Abstract
The medial entorhinal cortex (MEC) is known to contain spatial encoding neurons that likely contribute to encoding spatial aspects of episodic memories. However, little is known about the role MEC plays in encoding temporal aspects of episodic memories, particularly during immobility. Here, using a virtual “Door-Stop” task for mice, we show MEC contains a representation of elapsed time during immobility, with individual time encoding neurons activated at a specific moment during the immobile interval. This representation consisted of a sequential activation of time encoding neurons and displayed variations in progression speed that correlated with variations in mouse timing behavior. Time and spatial encoding neurons were preferentially active during immobile and locomotion periods, respectively, were anatomically clustered with respect to each other and preferentially encoded the same variable across tasks or environments. These results suggest the existence of largely non-overlapping sub-circuits in MEC encoding time during immobility or space during locomotion.
Introduction
Over the past 50 years, research from humans and animal models have implicated the medial temporal lobe, which includes the hippocampus and MEC, in the formation of personal memories of events that occur at specific places over a specific time interval1,2. While a vast amount of research has uncovered cellular substrates in the hippocampus and MEC that likely make up the spatial representation required for these episodic memories3–8, our understanding of the temporal representation is significantly less advanced and has focused mostly on the hippocampus9–11. Time-related neurons were first demonstrated in the hippocampus using studies where rodents were moving to some degree, either in a running wheel12, on a treadmill13 or in a small box14. Importantly, one study found hippocampal time-related activity during immobility15. These so-called hippocampal “time cells” fire briefly and consistently at specific times during the task such that behavioral time periods are tiled by a sequence of brief neuronal activations. Interestingly, specialized circuitry representing spatial information during immobility has also been demonstrated in the hippocampus16,17. This suggests that separate circuitry within the medial temporal lobe might be used to encode behaviorally relevant variables between mobile and immobile periods, though it is unclear from these studies if the representation of elapsed time maps onto a particular circuit(s).
In MEC, one study18 found that MEC grid cells can provide timing-related information during treadmill running and a separate study found MEC neurons that were more active at low instead of high running speeds during locomotion19. Inactivation of MEC during such mobile periods was found to produce deficits in encoding memories across trace periods20,21, produce deficits in a temporal memory task and cause instability in downstream hippocampal time cells22. These studies suggest that a code for elapsed time may exist in MEC during locomotion, but it is currently unknown if the neural circuity in MEC forms a representation of elapsed time during immobility, when sensory cues may not change in a temporally informative manner. Furthermore, if such a representation exists in MEC, it is unknown how the neural circuitry might be organized to generate it.
Results
To explore these ideas, we used our previously developed functional two-photon imaging methods23 to optically record from populations of layer II MEC neurons (Figure 1a, Supplementary Figure 1) during mouse navigation in a novel virtual “Door Stop” task. The Door Stop task combines both a locomotion dependent virtual navigation phase, and an explicit instrumental timing phase that was separated in time and location from reward delivery (Figure 1b, Supplementary Figure 2a). Mice were trained to run down a linear track to a specific location where they encountered an invisible door, which they could not run past, though they could still run on the treadmill. At the door location, the mice were required to stop and wait for at least 6 seconds (an auditory click signaled the start of the 6 second interval once the treadmill velocity fell below a threshold, see Methods); if the mice began running on the treadmill before the expiration of the 6 second interval, the mice could not progress past the closed door and the trial would start over (signaled by another click). After the 6 second interval, the door would open and the mice could run down the remaining length of the track to the reward zone. After 6–8 weeks of training, mice ran to the invisible door and stopped on their first attempt for the full 6 second wait period on 55.1% of trials (Figure 1c), referred to as “correct trials”. In order to easily compare neural activity during immobile timing periods and neural activity during locomotion periods, we excluded a transition zone between these periods and excluded the reward zone when behavior was more ambiguous (Figure 1e, Supplementary Figure 2a, see methods). During the wait periods, mice mostly sat immobile with essentially 0 velocity with small jerky movements occurring 12.9% of the wait period to maintain balance on the treadmill (mean velocity over wait periods = 0.33 ± 1.00 cm/sec (STD); Figure 1d,e). All of the data presented in Figures 2–4 using the (invisible door) Door Stop task come only from these correct trials (see Supplementary Figure 2b-f for velocity on all trials). Since the mice could not see the invisible door opening at the end of the 6 second interval, this Door Stop task therefore requires an internal temporal representation for efficient completion.
Figure 1:
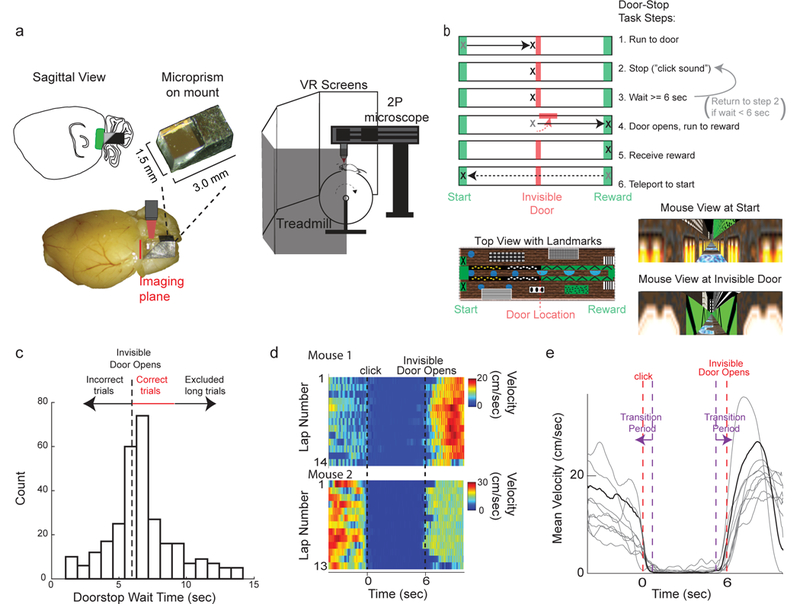
Head-fixed cellular resolution functional imaging during mouse navigation in a virtual Door Stop Task. a. MEC was imaged through a microprism (left) with two-photon microscopy in head-fixed mice navigating in a virtual “Door Stop” task (right). b. Virtual Door Stop task. c. Histogram of wait times at the invisible door from all mice (n=7) trained on 6-second Door Stop task. d. Mouse locomotion velocity leading into (time < 0 sec), during (0 sec < time < 6 sec) and after 6 second Door Stop wait interval for all correct trials from 2 different single sessions in 2 mice (top and bottom). e. Mean velocity leading into (time < 0 sec), during (0 sec < time < 6 sec) and after 6 second Door Stop wait interval across all correct trials of an example single session (grey). Mean velocity across all trials for example session shown in black. Purple dashed lines and arrows indicate Transition Period.
Figure 2:
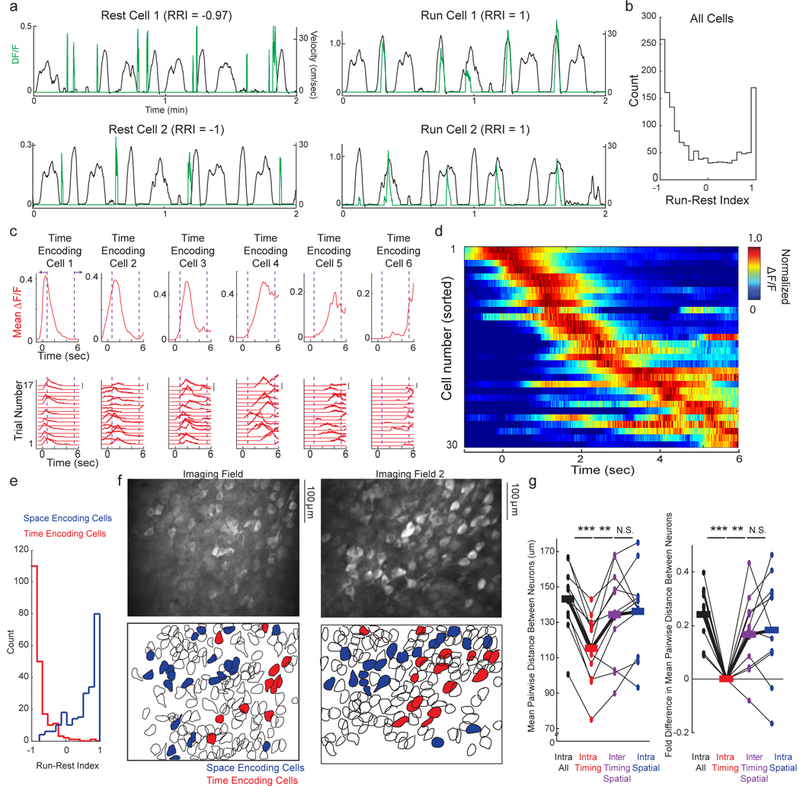
Functionally and anatomically clustered populations of neurons in MEC encode space during locomotion and elapsed time during immobile intervals of the Door Stop task. a. DF/F traces of significant transients (green traces) from individual example Rest Selective Cells (left) and Run Selective Cells (right) during running and resting periods (black traces) in Door Stop task. b. Histogram of RRI for all active cells, across all FOVs in all mice during Door Stop task; transition periods and reward zone excluded. c. Bottom, DF/F versus time for each correct trial of a single session for 6 individual neurons from the same FOV during the 6 second Door Stop wait interval. Scale bars indicate 100% DF/F. Top, Mean DF/F versus time across all correct trials. Purple dashed lines and arrows indicate transition period. d. Mean DF/F versus time across all correct trials in a single session for all neurons (each row represents single neuron mean DF/F) in a single FOV during the 6 second Door Stop wait interval. Mean DF/F normalized to peak for each neuron (each row). e. Histogram of RRI for all time encoding cells (red) and all spatial encoding cells (blue) across all FOVs in all mice during Door Stop task; transition periods and reward zone excluded. f. MEC FOVs of GCaMP6f labeled populations (top) colored red or blue to indicate cells encoding time or space (bottom), respectively. g. Mean pairwise distance (left) or fold-change (right) between neurons in various groups (x-axis). All spatial or time encoding cells from all mice in Door Stop task. Black lines connecting measures (dots) from same FOV, thick lines are mean across all FOVs. (n = 11 imaging fields from 7 mice; Repeated Measures ANOVA F = 11.8, P<0.0001; intra time-time vs intra all-all, P<0.001 Tukey’s post-test with Bonferroni Correction; intra time-time vs inter time-space, P<0.01 Tukey’s post-test with Bonferroni Correction.) Interestingly, spatial cells were not significantly clustered compared to all cells even though we previously found grid cells were clustered compared to non-grid cells (see reference 25). This difference is likely due to the heterogeneous spatial cell population defined here, which likely includes grid, border and spatially selective non-grid cells. *** indicate P<0.001. ** indicate P<0.01.
Figure 4:
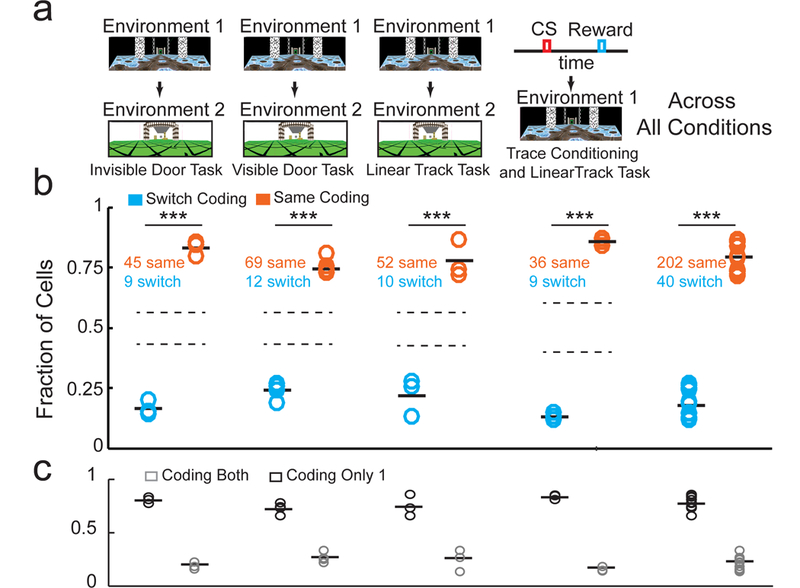
Subsets of neurons encoding time or space in one track (or task) are more likely than chance to encode the same variable in a different track (or task). a. Environment switches in the invisible door Door Stop task, visible door Door Stop task, linear track task, and task switch from classical trace conditioning to virtual linear track task (ordered from left to right). b. The fraction of cells that encoded the same variable (orange) or switched variables (blue) within the population of cells encoding a variable in both environments (or tasks). Mean for each group indicated by solid black bars. Standard deviation bounds for randomly shuffled distributions, for each type of environment or task switch, shown by dotted lines. (Across all conditions: n = 13 imaging fields; t = 13.8, df = 12, P<0.0001 two-sided Student’s Paired T-Test) c. The fraction of cells that encoded a variable in only one (black) or across both (grey) environments or tasks. Mean for each group indicated by solid black bars. N = 13 imaging fields. *** indicate P<0.001 for Shuffle Test for each track or task switch paradigm. *** indicates P<0.001 for paired Sign Rank Test for pool data across all track or task switch paradigms.
We first explored the possibility that MEC contains neurons active preferentially during immobility. We optically recorded from populations of layer II MEC neurons labeled with GCaMP6f (Figure 2, Supplementary Figure 3) during the Door Stop task (11 imaging sessions/fields of view (FOVs) across 7 Mice, FOV 414 ± 50 (STD) by 328 ± 40 microns, depth below surface = 89 ± 26 microns). Across the population (136 ± 54 (STD) active neurons/FOV, 1497 active neurons total, see Methods), active neurons exhibited calcium transients with different selectivity for running and immobile periods. This is consistent with previous reports of MEC neurons displaying positive and negative correlations to running speed19 (Supplementary Figure 3f). However, in addition to neurons that were negatively tuned to speed, we also observed neurons that were essentially silent during running periods and active selectively during immobility (when running speed was essentially zero, Supplementary Figure 2; see Methods). Therefore, in order to quantify this run/rest selectivity across the population, we developed a “Run-Rest Index” (RRI) that measures the run versus rest selectivity of a neuron, such that if all significant transients occurred during resting the RRI is −1 and if all occurred during running RRI = +1 (see Methods). The distribution of RRI values across the active MEC population appeared bimodal, with neurons active preferentially during rest/immobility periods or preferentially during run periods (Figure 2a,b; across all active neurons RRI = −0.24 ± 0.71, mean ± STD), though neurons with RRI values spanning the full range were observed. Thus, during the virtual Door Stop task, MEC contains subsets of run and (previously unknown) rest/immobility selective neurons.
We then sought to determine whether any of the rest specific neuronal activity might encode elapsed time during the immobile timing phase of the Door Stop task (11 FOVs in 7 mice). We identified all timing interval trials during single sessions and, for all active neurons, plotted the change in fluorescence (calcium transients) as a function of time since stopping. Across the different timing intervals, many neurons displayed calcium transients that occurred regularly at a specific time delay from the start of immobility (Figure 2c), with the mean activity over all trials forming significant timing fields in 18.3 ± 4.0 % (SEM) of all active neurons (P < 0.05 for Bootstrap Test, 11/11 FOVs had cells with timing fields, 22.5 ± 3.4 (SEM) number of cells had timing fields in each FOV, range of number of cells in each FOV [6 to 40], wait times between 6–9 seconds included). In single FOVs during single sessions, timing fields of different neurons across the population tiled the full timing interval (Figure 2d; similar results across all FOVs, see Supplementary Figure 3c). Therefore, during periods of immobility in a virtual Door Stop task, a subset of neurons in MEC form a representation of elapsed time through their sequential activation across the full wait interval.
During the locomotion phase of the task we found many neurons with significant spatial fields (17.9 ± 3.0 % (SEM) of all active neurons; P-Value < 0.05 for Bootstrap Test; 22.2 ± 11.9 (SEM) number of cells had spatial fields in each FOV, range of number of cells in each FOV [9 – 45]; Supplementary Figure 3d,e); this population likely contained many of the navigation encoding cells previously described (grid cells, border cells, velocity cells, etc)7,24,25. Across all 11 FOVs, the majority of space or time encoding cells had either spatial or timing fields (92.1 ± 1.8 % (SEM) were time or space encoding cells only), but not both (7.9 ± 1.8 % (SEM) had both timing and spatial fields; 3.1 ± 0.7 % (SEM) of all active cells) (t=7.9, df = 10, P<.00001, Student’s Paired T-Test). While the time encoding cells were most active during periods of rest (RRI = −0.86 ± 0.02 (median ± SEM)), the spatial encoding cells were most active during periods of locomotion (RRI = 0.69 ± 0.03 (median ± SEM)), with little overlap in the RRI distributions (Wilcoxon rank sum z-value = 17.3630; P<.00001) (Figure 2e). Interestingly, similar results suggesting functional bimodality within the MEC population were obtained using information theoretic metrics (Supplementary Figure 4). Thus, in a particular environment, the subset of neurons encoding elapsed time during immobility was largely non-overlapping with a separate subset of neurons encoding space during locomotion.
We then explored the anatomic organization of time and space encoding cells in MEC during the Door Stop task. Visual inspection of the anatomic location of the time and space encoding cells revealed that cells encoding similar information were often spatially clustered within individual imaging fields (Figure 2f). Across all 11 FOVs, time encoding cells were significantly clustered together in MEC compared to space encoding cells and compared to all active neurons (intra time-time distance: 115.0 ± 5.9 μm (SEM), intra space-space distance: 136.1 ± 7.4 μm, intra all-all neuron distance: 142.8 ± 5.6 μm, inter time-space distance: 134.2 ± 8.0 μm: Repeated Measures ANOVA F3,30 = 11.8, P<0.0001; intra time-time vs intra all-all, P<0.001 Tukey’s post-test with Bonferroni Correction; intra time-time vs inter time-space, P<0.01 Tukey’s post-test with Bonferroni Correction), resulting in a significant 24% and 17% difference in neuron-neuron distance between intra all neurons vs. intra time encoding cells and between inter time-space cells vs intra time encoding cells, respectively (Figure 2g). Together, the above results demonstrate the existence of different subsets of neurons in MEC during a navigation tasks: the canonical space encoding subset active during locomotion and a novel time encoding subset active during immobile timing intervals.
We observed that during correct trials in the Door Stop task, mice sometimes waited close to the ideal 6 seconds and sometimes waited significantly longer than 6 seconds (Figure 1c). Based on this observation, we next asked whether the temporal representation in MEC reflected this difference in timing behavior, as might be expected for such a representation and, if so, whether the sequential activation of time encoding cells advanced more slowly or whether additional time encoding cells were added to encode the additional wait time. For 4 sessions from 3 mice in which a relatively large number of trials containing a wide range of wait times was observed, we grouped trials into short wait times (6–7 seconds, mean= 6.5 ± 0.3 (STD) seconds) and long wait times (7–9.5 seconds, mean = 8.0 ± 0.7 (STD) seconds; Figure 3a). We observed an apparent slowing of the temporal representations between the short to long trials (in single trials, mean across trials from single fields and pooled over all fields; Figure 3b). To quantify this apparent slowing, we generated a trial-by-trial measure of the speed of sequence progression through the cells representing elapsed time during each wait interval (measured as the fitted slope in units of cell activations per second) and found that progression speed was significantly correlated to wait time (Spearman Rank Test rho = −0.52, df = 72, P<0.001; Figure 3b,c). Error trials in which mice waited between 4.5–6 seconds had the largest slopes on average (fastest sequence progression speeds), suggesting that the mice might run too early when the sequence progresses too quickly; when these trials were included with the correct trials, again progression speed was significantly correlated to wait time (Spearman Rank Test rho = −0.57, df = 85, P<0.001; Supplementary Figure 5a-b). Importantly, out of 796 active cells recorded across all 4 sessions, only 2 new time encoding cells formed with a field during the additional (long) wait time (1.5% of cells with significant time fields in either short or long trials). Therefore, the temporal representation formed by populations of time encoding cells in MEC reflected increases in mouse wait times through a decrease in the speed of sequence progression rather than by adding additional time encoding cells to the end of the sequence.
Figure 3:
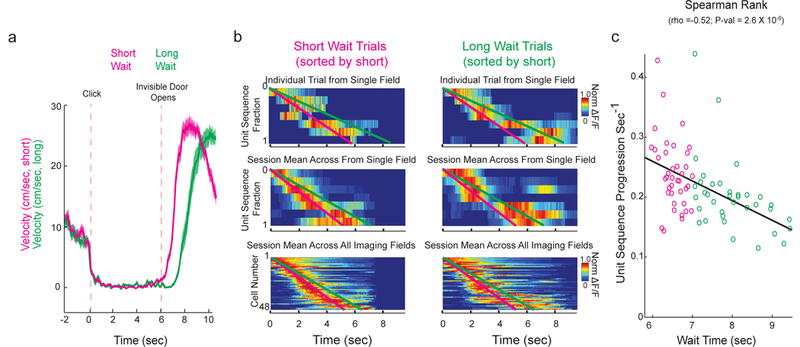
Sequence progression across time encoding MEC cells correlates with animal wait time. a. Velocity leading into (time < 0 sec), during (0 sec < time < 6 sec) and after 6 second Door Stop wait interval for all short wait (pink) and long wait (green) correct trials (mean in dark line, SEM in shaded color). b. Examples of normalized DF/F sequence for all cells from an individual trial (top, same cell ordering and same session in left and right; short wait = 6.1 sec, long wait = 8.0 sec), across all trials from an individual FOV (middle, same cell ordering and same session in left and right; mean short wait = 6.3 ± 0.3 sec (STD), mean long wait = 8.3 ± 0.7 sec (STD)) and for all cells across all FOVs (bottom, same cell ordering, includes multiple sessions in left and right; mean short wait = 6.5 ± 0.3 (STD), mean long wait = 8.0 ± 0.7 (STD)), short waits (6–7 seconds; left) and long waits (7–9.5 seconds; right). Cells were ordered according to each cell’s mean center of mass across all short wait trials (earliest mean center of mass at top, latest at bottom). Pink and green lines are linear fits of short (pink, left) and long (green, right) wait sequences. c. Plot of slopes (from linear fits of cell activations per second) as a function of animal wait time for all individual trials (each circle represents a single trial, as seen in b top). Cells were ordered according to each cell’s mean COM across all correct (6–9.5 sec) trials (earliest mean COM at top, latest at bottom). N=73 wait trials from 4 imaging fields in 3 mice.
The subsets of MEC neurons encoding space and time together formed a sequence of neuronal activations that encoded the full spatiotemporal extent of each trial of each task (from track start to end in the Door Stop task). These subsets could be recruited at random from the pool of all MEC neurons, suggesting MEC contains a single general and flexible circuit designed to generate sequences independent of the behavioral variable being encoded. Alternatively, the subsets could be recruited from largely non-overlapping pools of MEC neurons: one pool largely encoding time and a different pool largely encoding space. To distinguish between these possibilities, we asked if the subsets encoding time or space in one track (or task) were more likely than chance to encode the same variable in a different track (or task), suggesting largely non-overlapping pools, or were more likely to randomly switch their encoding between variables, suggesting a single large pool. We imaged the same population of MEC neurons and compared their space and time encoding properties across the following paradigms in which mice navigated across different (familiar) tracks or performed different (familiar) tasks: 1. Mice were switched between two (invisible door) Door Stop task tracks significantly different in visual appearance (Figure 4a-c, n=3 mice), 2. Mice were switched between two (visible door) Door Stop task tracks significantly different in visual appearance and door wait times (Supplementary Figure 6a,b; Figure 4a-c, n=3 mice), 3. Mice were switched between two different linear tracks (no Door Stop, time encoding during voluntary rest periods, Supplementary Figure 6c-f; see Methods; Figure 4a-c, n=3 mice) and 4. Mice were switched between an immobile classical trace conditioning task and the linear track navigation task (Supplementary Figure 6g-j; Figure 4a-c, n=3 mice). The tracks in environment switch paradigms 1–3 were sufficiently different in visual appearance (Figure 4a) to cause global remapping across populations of spatial encoding cells in MEC (mean spatial activity pattern correlations across environments not significantly different compared to chance in 7/9 mice, Pearson’s Correlation = 0.12 ± 0.05 (mean ± SEM), N = 9 mice; Supplementary Table 2A) and in place cells in the hippocampus26. Many cells with timing or spatial fields in one track (or task) did not have significant timing or spatial fields in the other track (or task), becoming largely inactive (77.3 ± 0.02% (SEM); Figure 4c; Supplementary Figure 6b). This observation leads to a refinement of the above two different possibilities: either two largely non-overlapping pools of MEC neurons exist, from which some fraction of cells can be recruited to encode time (from the time encoding pool) or space (from the space encoding pool) for any given environment and context, or a single large pool of MEC neurons exists, from which some fraction of cells can be recruited to encode time or space for any given environment and context. Importantly, we found that of the 22.7% of cells with timing or spatial fields in both tracks (or tasks), 83.7 ± 2.7% (SEM) encoded the same variable in both while 16.3 ± 2.7% (SEM) switched encoding (Figure 4b and Supplementary Table 1;t = 13.8, df = 12, P<0.0001 Student’s Paired T-Test), fractions highly unlikely to have arisen from chance (P < 0.001 for Shuffle Test for groups of timing cells and spatial cells in each of the 4 track or task switches). We also found that cells with significant timing or spatial fields present across multiple days were much more likely to encode the same variable rather than switch (Supplementary Figure 6k-m). Additionally, the RRI of the active cells in both tracks (paradigms 1–3) was similar across tracks (RRI difference for each cell between tracks = 0.30 ± 0.031 (SEM); Supplementary Figure 6n) and this difference was unlikely to have arisen from chance (Shuffled RRI difference for each cell between tracks = 0.864 ± 0.002 (SEM); P<0.001 for Shuffle Test). Together, the above results suggest the possible existence of largely non-overlapping pools of MEC neurons, one that preferentially encodes time during animal immobility and another encoding space during animal locomotion.
Next, we asked whether time and space encoding neurons within MEC were capable of forming temporal or spatial representations from the first moments of new spatio-temporal experiences, as might be expected if the representations arise from largely non-overlapping sub-circuits that specialize in encoding space or time. Alternatively, it is possible that the representations developed only after learning, as might be expected for non-specialized, flexible circuitry12,14,15. To answer this question, we used the virtual linear track task (no Door Stop) during which we observed a similar MEC temporal representation during periods of voluntary rest along the track (Supplementary Figure 6c-f). Mice were trained to familiarity in one linear track (familiar) and then switched to a second virtual linear track (novel), which was sufficiently different in visual appearance (Figure 5a) to cause global remapping across populations of spatial encoding cells in MEC (spatial activity pattern correlations across environments not significantly different compared to chance in 3/3 mice, Pearson’s Correlation = −0.07 ± 0.03 (mean ± SEM), N = 3 mice; Supplementary Table 2A) and in place cells in the hippocampus26. From the first moments of exploring the novel track, mice displayed periods of resting and running, similar to the behavior observed during navigation in a familiar linear track (Supplementary Figure 6d). Across all rest periods from single sessions in the novel track, we observed time encoding cells (Figure 5b) with similar properties to those described above (Figure 2 and Supplementary Figure 6). Importantly, time encoding cells very often were active in their respective timing fields during the very first rest period in the novel track (Figure 5b; mean time to first rest after transition from familiar to novel: 35.8 ± 14.1 (mean ± STD)seconds; 56% active on rest 1, 88% active by rest 3; Figure 5c,d), resulting in correlation values across all time encoding cells (between the calcium transients during each rest period and the mean timing field over all periods for each cell) that did not depend on exposure time in the novel track (Figure 5c). Furthermore, the fraction of trials during which a transient occurred within a cell’s timing field did not change between the first half and second half of the session (Figure 5e). Similar results were also observed for the spatial encoding subsets (Supplementary Figure 7a-e). Thus, the temporal (or spatial) representation formed by subsets of time (or space) encoding cells in MEC is present from the first moments of new experiences.
Figure 5:
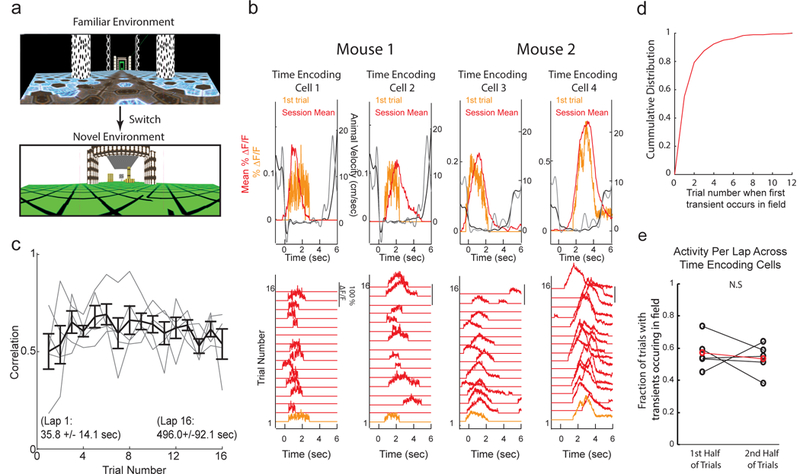
The temporal representation formed by populations of time encoding cells in MEC is present from the first moments of new experiences. a. Views of linear tracks mice navigated in during environment switch paradigm. b. Bottom, DF/F versus time for each voluntary rest period (wait trial) of a single session for 4 individual neurons from 2 different mice during first session in novel linear track (see 5a). Rest period 1 was the first time the mice ever stopped to rest in the novel track (orange trace). Top, Mean DF/F (red) and velocity (black) versus time across all rest periods; DF/F (orange) and locomotion velocity (grey) from first rest period. Note, negative deflection in mouse velocity trace reflects backwards movements on the treadmill. c. Pearson’s Correlation between the calcium transients during each rest period and the mean timing field over all periods (y-axis) as a function of number of wait periods in novel environment (x-axis); grey = mean across all cells in a single FOV in a single session; black = mean ± SEM across all cells in all sessions. N = 5 imaging fields across 3 mice. d. Cumulative distribution of the trial number on which a transient first occurred in the significant timing field in the novel session across all time encoding cells. e. Mean fraction of trials with transients occurring within the significant timing field across all cells for the first half of wait trials in the session versus the second half of wait trials in the session. (n = 5 imaging fields from 3 mice; P = 0.1875 two-sided Paired Wilcoxon Signed Rank Test).
Discussion
Altogether, our results establish the existence of a representation of elapsed time in MEC during immobility and further suggest the possible existence of largely non-overlapping functional sub-circuits in MEC that encode either time during animal immobility or space during animal locomotion. This later notion may differentiate MEC time encoding cells from previously described hippocampal “time cells” and time encoding neurons in other brain regions. The following data presented above is consistent with this notion. First, MEC time encoding cells were present from the first moments of exposure to novel environments, suggesting that learning was not required for their formation; “time cells” in the hippocampus12,14,15 and other time encoding cells in other brain regions27–33 have only been described in well-trained animals and thus it is not clear if learning is required for their formation. It is important to note that “time cells” in the hippocampus and MEC track time through the evolution of a neuronal ensemble through time; this differs from timing activity described in other brain regions, which is more often characterized by individual neuron firing patterns that ramp up or down in time after a cue27,34. Second, the subsets of neurons encoding time or space in one track (or task) were more likely than chance to encode the same variable in a different track (or task). This suggests that neurons encoding time or space during any given episode may be recruited from largely non-overlapping pools of neurons that may arise from specialized circuitry. Third, subsets of MEC time encoding cells formed temporal representations during different behavioral tasks, one with an explicit timing requirement during immobile periods of a navigation task (Door Stop), another with no explicit requirements during periods when the mice chose to rest during navigation (linear track) and a third non-navigation, immobile task with an explicit timing period defined (classic trace conditioning). This suggests that MEC contains circuitry that can be used to represent elapsed time given different tasks or contexts. Fourth, time encoding and spatial encoding cells were anatomically clustered, possibly suggesting that the separate circuits may have developed from separate precursor populations35 or developed to minimize wiring distance between their respective internal components36.
The neural basis for the generation of a temporal representation during immobility in MEC is thus far unknown, however previous research on the generation of grid cell firing patterns in this same brain region may provide insights since separate similarly organized circuits could be used to form a temporal representation. For example, circuit connectivity patterns capable of generating bumps of activity in continuous attractor networks have been described in MEC37,38. These networks could possibly generate sequential firing with appropriate input to move the bumps in one direction for timing cell networks, rather than in 2D for grid cell networks, or could be slightly rewired to form a 1D ring attractor for sequential timing cell activation. Additionally, beyond MEC, several general cortical or subcortical neural timing mechanisms have been proposed34,39–41. Our results here demonstrate that time encoding cells can exhibit fully formed timing fields from the first moment of exposure in a novel environment, suggesting that the mechanisms necessary for encoding elapsed time in MEC do not require learning. However, given our current data, it is not possible to determine whether the temporal encoding circuit arose through a developmental program or whether it could be formed through previous learning in related contexts.
The MEC timing representation described here could be used for online perception of elapsed time or could contribute to encoding temporal aspects of episodic memories, or both. While further research is required to discriminate between these possibilities, some insights can be drawn again from studies of grid cells. For example, grid cells appear to be important components of an online path integration system used during navigation7 and also for constructing context dependent spatial memories downstream in the hippocampus42–45. Therefore, it is possible that MEC timing circuitry is important for both the online perception used for interval timing46 and also for supporting the formation of episodic memories of temporally structured events in the hippocampus47. For the latter case, inactivation of “island cells” in MEC have demonstrated deficits in encoding memories across trace periods48 and distinct populations of neurons in CA2 of the hippocampus become active during periods of immobility16. Since strong and direct synaptic projections exist from superficial MEC to CA249, it is possible that distinct sub-networks within the hippocampus and MEC are recruited to encode elapsed time during immobility.
Similar to recent reports examining grid cells and spatially selective non-grid cells50, time encoding cells in MEC display a range of environment or context dependent selectivity, with the majority of cells being selective for only one of two environments or contexts. Results presented here suggest that temporal encoding and spatial encoding cells are predisposed to encode either time during immobility or space during locomotion, respectively, and may arise from sub-circuits that specialize in encoding these behavioral variables. However, several additional points should be considered for a rigorous interpretation of these findings. First, the majority of temporal and spatial encoding neurons (~77%) were active in only one of the two contexts during the environment (or task) switch experiments. This suggests that most of these cells are likely not part of “hard-wired” neural circuits, whereby the exact set of neurons are recruited to encode the same behavioral variable across all contexts. Second, although a large majority (~84%) of the neurons active in both contexts encoded the same behavioral variable (time or space), it is possible that this predisposition is caused by the animal encoding the contexts in a similar way, rather than through specialized sub-circuits. This possible explanation seems unlikely based upon the following observations indicating global remapping takes place across the context switches: 1) Of the neurons in MEC that encode time or space in at least one context, 77% encode only one of the two contexts in the environment or task switch experiments (~23% encode both) and 2) spatial encoding cells display signal correlations across environments not significantly different compared to chance in 7/9 mice (Supplementary Table 2A). Third, of the neurons in MEC that encode both contexts, a sub-set of neurons (~16%) switched from encoding time to encoding space (or vice versa) across environments or tasks. If specialized sub-circuits do indeed exist in MEC for encoding time during immobility and space during locomotion, then how might these sub-circuits be organized to account for such switching across contexts? One possibility is that given enough context switches, all MEC neurons could be eventually recruited to encode either time or space, which would indicate that any predisposition of sub-circuits to encode time or space is short-lived and continually evolving due to experience and plasticity. Alternatively, it is possible that within the time and space encoding subsets of MEC neurons, distinct sub-populations exist that reliably encode time or space across contexts (the ~20% of time or space encoding neurons here that encoded the same variable across contexts), while the rest of the population may be less tuned for one behavioral variable or the other. Consistent with this idea, a recent study showed that the most spatially selective grid cells encode space across environments and exhibit a coherent population phase shift with respect to their firing fields50, while spatially selective non-grid cells in MEC randomly turn off or turn on across environments, and as a population, exhibit global remapping. Here we found that in 3/9 mice, time encoding cells did exhibit temporal activity pattern correlations across environments (or tasks) that were above chance (Pearson’s Correlation = 0.33 ± 0.11 (mean ± SEM), N = 9 mice; Supplementary Table 3), though population coherence was not analyzed.
Finally, correlation between animal wait time and speed of sequence progression (Figure 3, Supplementary Figure 5) cannot speak to how the stretching between long and short wait trial neural sequences is read-out by downstream brain structures, nor to the animal’s “temporal perception” during long vs short wait trials. It is not possible to say whether the stretching is or is not a context-invariant neural representation of the time interval. For example, it is possible that the animal perceives the same amount of time during the short and long trials (egocentric timing perspective, no context difference). It is also possible that the animal does indeed perceive a difference in the amount of elapsed time (allocentric timing perspective, context difference). Our results only demonstrate a correlation between the wait time behavior of the animal and the speed of the neural sequence progression, suggesting that this temporally structured neural activity in MEC may play a role in the animal’s estimation of elapsed time.
Methods
Surgery and Behavior:
All experiments were approved and conducted in accordance with the Northwestern University Animal Care and Use Committee. Methods for MEC virus injection and microprism implant have been described previously23. Briefly, 12 male C57-BL6 mice (~p70) were anesthetized using 1–2% isoflurane. For virus injection, a small (~0.5 mm) craniotomy was made over the MEC centered at 3.1 mm lateral from bregma and 0.2 mm rostral from the rostral edge of the transverse sinus. Using a beveled pipette (1–2 MΩ), ~30nL of AAV1-Syn-GCaMP-6f was injected at each of three depths along the dorsal-ventral axis (1.0, 1.5 and 2.0 mm from the dorsal surface of the brain). In four mice, a second series of injections were made at 3.5 mm lateral from bregma and 0.2 mm rostral from the transverse sinus, using the same three depths along the dorsal-ventral axis as stated above. The mice then began water scheduling (receiving ~1mL of water/day) as described previously 51,52. 1–2 weeks after the viral injection, a surgery to chronically implant a microprism was performed. Mice were anesthetized and an approximately rectangular craniotomy was made over the dorsal surface of the cortex (above MEC) and cerebellum with corners positioned as follows: 1. ~2.1 mm lateral of bregma, ~4.5 mm caudal of bregma (~300–500 μm rostral of the transverse sinus), 2. ~4.5 mm lateral of bregma, ~4.5 mm caudal of bregma (~300–500 μm rostral of the transvers sinus), 3. ~2.1 mm lateral of bregma, ~7.75–8mm caudal of bregma (~3.25–3.5 mm caudal of the transverse sinus) and 4. ~4.5 mm lateral of bregma, ~7.75–8mm caudal of bregma (~3.25–3.5 mm caudal of the transverse sinus). After the skull was removed, a portion of the cerebellum was aspirated to expose the caudal surface of the cortex. A custom made hook and flat sharpened blade were then used to remove the tentorium separating the cerebellum and cortex, leaving the dura of the cortex completely intact. A microprism (right angle prism with 1.5 mm side length and reflective enhanced aluminum coating on the hypotenuse, Tower Optical) was mounted on a custom stainless-steel mount (using UV curable adhesive, Norland) and this assembly was then positioned by aligning the front face of the microprism parallel to the caudal surface of the MEC and aligning the top surface of the microprism perpendicular to the (eventual) axis of excitation light propagation. A thin layer of Kwik-Sil was applied to the caudal MEC surface prior to microprism implantation to fill the void between the brain and the front surface of the microprism. The microprism and mount were rigidly held in place and the craniotomy sealed by application of a thin layer of Metabond to all exposed sides of the microprism (except the top surface of the prism) and mount and on any exposed skull or brain. Subsequently, a titanium headplate (9.5 mm x 38 mm) was attached to the dorsal surface of the skull, centered upon and aligned parallel to the top face of the microprism. The headplate was used to head restrain the mouse as described previously 51,53. A titanium ring (27 mm outer diameter and 12.5 mm inner diameter, with a 3 mm high edge) was then attached to the top surface of the headplate, centered around the microprism, and the area between the craniotomy and the inner edge of the metal ring was covered with opaque dental cement (Metabond, Parkell, made opaque by adding 0.5 grams of carbon powder, Sigma Aldrich). The metal ring and opaque Metabond, combined with the loose fitting black rubber tube and tight fitting metal rings described previously51, were required to block stray light from the virtual reality screen. After the surgery, the mice recovered in their home cages for ~2–3 days and were regularly exposed to a large “playground” with running wheels and tunnels to encourage exploration and locomotion. Our virtual reality and treadmill system were the same as described previously23. Only mice that were able to perform the virtual reality tasks detailed below (reward rate >~1 reward/min) were included in this study.
Two-photon Imaging of MEC neurons:
The Moveable Objective Microscope and associated light shielding methods were the same as described previously54. Time-series movies (10000 frames, 1024×1024 pixels, 0.068ms/line; 20000 to 40000 frames, 1024×512 pixels, 0.068 ms/line; 40000–80000 frames, 1024 × 256 pixels, 0.068ms/line) were acquired at 14.4 Hz, 28.7 Hz and 57.5 Hz, respectively.
A Digidata1440A (Molecular Devices) data acquisition system was used to record (Clampex 10.3) and synchronize position in the linear track, timing interval, reward timing, lick timing and two-photon image frame timing.
Linear Track (no Door Stop) Virtual Reality Task:
Mouse locomotion speed on the treadmill was measured using a rotary encoder (E2–5000, US Digital). For the linear track task experiments, movement gain was set such that the full length of the virtual track was 3, 4 or 5 m of linear distance and the view angle in the virtual environment was fixed such that the mouse’s view was always straight down the center of the track. The rotational velocity of the treadmill (directly related to the mouse’s running speed on the treadmill) was linearly related to movement speed along the virtual track. Backward movement on the treadmill was also measured and used to update position (in the backward direction) in the virtual track, however mice rarely moved in the backward direction on the treadmill. Once the mice traversed the full length of the track, they received a small water reward (4 μL) in the track end zone. After the reward and a 2 second delay period, the mouse was “teleported” back to the start of the track to begin another traversal.
Approximately 7 days after surgery, behavior training on the virtual linear track began. Mice were trained ~1hr per day until they routinely ran ~1–3 track traversals/min. The number of days required to reach this criterion varied depending on the mouse and ranged from 2 days to 14 days. Once this criterion was reached, imaging experiments began.
During this behavior, trained mice displayed periods of spontaneous running and resting such that their locomotion velocity appeared bimodal and simple thresholding could easily distinguish between these two behavioral states (Supplementary Figure 6d; see “Defining temporal encoding and spatial encoding cells in the Door Stop and linear track tasks” below for thresholds used for defining rest and run periods). The rest periods often occurred near the reward zone (77.9 ± 19.4 % (STD) of rest periods) and were typically characterized by nearly complete immobility (mean velocity = 0.20 ± 0.84 cm/sec (STD) during rest; rest periods > 3 sec), while the running periods were characterized by locomotion (mean velocity = 21.4 ± 8.0 cm/sec (STD); run periods > 3 sec). The rest periods lasted several seconds (mean wait time = 9.1 ± 5.9 sec (STD)) (Supplementary Figure 6d) and the fraction of time spent resting vs. running was nearly equal (53% resting vs 47 % running). Time spent running vs. resting, location of resting, and locomotion velocities were all highly similar to the same metrics characterized in freely moving rodents navigating in real environments16,55. During these rest periods, MEC temporal encoding neurons were observed (Figure 4,5, Supplementary Figure 6c-f).
Invisible door Door Stop Virtual Reality Task (Figures 1–3 and Supplementary Figures 1,3–5):
Before the virus injection surgery, seven mice used for the invisible door Door Stop task were implanted with a headplate to allow for ~2–3 weeks of pre-training. During pre-training, mice were first exposed to a 3m linear track (no Door Stop) for ~1 hr per day until they ran ~2 laps/min. Once mice reached this criterion, they were transitioned to the “visible door” version of the Door Stop task. In this task, mice ran 1.5 m down a linear track on the cylindrical treadmill to a visible door. At the door, the mice were required to stop (locomotion velocity below threshold of either 5.2 cm/sec for 6 of 7 mice and 6.9 cm/sec for 1 of 7 mice) within 10 cm of the door location. An instrumental cue in the form of an auditory click was presented to inform the mouse that the Door Stop timing period had begun. Only once the mice had stopped for a given interval did the door open, at which point they could run forward through the open door another 1.5 m to the track end zone to gain a small water reward (4 μL). Because the treadmill was not fixed in place during the timing interval, the mice could begin running on the treadmill before the interval was complete. In such cases, the door did not open and the mice could not progress forward along the virtual track; once the mice stopped again, the interval started over with another auditory click sound. At the beginning of training on the visible door task, the timing interval was set to 2 seconds, and it was gradually increased over weeks of training to 6 seconds, as each mouse gained >~1 reward/min averaged over an entire 1 hr training session. During this training period on the visible door Door Stop task, the virus injection and MEC window implant surgeries described above were performed, with multiple days for recovery (without training) following each.
Once the mice were able to obtain ~1 reward/min on the 6 second interval visible door Door Stop task, the door was made invisible. This task was identical to the visible door version of the task, except the door was made completely invisible. Mice were therefore not able to visualize when the door was present or not, but when the door was present, it would block the forward progress of the mice down the track. Further, since the mice could not see the invisible door opening at the end of the 6 second interval, this Door Stop task therefore requires an internal temporal representation for efficient completion. If mice did not perform well on the invisible door task after switching from the visible door task, the timing interval was reduced to ~4–5 seconds until performance improved, at which point it was again increased to 6 seconds. Once mice were able to obtain ~1.5 rewards/min on the 6 second interval invisible door Door Stop task (referred to in the text as the “Door Stop” task, ~6–8 weeks of total training time), imaging experiments began.
During wait periods at the invisible door, jerky movements (for the animal to maintain balance on the treadmill) occurred intermittently (0.61 ± 0.25 Hz (STD)), aperiodically (randomly), and for short durations (lasting 0.23 ± 0.23 sec (STD) in duration, and occurred during 12.9 ± 8.6% of the rest periods (STD); mean amplitude 1.1 ± 1.5 cm/sec (STD); see Supplementary Figure 2b-f).
Linear track environment switch (Figure 4) and Novel Environment exposure (Figure 5 and Supplementary Figure 7, Supplementary Tables 1–3):
Before exposure to the novel linear track (Figure 5 and Supplementary Figure 7), mice ran on a different (familiar) linear track for 2–8 weeks. When transitioning to the novel track, mice either ran on the familiar environment first and were switched to the novel during the same recording session (3 transitions) or were exposed to familiar environment on one day and switched to the novel at the beginning of the session on the next day (2 transitions). Once the second environment became familiar, the same FOV of MEC neurons were imaged across the two familiar environments (Figure 4) in 3 mice.
Visible and Invisible Door Stop Task Environment Switch (Figure 4 and Supplementary Figure 6a, k-m, Supplementary Tables 1–3):
Three mice were trained on the Door Stop task with a visible door (described above) using two visually different environments: one had an 8 second wait interval and the other had a 6 second interval wait. Once mice were able to obtain ~1–2 rewards per minute over a 1 hour training session (averaged over trials in both environments) imaging experiments commenced. The same FOV was imaged as mice navigated one familiar Door Stop environment for the first half of the imaging session and the second familiar Door Stop environment for the second half of the imaging session. The same procedure was repeated for 3 mice trained and imaged in the invisible door Door Stop task switch paradigm, except in this case both environments had the same 6 second wait interval.
Classical Trace Conditioning Task and switch to virtual linear navigation task (Figure 4 and Supplementary Figure 6g-j):
3 mice were trained in an immobile head-fixed classical trace conditioning task. Mice were head-fixed in the dark on top of a cylindrical treadmill that was held in place (i.e. mice could not move the treadmill). Mice were presented with a conditioned stimulus (CS) in the form of an auditory click, followed by a fixed trace period, and subsequently an unconditioned stimulus in the form of a water reward (2uL). Inter-trial intervals consisted of a fixed 10 second baseline combined with a random time interval drawn from a uniform distribution from 0 to 20 seconds. Early in training the fixed trace period was set at 1 second, and as mice began to display predictive licking (Supplementary Figure 6j), this interval was gradually increased until mice displayed predictive licking during a 5.5 second trace interval. Once mice displayed predictive licking during the 5.5 second trace period, MEC imaging experiments began. Prior to this classical conditioning task, mice had been trained to run on the (no door) virtual linear track navigation task described above. Mice were imaged during the classical conditioning task and during the same imaging session, the 3 meter virtual linear track was presented upon previously dark screens around the head-fixed mouse and the treadmill was freed to allow for the mice to run and navigate along the track for water rewards at the end of the track. During navigation on the linear track, the same FOV was imaged as during the classical conditioning task.
Data Analysis
Imaging data was analyzed on a Dell Power Edge 720 Server using ImageJ (Version 1.47) and custom software written in MATLAB (2017a,b). No statistical methods were used to predetermine sample sizes. Sample sizes were based on reliably measuring experimental parameters while remaining in compliance with ethical guidelines to minimize the number of animals used. Spearman’s Rank tests, T tests, repeated measures ANOVA, Tukey’s post-test with Bonferroni Correction, Bootstrap tests, Shuffle tests and Wilcoxon rank tests were used to test for statistical significance when appropriate and all statistical tests were two-sided unless stated otherwise. For tests assuming normality, data distributions were assumed to be normal but this was not formally tested. Data collection and analysis were not performed blind to the conditions of the experiments. Data collection was not randomized and different experimental groups were not used or defined. All data in the text and figures are labeled as either mean ± s.d. or mean ± s.e.m. See Life Sciences Reporting Summary for more information.
Image processing, ROI selection and transient analysis:
Motion correction was performed using whole frame cross-correlation, as described previously23, and the motion corrected time-series was used for all subsequent analysis. Regions of interest (ROIs) were defined using Cell Sort 56 (threshold = 1.5–3; area limits = 150–4000 pixels; smoothing width = 0.75–1.0; mu = 0.5; Principal Components = 150) as described previously23 but with the following modification. The motion corrected time-series first was divided spatially into sub-regions of ~160 by ~200 microns (for example, a time series of a 320×400 micron field would be broken up into 4 time series, each on containing a 160×200 micron portion of the original field). Cell Sort was then applied to each sub-movie independently and all ROIs generated by Cell Sort were visually inspected to select for neuronal somata. The ROIs for the original field were then reconstructed using the ROIs defined on the subfields. If ROIs at borders were determined to be from the same cell split into different subfields then the ROIs were merged and the DF/F traces were averaged to generate a single cell and single DF/F trace. Cells were determined to be the same cell if at least 50% of each ROI’s pixels along the border were shared between ROIs and the Pearson’s correlation between the two ROI’s DF/F traces > 0.7. To avoid duplicate ROIs in any subfield, if 2 or more ROIs had centroids separated by less than 50 microns and the Pearson’s correlation between the two cell’s significant transient only traces was greater than 0.8, then only one of the ROIs was retained. ROIs that appeared to be dendrites were also removed from each data set. DF/F versus time traces from each ROI were then processed as previously described 51. Briefly, slow changes in the fluorescence traces were removed by examining the distribution of fluorescence in a 10.2 sec interval centered around each sample in the trace and normalized by the 8th percentile value. These baseline corrected somatic fluorescence traces were then subjected to the analysis of the ratio of positive to negative going transients of various amplitudes and durations described previously51,52,57. We used this analysis to identify significant transients with < 1% false positive error rates and generated the significant transient only traces that were used for all analysis in this research.
Defining Immobile Timing and Locomotion Dependent Behavioral Epochs in the Door Stop Task:
The immobile timing phase of the (visible and invisible) Door Stop tasks was defined as the middle 75% of the timing period (Figure 1e, Supplementary Figure 2a). The first 12.5% (0.75sec) and last 12.5% (0.75sec) of the 6 sec wait period were considered part of a transition period between locomotion and immobility and were removed from the analysis. The locomotion dependent navigation phase of the Door Stop task was defined as 58.3% (1.75m) of the 3m track; 12.5% of the track (37.5cm) before and after the door and 8.3% of the track (25 cm) before and after (beginning of track) the reward zone were considered part of a transition period between locomotion and immobility or transition when mice slow down and stop to consume the reward, respectively, and were removed from the analysis. These transitions regions were purposefully chosen to be large so as to include only clearly defined behavioral periods. These steps thus allowed us to exclude ambiguous behavioral periods and more clearly identify the small number of neurons with separate/isolated fields in both the immobile timing period and the locomotion dependent navigation period, rather than cells with single fields straddling the transition regions.
Defining Active Cells:
All active cells included in this study were defined as cells identified using Cell Sort56 segmentation software (see above), that had a mean ΔF/F > 0.7% in at least one of the two defined behavioral epochs (the immobile timing phase at the door location or the locomotion dependent navigation phase along the track).
Defining temporal encoding and spatial encoding cells in the Door Stop and linear track tasks:
Spatial fields were defined as described previously51,58, with minor changes. First, running periods were defined when mouse movement velocity along the virtual track first increased above 10 cm/sec and maintained at least 5 cm/sec for at least 25 cm of movement along the track (excluding transition zones). Note, these are the running periods used for calculating the Run-Rest Index (see below). For each cell, the mean somatic ΔF/F was calculated as a function of virtual track position for 35 (Door Stop) position bins and 50 (linear track) position bins (5 cm per bin). Potential spatial fields were first identified as contiguous regions of this plot in which all of the points were greater than 20% of the difference between the peak somatic ΔF/F value (for all bins) and the baseline value (mean of the lowest 25% somatic ΔF/F values). These potential spatial field regions then had to satisfy the following criteria: 1. The field must be >15 cm in width; 2. The field must have one value of at least 6% mean ΔF/F; 3. The mean in field ΔF/F value must be >2 times the mean out of field ΔF/F value; and 4. Significant calcium transients must be present >33.3 % of the time the mouse spent in the spatial field and each cell must have > 4 transients that occurred in the field. Potential spatial field regions that met these criteria were then defined as spatial fields if their P value from bootstrapping was <0.05, as described previously51,58 and their mean widths were <125 cm. The ΔF/F time series for each cell was broken into periods of individual transients and inter-transient intervals. The sequence of these events were then randomly shuffled to produce a random ΔF/F time series from which mean shuffled ΔF/F versus position plots were generated and subjected to the same criteria outlined above. This process was repeated 1,000 times and the P-value was defined as the ratio of the number of times out of 1,000 that the random shuffled trace generated a spatial field that met the above criteria.
Significant timing fields were defined using a similar method (to spatial fields) applied to calcium transients during the wait periods of either the Door Stop or linear track tasks. For each recording session in the Door Stop task, wait trials were included if the mouse stopped at the door location and maintained velocity below threshold (5.2 or 6.9 cm/sec) for the entire 6 second wait period and then began running (velocity>5.2 or 6.9 cm/sec) within 3 seconds from the door opening. Wait trials on which the mouse stopped at the door, and began running before the 6 second wait period, or trials when the animal waiting longer than 9 seconds were not included (except for error, short and long wait trial data included in Figure 3 (6 sec <= waits <= 9.5 sec) and Supplementary Figure 5a,b (4.5 sec <= waits <= 9.5 sec)). For each recording session in the linear track task, rest periods were included if the mouse went below 2.5 cm/sec and subsequently maintained velocity below 5 cm/sec for at least 5 seconds and not longer than 30 seconds. To measure the percentage of stopping periods at different locations along the track, the linear track was divided into 4 equal spatial regions. For each imaging session the number of stops in each region was divided by the total number of stops. Region 1 (start/reward location) contained the first 1/8 and last 1/8 of the track, regions 2 to 4 were spaced from 1/8 to 3/8 of the track, 3/8 to 5/8 of the track and 5/8 to 7/8 of the track, respectively. Since each mouse nearly always stopped at region 1 (77.9 ± 19.4 % (STD) of stops at region 1), timing cells were defined at this region only for the linear track task. Over all rest/wait periods (meeting the above criteria, and excluding transition periods) in each recording session (Door Stop and linear track tasks), the mean DF/F for each cell was calculated and binned using the imaging sample time (0.0174–0.0694 sec bins). Note, these are the rest periods used in the Run-Rest Index (see below). Potential timing fields were identified as a contiguous regions of this mean plot in which all of the points were greater than 50% of the difference between the peak somatic ΔF/F value and the baseline value (mean of the lowest 25% ΔF/F bins). These potential timing field regions then were required to satisfy the following criteria: 1. The field must persist > 0.5 sec; 2. The field must have one value of at least 6% mean ΔF/F; 3. The mean in field ΔF/F value must be >2 times the mean out of field ΔF/F value; and 4. Significant calcium transients must be present in the field >33.3 % of the trials. Potential timing field regions that met these criteria were then defined as timing fields if their P value from bootstrapping (see above) was <0.05, and their mean duration < 5 sec.
Time encoding cells were active almost exclusively during periods of immobility (Figure 2e), which is different from previous reports of negative velocity tuning in MEC neurons (see Hinman et al. 2016)19. In contrast to the time encoding cells shown here, the negatively tuned velocity cells presented in Hinman et al. 2016 were active during locomotion, but with firing rates that were negatively correlated with the animal’s velocity. Most temporal coding cells were active during periods of immobility (mean velocity = 0.33 ± 1.00 cm/sec (STD) for the wait periods in the Door Stop task), and much less active during locomotion, even at low velocities (Figure 2e, Supplementary Figure 3f).
In the (invisible door) Door Stop task, cells that had significant fields in both the temporal and spatial epochs made up 3.1 ± 0.7 % (SEM) of all active cells and 7.9 ± 1.8 % (SEM) of all cells with at least one significant field in the temporal or spatial phase of the Door Stop task. Data presented in Figure 2f,g are shown for exclusive timing selective cells or exclusive spatial selective cells.
For all pairwise measurements of cell-cell distance, the pairwise distance between neurons was measured as the Euclidean distance between the centroids of each neuron’s ROI. A repeated measures ANOVA (with Tukey’s post-test with Bonferroni Correction) across 11 unique (non-overlapping) imaging fields across 7 mice was used in order to compare the inter and intra distances between time encoding cells, spatial encoding cells and all cells.
To measure speed of sequence progression (Figure 3 and Supplementary Figure 5), we included only temporally selective cells with transients on at least five trials in each of the long and short wait trial epochs. The center of mass (COM) of each timing cell’s mean ΔF/F vs time trace was calculated as previously described58). For data presented in Figure 3b, cells were ordered according to each cell’s mean COM across all short wait trials (earliest mean COM at top, latest at bottom), for all cells in one FOV from a single imaging session, except for Figure 3b bottom which was sorted across all cells from all fields. Those same indices used to sort the cells on short wait trials, were then applied to sort cells on the long wait trials (Figure 3b). The trial-by-trial measure of the speed of sequence progression (unit sequence progression/sec) was computed for each trial as the slope of the linear fit of the sorted sequence of COMs for all time encoding cells active on that trial (see Figure 3b,c and Supplementary Figure 5a,b). For data presented in Figure 3c and Supplementary Figure 5a,b, cells were ordered according to each cell’s mean COM across all correct (6–9.5 sec) trials (earliest mean COM at top, latest at bottom), for all cells in one FOV from a single imaging session. To compare linear fits across FOVs (with different numbers of cells in each sequence), we normalized the position of each cell in the sorted sequence such that the kth cell in the ordered sequence had a position of k/N, where N is the total number of cells in the sequence. Therefore, for each trial, we computed the COM for each cell’s DF/F vs time trace on that trial, and left the ordering of the COMs according to each cell’s mean COM across all correct trials. A linear fit was then computed for the normalized cell sequence of COMs on each trial (example fits for error, short and long wait trials displayed as over-laid lines in Figure 3b and Supplementary Figure 5a). For each fit the intercept was fixed at (0,0) and cells that did not have transients on a given trial were not included in the fit.
In order to compare observed fractions of temporal and spatial encoding cells that either encode the same variable or switch coding across environments (or tasks) to the fraction expected by chance (Figure 4b), we performed a shuffle test as follows. For each environment (or task) switch experiment (1. visible door, 2. invisible door, 3. linear track and 4. Classical conditioning to linear track) we started by identifying the number of cells with timing or spatial fields in the first environment (or task) (say 50 cells with fields total, 30 timing and 20 spatial cells, as an example). We then found the number of these cells in the real data that encoded a variable (time or space) or did not encode a variable in the second environment (or task) (10 cells encoding a variable across both environments, 40 cells only encoding in the first environment, for the example 50 cells) and used these numbers to estimate variable encoding probability across the switch (20% chance for a cell encoding a variable in the first environment to encode a variable in the second, 80% chance to encode a variable only in the first environment). Across all of our datasets, we found 18% of active cells were time encoding cells and 18% were spatial encoding cells, implying that if a cell were randomly to be assigned to encode time or space in the second environment, it would have a 50% chance to be either. These probabilities were then used to generate a surrogate data set by randomly assigning each cell that had a timing or spatial field in the first environment (the 50 cells in our example) an identity in the second environment (80% chance to encode no variable, 10% chance to encode time and 10% chance to encode space). Among the sub-set of cells in the surrogate data set that encoded a variable in both environments, we calculated the fraction of cells that encoded the same variable, or switched variable encoding. This process was repeated 1,000 times and a P-value was defined as the ratio of the number of times out of 1,000 that the random shuffled fraction of cells that kept the same identity was at least as large as the observed fraction that kept the same identity in the real data. The standard deviation of this random shuffled fraction distribution, which is centered on 50%, is shown in Figure 4b (dashed lines).
Measuring temporal and spatial information for active cells in the (invisible door) Door Stop task (Supplementary Figure 4):
In order to measure temporal information during wait periods and spatial information during locomotion periods for each cell, we used information rate as defined in Skaggs et al. 1993 and recently applied to GCaMP6f [Ca2+] imaging data59,60. The only threshold used in this analysis was a minimum activity threshold that required neurons to have a mean DF/F > 0.7% during either locomotion periods along the track or immobile periods waiting at the door, which equates roughly to at least 5 calcium transients during one of these behavioral periods; the transition periods mentioned above were also excluded from analysis. Because the different parameters (spatial location vs wait time) occurred during different behavioral periods, we applied the below described information significance test separately to spatial (during locomotion periods) and temporal (during immobile periods) information for each cell.
During rest/wait (or long running) periods defined above, temporal (or spatial) information rate was computed for each cell as:
Where λ is the mean DF/F vs time across all wait trials (or vs position across all traversals across the track) excluding transition periods (or zones), λi is the mean DF/F at the ith wait time (or ith position along the track) and pi is the occupancy probability at the ith bin. The number of correct wait trials was less than the number of track traversals, therefore in order to compare the temporal and spatial information measures attributed to each cell directly, a random sub-set of the traversals along the track was included such that this number was equal the number of correct wait trials for each recording session. Cells with significant temporal or spatial information (Supplementary Figure 3b-e) were defined using a shuffle procedure. The ΔF/F time series during the rest/wait (or long running) periods for each cell was broken into periods of individual transients and inter-transient intervals. The sequence of these events were then randomly shuffled to produce a random ΔF/F time series from which a surrogate λ and λi were computed for the temporal (or spatial) period and these parameters were used to compute a surrogate temporal (or spatial) information rate. This process was repeated 1,000 times and a shuffle p-value was defined as the ratio of the number of times out of 1000 that the surrogate temporal (or spatial) information measure was greater than the temporal (or spatial) information rate measured in the real data. Cells were defined as having significant temporal (spatial) information if they had a P-value < 0.05.
The random shuffle distributions shown in Supplementary Figure 4c were generated by random shuffling. For all cells carrying significant spatial or temporal information, the spatial information values were randomly shuffled with respect to the temporal information values across the population. And a new histogram was generated for each random shuffle. K-Means clustering (Supplementary Figure 4e) was performed using the kmeans Matlab function with 2 clusters and Euclidian distance as the clustering parameter.
Defining the Run-Rest Index:
Running and resting periods included in the Run-Rest Index were defined using the same behavioral criteria used for defining spatial cells and temporal cells in the Door Stop tasks and linear track task (see above). The Run-Rest Index for the kth cell was computed as follows:
Where is the calcium transient DF/F value for the kth cell at the ith frame, is the set of all frames that occur during running periods, Rrest is the set of all frames that occur during rest periods. RRI was computed for all active cells (Figure 2b) and all significant temporal and spatial encoding cells (Figure 2e). For data shown in Supplementary Figure 3b, RRI was computed using the same equation, however in this case, Rrun is the set of all frames that occur during “stationary” running periods at the Door Stop location (i.e. running on the treadmill, but the door is closed so the mouse does not move in VR).
To generate a shuffle P-value to compare changes in RRI of cells across environments, in the real data, active cells were included if they had RRIs from either the top or bottom quartiles of the RRI distribution in the first environment (ie. run or rest selective cells). To generate the shuffle distribution, we randomly permuted the RRI values of these cells measured in environment 2, and measured the RRI difference across environments:
Where is the real RRI of the kth cell in environment 1 and is the shuffled RRI of the kth cell in environment 2. We then computed the mean RRI difference across all cells. This process was repeated 1,000 times and the P-value was defined as the ratio of the number of times out of 1,000 that the random shuffled mean RRI difference was smaller than the real mean RRI difference (Supplementary Figure 6n).
Identifying spatial and time encoding cells across days:
In order to identify with certainty, the same individual temporal or spatial encoding cells from recording sessions across days (see Supplementary Figure 6k-m), we compared the neuronal morphology for each ROI across days in FOVs that could be visually matched across days. FOVs were visually matched across days if both FOVs had a similar vasculature pattern, and a similar spatial organization and morphology of neuronal somata. Time or space encoding cells were then matched across days (FOVs) through visual inspection of their morphology (including both soma and dendrites) from frames averaged when the cells were active (see Supplementary Figure 6k,l). This criteria of requiring matching of dendritic morphology was conservative, but attempting to identify the same cells over days based only on the position of the somata often led to mis-identification (based on dendritic morphologies).
Histology
The surface vasculature seen across the micro-prism was recorded using epifluorescence microscopy (Supplementary Figure 1a). The location of each two-photon imaging field in MEC layer 2, with respect to the surface vasculature, was then recorded by comparing the surface vasculature patterned observed using two photon microscopy above (caudal to) the imaging field to the previously recorded epifluorescence pattern. Once imaging experiments in each mouse were complete, the mouse was anesthetized with 1–2% isoflurane and the headplate and prism/mount were removed to expose the caudal surface of MEC. A pin coated with Alexa594 was then inserted perpendicular to the caudal MEC surface at an identified region in the vascular pattern near to the two-photon imaging field locations. The mouse was then euthanized and the brain was removed and fixed in 4% PFA in 0.1M PBS for ~24 hours. The brains were then transferred into a 30% sucrose solution in 0.1M PBS for approximately 2 days until they sank in the solution. The tissue was sectioned in 50 micron sagittal slices using a freezing microtome. Free floating slices were then incubated 0.1M PBS with 0.1% Triton-X for 10 minutes, washing 3 times with 0.1M PBS and incubated for 1 hour in a 25 to 1 solution of 0.1M PBS to 435/455 blue fluorescent Nissl statin (Invitrogen). MEC was identified based the presence of lamina dissecans, the relative position of the post-rhinal border to the pin mark and the circular shape of the dentate gyrus shown at the medial-lateral position of the sagittal sections. Two photon imaging fields were then confirmed to be located in the identified MEC regions based on known distances from the pin mark sites (Supplementary Figure 1a-c).
Supplementary Material
Acknowledgements:
We thank M. Howe for developing the original version of the Door Stop task, J. Climer, M. Hasselmo, M. Howe and J. Issa for useful comments on this manuscript. We thank C. Woolley for use of the freezing microtome, and V. Jayaraman, R. Kerr, D. Kim, L. Looger, K. Svoboda from the GENIE Project (Janelia Farm, Howard Hughes Medical Institute) for GCaMP6. This work was supported by The McKnight Foundation (DAD), The Simons Collaboration on the Global Brain Post-Doctoral Fellowship (JGH), The Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (DAD), The NIH (1R01MH101297; DAD).
Footnotes
Competing Financial Interests Statement:
The authors declare no competing interests.
Code and data availability statement:
The data and custom code that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Scoville WB & Milner B LOSS OF RECENT MEMORY AFTER BILATERAL HIPPOCAMPAL LESIONS. J. Neurol. Neurosurg. Psychiatry 20, 11–21 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corkin S et al. H. M.’s medial temporal lobe lesion: findings from magnetic resonance imaging. J. Neurosci. 17, 3964–3979 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Keefe J & Dostrovsky J The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971). [DOI] [PubMed] [Google Scholar]
- 4.Taube JS, Muller RU & Ranck JB Head-direction cells recorded from the postsubiculum in freely moving rats. I. Description and quantitative analysis. J. Neurosci. 10, 420–35 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Keefe J & Recce ML Phase relationship between hippocampal place units and the EEG theta rhythm. Hippocampus 3, 317–330 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Skaggs WE & McNaughton BL Replay of Neuronal Firing Sequences in Rat Hippocampus During Sleep Following Spatial Experience. Science (80-. ). 271, 1870–1873 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Hafting T, Fyhn M, Molden S, Moser M-B & Moser EI Microstructure of a spatial map in the entorhinal cortex. Nature 436, 801–806 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Steffenach HA, Witter M, Moser MB & Moser EI Spatial memory in the rat requires the dorsolateral band of the entorhinal cortex. Neuron 45, 301–313 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Hasselmo ME A model of episodic memory: Mental time travel along encoded trajectories using grid cells. Neurobiol. Learn. Mem. 92, 559–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meck WH, Church RM & Matell MS Hippocampus, time, and memory—A retrospective analysis. Behav. Neurosci. 127, 642–654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichenbaum H Time cells in the hippocampus: a new dimension for mapping memories. Nat. Rev. Neurosci. 15, 732–744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastalkova E, Itskov V, Amarasingham A & Buzsaki G Internally Generated Cell Assembly Sequences in the Rat Hippocampus. Science (80-. ). 321, 1322–1327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus B, Robinson R, White J, Eichenbaum H & Hasselmo M Hippocampal ‘Time Cells’: Time versus Path Integration. Neuron 78, 1090–1101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald CJ, Lepage KQ, Eden UT & Eichenbaum H Hippocampal ‘time cells’ bridge the gap in memory for discontiguous events. Neuron 71, 737–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald CJ, Carrow S, Place R & Eichenbaum H Distinct Hippocampal Time Cell Sequences Represent Odor Memories in Immobilized Rats. J. Neurosci. 33, 14607–14616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kay K et al. A hippocampal network for spatial coding during immobility and sleep. Nature 531, 185–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arriaga M & Han EB Dedicated Hippocampal Inhibitory Networks for Locomotion and Immobility. J. Neurosci. (2017). doi: 10.1523/JNEUROSCI.1076-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus BJ et al. During Running in Place, Grid Cells Integrate Elapsed Time and Distance Run. Neuron 88, 578–589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinman JR, Brandon MP, Climer JR, Chapman GW & Hasselmo ME Multiple Running Speed Signals in Medial Entorhinal Cortex. Neuron 91, 666–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esclassan F, Coutureau E, Di Scala G & Marchand AR A Cholinergic-Dependent Role for the Entorhinal Cortex in Trace Fear Conditioning. J. Neurosci. 29, 8087–8093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh J, Rivest AJ, Nakashiba T, Tominaga T & Tonegawa S Entorhinal Cortex Layer III Input to the Hippocampus Is Crucial for Temporal Association Memory. Science (80-. ). 334, 1415–1420 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Robinson NTM et al. Medial Entorhinal Cortex Selectively Supports Temporal Coding by Hippocampal Neurons. Neuron (2017). doi: 10.1016/j.neuron.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heys JG, Rangarajan KV & Dombeck DA The functional micro-organization of grid cells revealed by cellular-resolution imaging. Neuron 84, 1079–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solstad T, Boccara CN, Kropff E, Moser M-B & Moser EI Representation of geometric borders in the entorhinal cortex. Science (80-. ). 322, 1865–1868 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kropff E, Carmichael JE, Moser M-B & Moser EI Speed cells in the medial entorhinal cortex. Nature 523, 419–424 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Sheffield MEJ, Adoff MD & Dombeck DA Increased Prevalence of Calcium Transients across the Dendritic Arbor during Place Field Formation. Neuron 96, 490–504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leon MI & Shadlen MN Representation of time by neurons in the posterior parietal cortex of the macaque. Neuron 38, 317–327 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Matell MS, Meck WH & Nicolelis MAL Interval timing and the encoding of signal duration by ensembles of cortical and striatal neurons. Behav. Neurosci. 117, 760–773 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Janssen P & Shadlen MN A representation of the hazard rate of elapsed time in macaque area LIP. Nat. Neurosci. 8, 234–241 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Jin DZ, Fujii N & Graybiel AM Neural representation of time in cortico-basal ganglia circuits. Proc. Natl. Acad. Sci. 106, 19156–19161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhlaghpour H et al. Dissociated sequential activity and stimulus encoding in the dorsomedial striatum during spatial working memory. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares S, Atallah BV & Paton JJ Midbrain dopamine neurons control judgment of time. Science (80-. ). 354, 1273–1277 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Narain D, Hosseini EA & Jazayeri M Flexible timing by temporal scaling of cortical responses. Nat. Neurosci. (2018). doi: 10.1038/s41593-017-0028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauk MD & Buonomano DV THE NEURAL BASIS OF TEMPORAL PROCESSING. Annu. Rev. Neurosci. 27, 307–340 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Donato F, Jacobsen RI, Moser M-B & Moser EI Stellate cells drive maturation of the entorhinal-hippocampal circuit. Science (80-. ). 355, eaai8178 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Chklovskii DB, Schikorski T & Stevens CF Wiring optimization in cortical circuits. Neuron 34, 341–347 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Burak Y & Fiete IR Accurate path integration in continuous attractor network models of grid cells. PLoS Comput. Biol. 5, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couey JJ et al. Recurrent inhibitory circuitry as a mechanism for grid formation. Nat. Neurosci. 16, 318–24 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Matell MS & Meck WH Neuropsychological mechanisms of interval timing behavior. BioEssays 22, 94–103 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Sussillo D & Abbott LF Generating Coherent Patterns of Activity from Chaotic Neural Networks. Neuron 63, 544–557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laje R & Buonomano DV Robust timing and motor patterns by taming chaos in recurrent neural networks. Nat. Neurosci. 16, 925–933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solstad T, Moser EI & Einevoll GT From grid cells to place cells: A mathematical model. Hippocampus 16, 1026–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Zhang S-J et al. Optogenetic Dissection of Entorhinal-Hippocampal Functional Connectivity. Science (80-. ). 340, 1232627–1232627 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Miao C et al. Hippocampal Remapping after Partial Inactivation of the Medial Entorhinal Cortex. Neuron 88, 590–603 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Kanter BR et al. A Novel Mechanism for the Grid-to-Place Cell Transformation Revealed by Transgenic Depolarization of Medial Entorhinal Cortex Layer II. Neuron 93, 1480–1492.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Buhusi CV & Meck WH What makes us tick? Functional and neural mechanisms of interval timing. Nat. Rev. Neurosci. 6, 755–765 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Eichenbaum H On the Integration of Space, Time, and Memory. Neuron (2017). doi: 10.1016/j.neuron.2017.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitamura T et al. Island cells control temporal association memory. Science 343, 896–901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hitti FL & Siegelbaum SA The hippocampal CA2 region is essential for social memory. Nature 508, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diehl GW, Hon OJ, Leutgeb S & Leutgeb JK Grid and Nongrid Cells in Medial Entorhinal Cortex Represent Spatial Location and Environmental Features with Complementary Coding Schemes. Neuron 94, 83–92.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dombeck DA, Harvey CD, Tian L, Looger LL & Tank DW Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 13, 1433–1440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harvey CD, Collman F, Dombeck DA & Tank DW Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dombeck DA, Graziano MS & Tank DW Functional clustering of neurons in motor cortex determined by cellular resolution imaging in awake behaving mice. J. Neurosci. 29, 13751–13760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heys JG, Rangaranjan KV & Dombeck DA The Functional Micro-organization of Grid Cells Revealed by Cellular-Resolution Imaging. Neuron 84, 1079–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giocomo LM et al. Grid cells use HCN1 channels for spatial scaling. Cell 147, 1159–1170 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Mukamel EA, Nimmerjahn A & Schnitzer MJ Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron 63, 747–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dombeck DA, Khabbaz AN, Collman F, Adelman TL & Tank DW Imaging Large-Scale Neural Activity with Cellular Resolution in Awake, Mobile Mice. Neuron 56, 43–57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheffield MEJ & Dombeck DA Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature 517, 200–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skaggs William E. and McNaughton Bruce L. and Gothard Katalin M. and Markus Etan J.. An Information-Theoretic Approach to Deciphering the Hippocampal Code. Proc. IEEE (1993). doi: 10.1109/PROC.1977.10559 [DOI] [Google Scholar]
- 60.Hainmueller T & Bartos M Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature 1 558, Ahead of eprint (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


