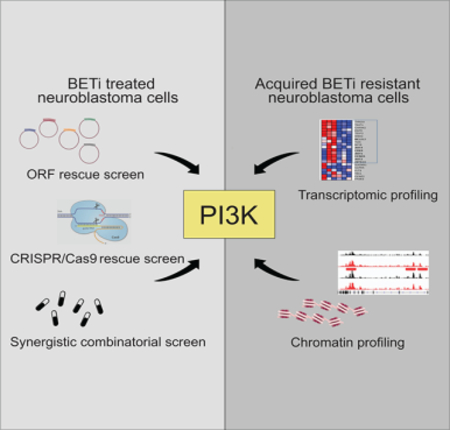
SUMMARY
Drug resistance represents a major challenge to achieving durable responses to cancer therapeutics. Resistance mechanisms to epigenetically-targeted drugs remain largely unexplored. We used BET inhibition in neuroblastoma as a prototype to model resistance to chromatin modulatory therapeutics. Genome-scale, pooled lentiviral open reading frame (ORF) and CRISPR knockout rescue screens nominated the PI3K pathway as promoting resistance to BET inhibition. Transcriptomic and chromatin profiling of resistant cells revealed that global enhancer remodeling is associated with upregulation of receptor tyrosine kinases (RTKs), activation of PI3K signaling and vulnerability to RTK/PI3K inhibition. Large-scale combinatorial screening with BET inhibitors identified PI3K inhibitors among the most synergistic upfront combinations. These studies provide a roadmap to elucidate resistance to epigenetic-targeted therapeutics and inform efficacious combination therapies.
IN BRIEF
Using functional screens, profiling of drug resistant cells, and drug combination screens in neuroblastoma, Iniguez et al. show that PI3K pathway activation via enhancer remodeling and transcriptional reprogramming confers resistance to BET inhibitors (BETi) and that PI3K inhibitors synergize with BETi.
Graphical Abstract
INTRODUCTION
The approval of targeted cancer therapeutics has initiated an age of precision medicine-based cancer treatment. Several tyrosine kinase inhibitors (TKIs) have seen remarkable success in the clinic, including imatinib, a BCR-ABL inhibitor, in chronic myeloid leukemia (Gambacorti-Passerini et al., 2011); lapatinib, a HER2 inhibitor, in HER2-amplified breast cancer (Geyer et al., 2006; Slamon et al., 2001); gefitinib and erlotinib, EGFR inhibitors, in EGFR-mutant cancers (Gridelli et al., 2010; Sridhar et al., 2003; Vecchione et al., 2011); crizotinib, an ALK inhibitor, in ALK-positive non-small cell lung cancer (Kwak et al., 2010); and vemurafenib, a BRAF inhibitor, in BRAF-mutant melanoma (Chapman et al., 2011). In order to optimize their clinical application, extensive preclinical studies were carried out to identify putative mechanisms of resistance to these TKIs. Shared resistance mechanisms included mutations in the drug target (most common), reactivation of the targeted pathway, and activation of compensatory pathways (Ramos and Bentires-Alj, 2015; Shah et al., 2002). Importantly, these preclinical studies of resistance accurately predicted resistance in human patients (Cools et al., 2004; Emery et al., 2009; Ercan et al., 2010; Johannessen et al., 2010).
The second wave of targeted agents is now entering clinical trials, and these molecules are focused on chromatin regulators. Epigenetic landscapes of tumors are frequently dysregulated, and mutations in several genes encoding epigenetic regulators have been identified, including genes involved in DNA methylation, chromatin remodeling complexes, and histone acetylation and methylation (Pfister and Ashworth, 2017). The mechanisms of resistance to many classes of epigenetic regulating drugs remain unknown.
BET (bromodomain and extra-terminal domain) protein inhibitors are a class of epigenetic inhibitors with several molecules being evaluated in clinical trials for the treatment of lymphoma, acute leukemia, and various solid tumors (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Mertz et al., 2011; Zuber et al., 2011). The BET family of proteins are epigenetic readers that regulate transcription through binding to acetylated lysine residues on histones (Zeng and Zhou, 2002). Previous work by our laboratories and others identified MYCN-amplified neuroblastoma as a disease sensitive to BET inhibitors (Henssen et al., 2016; Puissant et al., 2013; Wyce et al., 2013). Neuroblastoma is a good model system to study epigenetic-based resistance due to its low mutation frequencies with few genes mutated recurrently other than amplification of MYCN, mutations in ALK, and enhancer hijacking involving the telomerase reverse transcriptase (TERT) gene (Molenaar et al., 2012; Pugh et al., 2013; Sausen et al., 2013; Valentijn et al., 2015). The relatively stable genomes of these tumors implicate epigenetic dysregulation in the pathogenesis of this cancer.
Here, we present a roadmap for identifying mechanisms of resistance to diverse chromatin remodeling drugs in various cancers. We took an integrative approach using functional genomics screens, omic profiling of drug resistant cells and drug combination screens to nominate mechanisms of resistance to BET inhibitors in neuroblastoma and to discover efficacious drug combinations for preventing resistance.
RESULTS
Genome-scale lentiviral ORF and CRISPR rescue screens identify three major pathways that promote resistance to BET inhibitors
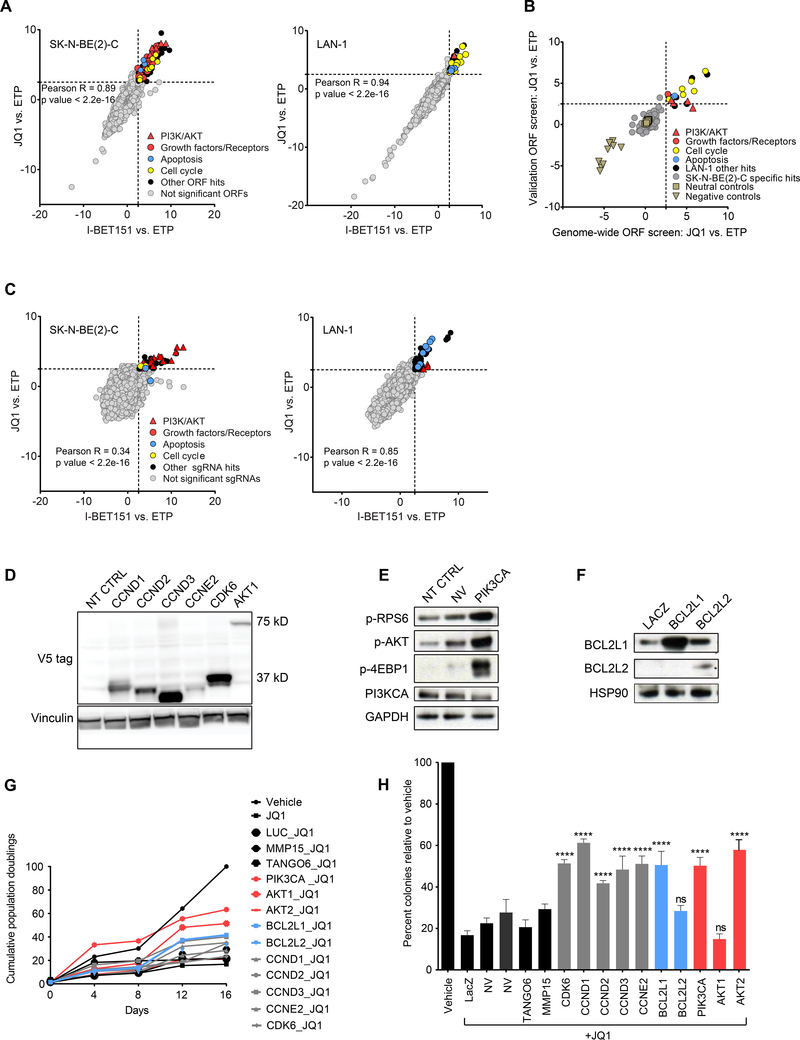
In anticipation of the testing of BET bromodomain inhibitors in neuroblastoma, we set out to elucidate mechanisms of acquired resistance to BET inhibitors to optimize their clinical application. We performed a genome-scale, lentiviral open reading frame (ORF) screen in two MYCN-amplified neuroblastoma cell lines sensitive to BET inhibitors (SK-N-BE(2)-C and LAN-1) with the BET inhibitors JQ1 and I-BET151. Cells were infected with a pooled lentiviral ORF library containing 17,255 barcoded ORFs, resulting in the individual overexpression of 10,135 distinct human genes with at least 99% nucleotide and protein match (Johannessen et al., 2013; Yang et al., 2011). Cells were then selected, passaged for 3–4 doublings, and an early time point (ETP) was harvested. ORF-expressing cells were passaged for three weeks in the presence of JQ1, I-BET151 or DMSO control (1 μM JQ1 and 5 μM I-BET151 for SK-N-BE(2)-C (Figure S1A) and 0.5 μM JQ1 and 5 μM I-BET151 for LAN-1). Cells were harvested and barcodes sequenced to compare log2 fold changes (log2(FC)) in ORF distribution under each treatment condition compared to the ETP to identify ORFs that were able to rescue the anti-viability effects of BET inhibition. ORF representation for JQ1 and I-BET151 treatment was strongly correlated in both cell lines (Figure 1A). ORFs were deemed hits if they had z-scores (standard deviations from the mean) for log2(FC) expression ≥ 2.5 with both JQ1 and I-BET151 vs. ETP. There were 154 ORF hits in the SK-N-BE(2)-C cell line, corresponding to 90 genes (Figure 1A and Table S1), and 34 ORF hits corresponding to 22 genes in the LAN-1 cell line (Figure 1A and Table S2). Importantly, top hits did not score in the DMSO control arm as promoting growth on their own over time in the SK-N-BE(2)-C cell line (Figure S1B). Top scoring hits in LAN-1 cells had a modest growth-promoting effect in the DMSO arm, which was strongly enhanced under selective drug pressure (Figure S1B). Genes that conferred resistance to both BET inhibitors were significantly enriched for three primary pathways: PI3K/AKT signaling, along with upstream growth factors and receptors, apoptosis, and cell cycle (Figure 1A and Figure S1C, D).
Figure 1: Genome-scale lentiviral ORF and CRISPR screens identify candidate drivers of BET inhibitor resistance in MYCN-amplified neuroblastoma.
A. Scatter plots of z-scores for log2 fold changes (log2(FC)) in ORF expression for JQ1 vs. ETP (y-axis) and I-BET151 vs. ETP (x-axis) in SK-N-BE(2)-C (left) and LAN-1 (right) cells. Genes with z-scores ≥ 2.5 with both BET inhibitors (dashed gray line) were nominated as candidate genes conferring resistance and classified as significant ORFs. B. Scatter plot showing the distribution of the JQ1 vs. ETP z-scores for the 150 ORFs included in the validation mini-ORF rescue screen in the LAN-1 cell line. C. Genome-scale pooled lenti-CRISPR screen in SK-N-BE(2)-C (left) and LAN-1 (right) cells under JQ1 and I-BET151 drug selection. Genes with z-scores ≥ 2.5 with both BET inhibitors (dashed gray line) were nominated as candidate sgRNAs conferring resistance and classified as significant sgRNAs. D-F. Western blots confirming overexpression of the indicated ORF hits with V5 antibody in cases where the V5 tag was expressed (D), or by antibodies directed against the ORF or downstream effectors (E, F) (p-AKT = pT308-AKT). G-H. Long-term viability assays (G) and colony formation assays (H) in SK-N-BE(2)-C cells overexpressing the indicated ORFs and treated with vehicle or 1 μM JQ1. Luciferase (LUC), LacZ, MMP15, and TANGO6 ORFs are included as negative controls. Data is presented as mean values of triplicate points ± standard deviation (SD), NT CTRL = non-targeting control ORF. NV= no virus. (* p value < 0.05, ** p value < 0.01, *** p value < 0.001, **** p value < 0.0001, Mann-Whitney nonparametric test). See also Figure S1 and Tables S1-S4.
To validate the genome-scale ORF screen, we conducted a secondary screen with 150 ORFs targeting 150 genes in the LAN-1 cell line and in an additional MYCN-amplified neuroblastoma cell line, CHP-212. The mini-pool lentiviral library included the top 90 ORFs that scored as hits with both BET inhibitors in the genome-scale screen in either SK-N-BE(2)-C or LAN-1 cells, as well as neutral control ORFs and negative control ORFs (Table S3). ORFs corresponding to genes regulating PI3K/AKT and cell cycle were validated as promoting JQ1 resistance in both LAN-1 and CHP-212 cells (Figure 1B, Figure S1E and Table S3).
In parallel, we performed a complementary genome-scale CRISPR rescue screen using the same two neuroblastoma cell lines and BET inhibitors as in the ORF screen. Cells were infected with the AVANA4 CRISPR, barcoded, pooled library containing 74,687 sgRNAs with ~4 sgRNAs/gene. Infected cells were then selected and passaged for one week. An ETP was collected and subsequently cells were treated with either JQ1 or I-BET151 for two weeks, collected, and sequenced to determine sgRNA distribution. There was a strong correlation between sgRNAs scoring with both BET inhibitors in both cell lines (Figure 1C). Genes were nominated as hits if they had a z-score ≥ 2.5 with both BET inhibitors vs. ETP. In the SK-N-BE(2)-C cell line, there were 39 high-scoring sgRNAs representing 19 genes (Figure 1C and Table S4). In the LAN-1 cell line, 50 sgRNAs scored corresponding to 31 genes (Figure 1C and Table S4). Strikingly, we found that sgRNAs against genes that repressed the three pathways nominated in the ORF screen scored very highly in the CRISPR screen: PI3K signaling, apoptosis and cell cycle.
To validate the resistance-promoting effects of each rescue ORF, we re-expressed ten top candidate genes in SK-N-BE(2)-C cells. We confirmed overexpression by V5 expression (Figure 1D) or by using individual antibodies against the target ORF (Figure 1E-F). We then treated these stably infected cells with either the vehicle control or JQ1. Overexpression of PIK3CA and AKT1 promoted resistance to the growth suppressive effects of JQ1 (Figure 1G) and did not promote growth in the absence of drug selection (data not shown). Additionally, eight ORFs rescued the effects of JQ1-mediated suppression of colony formation (Figure 1H). Furthermore, low-throughput suppression of PTEN via CRISPR/Cas9 mediated deletion also rescued the anti-viability effects of JQ1 treatment and conferred resistance to BET inhibition, confirming results of the CRISPR screen (Figure S1F, G).
Innate and acquired BET inhibitor resistance mechanisms in MYCN-amplified neuroblastoma
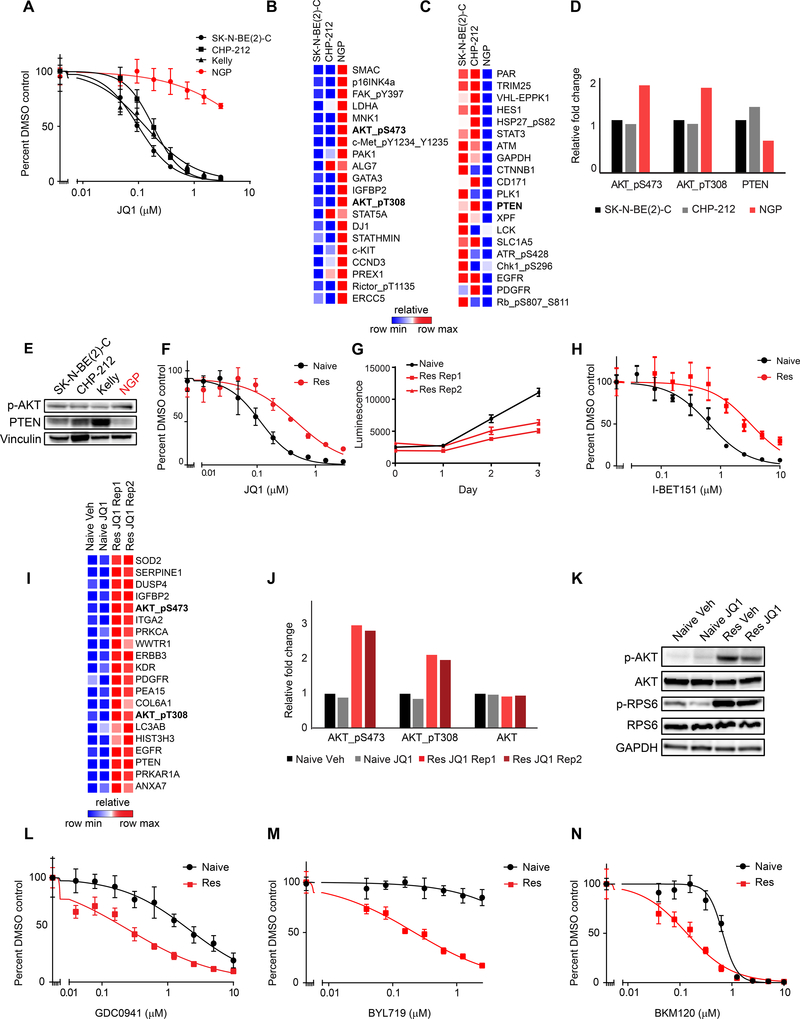
We next sought to identify innate resistance mechanisms to BET inhibition and determine whether these were related to top pathways scoring in the genetic resistance screens. We confirmed our previous observation (Puissant et al., 2013) and demonstrated that the MYCN-amplified neuroblastoma cell line NGP was innately resistant to BET inhibition (Figure 2A). We next performed proteomic and phosphoproteomic profiling of NGP cells and the JQ1 sensitive MYCN-amplified cell lines, SK-N-BE(2)-C and CHP-212, for comparison using a reverse phase protein array (RPPA) (Tibes et al., 2006). Among the top 20 upregulated proteins in NGP cells compared to SK-N-BE(2)-C and CHP-212 cells were two activating AKT phosphorylation sites: pS473-AKT and pT308-AKT (Figure 2B). Among the most downregulated proteins was PTEN, a lipid phosphatase and negative regulator of PI3K (Figure 2C, D, E). These findings suggest high PI3K pathway activation may underlie innate resistance to BET inhibitors in MYCN-amplified neuroblastoma.
Figure 2: Characterization of innate and acquired BET inhibitor resistant MYCN-amplified neuroblastoma cell lines.
A. Viability analysis of JQ1 treatment in four MYCN-amplified neuroblastoma cell lines. B-C. RPPA data demonstrating the top 20 upregulated (B) and downregulated (C) proteins and phosphoproteins in JQ1 resistant NGP cells compared to JQ1 sensitive SK-N-BE(2)-C and CHP-212 cells. D. Quantification of pS473-AKT, pT308-AKT, total AKT and PTEN expression levels based on RPPA data. E. Western blots for p-AKT and PTEN in neuroblastoma cell lines. (p-AKT = pT308-AKT). F. Effects of JQ1 treatment on the viability of naive and JQ1 resistant (Res) SK-N-BE(2)-C cells. G. Absolute growth rates of SK-N-BE(2)-C naive cells treated with vehicle control and two replicates of JQ1 resistant cells treated with 1 μM JQ1. H. Effects of I-BET151 treatment on the viability of naive and JQ1 resistant SK-N-BE(2)-C cells. I. RPPA data demonstrating the top 20 most differentially expressed proteins in JQ1 resistant vs. naive SK-N-BE(2)-C cells treated with vehicle (Veh) or JQ1. J. Quantification of pS473-AKT, pT308-AKT, and total AKT levels from data shown in (I). K. Western blot of PI3K pathway activity in JQ1 resistant and naive cells. (p-AKT = pT308-AKT). L-N. Effects of the PI3K inhibitors GDC0941 (L), BYL719 (M), and BKM120 (N) on the viability of JQ1 resistant vs. naive SK-N-BE(2)-C cells. Results are presented as representative dose response curves of three independent experiments. Data is presented as mean values of eight technical replicates ± SD. See also Figure S2.
To extend these findings to the context of naturally acquired BET inhibitor resistance, we generated JQ1 resistant Kelly and SK-N-BE(2)-C cells by treating chronically with 1 μM of JQ1 over several months. (Figure 2F and Figure S2A). Importantly, these resistant cells proliferated in the presence of 1 μM JQ1, albeit at a slower rate than untreated parental cells (Figure 2G and Figure S2B), and were cross-resistant to I-BET151 (Figure 2H and Figure S2C). Subsequently, we performed RPPA analysis of the SK-N-BE(2)-C resistant cells (Figure 2I) (Tibes et al., 2006). Similar to the innately JQ1-resistant NGP cells, SK-N-BE(2)-C cells with acquired JQ1 resistance activated PI3K signaling, indicated by increased levels of both activating phosphorylation marks on AKT, as well as compensatory increased levels of the negative regulator PTEN (Figure 2I, J). PI3K pathway activation in both JQ1 resistant cell line models was confirmed by western blot of downstream effectors of PI3K signaling (Figure 2K and Figure S2D).
Strikingly, genome-scale ORF and CRISPR rescue screens and the profiling of innate and acquired BET inhibitor resistant cell lines all converged on the PI3K pathway as promoting BET inhibitor resistance. As such, we hypothesized that BET inhibitor resistant cells would exhibit dependency on the PI3K pathway. Indeed, both JQ1 resistant SK-N-BE(2)-C and Kelly cells were markedly more sensitive to three distinct PI3K inhibitors compared to the naive cells from which they were derived (Figure 2L-N and Figure S2E-G). SK-N-BE(2)-C and Kelly JQ1 resistant cells were not cross-resistant to cytotoxic chemotherapy agents (Figure S2H-M). Rather, they were more sensitive to cisplatin and doxorubicin and retained similar sensitivity to etoposide (Figure S2H-M). Thus, cells with acquired resistance to BET inhibitors demonstrate resistance mechanisms that are specific to BET inhibitors.
Enhancer remodeling underlies transcriptional changes observed in the resistant state
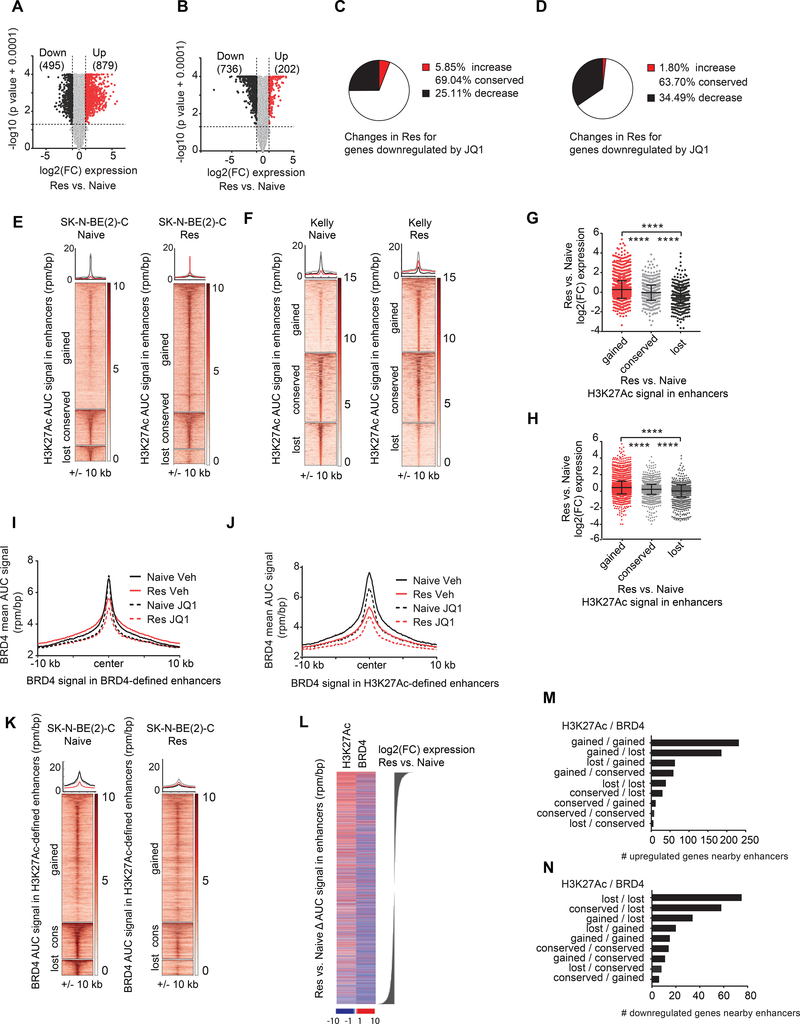
We next sought to elucidate the molecular basis of PI3K activation in resistance. Whole exome sequencing of JQ1 resistant SK-N-BE(2)-C and Kelly cells revealed no mutations in PI3K pathway members, and furthermore, there were no mutations in genes encoding BRD proteins (not shown). We thus performed RNA-sequencing of JQ1 resistant and naive SK-N-BE(2)-C and Kelly cells, treated with vehicle or JQ1 for 24 hours, to determine whether altered transcription was responsible for PI3K pathway upregulation in resistant cells. Expression of a large panel of housekeeping genes (Eisenberg and Levanon, 2013) was stable across conditions (Figure S3A-D). We also noted that JQ1 activity in the naive state downregulated more genes than it upregulated, consistent with the known repressive role of JQ1 on gene expression (Figure S3E, F). Our previously published gene signature of JQ1 treatment (Puissant et al., 2013) was strongly significantly enriched in JQ1-treated SK-N-BE(2)-C and Kelly naive cells (Figure S3G, H). We observed global differential gene expression in resistant vs. naive cells in both cell line models of BETi resistance (Figure 3A, B). Surprisingly, expression of the majority of genes downregulated by JQ1 in naive cells did not increase in resistant cells (Figure 3C, D).
Figure 3. Enhancer remodeling is associated with the transcriptional changes in the BET inhibitor resistant state.
A-B. Volcano plots highlighting the genes differentially expressed in resistant vs. naive SK-N-BE(2)-C (A) and Kelly (B) cells. The number of differentially expressed genes are shown in parentheses. C-D. Pie charts depicting the percent transcriptional changes in the resistant cells for genes downregulated by JQ1 in naive cells SK-N-BE(2)-C (C) and Kelly (D) cells. E-F. Heatmaps showing H3K27Ac binding among gained, conserved and lost enhancers in resistant vs. naive SK-N-BE(2)-C (E) and Kelly (F) cells. Regions are ranked by H3K27Ac binding signal in naive cells. Metaplots for average binding intensities across the gained (red), conserved (gray) and lost (black) enhancer regions are shown on top. G-H. Dot plots showing log2(FC) in expression for the genes associated with gained, conserved, and lost enhancers in SK-N-BE(2)-C (G) and Kelly (H) JQ1 resistant vs. naive cells. (**** p value < 0.0001 un-paired two sample Student t-test with Welch correction). Data are presented as mean values ± SD. I. Metaplot showing the average BRD4 binding signal (rpm/bp) on BRD4-defined enhancer regions +/− 10 kb in naive and resistant SK-N-BE(2)-C cells treated with vehicle control or JQ1. J. Metaplot showing the average BRD4 binding signal (rpm/bp) on H3K27Ac-defined enhancer regions +/− 10 kb in naive and resistant SK-N-BE(2)-C cells treated with vehicle control or JQ1. K. Heatmaps showing BRD4 binding on gained, conserved and lost H3K27Ac-defined enhancer regions in resistant vs. naive SK-N-BE(2)-C cells. Regions are ranked by BRD4 binding signal in naive cells. Metaplots for average binding intensities across the gained (red), conserved (gray) and lost (black) enhancer regions are shown on top. L. Heatmap showing ΔAUC for H3K27Ac and BRD4 signal in enhancers in resistant vs. naive SK-N-BE(2)-C cells ranked by log2(FC) expression changes. M-N. Barplots depicting the number of upregulated (M) and downregulated (N) genes nearby enhancers in resistant vs. naïve SK-N-BE(2)-C cells grouped according to gained, conserved or lost combinations of H3K27Ac and BRD4 levels in enhancer regions. For heatmaps, each row represents a single genomic region (+/− 10 kb) from the enhancer center. Genomic occupancy is shaded by binding intensity in units of reads per million per base pair (rpm/bp). See also Figure S3 and S4.
We hypothesized that gene expression changes observed in resistance are mediated by chromatin remodeling since JQ1 has been shown to preferentially repress genes marked by super-enhancers (SEs), regions in the genome that have high H3K27Ac and BRD4 binding (Loven et al., 2013). We therefore profiled genome-wide distribution of H3K27Ac by ChIP-sequencing of JQ1 naive and resistant cells across both cell line models (Fig S3I-P). In concordance with previous studies, SE-associated genes: HAND1/2, GATA3, and PHOX2B, were identified in naive cells (Boeva et al., 2017; Chipumuro et al., 2014; van Groningen et al., 2017, Durbin et al., 2018). SE-marked genes and typical enhancer (TE)-marked genes, defined by high H3K27Ac signal in enhancer regions, were preferentially repressed by JQ1 (Figure S4A, B). We found that enhancers were remodeled in the resistant vs. naive state (Figure 3E, F). In fact, there were 311 genes marked by “de novo” SEs and 1,271 genes marked by “de novo” TEs, in addition to 136 genes which lost a SE and 750 genes which lost a TE in the SK-N-BE(2)-C resistant cells (Figure S4C, D). Similar results were observed in the Kelly resistant vs. naive cells (Figure S4E, F). Enhancers gained in resistance were associated with increased transcriptional changes observed in resistance; whereas enhancers lost in resistance were associated with decreased transcriptional changes observed in resistance (Figure 3G, H and Figure S4G, H). Among genes transcriptionally upregulated in resistance and associated with a nearby enhancer, 69.2% and 55.87% gained H3K27Ac signal in these enhancer regions in SK-N-BE(2)-C and Kelly cells respectively (Figure S4I, J). Conversely, among genes transcriptionally downregulated in resistance and associated with an enhancer, the majority either lost H3K27Ac signal or showed no change in H3K27Ac signal in these enhancer regions (Figure S4I, J). Taken together, these results establish that transcriptional changes characterizing the resistant state are associated with global enhancer remodeling.
To further explore the mechanism of enhancer remodeling in resistance, we performed BRD4 ChIP-sequencing in SK-N-BE(2)-C naive and JQ1 resistant cells treated with vehicle or JQ1 for 24 hr. Consistent with the known activity of BET inhibitors, JQ1 treatment in naive cells preferentially repressed genes with high levels of BRD4 binding (i.e., SE and TE marked genes defined by levels of BRD4 AUC signal) (Figure S4K). BRD4 binding in the resistant state was suppressed at both BRD4-defined and H3K27Ac-defined enhancers and was further suppressed by JQ1 treatment (Figure 3I, J). BRD4 was globally repressed regardless of alterations (gained or lost) in H3K27Ac signal in the resistant state (Figure 3K). Among genes upregulated in resistance and nearby a BRD4-defined enhancer, BRD4 was gained in 45.09% of cases (Figure S4L). However, among the genes downregulated in resistance and nearby a BRD4-defined enhancer, BRD4 was lost in the majority, 61.60%, of cases (Figure S4L). Similar effects were observed for BRD4 signal restricted to H3K27Ac-defined enhancers. Among genes upregulated in resistance and nearby an H3K27Ac enhancer, BRD4 was gained in 51.9% of cases (Figure S4M). However, among genes downregulated in resistance and nearby an H3K27Ac-defined enhancer, BRD4 was lost in 80.9% of cases (Figure S4M).
We then assessed the combined effect of BRD4 and H3K27Ac chromatin remodeling in resistance. Gains in H3K27Ac signal in enhancer regions were strongly associated with transcriptional upregulation in resistance; whereas, losses of BRD4 signal in enhancer regions were strongly associated with transcriptional downregulation in resistance (Figure 3L). Among genes upregulated in resistance and nearby an enhancer, the vast majority either gained both H3K27Ac and BRD4 signal or gained H3K27Ac and lost BRD4 signal in resistance (Figure 3M). In contrast, among genes downregulated in resistance and nearby an enhancer, the vast majority either lost both H3K27Ac and BRD4 signal, or solely lost BRD4 signal and H3K27Ac remained conserved (Figure 3N). Thus, the global BRD4 loss observed in the resistant state is able to account for the majority of transcriptionally downregulated changes in resistance; whereas, H3K27Ac gains can account for the majority of transcriptionally upregulated changes in resistance.
Differential RTK reprogramming engenders therapeutic vulnerabilities in the resistant state
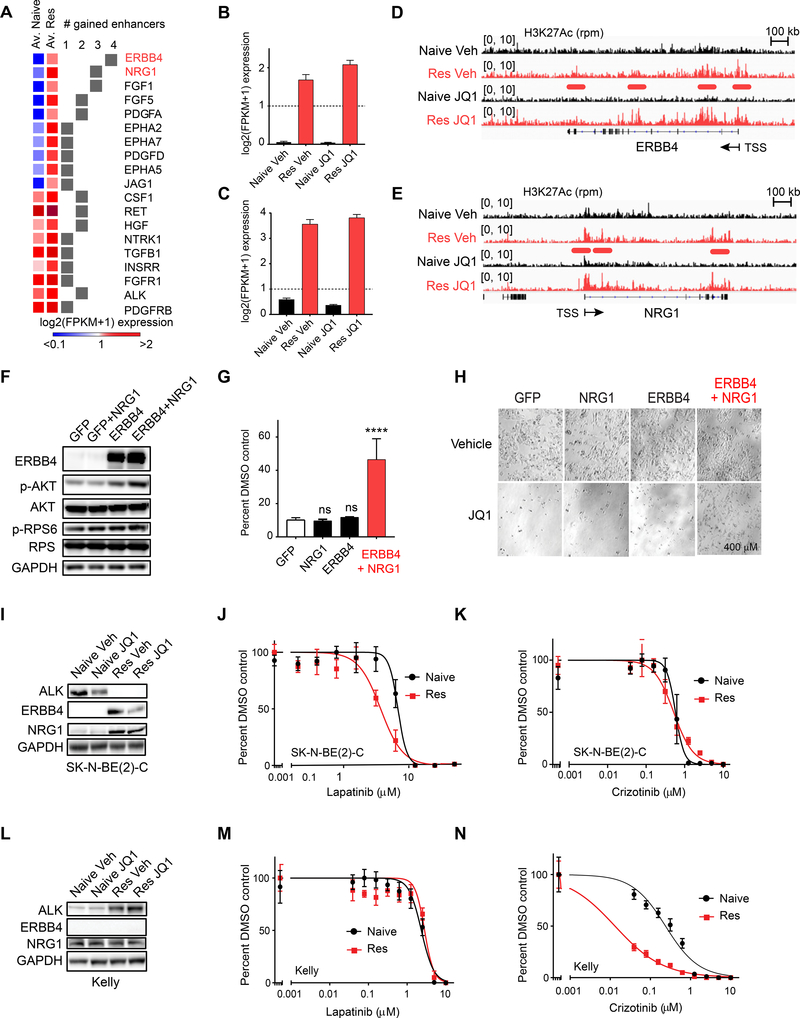
We hypothesized that the molecular basis of PI3K activation was mediated through activation of upstream growth factors and receptor tyrosine kinases. We queried the human kinome (Manning et al., 2002) and found that transcription of 19 RTK family genes were upregulated (log2(FC) expression > 1) and also associated with gained H3K27Ac-defined enhancers in the resistant vs. naive state in SK-N-BE(2)-C cells (Figure 4A). Interestingly, 10 out of the 19 RTK genes were newly expressed in the resistant vs. naive state, with log2(FPKM+1) expression in the naive state less than 1, and high expression in the resistant state. The EGFR family member, ERBB4, and its ligand NRG1, were the top scoring growth factors/RTKs transcriptionally activated in resistance (Figure 4B, C), and both genes encoding these proteins were associated with de novo enhancers in the resistant vs. naive state (Figure 4D, E). Co-overexpression of ERBB4 and NRG1 in naive cells was sufficient to activate PI3K signaling (Figure 4F) and to partially rescue JQ1-mediated cell death (Figure 4G, H). Importantly, overexpression of ERBB4 or NRG1 on their own was not sufficient to promote resistance to BET inhibition (Figure 4G, H), explaining why these genes did not score in the ORF rescue screen. We also performed similar analyses in the Kelly resistant model and found that ALK, RET and KITLG were transcriptionally upregulated (log2(FC) expression > 1) (Figure S5A-S5D) and also associated with gained enhancers in the resistant vs. naive state (Figure S5A-G). In the SK-N-BE(2)-C cell line, upregulation of ERBB4 and NRG1 were observed at the protein level in cells with acquired BET inhibitor resistance (Figure 4I). This upregulation engendered a vulnerability to the EGFR/ERBB4 inhibitor, lapatinib (Figure 4J). Importantly, ALK was not upregulated at a protein level in the resistant state in these cells (Figure 4I), and accordingly, the cells were not differentially sensitive to the ALK inhibitor, crizotinib (Figure 4K). Analogously, in the Kelly cell line, ALK was strongly upregulated in resistance, while ERBB4 and NRG1 were not (Figure 4L), engendering vulnerability to crizotinib but not to lapatinib (Figure 4M, N). Taken together, our data demonstrate that upstream regulators of PI3K signaling undergo enhancer remodeling associated with their overexpression, and subsequent activation of PI3K signaling in the resistant state, engendering vulnerability to agents that target these kinases.
Figure 4: Enhancer remodeling is associated with transcriptional upregulation of RTKs upstream of PI3K signaling engendering therapeutic vulnerabilities.
A. Heatmap demonstrating the average expression in naive and resistant cells for all RTK/GF genes associated with 1–4 gained enhancers and log2(FC) expression > 1 in resistant vs. naive cells. B-C. Average log2 FPKM expression for ERBB4 (B) and NRG1 (C) across JQ1 naive and resistant samples. Error bars represent SD. D-E. H3K27Ac ChIP-sequencing tracks for ERBB4 (D) and NRG1 (E). Enhancers gained in resistance are underlined in red. F. Western blot of SK-N-BE(2)-C cells engineered to overexpress GFP or ERBB4 and stimulated with vehicle (Veh) or recombinant NRG1 for 6 hr. Western blots are probed for downstream effectors of PI3K signaling. G. Long-term viability assays in SK-N-BE(2)-C cells overexpressing the indicated proteins and treated with vehicle (DMSO) or 1 μM JQ1. Data are presented as percent viable cells relative to the DMSO arm for each condition. Shown are mean values of quadruplicate points ± SD. (ns = not significant, **** p value < 0.0001, un-paired two sample Student t-test with Welch correction). H. Representative images of data presented in (G). I. Western blot analysis of naive and JQ1 resistant SK-N-BE(2)-C cells probed for ALK, ERBB4, and NRG1. Cells were treated with vehicle (Veh) or JQ1 for 24 hr. J-K. Effects of lapatinib (J) and crizotinib (K) treatment on viability in naive and JQ1 resistant SK-N-BE(2)-C cells. L. Western blot analysis of naive and JQ1 resistant Kelly cells treated with vehicle (Veh) or JQ1 for 24 hr. M-N. Effects of lapatinib (M) and crizotinib (N) treatment on viability in naive and JQ1 resistant Kelly cells. See also Figure S5.
Activation of PI3K signaling induces gene expression changes and enhancer remodeling associated with the drug resistant state
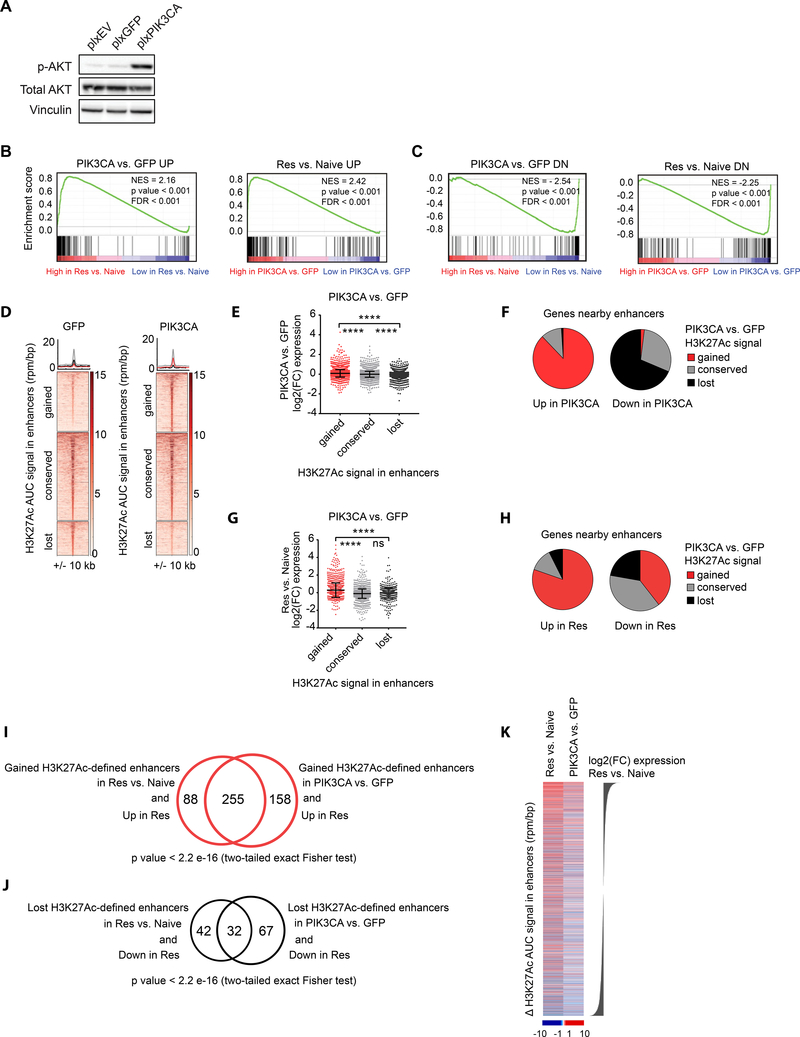
We next performed RNA-sequencing of SK-N-BE(2)-C cells engineered to overexpress either a GFP control or PIK3CA (Figure 5A) and found significant enrichment for genes upregulated in resistance among genes upregulated by PIK3CA overexpression and vice versa (Figure 5B). We similarly found a significant enrichment for downregulated genes (Figure 5C). We then performed H3K27Ac ChIP-sequencing of these engineered cells. We found that PIK3CA overexpression was associated with alterations in enhancers (Figure 5D), and genes that gain enhancers when PIK3CA was overexpressed were on average transcriptionally upregulated by PIK3CA overexpression (Figure 5E).
Figure 5: Transcriptomic analysis of BET inhibitor resistant cells reveals overexpression of PI3K signaling recapitulates enhancer remodeling and transcriptional changes characterizing the resistant state.
A. Western blot of SK-N-BE(2)-C cells engineered to overexpress an empty vector (plxEV), plxGFP or plxPIK3CA. B. GSEA demonstrating enrichment of genes upregulated in resistance among genes upregulated by PIK3CA overexpression (left) and vice versa (right). C. GSEA demonstrating enrichment of genes downregulated in resistance among genes downregulated by PIK3CA overexpression (left) and vice versa (right). D. Heatmaps showing H3K27Ac binding in gained, conserved and lost enhancer regions in PIK3CA vs. GFP samples. Each row represents a single genomic region +/− 10 kb from the enhancer center. Genomic occupancy is shaded by binding intensity in units of reads per million per base pair (rpm/bp). Regions are ranked by H3K27Ac binding signal in GFP cells. Metaplots for average binding intensities across the gained (red), conserved (gray) and lost (black) enhancer regions are shown on top. E. Dot plots showing log2(FC) in expression in PIK3CA vs. GFP cells for the genes associated with gained, conserved, and lost enhancers with PIK3CA overexpression (**** p value < 0.0001 un-paired two sample Student t-test with Welch correction). F. Pie charts showing the percentages of genes with gained, conserved or lost nearby enhancers with PIK3CA overexpression, among genes which are upregulated or downregulated by PIK3CA overexpression. G. Dot plots showing log2(FC) in expression in resistant vs. naive SK-N-BE(2)-C cells for the genes associated with gained, conserved, and lost enhancers with PIK3CA overexpression (**** p value < 0.0001, ns= not significant, un-paired two sample Student t-test with Welch correction). H. Pie charts showing the percentages of genes with gained, conserved and lost nearby enhancers with PIK3CA overexpression, among genes which are upregulated or downregulated in resistance. I. Venn-diagram showing the overlap of genes upregulated in resistant SK-N-BE(2)-C cells with nearby gained enhancers in resistance vs. genes upregulated in resistant SK-N-BE(2)-C cells nearby gained enhancers in PIK3CA overexpressing SK-N-BE(2)-C cells. Significance estimated based on two-tailed Fisher exact test. J. Venn-diagram showing the overlap of genes downregulated in resistant SK-N-BE(2)-C cells with nearby lost enhancers in resistant SK-N-BE(2)-C cells vs. genes downregulated in resistant SK-N-BE(2)-C cells with nearby lost enhancers in PIK3CA overexpressing SK-N-BE(2)-C cells. Significance estimated based on two-tailed Fisher exact test. K. Heatmaps showing ΔH3K27Ac AUC signal in enhancers for resistant vs. naive and PIK3CA vs. GFP samples ranked by log2(FC) expression in resistant vs. naive SK-N-BE(2)-C cells. Enhancers in this figure were defined by H3K37Ac binding. Dot plots in this figure are presented as mean values ± SD.
In addition, the large majority of genes upregulated by PIK3CA overexpression also gained an enhancer when PIK3CA was overexpressed; and the majority of genes downregulated by PIK3CA overexpression lost an enhancer when PIK3CA was overexpressed (Figure 5F). Importantly, genes associated with gained enhancers when PIK3CA was overexpressed were, on average, transcriptionally upregulated in JQ1 resistant cells (Figure 5G). Consistently, the majority of genes upregulated in resistance gained enhancers when PIK3CA was overexpressed and the majority of genes downregulated in resistance were associated with enhancers lost when PIK3CA was overexpressed (Figure 5H). Furthermore, there was significant overlap between genes upregulated transcriptionally in JQ1 resistance which gained an enhancer in JQ1 resistance and genes transcriptionally upregulated in resistance which gained an enhancer when PIK3CA was overexpressed (Figure 5I). There was a similarly significant overlap among genes downregulated in resistance with a lost enhancer in resistance and genes downregulated in resistance with a lost enhancer with PIK3CA overexpression (Figure 5J). Finally, H3K27Ac-defined enhancers gained in resistant vs. naive cells and PIK3CA vs. GFP cells were associated with strong transcriptional upregulation in resistance; whereas, H3K27Ac-defined enhancer losses were associated with strong transcriptional downregulation in the resistant state (Figure 5K). Collectively, these results demonstrate that PI3K overexpression can, in part, recapitulate both the enhancer remodeling and the transcriptional changes associated with BET inhibitor resistance.
Identification of upfront synergistic combination therapies
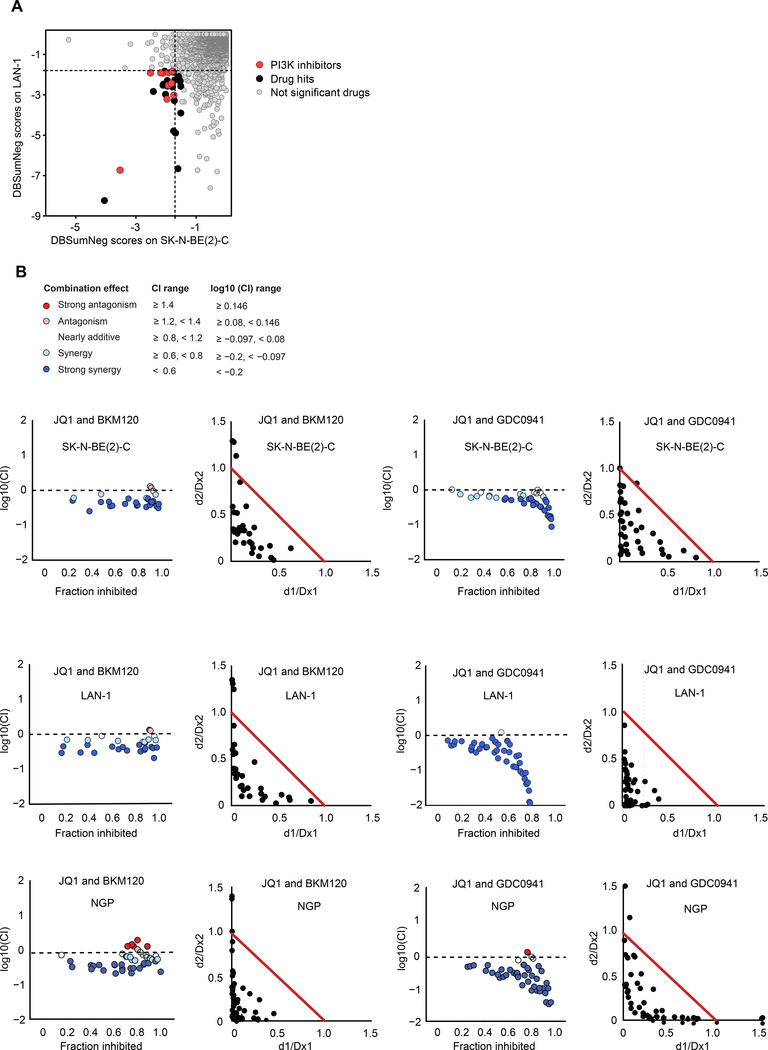
We next sought to systematically identify synergistic combinations of drugs with BET inhibitors in the naive cell state in order to identify effective combination strategies to block emergent resistance. We thus screened JQ1 against the Mechanism Interrogation PlatE (MIPE) library of ~1900 oncology focused compounds possessing diverse mechanisms of action (Mathews Griner et al., 2014) in the SK-N-BE(2)-C and LAN-1 cell lines (Figure 6A). PI3K inhibitors were enriched among the compounds that scored as synergistic with JQ1 in both cell lines (Figure 6A and Table S5). Validation of two PI3K inhibitors that scored in the chemical screen, BKM120 and GDC0941, demonstrated strong synergy with JQ1 based on the Chou-Talalay combination index (CI) model across a diverse panel of neuroblastoma cell lines (Figure 6B and Fig S6A-P) (Chou and Talalay, 1984). Importantly, in the innately BETi resistant NGP cells, treatment with PI3K inhibitors sensitized the cells to BET inhibition (Fig S6Q). Additionally, strong synergy was observed with PI3K and BET inhibitors in the NGP cell line (Figure 6B), strengthening the rationale to combine PI3K and BET inhibitors in the upfront setting. This provides support for the concept that studying adaptive mechanisms of resistance also allow for the identification of innate mechanisms of resistance. Furthermore, we screened I-BET151 across 58 compounds selected for validation from the primary 1900 compound, JQ1 sensitizer screen and used an apoptosis assay as the readout (Figure S6R). Two PI3K inhibitors scored among the top 9 synergistic combinations with I-BET151 (Figure S6R), further supporting the rationale for combining PI3K and BET inhibition.
Figure 6: Chemical combinatorial screening identifies PI3K inhibitors as highly synergistic with JQ1 in MYCN-amplified neuroblastoma.
A. JQ1 screened against the Mechanism Interrogation PlatE (MIPE) library in SK-N-BE(2)-C and LAN-1 MYCN-amplified neuroblastoma cell lines. Synergy was assessed using the Bliss model. DBSumNeg is defined as the sum of negative deviations from the Bliss model. Dotted black lines indicate threshold for synergy. B. Synergy was assessed by Chou-Talalay combination index (CI) for JQ1 and the PI3K inhibitors, BKM120 and GDC0941, across the indicated cell lines. For CI plots, the x-axis represents fraction inhibited and the y-axis represents log10(CI). Normalized isobolograms depict CI scores over a range of concentrations. The coordinates of the CI scores are d1/Dx1 and d2/Dx2, where Dx1 is the concentration of drug 1 (JQ1) that alone produces the fractional inhibition effect x, and Dx2 is the concentration of drug 2 (PI3Ki) that alone produces the fractional inhibition effect x. The red line displayed is the line of additivity.
See also Figure S6 and Table S5.
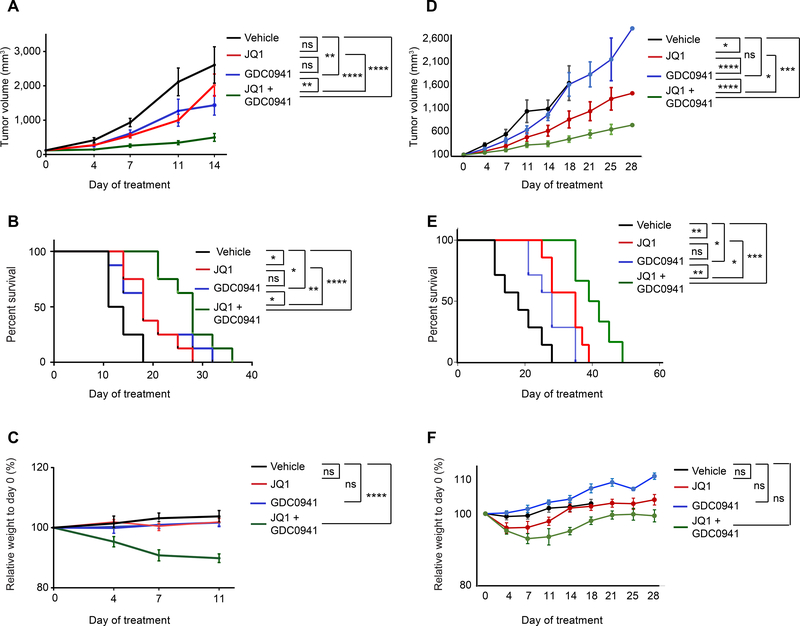
To assess the in vivo efficacy of combining PI3K and BET inhibitors, we performed a four-arm study in an aggressive SK-N-BE(2)-C xenograft mouse model of MYCN-amplified neuroblastoma. The combination of JQ1 and GDC0941 treatment delayed tumor progression and increased overall survival compared to either single agent alone (Figure 7A, B) with the combination incurring mild weight loss at 7–11 days of treatment (<15%) (Figure 7C). We performed additional preclinical studies with JQ1 and GDC0941 as single agents and in combination in a patient-derived xenograft (PDX) mouse model of MYCN-amplified neuroblastoma. In this model, JQ1 and GDC0941 strongly decreased tumor progression and increased survival to a greater extent in combination than as single agents (Figure 7D, E). Furthermore, although initial weight loss was observed at fourteen days of treatment, mouse weights increased and then plateaued after prolonged treatment. In fact, there was no statistical difference in weight among treatment groups at 28 days when treatment was ended (Figure 7F). Notably, our studies provide proof-of-concept that identification of resistance mechanisms to a drug can inform upfront synergistic combination therapies.
Figure 7. BET inhibitors and PI3K inhibitors are strongly synergistic in mouse models of MYCN-amplified neuroblastoma.
A. Tumor volume measurements for SK-N-BE(2)-C xenograft nude mice treated with vehicle control, 50 mg/kg JQ1 IP QD, 100 mg/kg GDC0941 PO QD, or the combination of JQ1 and GDC0941 for 14 days. Data for a given time point were plotted if >50% of mice in the group were alive. Data are plotted as mean values ± SD (n = 8). B. Kaplan-Meier survival curves for the experiment described in (A). C. Relative weight measurements of mice from experiment described in (A). Data are plotted as mean values ±SEM (n=8). Each treatment condition was compared to the vehicle treatment. D. Tumor volume measurements of a PDX mouse model of MYCN-amplified neuroblastoma treated with vehicle control, 50 mg/kg JQ1 IP QD, 100 mg/kg GDC0941 QD PO, or the combination of JQ1 and GDC0941 for 28 days. Data for a given time point were plotted if >50% of mice in the group were alive. Data are plotted as mean values ± SD (n = 7). E. Kaplan-Meier survival curves for the experiment described in (D). F. Relative weight measurements of mice from experiment described in (D). (ns = not significant, * p value < 0.05, ** p value < 0.01, *** p value < 0.001, **** p value < 0.0001). For tumor volume and weight measurements, significance was determined by 2-way ANOVA with Tukey post hoc test. For survival analysis, significance was determined by log-rank Mantel Cox test.
DISCUSSION
Systematic analysis of cancer genomes has revealed that pediatric cancers are among the most genetically stable tumors (Lawrence et al., 2013). Pediatric neuroblastoma tumors in particular harbor few recurrently mutated genes (Molenaar et al., 2012; Pugh et al., 2013; Sausen et al., 2013; Valentijn et al., 2015). The low mutation rates of many pediatric cancers suggest that these tumors are epigenetically dysregulated, making epigenetic regulators promising therapeutic targets. BET inhibitors are a class of epigenetic-targeting drugs being evaluated in clinical trials. Currently, there are 16 active clinical trials with BET inhibitors in various malignancies. Results from many of these trials have not been reported; however, OTX015, a BET protein inhibitor developed by OncoEthix, was tested in a phase I study for acute leukemia and three complete remissions in patients with refractory disease were documented (Berthon et al., 2016). Additionally, early reports of the BET inhibitor CPI-0610 have demonstrated anti-tumor effects in B-cell lymphoma and follicular lymphoma (Pfister and Ashworth, 2017). As initial findings of these trials support further development of BET inhibitors, studies of resistance mechanisms are needed in order to optimize their clinical application and to achieve durable responses to treatment. Here, we deployed a comprehensive genome-scale functional genomics approach to identify mechanisms of BET inhibitor resistance in neuroblastoma in an effort to prioritize upfront clinical combination therapies to prevent treatment failure and relapsed disease. The studies presented here provide a framework to identify mechanisms of resistance to diverse chromatin remodeling agents in varied cancer types.
Reported mechanisms of BET inhibitor resistance are distinct, implicating the importance of cellular context in understanding BET protein activity and resistance. BET inhibitor resistant AML cells have been shown to arise from leukemic stem cells driven by high Wnt signaling (Fong et al., 2015). Another study found that suppression of the PRC2 complex member, SUZ12, promoted BET inhibitor resistance in AML (Rathert et al., 2015). Furthermore, PRC2 suppression was able to restore expression of key target genes of BET inhibitors, such as MYC, through a WNT-dependent mechanism (Rathert et al., 2015). In triple negative breast cancer (TNBC), BET inhibitor resistant cells have hyperphosphorylated BRD4 as a result of decreased PP2A activity and bromodomain independent recruitment of BRD4 to chromatin (Shu et al., 2016). Consistent with our study, BET inhibitor resistant TNBC cells gain SEs, resulting in increased transcription of these SE-marked genes (Shu et al., 2016). Finally, adaptive kinome reprogramming in BET inhibitor resistant ovarian cancer cells was shown to activate several pro-survival compensatory kinases (Kurimchak et al., 2016). The molecular basis of kinome reprograming; however, remained unanswered. In the current study we analyzed proteomic, and epigenetic changes characterizing resistance, to explore the molecular basis for the adaptive kinome reprogramming observed. Our work demonstrates that altered enhancer remodeling is strongly associated with activation of PI3K signaling driving resistance. We intentionally focused on enhancer regulatory regions due to the known role of BETi in regulating these genomic locations. Future work is still needed to identify the contributions of other chromatin marks in defining the drug resistant state. Due to technical challenges, we have been unable to delete these enhancers via CRISPR to validate their function. Therefore, future work is also needed to effectively prove that the enhancer remodeling observed is sufficient to promote BET inhibitor resistance in this disease.
Importantly, we report that the combination of JQ1 and the PI3K inhibitor, GDC0941, significantly delayed tumor progression and extended survival of mice compared to either single agent alone in two aggressive mouse models of MYCN-amplified neuroblastoma. These studies nominate combination therapies that will enhance the efficacy of each drug and potentially prevent therapy resistance. Future work is needed to evaluate this combination in other molecular subtypes of neuroblastoma (namely, non-MYCN amplified tumors) as well as to evaluate the efficacy of clinical candidate molecules. Overall, our findings indicate that divergent chromatin states underlie resistance to BET inhibitors and engender vulnerabilities that can be exploited to block emergent resistance.
STAR METHODS
Contact for Reagent and Resource Sharing
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kimberly Stegmaier (kimberly_stegmaier@dfci.harvard.edu).
Experimental Model and Subject Details
Cell Culture
Human neuroblastoma cell lines, KELLY, CHP-212, and LAN-1 were provided by the Broad Institute. SK-N-BE(2)-C and SHSY5Y were purchased from ATCC. NGP cells were kindly provided by Dr. Rani George. EBC1 cells were kindly provided by Dr. A. Thomas Look. Cell line identities were confirmed by STR profiling at the Dana-Farber Cancer Institute.
Generation of JQ1 Resistant Cell Lines
JQ1 resistant neuroblastoma cell lines were established by exposing naive neuroblastoma cell lines to 1 μM of chronic JQ1 treatment over several months. Cells that eventually proliferated in the presence of 1 μM JQ1 were designated as JQ1 resistant cell lines.
In vivo Tumor Models
For cell line xenograft studies, eight-week old nude female mice were subcutaneously implanted with 1×10^6 SK-N-BE(2)-C cells in 30% matrigel/PBS. For PDX studies, a PDX mouse model of MYCN-amplified neuroblastoma was established from a tumor resected at time of diagnosis (COG-N-424X) from a patient with Stage 4 disease. Samples were obtained from the Children’s Oncology Group’s Childhood Cancer Repository. Informed consent was obtained and studies were approved by COG protocols. 1mm3 viably frozen tumor chunks were dipped in matrigel and implanted into the right flanks of eight-week old nude female mice via minor surgery. Once tumors engrafted and reached 100–150 mm3, treatment was initiated. Mice were randomized into four groups: vehicle, JQ1, GDC0941, or the combination of JQ1+GDC0941. JQ1 was delivered at 50 mg/kg QD IP and solubilized in 10% DMSO and 90% HPBCD. GDC0941 was delivered at 100 mg/kg QD PO and solubilized in 10% DMSO, 5% Tween20, 85% water. Mice were treated for 14 days or 28 days for the SK-N-BE(2)-C xenograft and PDX studies respectively, and followed for survival. Tumors were measured by calipers 2x week. Mice were weighed 2x week. Animals were sacrificed when tumors reached 2 cm in any one dimension. All dosing was performed at the Dana-Farber Cancer institute, and all animal protocols were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. Nude mice were maintained according to institutional guidelines.
Method Details Chemicals
JQ1 was synthesized by Dr. Jun Qi (Dana-Farber Cancer Institute). All other compounds for in vitro synergy experiments were obtained from Selleck. GDC0941 used for in vivo studies was obtained from ApexBio.
Western Blotting
Proteins were extracted using Lysis Buffer (Cell Signaling Technology) supplemented with Complete, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics). Protein samples were separated by SDS-PAGE and subsequently transferred to PVDF membranes, which were blocked in 5% BSA and incubated with primary antibodies against V5 (Life Technologies Cat. No. R960–25), GAPDH (Santa Cruz Cat. No. sc-137179), HSP90 (Abcam Cat. No. ab13492), Vinculin (Abcam Cat. No. 18058), pRPS6 (Cell Signaling Technology Cat. No. 2215), p-AKT (Cell Signaling Technology Cat. No. 9271), p-4EBP1 (Cell Signaling Technology Cat. No. 2855), total RPS6 (Cell Signaling Technology Cat. No. 2217), total AKT (Cell Signaling Technology Cat. No. 9272), total 4EBP1 (Cell Signaling Technology Cat. No. 9644), ALK (Cell Signaling Technology Cat. No. 3633), PTEN (Cell Signaling Technology Cat. No. 9552), ERBB4 (Cell Signaling Technology, Cat. No. 4795), NRG1 (Abcam Cat. No. ab180808), BCL2L1 (Cell Signaling Cat. No. 2762), and BCL2L2 (Sigma Cat. No. SAB4502627). Membranes were washed in TBS-T and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies. Signal was detected by enhanced chemi-luminescence (ThermoFisher Scientific).
Colony Formation Assays
Using a 16 gauge blunt end needle, 6 mL of ClonaCell-TCS Medium (Stemcell Cat. No. 03814) was added to 1.5 mL of neuroblastoma cells resuspended at 5,000 cells/mL. The solution was vortexed and 3 mL of methylcellulose/cells was added to a 6 cm dish in technical duplicate. Colonies were stained with MTT for 1 hr at 37°C and counted 10–14 days after plating when visible by the naked eye.
Lentiviral Infections
Lentivirus was generated by transfecting HEK-293T cells with the indicated vectors and the packaging plasmids, delta8.9 and VSVG, following the Fugene 6 protocol (Promega). Neuroblastoma cells were infected with 2 mL of virus and 8 μg/mL polybrene. Cells were selected with puromycin containing media 48 hours after infection.
Low-throughput JQ1 Rescue Experiments
Lentiviral infected cells were seeded onto 6 well plates in technical duplicate and were treated with DMSO or 1 μM JQ1. For ERBB4/NRG1 rescue experiments, cells were treated with 100 ng/mL recombinant human NRG1 (R&D systems) twice per day. Cumulative population doublings were calculated by manually counting cells every 3–4 days for a total of 14 days.
ORFeome and CRISPR Library Titration
Accurate virus volumes to use in large scale were determined in each cell line in order to achieve 30–40% infection efficiency, corresponding to a multiplicity of infection (MOI) of ~ 0.5–1. Spin-infections were performed in 12-well plate format with 3 × 106 and 1.5 × 106 cells each well for SK-N-BE(2)-C and LAN-1, respectively, with virus volumes (0,100, 200, 300, 400, 500 μL) with a final concentration of 8 μg/mL polybrene. Cells were spin-infected for 2 hours at 2000 rpm at 30 degrees. 24 hours later, cells were trypsinized and 2 × 105 SK-N-BE(2)-C cells and 3.5 × 105 LAN-1 cells from each infection, were seeded into duplicate wells in 6-well plates, each with complete medium and one treated with puromycin. 72–96 hours after selection, cells were counted to determine the amount of virus that yielded ~30 – 40% infection efficiency, and this amount was used for screening.
Genome-scale ORF Rescue Screens
The ORFeome barcoded library contains 17,255 barcoded ORFs overexpressing 10,135 distinct human genes with at least 99% nucleotide and protein match. Screening-scale infections of the ORFeome library were performed to achieve a representation of at least 1000 cells per ORF (~2 × 107 surviving cells containing 17,255 ORFs). Infections were performed with the optimized virus volume in 12-well format and pooled 24 hours post-infection. Approximately 24 hours after infection, all wells within a replicate were pooled and 48 hours after infection, cells were selected with puromycin. After selection was completed, 3 × 107 cells were divided into drug treated (1 μM JQ1 and 5 μM I-BET151 for SK-N-BE(2)-C and 0.5 μM JQ1 and 5 μM I-BET151 for LAN-1) and vehicle treated arms. Cells were passaged in fresh media containing drugs every 3–4 days, and throughout the screen we maintained an average representation of 1,000 cells per ORF construct. Cells were harvested 14–15 days after initiation of treatment. For both ORF screens, genomic DNA (gDNA) was isolated using Maxi (2 × 107–1 × 108 cells) or Midi (5 × 106 –3 × 107cells) kits according to the manufacturer’s protocol (Qiagen). PCR and sequencing were performed as previously described (Doench et al., 2016). We performed three replicates for the ETP and four replicates for the late time point for each cell line and each drug.
Genome-scale CRISPR Rescue Screens
CRISPR rescue screens were carried out using the CRISPR Avana library containing 74,687 guide RNAs corresponding to 4 gRNAs per gene (Doench et al., 2016). Infections were performed to achieve a representation of at least 500 cells per guide. The infection rate was confirmed to be ~30–50%. Approximately 24 hours post-infection, cells were pooled and selected in puromycin. Seven days later, cells were divided into drug treated arms (1 μM JQ1 and 5 μM of I-BET151 for SK-N-BE(2)-C and 0.5 μM JQ1 and 5 μM I-BET151). Cells were passaged in fresh media containing drugs every 3–4 days, and throughout the screen we maintained an average representation of 500 cells per sgRNA. Cells were harvested 14–15 days after initiation of treatment. Genomic DNA extraction and sequencing were carried out as in the ORF screen. We performed two replicates for the ETP and four replicates for the late time point for each cell line and each drug.
Reverse Phase Protein Array (RPPA)
RPPA data were generated by the RPPA core facility at the MD Anderson Cancer Center (Tibes et al., 2006). Cell lysates were serially diluted and arrayed on nitrocellulose-coated slides. Slides were probed with 287 unique antibodies by tyramide-based signal amplification and visualized by DAB (3,3’-diaminobenzidine) staining. Stained slides were quantified by Array-Pro Analyzer. All protein expression was normalized for protein loading before LogFCs were calculated between conditions.
RNA-sequencing
RNA was extracted from cells with the RNeasy Kit and on-column DNA digestion (Qiagen). For RNA-sequencing of SK-N-BE(2)-C cells, polyA mRNA was isolated and libraries were prepared using the TruSeq Stranded mRNA Kit (Illumina) according to the manufacturer’s protocol. For RNA-sequencing of Kelly cells, polyA mRNA was isolated using the NebNext PolyA mRNA magnetic isolation module (NEB #E7490) according to manufacturer’s instructions. Libraries were prepared using the NebNext Ultra II Directional RNA library prep kit for Illumina (NEB #E7760) and NEBNext Multiplex Oligos for Illumina (E7355) according to the manufacturer’s protocol. All samples were sequenced on a NextSeq500 instrument with single-end 75bp reads to a depth of 30–50M reads/sample.
RNA-sequencing Data Processing
Quality control tests for the mapped reads were performed using the FASTQC software (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were aligned to the GRCh37/hg19 human genes by using Tophat2 v2 (Kim et al., 2013). Quality control tests for the aligned reads and for the replicate consistency were performed by using the qualimap v2.2 (Garcia-Alcalde et al., 2012) and the SARTools (Hugo Varet, 2015) pipelines. The total number of reads for individual samples ranged from 35 to 54 Mb. The average percentage of uniquely mapped reads in the aligned data was 92.3%, with a standard deviation 1.8%.
Gene level reads and gene level expression estimated as log2(FPKM) scores were computed using the Feature Counts method implemented in the Bioconductor v3.2 RSubread package (Liao et al., 2014). The genome-wide expression data were projected onto a heatmap by using the Morpheus platform, (https://software.broadinstitute.org/morpheus). The overall significance of the differential expression between the control and treatment phenotypes was estimated by using the EdgeR method available from the Bioconductor v3.2 EdgeR library (Robinson et al., 2010) with the significance cut-offs: absolute fold change for log2(FPKM+1) expression ≥ 1, P ≤ 0.05, Benjamini-Hochberg false discovery rate (FDR) ≤ 0.05.
Gene Set Enrichment Analysis (GSEA)
GSEA v3.0 software (Mootha et al., 2003; Subramanian et al., 2005) was used to identify functional associations of the genome-wide molecular profiles. Significance cut-offs were assessed based on the GSEA standard recommendations: absolute Normalized Enrichment Score (NES) ≥ 1, p ≤ 0.05, Benjamini-Hochberg false discovery rate ≤ 0.25.
ChIP-sequencing
H3K27Ac ChIP-sequencing:
Neuroblastoma cells were plated in 15 cm dishes and grown to 70–80% confluence. Twenty million cells per condition were collected and washed twice in ice cold PBS. Cells were then crosslinked with 1% formaldehyde in cell culture media for 4 minutes and then quenched with 0.125 M glycine and 0.1M Tris HCl pH 7.5. Cells were lysed in 50 mM Tris-HCl pH 8.1, 5 mM EDTA, 0.5% SDS, 0.2% sodium azide and 100 mM NaCl supplemented with protease inhibitors (Roche). Debris was spun off. Cells were incubated in a 2:1 mixture of lysis buffer:dilution buffer (100 mM Tris HcL, pH 8.6, 100 mM NaCl, 5 mM EDTA, 0.2% sodium azide, and 5% Triton X-100). Chromatin was sheared to ~250 bp fragments using a Covaris instrument. Subsequently, 0.5% of the lysate was removed as the input control. The remaining lysate was incubated with 50 uL of protein A dynabeads (Thermofisher) and 5 μg of H3K27Ac antibody (ab4729, Abcam) at 4°C overnight (antibodies and beads were incubated together for 30 min at 4°C prior to the addition of cell lysate). The precipitated lysate was then washed sequentially in two washes of each of the following buffers: ice cold low salt buffer (20 mM Tris-HCl pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton-X100, 0.1% SDS), high salt buffer (20 mM Tris-HCl pH 8.1, 2 mM EDTA, 500 mM NaCl, 1% Triton-X100, 0.1% SDS), LiCl buffer (10 mM Tris-HCl pH 8.1, 0.25 M LiCl, 1 mM EDTA, 1% deoxycholic acid, 1% IGEPAL CA-630) and TE buffer (10 mM Tris-HCl pH 8.1, 1 mM EDTA). Samples were eluted in 1% SDS, 0.1M sodium bicarbonate for 15 min at RT. RNase A, proteinase K, and 0.2M NaCl was added to the elution buffer and samples were incubated at 65°C for >4 hours to reverse cross-linking. DNA was purified using AMPure XP beads (Agencourt). ChIP libraries were prepared using the Rubicon Genomics Thruplex DNA-seq prep kit according to the manufacturer’s protocol, and libraries were sequenced on a single-end 75bp NextSeq run.
BRD4 ChIP-sequencing:
Neuroblastoma cells were plated in 15 cm dishes and grown to 70–80% confluence. 100 million cells per condition were used. Cells were crosslinked in the dishes with 1% formaldehyde in cell culture media for 15 minutes, swirling every five minutes, and then quenched with ~0.2M Tris HCl pH 7.5. Cells were scraped off the plate and washed in ice cold PBS. 10 μg of BRD4 antibody (Bethyl, Cat. No. A301–985A100) was incubated with 100 μL of Protein G Dynabeads per condition overnight at 4°C. During the incubation, cells were lysed in Nuclei EZ lysis buffer (Sigma) for 5 min at 4°C. Debris was spun off and cells were incubated in buffer containing: Hepes-KOH, NaCl, EDTA (pH 8.0), EGTA (pH 8.0), Triton X-100, sodium deoxycholate and SDS and sonicated overnight on a Covaris instrument to shear chromatin to ~250–300 bp fragments. 2.5% of the lysate was removed as the input control. The antibody: bead solution was then added to the sonicated samples overnight at 4°C. The precipitated lysate was then washed sequentially with sonication buffer, high-salt sonication buffer, LiCl buffer, and TE. Samples were eluted and decrosslinked in 0.05 M Tris-HCl (pH 8.0), 1% SDS, and 10 mM EDTA at 65°C overnight. DNA was purified using Phenol:Chloroform:Isoamyl alcohol. ChIP libraries were prepared using the Rubicon Genomics Thruplex DNA-seq prep kit according to the manufacturer’s protocol, and libraries were sequenced on a single-end 75bp NextSeq run.
ChIP-sequencing Data Analysis
All of the ChIP-sequencing data sets were aligned using bwa-mem v0.7.15 (Li and Durbin, 2010) with the parameters -M -t 8, to the build version NCBI37/hg19 of the human genome. Quality control tests were performed based on the FastQC v.0.11.2 software (Babraham Bioinformatics, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and by using the ChIPQC library available from Bioconductor v3.2 (Carroll et al., 2014).
The ChIP-sequencing peaks for H3K27Ac and BRD4 were identified using MACS v1.4.3 (Feng et al., 2012). The significance cut-off for the p value was set < 1e-09 for all the H3K27Ac and BRD4 peaks, except for the H3K27Ac peaks on the SK-N-BE(2)-C naïve and JQ1 resistant cells for which the cut-off was set < 1e-05. The peaks were annotated by using the Annotate Peaks function available in the Homer v4.7 package (Heinz et al., 2010). Active enhancers were defined as significantly enriched regions which are outside promoters (i.e., not fully contained within ±2 kb region flanking the promoter) (Loven et al., 2013). The active enhancers within 12.5 kb of one another were stitched together, and the stitched regions spanning more than two promoters were excluded. The total ChIP occupancy signal at enhancers expressed in units of reads per million mapped reads per bp (rpm/bp) was computed by using the DeepTools v2.2.3 software (Ramirez et al., 2014) as the area under curve formula based on the H3K27Ac normalized signal with background input signal subtracted. The stitched active enhancers (“enhancers” for short) were ranked based on increasing total ChIP occupancy normalized signal and an inflection point in the signal distribution was geometrically detected. Super-enhancers (SEs) were defined as those stitched active enhancers with total ChIP occupancy signal above the inflection cut-off. Typical enhancers (TEs) were defined as those stitched active enhancers with total ChIP occupancy signal below the inflection cut-off. For the purpose of our study, we refer to SEs and TEs as “enhancers” for short.
Differential SE regions between resistant vs. naïve states were defined as in (Brown et al., 2014). Area under curve (AUC) for background subtracted H3K27Ac signal was calculated for all enhancer regions considered super in at least one condition. SEs were classified based on the log2 fold change signal: >1 gained, <−1 lost, and between −0.25 – 0.25 conserved. Differential TE regions between resistant vs. naïve states were defined similarly, based on the signal changes in non-SE, enhancer regions considered typical in at least one condition. Differential enhancer regions between resistant vs. naïve states were defined similarly, based on the signal changes in enhancer regions, with no other further requirement. Genes were annotated to enhancers based on the hg19 RefSeq basal+extension association rule: 5000 bp upstream, 1000 bp downstream, 100 kb max extension, curated regulatory domains included (McLean et al., 2010).
Cell Viability and Synergy Studies
Cells were resuspended at 25,000 cells/mL and seeded at 40 μl/ well onto 384-well plates. Cells were then treated with a single agent or combination of compounds and analyzed for cell viability on days 0, 3, 5 and 7 post-treatment using the Cell-TiterGlo luminescent assay kit (Promega) according to the manufacturer’s protocol. Luminescence was read on a Fluostar Omega Reader (BMG Labtech).
Synergy Analysis
To assess whether individual treatment combinations were synergistic, additive, or antagonistic we computed the combination index (CI) scores for Loewe Additivity based on the Chou-Talalay Median Effect model (Chou, 2006; Chou, 2010; Chou and Talalay, 1984) as implemented in CalcuSyn v2.11 (http://www.biosoft.com/w/calcusyn.htm). The median effect model states that the effect x of a dose of a drug is described by the equation x/(1-x) = (d/DIC50)m , where DIC50 is the IC50 dose of the drug, and m is a parameter that is estimated from the dose-effect curve kinetics of the drug: m = 1 (hyperbolic), m > 1 (sigmoidal), m < 1 (flat sigmoidal). Loewe additivity is a dose-effect model which states that additivity occurs in a two-drug combination if the sum of the ratios of the dose vs. the median-effect for each individual drug is d1/DIC50, 1 + d2/DIC50, 2 = 1, where d1, d2 are the doses for the two drugs, and DIC50, 1 and DIC50, 2 represent the IC50 concentrations (median-effect) for the two drugs, respectively. Chou and Talalay (Chou and Talalay, 1984) showed that Loewe equations are valid for enzyme inhibitors with similar mechanisms of action -- either competitive or non-competitive toward the substrate.
The Chou-Talalay Combination Index for Loewe Additivity model assigns a quantitative measure to any given effect x produced by the combination of dose d1 of drug 1 and dose d2 of drug 2: CI = d1/Dx1 + d2/Dx2, where Dx1 is the dose of drug 1 that alone produces the effect x and Dx2 is the dose of drug 2 that alone produces the effect x, as estimated from the median effect model. The drug combinations were analyzed based on two types of diagnostic plots:Effect-oriented plots (combination index plots): the effect x of the drug combination on the X axis, and CI = d1/Dx1 + d2/Dx2 on the Y axis, and Dose-oriented (isobologram) plots: d1/Dx1 on the X axis, d2/Dx2 on the Y axis, along with the Loewe additivity isobole line: d1/DIC50, 1 + d2/DIC50, 2 = 1. In the combination index plots, synergistic drugs have a CI < 1, whereas antagonistic combinations have a CI > 1. The C = 1 when the drugs are additive. For the isobolograms, the red line represents the line of additivity. Synergistic combinations fall below the line; whereas antagonistic combinations fall above the line.
Small-molecule library drug combination screening
A high-throughput screen was conducted in 1536-well white flat bottom plates (Corning) on a Kalypsys robotic system (Michael et al., 2008). Either LAN-1 or SK-N-BE(2)-C cells were dispensed at a density of 500 cells in 4 μL of media, seeded into plates pre-spotted with 2 μL of media (MultiDrop Combi, Thermofisher Scientific), making a total volume of 6 μL media per well. Following a 4 hour incubation, compounds were added using the EDC ATS100 acoustic dispenser (EDC) creating 6 by 6 (6×6) blocks using a method described previously (Mathews Griner et al., 2014). Briefly, a 6-concentration-point (including a zero concentration), 3-fold dilution of JQ1 was transferred to the screen plates (10 nL), and the library compounds were then transferred by acoustic dispensing (10 nL) into the same plates with each compound plated in six-point dilution series at five-fold serial dilutions. After 72 hours of incubation, 3 μL of Cell-TiterGlo reagent (Promega) was added to each well. Following 10 min incubation, luminescence was read using the Viewlux microplate reader (PerkinElmer). Plate data were normalized to in-plate controls (bortezomib as positive control, DMSO as negative control) and the normalized data was deconvoluted to individual dose combination matrices using in-house software. We employed the Bliss model of additivity (Bliss, 1956) to characterize the presence or absence of synergy for each combination. The synergistic activity of the in vitro combinations was assessed based on the Delta Bliss Sum Negative (DBSumNeg) score with the conservative synergy cutoff DBSumNeg < −3 (Mott et al., 2015). The DBSumNeg score was introduced as a new metric to quantify the synergy of the combination of two drugs across all dose combinations tested and was computed simply as the sum of the synergistic deviations from the Excess over Bliss model (Bliss, 1956). Combinations that demonstrated synergy based on the DBSumNeg scores in the primary screen were retested against both the SK-N-BE(2)-C and LAN-1 cell lines. The conditions for the confirmation screen were as described above, but a 10-concentration-point dose window was used for each agent, as a ‘10×10’ block. For medium-throughput screening with a second BET inhibitor, SK-N-BE(2)-C cells were treated with I-BET151 in 10×10 combination with 58 compounds showing prominent synergy in the primary screen. At 12, 18 and 24 hr post drug dispensing, 3 μL Caspase-Glo 3/7 (Promega) was dispensed, and plates were read to assess Caspase 3/7 activity.
Quantification and Statistical Analyses
GSEA v2.1.0, GraphPad PRISM 7, R 3.2.3 and Python 2.7.2 software packages were used to perform the statistical analyses. Statistical tests used are specified in the Figure legends. Errors bars represent standard deviation, unless otherwise stated. The threshold for statistical significance is p < 0.05, unless otherwise specified.
Supplementary Material
HIGHLIGHTS.
PI3K overexpression promotes resistance to BET inhibitors in neuroblastoma.
BET inhibitor resistance engenders dependency on PI3K signaling.
Enhancer remodeling is associated with transcriptional changes in BETi resistance.
PI3K and BET inhibitors are synergistic in vitro and in vivo in neuroblastoma.
SIGNIFICANCE.
Recent pan-cancer genome studies have revealed that pediatric cancers have markedly lower mutation rates than adult cancers. These low mutation rates suggest that epigenetic dysregulation plays an important role in pediatric cancer development. The BET family of proteins are epigenetic readers which regulate transcription through bromodomain motifs. Ongoing clinical trials in hematologic and solid tumors support the further development of BET inhibitors (BETi); however, modest and short-lived responses suggest that drug resistance remains a major clinical challenge to their optimization. Here, we demonstrated that BETi resistant cells undergo enhancer remodeling and transcriptional changes engendering tumor cell vulnerabilities that can be exploited for therapeutic benefit. Moreover, we demonstrate that PI3K and BET inhibitors are highly synergistic in the upfront setting.
ACKNOWLEDGEMENTS
We thank the members of the RPPA core facility at the MD Anderson Cancer Center for generating the RPPA data included in this manuscript. This facility is funded by NCI # CA16672. The authors thank the NCATS matrix team, including Sam Michael, Carleen Klumpp-Thomas, Paul Shinn, and Crystal McKnight for technical assistance.
GRANT SUPPORT
A.B.I. is a Damon-Runyon Fellow (DRSG 12–15). G.R. is supported by the Associazione Italiana per la Ricerca sul Cancro-AIRC. B.S. is supported by a DAAD (Deutscher Akademischer Austauschdienst) fellowship in the thematic network: Research for Rare Diseases and Personalized Medicine. This work was supported by a Hyundai Hope on Wheels grant (K.S.); P01CA217959 and R01NS088355 (K.S.; W.A.W); the St. Baldrick’s Foundation’s Robert J. Arceci Innovation Award (K.S.); Friends for Life (K.S.); and the NIH intramural research program (NCATS).
Footnotes
DECLARATION OF INTERESTS
K.S. participates in the DFCI/Novartis Drug Discovery Program which includes grant support for an unrelated project and previously included consulting and has consulted for Rigel Pharmaceuticals on a topic unrelated to this manuscript. R.B. and P.B. receive grant funding from Novartis Institute for Biomedical Research for unrelated projects. W.A.W. is founder of StemSynergy Therapeutics which works on targeting WNT signaling in colorectal cancer.
Data and Software Availability
The ChIP-sequencing and RNA-sequencing data for this study are available for download from the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/, accession no: GSE107708).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. (2016). Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. The Lancet Haematology 3, e186–195. [DOI] [PubMed] [Google Scholar]
- Bliss CI (1956). The calculation of microbial assays. Bacteriological reviews 20, 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugene C, Raynal V, Etchevers HC, Thomas S, Lermine A, Daudigeos-Dubus E, et al. (2017). Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nature genetics. [DOI] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, Bair S, Newton G, Lichtman A, Kung A, et al. (2014). NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Molecular cell 56, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TS, Liang Z, Salama R, Stark R, and de Santiago I (2014). Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Frontiers in genetics 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. (2011). Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine 364, 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. (2014). CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-C (2006). Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews 58, 621–681. [DOI] [PubMed] [Google Scholar]
- Chou TC (2010). Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446. [DOI] [PubMed] [Google Scholar]
- Chou TC, and Talalay P (1984). Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation 22, 27–55. [DOI] [PubMed] [Google Scholar]
- Cools J, Mentens N, Furet P, Fabbro D, Clark JJ, Griffin JD, Marynen P, and Gilliland DG (2004). Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer research 64, 6385–6389. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. (2011). Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, He S, Krill-Burger JM, Root DE, Vazquez F, et al. (2018). Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nature genetics 50, 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, and Levanon EY (2013). Human housekeeping genes, revisited. Trends in genetics : TIG 29, 569–574. [DOI] [PubMed] [Google Scholar]
- Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, et al. (2009). MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America 106, 20411–20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, Lifshits E, Brown A, Lee C, Christensen JG, et al. (2010). Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 29, 2346–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nature protocols 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. (2015). BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorti-Passerini C, Antolini L, Mahon FX, Guilhot F, Deininger M, Fava C, Nagler A, Della Casa CM, Morra E, Abruzzese E, et al. (2011). Multicenter independent assessment of outcomes in chronic myeloid leukemia patients treated with imatinib. Journal of the National Cancer Institute 103, 553–561. [DOI] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, Tarazona S, Dopazo J, Meyer TF, and Conesa A (2012). Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. [DOI] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, et al. (2006). Lapatinib plus capecitabine for HER2-positive advanced breast cancer. The New England journal of medicine 355, 2733–2743. [DOI] [PubMed] [Google Scholar]
- Gridelli C, Maione P, Bareschino MA, Schettino C, Sacco PC, Ambrosio R, Barbato V, Falanga M, and Rossi A (2010). Erlotinib in the treatment of non-small cell lung cancer: current status and future developments. Anticancer research 30, 1301–1310. [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen A, Althoff K, Odersky A, Beckers A, Koche R, Speleman F, Schafers S, Bell E, Nortmeyer M, Westermann F, et al. (2016). Targeting MYCN-Driven Transcription By BET-Bromodomain Inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 2470–2481. [DOI] [PubMed] [Google Scholar]
- Hugo Varet J-YC, Marie-Agnès Dillies (2015). SARTools: a DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. bioRxiv. [DOI] [PMC free article] [PubMed]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. (2010). COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, and Garraway LA (2013). A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, Gabbasov R, Fink LS, Li Y, Lounsbury N, et al. (2016). Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell reports 16, 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. (2010). Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine 363, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. (2013). Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, and Sudarsanam S (2002). The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, et al. (2014). High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America 111, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, and Sims RJ 3rd (2011). Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America 108, 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, and Simeonov A (2008). A robotic platform for quantitative high-throughput screening. Assay and drug development technologies 6, 637–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirguet O, Gosmini R, Toum J, Clement CA, Barnathan M, Brusq JM, Mordaunt JE, Grimes RM, Crowe M, Pineau O, et al. (2013). Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. Journal of medicinal chemistry 56, 7501–7515. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. (2012). Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Mott BT, Eastman RT, Guha R, Sherlach KS, Siriwardana A, Shinn P, McKnight C, Michael S, Lacerda-Queiroz N, Patel PR, et al. (2015). High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci Rep 5, 13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. (2010). Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, and Ashworth A (2017). Marked for death: targeting epigenetic changes in cancer. Nature reviews Drug discovery 16, 241–263. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. (2013). The genetic landscape of high-risk neuroblastoma. Nature genetics 45, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. (2013). Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer discovery 3, 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, and Manke T (2014). deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research 42, W187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P, and Bentires-Alj M (2015). Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene 34, 3617–3626. [DOI] [PubMed] [Google Scholar]
- Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, et al. (2015). Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA Jr., Papadopoulos N, et al. (2013). Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nature genetics 45, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, and Sawyers CL (2002). Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer cell 2, 117–125. [DOI] [PubMed] [Google Scholar]
- Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, et al. (2016). Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. (2001). Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine 344, 783–792. [DOI] [PubMed] [Google Scholar]
- Sridhar SS, Seymour L, and Shepherd FA (2003). Inhibitors of epidermal-growth-factor receptors: a review of clinical research with a focus on non-small-cell lung cancer. The Lancet Oncology 4, 397–406. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, and Kornblau SM (2006). Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics 5, 2512–2521. [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ, and Versteeg R (2015). TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nature genetics. [DOI] [PubMed] [Google Scholar]
- van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, Broekmans M, Haneveld F, Nowakowska NE, Bras J, et al. (2017). Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nature genetics 49, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Vecchione L, Jacobs B, Normanno N, Ciardiello F, and Tejpar S (2011). EGFR-targeted therapy. Experimental cell research 317, 2765–2771. [DOI] [PubMed] [Google Scholar]
- Wyce A, Ganji G, Smitheman KN, Chung CW, Korenchuk S, Bai Y, Barbash O, Le B, Craggs PD, McCabe MT, et al. (2013). BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PloS one 8, e72967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nature methods 8, 659–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, and Zhou MM (2002). Bromodomain: an acetyl-lysine binding domain. FEBS letters 513, 124–128. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.