Abstract
Many recent studies have now shown that even under healthy conditions, the bladder and urinary tract harbors its own microbial community, collectively known as the urinary microbiota. This contradicts the long held notion that urine is a sterile environment in the absence of an acute infection. Given this relatively new discovery, many basic questions which are critical for our understanding of the role that the urinary microbiota plays in human health and disease remain unanswered. As this is an emerging area of study, optimized techniques and protocols to identify microorganisms in the urinary tract are still being established. This is made more challenging for the urinary microbiota given its low microbial biomass. A clear understanding of the unique technical considerations of low microbial biomass samples, as well the impact of key elements of experimental design and computational analysis on downstream interpretation will improve the interpretability and comparability of results across methods and studies both for the urinary microbiota as well as other sites of low microbial abundance.
Introduction
The bladder and lower urinary tract have traditionally been considered a sterile environment where the presence of bacteria, revealed by culturing, is indicative of an infection. However, recent studies indicate that urine is actually not free of bacteria under healthy conditions1–8. These studies have sampled catheter-collected and clean-catch midstream urine as a reflection of the bladder and genitourinary microbiota (bacteria and other microbes) respectively and have identified a number of bacteria that are not detected by clinical microbiology but that can be detected using 16S rRNA gene sequencing2,5 and expanded culture techniques3,4,9. Understanding the normal microbial composition of the bladder and lower urinary tract in healthy individuals is essential so that microbial changes associated with bladder disorders such as overactive bladder syndrome, urgency urinary incontinence, interstitial cystitis, and recurrent urinary tract infections can be recognized.
In other regions of the body, resident microbes play a key role in maintaining homeostasis. Modulating these microbial communities through probiotics, prebiotics, or microbiota transplants has been explored as a therapeutic strategy for a variety of human diseases10. Likewise, mapping the healthy urinary microbiota and identifying deviations from normal may radically transform how we treat women and men with bladder disorders11,12. For example, literature from gut microbiota studies suggest that overuse, prolonged use, or incorrect use of antibiotics can alter microbial composition. This predisposes the host to colonization by pathogenic bacteria such a clostridium difficile13–15. Although a similar level of evidence does not yet exist in the genitourinary microbiota literature, a similar phenomenon may occur and explain why treatment with antibiotics for asymptomatic bacteriuria significantly increases the risk of recurrent UTI in young women16. It is possible that treatment for acute as well as recurrent UTI with oral and parenteral antibiotics may reduce the presence of beneficial bacteria that combat pathogenic ones. An understanding of the normal urinary microbiota may change our treatment approach, such that we focus more on rebuilding a more resilient microbiota that decreases host susceptibility to pathologic bacteria rather than antibiotic driven destruction of pathologic bacteria along with collateral damage to normal microbiota.
However, because this is a newly emerging field, optimal techniques to identify the microorganisms in the urinary tract and other sites of microbial scarcity that were previously thought to be sterile such as the lung and placenta, are not yet established. In contrast to other human body sites such as the gut and vagina, the urinary tract appears to harbor a relatively small amount of bacteria (less than 105 colony forming units [CFU] per milliliter compared to 1012 CFU that is typically found in gut microbiome samples)3,4. Thus, techniques for sample collection and data analysis need to account for issues that may arise due to the low microbial biomass environment. A consistent and high yield analysis pipeline is needed to help design studies appropriately and optimize the quality of our biological conclusions.
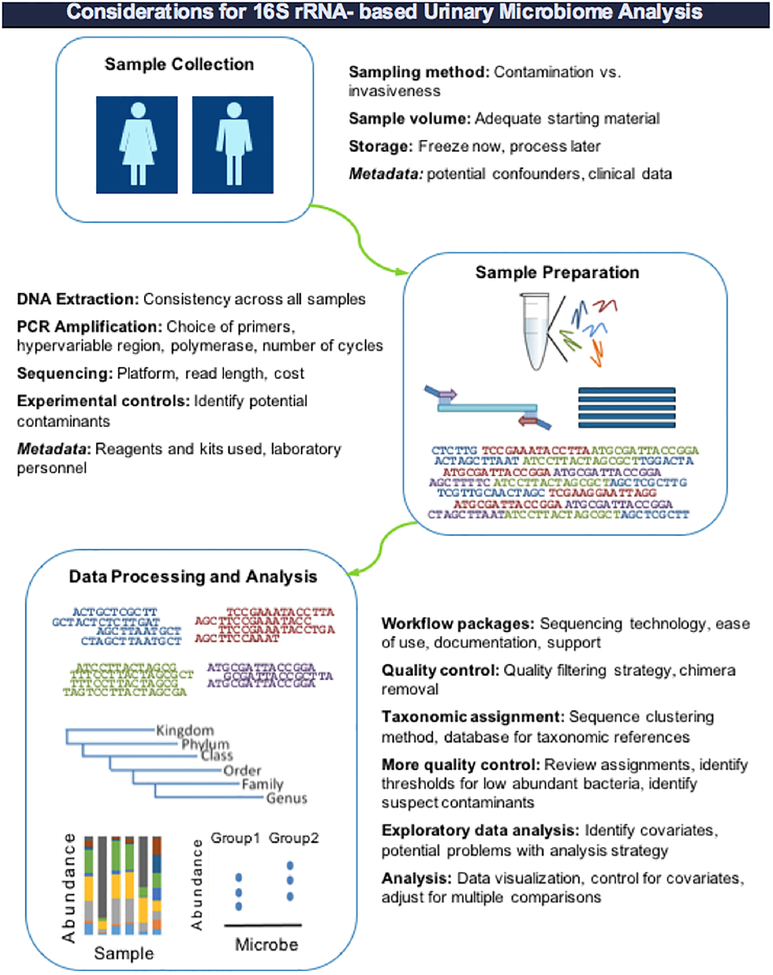
The purpose of this review is to give an overview of the methodology currently used to study the human microbiota and their genes (microbiome) in general, and to discuss elements of experimental design and data analysis that can have a large impact on studies of the urinary microbiota in particular (Figure 1). This information will be useful for researchers seeking to study the urinary microbiota as well as other sites of low microbial abundance. Our review is divided into four main sections: 1) considerations for sample collection, 2) considerations for laboratory methods to identify microbial communities, 3) sources of contamination and approaches to control for them, and 4) computational considerations for data processing and analysis.
Figure 1.
General overview of the workflow for 16S rRNA gene-based analysis of microbial communities, with specific considerations for urinary microbiome studies highlighted.
1. Considerations for sample collection
Bacteria isolated from urine are considered to be a reflection of the microbiota that resides in the bladder and lower urinary tract. While urine is relatively easy to collect, careful considerations must be made in order to minimize contamination of samples and maximize the microbial information that can be extracted from these samples. In addition, to minimize unintended variability, efforts should be made to collect specimens in a standardized manner throughout a study. Once collected, samples should be stored and transported under bacteriostatic conditions. Indeed, sample handling conditions are likely to profoundly impact the composition of a microbial community from its normal human environment. For example, oxygen exposure may promote the propagation of aerobes but induce cell death among strict anaerobes within minutes. Thus, variables such as temperature, pH and oxygen tension are all factors that should be controlled and standardized throughout sample collection and processing. To minimize contamination (discussed further below), samples should ideally be processed in a biosafety cabinet after specimen collection.
Specimen collection method.
When designing an experiment, the investigator should carefully weigh the benefits and drawbacks of each sampling method and determine if the sampling method chosen will appropriately address the scientific question. Methods for collecting urine specimens directly from the bladder include suprapubic aspirate and transurethral catheter. To investigate the genitourinary microbiota arising from the lower urinary tract and surrounding genital areas, midstream clean-catch voided specimens can be used.
Suprapubic aspirates, where a needle is used to puncture the skin and abdomen to aspirate urine from the bladder directly, provides the cleanest method of urine collection with minimal contamination1. Risks associated with this collection method include pain associated with the use of a needle, and the possibility of traversing intestinal contents. Thus, collecting urine in this way for research or clinical purposes may not be practical due to the invasiveness and risks of the procedure.
Collecting urine using a transurethral catheter is less invasive, and the bacterial communities identified using this sampling method have been shown to be consistent with samples collected by suprapubic aspirate in women1. Risks associated with this collection method include urethral irritation as well as discomfort during and following catheterization. There is also a small risk of acquiring a urinary tract infection. Despite these risks, this approach is generally well tolerated with minimal side effects in the research setting.
Obtaining a clean-catch midstream urine specimen is the least invasive and the easiest way to obtain urine from participants; however, it only allows characterization of the genitourinary microbiota including those found in the lower urinary tract and genital regions, not explicitly the bladder. Due to anatomical differences, urine collection from men and women generates different issues that must be considered to minimize contamination. One challenge with collecting midstream urine specimens from women is potential contamination from the surrounding vulvovaginal region, which has its own microbial environment. Wolfe et al. demonstrated that urine collected by midstream clean catch can be contaminated by vulvovaginal microbiota1. The bacterial profile of each individual’s clean-catch urine specimen was more similar to that individual’s vaginal bacterial profile than to the urine specimen profile collected from the same individual by suprapubic aspirate. Other approaches have been used to minimize vulvovaginal contamination of midstream urine samples from female participants. For example, Siddiqui et al. collected urine from women using clean-catch with labial separation under the supervision of a trained nurse in order to minimize contamination6,8. However, these studies did not compare the urinary microbiota profiles obtained to vulvar, vaginal, or catheter-collected specimens, so the success of this technique at minimizing vulvovaginal microbiome contamination is unknown. In general, obtaining a clean-catch midstream urine specimen that is free of microbiota from the vulvovaginal area is difficult. It may be possible to identify and computationally remove the signal from the vulva and/or vaginal microbiome prior to downstream analysis, but currently no standard analytical approach for this exists. If an approach were successfully developed, voided urine would provide a means to identify bacteria from the lower urinary tract with minimal contribution from surrounding tissue.
In men, there is a decreased risk of contamination from surrounding tissue. To date studies of the male urinary microbiota have primarily been from clean catch voided specimens17–19. However, it is currently unknown whether the male urethra has a microbial environment different from that of the urinary bladder. The male urinary microbiota has been found to be distinct from the microbiota inhabiting the coronal sulcus19, but similar to the microbiota identified from urethral swabs17,18. It is likely that, like women, the bladder of men has a distinct microbiome and future studies should be design to determine if the male bladder microbiome contributes to lower urinary tract disorders in men.
Sample volume.
Another consideration when collecting samples is the amount of urine needed to extract bacterial DNA from urine. One key factor is the use of voided versus catheter collected urine, since catheter collected urine will contain less amount of bacteria. Studies have had high success rates (>85%) of identifying bacteria from voided urine using 1–2mL20. However, studies investigating the bladder microbiome with catheter collected urine were only able to extract bacterial DNA from about half of the samples collected using 1–2 mL of urine.1,5,21 By increasing the amount of urine collected to at 30 – 50 mL, Karstens et al. have reported successful extraction of bacterial DNA from almost all (95%) samples with catheter-collected urine22. Thus, since urine has a low microbial biomass, there is likely more success when extracting and amplifying bacterial DNA from larger sample volumes. The ideal urine volume needed to achieve high yield is yet to be determined, but 30 – 50 mL has proven to be successful for studies of the bladder22 and genitourinary6,8 microbiota.
Specimen storage.
Specimens for sequencing should be frozen as soon as possible. Freezing samples upon collection allows for subsequent parallel processing of multiple samples, minimizing untraceable batch effects that can occur during sample processing. As of now, there is no published study examining the effect of sample handling and storage conditions on the downstream analysis of the urinary microbiota. Thus, no definitive guidelines currently exist. Extensive research regarding the influence of storage conditions on the microbial composition from other body sites, such as the vagina, skin, and gut, has been conducted. The effect of storage at −20C and −80C has been found to have little impact on microbial composition for these samples23,24. If immediate freezing and storage is not possible, freezing delay (e.g. storage either at room temperature or 4C) can introduce variation in sample composition. In other microbiota sites, these alterations have been minimized by use of different buffers25. However, it is currently unknown if these findings translate to urine specimens.
Sample metadata.
The human microbiome is dynamic and highly influenced by its host environment. In the gut, dietary changes26, aging27, and lifestyle28 amongst other factors have been found to affect microbial composition. In the vagina, sexual activity and the menstrual cycle have been associated with changes in the composition of the vaginal microbiome29. Such variations in the urinary microbiome are currently under-studied, but compositional differences that covary with age and sex have been identified in the genitourinary microbiome7. Therefore, information about study participants should include such parameters as age, sex, body mass index, menopausal status and medical history, in addition to information and questionnaires about the clinical condition being examined. Additional information such as dietary preferences, over the counter and prescription drug use including estrogen products or antibiotics, sexual activity, and use of genital wipes or lubricants should also be collected as they may influence the composition or function of the urinary microbiota. Additionally, detailed information about the sample collection such as time of day and study staff involved with sample collection and sample handling should be recorded. These additional metadata can be used for exploratory data analysis to identify unanticipated relationships with the composition of the microbiota (i.e. batch effects), which may need to be controlled for in the analyses.
2. Methods to identify microbial communities
Strategies for identifying microbes.
Once samples have been collected, there are a variety of complementary methods currently used to identify the microbial composition of a sample. In addition to conventional culture-based methods, there are DNA sequencing-based methods. Whole-genome shotgun sequencing (metagenomics) aims to sequence all of the bacterial genes in a given sample, while marker gene sequencing aims to sequence a specific gene region. Common marker genes are the 16S ribosomal RNA (rRNA) gene that is specific to bacteria and archaea; the 18S rRNA gene for eukaryotes; and the internal transcribed spacer (ITS) for fungi. Additional methods for investigating microbiota include expanded quantitative urine culture (EQUC), which uses a variety of media and incubation conditions to grow bacteria that do not grow under standard culturing conditions4,9,30; and quantitative PCR, which provides a targeted approach to quantify specific bacteria31. While certainly useful, these latter two methods may require a priori knowledge of physiological or DNA sequence properties of constituent bacteria and hence are inherently selective. The remainder of this review will focus on the major steps involved in microbial community profiling by 16S rRNA sequencing, since this technique is less intrinsically biased and currently the most widely adopted method for studying complex microbial communities.
The 16S rRNA gene is a highly conserved gene that exists in all bacteria and archaea, and has been used to study bacterial phylogeny since the late 1970’s32,33. These studies led to a major shift in how scientists classify microorganisms from phenotypic classification (e.g. Gram-positive bacteria) to taxonomic classification into various kingdoms, phyla, classes, orders, families, genera, and species. The 16S rRNA gene is approximately 1,550 base pairs long and is composed of multiple hypervariable regions that vary in sequence between species and that are flanked by highly conserved regions. The conserved regions allow the 16S rRNA gene to be targeted by primers for broad PCR amplification, while sequence variation in the targeted hypervariable region allows for taxonomic identification of the bacteria present in a sample. Importantly since the degree of variation of samples acts as an evolutionary chronometer, their (dis)similarity also reveals the relatedness of particular taxa, which also may give insight to shared biological properties. Most 16S rRNA gene sequencing studies target a specific hypervariable region or set of adjacent hypervariable regions, which typically allow for taxonomic identification down to the genus level. Because 16S rRNA sequencing cannot discriminate DNA from viable or non-viable bacteria, it is worth noting that most microbiota analyses represent a snapshot of both the living and the dead microbiota.
DNA extraction.
DNA extraction from bacteria involves two major steps of cell lysis and DNA purification. In the first step, the cell membrane is chemically lysed using detergents and surfactants. Heat and/or mechanical lysis (e.g. bead beating) can expedite this process and is often used for more complex or fibrous sample types such as stool. In addition, proteases and RNAases can be used to breakdown proteins and RNA respectively to improve DNA yield and limit non-DNA contamination. In the second step, DNA is purified from cellular debris, salts, proteins, and other remaining contaminants in addition to reagents used for cell lysis. DNA purification techniques include: ethanol/isopropanol precipitation; phenol-chloroform extraction, in which aqueous phase DNA is separated from phenol-denatured proteins that segregate to a distinct organic phase; or DNA adsorption to a solid phase (e.g. silica) minicolumn, in which DNA binding is modified by pH and salt concentration.
The DNA extraction technique chosen will significantly affect how faithfully the bacterial composition of the original sample is represented by the DNA extracted from it. Some bacteria, such as Gram-positive bacteria and Mycobacteria, may be more difficult to lyse than others in a microbial community, and hence become underrepresented34. On the other hand, if an extraction method is too harsh, the DNA from the easily lysed species may become sheared. Currently, there is no standard technique that works equally well for lysing all bacteria in a given sample35, though methods involving bead beating and/or mutanolysin are recommended to effectively lyse cells34,36. Future studies should consider comparing different DNA extraction methods to identify optimal methods for DNA extraction from urine specimens, as has been performed for a variety of samples with high and low microbial biomass such as feces37, cervicovaginal lavage samples,38 and marine biofilms39.
While there have not been comprehensive studies investigating optimal methods for DNA extraction from urine, there are some common practices amongst urinary microbiota researchers. In common with other high volume, low microbial biomass samples such as air particulates40, sea water or glacial ice41, microbes within a urine sample are typically concentrated through filtration and/or centrifugation prior to DNA extraction. After concentration, DNeasy blood and tissue kit (Qiagen, Valencia, CA) has been successfully used for urinary microbiome studies4,6,8,22
PCR amplification.
The use of a marker gene such as the 16S rRNA gene for community profiling increases the efficiency of sequencing effort, but at the cost of multiple sources of variability introduced during the PCR amplification, which is required to generate amplicons for sequencing. Examples, which have been extensively reviewed elsewhere42,43, include the fidelity of DNA polymerases (each with a distinct error profile); quantity of input DNA (too little or too much DNA may inhibit a PCR reaction); cycle number (a high number of PCR cycles significantly enhances the likelihood of amplifying contaminant DNAs); choice of primers (which may bias amplification efficiencies toward some species over others); and other PCR reaction conditions. Additionally, chimeric sequences may arise during PCR amplification, when two distinct DNA sequences anneal to form a new, non-biological sequence, which is subsequently amplified. The likelihood of generating chimeras increases with the number of PCR cycles used42, and these chimeric sequences need to be removed bioinformatically (discussed in further detail in Computational Considerations).
Another major consideration is the choice of hypervariable region targeted by the primers used for PCR44,45. Since no individual hypervariable region will universally distinguish all bacteria, targeting different hypervariable regions will result in different biases in the analysis. There has not yet been a comprehensive systematic comparison of how well different hypervariable regions capture the diversity and composition of the urinary microbiota. However, a variety of hypervariable regions have been used to study the urinary microbiota, including the V4 hypervariable region9,20,22, as well as V1–V32,7, V1–V2, and V6 regions6,8. This choice of variable region is particularly important for taxonomic identification, and may lead to the under- or over- representation of microbial taxa46 in addition to limiting the resolution of taxonomic identification possible47. This has also been observed for PCR reaction conditions such as annealing temperature48. While a direct comparison of the fidelity of representation by different variable regions has yet to be performed for urine samples, the V149 and V646 regions are reported to be inferior to other variable regions. We currently favor the V4 or V1–3 region. The former since this region is also used by the Earth Microbiome Project, the most comprehensive effort to date to characterize thousands of samples from a huge diversity of sample types50,51 The latter since this has been used by the vaginal microbiome project52.
Sequencing.
The development of high-throughput Next-Generation Sequencing (NGS) has revolutionized our ability to study microbial communities. NGS is used to identify the DNA sequences that represent the different types of microbiota in a sample when sequencing the 16S rRNA gene or performing metagenomics. Currently, popular NGS platforms are 454 pyrosequencing and Illumina’s HiSeq and MiSeq. Illumina sequencing is now becoming the norm due to its low cost and high speed51, as well as the withdrawal of the Roche 454 pyrosequencing platform in 2016. However, this is a rapidly developing field, and newer sequencing platforms are being developed, such as the Ion Torrent™ (Thermo Fisher Scientific), PacBio™ (Pacific Biosciences), and the MinION™ (Oxford Nanopore Technologies) sequencers.
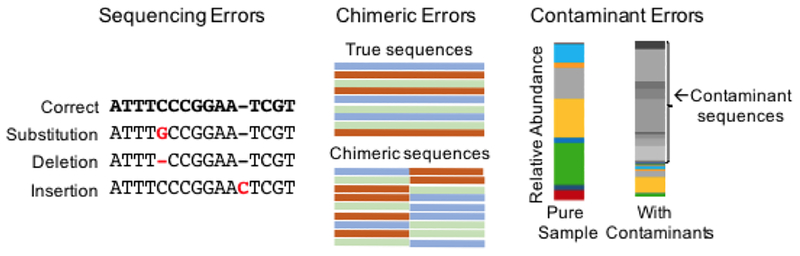
The accuracy of sequence-based microbial profiling is highly dependent on the quality and pre-processing of the sequencing data. Sequencing errors originate from the NGS technologies and result in incorrect base calls (Figure 2) in the output sequences. Sequencing errors make it difficult to distinguish technical variability from biological variability, and leads to inflated diversity measures, incorrect taxonomic classifications, and incorrect inferred function. The unique challenges and importance of correcting sequencing errors in 16S rRNA gene sequencing data53,54 have prompted development of bioinformatics tools specifically for 16S rRNA gene sequencing data. Sequencing errors and the approaches to correct them are dependent on the specific sequencing technology being used. Most 16S rRNA gene sequence correction tools were developed for use with pyrosequencing technology, which is characterized by insertions, deletions, and homopolymers (stretches of identical nucleotides)55 and are not applicable to Illumina sequencing data, where errors are primarily substitution mis-calls (e.g. an A is called instead of a C) and are not randomly distributed54. Sequencing replicates can allow for identification and removal of spurious errors and has been used in sequence-based urinary microbiome studies5,9,21. Other commonly used approaches for handling Illumina sequencing errors are quality trimming and merging overlapping paired-end reads54, though more sophisticated algorithms such as Illumina-specific denoising algorithms56 and DADA257 are being developed and used (See Computational Considerations).
Figure 2. Common errors in 16s rRNA sequencing-based microbiome studies.
Sequencing errors arise from incorrect base calling during sequencing and can be corrected when processing sequence reads with platform specific algorithms. Chimeric errors occur during the PCR amplification step and can be corrected for in the bioinformatics processing workflow. Contaminant errors arise from foreign bacteria being introduced to the sample throughout the sample collection and laboratory methods used to extract the bacterial DNA from the sample. Identifying and removing contaminant sequences is a particular challenge for samples with low microbial biomass.
3. Handling contamination
Unlike other human-associated microbial communities, urine, especially catheter-collected urine, typically yields a low microbial biomass and the true signal arising from the resident microbiota can easily be overshadowed by signal arising from technical variation and experimental errors. The accuracy of microbial profiling in low microbial biomass samples is highly dependent on appropriately identifying, controlling, and correcting these errors (Figure 2). The previous sections described sources of error arising from PCR amplification (chimeras) and NGS (sequencing errors). An additional key consideration for accurate representation of a microbial community is the exposure of a urine or other sample to environmental or ‘foreign’ nucleic acids, either during sample collection, DNA extraction, or PCR amplification. This is most relevant for samples with low microbial biomass.
Sources of contaminants.
Sources of foreign nucleic acids may include patients, healthcare staff, laboratory personnel, airborne bacteria, surface contaminants on laboratory instruments and plasticware (e.g. PCR tube lids), molecular biology reagents, and cross-sample contamination. Indeed, it has been reported that contaminating DNA is widespread in commercially available kits used for DNA extraction and may represent a significant source of error in community profiling of samples prepared with these kits58–60. Batch-to-batch variation in otherwise identical kits can produce distinct contamination profiles58, and as noted by Glassing et al, a DNA extraction kit itself can generate over 20,000 reads from dozens of bacterial genera.60 Taken together, this makes contaminant DNA a major problem for low microbial biomass samples that can be difficult to identify.
Control of contaminants.
Some level of laboratory contamination should be anticipated in studies involving urine or other low microbial biomass samples. Unfortunately, there is little consensus on how to best mitigate contamination of microbiome samples by exogenous bacterial DNA and there is currently no standard technique to remove these contaminants. Procedures can be taken to minimize the amount of exogenous DNA introduced to the samples during sample processing, such as using pretreating reagents60,61 and using DNA extraction kits designed specifically to minimize contamination. However, Glassing el al. concluded that techniques involving pretreating reagents may not be suitable for low microbial biomass samples60. Analytic approaches have also been developed to identify and remove contaminants from low microbial biomass samples by exploiting the relationship between the abundance of individual bacteria and each sample’s overall bacterial DNA concentration62. Another potential analytical approach for identifying contamination is by using a Bayesian approach implemented in SourceTracker63 if the contaminant sources are adequately defined. Further studies identifying which of these approaches are suitable for identification of the urinary microbiota are needed.
Salter et al and others have developed lists of common laboratory contaminants58,60. While such lists are useful, care should be used in how this information is handled during analysis since some of these identified contaminants may actually be biologically meaningful in a sample of interest. Hence, researchers should not simply disregard and remove all sequences mapping to bacteria in these lists, but should instead carefully monitor any results that include these bacteria and evaluate whether the result is most likely of biological relevance or a technical artifact.
Finally, not all contaminants in a PCR reaction arise from laboratory reagents and kits. For instance, cross-sample contamination can easily occur between PCR tubes, plate covers and pipette tips. This is particularly an issue for amplified DNA and hence the steps pre- and post- PCR amplification should be performed in dedicated spaces with their own equipment. Extra care should be taken to avoid cross contamination between low and high microbial biomass samples (i.e. do not process high and low microbial biomass samples at the same time).
Negative and positive control samples.
The use of both positive and negative controls is essential for evaluating the urinary microbiome or similar low microbial biomass samples. Negative controls can aid in the identification of the contaminants potentially introduced during lab processing and allow for identification of samples that only contain contaminant DNA and should not be used for downstream analyses. Ideally such controls are subject to every step of the DNA extraction procedure, are run in parallel to samples and incorporate use of original sample collection materials such as a ‘clean’ swab or rinse of sample collection materials. In addition to negative controls, positive controls are also necessary to verify that the experimental procedures worked as expected. A positive control can consist of a single known bacterial isolate or a mock community with a known polymicrobial profile. Mock communities can be created by mixing known proportions of pure cultures or be purchased commercially. Since the microbial community composition is known, the biases and accuracy of the experimental and computational procedure can be evaluated53, in addition to identifying contaminants in urine or other specimens from the laboratory methods used for sample preparation.
4. Computational considerations
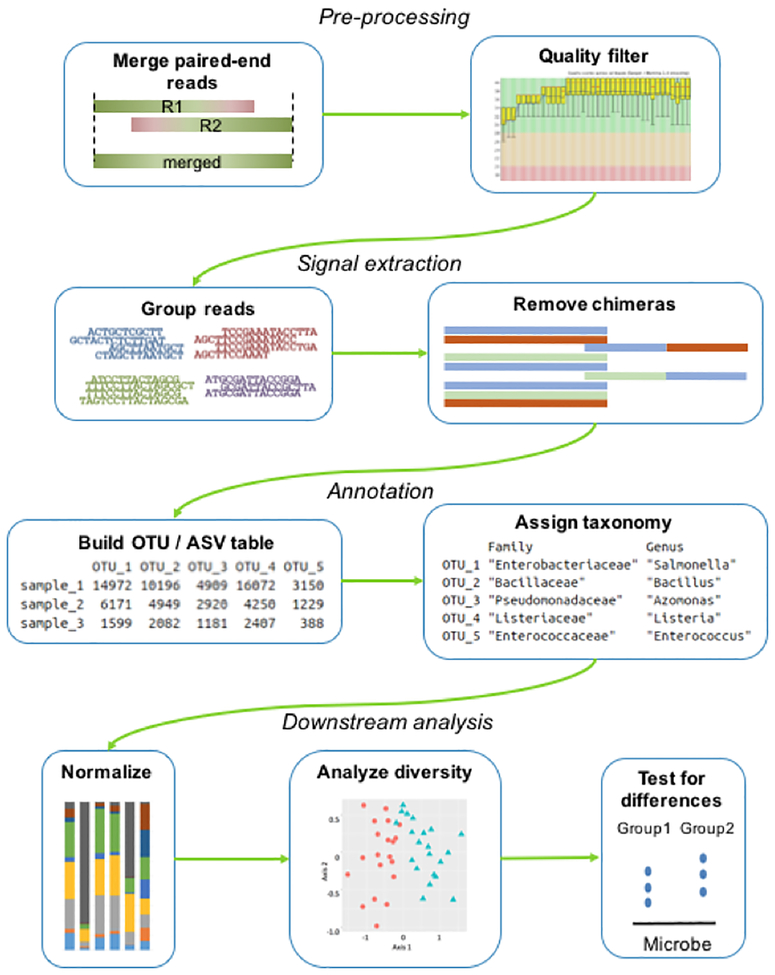
Next generation sequencing generates millions of sequencing reads that require manipulation to generate information about the microbial community of interest. Many bioinformatics methods and tools have been developed to process the sequenced reads. Individual bioinformatics tools are typically joined to form a bioinformatics pipeline, where the input is the raw sequencing reads and the output is a data table of counts of identified bacteria (e.g., an operational taxonomic unit table) used for downstream statistical analysis (Figure 3). The exact workflow selected depends on the sequencing technology employed, the data format received from the sequencing facility, the experimental design, and available computational resources. Developing a bioinformatics workflow for 16S rRNA gene sequence processing is important but challenging in large part because software is continuously emerging and each processing step presents multiple decision points.
Figure 3.
Typical bioinformatics pipeline for 16S rRNA sequencing based microbiome studies. Raw sequence reads are preprocessed typically by merging paired end reads and quality filtering to remove low quality sequences. The sequence reads passing this step are then grouped into operational taxonomic units (OTUs) or amplicon sequence variants (ASVs). This results in a data matrix of OTU/ASVs per sample that is annotated with taxonomic assignments. This table is used for downstream analysis to describe and understand the microbial communities.
We review the key processing and data analysis steps below to help inform readers of the techniques commonly used to analyze 16S rRNA gene sequencing data from urine and summarize the methods that have been used in urinary microbiome papers using 16S rRNA sequencing in Table 1. We also highlight current areas of development which should be considered for future studies involving the urinary microbiome. It should be noted that these steps are applicable to sequence data derived from a diversity of sample types including urine, vaginal swabs or feces.
Table 1.
Summary of bioinformatic methods used for processing 16S rRNA sequencing data from urinary microbiome studies
| Sequencing Technology | Workflow Package | OTU Clustering | Chimera Removal | Taxonomic Database | Taxonomic Classifier | Reference |
|---|---|---|---|---|---|---|
| Sanger | none | none | Bellerophon | RDP | RDP Classifier | 17 |
| Sanger | none | none | Bellerophon | unspecified, subset with Silva | RDP Classifier, subset with BLASTn | 19 |
| 454 Pyrosequencing | mothur | mothur (unspecified) | unspecified | unspecified | RDP Classifier | 103 |
| 454 Pyrosequencing | none | none | unspecified | unspecified | RDP Classifier | 18 |
| 454 Pyrosequencing | none | mothur (unspecified) | unspecified | custom | MEGAN | 6 |
| 454 Pyrosequencing | none | mothur (unspecified) | unspecified | custom | MEGAN | 8 |
| 454 Pyrosequencing | mothur | mothur (unspecified) | UCHIME | RDP | RDP Classifier | 7 |
| 454 Pyrosequencing | none | none | unspecified | unspecified, subset with Silva | RDP Classifier, subset with BLASTn | 19 |
| Illumina MiSeq | mothur | mothur (unspecified) | UCHIME | Silva | RDP Classifier | 4 |
| Illumina MiSeq | mothur | mothur (unspecified) | UCHIME | Silva | METAGENassist | 9 |
| Illumina MiSeq | mothur | mothur (unspecified) | UCHIME | Silva | RDP Classifier | 5 |
| Illumina MiSeq | QIIME | usearch | blast | Silva | RDP Classifier | 22 |
| Illumina MiSeq | QIIME | uclust | UCHIME | Greengenes | RDP Classifier | 98 |
| Illumina MiSeq | none | vsearch | unspecified | STIRRUPS | unspecified | 99 |
| Illumina MiSeq | QIIME | open reference uclust | unspecified | Greengenes | unspecified | 100 |
| Illumina MiSeq | mothur | mothur (unspecified) | UCHIME | Silva | RDP Classifier | 20 |
| Illumina MiSeq | QIIME | unspecified | unspecified | unspecified + Resphera Insight | unspecified | 102 |
| Illumina MiSeq | QIIME | open reference (unspecified) | unspecified | Greengenes | unspecified | 104 |
| Illumina MiSeq | QIIME | open reference (unspecified) | unspecified | Greengenes | unspecified | 105 |
| Illumina HiSeq | QIIME | not specified | uclust | unspecified | Resphera Insight | 106 |
| Ion Torrent | QIIME | unspecified | unspecified | Greengenes | unspecified, subset with BLASTn | 101 |
Footnotes: Unspecified is indicated when the specific method was not mentioned in the manuscript’s methods section. The bioinformatics methods will vary according to the sequencing platform used (e.g. the chimera method Bellerophon85 was used on Sanger sequences, but is not commonly used on short reads produced by Illumina).
Bioinformatics workflow software packages.
Currently, several software packages can implement the bioinformatics steps described below to process and analyze 16S rRNA gene sequencing data. Of these, Quantitative Insights Into Microbial Ecology (QIIME)64 and mothur65 are commonly used and have been used in urinary microbiome studies. Additional workflow software for 16S rRNA sequencing data exist and have been reviewed elsewhere66,67. These software packages enable users to create customized workflows without deep knowledge of the intricacies of each individual step. Furthermore, they can minimize problems that may arise when input and output formats differ between individual bioinformatics tools. QIIME and mothur are well documented and well supported through online communities, which are excellent resources for learning about the necessary steps for data processing and for troubleshooting when technical issues occur. When using these workflow packages such as QIIME or mothur, researchers should cite the package as well as specific methods used within the package.
Documentation.
Documenting the bioinformatics workflow specifics (including the method, version, and parameters used for each tool) is essential to ensure the fidelity of bioinformatics processing, as well as to promote analytical transparency and reproducibility in the urinary microbiome research field. Multiple options exist to support documentation and sharing data analysis workflows. Mothur users should generate a batch file to document each generated workflow. QIIME users may create Jupyter Notebooks to document the workflow and display key output or summaries, downstream analyses can be completed in R Markdown, which enables sharing key output and underlying code for analyses.
Preprocessing.
The first step in analyzing 16S rRNA sequences requires preprocessing the sequencing data so that it is in the proper format for subsequent processing steps. This commonly includes demultiplexing sequence reads if the samples are multiplexed, a common practice in which multiple microbiome samples are combined on a single sequencing run, and removing barcode and primer sequences. Preprocessing also includes quality filtering the sequences to avoid bias and minimize artifacts caused by PCR amplification and sequencing68. When sequencing produces overlapping paired-end reads, they can be merged prior to quality filtering, taking advantage of redundant base calls at overlapping nucleotide positions to generate improved posterior quality scores. Low quality reads are typically removed on the basis of quality scores, presence of ambiguous bases, expected errors, mismatched bases in the barcodes and primers, and sequence length. While this reduces the error rate by removing poor quality and low abundant sequences, a more rigorous, complementary solution (discussed further below) is to actively resolve sequencing errors using amplicon sequence variant algorithms that correct sequences, allowing for more data to be kept in downstream analyses. This is an active area of research, and new computational approaches to infer and correct Illumina sequencing errors are currently being developed.
OTU clustering.
Commonly, 16S rRNA gene sequencing workflows employ operational taxonomic unit (OTU) clustering methods that group sequences according to their similarity. The primary purpose of clustering sequences into OTUs is to render downstream computations less intensive and to minimize noise. The choice of clustering method and similarity threshold significantly impact downstream results69, and a similarity threshold of 97% is commonly used. Most OTU-picking algorithms can be grouped into 3 categories: de novo, closed-reference, and open-reference70. De novo approaches are data-driven methods that perform clustering based on the similarity of sequences in the data set to each other69,71. Closed-reference methods cluster sequences based on similarity to a reference database70, and therefore results will depend on the database used. This is problematic for urine or other understudied tissues which may have fewer constituent bacteria represented in existing databases. Finally, open-reference methods first perform closed-reference OTU picking and then perform de novo picking on the sequences that failed to cluster using the reference database70,72. Though all of the urinary microbiome studies to date have used OTU clustering methods, a major limitation of OTU clustering methods is that they tend to overinflate the number of bacteria present in a sample, typically identifying thousands of unique OTUs from mock communities which have under 100 true OTUs present71,73. This can lead to incorrect determination of ecological diversity in microbiota samples obtained from the bladder, urinary tract, or any other tissue site.
Alternatives to OTU clustering.
Recently, a number of alternative approaches to OTU clustering have been developed, which we refer to as amplicon sequence variant (ASV) methods. ASV methods attempt to achieve finer taxonomic resolution than traditional OTU clustering methods. These methods consider the frequency distribution of sequences, and may attempt to identify and resolve Illumina sequencing errors. Some ASV algorithms, such as UNOISE274 and Deblur75, employ a priori models of sequencer error profiles, while DADA257 uses experiment-specific, adaptive error models estimated from the data, and MED76 uses information theory to distinguish closely related taxa. Once the errors have been estimated and corrected or removed, all variation is considered to be biologically meaningful. Thus, in addition to removing noise, ASV methods offer finer taxonomic resolution than is possible with traditional OTU clustering methods73. Benefits thus include more accurate estimates of microbial diversity since a) closely related bacterial taxa that OTU methods may group into a single entity may be more successfully distinguished as ASVs57,73,76 and b) ASV methods avoid assigning sequence variants that arise due to error into distinct OTU clusters57,74,75. Additional benefits of ASV methods over OTU clustering methods are that ASVs are more reproducible and reusable across studies, and are independent of reference databases73. These benefits may prove particularly useful for urinary microbiome studies.
Chimera removal.
Removal of chimeric sequences, which are an artifact of PCR where two DNA sequences anneal to form a new, non-biological sequence and are amplified, is essential for 16S rRNA studies of any sample, but particularly low biomass or poorly characterized samples such as urine since the number of PCR cycles are usually high, and this has been shown to increase the formation of chimeras significantly42. Failure to remove chimeras can lead to errors such as inflated diversity and incorrect identification of novel taxa77. Chimera removal algorithms can be database-dependent (relying on a reference database of known sequences), which again may be problematic for urine, or database-independent (relying on the sequences in the dataset). Commonly used algorithms include ChimeraSlayer77, Perseus78, UCHIME79, which are included in the bioinformatics workflow packages QIIME and mothur.
Taxonomy assignment.
After sequences have been grouped into OTUs or ASVs, the representative sequences must be assigned a taxonomy to identify which microbes are present in the samples. In addition to choosing the appropriate database for taxonomy assignment, there are also various algorithms for assigning taxonomy to the individual sequence clusters, many of which are implemented in different bioinformatics workflow packages. Most methods use either a Bayesian k-mer matching approach (such as the RDP Classifier)80 or sequence matching based on similarity to a reference database. This is also a current area of active research and improved classifiers are being developed, such as the BLCA algorithm81, to improve taxonomic classification down to the species level.
The reference database chosen for taxonomy assignment will bias downstream results and must be taken into account when attempting to compare results from different studies. Currently, three major databases are commonly used: SILVA82, Ribosomal Database Project (RDP)83, and Greengenes84. All of these databases include taxonomy based on 16S rRNA, but take different approaches for obtaining names of associated organisms. Both SILVA and RDP offer contain taxonomy to the genus level and additionally contain information about additional marker genes (such as 18S and large subunit rRNAs) and eukaryotes (such as fungi). Greengenes is currently limited to 16S rRNA sequences, but contains some taxonomy down to the species level. Each database has several versions available to incorporate the generation of new information, and it is important to use the latest version for taxonomy assignment when possible. SILVA and RDP were both updated in September 2016. Greengenes, though widely used, has not been updated since 2013. While there has yet to be a study specifically on database performance for the urinary microbiome, SILVA is currently the most frequently used.
Site-specific taxonomic databases can overcome some limitations of general databases. Microbial site-specific and hand-curated databases have been created, such as databases specific to the vaginal microbiome52 (optimized for V1–V3 region of 16S rRNA), the oral microbiome86 and the gut microbiome87. This is especially important in microbial niches that are poorly represented in the currently available databases or where species level resolution is expected to be of importance, such is in the urinary microbiome. The cultured isolates from the bladder microbiota appear to be distinct from vaginal microbiota88, which supports the need for a urinary microbiome 16S rRNA database.
Data analysis.
After the above steps are completed, the processed 16S rRNA sequencing data takes the form of a large data matrix, where typically the columns represent individual samples and the rows contain the bacterial features at the taxonomic level of interest (Phylum, Class, Order, Family, Genus, or OTU/ASV). Each cell in the matrix thus indicates the number reads identified for a given bacterial feature and sample. There are several characteristics of these data that make downstream statistical analysis challenging. One issue is that there is variation in the total number of sequences obtained per sample (referred to as library size), which can vary by orders of magnitude. With Illumina MiSeq and HiSeq data, the number of sequences per sample can range from tens of thousands to millions of sequences, and with 454 pyrosequencers this range is from hundreds to tens of thousands of sequences. The variation in the number of reads per sample does not necessarily reflect biological information, but can be due to technical variation. Normalization attempts to correct the distribution of the data and minimize the effects of uneven sampling depth on downstream analysis. Many different normalization strategies have been proposed for 16S sequencing data89–91, but no one-size-fits-all approach exists. Another technical challenge is the sparseness of the data matrix. Due to the heterogeneity of the human microbiome, many cells of the full data matrix will contain zeros, especially at lower taxonomic rankings (genus, OTU, or ASV level). This sparseness can cause problems when attempting to apply certain statistical analyses, violating their assumptions or weakening their power.
Identifying the community structure.
The microbial communities of a given sample are commonly described by measures of alpha and beta diversity. Alpha diversity describes the within-sample diversity by taking into account how many different types of bacteria are in a sample (richness, commonly measured with the Chao1 estimator) and the distribution of these bacteria (evenness). Many popular alpha diversity metrics, such as the inverse Simpson index and Shannon index, account for both richness and evenness of a sample. Beta diversity refers to between-sample diversity and describes how individual samples relate to each other. Beta diversity measures are based on a distance or dissimilarity matrix, usually calculated by the Bray-Curtis method92 (non-phylogenetic) or UniFrac method93 (phylogenetic), and may account for information on the abundance of bacteria (weighted methods) or simply the presence or absence of bacteria (unweighted). Beta diversity is usually visualized with a Principal Coordinate Analysis plot (PCoA) or non-metric multidimensional scaling (NMDS). These are data reduction methods that summarize the multidimensional data to a new set of axes to represent the data in low-dimensional space. Such methods provide a concise overview of the dataset, and can be used to identify whether any covariates are contributing to microbial composition.
Identifying differentially abundant bacteria.
A common goal of microbiome studies is to identify bacteria that are different between groups of individuals (for example, between women with urgency urinary incontinence and healthy controls) in order to identify whether specific bacteria are associated with a disease or phenotype. Following the terminology of other -omic fields, this is referred to as differential abundance testing89,91.
Because 16S rRNA sequencing data is complex and does not follow a normal distribution, many studies have used non-parametric tests such as the Kruskal-Wallis rank sum or Wilcoxon rank sum tests to identify significant differences between groups. While these approaches do not require the data to come from a normal distribution, they do assume that the data come from identical distributions, and will result in erroneous conclusions if the data being compared come from different distributions or have unequal variances. Additionally, use of these tests when an appropriate parametric statistical test exists may result in a loss of power to detect differences. More recently, methods based on generalized linear models have been developed for identifying differentially abundant bacteria between groups. Some of these methods, such as DESeq294, have been adapted from approaches to identify differential expression from RNA-Seq data and repurposed to identify differentially abundant bacteria from 16S rRNA sequencing data. Additional methods such as ANCOM95 and metgenomeSeq91 have been specifically developed for microbiome analyses. These new methods hold promise and are growing in popularity, though some statistical expertise is needed to appropriately use them.
Scope and limitations of sequencing-based microbiome studies
Identifying the individual microbes and understanding the structure of a microbial community is an important first step in understanding a new microbial environment. However, it is only a first step and additional studies must be conducted in order to understand the complex biology of human-associated microbial communities such as those of the urinary tract. 16S rRNA gene sequencing is one approach to sample the microbiota of an environment, but it has limitations, such as its semi-quantitative nature and biases that are introduced through sample and data processing. Furthermore, 16S rRNA gene sequencing-based approaches currently used usually lack taxonomic resolution down to the species or strain level and do not give insight into the functional capabilities of the bacteria that are present, which would provide insight into how they contribute to host health. Functional microbial community profiling by shotgun metagenomics, transcriptomics, proteomics, and metabolomics can provide novel insights into the function of microbial communities and should thus be pursued as well12. Another limitation of sequencing-based approaches to understand human-associated microbes is that they only yield information about the microbial DNA in a sample. While this allows for identification of the types of bacteria present in a biological sample, it does not distinguish between bacteria that are live from those that are dead. The use of complementary techniques such as EQUC can identify live bacteria, though some may be missed if they are difficult to culture under these conditions.
Finally, though bacteria isolated from catheter-collected urine specimens are considered to be a reflection of the microbiota that resides in the bladder, sampling the microbiota from urine may not enable detection of all bladder-resident microbes. It is currently unknown how the urinary microbiome may differ from the urothelium-associated microbiota of the bladder wall. In light of the fact that the mucosal-associated microbiota of the intestinal tract are distinct from those found in stool96,97 such site-specific differences in the bladder are probable and will likely require more invasive sampling procedures such as biopsies or lavages.
Conclusions
We are only just now beginning to understand the clinical relevance of the urinary microbiota in lower urinary tract health and disease88. Since the discovery of the urinary microbiota in 201017 and bladder microbiota in 20121, several studies have confirmed that most adult women have microbial communities in their bladders2,3 and lower urinary tracts. Furthermore, these microbes are been identified as live and cultivatable under non-standard culturing conditions6,20. The urinary microbiota appear to be altered in women with bladder disorders such as urgency urinary incontinence4,5,9,22 and interstitial cystitis8. Characteristics of these microbial communities have also been associated with successful treatment of UUI21 and post-treatment risk of infection5. These studies provide strong evidence that the genitourinary microbiota are likely clinically relevant and warrant further investigation.
As we begin to elucidate the role of genitourinary microbiota in bladder disorders as well as their potential role for therapy, we must consider the technical and computational nuances for low microbial biomass environments. This review provides initial framework to help shape community guidelines to maximize the potential of this emerging and highly interdisciplinary field of study.
Key Points.
Like other areas of the human body, the urinary tract is inhabited by commensal microbes referred to as the urinary microbiota.
Characteristics of the urinary microbiota have been associated with response to treatments, risk of infection, and urological disorders.
Studying the urinary microbiota presents many challenges due to its low microbial biomass and proximity to other body sites with rich microbial environments.
Further research is needed to understand these microbial communities and their relationship to urological disorders are warranted and require careful planning from sample collection to data analysis to ensure conclusions drawn are robust and reproducible.
Acknowledgements
The authors thank R. Searles and L. Brubaker for helpful discussions and the three reviewers for constructive feedback that has greatly improved the manuscript. The authors also thank their funders who supported this work: L.K. is funded by the Oregon BIRCWH (Building Interdisciplinary Research Careers in Women’s Health) program (US NIH award number K12HD043488), the NIH (award number (K01DK116706) and the Rheumatology Research Foundation. M.A. is funded by the Rheumatology Research Foundation and the Spondylitis Association of America. J.T.R. is funded by the NIH (award number R01EY029266), Research to Prevent Blindness (RPB), the Rheumatology Research Foundation, the Stan and Madelle Rosenfeld Family Trust, the William and Mary Bauman Family Foundation and the Spondylitis Association of America. J.B. is funded by the NIH (award numbers P01DK46763, P30CA016042 and UL1TR001881). D.A.F. is funded by the NIH (award numbers R01MH115357, R01MH105538, R01MH096773 and R00MH091238). S.K.M. is funded by the NIH (award numbers UL1TR002369 and U24TR002306). R.N. is funded by the Society of Urodynamics, Female Pelvic Medicine & Urogenital Reconstruction (Overactive Bladder Syndrome Urgency Urinary Incontinence grant). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or any other funding agency.
Online Resources
QIIME: https://qiime2.org/
Mothur: https://mothur.org/
DADA2: https://benjjneb.github.io/dada2/index.html
Jupyter Notebook: http://jupyter.org/
R Markdown: https://rmarkdown.rstudio.com/
Earth Microbiome Project: http://www.earthmicrobiome.org/
Human Microbiome Project: https://hmpdacc.org/
References
- 1.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, FitzGerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE & Brubaker L Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol 50, 1376–1383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh M-J, Huang S-T, Ljungberg I, Sprague B, Lucas SK, Torralba M, Nelson KE & Groah SL Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med 10, 174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL & Malone-Lee J Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J. Clin. Microbiol 51, 2054–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ & Schreckenberger PC Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol 52, 871–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, Visco AG, Nygaard IE, Barber MD, Schaffer J, Moalli P, Sung VW, Smith AL, Rogers R, Nolen TL, Wallace D, Meikle SF, Gai X, Wolfe AJ & Brubaker L The female urinary microbiome in urgency urinary incontinence. Am. J. Obstet. Gynecol 213, 347.e1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui H, Nederbragt AJ, Lagesen K, Jeansson SL & Jakobsen KS Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol 11, 244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis D, Brown R, Williams J, White P, Jacobson SK, Marchesi JR & Drake MJ The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell. Infect. Microbiol 3, 41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL & Jakobsen KS Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol 12, 205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-white K, Fok C, Kliethermes S, Schreckenberger PC, Brubaker L, Gai X, Wolfe a. J. & Wolfe J The Female Urinary Microbiome: a Comparison of Women with and without Urgency Urinary Incontinence. MBio 5, e01283–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimee M, Citorik RJ & Lu TK Microbiome therapeutics — Advances and challenges. Adv. Drug Deliv. Rev 105, 44–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brubaker L & Wolfe AJ The new world of the urinary microbiota in women. Am. J. Obstet. Gynecol 213, 644–649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, Al KF, Chanyi RM, Whiteside S, Dewar M, Razvi H, Reid G & Burton JP Questions and challenges associated with studying the microbiome of the urinary tract. Ann. Transl. Med 5, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM & Sonnenburg JL Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaac S, Scher JU, Djukovic A, Jimenez N, Littman DR, Abramson SB, Pamer EG & Ubeda C Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J. Antimicrob. Chemother 72, 128–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Huang Y, Zhou Y, Buckley T & Wang HH Antibiotic Administration Routes Significantly Influence the Levels of Antibiotic Resistance in Gut Microbiota. Antimicrob. Agents Chemother 57, 3659–3666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai T, Mazzoli S, Mondaini N, Meacci F, Nesi G, D’Elia C, Malossini G, Boddi V & Bartoletti R The role of asymptomatic bacteriuria in young women with recurrent urinary tract infections: to treat or not to treat? Clin. Infect. Dis 55, 771–777 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, Sodergren E, Weinstock GM, Diao L & Fortenberry JD Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One 5, e14116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Q, Nelson DE, Toh E, Diao L, Gao X, Fortenberry JD & van Der Pol B The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS One 6, 1–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson DE, Dong Q, van der Pol B, Toh E, Fan B, Katz BP, Mi D, Rong R, Weinstock GM, Sodergren E & Fortenberry JD Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS One 7, 1–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas-White KJ, Kliethermes S, Rickey L, Lukacz ES, Richter HE, Moalli P, Zimmern P, Norton P, Kusek JW, Wolfe AJ & Brubaker L Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am. J. Obstet. Gynecol 216, 55.e1–55.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, Kliethermes S, Jacobs K, Zilliox MJ, Brincat C, Price TK, Kuffel G, Schreckenberger P, Gai X, Brubaker L & Wolfe AJ Incontinence medication response relates to the female urinary microbiota. Int. Urogynecol. J 27, 723–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karstens L, Asquith M, Davin S, Stauffer P, Fair D, Gregory WT, Rowsenbaum JT, McWeeney SK & Nardos R Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front. Cell. Infect. Microbiol 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai G, Gajer P, Nandy M, Ma B, Yang H, Sakamoto J, Blanchard MH, Ravel J & Brotman RM Comparison of Storage Conditions for Human Vaginal Microbiome Studies. PLoS One 7, e36934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauber CL, Zhou N, Gordon JI, Knight R & Fierer N Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett 307, 80–86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores R, Shi J, Yu G, Ma B, Ravel J, Goedert JJ & Sinha R Collection media and delayed freezing effects on microbial composition of human stool. Microbiome 3, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ & Turnbaugh PJ Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagi E, Candela M, Fairweather-Tait S, Franceschi C & Brigidi P Ageing of the human metaorganism: the microbial counterpart. Age (Omaha). 34, 247–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David L. a, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE & Alm EJ Host lifestyle affects human microbiota on daily timescales. Genome Biol 15, R89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UME, Zhong X, Koenig SSK, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ & Ravel J Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med 4, 132ra52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price TK, Dune T, Hilt EE, Thomas-white KJ, Kliethermes S, Brincat C, Brubaker L, Wolfe AJ, Mueller ER & Schreckenberger C The Clinical Urine Culture : Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. 54, 1216–1222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K & Tanaka R Quantitative PCR with 16S rRNA-Gene-Targeted Species-Specific Primers for Analysis of Human Intestinal Bifidobacteria. 70, 167–173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woese CR & Fox GE Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A 74, 5088–5090 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML & Pace NR Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci 82, 6955–6959 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan S, Cohen DB, Ravel J, Abdo Z & Forney LJ Evaluation of Methods for the Extraction and Purification of DNA from the Human Microbiome. PLoS One 7, e33865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parracho HMRT, Bingham MO, Gibson GR & McCartney AL Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J. Med. Microbiol 54, 987–91 (2005). [DOI] [PubMed] [Google Scholar]
- 36.O Cuiv P, Aguirre de Carcer D, Jones M, Klaassens ES, Worthley DL, Whitehall VLJ, Kang S, McSweeney CS, Leggett BA & Morrison M The effects from DNA extraction methods on the evaluation of microbial diversity associated with human colonic tissue. Microb. Ecol 61, 353–362 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Hart ML, Meyer A, Johnson PJ & Ericsson AC Comparative Evaluation of DNA Extraction Methods from Feces of Multiple Host Species for Downstream Next-Generation Sequencing. PLoS One 10, e0143334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill C, van de Wijgert JHHM, Blow F & Darby AC Evaluation of Lysis Methods for the Extraction of Bacterial DNA for Analysis of the Vaginal Microbiota. PLoS One 11, e0163148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcoll N, Osterlund T, Sinclair L, Eiler A, Kristiansson E, Backhaus T & Eriksson KM Comparison of four DNA extraction methods for comprehensive assessment of 16S rRNA bacterial diversity in marine biofilms using high-throughput sequencing. FEMS Microbiol. Lett 364, (2017). [DOI] [PubMed] [Google Scholar]
- 40.Jiang W, Liang P, Wang B, Fang J, Lang J, Tian G, Jiang J & Zhu TF Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc 10, 768–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yergeau E, Michel C, Tremblay J, Niemi A, King TL, Wyglinski J, Lee K & Greer CW Metagenomic survey of the taxonomic and functional microbial communities of seawater and sea ice from the Canadian Arctic. Sci. Rep 7, 42242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gohl Daryl M., Vangay Pajau, Garbe John, MacLean Allison, Hauge Adam, Becker Aaron, Gould Trevor J., Clayton Jonathan B., Johnson Timothy J., Hunter Ryan, Knights Dan Beckman KB Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol 1–11 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wu J-Y, Jiang X-T, Jiang Y-X, Lu S-Y, Zou F & Zhou H-W Effects of polymerase, template dilution and cycle number on PCR based 16 S rRNA diversity analysis using the deep sequencing method. BMC Microbiol 10, 255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barb JJ, Oler AJ, Kim H-S, Chalmers N, Wallen GR, Cashion A, Munson PJ & Ames NJ Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples. PLoS One 11, e0148047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar PS, Brooker MR, Dowd SE & Camerlengo T Target Region Selection Is a Critical Determinant of Community Fingerprints Generated by 16S Pyrosequencing. PLoS One 6, e20956 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, DeSantis TZ, Andersen GL & Knight R Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizrahi-Man O, Davenport ER & Gilad Y Taxonomic Classification of Bacterial 16S rRNA Genes Using Short Sequencing Reads: Evaluation of Effective Study Designs. PLoS One 8, e53608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sergeant MJ, Constantinidou C, Cogan T, Penn CW & Pallen MJ High-Throughput Sequencing of 16S rRNA Gene Amplicons: Effects of Extraction Procedure, Primer Length and Annealing Temperature. PLoS One 7, e38094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Z & Morrison M Comparisons of Different Hypervariable Regions of rrs Genes for Use in Fingerprinting of Microbial Communities by PCR-Denaturing Gradient Gel Electrophoresis. Appl. Environ. Microbiol 70, 4800–4806 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vazquez-Baeza Y, Gonzalez A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA & Knight R A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G & Knight R Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, Jefferson KK & Buck GA Species-level classification of the vaginal microbiome. BMC Genomics 13 Suppl 8, S17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amore RD, Ijaz UZ, Schirmer M, Kenny JG, Gregory R, Darby AC, Shakya M, Podar M, Quince C & Hall N A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics 17, 55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT & Quince C Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res 43, e37–e37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huse SM, Huber JA, Morrison HG, Sogin ML & Welch DM Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8, R143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tikhonov M, Leach RW & Wingreen NS Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 9, 68–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJ & Holmes SP DADA2 : High resolution sample inference from amplicon data. Nat. Method 13, 581–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ & Walker AW Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corless CE, Guiver M, Borrow R, Kaczmarski EB & Fox AJ Contamination and Sensitivity Issues with a Real-Time Universal 16S rRNA PCR. 38, 1747–1752 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ, Pace N, Mojzsis S, Burcelin R, et al. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog 8, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Champlot S, Berthelot C, Pruvost M, Bennett EA, Grange T & Geigl EM An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS One 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jervis-Bardy J, Leong LEX, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, Nosworthy E, Morris PS, O’Leary S, Rogers GB & Marsh RL Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome 3, 19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R & Kelley ST Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J & Knight R QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ & Weber CF Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Pop M, et al. Analysis of bacteria contaminating ultrapure water in industrial systems. PLoS One 8, 87 (2013). [Google Scholar]
- 67.Mysara M, Njima M, Leys N, Raes J & Monsieurs P From reads to operational taxonomic units: an ensemble processing pipeline for MiSeq amplicon sequencing data. Gigascience 6, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schloss PD, Gevers D & Westcott SL Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS One 6, e27310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westcott SL & Schloss PD De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ 3, e1487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Navas-Molina J. a., Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG & Knight R Advancing our understanding of the human microbiome using QIIME. Methods in Enzymology 531, 371–444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kopylova E, Navas-Molina JA, Mercier C, Xu ZZ, Mahé F, He Y, Zhou H-W, Rognes T, Caporaso JG & Knight R Open-Source Sequence Clustering Methods Improve the State Of the Art. mSystems 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rideout JR, He Y, Navas-Molina J. a, Walters W. a, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert J. a, Huse SM, Zhou H-W, Knight R & Caporaso JG Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Callahan BJ, McMurdie PJ & Holmes SP Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv (2016). doi: 10.1101/081257 [DOI] [Google Scholar]
- 75.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A & Knight R Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH & Sogin ML Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9, 968–979 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R & Birren BW Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quince C, Lanzen A, Davenport RJ & Turnbaugh PJ Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC, Haas BJ, Clemente JC, Quince C & Knight R UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Q, Garrity GM, Tiedje JM & Cole JR Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73, 5261–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao X, Lin H, Revanna K & Dong Q A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 18, 247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J & Glöckner FO The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41, 590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cole JR, Wang Q, Fish J. a., Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR & Tiedje JM Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 42, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P & Andersen GL Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huber T, Faulkner G & Hugenholtz P Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Chen T, Yu W-H, Izard J, Baranova OV, Lakshmanan A & Dewhirst FE The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, baq013–baq013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ritari J, Salojärvi J, Lahti L & de Vos WM Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics 16, 1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brubaker L & Wolfe AJ The Female Urinary Microbiota/Microbiome: Clinical and Research Implications. Rambam Maimonides Med. J 8, e0015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McMurdie PJ & Holmes S Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLOS Comput. Biol 10, e1003531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER & Knight R Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5, 27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paulson JN, Stine OC, Bravo HC & Pop M Differential abundance analysis for microbial marker-gene surveys. Nat Meth 10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bray JR & Curtis JT An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr 27, 325–349 (1957). [Google Scholar]
- 93.Lozupone C & Knight R UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mandal S, Treuren W Van, White R. a, Eggesbø M, Knight R & Peddada SD Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis 26, 27663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L & Sargent M Diversity of the human intestinal microbial flora. Science (80-.). 308, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G & Neufeld JD Bacterial biogeography of the human digestive tract. Sci. Rep 1, 170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu P, Chen Y, Zhao J, Zhang G, Chen J, Wang J & Zhang H Urinary Microbiome and Psychological Factors in Women with Overactive Bladder. Front. Cell. Infect. Microbiol 7, 488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gottschick C, Deng Z-L, Vital M, Masur C, Abels C, Pieper DH & Wagner-Dobler I The urinary microbiota of men and women and its changes in women during bacterial vaginosis and antibiotic treatment. Microbiome 5, 99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shoskes DA, Altemus J, Polackwich AS, Tucky B, Wang H & Eng C The Urinary Microbiome Differs Significantly Between Patients With Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls as Well as Between Patients With Different Clinical Phenotypes. Urology 92, 26–32 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Adebayo AS, Survayanshi M, Bhute S, Agunloye AM, Isokpehi RD, Anumudu CI & Shouche YS The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Negl. Trop. Dis 11, e0005826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abernethy MG, Rosenfeld A, White JR, Mueller MG, Lewicky-Gaupp C & Kenton K Urinary Microbiome and Cytokine Levels in Women With Interstitial Cystitis. Obstet. Gynecol 129, (2017). [DOI] [PubMed] [Google Scholar]
- 103.Plichta JK, Holmes CJ, Nienhouse V, Puszynski M, Gao X, Dong Q, Lin H, Sinacore J, Zilliox M, Toh E, Nelson DE, Gamelli RL & Radek KA Cutaneous Burn Injury Modulates Urinary Antimicrobial Peptide Responses and the Urinary Microbiome. Crit. Care Med 45, e543–e551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu F, Ling Z, Xiao Y, Yang Q, Wang B, Zheng L, Jiang P, Li L & Wang W Alterations of urinary microbiota in type 2 diabetes mellitus with hypertension and/or hyperlipidemia. Front. Physiol 8, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu F, Ling Z, Xiao Y, Yang Q, Zheng L, Jiang P, Li L & Wang W Characterization of the urinary microbiota of elderly women and the effects of type 2 diabetes and urinary tract infections on the microbiota. Oncotarget 8, 100678–100690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shrestha E, White JR, Yu S-H, Kulac I, Ertunc O, De Marzo AM, Yegnasubramanian S, Mangold LA, Partin AW & Sfanos KS Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J. Urol 199(1),161–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]