Summary
It is now well appreciated that the human microbiome plays a significant role in a number of processes in the body, significantly affecting its metabolic, inflammatory and immune homeostasis. Recent research has revealed that almost every mucosal surface in the human body is associated with a resident commensal microbiome of its own. While the gut microbiome and its role in regulation of host metabolism along with its alteration in a disease state has been well studied, there is a lacuna in understanding the resident microbiota of other mucosal surfaces. Among these, the scientific information on the role of lung microbiota in pulmonary diseases is currently severely limited. Historically, lungs have been considered to be sterile and lung diseases have only been studied in the context of bacterial pathogenesis. Recently however, studies have revealed a resilient microbiome in the upper and lower respiratory tracts and there is increased evidence on its central role in respiratory diseases.
Knowledge of lung microbiome and its metabolic fallout (local and systemic) is still in its nascent stages and attracting immense interest in recent times. In this review, we will provide a perspective on lung-associated metabolic disorders defined for lung diseases (e.g. COPD, Asthma, respiratory depression due to infection) and correlate it with lung microbial perturbation. Such perturbations may be due to altered biochemical or metabolic stress as well. Finally, we will draw evidence from microbiome and classical microbiology literature to demonstrate how specific lung morbidities associate with specific metabolic characteristics of the disease, and with the role of microbiome in this context.
Keywords: Lung, pulmonary microbiome, Metabolome, commensal flora, dysbiosis, mucosa, 16s rRNA sequencing
Introduction
A multitude of microbes inhabit various mucosal surfaces in the human body, with a majority residing in the gastrointestinal surface, followed by other mucosal surfaces, including lungs [1]. One of the major roles of the microbial ecosystem is the maturation of the host immune system in the postnatal period. In adults too, there is evidence of constant “tutoring” of the host immune system by the microbial ecosystem residing in various mucosal surfaces [2–4]. Hence, its perceivable that external factors influencing the commensal microbial ecosystem, like lifestyle and/or dietary changes, infections, aging etc. to name a few, would induce adaptive changes in the host immune system. Recent reports indeed suggest that external infection of the gut, fosters host systemic inflammation by out-competing the commensal flora and alternatively, systemic inflammation preferentially depletes beneficial gut flora to promote otherwise dormant commensal bacteria with potential pathogenic properties (pathobionts) [1,2,4–7]. Historically, lungs have been considered to be sterile and lung diseases have only been studied in the context of bacterial pathogenesis [8,9]. Recently, however, 16s ribosomal RNA (rRNA) and shotgun metagenomics studies have revealed a resilient microbiome in the upper and lower respiratory tracts and there is increased evidence of its central role in respiratory diseases [4,9–12]. While the upper respiratory tracts (trachea, upper bronchus) have microbiota resembling the oral cavity, lower respiratory tracts have unique microbial signatures. Compared to the oral cavity, lung harbors a significantly diminished (50-fold less, in terms of biomass) microbiome [13], and yet, the composition and homeostasis of both microbiota is modulated by the host immune system and vice-versa [4].
While gut microbiome and its role in influencing a vast majority of diseases has gained immense popularity over the last few years, there has not been much systematic study on the lung microbiome [9,14]. Cystic fibrosis is one of the most researched lung complication vis-à-vis microbial dysbiosis (see [15] and reviewed in [8]). Apart from that, several reports have implicated lung microbiome in pathogenesis of asthma/allergic diseases [16–18] and Chronic Obstructive Pulmonary Disease (COPD) [19–22]. In all these studies, microbial dysbiosis, characterized by expansion of specific microbial communities, has been a center-point for the disease type, accompanied by uncontrolled inflammation. However, evidence has been gathered over the years which supports direct and indirect role of gut microbiota in pulmonary diseases [23–26]. The specific composition of lung microbiome in healthy individuals and an altered scenario in disease, has attracted a plethora of human studies to define exact changes due to specific disease conditions, for example, in cystic fibrosis [27]. Clinical evidence suggest that restoration of gut microbiome has positive prognosis for lung microbial homeostasis and by association, lung immune homeostasis [14,27,28].
Most of the microbiome studies rely on an amplicon analysis of 16S rRNA gene. Though widely used, the limitations of the 16s rRNA sequencing technology prohibit exact identification of bacterial species. Therefore, clustering of collected sequences is the standard tactic of organizing 16s rRNA sequencing data [29–31]. The clusters represent groups of genetically similar bacteria, which are called operational taxonomic units (OTUs). Analysis of 16s sequencing is often a function of diversity of bacterial OTUs within an individual (alpha-diversity) and among individuals (beta-diversity)[32]. A microbiome that hosts a wide variety of bacterial species will have a high alpha-diversity. Alternatively, a microbiome that is present in multiple individuals, but varies in species presence between individuals will be have a high beta-diversity.
Here, we present a perspective on the role of lung microbiome in several well-studied lung morbidities, as compared to its homeostasis in health. We will discuss the considerations for the analysis of lung microbiome and the challenges involved. Finally, we will discuss the current state of knowledge in host metabolic perturbations due to lung microbial dysbiosis, which itself is a nascent and upcoming field of study.
The Respiratory Microbiome in health
Source and replenishment
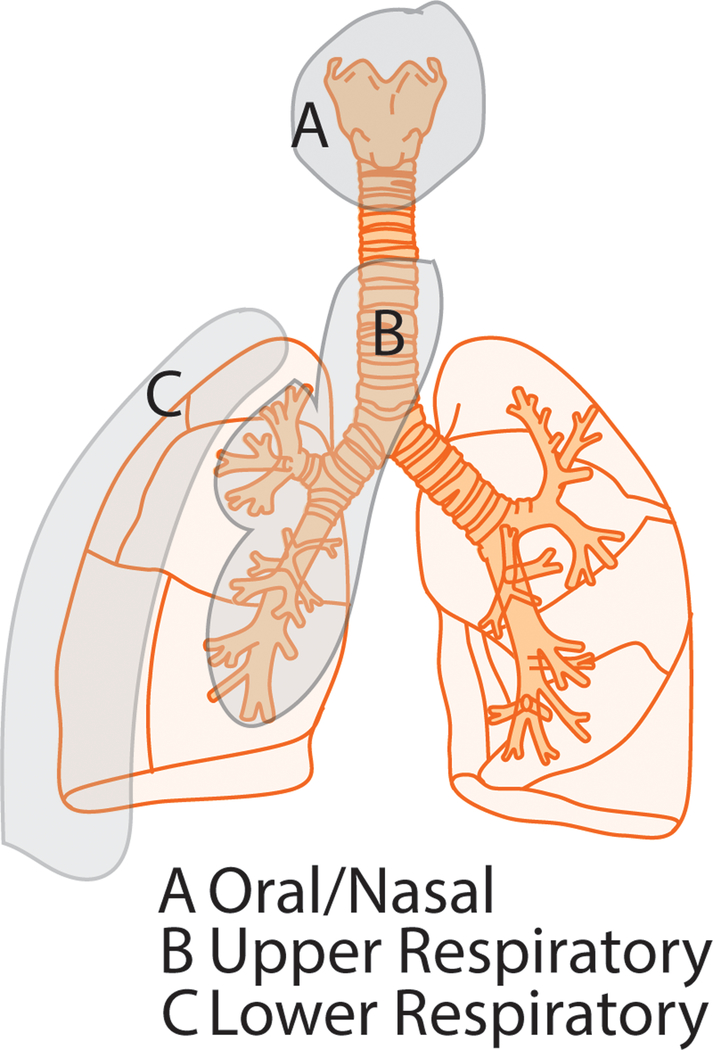
Current scientific literature divides the respiratory microbiome broadly into 3 major anatomical regions—oral/nasal, upper and lower respiratory microbiome (Fig 1-A, B and C respectively). Oral microbiome is heavily influenced by environmental factors, which in turn, shapes the composition of upper respiratory tract microbiome. The lower respiratory microbiome is unique, mildly influenced by microbiota from other mucosal surfaces, with the exception of diseased lung, where each of these compartments influences each other in a dysbiotic state. Major disparity pertains to the description of the source and resilience of lower airway microbiome is described in the literature. Some reports describe it as an independent unique community of microflora, relatively unperturbed due to fluctuations in oral microbiome due to environmental influences [4,8,33]. While others describe it as diminutive community requiring constant replenishment from upper airway and oral communities, hence heavily influenced by perturbation in those communities [34–36]. Since these reports are mostly based on clinical end-point studies, variability in collection methods and diverse disease and medicinal background of the subjects make it impossible to conclusively characterize the origin and sustainability of the lower airway microbiota. More studies need to be performed, preferably with small animals, utilizing all acquisition methods including lung tissues, in controlled environment, to obtain proof-of-concept in this aspect of respiratory microbiome. In this regard, strict demarcation has to be made with respect to the anatomical region from where the microbial sample is derived. As figure 1 describes, careful sampling from the disparate regions of the excised lung, especially in murine studies, may provide an interesting perspective for the dynamics of upper and lower airways in health and disease.
Figure 1: Different anatomical regions of the lung, representing unique microbial signatures.
The environment heavily influences oral/nasal microbiome (i), which in turn, influences the upper respiratory microbiota (ii). The resilience and uniqueness of the lower respiratory microbiome (iii) has been controversial due to challenges in sampling techniques and analysis pipelines.
Sampling and analysis pipelines
Until recently, the lungs were thought to be a sterile site within the body [9,37]. This idea was difficult to disprove due to the reliance of bacterial detection and identification on culture-dependent studies. The advent of culture-independent studies has identified the lung as having its own microbiome. In particular, the sequencing of 16s ribosomal RNA has been a critical tool in the description of microbiomes [38,39].
Assessment of the lung microbiome presents its own unique challenges. Sample contamination by bacteria from the upper respiratory tract presents a difficult hurdle in these studies [40]. Typically, these lung microbiome samples are collected via broncho-alveolar lavage (BAL). While colonization by bacteria from the upper respiratory tract is a common source of bacterial migration into the lung, not all bacteria that migrate this way can be expected to replicate and survive in the lower airways. Additionally, the process by which lung microbial samples are collected can introduce bacteria from the upper respiratory tract that never migrated to the lung. These problems can be overcome by refined collection methods to reduce contamination from the upper respiratory tract or analytical methods that assume and adjust for this contamination [36,40,41]. Recently, Bassis et al. have suggested that these sources of contamination may have less impact than previously understood [42]. In their study, bacterial population from an oral wash was compared to those of BAL taken from separate lung lobes without fully removing the bronchoscope past the epiglottis. Analysis of these samples showed that the BAL microbes did not differ significantly in their overall variance from the oral wash microbial population.
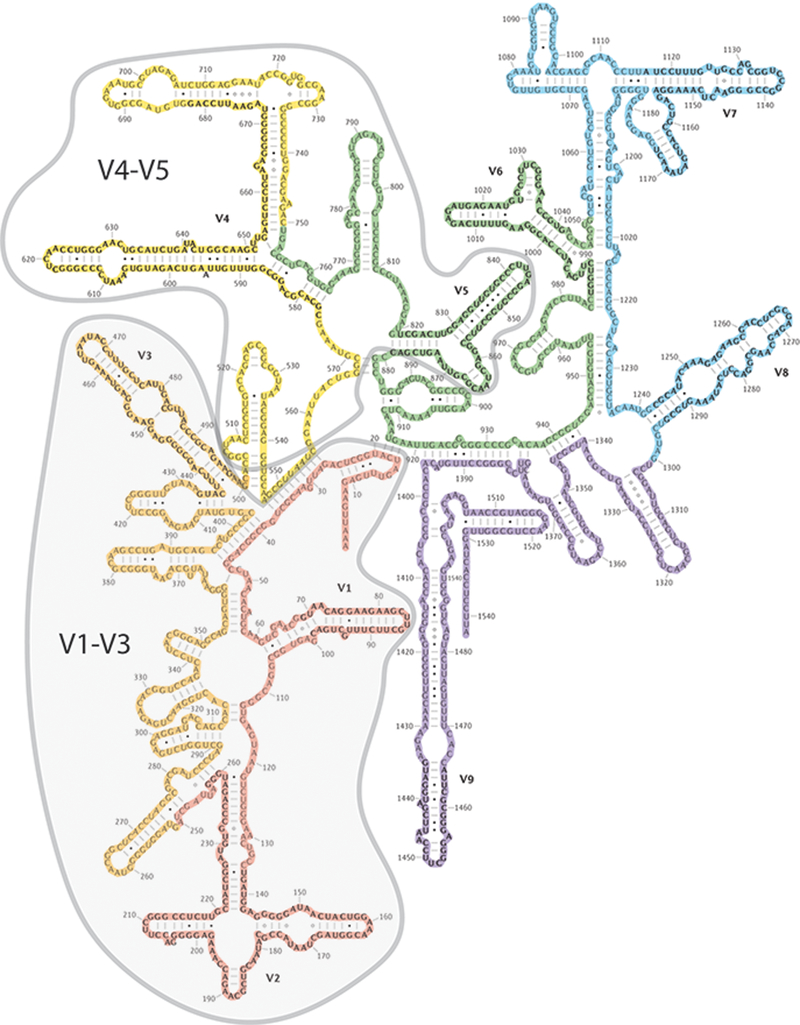
In terms of analysis pipeline, some controversy exists between the choices of 16s versus shotgun metagenomics. While discussions in the published reports mostly pertain to the analysis of gut microbiome, the arguments are valid for the analysis of microbiota from other mucosal surfaces, including the airways. Both 16s and shotgun metagenomics accord high level of resolution and throughput to the investigators, however, sampling and procedural bias for both methods have been reported [38,43]. Lung microbiome is several orders of magnitude smaller than gut microbiome and hence, presents several technical challenges. Major ways in which technical challenges can be mitigated are maintaining consistency in methods and facilities in obtaining data. The choice of the 16s region being amplified also plays a role in maintaining consistency (Figure 2). It is known that variable region (V) 1–3 amplification has better taxonomic coverage, while V4–5 region has higher specificity of detection [10,19]. As the field has advanced over years, preference for higher specificity (hence, V4-V5 amplicon) has gained more popularity over higher taxonomic coverage. With improvement in instrumentation, improvement in database curation and better protocols, it is gradually becoming easier to have better taxonomic coverage with high specificity. It is clear from the literature, that data emanating from various groups can be further reconciled if methodological and analytical consistency is maintained and agreed upon in the field. In this regard, the tools, methods and protocols in the Human Microbiome Project portal, which includes lung as an important mucosal surface, might serve as an exciting resource for researchers in the field.
Figure 2: 16s rRNA with V1-V3 and V4-V5 hyper variable regions.
16s rRNA has been extensively used for building molecular phylogeny of different bacterial species. Among the 9 hyper variable regions (V1-V9), V1-V3 and V4-V5 regions have remained a popular choice for constructing phylogeny of commensal bacteria in mammals. While V1-V3 region has wider taxonomic coverage, the V4-V5 region is known to provide better identification of bacterial genera.
Consensus communities at homeostasis
Multiple studies have attempted to define the microbiome of the healthy lung. By comparing BAL samples with oral wash samples from the same individual, a marked similarity has been observed between the lung and oral microbiome [40]. This is likely due to micro aspirations of bacteria-containing saliva into the lung [44,45]. The oral microbiome has thus been identified as a major contributor to the lung microbiome [46]. There is a lack of consensus on the basic composition of ‘healthy’ lung microbiome in human. On the phylum level, there are varied reports of dominance described: A. Bacteroidetes and Firmicutes dominated (BAL derived) [36]. B. Bacteroidetes and Proteobacteria dominated (Lung tissue derived) [22] and C. Diverse/no dominance (BAL derived)[10,21]. Methodologically, the consensus seems to favor BAL as the cleanest source for lung microbiota and Bacteroidetes and Firmicutes dominant composition [33].
Selection of resident microbial content on mucosal surfaces
Development of the microbiome is dependent on multiple factors. Exposure of a newborn to the microbiome of the mother is the source of initial inoculation. The birthing process exposes the newborn to vaginal, fecal, and skin microbiomes from the mother [47]. After birth, the mother continues to transfer bacteria to the child via breastfeeding. Children delivered vaginally have gut microbiomes that closely resemble that of their mother, while those born by cesarean section do not bear the same similarities [48].
The maturing fetus, once thought to develop in a sterile environment, has been shown to be exposed to maternal bacteria before birth [49–51]. Additionally, a recent study showed that the airways of preterm newborns have colonized airways as early as 6 hours after birth, including those born by cesarean section, and that those born by cesarean section have microbiomes that do not differ from those born vaginally [52]. This may imply that colonization of the lung microbiome may begin before birth, or lung colonization is independent and more environmentally acquired phenomenon, rather than maternally transferred one.
Microbiome status is adjusted and maintained by the host immune system, as well as environmental factors. Maintenance balances the populations of commensal bacterial species and discourages growth of pathogenic species. Hence, as a corollary, it is perceivable that changes in the microbiome would modulate host immunity and metabolism; the premise for all the interest and importance of the field.
Cross talk of resident microbial flora in mucosal surfaces
Cross talk between the microbial populations across different mucosal surfaces has been a point of interest in the field recently. Inflammatory bowel disease (IBD) has long been linked to pulmonary dysfunction and disease [53]. Lung dysfunction as a result of IBD has been reported as early as 1976 [54]. This study described severe pulmonary disease in six patients with IBD; four of which had no history of smoking. These patients demonstrated chronic bronchitis, bronchiectasis, and obstructive pulmonary dysfunction.
In a study of ulcerative colitis patients, impaired lung function presented in 57.6% of patients compared to healthy controls [55]. These manifested primarily as restrictive dysfunctions. Unfortunately, this study was unable to determine carbon monoxide diffusion capacity. Another study, however, did show a link between children with Crohn’s disease an impaired carbon monoxide diffusion capacity [56]. Additionally, a study of 314 Crohn’s disease patients demonstrated an increased risk of COPD [57]. Both IBD and COPD are diseases linked to alterations in relative microbial abundance.
Smoking is a major risk factor for the development and exacerbation of COPD. It is also a risk factor in the development of both ulcerative colitis and Crohn’s disease [58–60]. The hypothetical link between smoking and COPD is simple enough to see, considering the direct exposure of the lung to inhaled smoke. However, the link between smoking and IBD is less clear. When considering the potential of cigarette smoke to disrupt lung epithelium, a possible link emerges between the two diseases in the immune response to common commensal bacteria. This is supported by findings that smoking perturbs the gut microbiome [61]. One of the most interesting, yet complex interaction was recently reported by McFarlane et.al., [62] where they showed that intestinal helminth parasite infection and resulting gut microbial changes have a protective effect on symptoms arising from respiratory syncytial virus (RSV). Point to note here is that this study ensured that the helminth and RSV infections were confined to the gut and lungs respectively. Hence, ample evidence exists to suggest cross talk between the commensal flora in different isolated mucosal surfaces, however, studies need to be done to understand the micro-dynamics, e.g. whether the host immune system mediates this cross talk, or whether these are independent effects of a systemic insult in patients.
Environmental factors of microbial status
Smoking
Yu et al. performed a study of lung microbiota taken from non-malignant tissues of lung cancer patients. They found no correlation between lung microbiome and smoking intensity, years of smoking, or cigarettes per day. They did, however, note that alpha diversity of the lung microbiome increases with pack-years of tobacco smoking[63]. The authors hypothesize that the overall apparent lack of relation between smoking and lung microbiome status in this study may be explained by the lack of variability in smoking status between study subjects.
Smoking has been shown to disrupt the lung epithelial layer responsible for preventing migration of commensals from the lumen [64,65]. Previous studies on gut microbiome and disruption of the gut barrier have shown that this may lead to bacterial translocation and systemic immune activation [66]. This leads to the reasonable assumption that disruption of the lung epithelial barrier could lead to translocation of members of the lung microbiome.
An interesting piece of supporting evidence for this view was discovered by Jungnickel et al [67]. This study used a lung carcinoma model in which lung carcinoma cells were injected in mice. After exposure to both cigarette smoke and aerosolized nontypeable Haemophilus influenza (NTHi) bacterial infection, mice showed increased level of metastatic growth compared control counterparts that were only injected with lung carcinoma cells. Under this combined treatment, bacterial translocation to lung cancer tumors also occurred. This reinforces the previous findings of cigarette smoke-induced disruption of microbial barriers in the lungs. Additionally, the study asserts that bacterial migration to the tumor sites links inflammation caused by cigarette smoke to tumor proliferation.
Breast-Feeding
Breast milk plays two important roles in the establishment of the microbiome of a newborn [68]. First, the milk acts as a carrier of maternal bacteria. In fact, the human breast milk has been described as having its own microbiome, boasting over 200 species of bacteria. This collection of bacteria includes commensal staphylococci, streptococci, and lactobacilli [69]. These commensals have been shown to inhibit colonization and growth of pathogenic bacterial species in the host.
Second, the milk contains prebiotic oligosaccharides that nourish colonizing bacteria [70]. These oligosaccharides are resistant to digestion and absorption by the host and are available for bacterial utilization in the gut. Specifically, members of the Bifidobacterium and Bacteriodes bacterial genera are able to gain a competitive advantage from the presence of milk oligosaccharides. Human milk oligosaccharides also demonstrate antimicrobial properties, inhibiting adhesion and infection by pathogenic bacteria [71].
The above stated properties of breast milk have been demonstrated as influential in the establishment of the gut microbiome in infants. Additionally, cross-contamination of the lung microbiome from the gut has been hypothesized via micro-aspiration. It is conceivable that breast-feeding plays a role in the development of the lung microbiome, as well. Indeed, breast feeding has been shown to impact development of the nasopharygeal microbiome[68]. Breast feeding has been associated with microbial stability and early colonization of the upper respiratory tract by Moraxella, Corynebacterium, and Dolosigranulum [72]. These factors were also associated with reduced rates of respiratory infection.
Antibiotics
Antibiotic usage is a major factor in microbial homeostasis and can greatly reduce its diversity [73]. Antibiotics are often prescribed during pregnancy and can be found in both newborn blood and mother’s breast milk after birth. The impact from this on the microbiome is two-fold. First, bacteria are eliminated from the mother before they are passed on to the offspring. Second, the antibiotic present in breast milk can interfere with establishment and colonization by the microbes that escaped elimination in the mother and were passed down. Studies have shown that exposure to antibiotics early in life may be a causal factor in the development of asthma and chronic inflammation [44,47].
Diet
The effect of diet on lung microbiome has been a very scantily explored field. While there is some literature on how the maternal diet can influence the microbiome in an infant and may even be related to development of childhood asthma, there is not a lot of study to show that there is a direct influence of diet on lung microbiome [74].
A recent study by Cait et al in 2017 showed that mice treated with vancomycin have an altered microbiome and metabolite profile and exhibit exacerbated Th2 responses. These animals also showed more susceptibility to allergic lung inflammation. This study further showed that gut dysbiosis can aggravate allergic lung inflammation through both T cell- and DC-dependent mechanisms that are inhibited by bacterial short-chain fatty acids (SCFAs). However, even though this study showed that lung diseases may be affected by alteration of gut microbiome (and hence diet), it did not highlight if the lung microbiota was influenced [75]. It is evident from some studies that the gut microbiome has a profound influence on lung inflammation and its pathology (asthma, COPD) however whether it is mediated via change in the lung microbiome is not clear [76]. Further studies are required for the complete understanding of diet-induced lung microbial perturbations as exemplified in two independent reports from Bou Ghanem et.al., where they have shown that dietary α-tocopherol supplementation results in reversal of age-induced susceptibility to Streptococcus pneumoniae lung infection[77] and this is achieved through enhanced elastase activity of PMNs in the lung[78]. Considering that S. pneumoniae is a part of the lung commensal flora, the role of microbial dysbiosis or homeostasis cannot be discounted. This is further corroborated by the recent work by Brown et.al., where microbial role in controlling pulmonary infection via IL17A and GM-CSF has been established[79]. The authors demonstrate that microbiota-derived signals for controlling pulmonary infections, is not confined to lungs alone and could come from the intestinal microbes as well. Hence, diet-induced gut microbial changes can indeed translate into lung microbial perturbations and vice-versa.
Pollution
Pollutants in our environment have a direct effect on lung diseases like COPD and asthma. In the developing world, exposure to smoke during cooking, burning coal, and other biomass fuels have a direct consequence on lung health [80]. It is hypothesized that the respiratory microbiome acts as a first line of defense against the environmental pollutants. However, it is also equally likely that the members of the lung microbiome can be selectively killed or injured by the toxins released from these pollutants leading to an altered compromising microbiome. Disrupting the existing microbial community structure as a result of environmental pollutants can alter the balance of pro-inflammatory and anti-oxidant conditions leading to a diseased state [81].
Even though the lung microbiome research community firmly acknowledges the role of pollution in altering microbial composition in context of disease, there is not a lot of literature available in this field regarding the composition of the resident bacteria in the lung microbiome or how they may change upon exposure to environmental pollutants [82].
Lung microbiome and disease
Chronic obstructive pulmonary disease
Chronic obstructive pulmonary disease (COPD) is a highly morbid and potentially fatal lung disease. It presents a major burden of disease currently and has been projected to increase in this regard [83]. COPD development is marked by sudden, temporary worsening of the condition, called exacerbations [84], which typically cause significant, but temporary decrease in pulmonary function, with baseline lung function returning in roughly a month. However, rarely, baseline lung function is impaired longer or permanently[85].
Disease progression in COPD has been described by the vicious circle hypothesis [86]. According to this hypothesis, impaired innate immune response allows for bacterial infection. After initial infection, pathogenic bacteria induce inflammation and increased mucus production. This leads to insult of the respiratory epithelium, which impairs innate immune response. The impaired immune response leaves the lung vulnerable to subsequent infection by pathogenic bacteria, closing the vicious circle.
COPD is characterized by H. influenzae colonization and exacerbations of the disease are marked by its presence in the patient sputum [87]. Presence of the H. influenzae membrane protein, P6, has been shown to induce mucin production in COPD mice and cultured human epithelial cells [88]. During periods between exacerbations, H. influenzae is not present [87]. Interestingly, sputum samples from different exacerbation periods contain the same strain of H. influenzae. This may indicate that, even though the bacteria reach an undetectable level between exacerbations, the pathogens remain within the host and expand in population during or before exacerbations.
Cystic Fibrosis
Cystic Fibrosis (CF) is a genetic disease that is primarily linked to a mutation in the CFTR gene [89]. CF manifests symptoms in multiple organs throughout the body, but the most pronounced effects are localized to the lung. The hallmarks of the disease are the presence of secretions within the lung and increased rate of pulmonary bacterial infection. Unsurprisingly, these alterations to lung function have a significant impact on the lung microbiome.
It has long been understood that CF patients suffer from pulmonary infections of S. aureus, H. influenzae, B. cepacia, and P. aeruginosa [90]. More recently, S. maltophilia, MRSA, M. abscessus, and S. milleri has been identified as members of the CF lung microbiome. Lung tissue damage from bacterial infection often necessitates lung transplantation. A recent study showed that, post-transplant, P. aeruginosa rapidly invades the transplanted tissue [91]. The invading bacteria are suspected to have originated in the sinuses and adapted to the non-CF donor lung.
Cox et al. studied the differences in airway microbiota and lung function in young and old CF patients [92] and found that increasing age is associated with loss of pulmonary function, as well as, decreasing microbiotic richness, diversity, and evenness.
Asthma
The hygiene hypothesis states that exposure to microbes in early life can impact the development of asthma and similar diseases [93]. Development of asthma has been linked to early exposure to antibiotics. As previously stated, the use of antibiotics in pregnant mothers can reduce bacterial colonization [73]. Studies have shown that early exposure to the bacterial endotoxin, lipopolysaccharide (LPS), is protective against development of asthma. Studies in mice have shown that LPS exposure confers protection via expression of the A20 protein, which attenuates NF-kB activation.
The lung microbiomes of asthmatic individuals were compared to those of healthy controls and found to have increased levels of Proteobacteria, but decreased levels of Bacteriodetes [37]. Asthmatics were also found to have greater microbial abundance and diversity compared to healthy controls. This may be indicative of the lack of early life immune cell “tutoring” as mentioned earlier.
Pneumonia (idiopathic or ventilator)
With research on lung microbiome gaining impetus, the understanding of the pathology of pneumonia has changed from its conventional understanding. It is now believed that the dysbiosis of the lung microbial flora, not merely introduction of pathogenic bacteria, is the underlying cause of the disease [22,37,94].
Ventilator Associated Pneumonia (VAP):
Children under mechanical ventilation are at risk for a number of nosocomial infection like VAP. This leads to increased risk of mortality, prolonged hospitalization and extensive rehabilitation [94]. Recent research has shed light on how microbial organisms interact with each other and their environmental elements and the contribution of this to development of infection. VAP has been associated with viral lower respiratory tract infection along with bacterial infection. Among bacterial species, Staphylococcus aureus is the most prevalent. Airway samples collected from ventilated children exhibit a fairly diverse bacterial community. The studies also show that the diversity decreases in subsequent days and airways become rapidly dominated by pathogenic bacteria before diagnosis of VAP. In this study, Streptococcus flora was visibly increased in children with VAP compared to subjects without VAP [95].
Idiopathic pneumonia:
Analysis of the airway microbiome showed that the most important causative bacterial pathogen for lower respiratory tract infection like pneumonia was Streptococcus pneumoniae [96]. This is a common pathogen in the upper respiratory tract that exists asymptomatically with other bacterial community of pathogenic and commensal species. Exposure to environmental stimuli as well as immunological changes upset the “equilibrium” within the community, leading to dysbiosis and eventually infection [97]. A study comparing elderly pneumonia (to healthy elderly control subject) and adult pneumonia (to heathy adult subjects) is one of the very few, that have performed an in-depth study on the changes in microbial composition during the disease. The investigators observed a decrease in gram negative population like Prevotella, Veillonella and Leptotrichia and gram-positive genus Parascardovia in pneumonia patients [98]. Further, another study showed a strong correlation between the tongue microbiome with mortality associated with pneumonia in nursing home patients [99].
Lung cancer
Microbiome contributions to the development of different cancers have been previously studied[100]. Recently, studies describing the link between pulmonary microbiome and lung cancer have come to light [63,101]. In a recent study comparing patients diagnosed with lung cancer to those diagnosed with a benign lung mass, BAL was collected and microbiota were examined [101]. Compared to those with benign lesions, cancerous lungs had increased prevalence of the Fermicutes and TM7 phyla, as well as, the Veillonella and Megasphaera genera. The authors hypothesize that the microbiota of lung cancer patients may be altering the lung microenvironment.
A study using non-malignant lung tissue samples from 165 lung cancer patients described microbiota profiles associated with the disease [63]. In this study, the authors found that Proteobacteria was the dominant phylum. In patients with stage 3 or stage 4 cancers, the Thermus genus was relatively abundant. In patients who developed metastases, the Proteobacteria genus, Legionella, was relatively abundant. Both of these studies present an exciting possibility of alterations to the microbiome as a novel biomarker in lung cancer.
Human Immunodeficiency Virus (HIV) and AIDS
HIV has been shown to have a major effect on the condition of gut microbial status due to its interference with immune response and disruption of gut epithelium[102]. The literature available of the role of HIV infection in alteration of lung microbiome is less clear. A study of BAL from HIV-infected patients undergoing antiretroviral therapy and HIV-uninfected individuals found no clear difference between their microbiomes[34]. In patients with advanced HIV, decreased alpha diversity but greater beta diversity was observed in the lung microbial species[103]. After one year of treatment with antiretroviral therapy, these patients displayed increased abundance of Prevotella and Veillonella, which have been associated with lung inflammation. While abundant, yet inconclusive data is available from HIV patients vis-à-vis lung microbiome, this field would benefit from controlled prospective study, preferably on newly developed murine models, e.g. Humanized mice with human microbial engraftments. Outcomes from these controlled experiments could then be verified from patients, thereby furthering our knowledge with potentially therapeutic and non-invasive ways to control co-morbidities associated with HIV and lung inflammation.
Viral Infections of the Lung
Viruses are mainly responsible for acute respiratory infections (ARIs), which is common in children and adults[104]. Approximately 40% of healthcare-associated lung infections in children have been attributed to some form of ARI[105]. The main perpetrators of most viral ARIs include Influenza A virus (IAV), Respiratory syncytial virus (RSV), Metapneumovirus (MPV) and rhinoviruses (RV). From the perspective of ARI pathophysiology and the role of microbial homeostasis in the lungs, this presents a unique opportunity to study cross-kingdom interactions and whether respiratory microbiome can influence ARI for better or worse. In this context, IAV and RSV have recently been studied in detail. In a recent report, it has been shown that sublethal infection with a low-pathogenic variant of H5N1 IAV can change microbial composition in the lower respiratory tract (LRT)[106]. The authors show a Bacillus to Lactobacillus shift in the lung microbiome, and interestingly, gut microbiome depletion and disruption of gut barrier integrity and Type-1 interferon (IFN-1) response due to lung IAV infection. Another study also shows subtle changes in the microbiome, with enriched Streptococcus and depleted Pseudomonas within the respiratory microbiome[107]. This has significant implication not only in the context of pathophysiology of IAV infection, but also whether microbial changes contribute to the exacerbation of disease phenotype. Indeed, Bartley et.al.’s work has indicated that age-related microbial changes contribute to poor prognosis and IFN-1 responses due to IAV infection and this can be prevented to a large extent by maintaining the host’s microbial composition, in this case, by caloric restriction[108]. Most of the symptoms in ARIs are a result of excessive inflammation and hence, it is understandable that modulation in the microbiome due to viral infections can feed into this vicious cycle of inflammation and aberrant immune responses, leading to enhanced viral load[104]. For other ARI perpetrators, namely RV and RSV, distinct microbial signatures have been associated for each viral infection[62]. Here, the authors show a shift from a Staphylococcus dominated microbiome in uninfected infants to a Moraxella, Streptococcus, Corynebacterium, Haemophilus and Dolosigranulum dominated respiratory microbiome in RV and RSV infected patients. The predominance of Haemophilus in RSV infected lung has been corroborated by an independent study by Edervine et.al[109]. There are interesting points to be taken from these studies. It is apparent that respiratory microbial changes due to IAV infection is distinct from that due to RV or RSV infection. Presuming that these changes are early onset, there is an opportunity to use this information for early diagnosis in patients. Additionally, lung and/or gut microbial restoration could be potentially used as a therapeutic tool to modulate systemic immune response against viral perpetrators of ARI.
Lung Microbiome and the immune modulation of the host
It is well known the GI microbial flora plays a major role in modulating the host inflammatory status, specifically in the maturation of Th17 response in the mucosal immune system [110,111]. In lungs, the changes in the microbiome of healthy individuals are associated with a low-level inflammation [112]. In asthma patients, the microbial signatures associated with a Th17 phenotypes have been reported [113]. In the study by Huang et al, members of the Proteobacter taxa like Pasteurellaceae, Enterobacteraceae and Bacillaceae have been found to be associated with Th17 associated gene signature. This Th17 inflammatory phenotype may represent another pathway that is independent of Th2 response in asthma patients [113].
Furthermore, in a study by Yadava et. al., using an experimental mouse model, it was found that exposure to LPS and elastase led to a dysbiotic lung microbiome that resulted in an increase of IL-17A expression. This was due to an increase in γδ+ T cell phenotype [114]. This murine inflammatory phenotype is associated with airway abnormalities that was similar to human COPD. BAL obtained from these mice challenged with LPS and elastase demonstrated a decrease in α diversity and an increase of relative abundances of Lactobacillus, Pseudomonas and Chryseobacterium [114]. The researchers found that microbiota enhanced the production of IL-17A by γδ+ T-cells by using microbiota-depleted mice. Upon transferring the enriched microbiome from LPS/Elastase treated animals and concurrent challenge with LPS/Elastase in antibiotic treated mice, the authors were able to show an upregulation of IL-17A immunological phenotype. This study thus showed that the lung microbiome has a functional role on an immunological phenotype.
Th17 inflammatory pathway and IL-17 upregulation are not the only ones that may be upregulated in a host with lung dysbiosis. In a recent study, Richmond et al demonstrated that in pIgR deficient mice loss of mucosal immunity results in microorganism invasion into the epithelium [115]. Since the percentage of bacteria present in the lumen of the airways did not differ between the wild-type mice and pIgR deficient mice, the study showed that the effects were due to invasion of microorganisms alone. Mice with pIgR deficiency also had activation of the NF-κB pathway with associated NF-κB-dependent chemokine keratinocyte chemoattractant in BAL fluid. This mouse model of mucosal immunodeficiency is thus another pathway that is activated by the interaction between innate microorganisms in the lower airway [115].
Even though literature is scant in lung microbiome, the above studies indicate that the lung microbiome has a profound effect in modulating the immune system and the inflammatory pathways in the host.
The Respiratory Microbiome and Metabolism
The role of the gut microbiome in host metabolism is well-studied [116–118]. Despite this focus, we are still learning about the full role that both commensal and pathogenic bacteria play in modifying metabolism in the gut. Gut microbiota perform vital functions in bile acid, choline, and phenol metabolism [119–121]. The role of bacteria in metabolism within the lung has yet to be studied in depth, but there is some indication that pathogenic bacteria may play a role in host metabolism. E. coli and S. aureus are pathogenic bacteria in the context of the lung microbiome. They have also been linked to the episodic increases in proteases and protease inhibitors that play a role in infection and immune response [122].
A recent study, found metabolomic differences in BAL of HIV-infected individuals compared to healthy controls and hypothesized that these differences could originate from microbiome alterations [123]. A follow-up study by the same group identified a correlation between altered metabolite levels and the presence of pathogenic bacteria species in the lungs [124]. This study identified Caulobacteraceae, Staphylococcaceae, and Nocardioidaceae as the main contributors to altered metabolite levels. These bacteria are noteworthy for their roles in the pathogenesis of pneumonia in HIV patients. Bacterial metabolism by itself is known to modulate host pulmonary immunity. In a seminal study, it was shown that constant enrichment of lung microbiota with oral taxa (called supraglottic predominant taxa) creates an unique metabolic milieu, which promotes a Th17 and neutrophil mediated inflammation and suppresses innate responses[125]. Conversely, metabolites from activated immune cells, e.g. reactive nitrogen species have been shown to preferentially promote growth of facultative anaerobes (mostly Proteobacteria) on mucosal surfaces, thereby altering the microbial composition[13].
A number of different approaches are being explored to study the effect of the lung microbiome on the host metabolome. In a study by Garg et. al., the authors created a methodology in order to visualize human lung in 3D and applied this to mapping 16S rRNA sequencing and metabolomics data to patient specific reference space. This allowed a better visualization for correlation between microbial and molecular penetration. These visuals also revealed that local environments within the lung vary in terms of pathogen abundance, which can affect the metabolome in unique ways [126].
Microbial manipulations
The microbiome residing on various mucosal surfaces plays a central role in health and disease [1,58,127]. While much is known about gut microbiome, fecal microbial transplant (FMT) has been used only in the context of difficult gut infections like C. difficile, and that too in an effective, yet simple approach, where ‘normal’ microbiota is obtained from an identified and monitored cohort of individuals and cryopreserved for future FMT applications [128–132]. Only recently, identified species or communities of commensal bacteria are being considered for remedy in gut complications [133,134]. Consensus is gradually building on the concept that constant ‘tutoring’ of the host immune system, important for homeostasis, is not the function of the whole microbiome. Rather, this responsibility falls upon a few members of the community, where the majority of other microbes provide ‘buffering’ effect to keep these so called ‘pathobionts’ in check (For significantly changing bacteria, implicated in environmental and disease conditions, see Table 1). Disease is a state, where these pathobionts are able to overcome this buffering, and trigger a full-scale activation of host immune system-starting a vicious cycle, feeding into each other and manifesting the aforementioned morbidities and inflammation [4,134–137]. Hence, there is a huge potential in microbial restoration in the lungs, in the context of management of lung co-morbidities associated with different disease conditions as discussed in the previous sections.
Table 1:
Summary of documented lung microbial changes due to environmental factors and diseases.
| Disease or Environmental factor | Alterations to diversity | Increase in relative abundance (Phyla/ class/ genera) | Decrease in relative abundance (Phyla/ class/ genera) | Key genera | Reference |
|---|---|---|---|---|---|
| Asthma | Not reported | Proteobacteria | Bacteroidetes |
Haemophilus Moraxella Neisseria Prevotella Veillonella |
28 |
| Breast Feeding | Not reported |
Proteobacteria Actinobacteria Firmicutes |
Not reported |
Moraxella Corynebacterium Dolosigranulum |
65 |
| Diet | Decreased alpha diversity |
Streptococcus Klebsiella |
All other phyla |
S. pneumoniae K. pneumoniae |
77–79 |
| Smoking | Increased alpha diversity | Not reported | Not reported | Not reported | 56 |
| Pollutants | Not reported |
Firmicutes Verrucomicrobia |
Bacteroidetes | Not reported | 82 |
| COPD | Not reported |
Proteobacteria Bacteriodetes Actinobacteria Firmicutes |
Bacteroidetes |
Haemophilus Pseudomonas Prevotella Streptococcus Moraxella Acinetobacter Fusobacterium Neisseria |
28, 71 |
| Cystic Fibrosis | Decreased Diversity and Richness |
Firmicutes Proteobacteria Actinobacteria |
Not reported |
Streptococcus Staphylococcus Mycobacterium Hemophilus Burkholderia Pseudomonas Stenotrophomonas |
74, 76 |
| HIV | Decreased alpha diversity |
Proteobacteria Firmicutes Actinobacteria |
Not reported |
Prevotella Veillonella |
81, 87 |
| Lung Cancer | Not reported |
Proteobacteria Firmicutes TM7 |
Not reported |
Thermus Legionella Veillonella Megasphaera |
52, 79 |
| Influenza A virus | Decreased alpha diversity |
Lactobacillus Streptococcus |
Bacillus Pseudomonas |
Lactobacillus Streptococcus Bacillus Pseudomonas |
106,107 |
| Rhinovirus and SRV | Not reported |
Moraxella Streptococcus Corynebacterium Hemophilus Dolosigranulum |
Staphylococcus |
Moraxella Streptococcus Corynebacterium Hemophilus Dolosigranulum Staphylococcus |
62, 109 |
Future Directions in the Field
One of the many challenges in understanding the lung microbiome in context of diseases is the consensus among researchers regarding sample collection and analysis. Most of the reported studies on lung microbiome include from samples collected from upper respiratory tract, lower respiratory tract, bronchial lavage as well as lung lobectomy. This is a wide range. The upper respiratory tract bacteria often cross-talks with the oral microbiome thereby resulting in confounding results. On a positive note, the researchers in the field realize this. There has been an effort by National Institutes of Health to start a human microbiome project (https://hmpdacc.org/hmp/)). This portal acts as a repository for the existing data and streamlines protocols and analysis tools.
Improvement of our understanding of the lung microbiome will also require agreeing to common commensal bacterial species in the lung. Doing so will afford researchers with novel lines of inquiry by allowing them to target known commensals for study. It may also identify commensals that the lung microbiome shares with other microbial pools and help to clarify the relationships between host microbiome in different mucosal surfaces.
The limitations of lung as a microbiome reservoir reside in the fact that the bacterial species are not as abundant as in the gut. However, understanding the composition of this is more likely to have a profound effect on the host inflammatory status as the highly vascular nature of the lungs will expose the microbial components directly to the blood stream, eliciting a more immediate response from the immune system.
Given the importance of gut microbiota in host metabolism, role of commensal bacteria in the biochemical processes of the lung is another topic of future interest. Understanding the metabolic significance of commensal presence, or lack thereof, in disease states could be valuable in development of novel disease treatments. If these links are indeed established, the use of FMT as a treatment method, limited as it may be, could serve as a basis for similar treatments in lung disease using BAL-derived lung microbiome from healthy donors. Recent breakthroughs in the study of the gut microbiome and potential treatments derived from such studies hint at a wealth of opportunities in discovery within the lung microbiome.
Acknowledgements
The relevant and pertinent projects in author’s laboratories, some forming basis of this review are supported in part by the NIH grants R21HL125021 (SB), EY14801 and an unrestricted grant from Research to Prevent Blindness to University of Miami.
Footnotes
The authors declare no competing interests.
The authors declare no pertinent conflict of interest or competing interests.
References cited
- 1.Ferreira CM, Vieira AT, Vinolo MAR, Oliveira F a, Curi R & Martins FDS (2014) The central role of the gut microbiota in chronic inflammatory diseases. J. Immunol. Res 2014, 689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Power SE, O’Toole PW, Stanton C, Ross RP & Fitzgerald GF (2014) Intestinal microbiota, diet and health. Br. J. Nutr 111, 387–402. [DOI] [PubMed] [Google Scholar]
- 3.McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, Graeber TG, Sonnenburg JL, Horvath S, Huttenhower C, McGovern DP, Fornace AJ, Borneman J & Braun J (2013) Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs JP & Braun J (2014) Immune and genetic gardening of the intestinal microbiome. FEBS Lett [DOI] [PMC free article] [PubMed]
- 5.Kamada N, Chen GY, Inohara N & Núñez G (2013) Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol 14, 685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Hurtado I, Santacruz A, Peiró G, Zapater P, Gutiérrez A, Pérez-Mateo M, Sanz Y & Francés R (2011) Gut microbiota dysbiosis is associated with inflammation and bacterial translocation in mice with CCl4-induced fibrosis. PLoS One 6, e23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maier L, Diard M, Sellin ME, Chouffane E-S, Trautwein-Weidner K, Periaswamy B, Slack E, Dolowschiak T, Stecher B, Loverdo C, Regoes RR & Hardt W-D (2014) Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Salmonella Typhimurium Colitis. PLoS Pathog 10, e1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson R, Erb-Downward J & Huffnagle G (2013) The Role of the Bacterial Microbiome in Lung Disease. Expert Rev. Respir. Med 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP & Huffnagle GB (2015) The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLOS Pathog 11, e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM & Weinstock GM (2013) Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med 187, 1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourzac K (2014) The bacterial tightrope. Nature 516, S14–S16. [DOI] [PubMed] [Google Scholar]
- 12.Iii HDH, Sapkota AR, Rothman N, Rohan T, Hu W, Xu J, Vermeulen R, He X, White JR, Wu G, Wei F & Mongodin EF (2014) The Potential Role of Lung Microbiota in Lung Cancer Attributed to Household Coal Burning Exposures. Environ. Mol. Mutagen 00. [DOI] [PMC free article] [PubMed]
- 13.Scales BS, Dickson RP & Huffnagle GB (2016) A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J. Leukoc. Biol 100, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dwyer DNO, Dickson RP & Moore B (2016) The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol 196, 4839–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stressmann FA, Rogers GB, Klem ER, Lilley AK, Donaldson SH, Daniels TW, Carroll MP, Patel N, Forbes B, Boucher RC, Wolfgang MC & Bruce KD (2011) Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from American and British centers. J. Clin. Microbiol 49, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wills-karp M, Santeliz J & Karp CL (2001) Revisiting the Hygiene Hypothesis. Nat. Rev. Immunol 1, 69–75. [DOI] [PubMed] [Google Scholar]
- 17.Risnes KR, Belanger K, Murk W & Bracken MB (2011) Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am. J. Epidemiol 173, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKeever TM, Lewis SA, Smith C, Collins J, Heatlie H, Frischer M & Hubbard R (2002) Early exposure to infections and antibiotics and the incidence of allergic disease: A birth cohort study with the West Midlands General Practice Research Database. J. Allergy Clin. Immunol 109, 43–50. [DOI] [PubMed] [Google Scholar]
- 19.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ & Huffnagle GB (2011) Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabrera-Rubio R, Garcia-Nunez M, Seto L, Anto JM, Moya A, Monso E & Mira A (2012) Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J. Clin. Microbiol 50, 3562–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pragman AA, Kim HB, Reilly CS, Wendt C & Isaacson RE (2012) The Lung Microbiome in Moderate and Severe Chronic Obstructive Pulmonary Disease. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV., Cooper J, Sin DD, Mohn WW & Hogge JC (2012) The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 185, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huffnagle GB (2010) The microbiota and Allergies/Asthma. PLoS Pathog 6, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM & Souza DG (2012) Transient TLR Activation Restores Inflammatory Response and Ability To Control Pulmonary Bacterial Infection in Germfree Mice. J. Immunol 188, 1411–1420. [DOI] [PubMed] [Google Scholar]
- 25.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y & Weiser JN (2010) Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med 16, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, Vasson MP & Filaire E (2017) Desired Turbulence? Gut-Lung Axis, Immunity, and Lung Cancer. J. Oncol 2017. [DOI] [PMC free article] [PubMed]
- 27.Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E & Guarino A (2014) Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with lactobacillus gg: A randomised clinical trial. PLoS One 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L & Jenmalm MC (2012) Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol 129. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Cai Y, Huse SM, Knight R, Farmerie WG, Wang X & Mai V (2012) A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief. Bioinform 13, 107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD & Westcott SL (2011) Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol 77, 3219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, DeSantis TZ, Andersen GL & Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costello EEK, Lauber CCL, Hamady M, Fierer N, Gordon JI & Knight R (2009) Bacterial community variation in human body habitats across space and time. Science (80-. ) 326, 1694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN & Huffnagle GB (2014) Cell-associated bacteria in the human lung microbiome. Microbiome 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck JM, Schloss PD, Venkataraman A, Twigg H, Jablonski KA, Bushman FD, Campbell TB, Charlson ES, Collman RG, Crothers K, Curtis JL, Drews KL, Flores SC, Fontenot AP, Foulkes MA, Frank I, Ghedin E, Huang L, Lynch SV, Morris A, Palmer BE, Schmidt TM, Sodergren E, Weinstock GM & Young VB (2015) Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am. J. Respir. Crit. Care Med 192, 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly BJ, Imai I, Bittinger K, Laughlin A, Fuchs BD, Bushman FD & Collman RG (2016) Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD & Collman RG (2011) Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med 184, 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF & Cookson WOC (2010) Disordered Microbial Communities in Asthmatic Airways. PLoS One 5, e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL & Wong GK-S (2016) Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol 7, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janda JM & Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol 45, 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG & Bushman FD (2012) Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One 7, e42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethi S, Maloney J, Grove L, Wrona C & Berenson CS (2006) Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 173, 991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL & Huffnagle GB (2015) Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks JP, Edwards DJ, Harwich MD, Rivera MC, Fettweis JM, Serrano MG, Reris RA, Sheth NU, Huang B, Girerd P, Strauss JF, Jefferson KK, Buck GA, Lagier J-C, Million M, Hugon P, Armougom F, Raoult D, Knight R, Jansson J, Field D, Fierer N, Desai N, JA F, Pinto A, Raskin L, Hong S, Bunge J, Leslin C, Jeon S, Epstein S, Ahn J-H, Kim B-Y, Song J, Weon H-Y, Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Lee C, Herbold C, Polson S, Wommack K, Williamson S, IR M, Wu J-Y, Jiang X-T, Jiang Y-X, Lu S-Y, Zou F, Zhou H-W, Wu G, Lewis J, Hoffmann C, Chen Y-Y, Knight R, K B, Kanagawa T, Feinstein L, Sui W, Blackwood C, Whitehouse C, Hottel H, Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, EB H, Quince C, Lanzén A, Curtis T, Davenport R, Hall N, IM H, Kunin V, Engelbrektson A, Ochman H, Hugenholtz P, Huse S, Welch D, Morrison H, Sogin M, Haas B, Gevers D, Earl A, Feldgarden M, Ward D, Giannoukos G, Kembel S, Wu M, Eisen J, Green J, Dubourg G, Lagier J-C, Armougom F, Robert C, Hamad I, P B, Paulson J, Stine O, Bravo H, Pop M, Bergmann G, Bates S, Eilers K, Lauber C, Caporaso J, WA W, Lauber C, Hamady M, Knight R, Fierer N, Andreson R, Mols T, Remm M, Shinoda N, Yoshida T, Kusama T, Takagi M, Hayakawa T, T O, Kiviharju K, Leisola M, Eerikäinen T, Rispoli F, Shah V, Bautista-Gallego J, Arroyo-López F, Chiesa A, Duráin-Quintana M, Garrido-Fernández A, Harbi B, Chaieb K, Jabeur C, Mahdouani K, Bakhrouf A, Arroyo-López F, Bautista-Gallego J, Chiesa A, Durán-Quintana M, Garrido-Fernández A, Wang Q, Garrity G, Tiedje J, Cole J, Polz M, Cavanaugh C, Huber J, Morrison H, Huse S, Neal P, Sogin M, Welch D, Cornell J, Scheffé H, Goos P, Jones B, Fettweis J, Serrano M, Sheth N, Mayer C, Glascock A, JP B, Ravel J, Gajer P, Abdo Z, Schneider G, Koenig S, SL M, Harwich MJ, Serrano M, Fettweis J, Alves J, Reimers M, Wickham H, Liaw A & Wiener M (2015) The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol 15, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal LN & Blaser MJ (2014) A brave new world: The lung microbiota in an era of change. Ann. Am. Thorac. Soc 11, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gleeson K, Eggli DF & Maxwell SL (1997) Quantitative aspiration during sleep in normal subjects. Chest 111, 1266–72. [DOI] [PubMed] [Google Scholar]
- 46.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB & Schmidt TM (2015) Application of a neutral community model to assess structuring of the human lung microbiome. MBio 6, e02284–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller NT, Bakacs E, Combellick J, Grigoryan Z & Dominguez-Bello MG (2015) The infant microbiome development: mom matters. Trends Mol. Med 21, 109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N & Knight R (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A 107, 11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones HE, Harris KA, Azizia M, Bank L, Carpenter B, Hartley JC, Klein N & Peebles D (2009) Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS One 4, e8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S & Relman DA (2008) Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 3, e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J & Versalovic J (2014) The placenta harbors a unique microbiome. Sci. Transl. Med 6, 237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lal CV, Travers C, Aghai ZH, Eipers P, Jilling T, Halloran B, Carlo WA, Keeley J, Rezonzew G, Kumar R, Morrow C, Bhandari V, Ambalavanan N, Rogers GB, Segata N, Mu J, Arrieta MC, D’Angio CT, Basavegowda K, Avissar NE, Finkelstein JN, Sinkin RA, Ramsay PL, Watterberg KL, Demers LM, Scott SM, Murphy S, Viscardi RM, Dominguez-Bello MG, Aagaard K, Mourani PM, Harris JK, Sontag MK, Robertson CE, Abman SH, Lohmann P, Gritz EC, Bhandari V, Jobe AH, Cox MJ, Fujimura KE, Cope EK, Lynch SV, Pellaton C, Yun Y, Taft DH, Stout MJ, Fardini Y, Chung P, Dumm R, Joshi N, Han YW, Han YW, Han YW, Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS, Wassenaar TM, Panigrahi P, Barberan A, Justino PF, Mohamadzadeh M, Fernandez EM, Plaza-Diaz J, Sagar S, Simeoli R, Trompette A, Walsh MC, Mody K, Aghai ZH, Aghai ZH, Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD, Kumar R, Caporaso JG, Lozupone C, Hamady M, Knight R, Vazquez-Baeza Y, Pirrung M, Gonzalez A & Knight R (2016) The Airway Microbiome at Birth. Sci. Rep 6, 31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.EDWARDS FC & TRUELOVE SC (1964) THE COURSE AND PROGNOSIS OF ULCERATIVE COLITIS. III. COMPLICATIONS. Gut 5, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraft SC, Earle RH, Roesler M & Esterly JR (1976) Unexplained Bronchopulmonary Disease With Inflammatory Bowel Disease. Arch Intern Med 136, 454–459. [PubMed] [Google Scholar]
- 55.Mohamed-Hussein AA, Mohamed NA & Ibrahim M-EA (2007) Changes in pulmonary function in patients with ulcerative colitis ARTICLE IN PRESS. Respir. Med 101, 977–982. [DOI] [PubMed] [Google Scholar]
- 56.Munck A, Murciano D, Pariente R, Cezard J & Navarro J (1995) Latent pulmonary function abnormalities in children with Crohn’s disease. Eur. Respir. J 8. [DOI] [PubMed] [Google Scholar]
- 57.Jess T, Loftus EV, Harmsen WS, Zinsmeister AR, Tremaine WJ, Melton LJ, Munkholm P & Sandborn WJ (2006) Survival and cause specific mortality in patients with inflammatory bowel disease: a long term outcome study in Olmsted County, Minnesota, 1940–2004. Gut 55, 1248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keely S, Talley NJ & Hansbro PM (2012) Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol 5, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somerville KW, Logan RF, Edmond M & Langman MJ (1984) Smoking and Crohn’s disease. Br. Med. J. (Clin. Res. Ed) 289, 954–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cosnes J (2004) Tobacco and IBD: relevance in the understanding of disease mechanisms and clinical practice. Best Pract. Res. Clin. Gastroenterol 18, 481–496. [DOI] [PubMed] [Google Scholar]
- 61.Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, Steurer-Stey C, Frei A, Frei P, Scharl M, Loessner MJ, Vavricka SR, Fried M, Schreiber S, Schuppler M & Rogler G (2013) Smoking Cessation Induces Profound Changes in the Composition of the Intestinal Microbiota in Humans. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, Shankar J, Yooseph S, Nelson KE, Halpin RA, Moore ML, Anderson LJ, Peebles RS, Das SR & Hartert TV. (2016) Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J. Infect. Dis 214, 1924–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, Caporaso NE, Goedert JJ, Ravel J & Landi MT (2016) Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 17, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones JG, Lawler P, Crawley JCW, Minty BD, Hulands G & Veall N (1980) INCREASED ALVEOLAR EPITHELIAL PERMEABILITY IN CIGARETTE SMOKERS. Lancet 315, 66–68. [DOI] [PubMed] [Google Scholar]
- 65.Moazed F, Burnham EL, Vandivier RW, Kane CMO, Shyamsundar M, Hamid U, Abbott J, Thickett DR, Matthay MA, Mcauley DF & Calfee CS (2016) Cigarette smokers have exaggerated alveolar barrier disruption in response to lipopolysaccharide inhalation, 1130–1136. [DOI] [PMC free article] [PubMed]
- 66.Kelly JR, Kennedy PJ, Cryan JF & Dinan TG (2015) Breaking down the barriers : the gut microbiome, intestinal permeability and stress-related psychiatric 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jungnickel C, Wonnenberg B, Karabiber O, Wolf A, Voss M, Wolf L, Honecker A, Kamyschnikow A, Herr C, Bals R & Beisswenger C (2015) Cigarette smoke-induced disruption of pulmonary barrier and bacterial translocation drive tumor-associated inflammation and growth. Am. J. Physiol. - Lung Cell. Mol. Physiol 309. [DOI] [PubMed] [Google Scholar]
- 68.Gallacher DJ & Kotecha S (2016) Respiratory Microbiome of New-Born Infants. Front. Pediatr 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heikkila MP (2003) Inhibition of Staphylococcus aureus by the commensal bacteria of human milk, 471–478. [DOI] [PubMed]
- 70.Jost T, Lacroix C, Braegger C & Chassard C (2015) Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health 73, 426–437. [DOI] [PubMed] [Google Scholar]
- 71.Simon PM, Goode PL, Mobasseri A & Zopf D (1997) Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect. Immun 65, 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biesbroek G, Tsivtsivadze E, Sanders EAM, Montijn R, Veenhoven RH, Keijser BJF & Bogaert D (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med 190, 1283–1292. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J, Murray S, LiPuma JJ, Costello EK, Yatsunenko T, Kostic AD, Karlsson FH, Yoshimoto S, Arthur JC, Atarashi K, Koren O, Jansson J, Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK, Gajer P, Westermann AJ, Gorski SA, Vogel J, Sharma CM, Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA, Lemon KP, Armitage GC, Relman DA, Fischbach MA, Dethlefsen L, Relman DA, Jernberg C, Lofmark S, Edlund C, Jansson JK, Jakobsson HE, Jernberg C, Lofmark S, Edlund C, Jansson JK, Dethlefsen L, Huse S, Sogin ML, Relman DA, Zhao J, Konstan MW, Wagener JS, VanDevanter DR, Filkins LM, Nick JA, Cox MJ, Klepac-Ceraj V, van der Gast CJ, VanDevanter DR, LiPuma JJ, Schloss PD, Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ, Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R, Huse SM, Welch DM, Morrison HG & Sogin ML (2014) Modeling the Impact of Antibiotic Exposure on Human Microbiota. Sci. Rep 4, 1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray LEK, O’Hely M, Ranganathan S, Sly PD & Vuillermin P (2017) The maternal diet, gut bacteria, and bacterial metabolites during pregnancy influence offspring asthma. Front. Immunol 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, Reynolds LA, Hacker L, Mohr J, Finlay BB, Zaph C, McNagny KM & Mohn WW (2017) Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol [DOI] [PubMed]
- 76.Sokolowska M, Frei R, Lunjani N, Akdis CA & O’Mahony L (2018) Microbiome and asthma. Asthma Res. Pract 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bou Ghanem EN, Clark S, Du X, Wu D, Camilli A, Leong JM & Meydani SN (2015) The α-Tocopherol Form of Vitamin E Reverses Age-Associated Susceptibility to Streptococcus pneumoniae Lung Infection by Modulating Pulmonary Neutrophil Recruitment. J. Immunol 194, 1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bou Ghanem EN, Lee JN, Joma BH, Meydani SN, Leong JM & Panda A (2017) The Alpha-Tocopherol Form of Vitamin E Boosts Elastase Activity of Human PMNs and Their Ability to Kill Streptococcus pneumoniae. Front. Cell. Infect. Microbiol 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown RL, Sequeira RP & Clarke TB (2017) The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han MK, Postma D, Mannino DM, Giardino ND, Buist S, Curtis JL & Martinez FJ (2007) Gender and Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med 176, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dickson RP, Martinez FJ & Huffnagle GB (2014) The role of the microbiome in exacerbations of chronic lung diseases. Lancet 384, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adar SD, Huffnagle GB & Curtis JL (2016) The respiratory microbiome: an underappreciated player in the human response to inhaled pollutants? Ann. Epidemiol 26, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lopez AD, Shibuya K, Rao C, Mathers CD, Hansell AL, Held LS, Schmid V & Buist S (2006) Chronic obstructive pulmonary disease: Current burden and future projections. Eur. Respir. J 27, 397–412. [DOI] [PubMed] [Google Scholar]
- 84.Wedzicha JA & Donaldson GC (2003) Exacerbations of chronic obstructive pulmonary disease. Respir. Care 48, 1204–13; discussion 1213–5. [PubMed] [Google Scholar]
- 85.SEEMUNGAL TAR, DONALDSON GC, BHOWMIK A, JEFFRIES DJ & WEDZICHA JA (2000) Time Course and Recovery of Exacerbations in Patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med 161, 1608–1613. [DOI] [PubMed] [Google Scholar]
- 86.Sethi S & Murphy TF (2008) Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med 359, 2355–65. [DOI] [PubMed] [Google Scholar]
- 87.Murphy TF, Brauer AL, Schiffmacher AT & Sethi S (2004) Persistent Colonization by Haemophilus influenzae in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med 170, 266–272. [DOI] [PubMed] [Google Scholar]
- 88.Chen R, Lim JH, Jono H, Gu XX, Kim YS, Basbaum CB, Murphy TF & Li JD (2004) Nontypeable Haemophilus influenzae lipoprotein P6 induces MUC5AC mucin transcription via TLR2-TAK1-dependent p38 MAPK-AP1 and IKK??- I??B??-NF-??B signaling pathways. Biochem. Biophys. Res. Commun 324, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 89.Cutting GR (2015) Cystic fibrosis genetics: from molecular understanding to clinical application. HHS Public Access 16, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Høiby N (1988) Haemophilus Influenzae, Staphylococcus aureus, Pseudomonas cepacia, and Pseudomonas aeruginosa in Patients with Cystic Fibrosis. Chest 94, 97S–102S. [DOI] [PubMed] [Google Scholar]
- 91.Beaume M, Köhler T, Greub G, Manuel O, Aubert J-D, Baerlocher L, Farinelli L, Buckling A, van Delden C, Achermann R, Amico P, Baumann P, Beldi G, Benden C, Berger C, Binet I, Bochud P-Y, Boely E, Bucher H, Bühler L, Carell T, Catana E, Chalandon Y, Geest de S, Rougemont de O, Dickenmann M, Duchosal M, Fehr T, Ferrari-Lacraz S, Garzoni C, Soccal PG, Giostra E, Golshayan D, Good D, Hadaya K, Halter J, Heim D, Hess C, Hillinger S, Hirsch HH, Hofbauer G, Huynh-Do U, Immer F, Klaghofer R, Koller M, Laesser B, Lehmann R, Lovis C, Marti H-P, Martin PY, Martinolli L, Meylan P, Mohacsi P, Morard I, Morel P, Mueller U, Mueller NJ, Mueller-McKenna H, Müller A, Müller T, Müllhaupt B, Nadal D, Pascual M, Passweg J, Ziegler CP, Rick J, Roosnek E, Rosselet A, Rothlin S, Ruschitzka F, Schanz U, Schaub S, Seiler C, Stampf S, Steiger J, Stirnimann G, Toso C, Tsinalis D, Venetz J-P, Villard J, Wick M, Wilhelm M & Yerly P (2017) Rapid adaptation drives invasion of airway donor microbiota by Pseudomonas after lung transplantation. Sci. Rep 7, 40309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL & Lynch SV. (2010) Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Froidure A & Pilette C (2016) From the hygiene hypothesis to A20: The protective effect of endotoxins against asthma development. Clin. Exp. Allergy 46, 192–193. [DOI] [PubMed] [Google Scholar]
- 94.Foglia E, Meier MD & Elward A (2007) Ventilator-associated pneumonia in neonatal and pediatric intensive care unit patients. Clin. Microbiol. Rev 20, 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mourani PM & Sontag MK (2017) Ventilator-Associated Pneumonia in Critically Ill Children: A New Paradigm. Pediatr. Clin. North Am 64, 1039–1056. [DOI] [PubMed] [Google Scholar]
- 96.File TM (2003) Community-acquired pneumonia. Lancet 362, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M & Sanders E (2011) Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Steenhuijsen Piters WAA, Huijskens EGW, Wyllie AL, Biesbroek G, Van Den Bergh MR, Veenhoven RH, Wang X, Trzcinski K, Bonten MJ, Rossen JWA, Sanders EAM & Bogaert D (2016) Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J 10, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kageyama S, Takeshita T, Furuta M, Tomioka M, Asakawa M, Suma S, Takeuchi K, Shibata Y, Iwasa Y, Yamashita Y & Newman A (2017) Relationships of Variations in the Tongue Microbiota and Pneumonia Mortality in Nursing Home Residents. Journals Gerontol. Ser. A 00, 1–6. [DOI] [PubMed] [Google Scholar]
- 100.Vogtmann E & Goedert JJ (2016) Epidemiologic studies of the human microbiome and cancer. Br. J. Cancer 114, 237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, Chung KS, Kim EY, Jung JY, Kang YA, Kim YS, Kim SK, Chang J & Park MS (2016) Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 102, 89–95. [DOI] [PubMed] [Google Scholar]
- 102.Zilberman-Schapira G, Zmora N, Itav S, Bashiardes S, Elinav H & Elinav E (2016) The gut microbiome in human immunodeficiency virus infection. BMC Med 14, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Twigg HL, Knox KS, Zhou J, Crothers KA, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, Mi D, Katz BP, Sodergren E & Weinstock GM (2016) Effect of advanced HIV infection on the respiratory microbiome. Am. J. Respir. Crit. Care Med 194, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pichon M, Lina B & Josset L (2017) Impact of the Respiratory Microbiome on Host Responses to Respiratory Viral Infection. Vaccines 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harris J-AS (2006) Infection Control in Pediatric Extended Care Facilities. Infect. Control Hosp. Epidemiol 27, 598–603. [DOI] [PubMed] [Google Scholar]
- 106.Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B & Schmolke M (2018) Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Borges LG dos A, Giongo A, Pereira L de M, Trindade FJ, Gregianini TS, Campos FS, Ghedin E & da Veiga ABG (2018) Comparison of the nasopharynx microbiome between influenza and non-influenza cases of severe acute respiratory infections: A pilot study. Heal. Sci. Reports, e47. [DOI] [PMC free article] [PubMed]
- 108.Bartley JM, Zhou X, Kuchel GA, Weinstock GM & Haynes L (2017) Impact of Age, Caloric Restriction, and Influenza Infection on Mouse Gut Microbiome: An Exploratory Study of the Role of Age-Related Microbiome Changes on Influenza Responses. Front. Immunol 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, Timmerman HM, Huynen MA, van Hijum SAFT & de Jonge MI (2018) Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ivanov II, Frutos R de L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB & Littman DR (2008) Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, Montelaro RC, Metzger DW, Gulati AS & Kolls JK (2016) Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 44, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Segal LN, Alekseyenko A, Clemente JC, Berger K, Goldring R, Rom WN, Blaser MJ & Weiden MD (2014) Enrichment of Lung Microbiome with Supraglotic Microbes Is Associated with Increased Pulmonary Inflammation. Ann. Am. Thorac. Soc 11, S71–S71. [Google Scholar]
- 113.Huang YJ, Nariya S, Harris JM, Lynch SV., Choy DF, Arron JR & Boushey H (2015) The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol 136, 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, Von Garnier C, Nicod LP & Marsland BJ (2016) Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am. J. Respir. Crit. Care Med 193, 975–987. [DOI] [PubMed] [Google Scholar]
- 115.Richmond BW, Brucker RM, Han W, Du RH, Zhang Y, Cheng DS, Gleaves L, Abdolrasulnia R, Polosukhina D, Clark PE, Bordenstein SR, Blackwell TS & Polosukhin VV. (2016) Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat. Commun 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Nitert MD, Tremellen A, Tobin J, Wilkinson S, McSweeney C, O’Rourke P, Lingwood B, Kang A, Shanahan E, Fukuma N, Angel N & Foxcroft K (2016) Connections between the gut microbiome and metabolic hormones in early pregnancy in overweight and obese women. Diabetes 65, 2214–2223. [DOI] [PubMed] [Google Scholar]
- 117.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD & Pedersen O (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. [DOI] [PubMed] [Google Scholar]
- 118.Devaraj S, Hemarajata P & Versalovic J (2013) The human gut microbiome and body metabolism: Implications for obesity and diabetes. Clin. Chem 59, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Velasquez MT, Ramezani A, Manal A & Raj DS (2016) Trimethylamine N-oxide: The good, the bad and the unknown. Toxins (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Horáčková Š, Plocková M & Demnerová K (2017) Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol. Adv [DOI] [PubMed]
- 121.Duda-Chodak A, Tarko T, Satora P & Sroka P (2015) Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur. J. Nutr 54, 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davies PL, Spiller OB, Beeton ML, Maxwell NC, Remold-O’Donnell E & Kotecha S (2010) Relationship of proteinases and proteinase inhibitors with microbial presence in chronic lung disease of prematurity. Thorax 65, 246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cribbs SK, Park Y, Guidot DM, Martin GS, Brown LA, Lennox J & Jones DP (2014) Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res. Hum. Retroviruses 30, 579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L, Fitch A, Greenblatt RM, Kingsley L, Guidot DM, Ghedin E & Morris A (2016) Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Segal LN, Clemente JC, Tsay JJ, Koralov SB, Keller C, Wu BG, Li Y, Shen N, Ghedin E, Morris A, Diaz P, Huang L, Wikoff WR, Ubeda C, Artacho A & Weiden MD (2016) Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol 1, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, Protsyuk I, Bouslimani A, Lim YW, Wong R, Humphrey G, Ackermann G, Spivey T, Brouha SS, Bandeira N, Lin GY, Rohwer F, Conrad DJ, Alexandrov T, Knight R & Dorrestein PC (2017) Three-Dimensional Microbiome and Metabolome Cartography of a Diseased Human Lung. Cell Host Microbe 22, 705–716.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang H, Liu J-S, Peng S-H, Deng X-Y, Zhu D-M, Javidiparsijani S, Wang G-R, Li D-Q, Li L-X, Wang Y-C & Luo J-M (2013) Gut-lung crosstalk in pulmonary involvement with inflammatory bowel diseases. World J. Gastroenterol 19, 6794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khoruts A, Sadowsky MJ & Hamilton MJ (2015) Development of Fecal Microbiota Transplantation Suitable for Mainstream Medicine. Clin. Gastroenterol. Hepatol 13, 246–250. [DOI] [PubMed] [Google Scholar]
- 129.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ & Khoruts A (2014) Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol 306, G310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hamilton MJ, Weingarden AR, Sadowsky MJ & Khoruts A (2012) Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am. J. Gastroenterol 107, 761–7. [DOI] [PubMed] [Google Scholar]
- 131.Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, Gordon S, Gluck M, Hohmann EL, Kao D, Kao JY, McQuillen DP, Mellow M, Rank KM, Rao K, Ray A, Schwartz M a, Singh N, Stollman N, Suskind DL, Vindigni SM, Youngster I & Brandt L (2014) Fecal Microbiota Transplant for Treatment of Clostridium difficile Infection in Immunocompromised Patients. Am. J. Gastroenterol 109, 1065–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Khoruts a & Sadowsky MJ (2011) Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol 4, 4–7. [DOI] [PubMed] [Google Scholar]
- 133.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M & Honda K (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–6. [DOI] [PubMed] [Google Scholar]
- 134.Honda K & Littman DR (2012) The Microbiome in Infectious Disease and Inflammation. Annu. Rev. Immunol 30, 759–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jones ML, Tomaro-Duchesneau C & Prakash S (2014) The gut microbiome, probiotics, bile acids axis, and human health. Trends Microbiol 22, 306–8. [DOI] [PubMed] [Google Scholar]
- 136.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone C a., Lauber C, Clemente JC, Knights D, Knight R & Gordon JI (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Manor O, Levy R & Borenstein E (2014) Mapping the Inner Workings of the Microbiome: Genomic- and Metagenomic-Based Study of Metabolism and Metabolic Interactions in the Human Microbiome. Cell Metab [DOI] [PMC free article] [PubMed]