; Tamura G; Matsui M; Arima K; Yamaguchi I; Misato T Appl. Microbiol 1969, 18, 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang Y; Ma W; Zheng W Mol. Clin. Oncol 2013, 1, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Johnson TA; Sohn J; Inman WD; Estee SA; Loveridge ST; Vervoort HC; Tenney K; Liu J; Ang KK-H; Ratnam J; Bray WM; Gassner NC; Shen YY; Lokey RS; McKerrow JH; Boundy-Mills K; Nukanto A; Kanti A; Julistiono H; Kardono LBS; Bjeldanes LF; Crews PJ Nat. Prod 2011, 74, 2545–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tu Y; Jeffries C; Ruan H; Nelson C; Smithson D; Shelat AA; Brown KM; Li X-C; Hester JP; Smillie T; Khan IA; Walker L; Guy K; Yan BJ Nat. Prod 2010, 73, 751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bugni TS; Richards B; Bhoite L; Cimbora D; Harper MK; Ireland CM J. Nat. Prod 2008, 71, 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lang G; Ainy Mayhudin N; Mitova MI; Sun L; van der Sar S; Blunt JW; Cole ALJ; Ellis G; Laatsch H; Munro MH G. J. Nat. Prod 2008, 71, 1595–1599. [DOI] [PubMed] [Google Scholar]
- (18).Schmid I; Sattler I; Grabley S; Thiericke RJ Biomol. Screening 1999, 4, 15–25. [DOI] [PubMed] [Google Scholar]
- (19).Wagenaar MM Molecules 2008, 13, 1406–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Appleton DR; Buss AD; Butler MS Chimia 2007, 61, 327–331. [Google Scholar]
- (21).Eldridge GR; Vervoort HC; Lee CM; Cremin PA; Williams CT; Hart SM; Goering MG; O’Neil Johnson M; Zeng L Anal. Chem 2002, 74, 3963–3971. [DOI] [PubMed] [Google Scholar]
- (22).Abel U; Koch C; Speitling M; Hansske FG Curr. Opin. Chem. Biol 2002, 6, 453–458. [DOI] [PubMed] [Google Scholar]
- (23).Aliquots of some of the unprocessed crude extracts were not included in the library during early implementation of the prefractionation protocol.
- (24).This represents the total available prefractionated natural product samples at the time the NF1 screen was run.
- (25).Unavailable extracts were either reserved for other research groups or had insufficient mass to support chemical studies.
- (26).Crombie L; Lown JW J. Chem. Soc 1962, 775–781. [Google Scholar]
- (27).Dagne E; Yenesew A; Waterman PG Phytochemistry 1989, 28, 3207–3210. [Google Scholar]
- (28).Manjary F; Petitjean A; Conan JY; Martin MT; Frappier F; Rasoanaivo P; Ratsimamanga-Urverg S Planta Med. 1994, 60, 602. [DOI] [PubMed] [Google Scholar]
- (29).Lin Y-L; Kuo Y-H Heterocycles 1995, 41, 1959–1965. [Google Scholar]
- (30).Reilly KM; Tuskan RG; Christy E; Loisel DA; Ledger J; Bronson RT; Smith CD; Tsang S; Munroe DJ; Jacks T Proc. Natl. Acad. Sci. U. S. A 2004, 101, 13008–13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Peng XH; Karna P; O’Regan RM; Liu X; Naithani R; Moriarty RM; Wood WC; Lee HY; Yang L Mol. Pharmacol 2006, 71, 101–111. [DOI] [PubMed] [Google Scholar]
- (32).Li Z; Wu J; Wu C; Jiang J; Zheng X; Xu B; Li M Oncol. Lett 2012, 4, 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kang HW; Kim JM; Cha MY; Jung HC; Song IS; Kim JS Dig. Dis. Sci 2012, 57, 2873–2882. [DOI] [PubMed] [Google Scholar]
- (34).Jin Q; Feng L; Behrens C; Bekele BN; Wistuba II; Hong WK; Lee HY Cancer Res. 2007, 67, 11630–11639. [DOI] [PubMed] [Google Scholar]
- (35).Hail N Jr.; Lotan R. Apoptosis 2004, 9, 437–447. [DOI] [PubMed] [Google Scholar]
- (36).Ji BC; Yu CC; Yang ST; Hsia TC; Yang JS; Lai KC; Ko YC; Lin JJ; Lai TY; Chung JG Oncol. Rep 2012, 27, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).McCloud TG Molecules 2010, 15, 4526–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Scudiero DA; Shoemaker RH; Paull KD; Monks A; Tierney S; Nofziger TH; Currens MJ; Seniff D; Boyd MR Cancer Res. 1988, 48, 4827–4833. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.