Abstract
The formation of ordered nanostructures by molecular self-assembly of proteins and peptides represents one of the principal directions in nanotechnology. Indeed, polyamides provide superior features as materials with diverse physical properties. A reductionist approach allowed the identification of extremely short peptide sequences, as short as dipeptides, which could form well-ordered amyloid-like β-sheet-rich assemblies comparable to supramolecular structures made of much larger proteins. Some of the peptide assemblies show remarkable mechanical, optical, and electrical characteristics. Another direction of reductionism utilized a natural noncoded amino acid, α-aminoisobutryic acid, to form short superhelical assemblies. The use of this exceptional helix inducer motif allowed the fabrication of single heptad repeats used in various biointerfaces, including their use as surfactants and DNA-binding agents. Two additional directions of the reductionist approach include the use of peptide nucleic acids (PNAs) and coassembly techniques. The diversified accomplishments of the reductionist approach, as well as the exciting future advances it bears, are discussed.
Keywords: bionanotechnology, molecular materials, molecular recognition, peptide engineering, self-assembly, supramolecular chemistry
1. Introduction
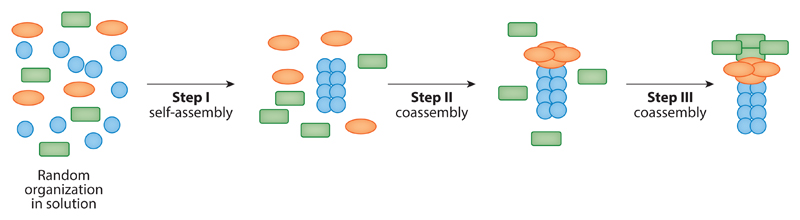
The use of molecular self-assembly is envisioned as one of the principal directions for advanced nanotechnological applications (1–6). The ability of small building blocks to organize spontaneously in a bottom-up manner is extremely attractive for the fabrication of functional devices without the need for top-down lithography methods and blueprint designs. Biological and bioinspired self-organizing units are especially attractive as all biological systems inherently feature the process of bottom-up assembly into ordered functional structures at the nanoscale (7–11). Biomolecules are indeed synthesized as discrete entities, which consequently form complex functional architectures by the processes of molecular recognition and self-assembly. This activity is a specific case of supramolecular chemical processes as most of the interactions that lead to the assembly of multifarious structures are noncovalent. Owing to these intrinsic properties, there is a continuous quest for the identification and characterization of adequate biological recognition and assembly elements that could be used for technological applications by mimicking natural self-organization processes. Furthermore, the process of self-organization could be hierarchal (Figure 1), and its mimicking is the ultimate goal of bioinspired nanotechnology. The initial attempts to achieve control and new architectures using coassembly are discussed below in Section 7.
Figure 1.
Hierarchal assembly of building blocks and the schematic representation of three molecular species that undergo a sequential self-organization process. Initially, the three types of molecules are randomly distributed. Then, one type of molecules undergoes self-assembly, and the two others are randomly distributed. In the next stage, a coassembled structure that contains two molecules is formed, and the third molecular species is in the solution. At the final stage, a three-component supramolecular structure is formed.
2. Molecular Self-Assembly of Proteins and Peptides
Proteins serve as a central source of materials for all biological systems (12–14). Ranging from rigid material such as keratin-based horns to the highly elastic resilin and elastin protein assemblies, polyamide chains, composed of natural or modified amino acids, are the preferred material for most natural functions both at the cellular and at the organismal level (15, 16). The organization of proteins into functional arrays can also result in strong and lightweight materials, such as spider silk or flexible assemblies including collagen-based fibrils in connective tissues (17–19).
Proteins are also very useful in the preparation of nanostructures of various architectures at various scales (20–26). The tendency of proteins to self-assemble into ordered architectures at the nanoscale can lead in many cases to different properties than those of the individual building blocks. This is an important aspect of the extensively studied fields of molecular self-assembly, supramolecular chemistry, nanoscience, and nanotechnology, which involve investigating the effects of molecule association, organization, orientation, and architecture on the physical and chemical properties of the resulting assemblies. The emerging physical properties of protein and peptide assemblies are also very useful for technological and medical applications. Such protein and peptide-based structures are remarkably applicable as scaffolds and molecular supports in applications ranging from tissue engineering and regeneration to drug delivery and controlled release of drugs.
2.1. Polyamides as Preferable Synthetic Materials
When proteins are discussed as preferred materials for technological applications, it should be noted that polyamides in general are also extremely useful as synthetic materials, including various forms of Nylon® (aliphatic polyamides), Kevlar® and Nomex®R (aromatic polyamides), and polyphthalamide. Proteins and synthetic polyamides, such as peptoids, share common structural features especially in the context of their organization as bulk materials (27–31). Other synthetic polyamide peptoids, discussed below in Section 8.1, are the bioinspired peptide nucleic acid (PNA) polymers, a family of peptide mimetics of nucleic acids with polyamide backbone and nucleobase side chains (32, 33), which were also recently explored for the production of self-assembled structures, showing unique architectural and physical characteristics (34, 35).
In spite of the usefulness of proteins as materials, these biomolecules are often complex to prepare, some of them are inherently unstable, and their production may be relatively costly and requires special settings. Therefore, there is a genuine scientific and industrial need for a mini-malistic approach to creating mimetic, functional protein materials using much simpler building blocks. Currently used both in the biological world and in synthetic settings, peptides offer the most straightforward alternative to protein-based materials and nanostructures.
2.2. Peptides Are Minimal Functional Units That Could Be Readily Synthesized
Peptides, as short protein fragments or stand-alone entities, serve as ideal building blocks for attaining protein-like properties in significantly simpler and much more robust systems (36–46). Ranging from the conjugation of two amino acids to a few tens of residues, peptides are readily produced by solid-phase or solution-phase (for short peptides) chemical synthesis, facilitating their efficient production (47–52). We currently witness an effort in the optimization of the speed of peptide synthesis similar to the revolution in DNA mass production. The current state-of-the-art for peptide synthesis is a total cycle time of 40 s per amino acid residue (53). These new developments may revolutionize the availability of the repertoire of peptides for technological applications as well as for basic research. Peptides are also used as ingredients in the food industry for which some of the peptides are produced in very large amounts, for example, the NH2-Phe-Asp-Me dipeptide (the artificial Aspartame® sweetener), which is remarkably similar to some of the building blocks discussed below (54). The annual production of this dipeptide was over 4 million tons a year at its peak with a production cost of only a few cents per gram. Therefore, if a practical commercial use is developed for dipeptide nanostructures, such as diphenylalanine mentioned below in Section 5, the cost of production should be economically practicable.
Automated peptide synthesizers are widely available in both academic and industrial settings, and the production of functional sequences following their biophysical and biochemical identification is remarkably simple. Another very important technological advancement facilitating the exploitation of functional peptide sequences is related to the production of thousands of peptides in the format of custom-made peptide arrays. An array of peptides can now be produced at a cost of less than 0.1 cent per peptide (e.g., PEPperCHIP® peptide microarray technology), resulting in the cost-effective production of thousands of peptides on a standard solid support chip (54–59). Peptide arrays allow the mapping of molecular recognition between peptide elements and the identification of interaction modules. We envision that this technology will revolutionize the field of peptide research, similar to the effect of DNA microarray technologies (known as DNA chip or biochip) on cellular and molecular biology research.
2.3. Beyond the Twenty Coded Amino Acids
The collection of explored synthetic peptides is not limited to the 20 coded amino acids used by the ribosomal synthesis of many peptides in living systems. Hundreds of nonnatural amino acid analogs are available for synthetic chemistry, many of them already in N-terminal protected forms that make them readily available for facile synthesis, thus remarkably increasing the available chemical space to be explored (60–62). This tactic is also employed by biological systems in which many peptides that contain noncoded amino acids are synthesized in enzymatic nonribosomal pathways (63–65). An additional approach to increasing chemical diversity is the use of posttranslational enzymatic modifications. For example, tyrosine residues are converted to 3,4-dihydroxyphenylalanine (L-Dopa) to allow a remarkably efficient surface adhesion observed in many marine organisms (66–70).
Moreover, modification for the efficient production of cyclic peptides can be applied to obtain various properties. The simplest cyclic peptides, cyclic dipeptides chemically denoted as diketo-piperazines, are utilized in various biological systems, as well as by peptide chemists, for the production of new materials with altered properties as compared to the linear peptides (71–73). Diketopiperazines are actually available in many types of natural food products, which suggests their safety and availability for human consumption (74). The ability to explore a vast chemical space together with the ease of synthesis make peptides ideal building blocks for various material-related applications. Moreover, peptides are usually much more stable than proteins under thermal or chemical stress. Therefore, the use of peptide building blocks may allow scientists to take advantage of the superb material properties of proteins but to utilize the minimal building blocks in a cost-effective manner for a large variety of applications.
2.4. Early Work on the De Novo Design Self-Assembly of Peptides
The availability of facile peptide synthesis and the ability to study self-assembly using spectroscopy, atomic force microscopy, and electron microscopy resulted in the intriguing exploration of peptide self-assembly in the 1990s and 2000s (75–83). It was shown that designed cyclic peptides with alternating D and L amino acids, a histidine-rich designed peptide, charge complementary peptides, and surfactant-like peptides could form well-ordered nanoassemblies with attractive architectures and functionalities. The most notable structures were those of nanotubes, which were of primary interest to the scientific community following the discovery of carbon nanotubes by Sumio Iijima in 1991 (84). This is a very important chapter in the field of peptide nanotechnology that is still relevant for current applications. However, this direction of research and development was extensively reviewed (e.g., 82, 83). Here, we limit our focus to the minimalistic-reductions approach for the discovery of increasingly shorter building blocks and for emerging future directions, including coassembly and the convergence of the DNA and peptide worlds, and the use of structurally hindered peptide motifs that form superhelical structures.
3. Amyloid Assemblies as a Generic Organization Motif
The β-sheet-rich amyloid fibrillary structures are key examples of generic protein organizations of ordered arrays of supramolecular structures. Originally identified in association with human diseases, the generic amyloid organization appears to represent a global energetic minimum for the aggregated state of proteins and peptides (85–91). The general arrangements of amyloid structures are fibrillary assemblies of 7–10 nm in diameter in which the polyamide chains are organized in cross-β secondary structures stabilized by dense networks of hydrogen bonds. The assemblies are remarkably ordered, as reflected by X-ray fiber diffraction and electron diffraction analyses, and show typical features, such as birefringence upon binding to the indicative Congo red dye and fluorescence following staining with thioflavin T. Amyloids also show high affinities to biological membranes, which may be useful for their biological activities.
The formation of amyloids was initially identified only for disease-related proteins and polypeptides, underlying the cytotoxic activity observed in neurodegenerative disorders such as Alzheimer’s disease and Parkinson’s disease. However, many proteins not related to any pathological state were later shown to self-assemble into amyloid fibrils that were structurally identical to those formed by the pathological building blocks. Dobson and coworkers pioneered the concept of amyloid structure as archetypical for protein and peptide assemblies in general based on studies from their group and others (92–95). The difference between the various amyloidogenic proteins appeared to be mainly related to the kinetics of formation rather than the structural and physical properties of the final products. Thus, amyloids serve as unique structural elements of generic properties and universal chemical organization. As most or all proteins eventually aggregate to produce the amyloidal state if a sufficient incubation time is provided, it seems that the soluble form of proteins actually only represents an energetically metastable state, and in time, the majority of proteins will adopt an amyloid structure (96, 97). Living cells actively prevent the unwanted association of proteins to form aggregated structures in a process called proteostasis (98–100).
3.1. Amyloids Are Also Natural Assemblies of Unique Physical Properties
Amyloids are also very interesting assemblies from a functional point of view. In various microorganisms, amyloids serve as structural material for the formation of a communal biofilm or the production of hyphae by a process of coordinated self-assembly (101). It appears that these organisms employ the remarkably efficient process of self-assembly for environmental fitness and protection against chemical and physical challenges. Another functional role of amyloid fibrils appears to be the production of melanin. Pigment cell-specific premelanosomal proteins undergo a process of assembly into amyloid structures that catalyze the formation of melanin in the melanosomes.
These functional amyloid assemblies, as well as the disease-related ones, were found to be extremely stable, showing notable mechanical rigidity (102) and label-free luminescence (103). These physical properties are of special interest as peptide nanostructures, which were discovered by the reductionist analysis of amyloid structures, possess properties similar to the protein assemblies, as described in Section 3.2 below.
3.2. The Formation of Amyloids by Self-Organization
The process of amyloid formation is based on a nucleation-growth mechanism, similar to the process of biomolecular crystallization into ordered three-dimensional arrays (104–108). In such a process, a set of thermodynamically unfavorable steps of molecular association occurs until a nucleus of a critical size is formed. This is followed by thermodynamically favorable steps of sequential noncovalent addition of new molecules to the preformed nucleus. The resulting structures are evidently supramolecular, and the organization of these stable assemblies does not depend on the formation of any new covalent bonds. The process is mainly mediated by a complex network of hydrogen bonds, although aromatic, hydrophobic, and electrostatic interactions can also contribute to the nucleation and growth steps.
4. Searching for the Smallest Recognition Modules
Aimed at identification of the shortest recognition module that can form typical amyloid fibrils, Kapurniotu and coworkers (109) performed a seminal work. The researchers demonstrated that a hexapeptide fragment of the islet amyloid polypeptide, a 37-amino acid amyloidogenic polypeptide associated with Type II diabetes, could form typical amyloid fibrils, showing the structural and biological properties of amyloids formed by much larger amyloidal polypeptides. The hexapeptide, Asn-Phe-Gly-Ala-Ile-Leu, forms typical amyloid fibrils that show the same morphology, cytotoxic activities, and dye-binding specificity as the full-length polypeptide. An even shorter tetrapeptide, Phe-Gly-Ala-Ile, can form unusually broad ribbons. The assemblies formed by both the hexapeptide and pentapeptide building blocks show a high amount of β-sheet secondary structure, as determined by analysis of the amide I vibration frequency using Fourier-transformed infrared spectroscopy.
4.1. Identification of Amyloidogenic Pentapeptides
In 2002, it was demonstrated that a pentapeptide fragment could form typical amyloid fibrils with structural and chemical properties as found in assemblies obtained by aggregation of much larger building blocks (110). The Asp-Phe-Asn-Lys-Phe peptide is part of the 31-amino acid calcitonin polypeptide, the organization of which into amyloid fibrils is associated with thyroid carcinoma. Identification of the fragment was based on the pH dependency of amyloid formation by the full-length polypeptide. The only charged amino acids in the entire peptide sequence were part of this pentapeptide, facilitating the truncation experiments that identified the pentapeptide motif as the shortest amyloid-forming element at that time. The organization of the peptide was recently determined by X-ray crystallography (111) and confirmed the earlier model for the organization of peptide building blocks.
Two other amyloidogenic pentapeptides were identified in another region of the Type II diabetes-associated islet amyloid polypeptide (112). The identification of two minimal self-associating sequences in the same polypeptide is quite intriguing and probably reflects the fact that this polypeptide is one of the most potent amyloid-forming molecules known so far. The Phe-Leu-Val-His-Ser and Asn-Phe-Leu-Val-His peptides, comprising a part of the central recognition module of the polypeptide, were identified by peptide array technology. The recognition module was identified by synthesis of a set of 28 overlapping decapeptides spanning the entire sequence of the islet amyloid polypeptide (i.e., peptide representation positions 1–10, 2–11, … 28–37 of the polypeptide). This array of peptides was incubated with the full-length 37-amino acid polypeptide, and the degree of binding was determined. The most active region in the binding process was then utilized for the systematic reductionist exploration of even the shortest elements within the decapeptide sequence that could self-assemble into amyloid-like structures.
4.2. Identification of Designed Amyloidogenic Tetrapeptides
Although the exploratory studies outlined above were conducted with peptide fragments of naturally occurring polypeptides, a later study using a designed peptide resulted in the identification of a tetrapeptide, Lys-Phe-Phe-Asp, that efficiently formed typical amyloid fibrils (113, 114). These assemblies were very similar to the fibrils formed by much larger polypeptides, and the driving forces seem to be aromatic interactions and electrostatic attraction.
Follow-up studies by several groups clearly established the formation of typical amyloid-like structures by very short peptides, ranging from tetrapeptides to hexapeptides and including both peptide fragments and designed peptides. Thus, very short peptides contain all the molecular information needed to mediate the process of molecular recognition and self-assembly to form ordered assemblies. Moreover, very short sequences are sufficient for the packing of polypeptide chains into regular structures.
4.3. The Role of Aromaticity in Amyloid Formation
Interestingly, all of these very short amyloid-forming peptides show high occurrence of aromatic amino acids, especially phenylalanine (115). It was therefore suggested by the author, in a hypothesis paper in 2002, that unlike the prevailing notion that amyloid formation stems merely from hydrophobic interactions, interactions between aromatic moieties may play a central role in the process of amyloid formation (115). Although aromaticity is not essential for amyloid formation per se, which is a backbone-related phenomenon, it appears to accelerate the process. This is especially crucial for very short peptide fragments in which the degree of hydrogen bonding between backbone elements is limited. It was suggested that the geometrically restricted interactions between aromatic side chains can contribute to the acceleration of the self-assembly process by providing order and directionality as well as binding energy (115).
The first high-resolution structural study of peptide amyloid fibrils using X-ray and electron diffraction techniques, performed in 2005 by Serpell and coworkers (116), clearly demonstrated the role of phenylalanine residues in the stabilization of intersheet packing via π-π stacking of the aromatic systems. Later studies demonstrated the same phenomenon in other short amyloidogenic peptide fragments (117–120).
The notion of the role of aromatic residues in the process of amyloid self-assembly was confirmed by a pioneering work by Dobson and coworkers (121). The authors mapped the aggregation propensity of all 20 naturally coded amino acids and discovered that the aromatic phenylalanine and tryptophan have the highest potential to promote amyloid formation. It was found that these two amino acids actually have more than three times the propensity to induce aggregation as compared to the highly hydrophobic and nonaromatic isoleucine amino acids (121). Furthermore, it was realized that some aliphatic amino acids, most notably leucine, actually have a negative propensity toward the self-assembly of amyloid fibrils (121). Thus, this comprehensive work further established that the aromatic model versus the hydrophobic model is the best way to understand the formation of ordered amyloid supramolecular assemblies by proteins and polypeptides.
5. Diphenylalanine as a Key Building Block
The diphenylalanine peptide is one of the most studied peptide building blocks in peptide nanotechnology. Its characterization was inspired by the study of amyloid self-assembly as described above in Section 3 (122). This peptide motif was also identified by a systematic, reductionist, non-biased exploration of an amyloid system to determine the most fundamental recognition unit that could form ordered structures at a nanoscale within the polypeptide. Diphenylalanine comprises the central recognition module of the β-amyloid polypeptide; an aggregation of this is associated with Alzheimer’s disease. The dipeptide was identified because it is the common denominator between a heptapeptide that forms amyloid fibrils and two pentapeptide inhibitors (122). It was found that the diphenylalanine peptide efficiently forms nanotubes in aqueous solution by a simple self-assembly process, very similar to that of proteins. Moreover, vibrational spectroscopy analysis demonstrated that the single amide bond in the formed structures had properties similar to the amide bonds in amyloidogenic proteins and peptides. The usefulness of the peptide as a template for inorganic material fabrication was demonstrated by self-assembly of the peptide nanostructures as a degradable cast to mold silver nanowires (122). Ionic silver was chemically reduced in the lumen of the nanotubes, followed by enzymatic degradation of the peptide, resulting in high-aspect ratio 20-nm silver wires (122). It was also demonstrated that similar nanotubes could be made with the self-assembly of a dipeptide made of the enantiomeric D-isomer amino acid building blocks.
Either solvent switch or temperature switch techniques are first applied to obtain the peptide in its monomeric form, followed by a remarkably efficient self-organization process to form ordered assemblies. Later studies revealed that under similar conditions the simpler nonnatural diphenylglycine analog forms spherical structures in aqueous solution with similarly remarkable efficiency and uniformity (123).
5.1. Formation of Amyloid-Like Assemblies by a Protected Dipeptide
Later studies on diphenylalanine analogs allowed the identification of an even smaller peptide, actually the simplest, that could form typical amyloid fibrils. It was found that acetylation of the N terminus and amidation of the C terminus of the diphenylalanine peptide result in the formation of fibrillary rather than tubular assemblies (124). In fact, the N- and C-capped analogs may represent the genuine recognition processes taking place within the polypeptide chains as the charge, present in the zwitterionic state of the peptide, does not play a role in the assembly process in the context of the polypeptide. Thus, Ac-Phe-Phe-NH2 is most likely the simplest amyloidogenic peptide element identified so far (124), which is consistent with the aromatic model that was hypothetically suggested (115) and computationally confirmed (121).
Other studies on the N-capped Boc-diphenylalanine peptide revealed the ability of this building block to form structures that undergo phase separation and its ability to evolve into diverse forms by a mechanism consistent with Ostwald’s rule of stages (125). It is therefore important to note that even very simple peptide sequences possess all the chemical information required to facilitate their organization into higher-order dynamic structures. Finally, modification of the peptide’s N terminus with the fluorenylmethyloxycarbonyl (Fmoc) protecting group resulted in its assembly into an ordered and rigid macroscopic hydrogel with nanoscale order (126, 127). Numerous research groups are currently exploring these hydrogels for various technological applications.
5.2. Physical Properties of the Peptide Assemblies
Careful characterization of the diphenylalanine structures revealed very unique physical properties. The tubular peptide assembly shows remarkable rigidity with Young’s modulus of 19–27 GPa as determined by point indentation and bending beam analysis (128, 129). The spherical assemblies have an even higher metallic-like stiffness with Young’s modulus of up to 300 GPa, as revealed by point indentation measurements (130). The assemblies were also found to act as semiconductors with a bandgap of several eV (131–133). The small size of the peptide building blocks actually allows the use of the quantum chemistry approach for the calculation of the bandgap and density function theory analysis for calculation of the bandgaps using first principles (133). Density function theory also helped explore the basis for the remarkable rigidity as was observed in peptide assemblies (134, 135). Further studies also demonstrated piezoelectricity comparable to lithium niobate, the best inorganic piezoelectric material (136). Moreover, the assemblies have clear pyroelectric properties, and electricity could be produced upon exposure of the nanostructures to heat (137). These observations promoted the field of peptide nanotechnology into a realm that was previously available only for inorganic or carbon-based materials, such as graphene, carbon nanotubes, and fullerenes (138).
The peptide assemblies also show clear luminescence at the visible range of the electromagnetic spectrum (139), remarkably similar to protein amyloids as described above in Section 3 (103). These features further demonstrate the structural and functional similarities between the dipeptide nanostructures and amyloid fibrils.
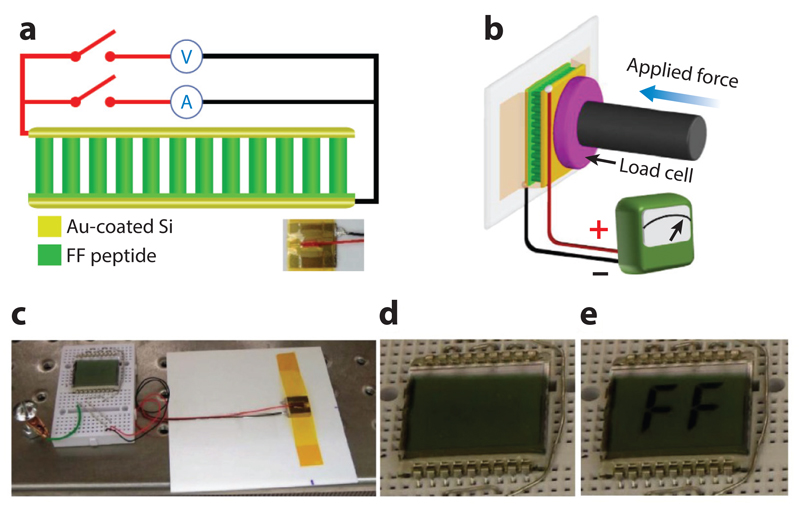
The use of diphenylalanine for various technological applications has been repeatedly demonstrated (reviewed in 138). Such applications include providing energy for the locomotion of a nanoboat (140), production of electricity using a piezoelectric array of peptide nanorods (141) (Figure 2), coating of medical devices (142), and fabrication of a thermometer that measures low physiological temperatures (143) and those near absolute zero (144). For a list of some examples of technological applications and their references see Table 1.
Figure 2.
The fabrication of a power-generating device based on a diphenylalanine array. (a) Schematic of the diphenylalanine peptide-based generator connected to the measurement equipment. Bottom-right inset: photograph of a real device. The functional device was produced by Professor Rusen Yang from the University of Minnesota and coworkers. (b) Schematic of the measurement setup in which a linear motor pushes with controlled forces on the top electrode in panel a. (c) Photograph of the generator as a direct power source for a liquid crystal display (LCD). (d,e) Photograph of the LCD before (d) and after (e) the generator in (c) was pressed by a human finger. Modified from Reference 141 under a Creative Commons CC-BY license.
Table 1. Technological applications of ultrashort peptide nanostructures.
| Physical property | Technological application | Reference(s) |
|---|---|---|
| Temperature-dependent luminescence | Thermal imaging and absolute temperature monitoring | 143, 144 |
| High quantum yield luminescence | Diagnostics of biological processes | 147 |
| Piezoelectricity | Energy harvesting devices | 136, 141 |
| Pyroelectricity | Energy harvesting devices | 137 |
| Capacitance | Supercapacitors | 145 |
| Chemical energy of assembly | Nanoboat locomotion | 140 |
| Electrochemical properties | Supersensitive sensor | 146 |
| Metal binding and drug loading | Drug-eluting stents | 142 |
| Light harvesting | Photosynthesis mimicking | 148 |
| Mechanical rigidity | Epoxy resin reinforcement | 149 |
6. Minimal Helical Motifs
Most of the work described above in Section 5 was based on β-sheet-rich protein amyloid assemblies and the organization of short peptides into a similar molecular organization. However, the other very common secondary structure of polypeptide chains, the α-helix, also plays a role in various structural proteins. The formation of these structures is based on the interactions between molecular interfaces in the organization of heptad repeats. Proteins comprising a coiled coil, one of the interesting helical motifs, contain several helical repeats that are arranged in tandem to mediate coordinated association into supramolecular structures.
Pioneering work of Woolfson & Ryadnov (150) and Woolfson (151, 152) allowed the use of peptide elements that are much shorter than the full-length coiled-coil forming proteins. Peptides composed of three to four natural amino acid heptad repeats were found stable enough to form supramolecular assemblies with structural and physical properties resembling the coiled structures formed by the natural proteins. These peptide assemblies were useful in various applications, including structural materials and the production of surfactants.
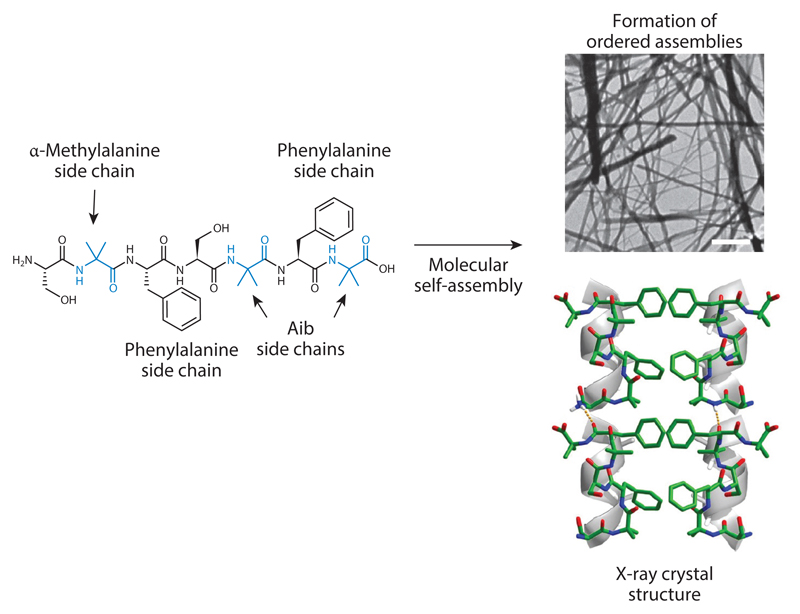
In spite of the usefulness of the short coiled-coil forming peptides, even shorter peptides that could still assemble into superhelical structures were required. The main limitation in a reductionist approach for exploring a peptide composed of the 20 coded amino acids is the inherent instability of short helical peptides. The solution for the limited stability of the short peptides was based on employing natural noncoded amino acids that are used for nonribosomal synthesis of short peptides, specifically the natural noncoded α-amino isobutyric acid (Aib) or α-methylalanine (Figure 3). This Cα-methylated amino acid is most likely one of the best helix inducer amino acids known in nature, as was established by Karle & Balaram (153). Aib is naturally used by microorganisms to stabilize short peptides, most notably in the structure of the channel-forming alamethicin peptide antibiotic produced by the Trichoderma viride fungus. In the context of peptide design to form ordered superhelical structures, it was established that the incorporation of Aib in a single heptad repeat allows the formation of superhelical assemblies that resemble the organization of the coiled coil (154–156). These assemblies were found to have functional properties, including the ability to act as a very efficient surfactant or to bind DNA molecules.
Figure 3.
Formation of superhelical supramolecular assemblies of a single heptad repeat with phenylalanines as interacting moieties stabilized by the incorporation of Aib, an α-methylated residue. The process of molecular self-assembly leads to the formation of ordered assemblies as observed using electron microscopy (scale bar is 100 nm). X-ray crystallography shows the organization into helical assemblies with the hydrophobic phenylalanines serving as interfaces between the helical elements. Modified from Reference 154 following author reuse guidelines.
7. Coassembly for Extension of the Structural Space
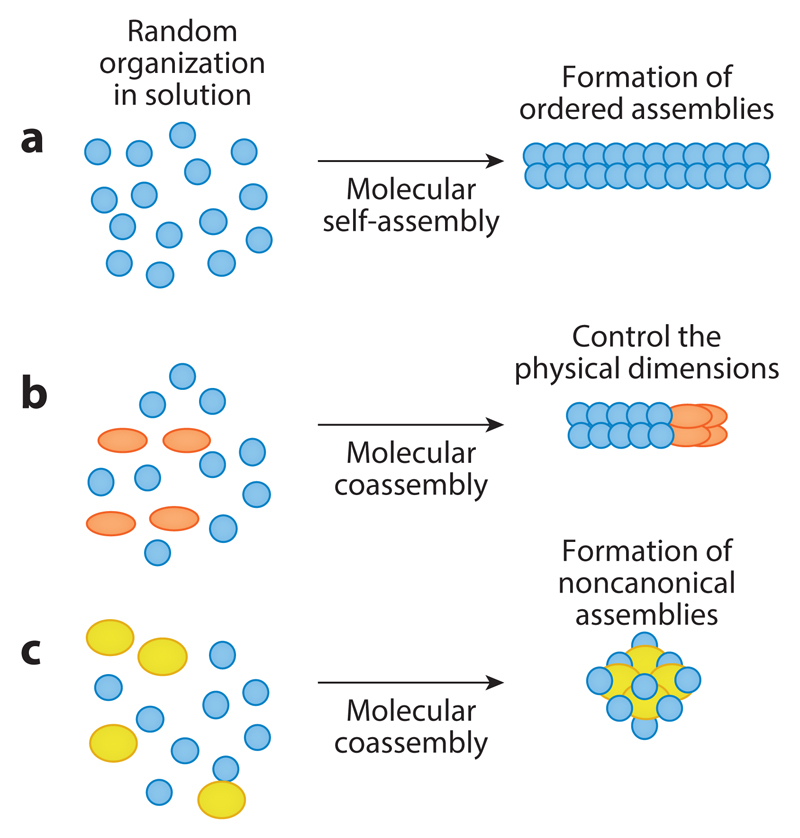
Molecular self-assembly is a very useful method for the formation of various nanoscale architectures, including nanotubes, nanospheres, nanoplates, and more. However, it also poses limitations for control of the physical dimensions (Figure 4b) of the assemblies and the formation of non-canonical morphologies (Figure 4c). Inspired by polymer chemistry and the extensive use of copolymers, it was demonstrated that the use of coassembly could allow manipulation of the self-assembly process (157–161), for example, by controlling the length and distribution of peptide nanotubes or by facilitating the formation of toroids (160).
Figure 4.
Coassembly as a strategy to tune dimensions and obtain new architectures. (a) Self-assembly of a single type of molecule forms elongated ordered assemblies. (b) The addition of a second species may control the assembly of the first type of molecule, resulting in structures of a more limited length. For example, the addition of Boc-diphenylalanine to diphenylalanine (as described in 161) led to the length of diphenylalanine nanotubes that were described in Section 5. (c) The addition of another type of molecular building block may lead to an alternative architecture. For example, the coassembly of diphenylalanine and triphenylalanine at a specific molar ratio leads to formation of noncanonical toroids (as described in 160).
The use of coassembly for the production of nanostructures is a relatively new innovation in peptide nanotechnology. The full potential of this approach still needs to be fully explored, and it will surely become an important research procedure in the years to come. Computational analysis using molecular dynamics has already been demonstrated as an effective tool in the design of such complex structures.
8. Extension of the Building Block Repertoire
8.1. Peptide Nucleic Acids
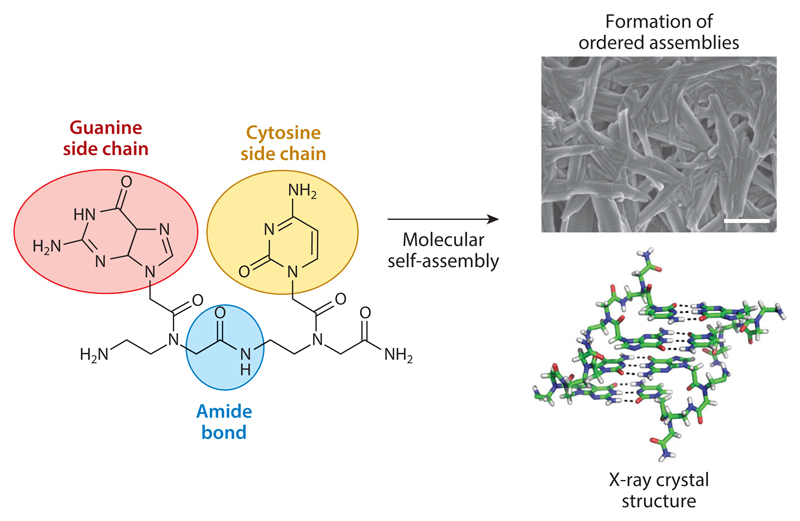
In parallel with the advancements in peptide and protein nanotechnology, significant work was performed on DNA nanotechnology. DNA has the advantage of intrinsic recognition between complementary bases that allows a precise organization of ordered assemblies. To take advantage of both peptide and DNA features, PNAs were studied because of their ability to form ordered nanoscale assemblies (35, 36). PNAs are DNA peptoid analogs that have a polyamide backbone and nucleobase side chains initially developed for the control of gene expression. When studied for their ability to form ordered structures at the nanoscale, it was revealed that guanine-containing PNA dipeptides could form well-ordered structures by molecular self-assembly (Figure 5). X-ray crystallography confirmed that indeed the structures consist of stacking interactions, such as a peptide and Watson-Crick hydrogen bonding between the bases as in DNA (Figure 5). Moreover, the PNA supramolecular assemblies show very interesting fluorescence properties, including the red-edge excitation shift. This is a lattice-related phenomenon that was previously reported in glassy materials and graphene oxide and has been observed in bioorganic self-assembled nanostructures.
Figure 5.
Formation of peptide nucleic acid (PNA)-based assemblies. The PNA building blocks contain an amide bond. This type of bond is present in proteins, peptides, and the synthetic polyamide polymers (such as Nylon® and Kevlar®), described in Section 2.1. However, the side chains in PNA consist of nucleobases as in DNA or RNA. The process of molecular self-assembly leads to the formation of ordered assemblies, observed using electron microscopy (scale bar is 10 μm). X-ray crystallography shows them in a stacked assembly similar to the dipeptide assemblies described in Section 5. However, in addition, there are Watson-Crick hydrogen bonds like those observed in nucleic acid structures. Modified from 34 following author reuse guidelines.
8.2. Assembly of Metabolites into Ordered Supramolecular Structures
A recent discovery that even a single phenylalanine could form amyloidal assemblies significantly extended the world of bioinspired nanotechnology (reviewed in 162). Although these are not peptide structures that share polypeptide organization and physicochemical properties, the assemblies did share the structural features and the dye binding specificities of protein- and peptide-amyloidal assemblies, and also revealed typical cytotoxicity, as observed for protein and peptide amyloids and fragment-based structures.
Follow-up studies demonstrated the ability of other amino acids to organize into clusters, similar to amyloidal oligomers, and their binding to membranes, another property that is observed for self-assembled amyloid structures (162). Although phenylalanine assemblies are clearly devoid of the amide bonds that occur in natural and synthetic polyamide, their organization into ordered structures remarkably resembles that of protein and peptide assemblies. The X-ray structure of the zwitterion form of phenylalanine clearly demonstrated a β-sheet-like organization stabilized by hydrogen bonding between layers of the amino acids and aromatic interactions between side chains. When a hypothetical polyphenylalanine strand, computationally confined to a β-strand structure, was overlaid on the amino acid crystal structure, a remarkable superposition was noted. Thus, it appears that even for the single amino acid organization, the structure is also very similar to that of the peptide assemblies, forming a type of supramolecular secondary structure resembling that of proteins and peptides.
Later studies demonstrated that not only amino acids but also nucleobases and other metabolites could form such typical amyloid-like fibrils (162). Thus, the formation of such layered β-sheet-like assemblies is most likely a generic property of a large variety of biological building blocks and is not limited only to the polyamides, proteins, and peptides. Although many of the current studies of metabolite amyloid formation are concerned with inborn errors of metabolism disorders in which the accumulation of various metabolites takes place, another line of research is devoted to the material properties of the metabolite assemblies. These include the ability of the metabolite to form peptide-like nanostructures and also the ability to form macroscopic gels with nanoscale order. Extensive investigations of this area are predicted for the future.
9. From Peptides Back to Proteins
The notable physical properties of the peptide assemblies raise the question of whether such properties could be observed in protein assemblies. The electric properties, including semiconductivity, piezoelectricity, and pyroelectricity, are intriguing. Piezoelectricity was previously reported in bones (163), but its physiological and pathological roles are not yet understood. It would be extremely interesting to test whether the electric properties of protein assemblies play a role in the biological world. One of the greatest innovations of the twentieth century was the use of inorganic semiconductors for the production of functional machines. It will be interesting to see whether proteins are naturally used in a similar way in biological systems.
10. Summary and Prospects
This review provides a detailed account of the ability of extremely short peptide sequences to form highly ordered and functional assemblies by the process of molecular recognition and self-organization. The formation of nano- and microstructures with unique physical properties by peptides, as short as dipeptides, clearly demonstrates the utility of the reductionist approach in the design and synthesis of technologically valuable peptide structures. Furthermore, the realization of the mechanism of assembly and the notable physical properties of the peptide structures are very useful for the development of diagnostic and therapeutic agents to treat amyloid-associated maladies.
As for the technological applications, peptides clearly represent a class of superior building blocks for the production of self-assembled materials of desirable and tunable physical and chemical properties. Even though the full potential of peptides is still not fully explored in the material world, in principle, the cost of peptides should be very low if a demand for them is formed. It should be emphasized that the notion of the inherent physical instability of proteins and peptides is incorrect. The polyamides that are being used by biological systems represent only a tiny part of the overall chemical space that could be obtained by the combinatorial arrangement of amino acids in a linear chain. When thermophilic organisms are studied, it is clear that functional proteins and peptides could be stable in temperatures over 100◦C. The use of synthetic polyamides in modern polymer science and the stability of such materials for tens of years provide vivid evidence of the usefulness of polyamide assemblies for technological applications and materials science.
The successful technological utilization of minimalistic peptide structures should drive production and common industrial applications. Important new directions include the introduction of building blocks that form superhelical assemblies, the use of PNA units, and the utilization of coassembly techniques. Finally, the ultimate reduction from very short peptides to small metabolites further extends the repertoire of building blocks for bioinspired self-assembly.
Future Issues.
In spite of vast advancements in the field, many questions remain to be resolved during the investigation of peptide-based nanotechnology in the coming years:
What is the minimal chemical information needed to achieve efficient self-assembly into ordered nanostructures?
What is the physiological significance of the semiconductivity of the assemblies? Does it have any role in the neurodegenerative processes observed in amyloid-related disorders?
Could the assemblies be integrated with biological systems for a better man-machine interface?
What is the minimal peptide length that allows the formation of functional helical assemblies?
What are the future uses of peptide and nucleic acid integration technologies? This is the biological process for the formation of highly complex machines such as the ribosome. Could peptide nucleic acid (PNA) provide the integration?
Are there other cases, in addition to melanin synthesis, in which amyloid assemblies play roles in natural biochemical synthesis pathways?
Did the formation of amyloid assemblies by very short peptides and the catalytic properties of the amyloids play roles in the origin of life? In 2005, this was hypothetically suggested (164), and it was very recently supported by remarkable experimental confirmation (165, 166).
Acknowledgments
The author thanks members of his group for many stimulating discussions and Dr. Sigal Rencus-Lazar for editing the wording of the manuscript.
Footnotes
Disclosure Statement
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Lehn JM. Perspectives in supramolecular chemistry—From molecular recognition towards molecular information processing and self-organization. Angew Chem Int Ed Engl. 1990;29:1304–19. [Google Scholar]
- 2.Whitesides GM, Mathias J, Seto C. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science. 1991;254:1312–19. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 3.Philp D, Stoddart JF. Self-assembly in natural and unnatural systems. Angew Chem Int Ed Engl. 1996;35:1154–96. [Google Scholar]
- 4.Stupp SI, LeBonheur V, Walker K, Li LS, Huggins KE, et al. Supramolecular materials: self-organized nanostructures. Science. 1997;276:384–89. doi: 10.1126/science.276.5311.384. [DOI] [PubMed] [Google Scholar]
- 5.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–21. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 6.Harada A, Kobayashi R, Takashima Y, Hashidzume A, Yamaguchi H. Macroscopic self-assembly through molecular recognition. Nat Chem. 2011;3:34–37. doi: 10.1038/nchem.893. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S. Emerging biological materials through molecular self-assembly. Biotechnol Adv. 2002;20:321–39. doi: 10.1016/s0734-9750(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21:1171–78. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 9.Stupp SI. Self-assembly and biomaterials. Nano Lett. 2010;10:4783–86. doi: 10.1021/nl103567y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habibi Y, Lucia LA, Rojas OJ. Cellulose nanocrystals: chemistry, self-assembly, and applications. Chem Rev. 2010;110:3479–500. doi: 10.1021/cr900339w. [DOI] [PubMed] [Google Scholar]
- 11.Stephanopoulos N, Ortony JH, Stupp SI. Self-assembly for the synthesis of functional biomaterials. Acta Mater. 2013;61:912–30. doi: 10.1016/j.actamat.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2:715–25. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez C, Arribart H, Guille MM. Biomimetism and bioinspiration as tools for the design of innovative materials and systems. Nat Mater. 2005;4:277–88. doi: 10.1038/nmat1339. [DOI] [PubMed] [Google Scholar]
- 14.Buehler MJ, Ackbarow T. Fracture mechanics of protein materials. Mater Today. 2007;10:46–58. [Google Scholar]
- 15.Steinert PM, Idler WW, Zimmerman SB. Self-assembly of bovine epidermal keratin filaments in vitro. J Mol Biol. 1976;108:547–67. doi: 10.1016/s0022-2836(76)80136-2. [DOI] [PubMed] [Google Scholar]
- 16.Qin G, Hu X, Cebe P, Kaplan DL. Mechanism of resilin elasticity. Nat Commun. 2012;3 doi: 10.1038/ncomms2004. 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keten S, Buehler MJ. Nanostructure and molecular mechanics of spider dragline silk protein assemblies. J R Soc Interface. 2010;7:1709–21. doi: 10.1098/rsif.2010.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. PNAS. 2006;103:12285–90. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy JG, Scheibel TR. Composite materials based on silk proteins. Prog Polym Sci. 2010;35:1093–115. [Google Scholar]
- 20.Hu X, Cebe P, Weiss AS, Omenetto F, Kaplan DL. Protein-based composite materials. Mater Today. 2012;15:208–15. [Google Scholar]
- 21.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–89. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 22.Silva NH, Vilela C, Marrucho IM, Freire CS, Neto CP, Silvestre AJ. Protein-based materials: from sources to innovative sustainable materials for biomedical applications. J Mater Chem B. 2014;2:3715–40. doi: 10.1039/c4tb00168k. [DOI] [PubMed] [Google Scholar]
- 23.DiMarco RL, Heilshorn SC. Multifunctional materials through modular protein engineering. Adv Mater. 2012;24:3923–40. doi: 10.1002/adma.201200051. [DOI] [PubMed] [Google Scholar]
- 24.Maretschek S, Greiner A, Kissel T. Electrospun biodegradable nanofiber nonwovens for controlled release of proteins. J Control Release. 2008;127:180–87. doi: 10.1016/j.jconrel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Wei G, Ma PX. Nanostructured biomaterials for regeneration. Adv Funct Mater. 2008;18:3568–82. doi: 10.1002/adfm.200800662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Marini DM, Hwang W, Santoso S. Design of nanostructured biological materials through self-assembly of peptides and proteins. Curr Opin Chem Biol. 2002;6:865–71. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 27.Bradbury EM, Elliott A. Infra-red spectra and chain arrangement in some polyamides, polypeptides and fibrous proteins. Polymer. 1963;4:47–59. [Google Scholar]
- 28.Tashiro K, Kobayashi M, Tadokoro H. Elastic moduli and molecular structures of several crystalline polymers, including aromatic polyamides. Macromolecules. 1977;10:413–20. [Google Scholar]
- 29.Jiang Y, Loos K. Enzymatic synthesis of biobased polyesters and polyamides. Polymers. 2016;8:243. doi: 10.3390/polym8070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Checco JW, Gellman SH. Targeting recognition surfaces on natural proteins with peptidic foldamers. Curr Opin Struct Biol. 2016;39:96–105. doi: 10.1016/j.sbi.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangloff N, Ulbricht J, Lorson T, Schlaad H, Luxenhofer R. Peptoids and polypeptoids at the frontier of supra- and macromolecular engineering. Chem Rev. 2016;116:1753–802. doi: 10.1021/acs.chemrev.5b00201. [DOI] [PubMed] [Google Scholar]
- 32.Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogs with an achiral peptide backbone. J Am Chem Soc. 1992;114:1895–97. [Google Scholar]
- 33.Gupta A, Bahal R, Gupta M, Glazer PM, Saltzman WM. Nanotechnology for delivery of peptide nucleic acids (PNAs) J Control Release. 2016;240:302–11. doi: 10.1016/j.jconrel.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger O, Adler-Abramovich L, Levy-Sakin M, Grunwald A, Liebes-Peer Y, et al. Light-emitting self-assembled peptide nucleic acids exhibit both stacking interactions and Watson-Crick base pairing. Nat Nanotechnol. 2015;10:353–60. doi: 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- 35.Berger O, Gazit E. Molecular self-assembly using peptide nucleic acids. Pept Sci. 2017;108:e22930. doi: 10.1002/bip.22930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gazit E. Self-assembled peptide nanostructures: the design of molecular building blocks and their technological utilization. Chem Soc Rev. 2007;36:1263–69. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- 37.Ulijn RV, Smith AM. Designing peptide based nanomaterials. Chem Soc Rev. 2008;37:664–75. doi: 10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 38.Williams BA, Lund K, Liu Y, Yan H, Chaput JC. Self-assembled peptide nanoarrays: an approach to studying protein-protein interactions. Angew Chem Int Ed Engl. 2007;119:3111–14. doi: 10.1002/anie.200603919. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Pan F, Xu H, Yaseen M, Shan H, et al. Molecular self-assembly and applications of designer peptide amphiphiles. Chem Soc Rev. 2010;39:3480–98. doi: 10.1039/b915923c. [DOI] [PubMed] [Google Scholar]
- 40.Hamley IW. Self-assembly of amphiphilic peptides. Soft Matter. 2011;7:4122–38. [Google Scholar]
- 41.Cui H, Webber MJ, Stupp SI. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Pept Sci. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briggs BD, Knecht MR. Nanotechnology meets biology: peptide-based methods for the fabrication of functional materials. J Phys Chem Lett. 2012;3:405–18. doi: 10.1021/jz2016473. [DOI] [PubMed] [Google Scholar]
- 43.Habibi N, Kamaly N, Memic A, Shafiee H. Self-assembled peptide-based nanostructures: smart nanomaterials toward targeted drug delivery. Nano Today. 2016;11:41–60. doi: 10.1016/j.nantod.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekiz MS, Cinar G, Khalily MA, Guler MO. Self-assembled peptide nanostructures for functional materials. Nanotechnology. 2016;27:402002. doi: 10.1088/0957-4484/27/40/402002. [DOI] [PubMed] [Google Scholar]
- 45.Yardeni JL, Amit M, Ashkenasy G, Ashkenasy N. Sequence dependent proton conduction in self-assembled peptide nanostructures. Nanoscale. 2016;8:2358–66. doi: 10.1039/c5nr06750b. [DOI] [PubMed] [Google Scholar]
- 46.Eskandari S, Guerin T, Toth I, Stephenson RJ. Recent advances in self-assembled peptides: implications for targeted drug delivery and vaccine engineering. Adv Drug Deliv Rev. 2017;110:169–87. doi: 10.1016/j.addr.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:2149–54. [Google Scholar]
- 48.Merrifield RB. Automated synthesis of peptides. Science. 1965;150:178–85. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- 49.Merrifield RB, Stewart JM, Jernberg N. Instrument for automated synthesis of peptides. Anal Chem. 1966;38:1905–14. doi: 10.1021/ac50155a057. [DOI] [PubMed] [Google Scholar]
- 50.Gausepohl H, Boulin C, Kraft M, Frank RW. Automated multiple peptide synthesis. Pept Res. 1992;5:315–20. [PubMed] [Google Scholar]
- 51.Mäde V, Els-Heindl S, Beck-Sickinger AG. Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J Org Chem. 2014;10:1197–212. doi: 10.3762/bjoc.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behrendt R, White P, Offer J. Advances in Fmoc solid-phase peptide synthesis. J Pept Sci. 2016;22:4–27. doi: 10.1002/psc.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mijalis AJ, Thomas DA, III, Simon MD, Adamo A, Beaumont R, et al. A fully automated flow-based approach for accelerated peptide synthesis. Nat Chem Biol. 2017;13:464–66. doi: 10.1038/nchembio.2318. [DOI] [PubMed] [Google Scholar]
- 54.Paul F, Auriol D, Monsan P. Direct enzymatic synthesis of aspartame. Ann NY Acad Sci. 1988;542:351–55. [Google Scholar]
- 55.Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol. 2001;12:59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 56.Cretich M, Damin F, Pirri G, Chiari M. Protein and peptide arrays: recent trends and new directions. Biomol Eng. 2006;23:77–88. doi: 10.1016/j.bioeng.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Schirwitz C, Loeffler FF, Felgenhauer T, Stadler V, Nesterov-Mueller A, et al. Purification of high-complexity peptide microarrays by spatially resolved array transfer to gold-coated membranes. Adv Mater. 2013;25:1598–602. doi: 10.1002/adma.201203853. [DOI] [PubMed] [Google Scholar]
- 58.Pai J, Hyun S, Hyun JY, Park SH, Kim WJ, et al. Screening of pre-miRNA-155 binding peptides for apoptosis inducing activity using peptide microarrays. J Am Chem Soc. 2016;138:857–67. doi: 10.1021/jacs.5b09216. [DOI] [PubMed] [Google Scholar]
- 59.Hansen CS, Østerbye T, Marcatili P, Lund O, Buus S, Nielsen M. ArrayPitope: automated analysis of amino acid substitutions for peptide microarray-based antibody epitope mapping. PLOS ONE. 2017;12:e0168453. doi: 10.1371/journal.pone.0168453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerr JM, Banville SC, Zuckermann RN. Encoded combinatorial peptide libraries containing non-natural amino acids. J Am Chem Soc. 1993;115:2529–31. [Google Scholar]
- 61.Hodgson DR, Sanderson JM. The synthesis of peptides and proteins containing non-natural amino acids. Chem Soc Rev. 2004;33:422–30. doi: 10.1039/b312953p. [DOI] [PubMed] [Google Scholar]
- 62.Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, et al. Structure-based design of non-natural amino acid inhibitors of amyloid fibrillation. Nature. 2011;475:96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuber P. Non-ribosomal peptide synthesis. Curr Opin Cell Biol. 1991;3:1046–50. doi: 10.1016/0955-0674(91)90127-k. [DOI] [PubMed] [Google Scholar]
- 64.Moutiez M, Schmitt E, Seguin J, Thai R, Favry E, et al. Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases. Nat Commun. 2014;5 doi: 10.1038/ncomms6141. 5141. [DOI] [PubMed] [Google Scholar]
- 65.Reimer JM, Aloise MN, Harrison PM, Schmeing TM. Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature. 2016;529:239–42. doi: 10.1038/nature16503. [DOI] [PubMed] [Google Scholar]
- 66.Waite JH, Tanzer ML. Polyphenolic substance of Mytilus edulis: novel adhesive containing L-Dopa and hydroxyproline. Science. 1981;212:1038–40. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- 67.Dalsin JL, Hu BH, Lee BP, Messersmith PB. Mussel adhesive protein mimetic polymers for the preparation of nonfouling surfaces. J Am Chem Soc. 2003;125:4253–58. doi: 10.1021/ja0284963. [DOI] [PubMed] [Google Scholar]
- 68.Lee BP, Dalsin JL, Messersmith PB. Synthesis and gelation of DOPA-modified poly(ethylene glycol) hydrogels. Biomacromolecules. 2002;3:1038–47. doi: 10.1021/bm025546n. [DOI] [PubMed] [Google Scholar]
- 69.Fichman G, Adler-Abramovich L, Manohar S, Mironi-Harpaz I, Guterman T, et al. Seamless metallic coating and surface adhesion of self-assembled bioinspired nanostructures based on di-(3,4-dihydroxy-l-phenylalanine) peptide motif. ACS Nano. 2014;8:7220–28. doi: 10.1021/nn502240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fichman G, Guterman T, Damron J, Adler-Abramovich L, Schmidt J, et al. Spontaneous structural transition and crystal formation in minimal supramolecular polymer model. Sci Adv. 2016;2:e1500827. doi: 10.1126/sciadv.1500827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prasad C. Bioactive cyclic dipeptides. Peptides. 1995;16:151–64. doi: 10.1016/0196-9781(94)00017-z. [DOI] [PubMed] [Google Scholar]
- 72.Fischer PM. Diketopiperazines in peptide and combinatorial chemistry. J Pept Sci. 2003;9:9–35. doi: 10.1002/psc.446. [DOI] [PubMed] [Google Scholar]
- 73.Martins MB, Carvalho I. Diketopiperazines: biological activity and synthesis. Tetrahedron. 2007;63:9923–32. [Google Scholar]
- 74.Borthwick AD, Da Costa NC. 2,5-Diketopiperazines in food and beverages: taste and bioactivity. Crit Rev Food Sci Nutr. 2017;57:718–42. doi: 10.1080/10408398.2014.911142. [DOI] [PubMed] [Google Scholar]
- 75.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. PNAS. 1993;90:3334–38. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature. 1993;366:324–27. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]
- 77.Ghadiri MR, Granja JR, Buehler LK. Artificial transmembrane ion channels from self-assembling peptide nanotubes. Nature. 1994;369:301–4. doi: 10.1038/369301a0. [DOI] [PubMed] [Google Scholar]
- 78.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, et al. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric β-sheet tapes. Nature. 1997;386:259–62. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 79.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–88. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 80.Vauthey S, Santoso S, Gong H, Watson N, Zhang S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. PNAS. 2002;99:5355–60. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banerjee IA, Yu L, Matsui H. Cu nanocrystal growth on peptide nanotubes by biomineralization: size control of Cu nanocrystals by tuning peptide conformation. PNAS. 2003;100:14678–82. doi: 10.1073/pnas.2433456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao X, Matsui H. Peptide-based nanotubes and their applications in bionanotechnology. Adv Mater. 2005;17:2037–50. doi: 10.1002/adma.200401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gazit E. Self-assembled peptide nanostructures: the design of molecular building blocks and their technological utilization. Chem Soc Rev. 2007;36:1263–69. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- 84.Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 85.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–32. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 86.Jahn TR, Radford SE. The Yin and Yang of protein folding. FEBS J. 2005;272:5962–70. doi: 10.1111/j.1742-4658.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 87.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 88.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–56. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 89.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 90.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–78. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adamcik J, Jung JM, Flakowski J, De Los Rios P, Dietler G, Mezzenga R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat Nanotechnol. 2010;5:423–28. doi: 10.1038/nnano.2010.59. [DOI] [PubMed] [Google Scholar]
- 92.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Amyloid fibril formation by an SH3 domain. PNAS. 1998;95:4224–28. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Litvinovich SV, Brew SA, Aota S, Akiyama SK, Haudenschild C, Ingham KC. Formation of amyloid-like fibrils by self-association of a partially unfolded fibronectin type III module. J Mol Biol. 1998;280:245–58. doi: 10.1006/jmbi.1998.1863. [DOI] [PubMed] [Google Scholar]
- 94.Chiti F, Webster P, Taddei N, Clark A, Stefani M, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. PNAS. 1999;96:3590–94. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fändrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165–66. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 96.Gazit E. The “correctly folded” state of proteins: Is it a metastable state? Angew Chem Int Ed Engl. 2002;41:257–59. doi: 10.1002/1521-3773(20020118)41:2<257::aid-anie257>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 97.Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–63. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 98.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–19. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 99.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 100.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 101.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–24. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 102.Knowles TP, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nat Nanotechnol. 2011;6:469–79. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 103.Pinotsi D, Buell AK, Dobson CM, Kaminski-Schierle GS, Kaminski CF. A label-free, quantitative assay of amyloid fibril growth based on intrinsic fluorescence. ChemBioChem. 2013;14:846–50. doi: 10.1002/cbic.201300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jarrett JT, Lansbury PT. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–58. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 105.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. PNAS. 1996;93:1125–29. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liang Y, Lynn DG, Berland KM. Direct observation of nucleation and growth in amyloid self-assembly. J Am Chem Soc. 2010;132:6306–8. doi: 10.1021/ja910964c. [DOI] [PubMed] [Google Scholar]
- 107.Knowles TP, White DA, Abate AR, Agresti JJ, Cohen SI, et al. Observation of spatial propagation of amyloid assembly from single nuclei. PNAS. 2011;108:14746–51. doi: 10.1073/pnas.1105555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gillam JE, MacPhee CE. Modelling amyloid fibril formation kinetics: mechanisms of nucleation and growth. J Phys Condens Matter. 2013;25 doi: 10.1088/0953-8984/25/37/373101. 373101. [DOI] [PubMed] [Google Scholar]
- 109.Tenidis K, Waldner M, Bernhagen J, Fischle W, Bergmann M, et al. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J Mol Biol. 2000;295:1055–71. doi: 10.1006/jmbi.1999.3422. [DOI] [PubMed] [Google Scholar]
- 110.Reches M, Porat Y, Gazit E. Amyloid fibril formation by pentapeptide and tetrapeptide fragments of human calcitonin. J Biol Chem. 2002;277:35475–80. doi: 10.1074/jbc.M206039200. [DOI] [PubMed] [Google Scholar]
- 111.Bertolani A, Pizzi A, Pirrie L, Gazzera L, Morra G, et al. Crystal structure of the DFNKF segment of human calcitonin unveils aromatic interactions between phenylalanines. Chem Eur J. 2017;23:2051–58. doi: 10.1002/chem.201604639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mazor Y, Gilead S, Benhar I, Gazit E. Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide. J Mol Biol. 2002;322:1013–24. doi: 10.1016/s0022-2836(02)00887-2. [DOI] [PubMed] [Google Scholar]
- 113.Tjernberg L, Hosia W, Bark N, Thyberg J, Johansson J. Charge attraction and β propensity are necessary for amyloid fibril formation from tetrapeptides. J Biol Chem. 2002;277:43243–46. doi: 10.1074/jbc.M205570200. [DOI] [PubMed] [Google Scholar]
- 114.Bellesia G, Shea JE. What determines the structure and stability of KFFE monomers, dimers, and protofibrils? Biophys J. 2009;96:875–86. doi: 10.1016/j.bpj.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gazit E. A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 116.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. PNAS. 2005;102:315–20. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jack E, Newsome M, Stockley PG, Radford SE, Middleton DA. The organization of aromatic side groups in an amyloid fibril probed by solid-state 2H and 19F NMR spectroscopy. J Am Chem Soc. 2006;128:8098–99. doi: 10.1021/ja0581898. [DOI] [PubMed] [Google Scholar]
- 118.Yang JH, Ho Y, Tzou DLM. A 13C solid-state NMR analysis of steroid compounds. Magn Reson Chem. 2008;46:718–25. doi: 10.1002/mrc.2235. [DOI] [PubMed] [Google Scholar]
- 119.Marshall KE, Morris KL, Charlton D, O’Reilly N, Lewis L, et al. Hydrophobic, aromatic, and electrostatic interactions play a central role in amyloid fibril formation and stability. Biochemistry. 2011;50:2061–71. doi: 10.1021/bi101936c. [DOI] [PubMed] [Google Scholar]
- 120.Genji M, Yano Y, Hoshino M, Matsuzaki K. Aromaticity of phenylalanine residues is essential for amyloid formation by Alzheimer’s amyloid β-peptide. Chem Pharm Bull. 2017;65:668–73. doi: 10.1248/cpb.c17-00203. [DOI] [PubMed] [Google Scholar]
- 121.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol. 2005;350:379–92. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 122.Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–27. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 123.Reches M, Gazit E. Formation of closed-cage nanostructures by self-assembly of aromatic dipeptides. Nano Lett. 2004;4:581–85. [Google Scholar]
- 124.Reches M, Gazit E. Self-assembly of peptide nanotubes and amyloid-like structures by charged-termini-capped diphenylalanine peptide analogues. Isr J Chem. 2005;45:363–71. [Google Scholar]
- 125.Levin A, Mason TO, Adler-Abramovich L, Buell AK, Meisl G, et al. Ostwald’s rule of stages governs structural transitions and morphology of dipeptide supramolecular polymers. Nat Commun. 2014;5 doi: 10.1038/ncomms6219. 5219. [DOI] [PubMed] [Google Scholar]
- 126.Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, et al. Nanostructured hydrogels for three-dimensional cell culture through self-assembly of fluorenylmethoxycarbonyl–dipeptides. Adv Mater. 2006;18:611–14. [Google Scholar]
- 127.Mahler A, Reches M, Rechter M, Cohen S, Gazit E. Rigid, self-assembled hydrogel composed of a modified aromatic dipeptide. Adv Mater. 2006;18:1365–70. [Google Scholar]
- 128.Kol N, Adler-Abramovich L, Barlam D, Shneck RZ, Gazit E, Rousso I. Self-assembled peptide nanotubes are uniquely rigid bioinspired supramolecular structures. Nano Lett. 2005;5:1343–46. doi: 10.1021/nl0505896. [DOI] [PubMed] [Google Scholar]
- 129.Niu L, Chen X, Allen S, Tendler SJ. Using the bending beam model to estimate the elasticity of diphenylalanine nanotubes. Langmuir. 2007;23:7443–46. doi: 10.1021/la7010106. [DOI] [PubMed] [Google Scholar]
- 130.Adler-Abramovich L, Kol N, Yanai I, Barlam D, Shneck RZ, et al. Self-assembled organic nanostructures with metallic-like stiffness. Angew Chem Int Ed Engl. 2010;49:9939–42. doi: 10.1002/anie.201002037. [DOI] [PubMed] [Google Scholar]
- 131.Amdursky N, Molotskii M, Gazit E, Rosenman G. Elementary building blocks of self-assembled peptide nanotubes. J Am Chem Soc. 2010;132:15632–36. doi: 10.1021/ja104373e. [DOI] [PubMed] [Google Scholar]
- 132.Hauser CA, Zhang S. Nanotechnology: peptides as biological semiconductors. Nature. 2010;468:516–17. doi: 10.1038/468516a. [DOI] [PubMed] [Google Scholar]
- 133.Akdim B, Pachter R, Naik RR. Self-assembled peptide nanotubes as electronic materials: an evaluation from first-principles calculations. Appl Phys Lett. 2015;106 183707. [Google Scholar]
- 134.Azuri I, Adler-Abramovich L, Gazit E, Hod O, Kronik L. Why are diphenylalanine-based peptide nanostructures so rigid? Insights from first principles calculations. J Am Chem Soc. 2014;136:963–69. doi: 10.1021/ja408713x. [DOI] [PubMed] [Google Scholar]
- 135.Zelenovskiy P, Kornev I, Vasilev S, Kholkin A. On the origin of the great rigidity of self-assembled diphenylalanine nanotubes. Phys Chem Chem Phys. 2016;18:29681–85. doi: 10.1039/c6cp04337b. [DOI] [PubMed] [Google Scholar]
- 136.Vasilev S, Zelenovskiy P, Vasileva D, Nuraeva A, Shur VY, Kholkin AL. Piezoelectric properties of diphenylalanine microtubes prepared from the solution. J Phys Chem Solids. 2016;93:68–72. [Google Scholar]
- 137.Esin A, Baturin I, Nikitin T, Vasilev S, Salehli F, et al. Pyroelectric effect and polarization instability in self-assembled diphenylalanine microtubes. Appl Phys Lett. 2016;109 142902. [Google Scholar]
- 138.Yan X, Zhu P, Li J. Self-assembly and application of diphenylalanine-based nanostructures. Chem Soc Rev. 2010;39:1877–90. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- 139.Amdursky N, Molotskii M, Aronov D, Adler-Abramovich L, Gazit E, Rosenman G. Blue luminescence based on quantum confinement at peptide nanotubes. Nano Lett. 2009;9:3111–15. doi: 10.1021/nl9008265. [DOI] [PubMed] [Google Scholar]
- 140.Ikezoe Y, Washino G, Uemura T, Kitagawa S, Matsui H. Autonomous motors of a metal-organic framework powered by reorganization of self-assembled peptides at interfaces. Nat Mater. 2012;11:1081–85. doi: 10.1038/nmat3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen V, Zhu R, Jenkins K, Yang R. Self-assembly of diphenylalanine peptide with controlled polarization for power generation. Nat Commun. 2016;7 doi: 10.1038/ncomms13566. 13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zohrabi T, Habibi N, Zarrabi A, Fanaei M, Lee LY. Diphenylalanine peptide nanotubes self-assembled on functionalized metal surfaces for potential application in drug-eluting stent. J Biomed Mater Res A. 2016;104:2280–90. doi: 10.1002/jbm.a.35764. [DOI] [PubMed] [Google Scholar]
- 143.Gan Z, Wu X, Zhang J, Zhu X, Chu PK. In situ thermal imaging and absolute temperature monitoring by luminescent diphenylalanine nanotubes. Biomacromolecules. 2013;14:2112–16. doi: 10.1021/bm400562c. [DOI] [PubMed] [Google Scholar]
- 144.Nikitin T, Kopyl S, Shur VY, Kopelevich YV, Kholkin AL. Low-temperature photoluminescence in self-assembled diphenylalanine microtubes. Phys Lett A. 2016;380:1658–62. [Google Scholar]
- 145.Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, et al. Self-assembled arrays of peptide nanotubes by vapour deposition. Nat Nanotechnol. 2009;4:849–54. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- 146.Yemini M, Reches M, Rishpon J, Gazit E. Novel electrochemical biosensing platform using self-assembled peptide nanotubes. Nano Lett. 2005;5:183–86. doi: 10.1021/nl0484189. [DOI] [PubMed] [Google Scholar]
- 147.Fan Z, Sun L, Huang Y, Wang Y, Zhang M. Bioinspired fluorescent dipeptide nanoparticles for targeted cancer cell imaging and real-time monitoring of drug release. Nat Nanotechol. 2016;11:388–94. doi: 10.1038/nnano.2015.312. [DOI] [PubMed] [Google Scholar]
- 148.Kim JH, Lee M, Lee JS, Park CB. Self-assembled light-harvesting peptide nanotubes for mimicking natural photosynthesis. Angew Chem Int Ed Engl. 2012;51:517–20. doi: 10.1002/anie.201103244. [DOI] [PubMed] [Google Scholar]
- 149.Even N, Adler-Abramovich L, Buzhansky L, Dodiuk H, Gazit E. Improvement of the mechanical properties of epoxy by peptide nanotube fillers. Small. 2011;7:1007–11. doi: 10.1002/smll.201001940. [DOI] [PubMed] [Google Scholar]
- 150.Woolfson DN, Ryadnov MG. Peptide-based fibrous biomaterials: some things old, new and borrowed. Curr Opin Chem Biol. 2006;10:559–67. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 151.Woolfson DN. Building fibrous biomaterials from α-helical and collagen-like coiled-coil peptides. Pept Sci. 2010;94:118–27. doi: 10.1002/bip.21345. [DOI] [PubMed] [Google Scholar]
- 152.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 153.Karle IL, Balaram P. Structural characteristics of α-helical peptide molecules containing Aib residues. Biochemistry. 1990;29:6747–56. doi: 10.1021/bi00481a001. [DOI] [PubMed] [Google Scholar]
- 154.Mondal S, Adler-Abramovich L, Lampel A, Bram Y, Lipstman S, Gazit E. Formation of functional super-helical assemblies by constrained single heptad repeat. Nat Commun. 2015;6 doi: 10.1038/ncomms9615. 8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mondal S, Gazit E. The self-assembly of helical peptide building blocks. ChemNanoMat. 2016;2:323–32. [Google Scholar]
- 156.Mondal S, Varenik M, Bloch DN, Atsmon-Raz Y, Jacoby G, et al. A minimal length rigid helical peptide motif allows rational design of modular surfactants. Nat Commun. 2017;8 doi: 10.1038/ncomms14018. 14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh P, Brar SK, Bajaj M, Narang N, Mithu VS, et al. Self-assembly of aromatic α-amino acids into amyloid inspired nano/micro scaled architects. Mater Sci Eng C. 2017;72:590–600. doi: 10.1016/j.msec.2016.11.117. [DOI] [PubMed] [Google Scholar]
- 158.Hnilova M, So CR, Oren EE, Wilson BR, Kacar T, et al. Peptide-directed co-assembly of nanoprobes on multimaterial patterned solid surfaces. Soft Matter. 2012;8:4327–34. [Google Scholar]
- 159.Maity S, Nir S, Reches M. Co-assembly of aromatic dipeptides into spherical structures that are similar in morphology to red and white blood cells. J Mater Chem B. 2014;2:2583–91. doi: 10.1039/c3tb21456g. [DOI] [PubMed] [Google Scholar]
- 160.Guo C, Arnon ZA, Qi R, Zhang Q, Adler-Abramovich L, et al. Expanding the nanoarchitectural diversity through aromatic di-and tri-peptide coassembly: nanostructures and molecular mechanisms. ACS Nano. 2016;10:8316–24. doi: 10.1021/acsnano.6b02739. [DOI] [PubMed] [Google Scholar]
- 161.Adler-Abramovich L, Marco P, Amon ZA, Creasey RC, Michaels TC, et al. Controlling the physical dimensions of peptide nanotubes by supramolecular polymer coassembly. ACS Nano. 2016;10:7436–42. doi: 10.1021/acsnano.6b01587. [DOI] [PubMed] [Google Scholar]
- 162.Gazit E. Metabolite amyloids: a new paradigm for inborn error of metabolism disorders. J Inherit Metab Dis. 2016;39:483–88. doi: 10.1007/s10545-016-9946-9. [DOI] [PubMed] [Google Scholar]
- 163.Fukada E, Yasuda I. On the piezoelectric effect of bone. J Phys Soc Jpn. 1957;12:1158–62. [Google Scholar]
- 164.Carny O, Gazit E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005;19:1051–55. doi: 10.1096/fj.04-3256hyp. [DOI] [PubMed] [Google Scholar]
- 165.Nanda J, Rubinov B, Ivnitski D, Mukherjee R, Shtelman E, et al. Emergence of native peptide sequences in prebiotic replication networks. Nat Commun. 2017;8:434. doi: 10.1038/s41467-017-00463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rout SK, Friedmann MP, Riek R, Greenwald J. A prebiotic template-directed peptide synthesis based on amyloids. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02742-3. 234. [DOI] [PMC free article] [PubMed] [Google Scholar]