Abstract
Rationale: The identification of causal variants responsible for disease associations from genome-wide association studies (GWASs) facilitates functional understanding of the biological mechanisms by which those genetic variants influence disease susceptibility.
Objective: We aim to identify causal variants in or near the FAM13A (family with sequence similarity member 13A) GWAS locus associated with chronic obstructive pulmonary disease (COPD).
Methods: We used an integrated approach featuring conditional genetic analysis, massively parallel reporter assays (MPRAs), traditional reporter assays, chromatin conformation capture assays, and clustered regularly interspaced short palindromic repeats (CRISPR)-based gene editing to characterize COPD-associated regulatory variants in the FAM13A region in human bronchial epithelial cell lines.
Measurements and Main Results: Conditional genetic association suggests the presence of two independent COPD association signals in FAM13A. MPRAs identified 45 regulatory variants within FAM13A, among which six variants were prioritized for further investigation. Three COPD-associated variants demonstrated significant allele-specific activity in reporter assays. One of three variants, rs2013701, was tested in the endogenous genomic context by CRISPR-based genome editing that confirmed its allele-specific effects on FAM13A expression and on cell proliferation, providing functional characterization for this COPD-associated variant.
Conclusions: The human GWAS association near FAM13A may contain independent association signals. MPRAs identified multiple functional variants in this region, including rs2013701, a putative COPD-causing variant with allele-specific regulatory activity.
Keywords: CRISPR editing, human bronchial epithelial cells, functional genomics, chromosome conformation capture, reporter assay
At a Glance Commentary
Scientific Knowledge on the Subject
Genome-wide association studies have conclusively demonstrated the association between an intragenic locus in FAM13A (family with sequence similarity member 13A) and susceptibility to chronic obstructive pulmonary disease (COPD). However, the specific genetic variant or variants responsible for COPD susceptibility are unknown, and there have been no systematic studies of COPD-related functional genetic variation in FAM13A.
What This Study Adds to the Field
Using high-throughput massively parallel reporter assay screening, we tested 606 common genetic variants spanning the full length of FAM13A to identify functional regulatory variants in bronchial epithelial cell lines. Additional functional validation was performed for six of these variants, and we demonstrate compelling evidence from both chromatin conformation capture and clustered regularly interspaced short palindromic repeats–based assays identifying rs2013701 as a functional variant likely to contribute to the development of COPD.
Chronic obstructive pulmonary disease (COPD) is a complex genetic disorder that arises from both cigarette smoke exposure and genetic susceptibility factors. A recent collaborative genome-wide association study (GWAS) identified 22 genome-wide significant COPD susceptibility loci (1), but candidate functional variants explaining the GWAS association at these loci have only been identified for the associations near HHIP (2) and AGER (3).
One of the earliest identified and strongest GWAS associations with COPD localizes to a >100 kb region within the FAM13A (family with sequence similarity member 13A) gene body (4). The broad association peak in this region complicates identification of the disease-causing genetic variants responsible for this GWAS signal, but notable progress has been made in characterizing the function of FAM13A. In murine models, we have demonstrated that Fam13a may promote COPD susceptibility by mediating the degradation of β-catenin, important for repair and regeneration of alveolar epithelial cells in COPD (5, 6); also, we have demonstrated that Fam13a−/− mice are resistant to smoke-induced emphysema and have increased levels of β-catenin in their lungs (7). However, the causal genetic variants influencing human COPD susceptibility in the FAM13A locus are unknown.
Most GWAS-identified loci exert their phenotypic effects through regulation of gene expression (8–10). In addition, allelic heterogeneity at GWAS loci may be common, with some GWAS signals arising from the cumulative effect of multiple causal variants (11). Thus, identification of the disease-causing variants within a GWAS locus often requires high-throughput functional screening, which can be achieved through a massively parallel reporter assay (MPRA). The MPRA leverages chip-based array technology to generate hundreds of thousands of synthetic oligonucleotides, each with its own unique genetic barcode sequence (12). An MPRA library is generated by cloning these oligonucleotides into reporter vectors. After transfection and RNA sequencing, the regulatory effects of each oligonucleotide on gene expression are quantified by high-throughput sequencing of genetic barcodes. Because the oligonucleotides are custom designed, MPRAs can be used to study the regulatory effects of various aspects of DNA sequences, including orientation and the effects of SNPs.
Recently, post-GWAS functional studies in complex diseases have suggested that functional variants tend to exert their regulatory effects across a long genomic distance (2). Such distal regulation through higher dimensional chromatin structure can be assessed by chromatin conformation capture (3C) assays (13). In this assay, a 3C library is generated from genomic DNA after cross-linking, enzymatic digestion, and diluted ligation. Then the physical proximity of DNA fragments is evaluated by 3C-PCR using unidirectional primers that can only amplify when a chromatin interaction exists between distant DNA segments.
We hypothesized that there may be at least one COPD-causing regulatory variant responsible for the GWAS association near FAM13A. To test this hypothesis and identify potential causal variants in this locus, we adopted a high-throughput functional screening strategy using MPRAs to comprehensively characterize the regulatory landscape near FAM13A in a human bronchial epithelial line (Beas-2B cells), and we performed comprehensive conditional genetic association analyses to identify potential secondary association signals. MPRAs identified multiple common regulatory variants in or near FAM13A, and conditional analyses identified two independent genetic association signals within the same 100-kb linkage disequilibrium (LD) block. Subsequently, we used reporter assays to confirm the allele-specific regulatory effects of selected MPRA SNPs, 3C-PCR assays to identify the interaction between SNP-containing genomic segments and the FAM13A promoter region, and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-9–based genome editing to confirm allele-specific regulatory effects of rs2013701 on FAM13A in the endogenous genomic context. All of these studies therefore support a COPD-related functional role for the rs2013701 variant in human bronchial epithelial cells.
Methods
Human Study Populations
Five study samples of smokers enriched for COPD were used for genetic association analysis (COPDGene [COPD Genetic Epidemiology], ECLIPSE [Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints], GenKOLS [Norway Genetics of Chronic Obstructive Lung Disease Study], NETT [National Emphysema Treatment Trial], and NAS [Normative Aging Study]). These study samples and genotyping methods have been previously described (14). For a detailed description of human populations, see online supplement.
Genotyping Data and Genetic Association Analysis
Genotyping and imputation for the study cohorts have been previously described (14). Genotype imputation to the 1,000 Genomes Phase 1 v3 EUR reference panel was performed with minimac (15). GWAS was performed for the five study populations using PLINK v1.9 (16). Genetic variants were related to COPD case-control status using logistic regression adjusting for age, sex, pack-years smoke exposure, and principal components of genetic ancestry calculated using the Eigenstrat software package (17). Conditional genetic association analyses were performed in PLINK using the –condition flag. R2 was calculated from COPDGene non-Hispanic white (NHW) study samples using PLINK, and haplotypes were defined with Haploview (18) using the criteria defined by Gabriel and colleagues (19).
Expression Quantitative Trait Locus Data
Expression quantitative trait locus (eQTL) SNPs within 250 kb of FAM13A were defined for inclusion in the MPRA panel using data from a previously published eQTL analysis performed in blood samples from 121 former smokers with COPD in the ECLIPSE Study (20). Lung tissue eQTL version 7 results from the Genotype-Tissue Expression (GTEx) project were accessed via the GTEx portal (https://www.gtexportal.org/home/) on June 4, 2018 (21).
Oligonucleotide Array Design and MPRA Library Construction
We designed MPRA oligonucleotides that included both biallelic variant alleles for all common SNPs in the FAM13A GWAS locus associated with COPD affection status with P < 0.02 in a COPD GWAS meta-analysis using 1000 Genomes imputed variants (14). To increase our likelihood of capturing regulatory variants in this region, we also included cis eQTL SNPs at FDR <0.01 for FAM13A mRNA levels in whole blood from subjects with COPD (20). MPRA oligonucleotides were synthesized on a 244K Agilent array and cloned into the pGL4.10M2 backbone MPRA library vector containing either a strong promoter (simian virus 40 [SV40] promoter) or a relatively weak promoter (minimal promoter [minP]) to test regulatory activities of 144-bp-length oligonucleotides containing potential functional genetic variants (12). Details on the oligonucleotide array design, cloning, cell cultures and transfection of library, MPRA data analysis, molecular cloning and reporter assays, 3C-PCR, CRISPR-based SNP modification, gene expression level detection, and cell proliferation rate measurement are included in the online supplement.
Results
MPRA Identifies Context-Specific Regulatory Variation near FAM13A
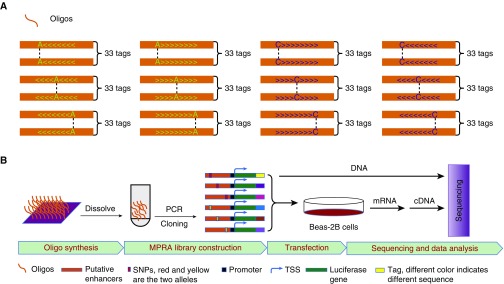
MPRA has been used effectively as a high-throughput screening method to identify disease-causing variants within GWAS loci (22). We used a similar screening approach to prioritize functional variants in the FAM13A GWAS locus. The overview of SNP selection for MPRA and subsequent follow-up characterization is shown in Figure E1 in the online supplement. To identify common regulatory variants in the FAM13A locus, we defined a set of SNPs to test in MPRA by selecting 606 SNPs within 250 kb of FAM13A that were either associated with COPD at P < 0.02 in a GWAS meta-analysis (14) or associated with FAM13A mRNA levels in blood gene expression from subjects with COPD (20). An overview of the MPRA procedure is shown in Figure 1. MPRA libraries were constructed by cloning oligonucleotides into two plasmid vectors, one with a weak promoter (minP) and another with a stronger promoter (SV40) to increase the ability to detect both positive (activating) and negative (repressing) regulatory elements. These libraries were subsequently transfected into Beas-2B cells in separate experiments, and mRNA was extracted for RNA sequencing.
Figure 1.
Overview of massively parallel reporter assay (MPRA) design. (A) Schematic illustration to indicate the design of oligonucleotides. (B) Experimental procedures of MPRA screening. TSS = transcription start site.
We analyzed the sequencing results of genetic barcodes included at the end of each synthesized sequence to understand the gene regulatory effects of each oligonucleotide. We first tested whether each oligonucleotide sequence acted as a regulatory element. We observed that 15.6% and 14.5% of oligonucleotide sequences showed significant regulatory effects in the minP and SV40 experiments, respectively (Table E1). These effects were highly consistent across the two experiments (Figure E2). Of these significant sequences, 42.3% and 34.1% were positive regulators for luciferase gene expression in the minP and SV40 experiments, respectively.
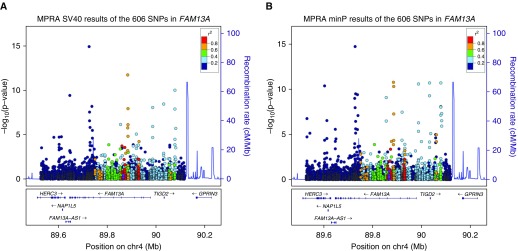
We also tested for allele-specific differences in regulatory activity as evidence for putative functional activity of tested SNPs, and we identified 85 oligonucleotides with significant allelic effects in at least one MPRA experiment (Table E2). These oligonucleotides correspond to 45 unique SNPs. Figure 2 shows the genomic positions, MPRA P values, and correlations with the previously published top COPD GWAS SNP (rs4416442) for the variants tested in MPRAs. The subset of these SNPs with significant COPD genetic associations (P <1 × 10−6) is shown in Table 1.
Figure 2.
Massively parallel reporter assay (MPRA) significant oligonucleotides with allele-specific effects shown by genomic position. Oligonucleotides representing 606 SNPs near and within FAM13A were tested in MPRA experiments with either a strong (A) or weak (B) promoter. The y-axis represents the P values for allele-specific expression from the MPRA experiment, and the x-axis represents the genomic position. Each dot represents one oligonucleotide sequence in the array. Different colors represent the relative correlation (linkage disequilibrium) with previously published GWAS SNP rs4416442 (marked in purple) as indicated by r2. The blue line shows the estimated recombination rate at each genomic location from HapMap data. FAM13A = family with sequence similarity member 13A; minP = minimal promoter; SV40 = simian virus 40 (stronger promoter).
Table 1.
SNPs near FAM13A with Significant Allele-Specific Effects in Massively Parallel Reporter Assays and Evidence of Genetic Association with Chronic Obstructive Pulmonary Disease Status
| SNP | Effect Allele* | GWAS OR† | GWAS P Value‡ | Orientation§ | minP Effect∥ | minP P Value¶ | SV40 Effect∥ | SV40 P Value¶ | R2 with rs4416442** | R2 with rs147089648** |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7674369 | A | 1.26 | 1.1 × 10−13 | L:F | −0.14 | 2.5 × 10−2 | −0.22 | 2.3 × 10−2 | 1.00 | 0.14 |
| rs2013701 | G | 0.80 | 1.5 × 10−12 | R:F | 0.89 | 3.8 × 10−3 | 0.47 | 5.0 × 10−1 | 0.63 | 0.08 |
| G | 0.80 | 1.5 × 10−12 | C:F | 0.52 | 1.1 × 10−5 | 0.36 | 2.0 × 10−4 | 0.63 | 0.08 | |
| G | 0.80 | 1.5 × 10−12 | R:RC | −2.4 | 6.0 × 10−1 | −4.78 | 2.7 × 10−3 | 0.63 | 0.08 | |
| G | 0.80 | 1.5 × 10−12 | L:F | 0.3 | 2.5 × 10−8 | 0.32 | 5.0 × 10−6 | 0.63 | 0.08 | |
| rs7671167 | C | 0.80 | 3.1 × 10−12 | L:F | −4.29 | 1.8 × 10−8 | −8.16 | 3.3 × 10−9 | 0.65 | 0.10 |
| C | 0.80 | 3.1 × 10−12 | L:RC | 0.36 | 6.1 × 10−1 | 0.89 | 3.8 × 10−4 | 0.65 | 0.10 | |
| C | 0.80 | 3.1 × 10−12 | C:F | −0.99 | 9.0 × 10−3 | −0.49 | 4.0 × 10−2 | 0.65 | 0.10 | |
| rs1964516 | C | 0.82 | 1.5 × 10−10 | L:F | −0.69 | 4.7 × 10−3 | −0.59 | 7.3 × 10−2 | 0.64 | 0.10 |
| rs1795739 | A | 1.21 | 2.5 × 10−9 | L:F | 1.98 | 3.3 × 10−2 | 1.66 | 2.2 × 10−1 | 0.78 | 0.13 |
| A | 1.21 | 2.5 × 10−9 | R:F | 1.64 | 7.6 × 10−2 | 1.91 | 7.6 × 10−3 | 0.78 | 0.13 | |
| rs7687539 | A | 1.21 | 9.6 × 10−8 | R:F | 1.72 | 8.9 × 10−3 | 2.48 | 1.5 × 10−2 | 0.46 | 0.30 |
| rs2464523 | C | 0.83 | 3.1 × 10−7 | C:F | 0.38 | 4.7 × 10−2 | 0.21 | 6.3 × 10−1 | 0.40 | 0.03 |
| C | 0.83 | 3.1 × 10−7 | L:RC | −0.66 | 3.0 × 10−1 | −1.55 | 2.6 × 10−1 | 0.40 | 0.03 | |
| rs78681184 | C | 0.73 | 8.2 × 10−7 | L:RC | −2.01 | 2.3 × 10−2 | −1.35 | 5.8 × 10−1 | 0.15 | 0.86 |
| C | 0.73 | 8.2 × 10−7 | C:RC | −0.95 | 4.8 × 10−1 | −2.21 | 1.5 × 10−2 | 0.15 | 0.86 |
Definition of abbreviations: C = center; F = forward; FAM13A = family with sequence similarity member 13A; GWAS = genome-wide association study; L = left; minP = minimal promoter; OR = odds ratio; R = right; RC = reverse complement; SV40 = simian virus 40.
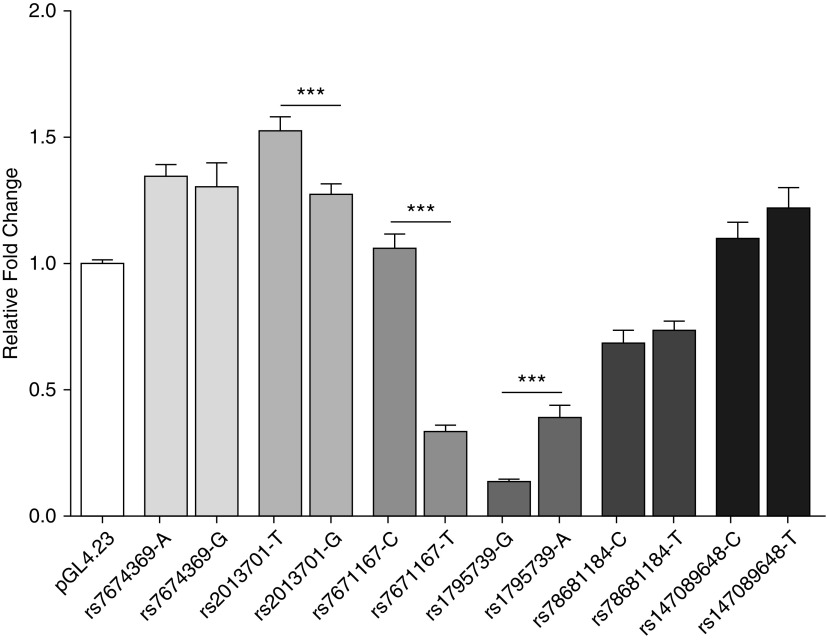
SNPs with significant massively parallel reporter assay results in either the minP or SV40 experiments and association with chronic obstructive pulmonary disease at P < 1 × 10−6 in the five cohort chronic obstructive pulmonary disease meta-analysis are shown. SNPs in bold are SNPs assessed by reporter assay in Figure 5.
Pertains to both GWAS ORs and massively parallel reporter assay results.
Chronic obstructive pulmonary disease GWAS OR.
P value from published chronic obstructive pulmonary disease GWAS meta-analysis (14).
SNP locations within oligonucleotides are shown. The oligonucleotide orientation is relative to the human genome reference.
Hodges-Lehmann provides a nonparametric estimate of the effect size, roughly equivalent to the difference of median allelic ratio between the effect allele and the other allele.
False discovery rate–adjusted P value for allelic expression effect in massively parallel reporter assay.
R2 values were calculated in COPDGene non-Hispanic white subjects.
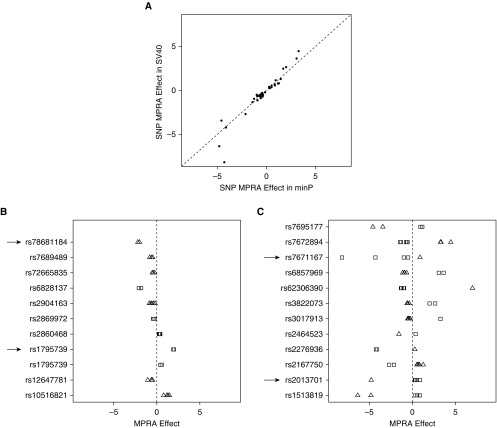
To gain additional insights into the regulatory effects of SNPs tested by MPRA, we examined factors that may impact the allele-specific activity of these 45 variants, including promoter type (strong or weak), SNP location within the oligonucleotide (left, center, or right), and oligonucleotide orientation (forward or reverse). We refer to SNP location and oligonucleotide orientation collectively as the oligonucleotide context for a given SNP. First, we observed that for 22 SNPs with significant effects in the same oligonucleotide context for both the minP and SV40 promoter, these effects were consistent in both experiments (Figure 3A). Thus, the choice of promoter did not alter the regulatory potential of these variants. Second, we examined SNPs with significant effects in multiple oligonucleotide contexts. For 11 SNPs that were significant in multiple positions but only one orientation, the effects were highly consistent, indicating that the SNP position within the oligonucleotide had little impact on allelic effects in our experiment (Figure 3B). For 12 SNPs with significant effects in both the forward and reverse orientations, these SNPs demonstrated opposite allelic effects in the two orientations. We refer to such orientation-dependent allelic effects as “divergent” SNP effects. Taken together, these observations demonstrate that the allelic effects observed in these MPRA experiments were highly reproducible, but a subset of SNPs showed opposite allelic effects dependent on oligonucleotide orientation.
Figure 3.
Allelic effects of significant massively parallel reporter assay (MPRA) SNPs with respect to promoter, flanking sequence, and oligonucleotide orientation. (A) Allelic gene regulatory effects are highly correlated for the same SNP within identical oligonucleotide sequences connected with either a weak (minimal promoter) or strong promoter (simian virus 40). The MPRA effect corresponds to the Hodges-Lehmann estimate of the difference between the input/output ratios comparing two SNP alleles. (B and C) Of the 23 SNPs that had significant MPRA results in more than one oligonucleotide context (orientation or position), 11 SNPs that were significant in one orientation showed consistent MPRA effects across different SNP positions within the oligonucleotide. (B) Twelve SNPs that were significant in both orientations had divergent effects, that is, opposite allelic effects in the forward and reverse orientations. (C) Each dot represents one significant oligonucleotide. Only significant MPRA results are displayed. Black arrows indicate SNPs that were chosen for reporter assay validation in Figure 5. Squares indicate forward orientation; triangles indicate reverse orientation. minP = minimal promoter; SV40 = simian virus 40.
Conditional Analysis Identifies Allelic Heterogeneity at the FAM13A Locus
To determine whether the set of 45 MPRA SNPs with allele-specific effects could explain the COPD genetic association at this locus, we performed conditional association analyses for each of these variants in 5,346 NHW subjects in COPDGene. The lead GWAS SNP in this region (rs4416442) was not significant in MPRAs, but it was included in the pairwise conditional analysis. We observed that conditioning on these variants, either singly (Table E3) or in comprehensive pairwise conditional tests for all 1,035 unique pairwise combinations of these 45 variants, did not completely attenuate the association signal (Figure E3), suggesting the presence of secondary association signals in this locus.
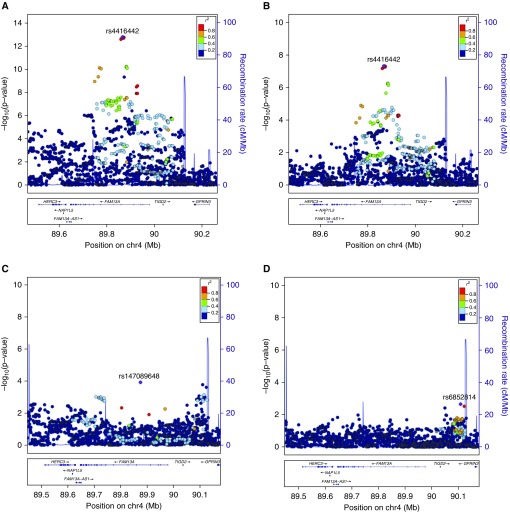
After conditioning on the lead SNP in the locus (rs4416442), we observed that the most significantly associated variant in this conditional analysis was rs147089648 (P = 1.3 × 10−4). Interestingly, this SNP was a low minor allele frequency (MAF) variant that was not included in the original multiethnic GWAS meta-analysis because of its low MAF in African Americans (1000 Genomes MAF 0.07 in European and 0.04 in African populations). This variant was in LD with the lead SNP (D′ = 1, r2 = 0.14). In a meta-analysis including all of the original populations, this variant had P = 1.1 × 10−6, but after excluding the African American subjects, it was associated with COPD at P = 3.9 × 10−9. Conditioning on both rs4416442 and rs147089648 resulted in attenuation of the association peak (Figure 4, minimum P = 2.2 × 10−3).
Figure 4.
Local chronic obstructive pulmonary disease (COPD) association plots near FAM13A (family with sequence similarity member 13A) for subjects of European descent only. (A) The unconditioned association peak at FAM13A. (B) Local association conditioned on the lead secondary association variant rs147089648. (C) Local association conditioned on rs4416442. (D) Conditioning on both rs4416442 and rs147089648 completely attenuates the association peak. Analyses were performed in COPD cases (n = 5,772) and smoking controls (n = 3,944) of European descent from five cohorts.
Analysis of LD patterns in this region indicated that rs4416442 and rs147089648 are located in an LD block spanning a 100-kb intragenic region in FAM13A (Figure E4). To prioritize a set of SNPs for additional functional investigation, we selected MPRA SNPs that were significant in both SV40 and minP experiments, that had genetic association with COPD (GWAS P <1 × 10−6), and that were in LD (r2 > 0.6) with either the primary or secondary association lead SNPs (rs4416442 or rs147089648). This resulted in five variants, and we also included the lead secondary association SNP, rs147089648, for assessment in subsequent reporter assays.
Allele-Specific Distal Enhancer Activities
We generated reporter constructs for these six SNPs to confirm allele-specific enhancer activity. Three out of six SNPs (rs2013701, rs7671167, and rs1795739) demonstrated significant allelic activity in 16HBE cells, a human bronchial epithelial cell line (Figure 5). Because these three SNPs are located >100 kb away from the promoter of FAM13A, we set out to determine whether genomic regions spanning these three SNPs have close contact with the promoter of FAM13A using 3C assays targeting the promoter of FAM13A in 16HBE cells. We observed interaction between the promoter of FAM13A with a genomic segment containing both rs2013701 and rs7671167, in contrast to rs1795739 (Figure E5).
Figure 5.
Reporter assay in 16HBE cells testing the allele-specific enhancer activity of massively parallel reporter assay SNPs. Results are given as ratios of firefly to Renilla luciferase activity. Mean ± SEM is shown from four to seven independent experiments with triplicate measurements in each repeat. ***P < 0.001, unpaired Student t test.
Based on our previous observations that Fam13a−/− mice are protected against CS-induced emphysema and that COPD risk alleles are associated with increased protein levels of FAM13A in human lung tissues (7), we expected that COPD risk alleles would be associated with increased enhancer activity relative to the non-risk allele, as we observed for rs2013701 (Figure 5).
CRISPR-based Genome Editing Targeting rs2017301 in 16HBE Cells
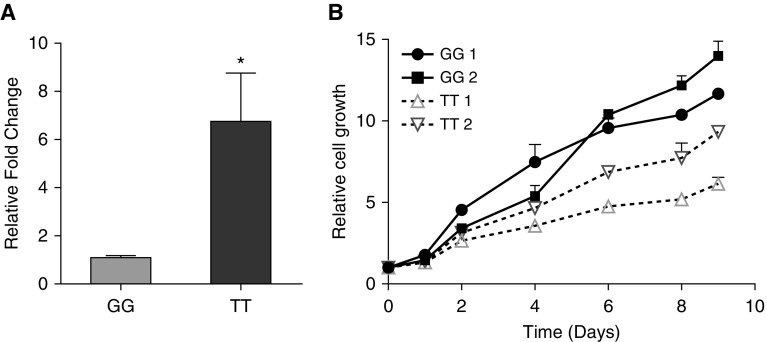
To confirm allele-specific enhancer activity of rs2013701 in the endogenous genomic context, we applied CRISPR/Cas-9 genome editing on rs2013701 in bronchial epithelial cells. We used CRISPR-based homology-directed repair to generate single clones homozygous for the TT genotype at rs2013701 from a GG homozygous 16HBE cell line (Figure E6). After single cell clone selection, we obtained two independent TT homozygous CRISPR lines for further studies. First, we observed that the rs2013701 G allele is associated with reduced expression of FAM13A, consistent with the reporter assay results (Figure 6A). Because we had previously observed that silencing FAM13A expression promotes cellular proliferation in 16HBE cell lines, we tested whether the allelic effects of rs2013701 were sufficient to alter cellular proliferation rates. We observed that GG clones demonstrated an increased rate of cellular proliferation relative to the TT clones, consistent with the anticipated direction of effect (Figure 6B).
Figure 6.
Expression levels of FAM13A (family with sequence similarity member 13A) and cellular proliferation assays in clustered regularly interspaced short palindromic repeats (CRISPR)-edited cell clones targeting rs2013701. (A) Expression levels of FAM13A were measured in 16HBE cell lines. After CRISPR editing, 16HBE cells homozygous for the G allele were altered to the homozygous T genotype of rs2013701. Means ± SEM are shown from two to three individual CRISPR single clones with three repeats for each line. (B) Cell growth curve in 16HBE CRISPR lines. Results from four single colonies with genotype GG or TT at rs2013701 are shown. Means ± SD are shown from triplicate wells. One representative experiment from four independent experiments is shown. *P < 0.05, unpaired Student’s t test.
Confirmation of rs2013701 Allelic Effects in GTEx Human Lung Tissue
To determine whether we could identify evidence to support the functional role of rs2013701 in human lung tissue, we queried publicly available eQTL data from the GTEx project, and we observed that rs2013701 is indeed associated with FAM13A expression in human lungs with the same directional effects as observed in our reporter assay and CRISPR-edited 16HBE cells (Figure E7).
Discussion
The identification of COPD-causal variants in the GWAS-identified region near FAM13A has been limited by the broad and complex nature of the association peak. Using MPRA screening, we interrogated 606 common variants near FAM13A and identified allele-specific effects for 45 variants in human bronchial epithelial cells lines. Integration of MPRA and fine mapping results prioritized six putative functional variants in a 100-kb LD block spanning an intragenic region of FAM13A. Follow-up reporter assays demonstrated allele-specific effects for rs2013701 consistent with previously postulated mechanisms of COPD susceptibility in murine models. Chromatin conformation capture suggested that the region containing this variant interacts with the FAM13A promoter in 16HBE cells, and CRISPR editing confirmed the allele-specific effects of this variant on expression of FAM13A in the endogenous genomic context.
The FAM13A gene was cloned in 2004 owing to interest in the association of this locus with bovine milk protein production (23). It is expressed in a wide range of human tissues, including club cells in the airway epithelium, type 2 alveolar epithelial cells, and alveolar macrophages. Until recently, little was known about FAM13A function. We have demonstrated through affinity purification and co-precipitation studies that FAM13A regulates protein phosphatase 2A/GSK3-β–mediated β-catenin degradation (7). In addition, Fam13a−/− mice are protected against the development of emphysema. These observations, paired with the robust replication of genetic association evidence near FAM13A in GWAS for lung function (24) and COPD (1), support a model in which a COPD risk allele in functional SNPs near FAM13A increases COPD risk by increasing expression of FAM13A.
In the present study, we conducted extensive conditional and pairwise conditional genetic association testing to examine the statistical evidence for association near FAM13A in more detail, and we identified novel, suggestive evidence for a secondary association in FAM13A for a low-frequency variant, rs147089648, located 7.6 kb from the lead GWAS SNP within an Alu insertion region. This variant, which was not included on our original MPRA screening panel, did not show statistically significant allelic enhancer activity in 16HBE reporter assays, but further investigation and replication of this secondary association is warranted. Of note, a recent GWAS analysis of population-based lung function identified a secondary association signal near FAM13A (secondary association lead SNP rs13110699) (24), although this secondary signal appears to be separate from the one we identified.
Both the primary and secondary COPD associations near FAM13A localize to a 100-kb LD block spanning the fourth intron of FAM13A. Whereas MPRAs and subsequent confirmatory assays identified multiple potentially functional variants within this LD block, only rs2013701 demonstrated upregulation of gene expression by the COPD risk allele and physical contact with the FAM13A promoter in bronchial epithelial cells. Analyses of CRISPR-edited clones confirm that this variant alters FAM13A expression in the endogenous genomic context and affects cellular proliferation, supporting rs2013701 as a functional, COPD-associated variant.
Previous MPRA studies have demonstrated that the regulatory activity of even short oligonucleotide sequences is dictated by complex interactions between multiple neighboring TF-binding regions (25), local GC content (26), and cell type–specific effects (27). In a focused MPRA study of allelic effects of single SNP and multi-SNP interactions in the rhodopsin promoter, the vast majority of allelic substitutions had functional effects, highlighting the extent to which gene expression is governed by the complex interplay between DNA-binding proteins even within a single promoter region (28). In our studies, we found that (1) different types of promoters, strong or weak, had consistent effects on enhancer activities (Figure 3A and Figure E2); and (2) context-specific allelic effects varied depending on the orientations of oligonucleotides but not SNP location (left, center, or right) (Figure 3B and 3C). This was also highlighted in a recent comprehensive review that suggested there are two types of enhancers with either loose or stringent context requirements (including genomic orientation) (29). A particular strength of this study was that MPRA screening was followed by comprehensive validation using traditional reporter assays, 3C, CRISPR-based editing, and cellular phenotype assessments in CRISPR-edited cell lines. These studies support a role for rs2013701 influencing COPD susceptibility through increased FAM13A expression in the bronchial epithelium. Despite divergent effects of this SNP shown in the MPRA analysis (Figure 3C), the direction of effects in subsequent reporter assays and CRISPR lines is consistent with human genetic discoveries and previous mouse models. Our data highlight the importance of multipronged validation approaches following MPRA screening and also warrant a degree of caution when drawing conclusions about allele-specific effects within a complex regulatory context.
In this study, we applied CRISPR-based genome editing targeting rs2013701 in the 16HBE cell line due to its single clonal expansion capability that was not feasible in normal bronchial epithelial cells. Admittedly, such a submerge growth condition of 16HBE cell line is distinct from a differentiated airway epithelial cell growth condition in vivo, which may complicate the SNP-mediated gene regulation. We attempted to grow CRISPR-edited 16HBE cell lines at the air–liquid interface, but we observed that these cells failed to form adequate tight junctions. Thus, as an alternative validation strategy we queried human lung eQTL data in GTEx and observed that rs2013701 is significantly associated with expression of FAM13A in human lung tissues (Figure E7), which confirmed allele-specific regulation of FAM13A by rs2013701 in 16HBE cells and provided compelling evidence for a causal role for rs2013701 in COPD.
Although we identified dozens of functional variants near FAM13A in bronchial epithelial cells, this study does not comprehensively identify variants that may be active in other cell types, in response to specific exposures, or at specific periods of development. We also recognize that functional variants that impact splicing and transcript stability may fall beyond our detection limit.
In summary, comprehensive screening for regulatory variants near FAM13A identified functional regulatory variation within a 250-kb window of FAM13A in human bronchial epithelial cell lines. Focused evaluation of a subset of these variants in strong linkage disequilibrium with the lead SNPs in this region prioritized rs2013701, with allele-specific effects on FAM13A regulation. Further studies in additional cell types and genomic contexts will be useful to further characterize the complex pattern of genetic associations in this locus. This approach can be generalized for functional variant identification in GWAS noncoding regions associated with other complex diseases.
Supplementary Material
Footnotes
Supported by U.S. NIH grants R01HL127200, R01HL137927, and R33 HL120794 (X.Z.); R01HL124233 and R01HL126596 (P.J.C.); P01 HL114501, R01HL137927, and P01 HL105339 (E.K.S.); R01HL113264, R01HL137927, and R01HL135142 (M.H.C.); and K01 HL129039 (D.Q.). The project was also supported by National Human Genome Research Institute grant 5R01HG006785 (T.S.M.).
Author Contributions: E.K.S. and X.Z. designed and supervised the overall study. M.H.C. and P.J.C. selected SNPs in MPRA screening. T.S.M. designed the oligonucleotide array, supervised MPRA library cloning, and helped with data interpretation. P.J.C. and D.Q. performed MPRA data analysis, conditional analysis, and fine-mapping. F.D. performed CRISPR/Cas-9–based genome editing. F.G. performed MPRA library transfection and 3C assays. B.P, Y.L., and Z.Z.C.N. performed all experimental work. F.G. and X.Z. supervised all experimental validation. P.J.C., E.K.S., F.G., D.Q., M.H.C., and X.Z. wrote the manuscript, which all authors reviewed and approved.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201802-0337OC on August 4, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21:1325–1335. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SJ, Kleffmann T, Hessian PA. The G82S polymorphism promotes glycosylation of the receptor for advanced glycation end products (RAGE) at asparagine 81: comparison of wild-type rage with the G82S polymorphic variant. J Biol Chem. 2011;286:21384–21392. doi: 10.1074/jbc.M111.241281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skronska-Wasek W, Mutze K, Baarsma HA, Bracke KR, Alsafadi HN, Lehmann M, et al. Reduced frizzled receptor 4 expression prevents WNT/β-catenin-driven alveolar lung repair in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;196:172–185. doi: 10.1164/rccm.201605-0904OC. [DOI] [PubMed] [Google Scholar]
- 6.Baarsma HA, Skronska-Wasek W, Mutze K, Ciolek F, Wagner DE, John-Schuster G, et al. Noncanonical WNT-5A signaling impairs endogenous lung repair in COPD. J Exp Med. 2017;214:143–163. doi: 10.1084/jem.20160675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Z, Lao T, Qiu W, Polverino F, Gupta K, Guo F, et al. A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of β-catenin. Am J Respir Crit Care Med. 2016;194:185–197. doi: 10.1164/rccm.201505-0999OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ.Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWASPLoS Genet20106e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hormozdiari F, Zhu A, Kichaev G, Segre AV, Ju CJT, Joo JW, et al. Widespread allelic heterogeneity in complex traits. Am J Hum Genet. 2017;100:789–802. doi: 10.1016/j.ajhg.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Laat W, Dekker J. 3C-based technologies to study the shape of the genome. Methods. 2012;58:189–191. doi: 10.1016/j.ymeth.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho MH, McDonald M-LN, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 20.Castaldi PJ, Cho MH, Zhou X, Qiu W, Mcgeachie M, Celli B, et al. Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum Mol Genet. 2015;24:1200–1210. doi: 10.1093/hmg/ddu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen M, Reichenstein M, Everts-van der Wind A, Heon-Lee J, Shani M, Lewin HA, et al. Cloning and characterization of FAM13A1--a gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics. 2004;84:374–383. doi: 10.1016/j.ygeno.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Understanding Society Scientific Group; Geisinger-Regeneron DiscovEHR Collaboration. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49:416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci USA. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci USA. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J, Melnikov A, Zhang X, Wang L, Rogov P, Mikkelsen TS, et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat Biotechnol. 2016;34:1180–1190. doi: 10.1038/nbt.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci USA. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.