Abstract
High-throughput 16S rRNA gene sequencing has been used to identify the intestinal microbiota of many animal species, but that of marine invertebrate organisms remains largely unknown. There are only a few high-throughput sequencing studies on the intestinal microbiota of echinoderms (non-vertebrate Deuterostomes). Here we describe the intestinal microbiota of the sea cucumber Holothuria glaberrima, an echinoderm, well-known for its remarkable power of regeneration. We characterized the microbiota from the anterior descending intestine, the medial intestine (these two comprise the small intestine) and the posterior descending intestine (or large intestine), using pyrosequencing to sequence the V4 region of the 16S rRNA gene. We compared animals in their natural marine environment and in sea-water aquaria. A total of 8,172 OTU’s were grouped in 10 bacterial phyla, 23 classes, 44 orders, 83 families, 127 genera and 1 group of unknown bacteria, present across the digestive tract of 10 specimens. The results showed that the anterior intestine is dominated by Proteobacteria (61%) and Bacteroidetes (22%), the medium intestine is similar but with lower Bacteroidetes (4%), and the posterior intestine was remarkably different, dominated by Firmicutes (48%) and Bacteroidetes (35%). The structure of the community changed in animals kept in aquaria, which had a general dominance of Firmicutes and Bacteroidetes, regardless the intestinal segment. Our results evidence that in the natural sea environment, there is intestinal segment differentiation in the microbiota of H. glaberrima, which is lost in artificial conditions. This is relevant for physiological studies, such as mechanisms of digestive regeneration, which might be affected by the microbiota.
Introduction
The microbiome refers to the genome of microbial life forms inhabiting a living host, and their interactions with the host [1]. The term was first suggested by Joshua Lederberg to describe the collective genome of our indigenous microbes and to introduce the idea that a genetic view of humans should include the microbial genes [2]. They play significant roles in the metabolism of the host. Among these the most studied have been the hydrolysis of ingested molecules, the synthesis of vitamins [3] and the stimulation of the immune system [4,5]. Other microbiota studies addressed the development of obesity [6,7]), the integrity of the intestinal mucosal barrier ([8–10], the proliferation and differentiation of epithelial lineages during intestinal development [11,12]. and the activity of the enteric nervous system [13,14], changes in the host behavior [15,16–19] and the microbiome associated to diseases, such as cancer [20,21].
The current knowledge of the gastrointestinal microbiome and its benefits are mainly focused on vertebrates particularly on mammals. Among marine animals, two of the groups most studied in terms of their microbiota are sponges and corals [22–24], however there are few investigations of other marine invertebrates.
Members of the phylum Echinodermata comprise some of the most important marine invertebrates. They are found in all marine environments, from coastal to benthic and from the tropics to the polar regions. In some of these they constitute the majority of biomass present [25]. Echinoderms include five different classes: Asteroidea (sea stars), Echinoidea (sea urchins and sand dollars), Crinoidea (crinoids or sea lilies), Ophiuroidea (brittle stars) and Holothuroidea (sea cucumbers). Culture-dependent studies of the microbial composition in the intestine of adult holothurians (and other echinoderms) have shown that they have a great diversity of microorganisms, such as bacteria, viruses, protozoa, and fungi that colonize the intestine [26]. Studies have shown the presence of bacteria inhabiting the guts in echinoids [27,28], holothuroids [29–33], and ophuiroids [29]. Some studies have focused on the bacteria found in specific compartments of the digestive tract, particularly in the foregut [34], intestine [31, 35–37], hindgut ([34], and cecum [27]. The characterization of bacteria in the gut showed that ~50% of the isolates were related to members of the genus Vibrio and neighboring taxa. Other isolates, included members of the genus Bacillus, the alpha and gamma subclasses of the Proteobacteria, the Cytophaga-Flavobacterium-Bacteroides lineage, and the order Actinomycetales [38]. In addition, it was found that gut microbiota of two species, A. japonicus and Holothuria leucospilota, are involved in the breakdown of indigestible products during intestinal metabolism [39–41].
Here we studied the microbiota of the sea cucumber Holothuria glaberrima, and determined the differences between individuals from natural and aquarium environments. This study is important for two different reasons. First, there is limited information on echinoderm microbiotas with only one study on the microbiota of holothurians [33] and two in sea urchins [42,43]. Our study contributes information on a holothurian species from a different ecological niche. H. glaberrima lives in the coastal rocky shore feeding on organic matter brought by the continuous wave action. Particulate matter, including algae, sand, mud, organic and inorganic debris, etc. are picked by the animals tentacles and introduced into the mouth.
The second, and most important reason (from our laboratory perspective), H. glaberrima has become an important model system to study intestinal regeneration [44–46]. This study provides the fundamental information on the microbiota of this species in natural and aquarium environments, thus paving the way for future studies on the changes in bacterial compositions associated with the intestinal regeneration process.
Materials and methods
This research deals only with invertebrate animals, thus the University of Puerto Rico IACUC waives ethical approval of research performed on invertebrates. Animals were sacrificed by sectioning the anterior part of the animal close to the oral nerve ring, which accounts for the principal nervous component.
Sample collection
Ten adult animals were captured from their natural habitat in Playa Piñones, Puerto Rico. Permission is not required for their capture since these animals are not either endangered or protected. The coastal area where they were collected is not private property and is considered public property. Five of the animals were dissected in situ while the remaining five were transported to the lab and placed in seawater aquarium.
Intestines that were dissected in situ were filled with the usual sand, algae, organic matter and other debris that the animals acquired by capturing from their surroundings with their tentacles and inserting them into their esophagus. These intestines were rinsed in seawater to remove most of the content. Each intestine was divided into three segments ranging from 5 to 7 cm: the anterior segment, which extends from the esophagus to the first descending intestine; the medial segment, which encompasses the ascending small intestine; and the posterior segment, which is the second descending or large intestine that ends in the cloacae (Fig 1). Dual cotton swabs (BD Diagnostics, BD-220135, Franklin Lakes, NJ USA) were used to collect the microbial sample from the luminal epithelium of each segment, and the samples were stored in a 1.5 mL centrifuge-tube containing 200 μl of sucrose lysis buffer (20mM EDTA, 400 mM NaCl, 0.75 M Sucrose, 50 mM Tris-HCl, pH 9.0)(Suppl. Venter et al. 2011). All samples were immediately frozen in dry ice at -78°C, and were transported to the University of Puerto Rico where the DNA was extracted. In addition, two liters of seawater were transported to the lab, and used as a control to determine what microorganisms were present in the surrounding environment. To obtain the microbial sample from the seawater, the two liters of water were filtered through 11 mm sterile filter paper (Qualitative 1, Whatman Filter Paper) to remove large particles from the water. The water was again filtered through Millipore membrane filters (0.45 μm pore size), and then filtered through another Millipore membrane filter (0.22 μm pore size) to obtain the bacterial cells. The two Millipore membranes (0.45 μm and 0.22 μm) were removed from their respective filter and were transferred to a sterile 15 mL centrifuge-tube with 10 mL of sucrose lysis buffer and stored at -20°C until DNA extraction.
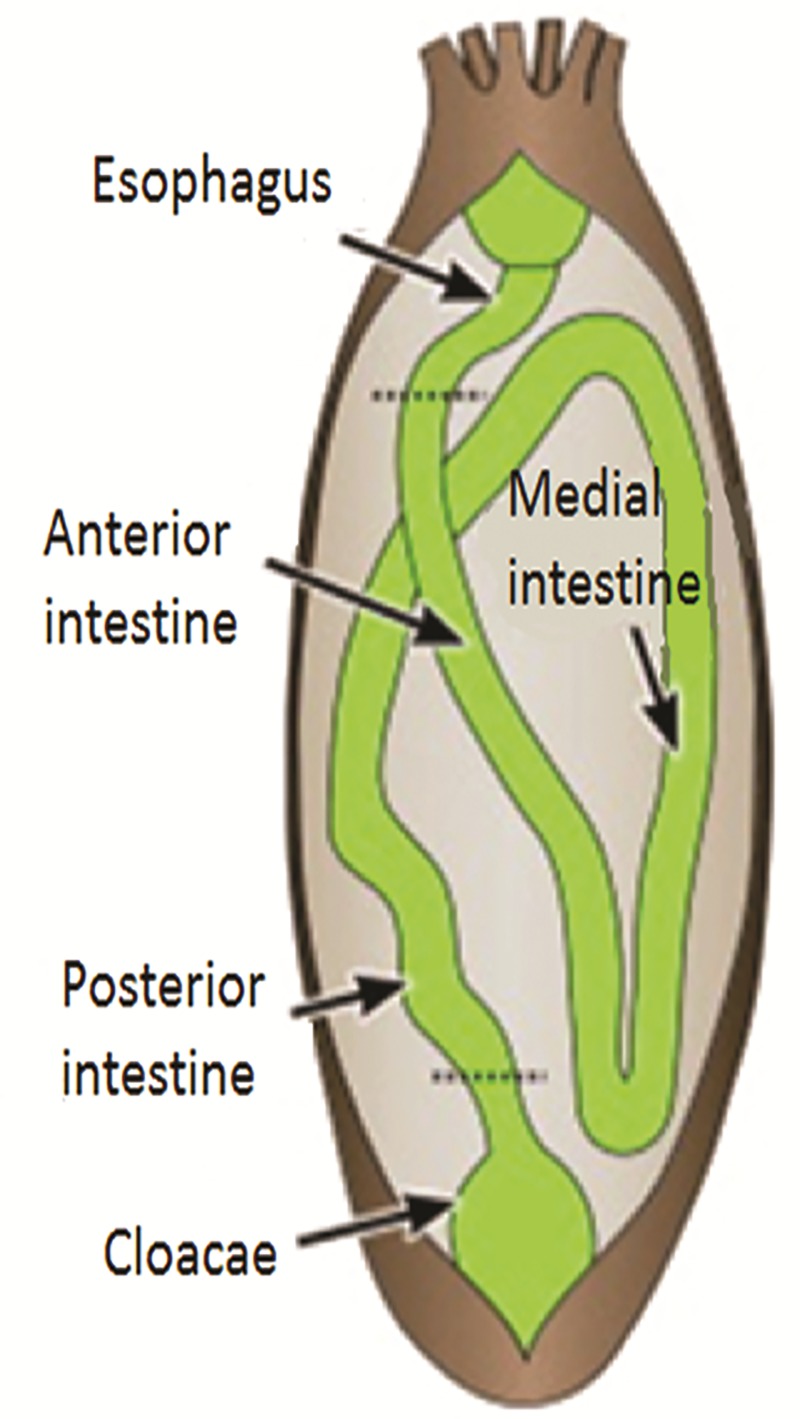
Fig 1. Anatomy of the digestive tract of the sea cucumber H. glaberrima.
The digestive tract of H. glaberrima is formed by a continuous tube that begins at the mouth, forming a short esophagus which is attached to a long descending small intestine (anterior) and a long ascending small intestine (medial). The final segment of the tube is a descending large intestine (posterior) that ends in the cloacae. (Diagram obtained from Mashanov et al. 2012. Adapted by MPJ).
The five animals that were transported to the lab were kept in a sea water aquarium for 24 hours. The sea water used in the aquaria was sea water brought from the animals’ natural environment. During this time the animals eliminated most of their intestinal content via the cloaca, and once this occurred, specimens were transferred to a sea water aquarium with fresh natural sea water to minimize the amount of digestive tract material present. These animals were dissected after three days in the aquarium, and digestive tract samples were taken using the same protocol as for the in situ dissections described above. By keeping animals in the aquaria for 3 days we reproduced the conditions that are used in regeneration experiments [44–46]. Therefore, the acquired data, and the comparison to natural environments will be important for future experiments on the association of bacteria with regenerative events taking place in the laboratory aquaria.
DNA extraction from intestine and water samples
DNA extraction was performed according to the manufacturer protocol (Qiagen's QIAamp DNA Mini Kit) (#51306, Valencia, CA US). For the DNA extraction of seawater bacteria, we removed the membranes and the remaining sucrose lysis buffer contained in the 15mL centrifuge-tube was centrifuged at 10,000 rpm for 30 minutes. The supernatant was discarded, and the pellet was dissolved in 1–2 mL NaCl (0.9%) to perform the DNA extraction following the same protocol as for the intestinal samples. DNA quantification was determined by absorbance measurements using a NanoDrop (1000 Spectrophotometer, Thermo Fisher Co.) device. The amount of DNA per sample varied from 2.6 mg/μl to 22.2 mg/μl. All samples were stored frozen at -20°C until used.
Sample preparation for pyrosequencing of 16S rRNA genes
PCR for multiplexing pyrosequencing was performed using universal bacterial barcoded primers. A set of primers was designed by adding a 12-nucleotide barcode to the forward primer 515F (5’-GAGTGCCAGCMGCCGCGGTAA). The reverse primer (not barcoded) was 806R (5’-CCGGACTACHVGGGTWTCTAAT). These primers targeted the V4-V5 regions of the 16S gene of bacteria for amplification. PCR was performed with a thermal cycler (PTC 100, Bio-Rad) under the following conditions: initial denaturation at 94°C for 3 min; 35 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 1 min; and a final extension at 72°C for 10 min. The PCR preparation consisted of 5μl of DNA, 2.5μl of barcoded primers and 10μl of Master Mix (Promega #M7502). PCR products were purified using Ultra Clean PCR Clean-Up (MoBio #12500) and were quantified with Quant-IT PicoGreen dsDNA Assay Kit (Invitrogen Cat # P11496). A mixture of PCR products was prepared and then was pyrosequenced using the Roche 454 FLX Titanium platform at the Sequencing and Genotyping Facility of University of Puerto Rico, according to the manufacturer’s instructions.
Taxonomic assignments and species richness of pyrosequencing reads
Statistical and bioinformatic analyses of bacterial 16S amplicons were done using QIIME pipeline to process data from high-throughput 16S rRNA sequencing studies [47]. Multiplexed and trimmed sequence reads (300bp) were clustered into OTU’s (Operational Taxonomic Units) at 97% sequence identity using UCLUST to estimate richness. The alignment of the sequences was done by PyNAST against the Greengenes core set. The OTU classification was done using RDP (Ribosomal Database Project)-classifier [48]. FastTree was used for building a phylogenetic tree [47]. Prior to phylogenetic tree building, the alignment was filtered to remove positions with gaps.
Comparison of microbial communities
Beta diversity metrics were calculated for each sample and the types of communities were compared using the taxonomic and phylogenetic assignments. UPGMA and PCoA plots were generated to visually depict the differences between the samples [47]. Beta significances were calculated as an “unweighted and weighted unifrac” which performs randomizations of sample/sequence assignments, and records the probability that one sample is phylogenetically different from the other samples, using Permutational multivariate analysis of variance (PermANOVA) test.
Results
A total of 138,029 V4-V5 16S rRNA gene sequences (~300bp) were obtained. The sequences were binned into 8,172 OTU’s (threshold cutoff for each OTU, 97% nucleotide sequence identity using UCLUST). The OTU classification was done using the RDP-classifier and we obtained 10 bacterial phyla, 23 classes, 44 orders, 83 families and 127 genera, that were present along the sea cucumber digestive tract. In terms of microbe relative abundance, the most abundant phyla were the Firmicutes (39.1%), Bacteroidetes (24.4%) and Proteobacteria (23.8%), followed by the Fusobacteria (4.2%) and Actinobacteria (1.3%). Unknown bacterial phyla represented 6.5% of OTU’s.
Within the Bacteroidetes, the most abundant genera included Bacteroides (2.5%) and Lewinella (1.3%); the families Porphyromonadaceae (8.9%) and Bacteroidaceae (2.5%); and the order Bacteroidales (3.2%). The most abundant groups in the Firmicutes were the genus Lactobacillus (2.1%); families, Lachnospiraceae (13.2%) and Ruminococcaceae (2%); and order Clostridiales (18.6%). The most abundant groups in the Proteobacteria were genera Vibrio (11.7%), Shimia (1.1%) and Helicobacter (1.2%); and order Oceanospirillales (2%). Within Fusobacteria the most important group was family Fusobacteriaceae (10%), and from Actinobacteria was the genus Corynebacterium (7%).
Bacterial distribution along the three segments of the intestine in the natural coastal environment
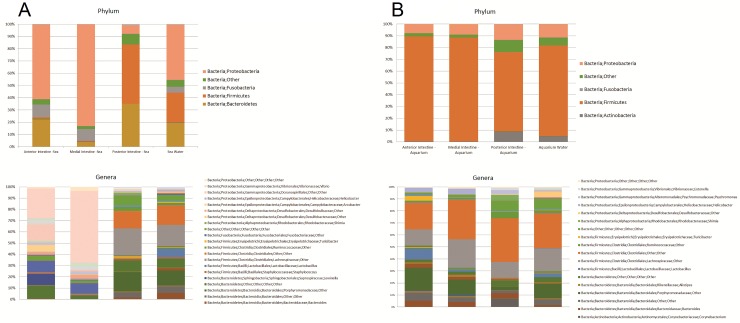
The microbiota found in different areas of the digestive tract of animals in their natural environment was similar at the phylum level. However, the relative representation differed remarkably. The results showed that the anterior and the medial (small) intestines are more similar between them when compared to the posterior (large) intestine. The former showed a greater proportion of Proteobacteria (61% in the anterior and 83% in the medial) and Fusobacteria (10%) and a smaller proportion of Firmicutes when compared to the posterior intestine (Fig 2). Notwithstanding, there were also differences between the two small intestinal segments, where the anterior intestine had a greater proportion of Bacteroidetes (22%), while the medial intestine only showed 4%. The posterior intestine was very different, with the most abundant phylum being the Firmicutes (48%), followed by Bacteroidetes (35%). In contrast with the anterior and medial intestine, the posterior intestine showed greatly reduced percentages of Proteobacteria (7%) while no Fusobacteria could be detected. Finally, the seawater sample reflects the taxonomy found in the three intestinal segments. Proteobacteria group 45%, Firmicutes 24%, Bacteroidetes 19%, and Fusobacteria 5% of the bacterial relative abundance. Similar to the digestive tract, a small number of bacteria (5%) could not be classified.
Fig 2. Bacterial taxa distribution in the intestinal system of H. glaberrima.
(A) Phylum and Genera of intestinal bacterial OTUs of animals in the sea (natural environment). B) Phylum and Genera of intestinal bacterial OTUs of animals in seawater tanks (aquarium environment).
At more specific levels, the anterior and medial intestines are dominated by the genus Vibrio (26% and 64% respectively), the families Fusobacteriaceae (10%) and Desulfobulbaceae (2% and 3% respectively), and other Bacteroidetes (11% and 3% respectively). In addition, the anterior intestine is dominated by the order Oceanospirillales (15%), and the genera Lewinella (10%) and Arcobacter (6%), whereas in the medial intestine these groups appear to be displaced by the genus Vibrio. On the other hand, the posterior intestine is different from the other two intestinal segments and is more similar to the seawater sample, where the Firmicutes and Bacteroidetes are dominant. Among the Firmicutes bacteria in the posterior intestine and seawater, the most abundant groups are: the Lachnospiraceae family (24% and 19% respectively), the order Clostridiales (15% and 17% respectively), and from this order, the Ruminococcaceae family (4% and 2% respectively). Secondly, the Bacteroidetes phylum is highly represented by the family Porphyromonadaceae (17% and 13% respectively), the order Bacteroidales (5% and 6% respectively) and others Bacteroidetes (10% and 8% respectively). In addition, we found a low representation of other Proteobacteria in both samples, except for the peculiar finding that the genus Shimia (2%) is found only in the posterior intestine segment.
Bacterial distribution along the three segments of the intestine in the aquarium environment
Many of the experiments performed in our laboratory require that animals be maintained in indoor seawater aquaria for prolonged periods of time. It is possible that the microbiota of animals in these conditions varies from that of animals in their normal habitats. To determine the microbiota of animals within the aquaria, we analyzed the bacterial taxonomy from intestinal samples after 3 days in the aquaria. Our results show that the microbiota of the digestive tract of animals in the aquaria was similar among the three different segments in terms of taxonomy and relative abundance. Their bacterial composition showed a large proportion of Firmicutes and Bacteroidetes. The Proteobacteria group is the least represented in the digestive tract of animals in the aquarium environment (Fig 2). On the other hand, we observed that the posterior intestine and aquarium water samples have representatives of Actinobacteria, a group of bacteria not found in the other samples.
At more specific levels all the samples have a similar taxonomic distribution. The dominant groups in the Bacteroidetes are the genera Bacteroides and Alistipes, and the family Porphyromonadaceae. For the Firmicutes, the dominant groups are the genera Lactobacillus, Turicibacter and Helicobacter; the families Lachnospiraceae and Ruminococcaceae; and the order Clostridiales.
On the other hand, as mentioned above, the posterior intestine has a representation of Actinobacteria that is dominated by the genus Corynebacterium (7%). Similar to animals in their natural environment, the posterior intestine from aquarium environment is the only sample that contains the genus Shimia (4%).
Beta-diversity of bacterial communities among water and intestinal samples
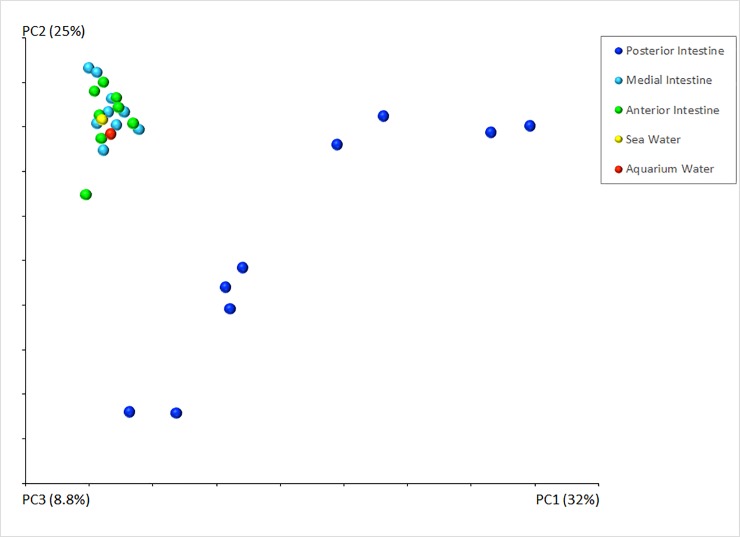
The weighted PCoA revealed that the anterior, medial and posterior intestine bacterial communities formed three significantly different clusters (P = 0.005). We compared the three intestinal segments, and the resulting graph showed a separation of the posterior intestine segment from both anterior and medial intestine segments (Fig 3) with a significance difference (p = 0.001). These results showed a concordance with the bacterial richness at phylogenetic levels (phylum), where the anterior and medial intestine segments shared greater similarity (No significant differences were found between anterior vs. medial and posterior intestine, or between medial vs anterior and posterior intestine. In general, both the aquarium water and the seawater samples were more similar to the anterior and medial intestine than with the posterior intestinal segment (Fig 3).
Fig 3. Principal Coordinate Analyses (PcoA) of bacterial communities in the intestine of H. glaberrima.
Samples clustered using PcoA of weighted UniFrac distance matrices that reflect the beta-diversity of the bacterial communities. The graph shows the UniFrac distance of bacterial communities from the anterior, medial and posterior intestine of the sea cucumber H. glaberrima.
Beta-diversity of bacterial communities between the host environments
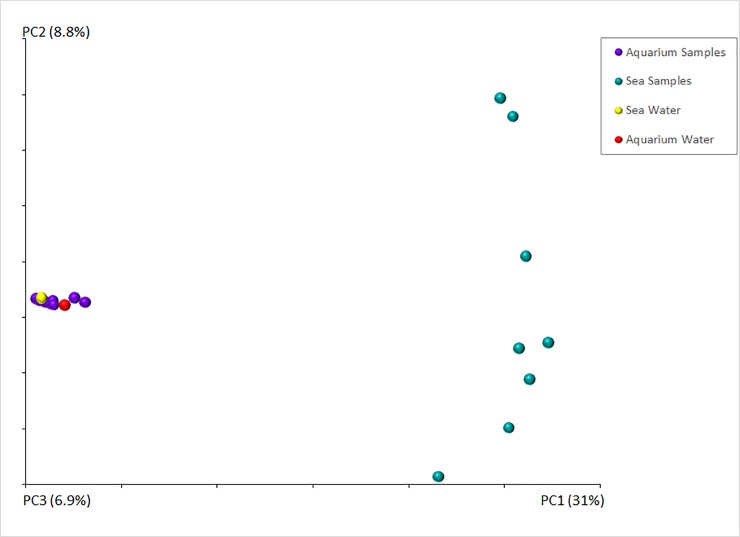
The UniFrac metric revealed that the samples from sea and aquaria formed two significantly different clusters (P = 0.001) based on the origin of the samples (Fig 4). Therefore the bacterial compositions of the host in the two environments are significantly different. Moreover, the seawater sample clustered with the aquarium intestinal samples (Fig 4).
Fig 4. Principal Coordinate Analyses (PcoA) of bacterial communities in the intestine of H. glaberrima in two different environmental settings.
The PcoA analysis reflects the beta-diversity of the bacterial communities from the various intestinal segment samples that originate from two environments; the animals collected in their natural sea environment and animals kept in seawater aquaria for 3 days.
Discussion
In accordance to many other studies where 16S data is used to determine microbial diversity, the number of genera identified by our study is much larger than the ~20 genera that have been identified in the digestive tract of several holothurian species using culture-dependent methods [38, 49]. This confirms the general finding that the use of “culture-dependent” methods to assess microbial diversity only detect a limited group of microorganisms; therefore, they cannot be used to define the entire microbiota within the intestine [50].
H. glaberrima intestinal microbiota comparison with other organisms
In this study, the characterization of intestinal bacteria of H. glaberrima revealed a dominance of Bacteroidetes, Firmicutes and Proteobacteria. Bacteroidetes and Firmicutes are typical dominant members of the vertebrate gut, particularly in mammals [51, 52], where the Bacteroidetes phylum is highly represented by the genus Bacteroides, and Firmicutes is mostly represented by the genera Clostridium, Ruminococcus, and Lactobacillus [1], this representation is similar to the most abundant taxonomy found in our study. Many studies have demonstrated that bacterial members of these two phyla are important for the normal intestinal physiology and homeostasis of vertebrate host [6, 7, 53]. Thus, our findings suggest that the gastrointestinal tract of marine and terrestrial deuterostomes share common microbial groups that can influence in the gastrointestinal metabolism of the host. The phylum Proteobacteria, the third most abundant phyla in H. glaberrima, has been found as a common member of the gut microbiota in adult zebrafish [54]. Both animals inhabit aquatic environments and it has been shown that in oceans and aquatic environments, Proteobacteria is the most abundant phylum comprising 79% of the bacterial biomass in deep sea, 64% in the sea surface, and 40% in fresh water [55].
The predominance of the phylum Proteobacteria is consistent with previous studies of the bacterial gut composition of other marine invertebrates [56,57]. Studies in the guts of Crustacea [Macrobrachium rosenbergii [58]; Palaemon paucidens [59]; Penaeus aztecus [60]; Mollusca [Donax gouldii [61], and Echinodermata [Echinus esculentus [37] reveal that genera members of Proteobacteria, such as, Vibrio and Pseudomonas, are commonly isolated in the three invertebrates phyla [57]. Vibrio is the most abundant genus found in H. glaberrima and it is consistent with studies in other echinoderms such as the sea urchins Strongylocentrotus droebachiensis and Tripneustes ventricosus [28], and the ophiuroid Ophionema sp, that suggest that echinoderms have a high population of Vibrio spp. in the gut, that may serve as reservoirs for the bacteria [24]. In addition, the phylum Proteobacteria has been found in high abundance in culture-dependent studies of members of the Holothuroidea: Benthodytes sp. [29], Stichopus japonicus [39], Holothuria atra [38], Holothuria leucospilota [40] and Apostichopus japonicus [33].
Taxonomic comparison between H. glaberrima microbiota and those of other echinoderms
Studies of the gut microbiota in echinoderms have been few, moreover high throughput sequencing studies are scarce. Using 454 pyrosequencing, Gao and colleagues [33], detected a higher bacterial diversity than previously described in the gut of sea cucumbers. They described 37 different phyla in the gut of A. japonicus, when previously only two phyla were reported. Similar findings have been done in two sea urchin species P. lividus and L variegatus [42,43].
When these studies are compared to our results, some interesting findings appear. The sea urchin L variegatus presents an almost exclusive abundance of Proteobacteria in the gut, and of these most belong to the Campylobacteraceae family [43]. This decreased biodiversity can be due to a proposed compartmentalization of gut bacteria that is separated from those in the ingesta pellet as proposed by the authors or to the specialized feeding strategy of the animals that depend mainly on sea grass for their nutrition.
More interesting is the comparison with A. japonicus. The gut content of both holothurian species show a high representation of Proteobacteria. However, while in A. japonicus the phylum Proteobacteria was the predominant group, our results in H. glaberrima show it as being one of three main groups represented. A. japonicus did not have an abundance of the Bacteroidetes and Firmicutes phyla. Moreover, A. japonicus also showed an abundance of Acidobacteria, Actinobacteria, Planctomycetes, and Chloroflexi that were not present (or present lower abundance) in our study. Interestingly, both pyrosequencing studies detected a high number of unknown bacteria that could not be classified by the database, making it possible that future studies could be directed to the identification of bacteria that have not yet been discovered.
There are many important differences between the two species that might influence their microbial diversity. H. glaberrima is a tropical and semi-tropical species in the Atlantic Ocean that is suspension-feeder and a detritivore, an animal that feeds on organic matter and detritus that comes from the action of waves breaking on the rocks that serve as the animal’s habitat [62, 63]. A japonicus is an epibenthic deposit-feeder that ingests sediments directly from the bottom floor, mainly found in temperate climates of the northern-western Pacific Ocean [33]. Although the main food sources of both are bacteria, microalgae, meiofauna, and dead organic matter of plant and animal origin [62, 64–67] the specific environment or food availability might be key to defining their microbiota.
Taxonomic, bacterial proportions and community structure among the three segments of the intestine of H. glaberrima
Our findings in H. glaberrima show that the distribution of microbes throughout the intestine is not homogenous. The anterior and medial intestines share a similar bacterial composition of Proteobacteria and Fusobacteria as predominant groups, while the posterior intestine (hindgut) has a higher diversity of microorganisms: Bacteroidetes, Firmicutes and Proteobacteria as predominant groups. The weighted PCoA plot showed that microbial communities of the anterior and medial intestines clustered together, while the posterior intestine was significantly different, this indicates similarities in the diversity and abundance of their microbial community. Our finding agreed with the results in A. japonicus that also reveal differences in bacterial communities between intestinal segments: in their case the anterior and posterior intestinal segments [33]. In the anterior gut content the most abundant phyla were Acidobacteria, Actinobacteria, Planctomycetes, Chloroflexi, and Proteobacteria, being the latter less abundant. On the other hand, the posterior gut content showed an abundance of Proteobacteria, and a low abundance of other phyla [33]. These results contrast with those obtained in H. glaberrima. As described above, the bacterial community of anterior and medial intestine of H. glaberrima showed an abundance of Proteobacteria. The posterior intestine (H. glaberrima) also reflected a difference in bacterial community. The most predominant groups were Firmicutes, Bacteroidetes and a low abundance of Proteobacteria (Fig 3). Despite these differences, at genera level we found some similarities between the two sea cucumbers: the genus Vibrio, the family Desulfobulbaceae and the class Gammaproteobacteria were dominant in the anterior parts of both animals. Moreover, although not abundant, both animals shared the presence of the genera Lactobacillus and Vibrio in posterior gut contents.
Comparison of bacterial taxonomy of the digestive tract between natural and aquarium environments
Our results showed a notable bacterial difference between the holothurian intestinal microbiota obtained from a natural coastal environment and those kept in indoor aquaria. It might be suggested that these differences in bacterial composition occur due to the intake of food available within the sea cucumber’s environment. In the still waters of the aquarium environment, H. glaberrima specimens do not have the ability to feed as they do in the ocean, and their digestive tract is usually empty of the detritus, organic and inorganic matter that can be found within animals in natural conditions. (Animals can be kept in the aquarium for over 2 months. It is not certain if these “unfed” animals are obtaining nutrients from other sources, such as aquarium bacteria”. Nonetheless, “unfed” animals, serve as controls for animals that have eviscerated their digestive tract and are in the process of regeneration, since the latter lack a functional digestive tracts for at least two weeks.) Our data suggest that bacterial groups found in the anterior digestive tract of animals in natural environments but not of those in aquaria, such as Fusobacteria and the Proteobacteria-Vibrio, could be originating from the food intake. For example, it was found that differences in bacterial communities in the foregut (anterior intestine) may be caused by the selective feeding of the sea cucumber [33, 68–70]. These animals may use the bacteria directly as food source or they can use the bacteria indirectly to provide them with essential nutrients [31,32,71]. In addition, it has been suggested that the variation in the bacterial composition could be due to the food source of the sea cucumber, because it is known that the process of succession (the progressive replacement of one community by another until a climax community is established) can be caused by host external factors such as exposition to new microbes that enter the gastrointestinal tract through food [72].
H. glaberrima core microbiota
Our finding that the posterior intestinal segment of animals in the sea environment was similar to the posterior segment of intestine of animal from the aquarium environment suggests that this segment was less susceptible to changes in its microbial composition despite changes in environment. It is known that of the gut regions of invertebrates, the most susceptible to harboring an indigenous microbiota is the posterior intestine [56,57]. Bacteria in this region have access to leftover digesta and are not competing directly with their host for uptake of digested compounds. Furthermore, the posterior intestine function is to eliminate waste material from the body [73], therefore, it is expected that the bacterial composition of this segment could help to carry out this function, after the food has been digested.
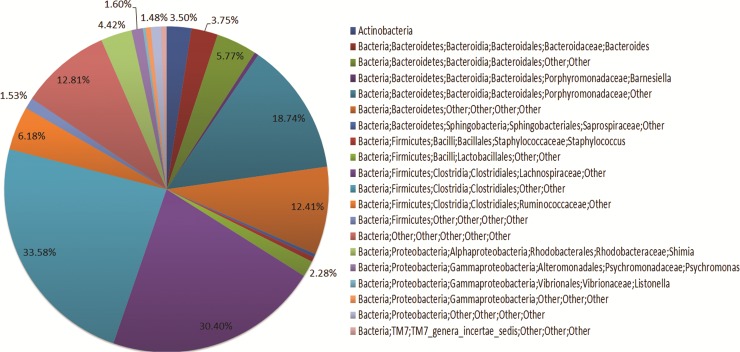
Based on the results and analysis of the taxonomy and the study of the different environments we can propose the bacterial community found in the posterior intestine represents the intestinal core microbiota for H. glaberrima (Fig 5). The most abundant groups would be from the phylum Bacteroidetes: the family Porphyromonadaceae (18.74%), and others Bacteroidetes (12.41%); and from the phylum Firmicutes: the family Lachnospiraceae (30.40%), the order Clostridiales (33.58%) and the family Ruminococcaceae (6.18%). The phylum Proteobacteria would be less abundant, with a representation of the genera Shimia (4.42%), Psychromonas (1.60%) and Listonella (0.38%), the class Gammaproteobacteria (0.73%) and other Proteobacteria (1.48%).
Fig 5. Proposed endogenous bacterial community of H. glaberrima intestine.
We propose that the microbiota found in the posterior intestine of the sea cucumber, either in their natural habitat (ocean) or aquarium environments represents the endogenous microbiota of these animals.
At genera level, there are six (6) specific members of the core microbiota of H. glaberrima. These are: Bacteroides, Barnesiella, Staphylococcus, Shimia, Psychromonas and Listonella.
A particularly interesting case is the presence of Shimia as part of the holothurian microbiota. Shimia is a novel rod-shaped marine proteobacterium isolated from a biofilm in a coastal fish farm [74] and from the gut of abalone [75]. It is motile and grows on marine agar as colorless or beige colonies [74]. In our study, this genus was found in the posterior intestine sample for both environments, and these are the only regions of the sea cucumber intestine that contains Shimia in high proportions compared to the other segments of intestine.
The finding that the microbiota of the posterior intestinal segment is similar between animals in the natural and the aquarium environments is of importance to our future regeneration studies. It provides a baseline comparative value that can be reproduce in the laboratory and analyzed to determine the possible changes taking place during intestinal regeneration.
In conclusion, this is the first high-throughput study characterizing the microbiota of the intestine of H. glaberrima, which can be used, along with other echinoderm microbiome studies as a base for understanding the microbial ecology of these marine invertebrates. We also present here the first study that compares the bacterial composition of different segments of intestine in two different environments, which can shed a clearer view of the core microbial community of this organism, and provides important changes in the microbiota of this animal that should be taken into account when performing other studies in aquaria.
Data Availability
All data from this study is publicly available on the European Nucleotide Archive study number PRJEB30491 (https://www.ebi.ac.uk/ena/data/view/ERP112954).
Funding Statement
This work was supported by the Puerto Rico Science and Technology Research Trust (http://prsciencetrust.org); University of Puerto Rico Sequencing Genomic Facility (http://www.mcc.com.pr/facilities/sequencing-and-genotyping-facility-sgf-2/).
References
- 1.Walter J, Ley R. The human gut microbiome ecology and recent evolutionary changes. Annu Rev Microbiol 2001; 65: 411–29 [DOI] [PubMed] [Google Scholar]
- 2.Hooper L, Gordon JI. Commensal host-bacterial relationship in the gut. Science 2001; 292: 1115–8 [DOI] [PubMed] [Google Scholar]
- 3.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomics analysis of the human distal gut microbiome. Science 2006; 312:1355–9 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corthier G, Doré J. A new era in gut research concerning interactions between microbiota and human health. Gastroenterol Clin Biol. 2010; 34 Suppl 1: S1–6 [DOI] [PubMed] [Google Scholar]
- 5.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010; 328:228–31 10.1126/science.1179721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA 2006; 103:10011–6 10.1073/pnas.0602187103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harley ITW, Karp CL. Obesity and the gut microbiome: Striving for causality. Molecular Metabolism 2012. p. 1–11 10.1016/j.molmet.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol 2003; 4:269–73 10.1038/ni888 [DOI] [PubMed] [Google Scholar]
- 9.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host–microbial relationships in the intestine. Science 2001; 291: 881–4 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- 10.Vindigni SM, Zisman TL, Suskind DL, Damman CJ. The intestinal microbiome, barrier function, and immune system in inflammatory bowel disease: a tripartite pathophysiological circuit with implications for new therapeutic directions. Therap Adv Gastroenterol. 2016; 9:606–25 10.1177/1756283X16644242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uribe A, Alam M, Midtvedt T, Smedfors B, Theodorsson E. Endogenous prostaglandins and microfiora modulate DNA synthesis and neuroendocrine peptides in the rat gastrointestinal tract. Scand J Gastroenterol 1997; 32:691–9 [DOI] [PubMed] [Google Scholar]
- 12.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of hostmicrobial interactions in an open mammalian ecosystem. Science 1996; 273:1380–3. [DOI] [PubMed] [Google Scholar]
- 13.Husebye E, Hellström PM, Midtvedt. The intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of the migrating myoelectric complex. Dig Dis Sci 1994; 39:946–56. [DOI] [PubMed] [Google Scholar]
- 14.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015; 161; 264–76 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci. USA 2011; 108:3047–52 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013; 18: 666–73 10.1038/mp.2012.77 [DOI] [PubMed] [Google Scholar]
- 17.Selkrig J, Wong P, Zhang X, Pettersson S. Metabolic tinkering by the gut microbiome: Implications for brain development and function. Gut Microbes 2014; 5:369–80 10.4161/gmic.28681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savignac HM, Kiely B, Dinan TG, Cryan JF. Bifidobacteria exert strain-specific effects on stress-related behavior and physiology in BALB/c mice. Neurogastroenterol. Motil. 2014; 26:1615–27 10.1111/nmo.12427 [DOI] [PubMed] [Google Scholar]
- 19.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav. Brain Res. 2015; 287:59–72 10.1016/j.bbr.2015.02.044 [DOI] [PubMed] [Google Scholar]
- 20.Zitvogel L, Galluzzi L, Viaud S, Vetizou M, Daillere R, Merad M, et al. Cancer and the gut microbiota: An unexpected link. Sci Transl Med 2015; 7(271): 271ps1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Intl J Mol Sci 2017; 18:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-Associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc R Soc B 2012; 280:20122328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 2016; 70:317–40 10.1146/annurev-micro-102215-095440 [DOI] [PubMed] [Google Scholar]
- 24.Webster NS, Thomas T. The sponge hologenome. mBio 2016; 7(2) e00135–16. 10.1128/mBio.00135-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClintock JB. Trophic biology of antarctic shallow-water echinoderms. Mar Ecol Prog Ser. 1994; 111:191–202 [Google Scholar]
- 26.Hirimuthugoda NY, Zhenming C, Zhu KL. Probiotics and sea cucumber farming. SPC Beche-de-mer Information Bulletin 2006; 24:45–8 [Google Scholar]
- 27.De Ridder C, Jangoux M, De Vos L. Description and significance of a particular intradigestive symbiosis between bacteria and deposit-feeding echinoid. J. Exp Mar Biol Ecol 1985; 91:65–76 [Google Scholar]
- 28.Guerinot ML, Patriquin DG. N2 –fixing vibrios isolated from the gastrointestinal tract of sea urchins. Can J Microbiol 1981; 27:311–7 [DOI] [PubMed] [Google Scholar]
- 29.Dilmore LA, Hood MA. Vibrios of some deep-water invertebrates. FEMS Microb Letters 1986; 35:221–4 [Google Scholar]
- 30.Odintsov VS. Nitrogen fixation (acetylene reduction) in the digestive tract of some echinoderms from Vostok Bay in the sea of Japan. Mar Biol Lett 1981; 2:259–63 [Google Scholar]
- 31.Deming JW, Colwell RR. Barophilic bacteria associated with digestive tracts of abyssal holothurians. Appl Environ Microbiol 1982; 44:1221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deming JW, Tabor PS, Colwell RR. Barophilic growth of bacteria from intestinal tracts of deep-sea invertebrates. Microb Ecol 1981; 7:85–94. 10.1007/BF02010480 [DOI] [PubMed] [Google Scholar]
- 33.Gao F, Li F, Tan J, Yan J, Sun H. Bacterial Community Composition in the Gut Content and Ambient Sediment of Sea Cucumber Apostichopus japonicus revealed by 16S rRNA Gene Pyrosequencing. PLOS ONE 2014; 9(6):e100092 10.1371/journal.pone.0100092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweijd NA, Pillay D, McQuaid CD, Bandu VH, Baecker AAW. Filamentous structures associated with the gut mucosa of the sea urchin Parechinus angulosus. Electron Microsc Soc SA 1989; 19:99–100 [Google Scholar]
- 35.Lynch JE. Studies on the ciliates from the intestine of Strongylocentrotus. I. Entorhipodium gen nov. Univ Calif Publ Zool 1929; 33:27–56 [Google Scholar]
- 36.Lynch JE. Studies on the ciliates from the intestine of Strongylocentrotus. II. Lechrioiphyla mystax, Gen nov., sp. nov. Univ Calif Publ Zool 1930; 33: 307–50 [Google Scholar]
- 37.Unkles SE. Bacterial flora of the sea urchin Echinus esculentus. Appl Environ Microbiol 1977; 34:347–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward-Rainey N, Rainey FA, Stackebrandt E. A study of the bacterial flora associated with Holothuria atra. J Exp Mar Biol Ecol 1996; 203: 11–26 [Google Scholar]
- 39.Dou SYC. The microbial composition of Stichopus japonicus and its physiological property. Oceanol Limnol Sinica 1989; 20:300–7 [Google Scholar]
- 40.Zhang X, Nakahara T, Miyazaki M, Nogi Y, Taniyama S. Diversity and function of aerobic culturable bacteria in the intestine of the sea cucumber Holothuria leucospilota. J Gen Appl Microbiol 2012; 58: 447–56 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Nakahara T, Murase S, Nakata H, Inoue T. Physiological characterization of aerobic culturable bacteria in the intestine of the sea cucumber Apostichopus japonicus. J Gen Appl Mic 2013; 59(1);1–10. [DOI] [PubMed] [Google Scholar]
- 42.Becker P, Egea E, Eeckhaut I. Characterization of the bacterial communities associated with the bald sea urchin disease of the echinoid Paracentrotus lividus. J Invertebr Pathol 2008; 98:136–47 10.1016/j.jip.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 43.Hakim JA, Koo H, Kumar R, Lefkowitz EJ, Morrow CD, Powell ML, et al. The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiol Ecol. 2016; 92(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Arrarás J, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrina Selenka (Holothuroidea: Echinodermata). J. Exp. Biol. 1998; 281:288–304 [DOI] [PubMed] [Google Scholar]
- 45.Mashanov VS, Zueva OR, García-Arrarás JE. Postembryonic organogenesis of the digestive tube: why does it occur in worms and sea cucumbers but fail in humans? Curr. Topics Dev. Biol. 2014; 108:185–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García-Arrarás JE, Lázaro-Peña MI, Díaz-Balzac CA. Holothurians as a model system to study regeneration. Results Probl Cell Differ. 2018; 65:255–83 10.1007/978-3-319-92486-1_13 [DOI] [PubMed] [Google Scholar]
- 47.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010; 7: 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. NuclAcids Res (2008) 37:141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu-feng, Tian L, Duzong J, Dong S. Comparative study on seasonal variation of heterotrophic bacteria flora associated with intestine of sea cucumber in feeding and unfeeding ponds. J Anhui Agricul Sc 2009; 27: 13113–17 [Google Scholar]
- 50.Dyall-Smith M, Aharon O. Culture-Dependent Study of Microbial Diversity of Lake Chaka. Appl Environ Microbiol. 2006; 72:7427 10.1128/AEM.01401-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The Human Microbiome Project. Nature 2007; 449:804–10 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009; 106:5859–64. 10.1073/pnas.0901529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 2007; 104:979–84 10.1073/pnas.0605374104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–608 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battistuzzi FU, Hedges SB. A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol 2009; 26:335–43 10.1093/molbev/msn247 [DOI] [PubMed] [Google Scholar]
- 56.Lau WW, Jumars PA, Armbrust EV. Genetic diversity of attached bacteria in the hindgut of the deposit-feeding shrimp Neotrypaea (formerly Callianassa) californiensis (decapoda: thalassinidae). Microb Ecol.2002; 43:455–66 10.1007/s00248-001-1043-3 [DOI] [PubMed] [Google Scholar]
- 57.Harris JM. The presence, nature and role of gut microflora in aquatic invertebrates: a synthesis. Microb Ecol 1993; 25:195–231 10.1007/BF00171889 [DOI] [PubMed] [Google Scholar]
- 58.Colorni A. A study on the bacterial flora of giant prawn Macrobacterium rosenbergii, larvae fed with Artemia salina nauplii. Aquaculture 1985; 49:1–10 [Google Scholar]
- 59.Sugita H, Takahashi T, Kanemoto Fl, Deguchi Y. Aerobic bacteria flora in the digestive tracts of freshwater shrimp Palaemon paucidens acclimated with seawater. Nipp Suisan Gakkaishi 1987; 53:511 [Google Scholar]
- 60.Dempsey AC, Kitting CL. Characteristics of bacteria isolated from penaeid shrimp. Crustaceana 1987; 2:90–4 [Google Scholar]
- 61.Beeson RJ, Johnson PT. Natural bacterial flora of the bean clam, Donax gouldi. J Invert Pathol 1967; 9:104–10 [Google Scholar]
- 62.Massin C. Food and feeding mechanisms: Holothuroidea In Jangoux M. and Lawrence J. M. (eds) Echinoderm nutrition. Rotterdam, A. A. Balkema: 1982; 43–55 [Google Scholar]
- 63.Michio K, Kurata K. Effects of deposit feeders Stichopus japonicus on algal bloom and organic matter contents of bottom sediments of the enclosed sea. Mar. Poll. Bull., 2003; 47:118–25 [DOI] [PubMed] [Google Scholar]
- 64.Yingst YI. The utilization of organic matter in shallow marine sediments by an epibenthic deposit-feeding holothurians. J Exp Mar Biol Ecol 1976; 23:55–69 [Google Scholar]
- 65.Moriarty DJW. Feeding of Holothuria atra and Stichopus chloronotus on bacteria, organic carbon and organic nitrogen in sediments of the Great Barrier Reef. Aust. J. mar. Freshwat. Res. 1982; 33:255–63 [Google Scholar]
- 66.Uthicke S. Nutrient regeneration by abundant coral reef holothurians. J Exp Mar Biol Ecol 2001; 265:153–70 [Google Scholar]
- 67.Lopez GL, Levinton JS. Ecology of deposit-feeding animals in marine sediments. Quart Rev Biol 1987; 62: 235–60 [Google Scholar]
- 68.Sloan NA, von Bodungen B. Distribution and feeding of the sea cucumber Isostichopus badionotus in relation to shelter and sediment criteria of the Bermuda Platform. Mar Ecol Prog Ser 1980; 2:257–64 [Google Scholar]
- 69.Uthicke S, Karez R. Sediment patch selectivity in tropical sea cucumbers (Holothuroidea: Aspidochirotida) analyzed with multiple choice experiments. J Exp Mar Biol Ecol 1999; 236:69–87 [Google Scholar]
- 70.Navarro PG, García-Sanz S, Barrio JM, Tuya F. Feeding and movement patterns of the sea cucumber Holothuria sanctori. Mar Biol 2013; 160(11):2957–66. [Google Scholar]
- 71.Amaro T, Witte H, Herndl GJ, Cunha MC, Billett DSM. Deep-sea bacterial communities in sediments and guts of deposit-feeding holothurians in Portuguese canyons 10 (NE Atlantic), Deep-Sea Res. I, 2009; 56: 1834–43 [Google Scholar]
- 72.Mackie RJ, Abdelghani S, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Amer. J. Clin Nutr Suppl. 1999; 69:1035S–45S. [DOI] [PubMed] [Google Scholar]
- 73.Sandle G. Salt and water absorption in the human colon: a modern appraisal. Gut 1998; 43:294–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi Dong H, Buyng Cho. Shimia marina gen. nov., a novel bacterium of the Rosebacter clade isolated from biofilm in a coastal farm fish. Int J Syst Evol Microbiol 2006; 56:1869–73 10.1099/ijs.0.64235-0 [DOI] [PubMed] [Google Scholar]
- 75.Hyun DW, Kim MS, Shin NR, Kim JY, Kim PS, Whon TW, et al. Shimia haliotis sp. nov., a bacterium isolated from the gut of an abalone, Haliotis discus hannai. Int J Syst Evol Microbiol, 2013; 63: 4248–53. 10.1099/ijs.0.053140-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from this study is publicly available on the European Nucleotide Archive study number PRJEB30491 (https://www.ebi.ac.uk/ena/data/view/ERP112954).