Abstract
Amino acids are the simplest biological building blocks capable of forming discreet nanostructures by supramolecular self-assembly. The understanding of the process of organization of amino acid nanostructures is of fundamental importance for the study of metabolic diseases as well as for materials science applications. Although peptide self-assembled structures have been the topic of many review articles, much less attention has been devoted to the ability of amino acid building blocks, both natural and synthetic, to form ordered assemblies with defined architectures and notable physical properties, by the process of self-association. Herein, we try to shed light on amino acid based nanostructures, their fabrication and implications. We discuss self-assembled nanostructures, including hydrogels with nanoscale order, obtained from both modified and unmodified single amino acids. We also envision some future prospects in this emerging field.
Keywords: nanostructures, amino acids, self-assembly, phenylalanine, hydrogels
1. Introduction
Assemblies of bonded atoms with a dimension between 1–100 nm are typically defined as “nanostructures”.[1] Nanostructures are highly ubiquitous in biological systems, with prominent examples including protein assemblies, viruses, lipid vesicles, and cellular organelles. These natural nanostructures are generated by molecular self-assembly, the underlying mechanism of numerous processes, such as, phospholipid membrane formation, DNA double helix organization, formation of amyloid fibrils (both functional and pathological), etc. During the process of molecular self-assembly, individual molecules (monomers) undergo independent organization via a number of non-covalent interactions, including hydrogen bonding, electrostatic attraction, van der Waals forces and aromatic stacking,[2] thereby forming thermodynamically stable, ordered, hierarchical nanostructures.[3] Specifically, the self-assembly of proteins plays a crucial role in the formation of biological scaffold materials,[4,5] giving rise to the actin cytoskeleton, microtubules, viruses, and other key nanostructures.[6] Some macroscopic constructs, like collagen in the skin, keratin in nails and hairs, and silk which has unique mechanical properties, are also composed of protein building blocks.
Peptides are fragments of proteins which may bear the functional and structural characteristics of proteins.[7] The self-assembly of peptides has been a subject of extensive research owing to the intrinsic biocompatibility of these building blocks, ease of synthesis, biodegradability and recyclability.[8] Applying the “bottom up” approach, in which self-assembly occurs at a molecular (or even atomic) level, allowed the design of biomaterials with unique properties that mimic the complexity and dimensions of biological systems.[4] The self-assembly of various peptide building blocks resulted in different nanostructured morphologies, including fibrils, nanotubes, spheres, vesicles and hydrogel matrices. Such nanostructured assemblies have diverse applications ranging from biology to nanotechnology.[9] The formation of nanostructures by short peptides was first reported 25 years ago by Ghadiri et al., who demonstrated the formation of nanotubular structures by the self-assembly of a cyclic octapeptide with alternating L and D amino acids.[10] Later, in 2003, applying a reductionist approach, an ultrashort dipeptide, diphenylalanine (Phe-Phe), which is the core recognition motif of Aβ, was demonstrated to be the simplest building block for peptide self-organization composed of coded amino acid, forming discreet nanotubes.[11] Later studies have confirmed that other dipeptides, including the simplest aromatic one, diphenylglycine, could form ordered assemblies.[12] Thus, for a long time it was assumed that the dipeptide building blocks represent the smallest recognition units stemming from the unique properties of the amide bond and its partial planarity due to resonance stabilization.
However, a very surprising observation reported in 2012 revealed that a single aromatic amino acid, phenylalanine (Phe), could also form ordered fibrillar assemblies with amyloid-like properties.[13] Follow-up studies had confirmed the ability of other amino acids, as well as additional metabolites, to form ordered supramolecular fibrillar assemblies.[14] X-ray crystallography of Phe in its zwitterionic state, under conditions allowing its fibrillization, confirmed the organization of the amino acid into an ordered β-sheet-like layered organization, which was stabilized by a network of hydrogen bonds and aromatic interactions.[15]
Amino acids are the building blocks of proteins, with their composition and sequence controlling the explicit characteristics of these polyamide structures. Amino acid based nanostructures are formed by self-assembly of the simplest building blocks in the milieu of biological systems, serving as the smallest member of the “bottom up” approach. Even though many comprehensive reviews have focused on nanostructures by peptide self-assembly,[5,16] nanostructures fabricated by self-assembly of amino acids has so far not been appraised.
In this focus review, the formation of amino acids nanostructures by supramolecular self-assembly is discussed. We depict different approaches for the design of such nanostructures from both modified and unmodified amino acids, along with their implications, both from diseases and material science perspective. We also portray a brief picture of the challenges and future prospects of this field.
2. Amino Acid Based Nanostructures
The self-assembly of both unmodified and modified amino acids to form discreet nanostructure has recently emerged as an intriguing field of research, with exciting potential applications. In some cases, the assembly forms an entangled fibrillar 3D network of nanostructures which can entrap solvent molecules to form physical gels. Here, we present a concise view of these self-assembled nanostructures and physical gels.
2.1. Self-assembled Nanostructures
Amino acid self-assembled nanostructures can be subdivided according to their constituent building blocks, i. e. modified and unmodified amino acid-based building blocks.
2.1.1. Modified Amino Acid Self-assembly
Self-assembled structures of modified amino acids were presented in 1984 when Onai and co-workers synthesized N-(2-hydroxydodecyl) amino acids and found that N-(2-hydroxydodecyl) valine formed fibers in volatile organic solvents (acetone or diethyl ether) with helicity which altered according to the chirality of valine.[17] Similar properties were observed for leucine (Leu) and alanine (Ala), while the tryptophan (Trp) derivative did not exhibit helical fiber formation. Later it was observed that N-acyl-L-aspartic acids (CnAsp, n=12–18) could form gel-like aggregates in aqueous solutions at medium pH and low temperatures.[18] The morphologies of these assemblies varied from vesicles to helical fibers, depending on the length of the hydrocarbon chains and/or the pH of the solution. It was also observed that C12Glu did not form fibers, whereas C12Ala assembled into cylindrical fibers without helicity.
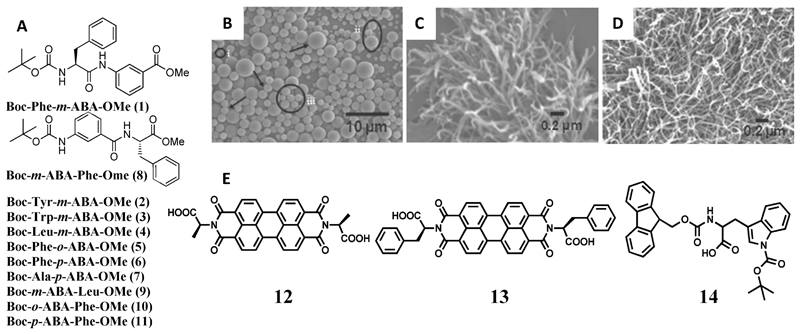
Pramanik and co-workers synthesized a series of derivatives of hydrophobic amino acids (Figure 1A, 1–11), including Phe, Trp, tyrosine (Tyr) and Leu by introducing an aminobenzoic acid (ABA) moiety and protecting the N and C termini by –Boc and –OMe groups, respectively.[19] The derivatives formed vesicles in methanol, whereas fibers or nanorods were obtained in less polar solvents (Figure 1B, C, D). However, derivatives of less hydrophobic amino acids, such as glycine (Gly), Ala, βAla, and proline (Pro), failed to produce nanostructures. Interestingly, a change in morphology was also observed with the positional isomerism of the ABA moiety. Single crystals were grown for the compounds 5, 7 and 8 (Figure 1A) and the crystal packing revealed the formation of β sheet layers. The incorporation of ABA improved the proteolytic stability of the nanostructures. Moreover, silver nanoparticles were also grown on both the inner and outer surfaces of the nanostructures. Another important application of these vesicles was the encapsulation and release of curcumin by disrupting them using a biocompatible metal ion (K+).[19]
Figure 1.
(A) Chemical structures of the amino acid derivatives (1-11). (B–D) Scanning electron microscopy (SEM) images of the nanostructures formed by compound (1) in (B) methanol, (C) chloroform + petroleum ether (3 : 7), and (D) toluene (10 mM).[19] (E) Molecular structures of the modified amino acids (12, 13, 14). Figures reproduced with permission from reference.
Applying another interesting strategy, Ala and Phe were utilized to functionalize 3,4,9,10-Perylenetetracarboxyl dianhydride, and the resultant imide derivatives (12, 13 in Figure 1E) were used to engineer composites with polyaniline.[20] The imide derivatives are electron dearth centers and thus can function as electron acceptors. A composite of these derivatives and polyaniline (acting as a donor) generated nanostructures that were deployed for dye sensitized solar cell applications. An improved power conversion efficiency was achieved using the Phe derivative, as compared to the Ala derivative, because of the better π-stacking interactions of polyaniline with the former through the aromatic phenyl rings, which enhances the electron mobility, and hence the power conversion efficiency.
Recently, Panda and co-workers depicted the formation of nanoparticles from a Trp derivative, N-alpha-(9-fluorenylmethyloxycarbonyl)-N(in)-tert-butyloxycarbonyl-l-tryptophan (Fmoc-Trp(Boc)-OH) (14 in Figure 1E).[21] The self-assembled nanoparticles (diameter: 489.4 ± 80.2 nm) were prepared by diluting a HFIP solution of Fmoc-Trp(Boc)-OH in water to a final concentration of 2 mM.
Taking a de novo approach, Musah and co-workers synthesized a boron cluster amino acid, 2-amino-3-(1,7-dicarbaclosododecacarboranyl-1-thio)-propanoic acid, by substituting the thiol proton in cysteine with m-carborane.[22] This modified amino acid formed floret like discs in ethanol and fibers in aqueous solutions. Crystal structures revealed the formation of bilayers via H-bonding interactions.
2.1.2. Unmodified Amino Acid Self-assembly
The self-assembly of nanostructures by unmodified amino acids is facilitated only when using the aromatic amino acids Phe, Tyr, Trp, Histidine(His), excluding one exception, in which Gly nanostructures were observed.
2.1.2.1. Phenylalanine Self-assembly and its Association with Phenylketonuria (PKU)
The most studied single amino acid capable of forming nanostructures is Phe. Accumulation of Phe due to genetic mutation underlies the etiology of PKU, a congenital metabolic disorder characterized by high levels of Phe in different portions of the brain, cerebrospinal fluid, and plasma, thereby mitigating brain development.[23–25] Understanding the self-assembly process of Phe is thus expected to shed light on the underlying etiology of PKU. Furthermore, developing novel approaches to inhibit Phe self-assembly may lay the basis for new therapeutics.
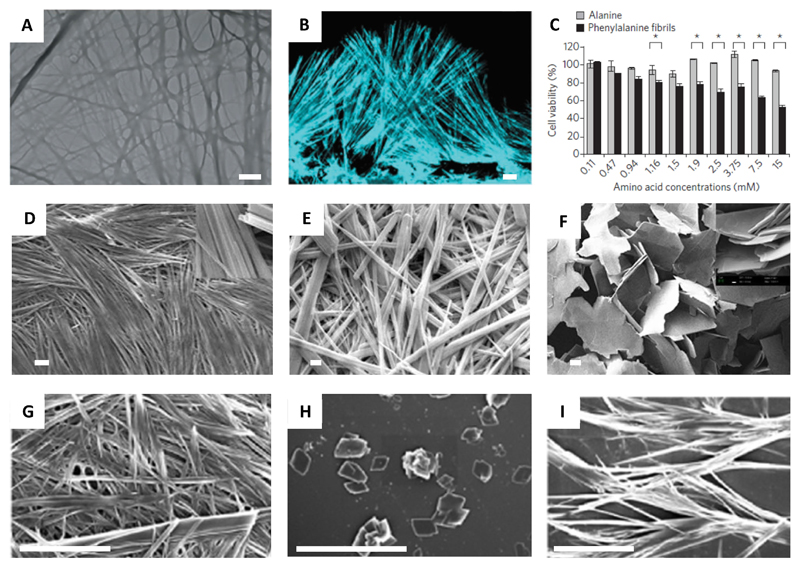
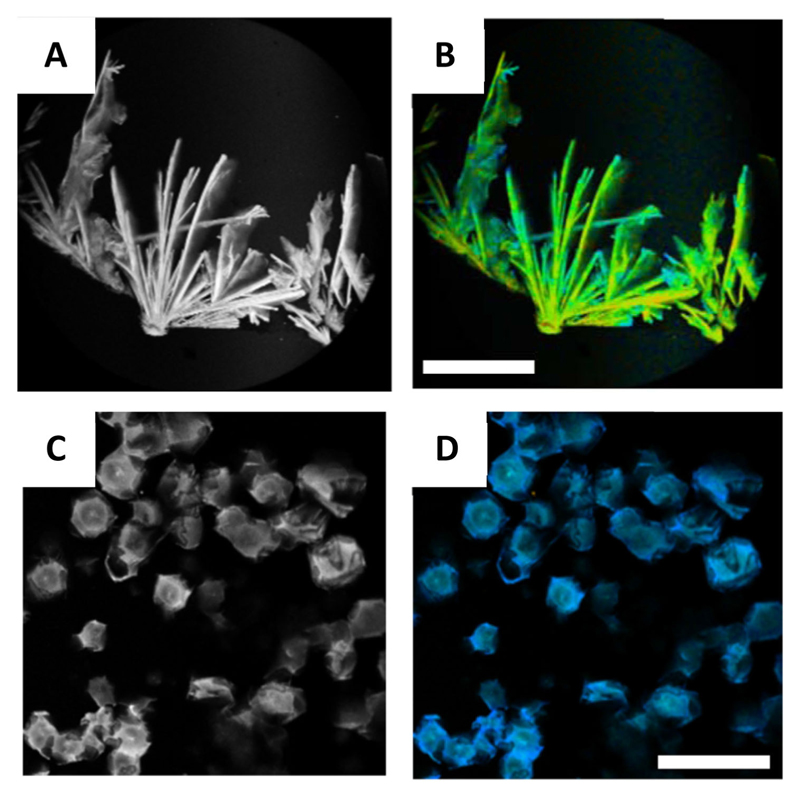
In 2012, we presented the first account of self-assembly of an unmodified amino acid (Phe) and its correlation with PKU. We have shown that Phe self-assembles in water forming well-ordered fibrillar aggregates at millimolar concentrations (Figure 2A).[13] The fibers exhibited typical birefringence upon staining with Congo red, clear electron diffraction patterns, Thioflavin T (ThT) fluorescence patterns (Figure 2B), and strong cytotoxicity (Figure 2C), all canonical characteristics of protein amyloids. For cytotoxicity measurements, PC12 cells were cultured in 96-well micro plates and were incubated overnight at 37 °C. Phe, dissolved in Dulbecco’s Modified Eagle Medium (DMEM) at various concentrations, were added to each well. After incubation for 6 hours at 37°C, cell viability was estimated using the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay. Furthermore, these structures were specifically recognized by antibodies generated upon their injection into rabbits. Binding by the antibodies significantly reduced the cytotoxicity of the assemblies. The hippocampus of model mice, as well as parietal cortex brain tissue from individuals with PKU, were found to contain these Phe assemblies. These findings indicate that PKU resembles amyloid-like diseases and may result from a similar underlying mechanism.
Figure 2.
(A) TEM image of Phe fibrils (scale bar 1 μm).[13] (B) Confocal microscopy image of Phe fibers dyed with ThT (scale bar 10 μm).[13] (C) Cytotoxicity of the Phe fibers (Ala as control) towards PC12 cell line determined using MTT assay.[13] (D–F) SEM images of (d) 300 mM L-Phe (top window 10 k× zoom) (scale bar 10 μm), (E) D-Phe (scale bar 2 μm), (F) DL-Phe (scale bar 20 μm) (top window 5 k×zoom). [27] (G) FESEM image of Phe fibrils (scale bar 4 μm).[28] (H–I) Phe fibril inhibition by 6 mM of (H) 18C6 (scale bar 20 μm) and (I) 15C5 (scale bar 10 μm) after 48 hours.[28] Figures reproduced with permission from respective references.
Phe fibrils were later demonstrated to cross-seed protein aggregation.[26] These fibrils could trigger in the self-assembly of proteins under physiological conditions and transform native protein structures to β-sheet assemblies. Interestingly, it was observed that both Phe fibrils, and, Phe-induced protein aggregates, exhibited hemolytic effect, which along with protein aggregation is a characteristic symptom usually found in PKU patients. These findings further reinforced the role of Phe aggregation in the disease mechanism of PKU.
Interestingly, D-Phe has been projected as a therapeutic molecule for PKU.[27] Thakur and co-workers reported the modulation of the aggregation behavior of L-Phe using D-Phe. It was observed that both L-Phe and D-Phe separately form hydrogels with fibrillar structure in water and in 300 mM phosphate buffer saline (PBS) (Figure 2D, E). Interestingly, an equimolar mixture of L-Phe and D-Phe did not produce gels, rather forming flake-like structures which phased out from solution (Figure 2F). The formation of flakes was also observed at a concentration ≥ 8% of D-Phe when L-Phe was titrated with increasing amounts of D-Phe.
Sarkar and co-workers applied another innovative strategy to arrest the fiber formation of L-Phe (Figure 2G).[28] Taking a supramolecular approach, they established that two crown ethers, 15-Crown-5 (15C5) and 18-Crown-6 (18C6) could inhibit the fiber formation of L-Phe, although 18C6 was found to be more effective (Figure 2H–I). This inhibition was ascribed to the favorable host-guest interaction of the −NH3+ group of Phe with 18C6. They further discovered that lanthanide cations also have the competence to arrest Phe fibrils with an efficacy order of Tb3+ < Sm3+ < Eu3+.[29]
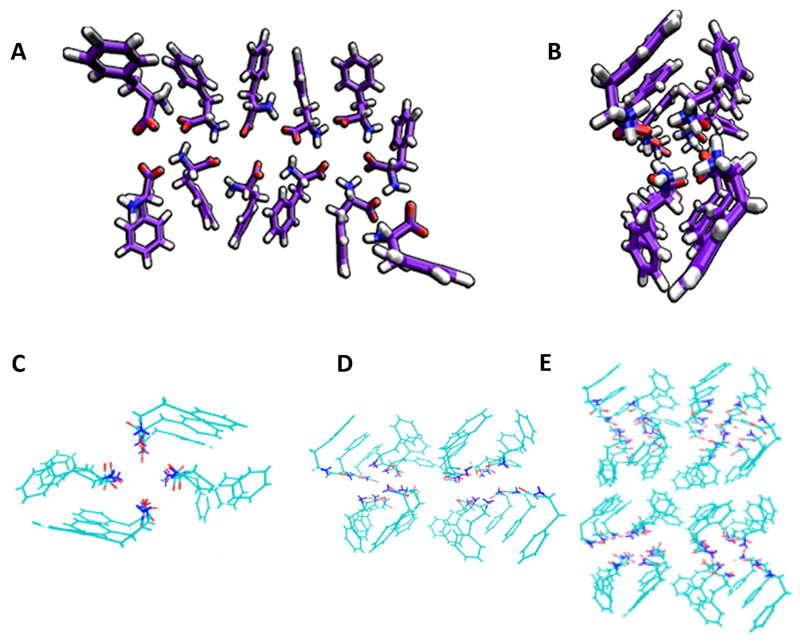
Aiming to decipher the structure of Phe assemblies, we performed molecular dynamics simulation of 27 Phe molecules, suggesting the formation of ladder-like structures at high pH (neutral N-terminal end groups) (Figure 3A, B).[13] This structure was quite different from that of the previously reported crystal structures of Phe.[30] Hansmann and co-workers studied the Phe self-assembly process by molecular dynamics at pH 7, in which both end groups are charged. Surprisingly, in contrary to the previously presented crystal structures,[30] or the ladder-like structures we have reported,[13] they found that tubular structures, in which each layer consisted of four Phe molecules, were the dominant assemblies.[31] Bowers and co-workers employed ion-mobility mass spectrometry to inspect the structure of the Phe oligomers (up to n = 60) produced during the early stages of the assembly process.[32] They found a signature of singly and multiply charged oligomers in a mass spectra of 6 mM Phe in water (pH 6.5). Based on the model proposed by Hansmann, postulating that Phe assembles in layers of four monomers,[30] they constructed single tube, double tube, and tetra tube pore-like models which were in agreement with the experimental data (Figure 3C–E). They also observed the presence of sideways π-stacking interactions between the tubes (T-shaped π-stacking), along with π-stacking along the fibril axis within the tubes. These structures could also explain the cell toxicity of Phe fibers. The hydrophobic outer surface assists in penetrating the cell membrane whereas the hydrophilic core, comprised of zwitterionic ends, causes ion leakage from the cells, ultimately damaging them.
Figure 3.
Molecular dynamics showing (A) ladder-like and (B) four-fold dodecamer structures, containing three four-monomer layers of Phe oligomers.[32] (C–E) (c) Single, (D) double and (E) tetra tube models of Phe oligomers.[32] Figures reproduced with permission from reference.
2.1.2.2. Tyrosine Self-assembly
Khushalani and co-workers first documented the self-assembly of Tyr to form self-assembled nanostructures.[33] They obtained straight fibers with a flat ribbon-like textures from a 1 mM solution of Tyr in deionized water. However, they did not find any signature of secondary structural characteristics.
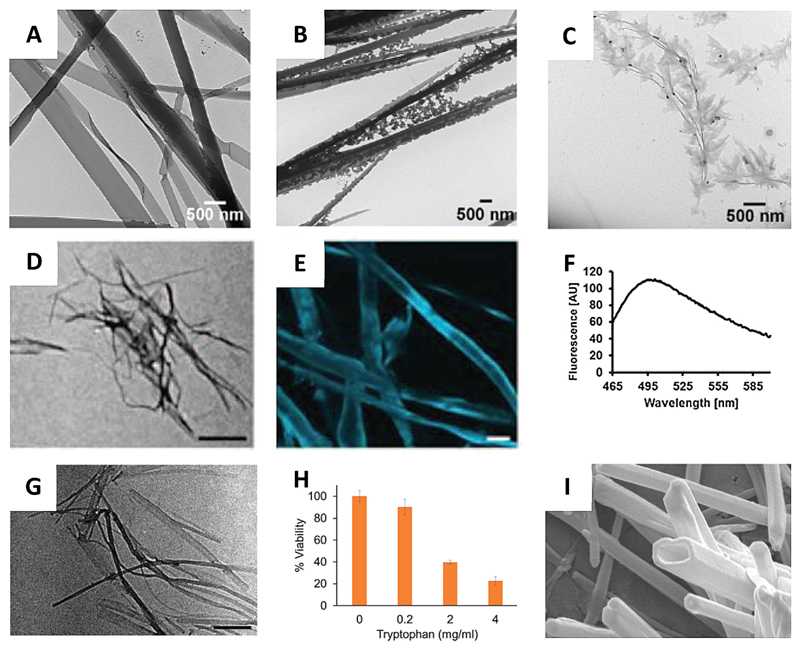
Studying the self-assembly of Tyr in deionized water, Verma, Bianco and co-workers observed different morphological features depending on the concentration of Tyr.[14a] At a concentration of 4.0–5.5 mM, fibers with a cross-section of a few microns were observed, whereas a decrease in concentration (~ 2.8 mM) provided nanoribbon strip-like nanostructures. At a further lower concentration (0.8 mM), small particles appeared at the end of the fibers. Interestingly, formation of dendritic structures from the tips of the nanoribbons were observed upon aging of a ~ 4 mM solution of Tyr (Figure 4A–C). No resemblance to amyloid-like morphology was observed for the aged Tyr assemblies.
Figure 4.
Tyr and Trp self-assembly. (A–C) TEM micrographs of Tyr assemblies at ~ 4 mM at various time points: (A) 6 days, (B) 14 days, (C) 22 days.[14a] (D–F) Amyloid nature of Tyr fibers. (D) TEM image (scale bar 500 nm), (E) ThT microscopy (scale bar 20 μm) and (F) fluorescence spectrum of Tyr fibers.[14b] (G) TEM image of Trp fibers (scale bar 500 nm).[14c] (H) Cytotoxicity of Trp fibers towards SH-SY5Y cells.[14c] (I) FESEM images of Trp nanotubes (scale bar 200 nm).[35] Figures reproduced with permission from respective references.
However, we observed that upon dissolving in PBS at a much higher concentration (~ 22 mM) by heating to 90 °C, Tyr could form amyloid resembling fibers (Figure 4D–F).[14b] These fibers also exhibited cytotoxicity and induced apoptotic cell death. This work further extended the hypotheses of amyloid formation by building blocks simpler than proteins and peptides. Moreover, it also strengthened the correlation of metabolic disorders and amyloid diseases.
Sarkar and co-workers shed light on the mechanism of Tyr fiber formation, presenting a self-assembly process different from that of Phe.[34] While inspecting the effect of 18C6 on the Tyr fibrils, they did not find any inhibition of the fibrillary morphology, contrary to previous observations for Phe.[28] As it was earlier established that host-guest interaction of the −NH3+ group of Phe with 18C6 resulted the Phe fiber arrest by 18C6, [28] it was apparent that −NH3+ was not involved in the fiber formation of Tyr, and the −OH and −COO- groups were the H-bonding partners.
2.1.2.3. Tryptophan Self-assembly
We have extended these earlier findings and shown that Trp could also form amyloid fibrils.[14c] At a concentration of ~ 19.5 mM in PBS, Trp formed fibrillar structures 5–20 nm in diameter (Figure 4G). As a typical signature of amyloid aggregates, these fibrils exhibited positive ThT staining and substantial cytotoxicity by inducing apoptotic cell death (Figure 4H).
Interestingly, in ethanol, Trp was found to form discreet nanotubes at a concentration of ~ 2.4 mM (Figure 4I). [35] These nanotubes exhibited fluorescence when excited at 385 (blue), 488 (yellow) and 561 (red) nm.
Wangoo, Sharma and co-workers studied the self-assembly and nanostructure formation of the aromatic amino acids Phe, Tyr, Trp and His in water/methanol mixtures.[36] Increasing the proportion of methanol, thereby decreasing dielectric constant of the solvent mixture resulted in different nanostructures. Phe formed more ordered fibers with a smaller diameter when methanol content was increased. In a 1 : 1 water: methanol ratio, Tyr formed rod like microstructures rather than nanoribbons in water. In the same solvent ratio, Trp was found to assemble into porous networks of twisted nanosheets. His was also reported to form dendritic structures in water: methanol (1 : 1), while increasing the methanol content to 90% v/v resulted in fused fibril formation.
2.1.2.4. Glycine Self-assembly
Amongst the non-aromatic amino acids, only Gly has been reported to form fibrous nanostructures which were probed using 2,2′-bipyridine-3,3′-diol (BP(OH)2) using the fluorescence lifetime imaging microscopic (FLIM) technique (Figure 5A, B).[14d] As in the case of Phe fibrils,[28] 18C6 inhibited the fiber formation of Gly by generating flake-like morphology (Figure 5C, D), whereas 15C5 showed no effect. Gly fibers are related with numerous genetic disorders, like nonketotic hyperglycinemia, D-glyceria aciduria, and iminoglycinuria, [14b] emphasizing the significance of these findings.
Figure 5.
Glycine self-assembled fibers and their disruption by 18C6.[14d] (A–B) Gly fibers images showing (a) fluorescence intensity and (b) lifetime images (scale bar 25 μm). (C–D) Gly flake-like structures formed in the presence of 18C6 showing (C) fluorescence intensity and (D) lifetime images (scale bar 25 μm). Figure reproduced with permission from reference.
2.2. Hydrogels
Single amino acids have also been demonstrated to form 3D networks of nanostructures which could entrap solvent molecules forming supramolecular gels. In this section we discuss different strategies to prepare these gels and their implications.
2.2.1. Enzymatic Approach
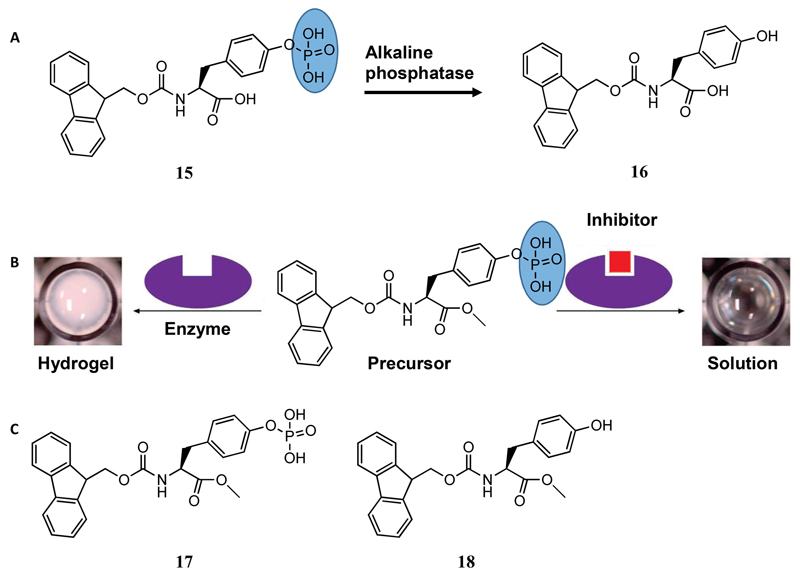
Being the first to apply the enzymatic approach to prepare amino acid-based hydrogels, Xu and co-workers used alkaline phosphatase, which removes the hydrophilic phosphate group from Fmoc tyrosine phosphate (15 in Figure 6A) and converts it to hydrophobic Fmoc-Tyr (16 in Figure 6A).[37] This change in solubility triggered the supramolecular gelation of Fmoc-Tyr consisting of nanofibrillar morphology. This strategy of enzymatic sol-gel transition was later utilized to visually screen the activity of inhibitors for a particular enzyme.[38] Pamidronate disodium, Zn2+, and sodium orthovanadate (Na3VO4) were examined as inhibitors and their minimum inhibitory concentrations were measured by monitoring the sol-gel phase transition (Figure 6B). Yang and co-workers also reported hydrogelation using the enzymatic approach where a “nonhydrogelator” Fmoc-L-Tyr(PO(OH)2-OMe (17 in Figure 6C) was converted into a hydrogelator Fmoc-L-Tyr-OMe (18 in Figure 6C) after treatment with phosphatase.[39] They proposed that the nanofibers of the hydrogel were principally composed of the hydrogelator, but were doped with the hydrophilic precursor, conferring their stability in aqueous medium.
Figure 6.
(A) Transformation of the precursor (15) to the hydrogelator (16) by alkaline phosphatase enzyme. (B) Schematic representation of utilizing enzymatic supramolecular hydrogelation for screening inhibitors of alkaline phosphatase.[38] (C) Chemical structures of the precursor (17) and hydrogelator (18). Figure reproduced with permission from reference.
2.2.2. pH Switching
Adams and co-workers used the pH switching method to prepare hydrogels of Fmoc-Phe and Fmoc-Tyr.[40] The amino acids were first dissolved in NaOH solutions, which were then added to required quantities of glucono-δ-lactone (GdL). GdL hydrolyzes in water to yield gluconic acid, resulting in a uniform change of the pH, thereby causing the formation of homogeneous gels. They also observed the formation of Fmoc-Phe crystals when HCl was used instead of GdL. As the Fmoc-Tyr hydrogels, which were mechanically stronger, could restrict the diffusion of larger dyes encapsulated in them. In contrast, the Fmoc-Phe hydrogels, being mechanically weak, were not able to retain even large dyes. This system can be potentially used as a controlled release system.
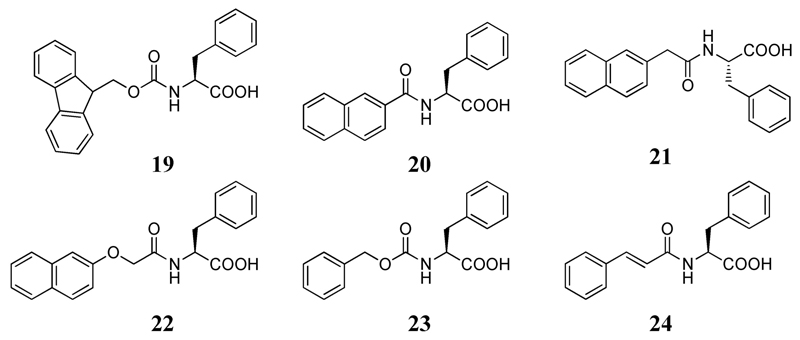
Xu and co-workers studied the gelation behavior of six Phe derivatives (19–24 in Figure 7).[41] The compounds were first dissolved in 1 N NaOH solution and then the pH of the solution was decreased. All the compounds, except 20 and 23, formed hydrogels. Interestingly, cis-trans isomerization was observed in 24, which caused a phase separation in the corresponding hydrogel.
Figure 7.
Chemical structures of the gelators used in reference 41.
2.2.3. Solvent Switching
Nilsson and co-workers studied the gelation properties of Fmoc protected pentafluorophenylalanine (Fmoc-F5-Phe) by applying the solvent switching method.[42] A solution of the peptide in DMSO (210 mM) was diluted with water to a final concentration of 4.2 mM (0.2%) yielding an optically transparent hydrogel within minutes. They also observed that Fmoc-Phe and Fmoc-4-CF3-Phe precipitated from the solution, whereas Fmoc-Tyr formed hydrogels after ~ 30 minutes. The Fmoc-F5-Phe hydrogels exhibited superior mechanical properties and lower gelation concentrations compared to the Fmoc-Tyr hydrogels. This work demonstrated that the physical properties of Fmoc-Phe based gelators could be tuned by side chain π-π stacking, which, by electronic contribution, dictates the assembly. To shed further light on the effect of electronegative groups/atoms on the π-π stacking, and consequently on the hydrogelation of Fmoc-Phe derivatives, single halogen substituents (F, Cl, Br) were introduced in ortho, meta and para positions of the benzyl side groups in Fmoc-Phe.[43] The mechanical properties and self-assembly rates of the hydrogels were found to vary with the halogen identity and position. Furthermore, polyethylene glycol (PEG) chains were introduced into Fmoc-F5-Phe to increase the solubility of the fibers which could lead to shear-responsiveness.[44] Indeed, co-assembly of Fmoc-F5-Phe and Fmoc-F5-Phe-PEG yielded hydrogels with adequate mechanical rigidity and ideal shear-responsiveness. Later, the C-terminal carboxylic acid groups of Fmoc-F5-Phe and Fmoc-3-F-Phe were modified to amide and methyl esters.[45] Interestingly, while the ester derivatives did not form hydrogels, the amide derivative formed hydrogels faster than the parent carboxylic acid derivatives. However, the lower solubility of the amide derivatives made them non-responsive to shear.
Our group reported the engineering of a two-component hydrogel composed of Fmoc-3,4-dihydroxyphenylalanine (Fmoc-DOPA) and Fmoc-Tyr by the solvent switching approach.[46] This hydrogel combined the physical properties of the Fmoc-Tyr gels with functionalities of the catechol groups of DOPA. This hydrogel could reduce ionic silver and form silver nanoparticles within the hydrogel matrix upon exposure to ambient light. This ability originated from Fmoc-DOPA, as Fmoc-Tyr did not exhibit nanoparticle formation under similar conditions.
2.2.4. Heating-cooling
Banerjee and co-workers obtained a hydrogel of Fmoc-Phe with a low gelation concentration (0.1%) in physiological pH (7.46) by the heating-cooling technique.[47] They observed that silver ions could be complexed with the gelator and consequently reduced to form fluorescent silver nanoclusters under diffused sunlight. They confirmed that the clusters were composed of very few silver atoms (Ag4). Functionalized single-walled carbon nanotubes were integrated with this hydrogel, resulting in improved mechanical properties, thermal stability, and conductivity.[48] It was shown that the carbon nanotubes were aligned in a 1D pattern along the hydrogel nanofibers.
A pyrene-conjugated Phe-based hydrogelator pyrene-Phe-OH, which forms thixotropic hydrogels in a wide pH range of 7.46–14, was also reported. [49] Interestingly, hydrogels were produced at a low gelation concentration of 0.037% in pH 7.46, indicating pyrene-Phe-OH to be a supergelator. Using the thixotropic property, vitamin B12 and doxorubicin, an anticancer drug, were incorporated into the hydrogels and their sustained release was studied. The thixotropic property allowed the incorporation of drug molecules inside the hydrogels, thereby avoiding the heating-cooling cycles which can potentially damage the drug activity.
Taking advantage of the co-assembly approach, an antibacterial amino acid-based building block, Fmoc-Leu, was co-assembled with Fmoc-Phe to prepare hydrogels with antimicrobial activity.[50] The hydrogels exhibited selective activity against gram-positive bacteria by disrupting the cell wall and membrane. The hydrogels were also found to be biocompatible with mammalian cells.
The hydrogelation ability of a Trp derivative was reported by Nandi and co-workers, who reported the gelation of Fmoc-Trp at pH 7.[51] The hydrogels were thixotropic in nature and exhibited strain recovery properties. To improve their mechanical properties, PEG was incorporated inside the hydrogels. The composite hydrogels indeed exhibited improved mechanical strength, higher strain recovery and enhancement of thixotropy. Utilizing the last property, doxorubicin was incorporated within the hydrogels and its release was tuned by changing the amount of PEG within the Fmoc-Trp networks. Recently, this Fmoc-Trp hydrogel was combined with a double network polyacrylamide/poly(vinyl alcohol) (PAAm/PVA) hydrogel.[52] The resulting hybrid hydrogels offered a reduction in friction coefficients on the hydrogel surface under application of shear.
Hanabusa and co-workers worked extensively on lysine (Lys) based hydrogels. They converted lysine based organogelators to hydrogelators by incorporating charged or non-charged hydrophilic groups into the organogelators. To create positively charged hydrogelators, pyridinium or imidazolium cations were incorporated on a terminal or pendant group,[53] whereas, negatively charged hydrogelators were prepared by de-esterification in alkaline condition.[54] Moreover, a non-ionic water soluble functional group, aldonamide, was introduced to produce non-ionic hydrogelators.[55]
New reports of amino acid based composite hydrogels are often published. A conducting hydrogel of Fmoc-Phe and polyaniline was reported, in which, a metastable hydrogel of Fmoc-Phe and aniline could be transformed into a stable, conducting hydrogel by polymerizing aniline inside the gel network.[56] Expectedly, the polymerized hydrogels exhibited much higher conductivity and capacitance, compared to the unpolymerized hydrogels.
There are also reports on amino acid based organogels. Nandi and co-workers reported the gelation of Fmoc-Phe in toluene and its co-assembly with 2-aminoanthracene (AA) and 2-aminonaphthalene.[57] The co-assembled gels of Fmoc-Phe and AA exhibited highly improved mechanical properties, compared to the Fmoc-Phe gels. Interestingly, Fmoc-Phe (acceptor) formed a charge transfer complex with AA (donor) in the gel state, accompanied by a color change of the gels from greenish-brown to deep-green. Recently, Lloyd and co-workers reported the gelation of unprotected Phe in DMSO.[58] They found that Phe could form gels at a much lower concentration in DMSO, as compared to water. Noticeably, the DMSO gels were transparent, unlike the opaque hydrogels.
3. Conclusions and Future Outlook
Amino acids have emerged as the simplest bio-inspired building blocks for producing nanostructures. As discussed in this focus review, both modified and unmodified amino acids are observed to form nanostructures with diverse morphologies, including fibers, vesicles, nanorods, nanoflakes, nanotubes, etc. The fibers could also physically cross-link to create entangled 3D network structures capable of immobilizing solvent molecules thus forming gels. These nanostructures are of paramount importance from both disease and materials science point of view. The fibers produced from the self-assembly of unprotected amino acids, such as Phe, Tyr and Trp, resemble typical protein amyloid assemblies. These discoveries reinforce the connection of metabolic disorders and amyloid diseases, and the understanding of the amino acid aggregation mechanism could shed light on the underlying etiology of these diseases. Moreover, methods to arrest the fiber formations potentially provide new paradigms for therapeutic intervention in these diseases. On the other hand, modified amino acids yield nanostructures/hydrogels for various applications, including drug encapsulation and release, electron acceptors for solar cells, formation and stabilization of silver nanoparticles in hydrogel matrix, antibacterial materials, lubrication under shear, etc.
As this field of research is comparably new, future discoveries are expected to further promote our understanding of disease etiology, as well as the development of novel materials for different applications. Thus, the ability of other unprotected amino acids to form ordered amyloid structures and their correlation with metabolic disorders may provide new insights into the underlying mechanisms of these ailments. Moreover, the design of de novo stimuli responsive assemblies of modified amino acids is thought-provoking, as they are of fundamental importance in drug delivery. Amino acid based hydrogels could be integrated with covalent polymers to improve their mechanical properties. Hybrids of hydrogels with graphene quantum dots/carbon dots are also fascinating, potentially combining the soft nature and intrinsic bio-compatibility of hydrogels with the electronic properties of graphene quantum dots/carbon dots.
Acknowledgements
This work was partially supported by grants from the European Research Council under the European Union’s Horizon 2020 research and innovation program (BISON, Advanced ERC grant, no. 694426) (E. G.). P.C. gratefully acknowledges the Center for Nanoscience and Nanotechnology of Tel Aviv University for financial support. The authors thank Sigal Rencus-Lazar for linguistic editing and all the members of the Gazit laboratory for helpful discussions.
Biographies
 Prof. Ehud Gazit is the incumbent Chair for Biotechnology of Neurodegenerative Diseases at Tel Aviv University. He received his BSc (summa cum laude) after completing his studies at the Special University Program for Outstanding Students at Tel Aviv University and his PhD (with highest distinction) from the Weizmann Institute of Science. He has been a faculty member at Tel Aviv University since 2000, following the completion of his postdoctoral studies at Massachusetts Institute of Technology (MIT). He is a member of the European Molecular Biology Organization (EMBO) and a Fellow of the Royal Society of Chemistry (FRSC). In 2015, he was knighted by the Italian Republic for his service to science and society. Prof. Gazit has received numerous awards including the Landau Research Award, the Dan David Scholarship Award, the Herstin Award for a leading scientist under the age of 44 in 2009, the Research Prize Award named after Teva Founders in 2013, and the Kadar Family Award in 2015.
Prof. Ehud Gazit is the incumbent Chair for Biotechnology of Neurodegenerative Diseases at Tel Aviv University. He received his BSc (summa cum laude) after completing his studies at the Special University Program for Outstanding Students at Tel Aviv University and his PhD (with highest distinction) from the Weizmann Institute of Science. He has been a faculty member at Tel Aviv University since 2000, following the completion of his postdoctoral studies at Massachusetts Institute of Technology (MIT). He is a member of the European Molecular Biology Organization (EMBO) and a Fellow of the Royal Society of Chemistry (FRSC). In 2015, he was knighted by the Italian Republic for his service to science and society. Prof. Gazit has received numerous awards including the Landau Research Award, the Dan David Scholarship Award, the Herstin Award for a leading scientist under the age of 44 in 2009, the Research Prize Award named after Teva Founders in 2013, and the Kadar Family Award in 2015.
 Dr. Priyadarshi Chakraborty obtained his PhD degree in 2016 from Indian Association for the Cultivation of Science, India. Subsequently, he joined the Prof. Ehud Gazit’s research group as a postdoctoral fellow under the Center for Nanoscience and Nanotechnology Post-Doctoral Fellowship program, Tel Aviv University. His research interests include bio-inspired, conducting, peptide-based hydrogels for electronic and biological applications.
Dr. Priyadarshi Chakraborty obtained his PhD degree in 2016 from Indian Association for the Cultivation of Science, India. Subsequently, he joined the Prof. Ehud Gazit’s research group as a postdoctoral fellow under the Center for Nanoscience and Nanotechnology Post-Doctoral Fellowship program, Tel Aviv University. His research interests include bio-inspired, conducting, peptide-based hydrogels for electronic and biological applications.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
This manuscript is part of a Special Issue on Supramolecular Nanostructures. A link to the Table of Contents will appear here once the Special Issue is assembled.
References
- [1].Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312–1318. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- [2].Whitesides GM, Grzybowski B. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- [3].Lehn JM. Science. 1993;260:1762–1763. doi: 10.1126/science.8511582. [DOI] [PubMed] [Google Scholar]
- [4].Zhang S. Nat Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- [5].Gazit E. Chem Soc Rev. 2007;36:1263–1269. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- [6].Liu Z, Qiao J, Niu Z, Wang Q. Chem Soc Rev. 2012;41:6178–6194. doi: 10.1039/c2cs35108k. [DOI] [PubMed] [Google Scholar]
- [7].Yan X, Zhua P, Li J. Chem Soc Rev. 2010;39:1877–1890. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- [8].a) Fichman G, Gazit E. Acta Biomater. 2014;10:1671–1682. doi: 10.1016/j.actbio.2013.08.013. [DOI] [PubMed] [Google Scholar]; b) Hamley IW. Angew Chem Int Ed. 2014;53:6866–6881. doi: 10.1002/anie.201310006. Angew. Chem2014, 126, 6984–7000. [DOI] [PubMed] [Google Scholar]; c) Stephanopoulos N, Ortony JH, Stupp SI. Acta Mater. 2013;61:912–930. doi: 10.1016/j.actamat.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Du X, Zhou J, Shi J, Xu B. Chem Rev. 2015;115:13165–13307. doi: 10.1021/acs.chemrev.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nagy-Smith K, Moore E, Schneider J, Tycko R. Proc Natl Acad Sci USA. 2015;112:9816–9821. doi: 10.1073/pnas.1509313112. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pappas CG, Sasselli IR, Ulijn RV. Angew Chem Int Ed. 2015;54:8119–8123. doi: 10.1002/anie.201500867. Angew. Chem. 2015, 127, 8237–8241. [DOI] [PubMed] [Google Scholar]; d) Knowles TPJ, Oppenheim TW, Buell AK, Chirgadze DY, Welland ME. Nat Nanotechnol. 2010;5:204–207. doi: 10.1038/nnano.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]; f) Luo Z, Zhang S. Chem Soc Rev. 2012;41:4736–4754. doi: 10.1039/c2cs15360b. [DOI] [PubMed] [Google Scholar]; g) Ghosh S, Reches M, Gazit E, Verma S. Angew Chem Int Ed. 2007;46:2002–2004. doi: 10.1002/anie.200604383. Angew. Chem. 2007, 119, 2048–2050. [DOI] [PubMed] [Google Scholar]; h) Zhang S, Greenfield MA, Mata A, Palmer LC, Bitton R, Mantei JR, Aparicio C, de La Cruz MO, Stupp SI. Nat Mater. 2010;9:594–601. doi: 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Ikezoe Y, Washino G, Uemura T, Kitagawa S, Matsui H. Nat Mater. 2012;11:1081–1085. doi: 10.1038/nmat3461. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G, Gazit E. Nat Nanotechnol. 2009;4:849–854. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- [10].a) Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Nature. 1993;366:324–327. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]; b) Hartgerink JD, Granja JR, Milligan RA, Ghadiri MR. J Am Chem Soc. 1996;118:43–50. [Google Scholar]
- [11].Reches M, Gazit E. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- [12].Reches M, Gazit E. Nano Lett. 2004;4:581–585. [Google Scholar]
- [13].Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E. Nat Chem Biol. 2012;8:701–706. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- [14].a) Mànard-Moyon C, Venkatesh V, Krishna KV, Bonachera F, Verma S, Bianco A. Chem Eur J. 2015;21:11681–11686. doi: 10.1002/chem.201502076. [DOI] [PubMed] [Google Scholar]; b) Shaham-Niv S, Adler-Abramovich L, Schnaider L, Gazit E. Sci Adv. 2015;1:e1500137. doi: 10.1126/sciadv.1500137. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shaham-Niv S, Rehak P, Vuković L, Adler-Abramovich L, Kràl P, Gazit E. Isr J Chem. 2017;57:729–737. [Google Scholar]; d) Banik D, Roy A, Kundu N, Sarkar N. J Phys Chem B. 2016;120:11247–11255. doi: 10.1021/acs.jpcb.6b07524. [DOI] [PubMed] [Google Scholar]
- [15].Mossou E, Teixeira SCM, Mitchell EP, Mason SA, Adler-Abramovich L, Gazit E, Forsyth VT. Acta Crystallogr C Struct Chem. 2014;70:326–331. doi: 10.1107/S2053229614002563. [DOI] [PubMed] [Google Scholar]
- [16].a) Adler-Abramovich L, Gazit E. Chem Soc Rev. 2014;43:6881–6893. doi: 10.1039/c4cs00164h. [DOI] [PubMed] [Google Scholar]; Yan X, Zhua P, Li J. Chem Soc Rev. 2010;39:1877–1890. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- [17].Hidaka H, Murata M, Onai T. J Chem Soc Chem Commun. 1984:562–564. [Google Scholar]
- [18].Imae T, Takahashi Y, Muramatsu H. J Am Chem Soc. 1992;114:3415–3419. [Google Scholar]
- [19].Koley P, Pramanik A. Adv Funct Mater. 2011;21:4126–4136. [Google Scholar]
- [20].a) Chal P, Shit A, Nandi AK. J Mater Chem C. 2016;4:272–285. [Google Scholar]; b) Chal P, Shit A, Nandi AK. J Mater Chem A. 2016;4:16108–16118. [Google Scholar]
- [21].Dube T, Mandal S, Panda JJ. Amino Acids. 2017;49:975–993. doi: 10.1007/s00726-017-2403-8. [DOI] [PubMed] [Google Scholar]
- [22].He T, Misuraca JC, Musah RA. Sci Rep. 2017;7:16995. doi: 10.1038/s41598-017-16926-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Blau N, van Spronsen FJ, Levy HL. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- [24].Kaufman S. Proc Natl Acad Sci USA. 1999;96:3160–3164. doi: 10.1073/pnas.96.6.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Williams RA, Mamotte CD, Burnett JR. Clin Biochem Rev. 2008;29:31–41. [PMC free article] [PubMed] [Google Scholar]
- [26].Anand BG, Dubey K, Shekhawat DS, Kar K. Sci Rep. 2017;7 doi: 10.1038/srep40744. 11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Singh V, Rai RK, Arora A, Sinha N, Thakur AK. Sci Rep. 2014;4:3875. doi: 10.1038/srep03875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Banik D, Dutta R, Banerjee P, Kundu S, Sarkar N. J Phys Chem B. 2016;120:7662–7670. doi: 10.1021/acs.jpcb.6b05209. [DOI] [PubMed] [Google Scholar]
- [29].Banik D, Kundu S, Banerjee P, Dutta R, Sarkar N. J Phys Chem B. 2017;121:1533–1543. doi: 10.1021/acs.jpcb.6b12220. [DOI] [PubMed] [Google Scholar]
- [30].a) Weissbuch I, Frolow F, Addadi L, Lahav M, Leiserowitz L. J Am Chem Soc. 1990;112:7718–7724. [Google Scholar]; b) King MD, Blanton TN, Korter TM. Phys Chem Chem Phys. 2012;14:1113–1116. doi: 10.1039/c1cp22831e. [DOI] [PubMed] [Google Scholar]
- [31].German HW, Uyaver S, Hansmann UHE. J Phys Chem A. 2015;119:1609–1615. doi: 10.1021/jp5077388. [DOI] [PubMed] [Google Scholar]
- [32].Do TD, Kincannon WM, Bowers MT. J Am Chem Soc. 2015;137:10080–10083. doi: 10.1021/jacs.5b05482. [DOI] [PubMed] [Google Scholar]
- [33].Perween S, Chandanshive B, Kotamarthi HC, Khushalani D. Soft Matter. 2013;9:10141–10145. [Google Scholar]
- [34].Banik D, Kundu S, Banerjee P, Dutta R, Sarkar N. J Phys Chem B. 2017;121:1533–1543. doi: 10.1021/acs.jpcb.6b12220. [DOI] [PubMed] [Google Scholar]
- [35].Babar DG, Sarkar S. Appl Nanosci. 2017;7:101–107. [Google Scholar]
- [36].Singh P, Brar SK, Bajaj M, Narang N, Mithu VS, Katare OP, Wangoo N, Sharma RK. Mater Sci Eng C. 2017;72:590–600. doi: 10.1016/j.msec.2016.11.117. [DOI] [PubMed] [Google Scholar]
- [37].Yang ZM, Gu HW, Fu DG, Gao P, Lam JK, Xu B. Adv Mater. 2004;16:1440–1444. [Google Scholar]
- [38].Yang ZM, Xu B. Chem Commun. 2004:2424–2425. doi: 10.1039/b408897b. [DOI] [PubMed] [Google Scholar]
- [39].Gao J, Wang H, Wang L, Wang J, Kong D, Yang Z. J Am Chem Soc. 2009;131:11286–11287. doi: 10.1021/ja9042142. [DOI] [PubMed] [Google Scholar]
- [40].Sutton S, Campbell NL, Cooper AI, Kirkland M, William Frith J, Adams DJ. Langmuir. 2009;25:10285–10291. doi: 10.1021/la9011058. [DOI] [PubMed] [Google Scholar]
- [41].Shi J, Gao Y, Yang Z, Xu B. Beilstein J Org Chem. 2011;7:167–172. doi: 10.3762/bjoc.7.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ryan DM, Anderson SB, Senguen FT, Youngman RE, Nilsson BL. Soft Matter. 2010;6:475–479. [Google Scholar]
- [43].Ryan DM, Anderson SB, Nilsson BL. Soft Matter. 2010;6:3220–3231. [Google Scholar]
- [44].Ryan DM, Doran TM, Nilsson BL. Chem Commun. 2011;47:475–477. doi: 10.1039/c0cc02217a. [DOI] [PubMed] [Google Scholar]
- [45].Ryan DM, Doran TM, Anderson SB, Nilsson BL. Langmuir. 2011;27:4029–4039. doi: 10.1021/la1048375. [DOI] [PubMed] [Google Scholar]
- [46].Fichman G, Guterman T, Adler-Abramovich L, Gazit E. CrystEngComm. 2015;17:8105–8112. [Google Scholar]
- [47].Roy S, Banerjee A. Soft Matter. 2011;7:5300–5308. [Google Scholar]
- [48].Roy S, Banerjee A. RSC Adv. 2012;2:2105–2111. [Google Scholar]
- [49].Nanda J, Biswas A, Banerjee A. Soft Matter. 2013;9:4198–4208. [Google Scholar]
- [50].Irwansyah I, Li Y-Q, Shi W, Qi D, Leow WR, Tang MBY, Li S, Chen X. Adv Mater. 2015;27:648–654. doi: 10.1002/adma.201403339. [DOI] [PubMed] [Google Scholar]
- [51].Chakraborty P, Mondal S, Khara S, Bairi P, Nandi AK. J Phys Chem B. 2015;119:5933–5944. doi: 10.1021/acs.jpcb.5b02424. [DOI] [PubMed] [Google Scholar]
- [52].Zhang X, Wang J, Jin H, Wang S, Song W. J Am Chem Soc. 2018;140:3186–3189. doi: 10.1021/jacs.7b12886. [DOI] [PubMed] [Google Scholar]
- [53].a) Suzuki M, Yumoto M, Kimura M, Shiraib H, Hanabusa K. Chem Commun. 2002:884–885. doi: 10.1039/b200242f. [DOI] [PubMed] [Google Scholar]; b) Suzuki M, Yumoto M, Kimura M, Shirai H, Hanabusa K. Chem Eur J. 2003;9:348–354. doi: 10.1002/chem.200390030. [DOI] [PubMed] [Google Scholar]; b) Suzuki M, Yumoto M, Kimura M, Shirai H, Hanabusa K. Helv Chim Acta. 2003;86:2228–2238. [Google Scholar]
- [54].a) Suzuki M, Yumoto M, Kimura M, Shirai H, Hanabusa K. Helv Acta Chim. 2004;87:1–10. [Google Scholar]; b) Suzuki M, Nanbu M, Yumoto M, Shirai H, Hanabusa K. New J Chem. 2005;29:1439–1444. [Google Scholar]
- [55].Suzuki M, Owa S, Shirai H, Hanabusa K. Tetrahedron. 2007;63:7302–7308. [Google Scholar]
- [56].Chakraborty P, Bairi P, Mondal S, Nandi AK. J Phys Chem B. 2014;118:13969–13980. doi: 10.1021/jp5086137. [DOI] [PubMed] [Google Scholar]
- [57].Bairi P, Roy B, Routh P, Sen K, Nandi AK. Soft Matter. 2012;8:7436–7445. [Google Scholar]
- [58].Nartowski KP, Ramalhete SM, Martin PC, Foster JS, Heinrich M, Eddleston MD, Green HR, Day GM, Khimyak YZ, Lloyd GO. Cryst Growth Des. 2017;17:4100–4109. [Google Scholar]