Abstract
Rationale: Obstructive sleep apnea (OSA) is associated with recurrent obstruction, subepithelial edema, and airway inflammation. The resultant inflammation may influence or be influenced by the nasal microbiome.
Objectives: To evaluate whether the composition of the nasal microbiota is associated with obstructive sleep apnea and inflammatory biomarkers.
Methods: Two large cohorts were used: 1) a discovery cohort of 472 subjects from the WTCSNORE (Seated, Supine and Post-Decongestion Nasal Resistance in World Trade Center Rescue and Recovery Workers) cohort, and 2) a validation cohort of 93 subjects rom the Zaragoza Sleep cohort. Sleep apnea was diagnosed using home sleep tests. Nasal lavages were obtained from cohort subjects to measure: 1) microbiome composition (based on 16S rRNA gene sequencing), and 2) biomarkers for inflammation (inflammatory cells, IL-8, and IL-6). Longitudinal 3-month samples were obtained in the validation cohort, including after continuous positive airway pressure treatment when indicated.
Measurements and Main Results: In both cohorts, we identified that: 1) severity of OSA correlated with differences in microbiome diversity and composition; 2) the nasal microbiome of subjects with severe OSA were enriched with Streptococcus, Prevotella, and Veillonella; and 3) the nasal microbiome differences were associated with inflammatory biomarkers. Network analysis identified clusters of cooccurring microbes that defined communities. Several common oral commensals (e.g., Streptococcus, Rothia, Veillonella, and Fusobacterium) correlated with apnea–hypopnea index. Three months of treatment with continuous positive airway pressure did not change the composition of the nasal microbiota.
Conclusions: We demonstrate that the presence of an altered microbiome in severe OSA is associated with inflammatory markers. Further experimental approaches to explore causal links are needed.
Keywords: microbiome, inflammation, chronic rhinosinusitis, biomarkers
At a Glance Commentary
Scientific Knowledge on the Subject
Obstructive sleep apnea (OSA) is a disease characterized by intermittent obstruction of the upper airway, which may be associated with increased inflammation. Nasal microbiota interacts with host immune response in the airway mucosa and may have a significant role on mucosal immune tone. Host immune tone, in turn, may subject nasal microbiota to selection pressure.
What This Study Adds to the Field
We identified nasal microbiota signatures associated with OSA that were consistent across two separate large cohorts. These microbiota signatures were associated with increased inflammatory biomarkers and did not change after treatment with continuous positive airway pressure.
Obstructive sleep apnea (OSA) is characterized by recurrent episodes of upper airway collapse during sleep and has an estimated prevalence ranging from 34% to 50% in men and 17% to 23% in women (1, 2). The pathology of OSA arises from the physiological effects of sleep on the upper airway and may include derangements of the upper airway mucosa, tissue, and oropharyngeal muscles (3–7). Both local and systemic inflammation have been reported in OSA, as well as immune response changes. In OSA, local inflammation of the upper airway could lead to increased airway resistance (7–9); for example, the repetitive closure of the narrowed upper respiratory airway can lead to increased subepithelial edema, serum inflammatory markers, and inflammatory cell infiltration (10–12). The nasal mucosa may also play an important role in OSA, as chronic nasal obstruction and congestion are risk factors for snoring and OSA (13).
The nasal cavity is a microbial–host interface that affects the mucosal immunological tone (14). For example, the colonization by Staphylococcus epidermidis in healthy hosts enhances host defense mechanisms against infection with influenza (15), and interspecies competition drives immune responses with complement-dependent phagocytic killing of Streptococcus pneumoniae (16). Furthermore, host immune factors, including antimicrobial peptides, provide differential selection pressures on the nasal microbiota, thereby potentially affecting its composition (17, 18), and host genetic factors are associated with changes to the nasal microbiome (19).
We therefore hypothesize that differences in host–microbial associations will be identified among subjects with OSA. To test this, we evaluated the association between OSA, the nasal microbiome, and inflammatory markers in a discovery multicenter cohort and confirmed the results using a validation cohort. In addition, we assessed longitudinal changes in the nasal microbiome in the validation cohort, including a subgroup of subjects with OSA treated with continuous positive airway pressure (CPAP) for 3 months.
Methods
Study Design and Participants
For this investigation, we used two prospective cohorts in whom sleep studies were conducted to diagnose OSA and nasal lavages were obtained. The discovery cohort was recruited from two sites of the World Trade Center (WTC) Health Program, New York University (NYU) and Rutgers, where 472 subjects exposed to World Trade Center dust in 2001 were enrolled (20). The research protocols were independently approved by the NYU and Rutgers University Institutional Review Boards (NYU IRB# i12–02578 and Rutgers IRB# Pro2012002164). The validation cohort consisted of 93 consecutive subjects with suspected OSA referred to the sleep center at the Hospital Miguel Servet (Zaragoza, Spain) (21). No patient was treated with positive airway pressure at baseline. Study procedures were approved by the Ethics and Clinical Research Committee of the Aragón Institute of Health Sciences (protocol #10/231), and informed consent was obtained from each participant. Detailed inclusion and exclusion criteria are provided in the online supplement.
Diagnosis of OSA
All subjects underwent home sleep testing. For the discovery cohort, subjects were issued an ARES Unicorder to take home and wear for two nights (SleepMed, Inc.). The validation cohort used the BITMED NGP 140 (Meditel Ingeniería Médica) for one night (22). Trained personnel manually scored polygraph data in accordance with American Academy of Sleep Medicine guidelines (23). Severity of sleep apnea was categorized by standard American Academy of Sleep Medicine criteria: no OSA (apnea–hypopnea index [AHI4] < 5 events/h), mild OSA (AHI4, 5–14 events/h), moderate OSA (AHI4, 15–29 events/h), and severe OSA (AHI4 ≥ 30 events/h) (24).
Nasal Lavage
From both cohorts, nasal lavage samples were collected by trained personnel. For the discovery cohort, a total of 8 ml of sterile saline was instilled, and returned nasal lavage was collected (25). The sample was immediately placed on ice and processed within 2 hours. For the validation cohort, nasal lavage was collected by instilling 10 ml of sterile saline. This was repeated for a total of three times, and each lavage was collected into the same sterile container (see online supplement for further details).
Longitudinal Samples in the Validation Cohort
Longitudinal nasal lavage samples were obtained in all subjects from the validation cohort 3 months apart from the baseline. A subgroup of subjects with OSA (1 mild, 4 moderate, and 17 severe OSA) were treated during those 3 months with CPAP.
Measurement of Inflammatory Markers in Nasal Lavage Fluid
For the discovery cohort, nasal lavage fluid was filtered through a 40-μm nylon mesh syringe filter to remove larger particles. An aliquot of whole nasal lavage fluid was separated for microbiome analysis. The remaining sample was then centrifuged at 500g for 10 minutes at 4°C. The cell-free supernatants were aliquoted and stored at −80°C for later analyses of soluble markers. Cells from the pellet were resuspended in 1 ml of a buffered salt solution. Cell counts were performed on a hemocytometer. For the differential cell counts, cytocentrifuge slides were prepared, fixed, and stained with a Wright-Giemsa stain. The cell-free supernatants were aliquoted and stored at −80°C for further analyses of inflammatory cytokines. Cytokines (IL-8 and IL-6) were measured using high-sensitivity ELISA (BD Biosciences; Cat #BDB550999 and Cat #BDB550799, respectively), and values were expressed in pg/ml. For the validation cohort, nasal lavage fluid was spun down (10 min, 1,700 rpm) to separate cell-free fluid from the cell pellet. The cell pellet was resuspended into sterile saline and recentrifuged (5 min, 800 rpm). Lymphocytes were counted by flow cytometry using anti-CD3 antibody (fluorescein isothiocyanate; BD Biosciences), and images were acquired in a FacsCanto (BD Biosciences). In addition, IL-8 and IL-6 concentrations were measured in the cell-free nasal lavage fluid using Singleplex Luminex Protein Assays (Affymetrix; Cat #EPX010-10204-901 and Cat #EPX010-10213-901, respectively; see online supplement for more details).
Bacterial 16S rRNA Gene Marker Quantitation and Sequencing
For the discovery cohort, whole nasal lavages from all 472 subjects and 16 sterile saline samples from different sterile saline batches used to perform nasal lavages were collected. For the validation cohort, whole nasal lavages (paired baseline and post 3 mo) from all 93 subjects and 10 sterile saline sample control samples were collected. For both cohorts, samples were sent to NYU, where they were processed and sequenced in pool.
The 16S rRNA gene sequences were analyzed using the Quantitative Insights into Microbial Ecology (QIIME version 1.9.1) (26). Sequences were clustered into operational taxonomic units using a 97% similarity threshold with UCLUST (27) and the Greengenes 16S reference dataset (28). Sequence data are available in Sequence Read Archive (SRA, discovery cohort = PRJNA419002, validation cohort = PRJNA419003; see online supplement for more details).
Statistical Analysis
For association with discrete factors, we used the Mann-Whitney test (in the case of two categories) or the Kruskal-Wallis ANOVA (in case of more than two categories). Paired nonparametric statistics (Wilcoxon rank sum test) was used for comparison between longitudinal samples (baseline and post 3 mo) obtained in the validation cohort. For distribution of frequencies we used chi-square analysis to assess for differences in the presence or absence of population differences in our cohorts. Multivariate linear regression using clinical covariates (e.g., age, body mass index [BMI], sex, AHI4, and smoking status) was performed using microbiota signatures identified as associated with OSA as outcome (SPSS version 23.0; IBM). For tests of association with continuous variables, we used nonparametric Spearman (rho) correlation tests. To evaluate differences in community composition between groups on the basis of 16S data, we used permutational multivariate ANOVA (PERMANOVA). To evaluate for taxonomic differences between groups, we used linear discriminant analysis combined with effect size (LEfSe) (29). Correlations between taxa were calculated using SparCC (30). Clinical and inflammatory variables were then correlated with the taxa in the cooccurrence network and displayed using Cytoscape v3.6.0 (31). Thus, the cooccurrence network is presenting information on how different bacteria, biomarkers, and AHI4 are correlated: solid lines represent positive correlations (blue: among bacteria; yellow: between bacteria and clinical/inflammatory markers), and dashed lines are negative correlations.
Results
Description of the Discovery and Validation Cohorts
A total of 472 subjects were enrolled in the discovery cohort and 93 subjects in the validation cohort. Table 1 shows demographics and clinical characteristics of both of these cohorts. In the discovery cohort, there was high prevalence of OSA (304 of 472 subjects; 65.8%), with 9.5% of these subjects categorized as having severe OSA. As expected, OSA was associated with older age, male sex, and higher BMI (see Table E1 in the online supplement). However, in the validation cohort, there was higher prevalence of OSA (68 of 93 subjects; 73.1%), as expected for subjects being enrolled from a sleep consultation clinic. Forty-five percent of the subjects in the validation cohort were categorized as having severe OSA. In the validation cohort, the subjects were younger and had lower BMI than the discovery cohort (Table 1). As per design, no subject in the validation cohort were smokers or on medications. The discovery cohort had more subjects with mild OSA (P = 0.002), and the validation cohort had subjects with more severe OSA (Table 1; P < 0.0001).
Table 1.
Demographics and Clinical Characteristics in Nasal Lavage of the Discovery and Validation Cohorts
| Discovery Cohort | Validation Cohort | P Value | |
|---|---|---|---|
| n | 472 | 93 | |
| Age, yr | 52 (47–58) | 45 (35–54) | <0.0001* |
| Sex, male | 388 (82.2) | 71 (76.3) | ns† |
| BMI, kg/m2 | 29 (26–32) | 28 (25–30) | 0.004* |
| Smokers | 46 (9.7) | 0 (0) | 0.0017† |
| Medications | |||
| Nasal steroid | 45 (9.5) | 0 (0) | 0.0019† |
| Inhaled steroid | 17 (3.6) | 0 (0) | ns† |
| Oral steroid | 6 (1.2) | 0 (0) | ns† |
| Antihistamine | 36 (7.6) | 0 (0) | 0.005† |
| Bronchodilator | 59 (12.5) | 0 (0) | 0.0003† |
| PPI | 95 (20.1) | 0 (0) | <0.0001† |
| OSA data | |||
| No OSA | 168 (35.6) | 25 (26.9) | ns† |
| Mild OSA | 172 (36.4) | 19 (20.4) | 0.002† |
| Moderate OSA | 87 (18.4) | 18 (19.4) | ns† |
| Severe OSA | 45 (9.5) | 31 (33.3) | <0.0001† |
Definition of abbreviations: BMI = body mass index; IQR = interquartile range; ns = not significant; OSA = obstructive sleep apnea; PPI = proton pump inhibitors.
All values are expressed as median (IQR) or total count (% of column total).
Mann-Whitney calculations.
Chi-square calculations.
Microbiota Signatures in OSA
The overall bacterial burden of nasal lavages from the discovery and validation cohorts was evaluated using quantitative PCR for the 16S rRNA gene. No differences in bacterial load were noted between subjects with no OSA and subjects with various degrees of OSA severity (Figure E1; Kruskal-Wallis P = not significant [ns]).
In the discovery cohort, evaluation of the microbiota present in the sterile saline used to perform the nasal lavages at NYU and Rutgers showed no significant β diversity differences between sites or differential clustering (Figure E2). Figure E3 shows a heatmap based on most abundant taxa (relative abundance > 2% in at least 10% of the samples) in nasal lavage samples. In this cohort, nasal microbiota frequently had high relative abundance of Staphylococcus and Corynebacterium, consistent with prior studies of nasal microbiota (32). Subclusters were enriched with Streptococcus, Prevotella, Neisseriaceae (undefined genus [u.g.]), and Veillonella. Similarly, evaluation of the nasal microbiota in the validation cohort demonstrated that most samples had high relative abundance of Staphylococcus and Corynebacterium (Figure E4), and a distinct cluster of samples were enriched with Streptococcus, Prevotella, and Veillonella.
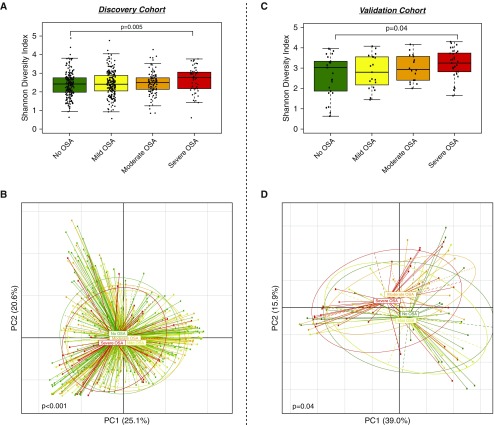
We then evaluated for differences in microbiome between subjects with no OSA compared with subjects with different OSA severity (defined by AHI4). Analysis of α diversity showed that the severe OSA group had increased Shannon diversity index (SDI) compared with the no OSA group (Figure 1A; Mann-Whitney P = 0.005). SDI is a marker of richness (the number of taxa) and evenness (the distribution of taxa) in a sample. Analysis of β diversity on the basis of weighted UniFrac distance demonstrated significant differences between OSA severity groups (Figure 1B; PERMANOVA P < 0.001). Weighted UniFrac distance is one marker to describe degree of compositional similarity between different samples (the fewer shared taxa between samples, the less likely they will cluster together, and therefore significantly different). Similarly, in nasal lavage samples from this cohort, there was a significantly higher SDI among subjects with severe OSA and significant compositional differences in β diversity (Figures 1C and 1D).
Figure 1.
Similar trends in α and β diversity parameters for obstructive sleep apnea (OSA) diagnosis in the discovery and validation cohorts. (A) Shannon diversity index (SDI) differences between subjects with mild OSA, moderate OSA, severe OSA, and no OSA in the discovery cohort. Higher α diversity was noted in the group with severe OSA compared with subjects with no OSA (Mann-Whitney P = 0.005). (B) Principal coordinate analysis on the basis of weighted UniFrac distances for groups of severity of OSA and no OSA subjects (permutational multivariate ANOVA [PERMANOVA] P < 0.001) in the discovery cohort. (C) α diversity differences (based on SDI) in the validation cohort. The severe OSA group had higher α diversity than subjects with no OSA (Mann-Whitney P = 0.04). (D) Principal coordinate analysis on the basis of weighted UniFrac shows significant differences between subjects with different severity of OSA and subjects with no OSA (PERMANOVA P = 0.04) in the validation cohort. PC = principal coordinate.
Taxonomic Differences among OSA Groups
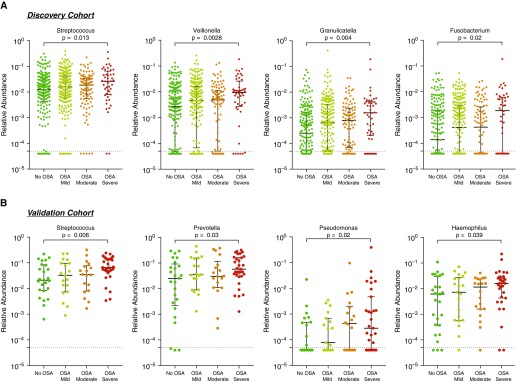
Taxonomic differences between OSA groups versus subjects with no OSA were explored. LEfSe identified multiple taxonomic differences in the nasal microbiota of subjects with mild, moderate, and severe OSA (Figure E5) compared with the nasal microbiota of subjects with no OSA in both discovery and validation cohorts. In the discovery cohort, the nasal microbiota of severe OSA was enriched with Streptococcus, Veillonella, Granulicatella, and Fusobacterium (Figure 2A), taxa commonly considered oral commensals. Similarly, in the validation cohort, the nasal microbiota of subjects with severe OSA was enriched with oral commensals (e.g., Streptococcus, Prevotella, Pseudomonas, and Haemophilus) compared with the nasal microbiota of subjects with no OSA (Figure 2B).
Figure 2.
Taxonomic differences between severe obstructive sleep apnea (OSA) diagnosis and subjects with no OSA in the discovery and validation cohorts show oral commensal enrichment. (A) Comparison of relative abundance of top differentially enriched taxa in the discovery cohort. The nasal microbiota from subjects with severe OSA was enriched with Streptococcus, Veillonella, Granulicatella, and Fusobacterium when compared with samples from subjects with no OSA (Mann-Whitney). (B) Comparison of relative abundance of top differentially enriched taxa in the validation cohort. The nasal microbiota from subjects with severe OSA was enriched with Streptococcus, Prevotella, Pseudomonas, and Haemophilus when compared with samples from subjects with no OSA (Mann-Whitney). Samples with taxa less than 5.0 × 10−5 relative abundance of specific taxa were considered below the lower limit of detection (dotted line).
Multivariate Analysis for Microbiota Signatures Identified in Severe OSA
To evaluate the independent strength of the associations between OSA and the microbiota signatures identified above, we performed multivariate linear regression analysis adjusting for age, BMI, sex, and smoking status (Table E2). Of note, significant collinearity can be identified between AHI4, age, sex, and BMI (data not shown). In the discovery cohort, AHI4 predicted increases in the SDI and relative abundance of Streptococcus independent of the effects of age, BMI, smoking, and sex. In the validation cohort, higher AHI4 was associated with higher relative abundance of Streptococcus and Haemophilus independent of the effects of age, sex, and BMI.
Inflammatory Biomarkers and Nasal Microbiota
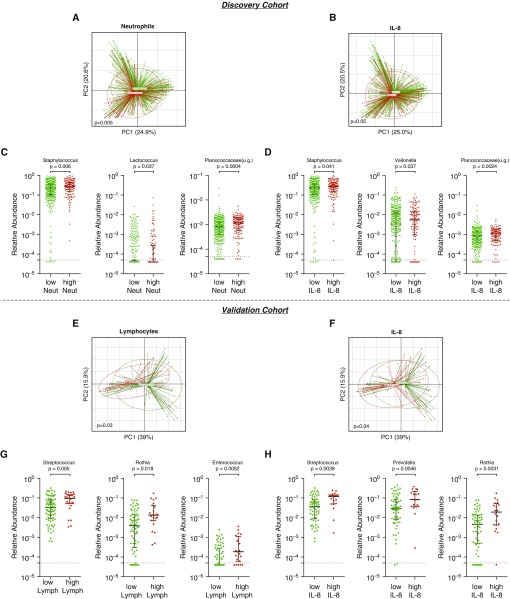
Inflammatory biomarkers were measured in the nasal lavages of both cohorts. Table E3 depicts these levels of biomarkers among different OSA categories. In the discovery cohort, levels of IL-8 and IL-6 in nasal lavage were higher among subjects with OSA than subjects without OSA (P < 0.01). Levels of IL-8 significantly correlated with concentration of neutrophils in the nasal lavages (Spearman rho = 0.609, P < 0.0001). Then, we evaluated microbiota changes associated with inflammatory biomarkers by grouping samples with high and low values of these biomarkers. In the discovery cohort, there were no significant differences in SDI between high versus low neutrophils, IL-6, or IL-8 levels (data not shown). Principal coordinate analysis based on weighted UniFrac showed significant differences in nasal microbiota composition between high versus low neutrophils, IL-8, and IL-6 levels (Figures 3A, 3B, and E7A, respectively; PERMANOVA P < 0.05 for all comparisons). LEfSe analysis identified top differentially enriched taxa (Figure E6). The nasal microbiota of subjects with high neutrophil count was enriched with Staphylococcus, Planococcaceae(u.g.), and Lactococcus, whereas high IL-8 levels were associated with enrichment with Staphylococcus, Veillonella, and Planococcaceae(u.g.) (Figures 3C and 3D). The nasal microbiota of subjects with high IL-6 was enriched with Moraxella (Figure E7A).
Figure 3.
Changes in nasal microbiota associated with levels of inflammatory markers in the discovery and validation cohorts. (A) β diversity differences between high and low neutrophils in the discovery cohort (permutational multivariate ANOVA [PERMANOVA] P < 0.005). (B) β diversity differences between high and low IL-8 levels in the discovery cohort (PERMANOVA P < 0.05). (C) The nasal microbiota of lavages with high neutrophils was enriched with Staphylococcus, Lactococcus, and Planococcaceae(u.g.) (Mann-Whitney). (D) The nasal microbiota of lavages with high IL-8 was enriched with Staphylococcus, Veillonella, and Planococcaceae(u.g.) (Mann-Whitney). (E) β diversity differences between high and low lymphocyte levels in the validation cohort (PERMANOVA P < 0.03). (F) Significant differences were noted in β diversity when comparing high versus low IL-8 (PERMANOVA P = 0.04). (G) The nasal microbiota of lavages with high lymphocytes was enriched with Streptococcus, Rothia, and Enterococcus (Mann-Whitney). (H) The nasal microbiota of lavages with high IL-8 was enriched with Streptococcus, Prevotella, and Rothia (Mann-Whitney). Samples with taxa less than 5.0 × 10−5 relative abundance were considered below the lower limit of detection (dotted line). lymph = lymphocytes; neut = neutrophils; PC = principal coordinate; u.g. = undefined genus.
In the validation cohort, lymphocyte count and IL-8 levels in nasal lavage were higher among subjects with OSA (Table E3; P < 0.01). Levels of IL-8 were significantly correlated with lymphocytes found in nasal lavage via flow cytometry (Spearman rho = 0.328; P = 0.003). In this cohort, α diversity (SDI) of nasal microbiota of subjects with high lymphocyte percentage was higher than nasal microbiota of subjects with low lymphocyte percentage (Figure E9; Mann-Whitney P = 0.005). Significant differences were noted in β diversity of the nasal microbiota of subjects with high versus low lymphocyte percentage (Figure 3E; PERMANOVA P = 0.03) and IL-8 levels (Figure 3F; PERMANOVA P = 0.04). No significant difference in α or β diversity was noted between samples with high versus low IL-6 (data not shown). LEfSe analysis shows differential enrichment in microbiota between groups of high versus low lymphocyte count (Figure E6C) and high versus low IL-8 (Figure E6D). In nasal lavage samples from this cohort, analysis showed that samples with high lymphocyte count were enriched with Streptococcus, Rothia, and Enterococcus (Figure 3G). The validation cohort samples with high IL-8 were enriched with Streptococcus, Prevotella, and Rothia (Figure 3H). The high IL-6 nasal lavage samples were enriched with Brochothrix (Figures E7B and E8B). Thus, the observations made across the discovery and validation cohorts suggest an association between enrichment of oral commensal taxa in the nasal microbiota, inflammation, and OSA.
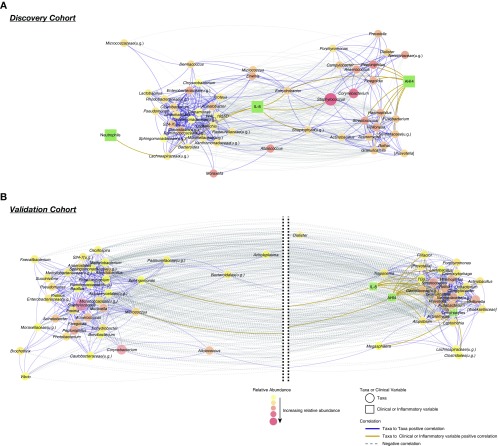
We then built a cooccurrence network of most abundant taxa (at the genus level), inflammatory markers, and AHI4 (Figure 4). Essentially, this multidimensional analysis explores how different bacteria, biomarkers, and AHI4 are correlated. The graphical representation is such that highly positive correlated features (either taxa, inflammatory markers, or AHI4) will be displayed in close proximity. In contrast, features highly negatively correlated will be displayed farther apart. In the discovery cohort, this network analysis identified taxa that tend to cooccur, defining metacommunities within the nasal microbiota (Figure 4A). One such metacommunity was characterized by cooccurrence between Acinetobacter, Moraxella, Lactobacillus, and Pseudomonas. A separate metacommunity was characterized by cooccurrence of oral commensals such as Streptococcus, Veillonella, Prevotella, Rothia, Porphyromonas, and Campylobacter. Among the significant correlations with inflammatory biomarkers (false discovery rate < 0.2), IL-8 were correlated with Lachnospiraceae(u.g.), Bacteroides, and Staphylococcus, whereas neutrophils correlated with Lachnospiraceae(u.g.). AHI4 correlated with multiple oral commensals such as Streptococcus, Veillonella, Fusobacterium, and Granulicatella. A similar cooccurrence network built using data from the validation cohort supported observations that oral commensals were associated with AHI4 and included significant associations with lymphocytes and IL-8 (Figure 4B).
Figure 4.
The cooccurrence network between taxa, apnea–hypopnea index (AHI4), and inflammatory biomarkers from the discovery and validation cohorts. Cooccurrence network for the top 50 genus-level taxa (>2% relative abundance in at least 10% of the samples) built using SparCC using data from both cohorts. Genera (circles) were correlated with AHI4 and inflammatory biomarkers (squares), and significantly correlated variables were kept in the network (false discovery rate < 0.20). Relative abundance of genera is represented by the color and size of the circles. Positive correlations between taxa are represented by solid blue edges and the length of edges calculated as 1 − rho. Negative correlations are dotted gray edges, and the length of the edges calculated as the absolute rho. Therefore, nodes in close proximity tend to cooccur in the community. Correlations of genera with AHI4 and inflammatory biomarkers are represented by gold edges. Cytoscape 3.6.0 was used to visualize the network with a prefuse force-directed layout. (A) The cooccurrence network using data from the discovery cohort shows differential clustering of taxa. Oral commensals, such as Streptococcus, Fusobacterium, Rothia, Veillonella, and Prevotella, tended to cooccur and correlate with AHI4. (B) The cooccurrence network using data from the validation cohort demonstrates similar cooccurrence pattern. This network showed two well-defined large clusters (note that break between the two clusters is represented by dotted black lines and was introduced to facilitate visualization). Oral commensals cooccurred with AHI4 and inflammatory markers (lymphocytes and IL-8). u.g. = undefined genus.
Effects of Treatment of OSA with CPAP on the Nasal Microbiota
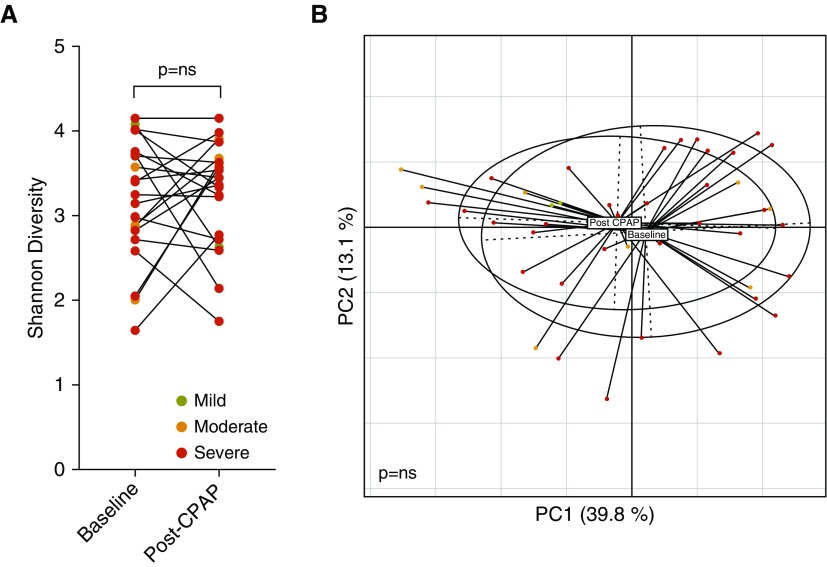
To evaluate this, we used the validation cohort, where nasal lavage sampling was repeated after 3 months in both subjects with no OSA and the subjects with OSA. A subgroup of subjects with OSA underwent treatment with CPAP. Evaluation of the longitudinal change in nasal microbiota for subjects with no OSA and three groups of OSA severity showed no statistically significant differences in α or β diversity (Wilcoxon rank sum P = ns and PERMANOVA P = ns, respectively; Figures E10 and E11). The subgroup of subjects who underwent CPAP treatment also did not show statistically significant differences in either α or β diversity (Figure 5A,Wilcoxon rank sum P = ns, and Figure 5B, PERMANOVA P = ns, respectively).
Figure 5.
Treatment with continuous positive airway pressure (CPAP) does not significantly alter the nasal microbiome. Longitudinal change of nasal microbiota comparing baseline samples with samples obtained after 3 months on treatment with CPAP showed no significant difference in (A) α diversity (Wilcoxon rank test P = ns), and (B) β diversity (permutational multivariate ANOVA P = ns). ns = not significant; PC = principal coordinate.
Discussion
This study shows that the nasal mucosa of subjects with severe OSA contains distinct microbiota and inflammatory signatures. This study was conducted using two well-characterized cohorts: one involving a large number of subjects without a history of snoring before exposure to World Trade Center dust used as a discovery cohort and a second validation cohort characterized by subjects without significant comorbidities who were sent for a sleep consultation. These cohorts represent the largest study of nasal microbiome to date. In both cohorts, sampling the nasal mucosa using nasal lavages identified microbiota changes associated with OSA and with inflammatory markers. The microbes identified as associated with severe OSA were frequently human oral commensals, such as Streptococcus, Prevotella, Veillonella, and Porphyromonas. The association between nasal microbiota, local inflammation, and OSA was corroborated in the validation cohort, reinforcing the generalizability of this study.
The anterior nares are a microbial niche characterized by keratinized epithelium in close proximity to the nasopharynx and skin. The mucosa in the posterior nares is characterized by a pseudostratified epithelium that shares multiple features with the lower respiratory airways (33). The nasal cavity is the first interface between the external environment and the airway mucosa, receiving first-hand the exposure to airborne toxins, particulate matter, and airborne microbes. Prior reports have shown changes in the nasal microbiota, evaluated by culture-dependent and -independent methods, are associated with the inflammatory tone in the host (34–36). Nasal colonization with Moraxella catarrhalis, Corynebacterium pseudodiphtheriticum, Streptococcus agalactiae, and S. pneumoniae are associated with increased risk of chronic rhinosinusitis (37–39). Surgical brushings show taxonomically and functionally unique microbial composition that is associated with increased proinflammatory T-helper cell type 1 response and nasal polyps (40). Newer longitudinal data demonstrate that in infants the composition of the microbiota of the nasopharynx is associated with increased susceptibility to respiratory tract infections (41), thus supporting the important role of the nasal microbiota and host immunological tone.
In our investigation, OSA was associated with changes in nasal microbiota found in α and β diversity, most notably in the severe OSA group. In these subjects, the nasal microbiota was enriched with several taxa that represent commensals frequently found in the oral cavity, such as Streptococcus, Veillonella, Granulicatella, Gemella, and Prevotella. Some of the taxa were also associated with biomarkers of increased inflammation in the nasal lavage. The detection of oral commensals in the nasal lavage of subjects with severe OSA could be due to recurrent obstruction during sleep causing reflux of oropharyngeal secretions that otherwise would be swallowed in a healthy subject. Also, biofilms constitute a hypoxic environment across multiple sites of the aerodigestive tract and have long been described in the sinus and nasal cavity (42–44). Thus, it is possible that biofilms provide a distinct environment that may favor facultative anaerobes, such as some of the taxa identified in the severe OSA group (42). In addition, the nasal mucociliary system is significantly deteriorated in patients with severe OSA (45) and may play a role in the nasal microbiome changes seen in these subjects.
It is possible that differences in the inflammatory tone present in OSA exert selection pressure on resident microbiota in the nasal mucosa. Alternatively, whether the dysbiotic signature identified (enrichment with oral commensals in the nasal lavage) causes mucosal inflammation remains to be tested, but both scenarios would be consistent with an increased inflammatory tone reported in OSA. Increased nasal inflammatory changes have been well described in OSA, such as the increase in nitric oxide, lipid peroxidation, nuclear factor-κB activation, and neutrophils in the nasal passages (46–48). Our investigation also found an association between OSA and increased inflammatory biomarkers: neutrophils, lymphocytes, IL-6, and IL-8. In both discovery and validation cohorts, several of these inflammatory biomarkers were also associated with a unique microbiome profile (although not identical between the two cohorts). Furthermore, colonization of the upper airway with these organisms may contribute to the inflammatory process (37) and to subepithelial edema (49), perpetuating a cycle of colonization, upper airway obstruction during sleep, reflux, and worsening inflammation.
Microbiome alterations can occur in different mucosae in OSA and have systemic “spill-over” effects that may contribute to comorbidities prevalent in this disease. For example, in a rat model, gut dysbiosis induced by changes in diet causes hypertension (50). Intermittent hypoxia and hypercapnia, prominent in OSA, may accelerate atherosclerosis by increasing levels of gut microbial metabolites such as trimethylamine oxide (51, 52). The upper airway dysbiosis identified in the current investigation could be a marker of dysbiosis occurring in different mucosae. Further investigation should focus on the systemic effects of upper airway dysbiosis on the systemic inflammatory tone and on the spill-over of microbial metabolites into systemic circulation in OSA.
Other factors that could affect the nasal microbiome in OSA include treatment with PAP either through pressure or contact with the mask interface (53). In the validation cohort, we were able to assess the effects of CPAP treatment on the nasal microbiota, and we did not identify significant differences in α or β diversity. Differences in delivery systems may be relevant. For example, most patients used nasal masks and not nasal pillows, and none were on full face mask. Given the small number of subjects, it is possible that the study is not powered to dissect differences within subgroups. It is also possible that 3 months of treatment is not sufficient to affect the nasal microbiota. Alternatively, CPAP may improve the mechanical obstruction but may not alter the inflammatory tone in the nasal cavity (such as what has been shown in systemic circulation) (54) or may even represent a “second hit,” where the pressure in the upper airway system may lead to a shear stress on the nasal mucosa and actually contribute to the inflammatory milieu affecting the nasal microbiota. Future investigations should evaluate the effects of CPAP on the upper airway microbiota by sampling both the environment (e.g., water reservoir, nasal mask) and the upper airways before and after initiation of treatment. Last, it is possible that other confounders might affect the observations found in the study. In this investigation, we attempted to address this with some of the confounders (age, sex, BMI, and smoking) that we could adjust for in a multivariate analysis. The collinearity existing between these covariates likely weakened the strength of the associations. However, the independent nature of the association between OSA (as determined by AHI4) and microbiota signatures could be demonstrated for SDI and Streptococcus. In addition, some of these possible confounders were also present in the mild OSA and moderate OSA groups, where the microbiota signatures associated with severe OSA were not present.
The strengths of this study include the use of two large well-characterized cohorts, the consistent diagnosis of OSA, and quantification of inflammatory biomarkers. However, there are several limitations to note. For the discovery cohort, we used a convenience cohort of subjects with high prevalence of OSA who had been previously exposed to WTC dust more than a decade ago. This cohort did not include subjects without WTC dust exposure. However, in the validation cohort, we observed similar associations between microbiota, OSA, and inflammation; thus, we believe that the observations made in this investigation are generalizable. The use of nasal lavage may be a limited way to sample the upper airway, where multiple microbial niches, such as the anterior nasopharynx, posterior nasopharynx, oral cavity, and supraglottic space, exist in close proximity. The nasal lavage approach used in this investigation may preferentially sample the anterior nasopharynx. Other upper airway mucosae may have distinct microbiota signatures associated with OSA, and future investigations will require a more comprehensive topographical approach that considers crosspollination of microbes between different mucosae. Furthermore, prior studies have shown that the nasal mucosa hosts a complex viral and fungal community (55); thus, evaluation of the virome and mycobiome should be considered in future studies. Differences between the sampling methods between the two cohorts may have influenced our findings. However, despite these sampling differences, similar microbiota signatures were identified associated with severe OSA. The microbiota signatures associated with severe OSA had some differences between the cohorts. It is likely that the associations between specific bacteria genera, inflammation, and OSA severity are affected by factors that are geographically specific (e.g., environmental exposures, diet, genotype, etc.). Finally, treatment with CPAP did not significantly affect α or β diversity. It is possible that, to restore the nasal microbiota, 3 months of treatment is not enough or that it requires complete resolution of the upper airway obstruction (which we were not able to assert with the available data from this cohort). Alternatively, dysbiotic signatures remaining after CPAP treatment may be relevant to consider for development of therapies that, by impacting the nasal microbiota, may affect the inflammatory process in the upper airways.
In conclusion, the current study provides evidence that changes in nasal microbiota are associated with OSA and inflammation. The associations between distinct taxa, OSA, and inflammation require further longitudinal, interventional, and experimental investigations to evaluate causal relationships. This study also provides a possible target in those with severe OSA to evaluate for the role of reflux of oral contents into the nasopharynx as a contributor to the pathological derangements occurring in the upper airways of these subjects.
Supplementary Material
Footnotes
Supported by NIH grants K23 AI102970 (L.N.S.), T32 CA193111 (B.G.W.), UL1TR001445 (B.G.W.), and K24HL109156 (I.A.); Flight Attendant Medical Research Institute Young Clinical Scientist Award (B.G.W.); Stony Wold Herbert Foundation Fellowship (B.G.W.); Centers for Disease Control and Prevention/National Institute for Occupational Safety and Health grant U01OH010415; the Instituto de Salud Carlos III grants PI10/02696, PI12/02175, and PI15/01940; and the Instituto de Salud Carlos III and European Regional Development Fund, Madrid, Spain (J.M.M. and E.V.).
Author Contributions: Conception and design: B.G.W., D.M.R., E.V., J.M.M., J.S., I.A., and L.N.S.; acquisition of data: I.S., Y.L., R.J.L., S.-E.L., I.U., D.M.R., O.L.-H., A.P., S.A., K.B., M.P., A.T., H.S., A.W., D.H., N.C., E.V., J.M.M., J.S., I.A., and L.N.S.; analysis and interpretation of data: B.G.W., J.W., N.S., J.C.C., R.J.L., K.B., P.M., B.K., B.D.S., A.W., C.B., M.D.W., and L.N.S.; drafting or revising of article: B.G.W., J.W., R.J.L, K.B., M.D.W., D.M.R., J.M.M., J.S., I.A., and L.N.S.; final approval of the manuscript: B.G.W., J.W., D.M.R., J.S., I.A., and L.N.S.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201801-0119OC on July 3, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Heinzer R, Vat S, Marques-Vidal P, Marti-Soler H, Andries D, Tobback N, et al. Prevalence of sleep-disordered breathing in the general population: the HypnoLaus study. Lancet Respir Med. 2015;3:310–318. doi: 10.1016/S2213-2600(15)00043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peppard PE, Young T, Barnet JH, Palta M, Hagen EW, Hla KM. Increased prevalence of sleep-disordered breathing in adults. Am J Epidemiol. 2013;177:1006–1014. doi: 10.1093/aje/kws342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan CF, Lowe AA, Li D, Fleetham JA. Magnetic resonance imaging of the upper airway in obstructive sleep apnea before and after chronic nasal continuous positive airway pressure therapy. Am Rev Respir Dis. 1991;144:939–944. doi: 10.1164/ajrccm/144.4.939. [DOI] [PubMed] [Google Scholar]
- 4.Paulsen FP, Steven P, Tsokos M, Jungmann K, Müller A, Verse T, et al. Upper airway epithelial structural changes in obstructive sleep-disordered breathing. Am J Respir Crit Care Med. 2002;166:501–509. doi: 10.1164/rccm.2109099. [DOI] [PubMed] [Google Scholar]
- 5.Dempsey JA, Veasey SC, Morgan BJ, O’Donnell CP. Pathophysiology of sleep apnea. Physiol Rev. 2010;90:47–112. doi: 10.1152/physrev.00043.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies RJ, Stradling JR. The relationship between neck circumference, radiographic pharyngeal anatomy, and the obstructive sleep apnoea syndrome. Eur Respir J. 1990;3:509–514. [PubMed] [Google Scholar]
- 7.Isono S, Remmers JE, Tanaka A, Sho Y, Sato J, Nishino T. Anatomy of pharynx in patients with obstructive sleep apnea and in normal subjects. J Appl Physiol (1985) 1997;82:1319–1326. doi: 10.1152/jappl.1997.82.4.1319. [DOI] [PubMed] [Google Scholar]
- 8.Anch AM, Remmers JE, Bunce H., III Supraglottic airway resistance in normal subjects and patients with occlusive sleep apnea. J Appl Physiol. 1982;53:1158–1163. doi: 10.1152/jappl.1982.53.5.1158. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick MF, McLean H, Urton AM, Tan A, O’Donnell D, Driver HS. Effect of nasal or oral breathing route on upper airway resistance during sleep. Eur Respir J. 2003;22:827–832. doi: 10.1183/09031936.03.00047903. [DOI] [PubMed] [Google Scholar]
- 10.Hatipoğlu U, Rubinstein I. Inflammation and obstructive sleep apnea syndrome: how many ways do I look at thee? Chest. 2004;126:1–2. doi: 10.1378/chest.126.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Anastassov GE, Trieger N. Edema in the upper airway in patients with obstructive sleep apnea syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1998;86:644–647. doi: 10.1016/s1079-2104(98)90197-4. [DOI] [PubMed] [Google Scholar]
- 12.Nadeem R, Molnar J, Madbouly EM, Nida M, Aggarwal S, Sajid H, et al. Serum inflammatory markers in obstructive sleep apnea: a meta-analysis. J Clin Sleep Med. 2013;9:1003–1012. doi: 10.5664/jcsm.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young T, Finn L, Palta M. Chronic nasal congestion at night is a risk factor for snoring in a population-based cohort study. Arch Intern Med. 2001;161:1514–1519. doi: 10.1001/archinte.161.12.1514. [DOI] [PubMed] [Google Scholar]
- 14.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HW, Liu PF, Liu YT, Kuo S, Zhang XQ, Schooley RT, et al. Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci Rep. 2016;6:27870. doi: 10.1038/srep27870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Peters-Hall JR, Ghimbovschi S, Mimms R, Rose MC, Peña MT. Glandular gene expression of sinus mucosa in chronic rhinosinusitis with and without cystic fibrosis. Am J Respir Cell Mol Biol. 2011;45:525–533. doi: 10.1165/rcmb.2010-0133OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanathan M, Jr, Lee WK, Spannhake EW, Lane AP. Th2 cytokines associated with chronic rhinosinusitis with polyps down-regulate the antimicrobial immune function of human sinonasal epithelial cells. Am J Rhinol. 2008;22:115–121. doi: 10.2500/ajr.2008.22.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal LN, Sunderram J, Kipen H, Laumbach RJ, Lu S-E, Udasin I, et al. Evaluation of the nasal microbiome in World Trade Center dust exposed subjects [abstract] Am J Respir Crit Care Med. 2016;193:A2542. [Google Scholar]
- 21.Vicente E, Marin JM, Carrizo SJ, Osuna CS, González R, Marin-Oto M, et al. Upper airway and systemic inflammation in obstructive sleep apnoea. Eur Respir J. 2016;48:1108–1117. doi: 10.1183/13993003.00234-2016. [DOI] [PubMed] [Google Scholar]
- 22.Candela A, Hernández L, Asensio S, Sánchez-Payá J, Vila J, Benito N, et al. Validation of a respiratory polygraphy system in the diagnosis of sleep apnea syndrome [in Spanish] Arch Bronconeumol. 2005;41:71–77. doi: 10.1016/s1579-2129(06)60400-x. [DOI] [PubMed] [Google Scholar]
- 23.Kapur VK, Auckley DH, Chowdhuri S, Kuhlmann DC, Mehra R, Ramar K, et al. Clinical practice guideline for diagnostic testing for adult obstructive sleep apnea: an American Academy of Sleep Medicine clinical practice guideline. J Clin Sleep Med. 2017;13:479–504. doi: 10.5664/jcsm.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Hoz RE, Aurora RN, Landsbergis P, Bienenfeld LA, Afilaka AA, Herbert R. Snoring and obstructive sleep apnea among former World Trade Center rescue workers and volunteers. J Occup Environ Med. 2010;52:29–32. doi: 10.1097/JOM.0b013e3181c2bb18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laumbach RJ, Fiedler N, Gardner CR, Laskin DL, Fan ZH, Zhang J, et al. Nasal effects of a mixture of volatile organic compounds and their ozone oxidation products. J Occup Environ Med. 2005;47:1182–1189. doi: 10.1097/01.jom.0000183338.95778.f0. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chouake J, Friedman A. The outer aspect of our inner anxiety: the skinny on stress. J Drugs Dermatol. 2012;11:883–884. [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BP, Jensen BJ, Ransom EM, Heinemann KA, Vannatta KM, Egland KA, et al. Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA. J Bacteriol. 2009;191:5563–5565. doi: 10.1128/JB.01226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles MR, Carson JL, Collier AM, Gatzy JT, Boucher RC. Measurements of nasal transepithelial electric potential differences in normal human subjects in vivo. Am Rev Respir Dis. 1981;124:484–490. doi: 10.1164/arrd.1981.124.4.484. [DOI] [PubMed] [Google Scholar]
- 34.Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoggard M, Biswas K, Zoing M, Wagner Mackenzie B, Taylor MW, Douglas RG. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7:230–239. doi: 10.1002/alr.21871. [DOI] [PubMed] [Google Scholar]
- 36.Hoggard M, Wagner Mackenzie B, Jain R, Taylor MW, Biswas K, Douglas RG. Chronic rhinosinusitis and the evolving understanding of microbial ecology in chronic inflammatory mucosal disease. Clin Microbiol Rev. 2017;30:321–348. doi: 10.1128/CMR.00060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, Sanford T. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2013;139:1328–1338. doi: 10.1001/jamaoto.2013.5465. [DOI] [PubMed] [Google Scholar]
- 38.Boase S, Foreman A, Cleland E, Tan L, Melton-Kreft R, Pant H, et al. The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infect Dis. 2013;13:210. doi: 10.1186/1471-2334-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brook I. Microbiology of sinusitis. Proc Am Thorac Soc. 2011;8:90–100. doi: 10.1513/pats.201006-038RN. [DOI] [PubMed] [Google Scholar]
- 40.Cope EK, Goldberg AN, Pletcher SD, Lynch SV. Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome. 2017;5:53. doi: 10.1186/s40168-017-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, Chu MLJN, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences: a prospective cohort study. Am J Respir Crit Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 42.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 43.Al-Mutairi D, Kilty SJ. Bacterial biofilms and the pathophysiology of chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2011;11:18–23. doi: 10.1097/ACI.0b013e3283423376. [DOI] [PubMed] [Google Scholar]
- 44.Frederick J, Braude AI. Anaerobic infection of the paranasal sinuses. N Engl J Med. 1974;290:135–137. doi: 10.1056/NEJM197401172900304. [DOI] [PubMed] [Google Scholar]
- 45.Deniz M, Gultekin E, Ciftci Z, Alp R, Ozdemir DN, Isik A, et al. Nasal mucociliary clearance in obstructive sleep apnea syndrome patients. Am J Rhinol Allergy. 2014;28:178–180. doi: 10.2500/ajra.2014.28.4094. [DOI] [PubMed] [Google Scholar]
- 46.Greenberg H, Ye X, Wilson D, Htoo AK, Hendersen T, Liu SF. Chronic intermittent hypoxia activates nuclear factor-kappaB in cardiovascular tissues in vivo. Biochem Biophys Res Commun. 2006;343:591–596. doi: 10.1016/j.bbrc.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Rubinstein I. Nasal inflammation in patients with obstructive sleep apnea. Laryngoscope. 1995;105:175–177. doi: 10.1288/00005537-199502000-00012. [DOI] [PubMed] [Google Scholar]
- 48.Hamada S, Tatsumi S, Kobayashi Y, Yasuba H. Nasal nitric oxide improved by continuous positive airway pressure therapy for upper airway inflammation in obstructive sleep apnea. Sleep Breath. 2017;21:405–410. doi: 10.1007/s11325-016-1431-z. [DOI] [PubMed] [Google Scholar]
- 49.Min YG, Oh SJ, Won TB, Kim YM, Shim WS, Rhee CS, et al. Effects of staphylococcal enterotoxin on ciliary activity and histology of the sinus mucosa. Acta Otolaryngol. 2006;126:941–947. doi: 10.1080/00016480500469016. [DOI] [PubMed] [Google Scholar]
- 50.Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension. 2016;67:469–474. doi: 10.1161/HYPERTENSIONAHA.115.06672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue J, Zhou D, Poulsen O, Imamura T, Hsiao YH, Smith TH, et al. Intermittent hypoxia and hypercapnia accelerate atherosclerosis, partially via trimethylamine-oxide. Am J Respir Cell Mol Biol. 2017;57:581–588. doi: 10.1165/rcmb.2017-0086OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffin JL, Wang X, Stanley E. Does our gut microbiome predict cardiovascular risk? A review of the evidence from metabolomics. Circ Cardiovasc Genet. 2015;8:187–191. doi: 10.1161/CIRCGENETICS.114.000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipin RB, Deshpande A, Wise SK, DelGaudio JM, Patel ZM. Chronic rhinosinusitis and obstructive sleep apnea: CPAP reservoir bacterial colonization is not associated with sinus culture positivity. Sinusitis. 2016;1:44–48. [Google Scholar]
- 54.Koutsourelakis I, Vagiakis E, Perraki E, Karatza M, Magkou C, Kopaka M, et al. Nasal inflammation in sleep apnoea patients using CPAP and effect of heated humidification. Eur Respir J. 2011;37:587–594. doi: 10.1183/09031936.00036910. [DOI] [PubMed] [Google Scholar]
- 55.Mahdavinia M, Keshavarzian A, Tobin MC, Landay AL, Schleimer RP. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS) Clin Exp Allergy. 2016;46:21–41. doi: 10.1111/cea.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.