Abstract
Background
Long noncoding RNAs (lncRNAs) are important regulators in human disease, including cancers. LncRNA MIR22HG has been shown to inhibit the progression of endometrial carcinoma, lung cancer, and hepatocellular carcinoma. Its role in gastric cancer is unclear. This study investigated MIR22HG effects on gastric cancer.
Material/Methods
Gastric cancer tissues (n=43) and adjacent normal tissues (n=21) were collected. Patients’ 5-year overall survival rate was analyzed. Human normal gastric mucosal cell line (GES-1) and gastric cancer cell lines (MKN-45, AGS, SGC-7901) were cultured. AGS and MKN-45 cells were transfected by pcDNA3 empty vector, pcDNA3-MIR22HG overexpression vector, MIR22HG siRNA and its negative control, NOTCH2 siRNA and its negative control, respectively. Proliferation was explored by CCK-8 assay. Migration and invasion were explored by Transwell. qRT-PCR and western blot were used to investigate mRNA and proteins expression, respectively.
Results
MIR22HG expression was decreased in gastric cancer tissues and cells (P<0.05). Low MIR22HG expression indicated lower 5-year overall survival rate (P<0.05). Upregulation of MIR22HG inhibited AGS and MKN-45 cell proliferation, migration and invasion (all P<0.05). Downregulation of MIR22HG elevated AGS and MKN-45 cell proliferation, migration, and invasion (all P<0.05). MIR22HG negatively regulated NOTCH2 signaling. Silencing MIR22HG elevated HEY1 and nucleus NOTCH2 expression. Silencing of NOTCH2 suppressed AGS and MKN-45 cells proliferation, migration and invasion (all P<0.05).
Conclusions
LncRNA MIR22HG suppressed gastric cancer progression through attenuating NOTCH2 signaling.
MeSH Keywords: Cell Proliferation; Receptor, Notch2; RNA, Long Noncoding; Stomach Neoplasms
Background
Similar to most other tumors, gastric cancer is also a disease characterized by excessive cell proliferation and infinite growth [1,2]. Oncogene activation as well as tumor suppressor gene inactivation is the main cause of cancer induction [3]. Currently, the treatment options for gastric cancer are mainly surgery, radiotherapy, chemotherapy [4,5]. However, these treatment methods also damage normal tissues and cells while destroying the tumor. Moreover, gastric cancer cells are more and more tolerant to drug after chemotherapy, which is a main cause of recurrence after chemotherapy [6,7]. Given the serious life threat that gastric cancer poses to patients, it is urgently needed to find effective targets for the therapy of gastric cancer.
It is well known that deregulation of coding genes exerts a crucial role in gastric cancer progression [8]. Small non-coding RNAs (miRNAs) are also recently found to be involved in the occurrence and development of gastric cancer through regulating some other gene expression [9]. Currently, long-chain non-coding RNAs (lncRNAs), another type of non-coding RNAs consisting of more than 200 nucleotides [10], have become a research hotspot for tumor-targeted therapy, including for gastric cancer [11]. LncRNAs are found to be abnormally expressed in several cancers and stimulate cancer progression by regulating the expression of other coding oncogenes, tumor suppressor genes or non-coding miRNAs [12,13]. A meta-analysis of 40 related studies indicated that, in hepatocellular carcinoma patients with poor prognosis, 27 types of lncRNAs are abnormally up-regulated and 18 types of lncRNAs are remarkably down-regulated [14]. In gastric cancer, existing literature findings demonstrated that TINCR, CCAT2, AOC4P, BANCR and LINC00857 are all associated with tumor size, advanced tumor stages as well as lymphatic metastasis, which might be novel diagnostic biomarkers for gastric cancer [15]. Furthermore, MALAT1 acts as an oncogene in gastric cancer, whose aberrantly up-regulation increases gastric cancer aggressiveness by regulating HMGB2 [16]. lncRNA TCONS_00068220 also suppresses gastric cancer cell apoptosis rate and it might be involved in the pathogenesis of gastric cancer [17]. These biological targets provide great possibilities for the early diagnosis and effective treatment strategy of gastric cancer. Thus, we presented a new diagnostic and therapeutic target for gastric cancer in this study, namely lncRNA MIR22HG. The mechanism of MIR22HG in affecting gastric cancer progression has been further explored.
Material and Methods
Tissue samples collection
From March 2010 to May 2012 gastric cancer tissues of 43 patients who were diagnosed with gastric cancer for the first time and underwent surgery therapy in our hospital were collected. Adjacent normal tissues of 21 cases were also obtained. Of these patients, 24 cases were male, and 19 cases were female. All patients average age was 54.2±9.1 years. All patients were not complicated with other severe organic lesions, and lactating and pregnant women were not allowed to join the study. All patients were followed-up for at least 5 years to calculate their 5-year overall survival rate by Kaplan-Meier survival analysis. With the informed consent of all participants, this study has been approved by the ethics committee of our hospital.
Cell culture
Human normal gastric mucosal cell line (GES-1) and gastric cancer cell lines (MKN-45, AGS, SGC-7901), bought from Institute of Digestive Surgery, Shanghai Jiaotong University, China, were cultured in RPMI1640 medium containing 10% fetal bovine serum (FBS), 50 U/mL penicillin and 50 μg/mL streptomycin. All cells were maintained in an incubator at 37°C, 5% CO2 and were subcultured every 3 days. Cells of the third generation were collected at 80–90% confluence for follow-up studies.
Cell transfection
AGS and MKN-45 cells were seeded in 6-well plates with a density of 1×105 cells per well by serum-free RPMI1640 medium. pcDNA3 empty vector and pcDNA3-MIR22HG overexpression vector, constructed by Shanghai Jima Pharmaceutical Technology Co., Ltd., China, were used to transfect AGS and MKN-45 cells (named pcDNA3 vector group and pcDNA3-MIR22HG group, respectively). Furthermore, AGS and MKN-45 cells were also transfected by MIR22HG siRNA and its negative control, as well as NOTCH2 siRNA and its negative control (all provided by Shanghai Jima Pharmaceutical Technology Co., Ltd., China.). AGS and MKN-45 cells transfected with MIR22HG siRNA and its negative control were served as si-MIR22HG group and si-NC group, respectively, while those transfected by NOTCH2 siRNA and its negative control were used as si-NOTCH2 group and NC group. Twenty-four hours later, normal RPMI1640 medium containing 10% FBS was used to replace the residual medium of each well for 48 hours of incubation at 37°C, 5% CO2.
CCK-8 assay
AGS and MKN-45 cells of each group were inoculated in 96-well plates with 2.5×103 cells each well and were incubated at 37°C, 5% CO2. Cells of each group were set up in parallel with 6 duplicate holes. At 24 hours, 48 hours and 72 hours respectively, 10 μL CCK-8 solutions was added into each well for additional 4 hours of incubation at 37°C. The absorbance value of all wells was detected at 450 nm wavelength (OD450 value) by an automatic labeling instrument.
Transwell assay
The diluted Matrigel was spread into 6-well Transwell chamber with a volume of 70 μL. Cells of each group were prepared as single cell suspensions using serum-free RPMI1640 medium with a density of 1×106/mL. A total of 200 μL cell suspensions were added into the upper chamber and 1 mL RPMI1640 medium containing 10% FBS was added into the lower chamber. All Transwell chambers were incubated at 37°C, 5% CO2 for 24 hours. Then cells passed through the membrane were fixed with 10% formaldehyde for 20 minutes and stained with 0.1% purple crystal for 15 minutes. Before being fixed by formaldehyde, cells that failed to penetrate the membrane were gently scraped off using a cotton swab. Number of invasion cells was counted under an inverted microscope. Cells migration was also measured according to the above method without Matrigel. In addition, migration and invasion of normal AGS and MKN-45 cells without transfection were also detected. Relative migration or invasion of cells in each group was calculated according to the following formula: Relative migration or invasion (%)=(Number of migration or invasion cells of each group/Number of migration or invasion cells of normal cells)×100%.
qRT-PCR
Total RNA in tissues and cells were isolated by TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) as described before [18]. Totally 200 μL of each RNA sample was subjected to reverse transcription through application of cDNA Reverse Transcription Kit (Takara Bio Company, Shiga, Japan). All RNA samples were amplified by using the Fast Start Universal SYBR Green Mastermix (Roche, USA). Premiers sequences (synthesized by Guangzhou RiboBio Co., Ltd., China.) were as follows: MIR22HG, forward, 5′-CGGACGCAGTGATTTGCT-3′, reverse, 5′-GCTTTAGCTGGGTCAGGACA-3′. U6, forward, 5′-GCTTCG GCAGCACATATACTAAAAT-3′, reverse, 5′-CGCTTCACGAA TTTGCGTGTCAT-3′. NOTCH2, forward, 5′-TACCTGCCGATGTGCT-CC-3′, reverse, 5′-CGAGAGTGAAGTCGCCAGTC-3′. HES6, forward, 5′-GGATGCTAATGAGGACGAGCG-3′, reverse, 5′-CAGCTTCATCTGCGTGTCGC-3′. HEY1, forward, 5′-TGAGCTGA GAAGGCTGGTAC-3′, reverse, 5′-ACCCCAAACTCCGATAGTCC-3′. GAPDH, forward, 5′-GTCGATGGCTAGTCGTAGCATCGAT-3′, reverse, 5′-TGCTAGCTGGCATGCCCGATCGATC-3′. MIR22HG relative expression was normalized to U6, and other candidate genes were normalized to GAPDH. The 2−ΔΔCT method was selected to analyze data.
Western blot
Cells were washed 3 times with pre-cooled phosphate buffered saline (PBS) at 4°C. Total proteins were isolated by using cell lysates and quantified by BCA methods. In addition, nucleus proteins were also determined by BCA methods after being obtained using nuclear protein extraction kit (Beyotime, Jiangsu, China). After separation by sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transfer onto PVDF membranes and incubated with 50 μg/L skim milk powder for 1 hour. TBST solution were used to wash membranes 3 times. Rabbit anti-mouse NOTCH2 and HEY1 primary antibodies (Abcam, USA, 1: 1000) were subsequently added to membranes, respectively. After 12 hours incubation at 4°C, all membranes were subjected to 3 times washing with TBST solution and goat anti-rabbit anti-IgG antibodies (1: 2000, Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., China) were used to incubate membranes for 2 hours at room temperature. Three times washing with TBST were carried out once again. Finally, the blots were visualized by enhanced chemiluminescence Plus and integrate optical density was measure through software Lab Works 4.5. In this research, GAPDH was served as internal reference of total NOTCH2 and HEY1 proteins, while Histone H3 was set as the internal reference of nucleus NOTCH2 protein.
Statistical analysis
All experiments were conducted in triplicate in this research. Data were presented as mean ± standard deviation and were analyzed by using SPSS 19.0 software. Student t-test as well as one-way ANOVA was used to perform differential comparisons. The statistically significant threshold was P<0.05.
Results
Low MIR22HG expression indicated poor prognosis of gastric cancer patients
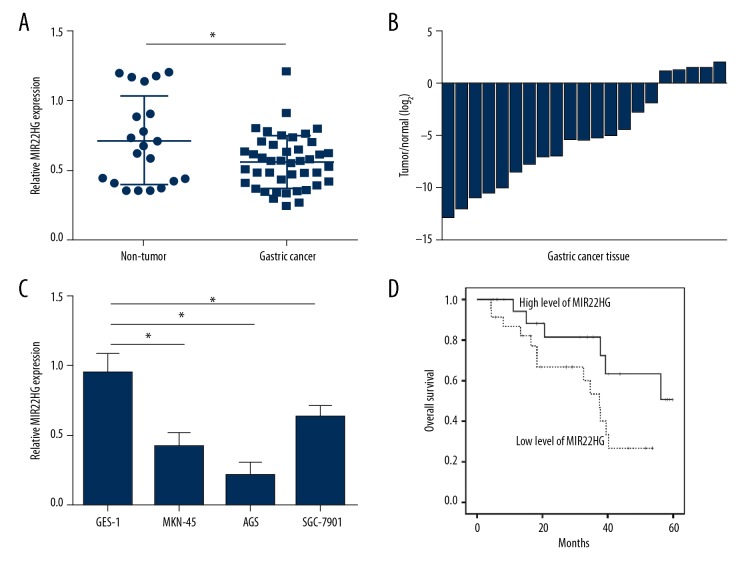
Relative MIR22HG expression in gastric cancer tissues (n=43) and adjacent non-tumor tissues (n=21) were measured by qRT-PCR. As shown in Figure 1A, gastric cancer tissues had much lower relative MIR22HG expression than that of non-tumor tissues (P<0.05). Meanwhile, the relative expression of MIR22HG in gastric cancer tissues and 21 corresponding non-tumor tissues were also compared. 17 cases were occurred lower MIR22HG expression in cancer tissues than that in non-tumor tissues (Figure 1B). In addition, human normal gastric mucosal cell line (GES-1) and gastric cancer cell lines (MKN-45, AGS, SGC-7901) were cultured to detect MIR22HG expression. Compared with GES-1 cells, relative MIR22HG expression in MKN-45, AGS and SGC-7901 cells was all much declined (P<0.05). Among three gastric cancer cell lines, AGS and MKN-45 cells had relatively lower MIR22HG expression than SGC-7901 cells. Therefore, AGS and MKN-45 cells were selected for follow-up studies (Figure 1C). More importantly, patients’ 5-year overall survival rate was analyzed by Kaplan-Meier survival analysis. For patients with high level of MIR22HG, they had obviously higher overall survival rate than those with low expression of MIR22HG (P<0.05) (Figure 1D). Thus, MIR22HG expression was declined in gastric cancer tissues and cells, and low MIR22HG expression indicated poor prognosis of gastric cancer patients.
Figure 1.
Low MIR22HG expression indicated poor prognosis of gastric cancer patients. (A) Obviously declined relative MIR22HG expression was found in gastric cancer tissues when compared with adjacent non-tumor tissues. (B) Among 21 gastric cancer patients, 17 cases were occurred lower MIR22HG expression in cancer tissues than that in non-tumor tissues. (C) Compared with GES-1 cells, relative MIR22HG expression in MKN-45, AGS, and SGC-7901 cells was all much declined. (D) Patients with high level of MIR22HG had obviously higher overall survival rate than those with low expression of MIR22HG. * P<0.05.
Upregulation of MIR22HG inhibited AGS and MKN-45 cells proliferation, migration and invasion ability
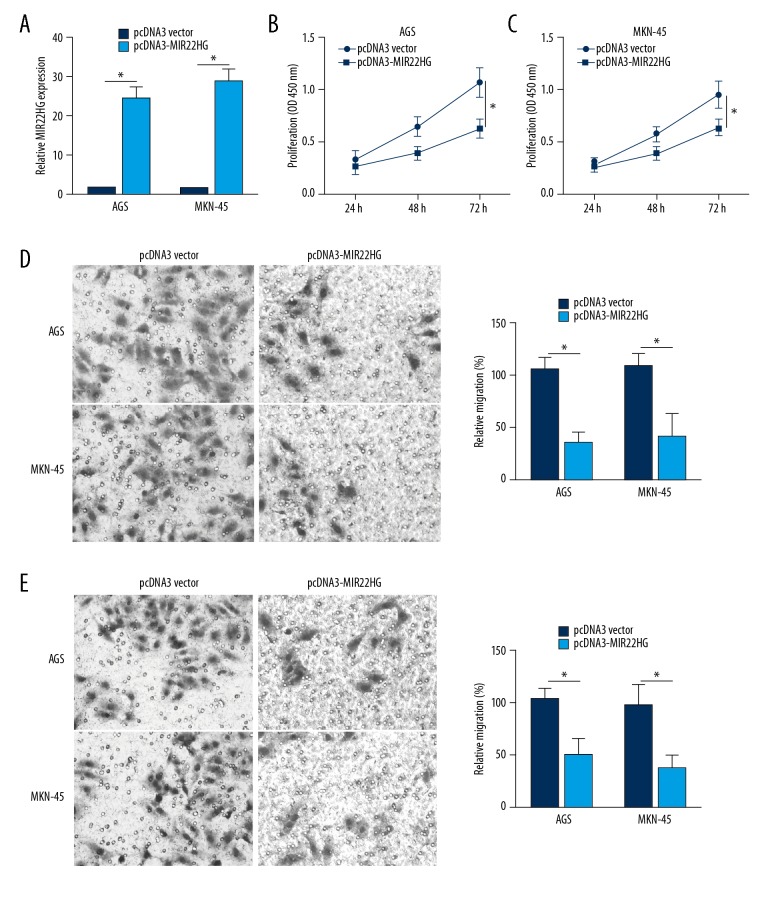
Relative MIR22HG expression in AGS and MKN-45 cells was significantly increased by transfection with pcDNA3-MIR22HG overexpression vector (P<0.05) (Figure 2A). According to the CCK-8 assay, at 72 hours, the OD450 value of AGS and MKN-45 cells of pcDNA3-MIR22HG group was remarkably lower than those of pcDNA3 vector group (P<0.05) (Figure 2B). Cell migration and invasion ability was also detected by Transwell assay, and the results revealed decreased relative migration and invasion of AGS and MKN-45 cells in pcDNA3-MIR22HG group when compared with pcDNA3 vector group (P<0.05) (Figure 2C, 2D). These results indicated that up-regulation of MIR22HG inhibited AGS and MKN-45 cells proliferation, migration and invasion ability
Figure 2.
Upregulation of MIR22HG inhibited AGS and MKN-45 cells proliferation, migration and invasion ability. (A) Relative MIR22HG expression in AGS and MKN-45 cells was increased by transfection with pcDNA3-MIR22HG overexpression vector. Upregulation of MIR22HG inhibited AGS and MKN-45 cells proliferation (B), migration (C), and invasion (D) ability. * P<0.05.
Downregulation of MIR22HG promoted AGS and MKN-45 cells proliferation, migration and invasion ability
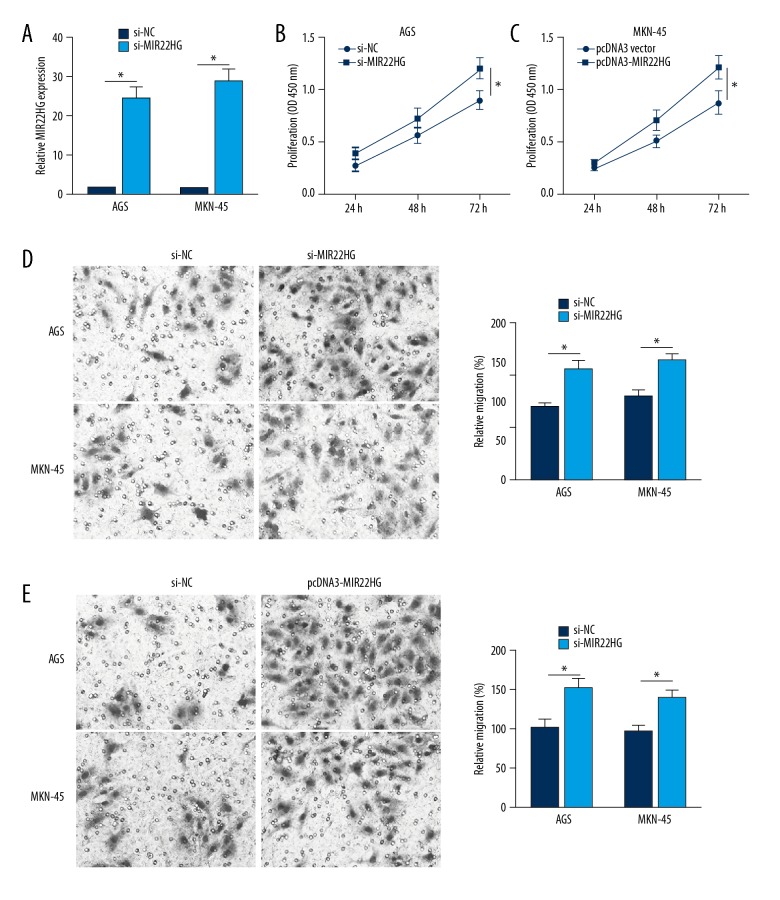
In this study, MIR22HG siRNA and its negative control were used to transfect AGS and MKN-45 cells. It could be observed that, compared with AGS and MKN-45 cells of si-NC group, MIR22HG relative expression in AGS and MKN-45 cells of si-MIR22HG group was significantly reduced (P<0.05) (Figure 3A). Meanwhile, dramatically enhanced OD450 value, relative migration and invasion of AGS and MKN-45 cells in si-MIR22HG group were found when compared with those in si-NC group (P<0.05) (Figure 3B–3D). The above data revealed that downregulation of MIR22HG promoted AGS and MKN-45 cells proliferation, migration and invasion ability.
Figure 3.
Downregulation of MIR22HG promoted AGS and MKN-45 cells proliferation, migration and invasion ability. (A) MIR22HG relative expression in AGS and MKN-45 cells was decreased by MIR22HG siRNA transfection. Downregulation of MIR22HG inhibited AGS and MKN-45 cells proliferation (B), migration (C), and invasion (D) ability. * P<0.05.
MIR22HG negatively regulated NOTCH2 signaling
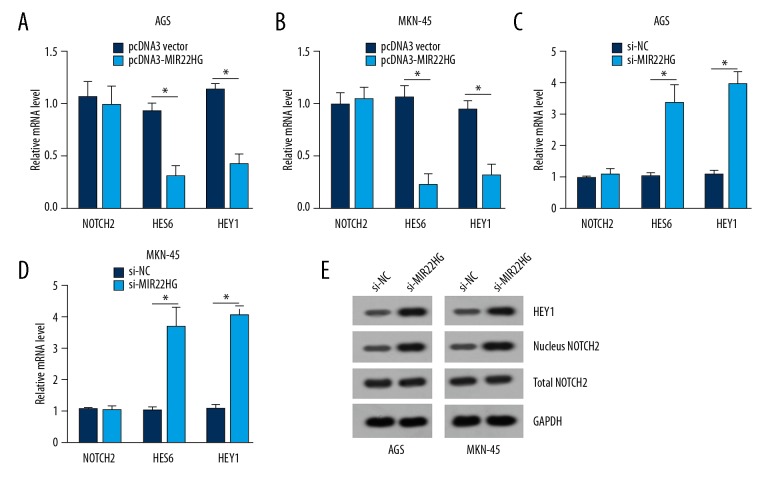
From Figure 4A and 4B, it could be noticed that, compared with pcDNA3 vector group, HES6 and HEY1 mRNA relative expression were both markedly decreased in AGS and MKN-45 cells of pcDNA3-MIR22HG group (P<0.05). At the same time, when MIR22HG was being silenced, HES6 and HEY1 mRNA relative expression were both dramatically higher in AGS and MKN-45 cells of si-MIR22HG group than those of si-NC group (P<0.05) (Figure 4C, 4D). However, over-expression or silencing of MIR22HG had no obviously effects on NOTCH2 mRNA relative expression in AGS and MKN-45 cells. Furthermore, western blot results showed that, silencing of MIR22HG did not affect total NOTCH2 protein expression in AGS and MKN-45 cells. However, after MIR22HG was silenced, HEY1 and nucleus NOTCH2 proteins expression were both elevated in AGS and MKN-45 cells (Figure 4E), indicating MIR22HG suppresses the activation of NOTCH2 pathway.
Figure 4.
MIR22HG negatively regulated NOTCH2 signaling. Upregulation of MIR22HG markedly decreased HES6 and HEY1 mRNA relative expression in AGS (A) and MKN-45 (B) cells. Downregulation of MIR22HG dramatically increased HES6 and HEY1 mRNA relative expression in AGS (C) and MKN-45 (D) cells. (E) Silencing of MIR22HG elevated HEY1 and nucleus NOTCH2 proteins expression in AGS and MKN-45 cells. * P<0.05.
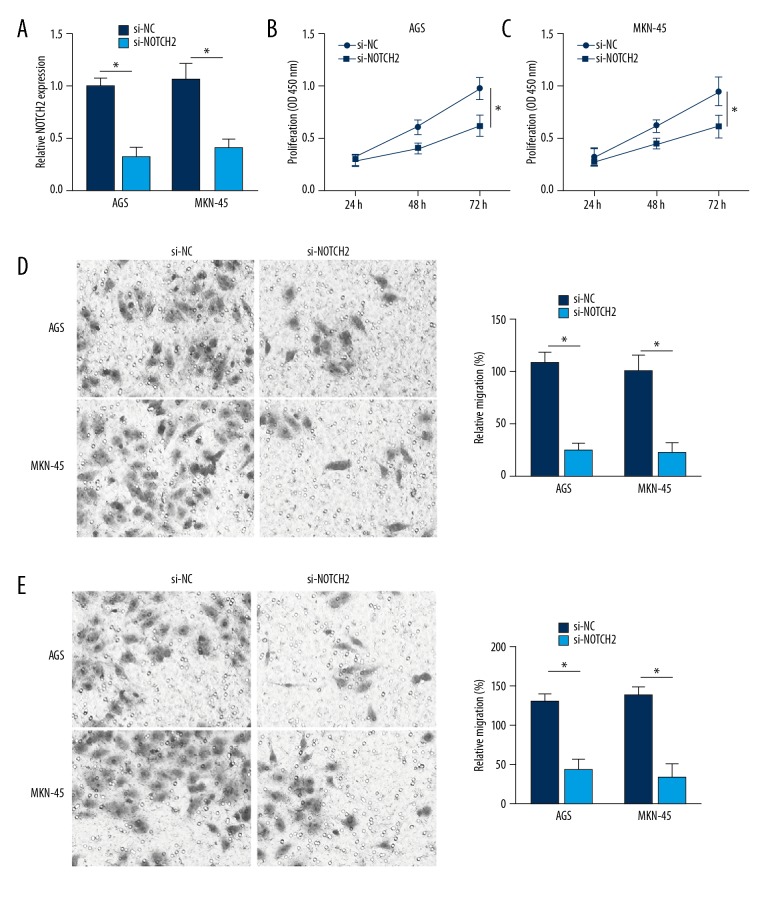
Silencing of NOTCH2 suppressed AGS and MKN-45 cells proliferation, migration and invasion
The relative expression of NOTCH2 mRNA in AGS and MKN-45 cells was successfully inhibited by NOTCH2 siRNA transfection (P<0.05) (Figure 5A). At 72 hours after transfection, the OD450 value of AGS and MKN-45 cells in si-NOTCH2 group was much declined when compared with NC group (P<0.05) (Figure 5B). In addition, relative migration and invasion of AGS and MKN-45 cells in si-NOTCH2 group was also significantly lower than that of NC group (P<0.05) (Figure 5C, 5D). Therefore, silencing of NOTCH2 suppressed AGS and MKN-45 cells proliferation, migration and invasion.
Figure 5.
NOTCH2 silencing suppressed AGS and MKN-45 cells proliferation, migration and invasion. (A) NOTCH2 mRNA relative expression in AGS and MKN-45 cells was successfully inhibited by NOTCH2 siRNA transfection. Silencing of NOTCH2 suppressed AGS and MKN-45 cells proliferation (B), migration (C), and invasion (D) ability. * P<0.05.
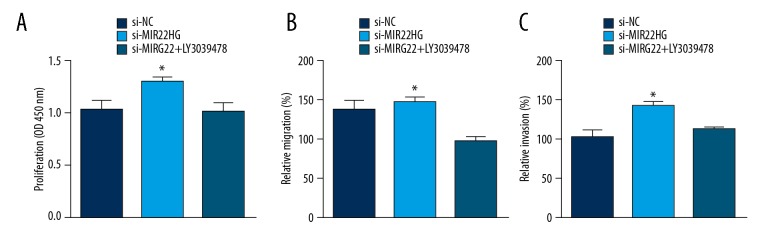
Inhibition of NOTCH2 pathway abrogated the effects of MIR22HG silencing
To further prove the essential role of NOTCH2 pathway in MIR22HG-mediated gastric cancer, we suppressed NOTCH2 pathway using a commercial inhibitor (LY3039478) in MIR22HG-depleted AGS cells. Through CCK-8 and Transwell assays, we found that MIR22HG silencing promoted proliferation, migration and invasion of tumor cells (Figure 6A–6C). However, inhibition of NOTCH2 pathway in the meantime abrogated these effects (Figure 6A–6C), indicating NOTCH2 pathway is essential for the role of MIR22HG in gastric cancer.
Figure 6.
Inhibition of NOTCH2 pathway abrogated the effects of MIR22HG silencing. Inhibition of NOTCH2 pathway suppressed proliferation (A), migration (B), and invasion (C) ability in MIR22HG-depleted AGS cells. * P<0.05.
Discussion
Gastric cancer was one of the most common cancers, which caused severe death worldwide with only 20% survival rate by existing treatments. Most patients were identified as advanced or metastatic gastric cancer in the first diagnosis and the 5-year survival rate was no more than 15% for advanced gastric cancer patients [19]. The emergence of novel therapeutic targets was of important significance for improving the early diagnosis and survival of gastric cancer. In this article, lncRNA MIR22HG was identified as a novel target of gastric cancer, which might inhibit the progression of gastric cancer by attenuating NOTCH2 signaling.
MIR22HG belongs to lncRNAs that have recently been discovered and studied. Currently, limited existing literatures have researched the effect of MIR22HG on tumor. MIR22HG was first discovered to be involved in hepatocellular carcinoma. In 2016, Liu et al. [20] firstly pointed out that MIR22HG damaged the invasion of hepatocellular carcinoma cells through deriving miR-22 and targeting HMGB1. The next year, Dong et al. [21] demonstrated in their retrospective study that MIR22HG expression was reduced in tumor tissues of 127 hepatocellular carcinoma cases and low MIR22HG level was closely associated with microvascular invasion, tumor encapsulation as well as TNM stage. In addition, after MIR22HG was knocked down, the proliferative capacity of several hepatocellular carcinoma cells was impaired. MIR22HG was also considered to be one of the independent risk factors of hepatocellular carcinoma patients’ prognosis. Cui et al. [22] indicated declined MIR22HG expression in endometrial cancer. Elevated MIR22HG expression level impaired endometrial cancer cell proliferative capacity and stimulated endometrial cancer cell apoptosis by arresting them in G0/G1 phase. Further data revealed that MIR22HG played tumor suppressive roles in endometrial cell cancer by interfering with miR-141-3p/DAPK1 axis. According to the results of 918 cases with lung cancer, reduced MIR22HG expression was associated with low survival rate. Decreased MIR22HG could elevate the expression of oncogenes, such as MET, YBX1 and p21, which blocked the transmission of cell death signals. In this paper, reduced MIR22HG expression was also observed in gastric cancer tissues and cells, which indicated poor outcomes of patients with gastric cancer.
NOTCH2 signaling was found to be involved in and regulated the development of multiple diseases, especially in tumors [23,24]. Results from this research suggested that silencing MIR22HG might enhance gastric cancer cells proliferation, migration and invasion by increasing HES6, HEY1 and nucleus NOTCH2 expression. HES6, a member of the hairy-enhancer of the split homolog family, was found to be involved in multiple tumors [25]. It was reported that elevated HES6 expression of patients with hepatocellular carcinoma exhibited adverse prognosis [26]. Carvalho et al. [27] still demonstrated that HES6 was increased in prostate cancer, and reduction in tumor cells invasion and colony formation ability was occurred by reduced HES6 expression. HEY1 was a direct target gene of NOTCH signaling pathway [28]. Xu et al. [29] data revealed that HEY1 was involved in the development of pancreatic cancer and it might be one of the potential therapeutic targets for patients with pancreatic cancer. In addition, elimination of HEY1 expression could inhibit proliferation and migration of hepatocellular carcinoma cells [30]. NOTCH2 was one of the important members of NOTCH signaling [31]. Litten et al. [32] declared overexpressed NOTCH2 in hepatoblastoma, which provided potential theoretical implications for targeted therapy of hepatoblastoma. We noticed in this research that declined MIR22HG had no effect on NOTCH2 expression. However, expression of NOTCH2 in the nucleus was significantly increased by silencing MIR22HG. We speculated that overexpression of NOTCH2 in the nucleus might interfere with other tumor suppressor genes or oncogenes expression, thereby promoting the proliferation, migration and invasion of gastric cancer cells. This would also be the focus of our future researches.
Conclusions
Collectively, this article researched lncRNA MIR22HG effects on gastric cancer and decreased MIR22HG expression was noted in gastric cancer tissues and cells. In-depth studies indicated that MIR22HG was a novel potential diagnostic and therapeutic target for gastric cancer, which might suppress gastric cancer cells proliferation, migration and invasion by attenuating NOTCH signaling.
Footnotes
Source of support: Departmental sources
Conflict of Interests
None.
References
- 1.Shi Q, Wang W, Jia Z, et al. ISL1, a novel regulator of CCNB1, CCNB2 and c-MYC genes, promotes gastric cancer cell proliferation and tumor growth. Oncotarget. 2016;7:36489–500. doi: 10.18632/oncotarget.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Sun Y. Overexpression of myosin phosphatase target subunit 1 (MYPT1) inhibits tumor progression and metastasis of gastric cancer. Med Sci Monit. 2018;24:2508–17. doi: 10.12659/MSM.906852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai T, Sowa Y. Molecular-targeting therapies against quantitative abnormalities in gene expression with malignant tumors. Cancer Sci. 2017;108:570–73. doi: 10.1111/cas.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim BH, Eom KY, Kim JS, et al. Role of salvage radiotherapy for regional lymph node recurrence after radical surgery in advanced gastric cancer. Radiat Oncol J. 2017;31:147–54. doi: 10.3857/roj.2013.31.3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Nong Z, Lu G. Correlation between nuclear factor E2-related factor 2 expression and gastric cancer progression. Med Sci Monit. 2015;21:2893–99. doi: 10.12659/MSM.894467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida K, Ito C, Ohmori Y, et al. Inhibition of PI3K suppresses propagation of drug-tolerant cancer cell subpopulations enriched by 5-fluorouracil. Sci Rep. 2017;7:2262. doi: 10.1038/s41598-017-02548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Pang X, Ji L, et al. Reduced expression of deubiquitinase USP33 is associated with tumor progression and poor prognosis of gastric adenocarcinoma. Med Sci Monit. 2018;24:3496–505. doi: 10.12659/MSM.908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu CG, Cui XL, Wei ZG, Guo JS. High expression of the ANKRD49 protein is associated with progression and poor prognosis of gastric cancer. Cancer Biomark. 2018;22:649–56. doi: 10.3233/CBM-171074. [DOI] [PubMed] [Google Scholar]
- 9.Irmakyazicioglu MB. Mechanisms of microRNA deregulation and microRNA targets in gastric cancer. Oncol Res Treat. 2016;39:136–39. doi: 10.1159/000443224. [DOI] [PubMed] [Google Scholar]
- 10.Ye B, Liu B, Yang L, et al. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018;37(8) doi: 10.15252/embj.201797174. pii: e97174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Cheng G, Yang H, et al. Reciprocal regulation of long noncoding RNAs THBS4-003 and THBS4 control migration and invasion in prostate cancer cell lines. Mol Med Rep. 2016;14:1451–58. doi: 10.3892/mmr.2016.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu P, Wang Y, Wu J, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu P, Wang Y, Huang G, et al. Lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23:631–39. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 14.Zheng C, Liu X, Chen L, et al. LncRNAs as prognostic molecular biomarkers in hepatocellular carcinoma: A systematic review and meta-analysis. Oncotarget. 2017;8:59638–47. doi: 10.18632/oncotarget.19559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang K, Shi H, Xi H, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics. 2017;7:213–27. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17:44. doi: 10.1186/s12935-017-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Xue J, Song Y, et al. Downregulated long non-coding RNA TCONS_00068220 upregulates apoptosis in gastric cancer cells. Oncol Lett. 2017;14(5):6143–50. doi: 10.3892/ol.2017.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Cheng Q, Liu J, Dong M. Leukemia stem cell-released microvesicles promote the survival and migration of myeloid leukemia cells and these effects can be inhibited by MicroRNA34a overexpression. Stem Cells Int. 2016;2016 doi: 10.1155/2016/9313425. 9313425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uprak TK, Attaallah W, Çelikel ÇA, et al. HER-2 incidence in gastric cancer, its association with prognosis and clinicopathological parameters. Ulus Cerrahi Derg. 2015;31(4):207–13. doi: 10.5152/UCD.2015.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Zhang D, Wu D. 748 Long non-coding RNA miR22HG represses hepatocellular carcinoma cell invasion by deriving miR-22 and targeting HMGB1. Gastroenterology. 2016;150:S1045–46. [Google Scholar]
- 21.Dong Y, Yan W, Zhang SL, et al. Prognostic values of long non-coding RNA MIR22HG for patients with hepatocellular carcinoma after hepatectomy. Oncotarget. 2017;8:114041–49. doi: 10.18632/oncotarget.23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Z, An X, Li J, et al. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–28. doi: 10.1016/j.biopha.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 23.Qu J, Song M, Xie J, et al. Notch2 signaling contributes to cell growth, invasion, and migration in salivary adenoid cystic carcinoma. Mol Cell Biochem. 2016;411:135–41. doi: 10.1007/s11010-015-2575-z. [DOI] [PubMed] [Google Scholar]
- 24.Hubmann R, Sieghart W, Schnabl S, et al. Gliotoxin targets nuclear NOTCH2 in human solid tumor derived cell lines in vitro and inhibits melanoma growth in xenograft mouse model. Front Pharmacol. 2017;8:319. doi: 10.3389/fphar.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam SM, Kim YN, Kim JW, et al. Hairy and enhancer of Split 6 (Hes6) deficiency in mouse impairs neuroblast differentiation in dentate gyrus without affecting cell proliferation and integration into mature neurons. Cell Mol Neurobiol. 2015;36:57–67. doi: 10.1007/s10571-015-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu YC, Liang CJ, Zhang DX, et al. LncSHRG promotes hepatocellular carcinoma progression by activating HES6. Oncotarget. 2017;8:70630. doi: 10.18632/oncotarget.19906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho FL, Marchionni L, Gupta A, et al. 847 HES6 promotes prostate cancer aggressiveness and is independent of Notch signalling. European Urology Supplements. 2014;13:e847. [Google Scholar]
- 28.Zamurovic N, Cappellen D, Rohner D, Susa M. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279:37704. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Shan X, Cai Y, Liu L. Qingyihuaji formula inhibits pancreatic cancer and prolongs survival by downregulating Hes-1 and Hey-1. Evid Based Complement Alternat Med. 2015;2015 doi: 10.1155/2015/145016. 145016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zhou X, Wu WN, Zeng AL, et al. MicroRNA-141-3p promotes glioma cell growth and temozolomide resistance by directly targeting p53. Oncotarget. 2017;8:71080–94. doi: 10.18632/oncotarget.20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen WC, Fischer MM, Axelrod F, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–95. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 32.Litten JB, Chen TT, Schultz R, et al. Activated NOTCH2 is overexpressed in hepatoblastomas: an immunohistochemical study. Pediatr Dev Pathol. 2011;14:378–83. doi: 10.2350/10-09-0900-OA.1. [DOI] [PubMed] [Google Scholar]