Abstract
Peptide nucleic acids (PNAs) are extensively studied for the control of genetic expression since their design in the 1990s. However, the application of PNAs in nanotechnology is much more recent. PNAs share the specific base-pair recognition characteristic of DNA together with material-like properties of polyamides, both proteins and synthetic polymers, such as Kevlar and Nylon. The first application of PNA was in the form of PNA-amphiphiles, resulting in the formation of either lipid integrated structures, hydrogels or fibrillary assemblies. Heteroduplex DNA–PNA assemblies allow the formation of hybrid structures with higher stability as compared with pure DNA. A systematic screen for minimal PNA building blocks resulted in the identification of guanine-containing di-PNA assemblies and protected guanine-PNA monomer spheres showing unique optical properties. Finally, the co-assembly of PNA with thymine-like three-faced cyanuric acid allowed the assembly of poly-adenine PNA into fibers. In summary, we believe that PNAs represent a new and important family of building blocks which converges the advantages of both DNA- and peptide-nanotechnologies.
1. Introduction
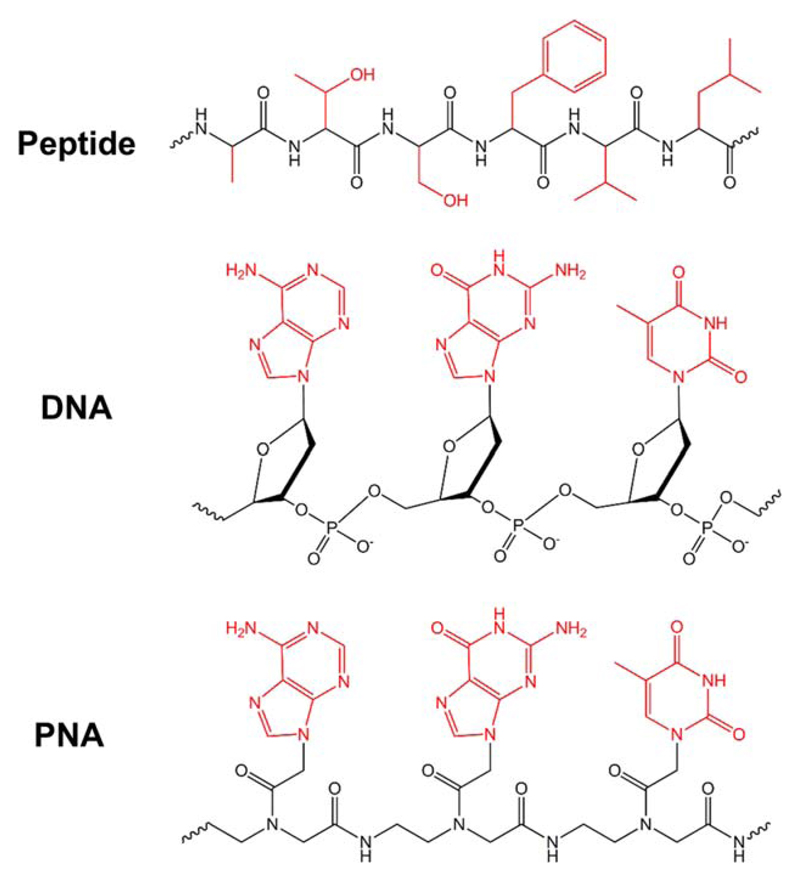
Designed in the early 1990s, peptide nucleic acids (PNAs) were presented as a novel strategy to target double-stranded DNA by strand displacement to achieve therapeutic gene modulation and to construct artificial restriction enzymes.[1–4] PNAs are synthetic DNA analogs in which the negatively charged sugar-phosphate backbone is replaced by uncharged repeats of N-(2-aminoethyl)glycine units linked by peptide bonds, to which the four primary nucleobases are attached as side-chains. For chemical structure see Figure 1. PNAs exhibit many attractive properties, including high affinity to DNA and RNA, compared with natural oligonucleotides, a significantly lower dependency on ionic strength, high sequence specificity for hybridization, chemical stability, and resistance to both nucleases and proteases. Furthermore, the peptide backbone allows relatively simple modifications and conjugation of various chemical groups. Owing to these remarkable features, PNAs have been used in a variety of biological and biomedical applications, mainly antisense therapy, and functional genomics research methods, such as enhanced polymerase chain reaction (PCR), diagnosis, detection, and biosensing.[5–14] To date, over 1,500 research papers and dozens of reviews have already been published on the use of PNAs in molecular biology and drug discovery.
Figure 1.
Representative chemical structures of peptide, DNA, and PNA. The side-chains which can be replaced by any amino acid residue in peptides, or nucleobase in the case of DNA and PNA, are highlighted in red
Much like in the case of DNA, PNAs were also utilized for applications beyond molecular biology in the larger context of nucleic acid nanotechnology. It was first suggested in the early 1980s that nucleic acids can be used as building blocks for materials science and nanotechnology, rather than merely as the carriers of genetic information.[15] This new field of science, which is commonly termed “structural DNA nanotechnology,” has led to the research and development of a vast array of highly defined self-assembled architectures of two and even three dimensions, including Holliday junctions, polyhedra, Borromean rings[16–20] and more elaborate shapes designed by computational methods.[21–23] Since PNA shares the specific molecular recognition employed by DNA via the precise Watson–Crick base pairing, as well as offers the advantages of peptides, including chemical versatility, architectural flexibility, and structural complexity, it was not long before researchers recognized its potential for nanotechnology. The polyamide nature of PNA is also compatible with solid material applications, as exploited in synthetic poly-amides (such as Nylon and Kevlar) and structural proteins (such as keratin and silk).
Owing to the superior hybridization properties of PNA, numerous studies have focused on the decoration of various nano-structured materials with PNA strands to develop tools for DNA detection.[24,25] A few notable examples include gold surfaces,[26] single-layered sheets of graphene oxide,[27] single-walled carbon nanotubes,[28] and nanodiamonds.[29] In this mini-review, we describe advances made in the last decade in the field of PNA nanotechnology, focusing on self-assembly of PNA-based structures (Figure 2).
Figure 2.
Schematic illustrations of the four main approaches to molecular self-assembly using PNA. (A) A PNA amphiphile is composed of a PNA strand (red) conjugated to a hydrophobic domain (yellow). When assembled in solution nanostructures such as spheres or fibers with a hydrophobic core are formed and the PNA are displayed on their surface in high density. (B) PNA can bind complementary sequences of DNA to form hybrid duplexes. These hybrids can then be used to generate typical nucleic acid assemblies such as G-quadruplex, or more elaborated designs using structural DNA nanotechnology methods. (C) PNA molecules as short as di-PNAs, have been shown to assemble through a combination of Watson–Crick interactions and aromatic stacking into different nanostructures with unique physical properties. (D) The co-assembly of PNA with cyanuric acid, a small molecule exhibiting three thymine-like faces, allows the organization of poly-adenine PNA into fibers
1.1. PNA amphiphiles
Peptide amphiphiles are a class of molecules composed of a hydrophobic domain conjugated to one terminus of a short peptide sequence that is relatively hydrophilic. The amphiphilic nature of these molecules allows assembly into supra-molecular nanostructures exhibiting attributes of surfactants combined with the biological function of the peptides embedded in the complex. This kind of assemblies is of great interest in tissue engineering, drug delivery, and other biomedical applications, as they can be endocytosed by cells, exhibit a high density of bioactive peptides and can be constructed as hydrogels, fibers, and micelles with adjustable dimensions.[30–32] Through the incorporation of PNA as part of the hydrophilic block, a nucleic acid binding dense surface can be achieved.
A series of works by Schneider and co-workers described the synthesis of different PNA sequences of up to 10-bases linked to alkanes of varying length, as well as their assembly and the stability of their duplexes with DNA oligomers.[33–36] The PNAs were also incorporated in phospholipid liposomes by their conjugation to a synthetic di-alkyl lipid tail. Naturally charged amino acids and a spacer composed of glycines were added to the sequence to confer sufficient head group hydration. These modifications resulted in stable liposomes that present the PNA domain and hence bind complementary DNA strands.[33,34] Characterization by small-angle X-ray scattering (SAXS) and dynamic light scattering (DLS) verified the formation of nanometric ellipsoidal micelles.[35] The PNA amphiphiles were shown to detect single stranded PCR products of varying lengths, and discriminated over other oligomers of same length, to give a novel gel-free DNA analysis method by separation with micellar electrokinetic chromatography.[36]
Stupp and co-workers have described a PNA–peptide amphiphile conjugate that self-assembles into nanofibers and binds to oligonucleotides with high affinity and specificity.[37] This PNA segment is a poly-thymine heptamer followed by the peptide sequence KGGGAAAK which promotes β-sheet formation to give nanofibers as previously reported by the same group.[38] Thermal denaturation experiments showed stronger binding of a poly-adenine DNA by the PNA–peptide amphiphile fibers compared with poly-thymine DNA and RNA, as well as a free heptathymine PNA molecule. Moreover, when a single thymine was introduced into the hepta-adenine DNA strand substrate, the melting temperature (Tm) was lower by 16°C, indicating high binding specificity. The authors suggested that these fibers can be highly attractive for application, including translation regulation studies, nucleic acid purification, and biosensors.
In a more recent work, Gianneschi and co-workers described the preparation, assembly and DNA hybridization properties of PNA-based amphiphilic polymer brushes.[39] These assemblies are comprised of a 10-base long single stranded PNA sequence formulated into high-density brushes via graft-through polymerization using ring-opening metathesis polymerization (ROMP). The resulting materials self-assemble into spherical nanoparticles with a hydrodynamic radius of 25 nm when dialyzed from DMSO into an aqueous solution, as indicated by electron microscopy and DLS and supported by molecular dynamics simulations. The PNA brush polymers show hybridization to complementary DNA sequences. As demonstrated by Tm measurements, hybridization to the nanoparticles is stronger than that of the unpolymerized PNA by a difference of 8°C. Potential applications for these assemblies, as proposed by the authors, are nucleic acid delivery into cells and DNA purification.
1.2. PNA–DNA duplexes
A different approach for the self-assembly of PNAs into nanoparticles is by inserting a PNA molecule into an array of self-assembled DNA building blocks. Hybrid duplexes would benefit from the advantages available to PNAs as discussed above, in particular improved chemical and thermal stability, while making minimal changes in the assembled DNA design.
Seeman and co-workers demonstrated this hetero-duplex strategy by the incorporation of PNA into a DNA double crossover (DX) molecule.[40] DX molecules, which normally consist solely of DNA, contain two crossover links between two double helical domains. These molecules can be expanded further by adding a secondary structure motif between the two crossovers. The researchers used two DX DNA tiles, one of which appended with a DNA hairpin perpendicular to the molecule plane between the crossovers, and both terminated with sticky ends. When co-assembled, these two tiles form a two-dimensional array displaying the hair-pins that appear as stripes at constant distances when imaged using atomic force microscopy (AFM). The unmodified tile was changed so it contained short segments of PNA instead of only DNA strands. Assembly of the hetero-polymer tiles showed arrays similar to those generated by the DNA-based molecules. Furthermore, the study details the use of these arrays as tools to measure the helical repeat of oligonucleotides by the periodic features identified in the AFM images.
Another well-studied DNA architecture that has been the subject of several studies with the goal of turning it into a hetero-duplex is the guanine (G) quadruplex. G-quadruplex is a guanine-rich sequence motif naturally occurring in the genome that adopts a distinct four-stranded structural topology.[41] It has been reported that designed PNA oligomers are also capable of forming hybrid-quadruplexes with DNA. Armitage and co-workers targeted a dimeric G-quadruplex telomeric sequence with a G-rich PNA probe.[42] Melting curves recorded by UV and circular dichroism (CD) spectroscopy revealed the formation of a stable hybrid containing G-quartets. The resulting hetero-quadruplex exhibits very high thermodynamic stability of −36.1 kcal/mole at low salt concentration, comparable with the remarkable stability of a four-stranded DNA quadruplex of a similar sequence. These quadruplexes can be used either as therapeutic targets owing to their high biological stability or as telomeres imaging agents.[42–44]
Different hybrid G-quadruplex forming PNA molecules were studied by Appella and co-workers. A set of short G-rich γ-substituted PNA constructed from L-lysine (LKγ) with varying number of γ-modifications was synthesized. This family of molecules has been shown previously by the same group to serve as a versatile scaffold to introduce a range of functional groups onto PNA without hindering the hybridization of complementary sequences.[45] While unmodified G-rich PNAs can generate quadruplex structures alone, these LKγ-PNAs form hybrids only with target human telomeric DNA sequences, making them highly selective. This level of specificity is suggested to be useful for the development of telomer targeting ligands.[46]
In a latter work, Balasubramanian and co-workers described the first PNA quadruplex formed with no DNA template.[47] The quartet-forming molecule is a tetra-PNA composed of one thymine residue followed by three guanines. A lysine was added to the C-terminus to enhance water solubility. Formation of quadruplexes was identified by mass spectrometry and UV melting curves. Analysis by CD spectroscopy revealed a pattern similar to that of antiparallel DNA quadruplexes. As the biological DNA-based G-quadruplexes are being employed as molecular wires and devices in nanotechnology, the authors envisioned the more chemically and thermally stable new structures to be excellent candidates for similar uses.
PNA–DNA interactions can also be utilized for the precise assembly of peptides and proteins as demonstrated in works by Fromme and co-workers. The researchers have used PNA–protein conjugates and introduced them into a DNA nanocage, four DNA sequences folded into a tetrahedral shape, through hybridization with the PNA linker.[48] PNA conjugates with either cytochrome c or azurin were assembled into the nanocages within merely 2 min at room temperature. Moreover, the catalytic activity and secondary structure of cytochrome c were analyzed and found to be unaffected by the assembled construct.[49] The authors suggested that the flexible design could be expanded to incorporate up to six different functional proteins that can be studied in defined sites within the DNA framework.
1.3. Minimalistic PNA
The shortest PNA building blocks that were shown to self-organize into ordered architectures are di-PNAs, i.e. PNA sequences of only two side-chains. All the 16 possible combinations of di-PNAs were screened, rising from the four PNA basic monomers (A, C, G, and T). It was demonstrated that certain guanine-containing molecules can self-assemble into ordered structures, while di-PNAs lacking a guanine base did not form any organized architecture.[50] X-ray diffraction analysis of the GC di-PNA revealed a highly organized structure stabilized by Watson–Crick interactions between guanines and cytosines of neighboring molecules, as well as inter- and intra-molecular aromatic π-stacking between the bases, as was also observed for short aromatic dipeptide assemblies.[51,52] This work, which was the first to describe the self-assembly of an unmodified PNA with no other additives, illustrated the duality of the PNA molecule. The resulting elongated structures were shown to exhibit excitation-dependent emission over the entire visible spectrum, a property that was not previously demonstrated for assemblies of either peptides or DNA and can be exploited for the fabrication of organic optical devices.
Following these results, an even smaller PNA-based molecule capable of self-association was discovered in a non-biased search.[53] All four unmodified PNA monomers, as well as their commercially available Fmoc- and Bhoc-protected forms, were tested for their self-association potential. Only the guanine-based compound with both protecting groups showed propensity to form ordered structures. DLS measurements and electron microscopy revealed spherical assemblies of highly uniform dimensions, resembling inorganic particles such as silica. These spheres underwent a secondary process of assembly at the interface of the solution and the air to generate a tightly packed monolayer arranged in hexameric symmetry. A similar array of nanocrystals is found in the skin of certain species of chameleon. Through changes in osmolarity of its tissues, this camouflaging animal is able to alter the spacing of the crystals, thereby reflecting a different wavelength, resulting in an active color change.[54] Since the crystals found in the skin of the chameleon are composed of guanine, the synthetic guanine-based PNA system was tested under different salt concentrations and adjustment of the reflected wavelength was indeed demonstrated. The change in color can be terminated by deposition of the layer on a solid substrate. The photonic crystals formed by this method are attractive for a variety of optics and photonic applications.
Notably, in both works only guanine-based PNA molecules were found to self-assemble.[50,55] This is interesting since guanine is a key component in the assembly of natural DNA structures such as the G-quadruplex.[56] Moreover, in addition to the arrays of guanine crystals in chameleons mentioned above, other nanometric architectures made of guanine crystals give rise to various physical colors in fish and in reflectors in reptile eyes (tapeta lucida).[57,58] Among the canonical nucleobases, guanine is known as the most potent in the field of supramolecular chemistry owing to its remarkably versatile chemical structure and the variety of non-covalent interactions it can form including hydrogen bonding, electrostatic interactions, hydrophobic interactions, metal coordination, and aromatic stacking.[56]
1.4. Modification of PNA assembly by a small molecule
A new approach to the self-assembly of PNAs is presented in a very recent work. While synthetic biologists are trying to expand the four-letter genetic alphabet, that allows for only two kinds of base-pairs, by incorporating unnatural bases into DNA, Sleiman and co-workers cleverly used a small molecule that possesses three thymine-like faces to alter the structure of poly-adenine nucleic acids.[59] This small molecule, termed cyanuric acid, is known to form hydrogen bonds with molecules that display complementary acceptor-donor motifs, such as adenine, melamine, and diaminotriazine. Upon addition of cyanuric acid to hepta-adenine PNA, assembly was monitored by a hypochromic effect and a red shift at the UV spectrum. AFM imaging revealed fibers that are 100 nm long and approximately 2 nm in height. This phenomenon was observed for poly-adenine DNA and RNA sequences, resulting in micron-long fibers. Since the structures can be generated in large quantities and the components are inexpensive and non-toxic, the authors suggested the fibers may have potential uses in tissue engineering and drug delivery.
2. Concluding Remarks
Nature has long served as an inspiration for scientists who desire to design functional synthetic systems with a high level of precision and efficiency. Two key directions for the development of bio-inspired nanomaterials are based either on the use of protein and peptide building blocks, or alternatively, on the use of DNA. Polypeptide structures are superior in terms of structural integrity and robustness, while nucleic acids have the advantage of specific molecular recognition between complementary bases. PNA is an artificially designed polymer that combines the rational design of DNA with the chemical versatility of peptides. Although originally developed for applications in molecular biology and drug design, it is emerging as a potent candidate for materials science and nanotechnology uses.
This mini-review highlights the four main approaches toward the generation of nanometric PNA-based supramolecular assemblies. While initial studies were based on the conjugating of PNA segments to self-assembling moieties to gain hybridization properties, the field is now expanding into exploitation of the nucleic acid nature of the polymer to achieve further rationally designed and well-organized architectures. Given the versatile characteristic of PNAs together with the ease of synthesis, new strategies toward the engineering of nanostructures for applications beyond nucleic acids binding are surely to rise in the future.
Acknowledgments
This work was supported in part by grants from the Israeli National Nanotechnology Initiative and Helmsley Charitable Trust for a focal technology area on Nanomedicine for Personalized Theranostics. The authors thank members of the Gazit laboratory for helpful discussions.
References
- [1].Egholm M, Buchardt O, Nielsen PE. J Am Chem Soc. 1992;114:1895. [Google Scholar]
- [2].Nielsen PE, Egholm M, Berg R, Buchardt O. Science. 1991;254:1497. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- [3].Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- [4].Wittung P, Nielsen PE, Buchardt O, Egholm M, Nordén B. Nature. 1994;368:561. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- [5].Nielsen PE, Egholm M, Buchardt O. Bioconjug Chem. 1994;5:3. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- [6].Nielsen PE, Haaima G. Chem Soc Rev. 1997;26:73. [Google Scholar]
- [7].Wang J. Biosens Bioelectron. 1998;13:757. doi: 10.1016/s0956-5663(98)00039-6. [DOI] [PubMed] [Google Scholar]
- [8].Nielsen PE. Acc Chem Res. 1999;32:624. [Google Scholar]
- [9].Larsen HJ, Bentin T, Nielsen PE. Biochim Biophys Acta Gene Struct Exp. 1999;1489:159. doi: 10.1016/s0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- [10].Ray A, Nordén B. FASEB J. 2000;14:1041. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- [11].Nielsen PE. Curr Opin Biotechnol. 2001;12:16. doi: 10.1016/s0958-1669(00)00170-1. [DOI] [PubMed] [Google Scholar]
- [12].Braasch DA, Corey DR. Biochemistry. 2002;41:4503. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- [13].Koppelhus U, Nielsen PE. Adv Drug Deliv Rev. 2003;55:267. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- [14].Shakeel S, Karim S, Ali A. J Chem Technol Biotechnol. 2006;81:892. [Google Scholar]
- [15].Seeman NC. J Theor Biol. 1982;99:237. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- [16].Seeman NC. Annu Rev Biophys Biomol Struct. 1998;27:225. doi: 10.1146/annurev.biophys.27.1.225. [DOI] [PubMed] [Google Scholar]
- [17].Seeman NC. Chem Biol. 2003;10:1151. doi: 10.1016/j.chembiol.2003.12.002. [DOI] [PubMed] [Google Scholar]
- [18].Seeman NC. Mol Biotechnol. 2007;37:246. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Seeman NC. Nano Lett. 2010;10:1971. doi: 10.1021/nl101262u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pinheiro AV, Han D, Shih WM, Yan H. Nat Nanotechnol. 2011;6:763. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- [22].Dietz H, Douglas SM, Shih WM. Science. 2009;325:725. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bonifazi D, Carloni L, Corvaglia V, Delforge A. Artif DNA PNA XNA. 2012;3:112. doi: 10.4161/adna.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Anstaett P, Gasser G. CHIMIA. 2014;68:264. doi: 10.2533/chimia.2014.264. [DOI] [PubMed] [Google Scholar]
- [26].Lioubashevski O, Patolsky F, Willner I. Langmuir. 2001;17:5134. [Google Scholar]
- [27].Park JS, Baek A, Park IS, Jun BH, Kim DE. Chem Commun. 2013;80:9203. doi: 10.1039/c3cc45750h. [DOI] [PubMed] [Google Scholar]
- [28].Williams KA, Veenhuizen PTM, Torre BG, Eritja R, Dekker C. Nature. 2002;420:761. doi: 10.1038/420761a. [DOI] [PubMed] [Google Scholar]
- [29].Gaillard C, Girard HA, Falck C, Paget V, Simic V, Ugolin N, Bergonzo P, Chevillard S, Arnaulta JC. RSC Adv. 2014;4:3566. [Google Scholar]
- [30].Cui H, Webber MJ, Stupp SI. Biopolymers. 2010;94:1. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao X, Pan F, Xu H, Yaseen M, Shan H, Hauser CAE, Zhang S, Lu JR. Chem Soc Rev. 2010;39:3480. doi: 10.1039/b915923c. [DOI] [PubMed] [Google Scholar]
- [32].Hartgerink JD, Beniash E, Stupp SI. Proc Natl Acad Sci. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vernille JP, Kovell LC, Schneider JW. Bioconjug Chem. 2004;15:1314. doi: 10.1021/bc049831a. [DOI] [PubMed] [Google Scholar]
- [34].Marques BF, Schneider JW. Langmuir. 2005;21:2488. doi: 10.1021/la047962u. [DOI] [PubMed] [Google Scholar]
- [35].Lau C, Bitton R, Bianco-Peled H, Schultz DG, Cookson DJ, Grosser ST, Schneider JW. J Phys Chem B. 2006;110:9027. doi: 10.1021/jp057049h. [DOI] [PubMed] [Google Scholar]
- [36].Grosser ST, Savard JM, Schneider JW. Anal Chem. 2007;79:9513. doi: 10.1021/ac7016376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guler MO, Pokorski JK, Appella DH, Stupp SI. Bioconjug Chem. 2005;16:501. doi: 10.1021/bc050053b. [DOI] [PubMed] [Google Scholar]
- [38].Behanna HA, Donners JJJM, Gordon AC, Stupp SI. J Am Chem Soc. 2005;127:1193. doi: 10.1021/ja044863u. [DOI] [PubMed] [Google Scholar]
- [39].James CR, Rush AM, Insley T, Vuković L, Adamiak L, Král P, Gianneschi NC. J Am Chem Soc. 2014;136:11216. doi: 10.1021/ja503142s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lukeman PS, Mittal AC, Seeman NC. Chem Commun. 2004;15:1694. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
- [41].Davis JT. Angew Chem Int Ed. 2004;43:668. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- [42].Datta B, Schmitt C, Armitage BA. J Am Chem Soc. 2003;125:4111. doi: 10.1021/ja028323d. [DOI] [PubMed] [Google Scholar]
- [43].Datta B, Bier ME, Roy S, Armitage BA. J Am Chem Soc. 2005;127:4199. doi: 10.1021/ja0446202. [DOI] [PubMed] [Google Scholar]
- [44].Gupta A, Lee LL, Roy S, Tanious FA, Wilson WD, Ly DH, Armitage BA. ChemBioChem. 2013;14:1476. doi: 10.1002/cbic.201300263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Englund EA, Appella DH. Angew Chem Int Ed Engl. 2007;46:1414. doi: 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]
- [46].Gupta P, Rastede EE, Appella DH. Bioorg Med Chem Lett. 2015;25:4757. doi: 10.1016/j.bmcl.2015.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Krishnan-Ghosh Y, Stephens E, Balasubramanian S. J Am Chem Soc. 2004;126:5944. doi: 10.1021/ja031508f. [DOI] [PubMed] [Google Scholar]
- [48].Flory JD, Shinde S, Lin S, Liu Y, Yan H, Ghirlanda G, Fromme P. J Am Chem Soc. 2013;135:6985. doi: 10.1021/ja400762c. [DOI] [PubMed] [Google Scholar]
- [49].Flory JD, Simmons CR, Lin S, Johnson T, Andreoni A, Zook J, Ghirlanda G, Liu Y, Yan H, Fromme P. J Am Chem Soc. 2014;136:8283. doi: 10.1021/ja501228c. [DOI] [PubMed] [Google Scholar]
- [50].Berger O, Adler-Abramovich L, Levy-Sakin M, Grunwald A, Liebes-Peer Y, Bachar M, Buzhansky L, Mossou E, Forsyth VT, Schwartz T, Ebenstein Y, et al. Nat Nanotechnol. 2015;10:353. doi: 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- [51].Reches M, Gazit E. Science. 2003;300:625. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- [52].Görbitz CH. Chem Commun. 2006;22:2332. doi: 10.1039/b603080g. [DOI] [PubMed] [Google Scholar]
- [53].Berger O, Yoskovitz E, Adler-Abramovich L, Gazit E. Adv Mater. 2016;28:2195. doi: 10.1002/adma.201504160. [DOI] [PubMed] [Google Scholar]
- [54].Teyssier J, Saenko SV, van der Marel D, Milinkovitch MC. Nat Commun. 2015;6:6368. doi: 10.1038/ncomms7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ulijn RV. Nat Nanotechnol. 2015;10:295. doi: 10.1038/nnano.2015.59. [DOI] [PubMed] [Google Scholar]
- [56].Davis JT, Spada GP. Chem Soc Rev. 2007;36:296. doi: 10.1039/b600282j. [DOI] [PubMed] [Google Scholar]
- [57].Ollivier FJ, Samuelson DA, Brooks DE, Lewis PA, Kallberg ME, Komáromy AM. Vet Ophthalmol. 2004;7:11. doi: 10.1111/j.1463-5224.2004.00318.x. [DOI] [PubMed] [Google Scholar]
- [58].Arnott HJ, Maciolek NJ, Nicol JA. Science. 1970;169:478. [PubMed] [Google Scholar]
- [59].Avakyan N, Greschner AA, Aldaye F, Serpell CJ, Toader V, Petitjean A, Sleiman HF. Nat Chem. 2016;8:368. doi: 10.1038/nchem.2451. [DOI] [PubMed] [Google Scholar]