Abstract
Bacterial diseases cause high mortality in Penaeus (Litopenaeus) vannamei postlarvae. Therefore, appropriate application of efficient therapeutic products is of vital importance for disease control. This study evaluated through in vitro analyses the antimicrobial effectiveness of commercial therapeutic products used for P. vannamei bacterial diseases and antibiotics against pathogenic Vibrio strains circulating in Ecuadorian hatcheries. Twenty strains were isolated from 31 larvae samples with high bacterial counts from 10 hatcheries collected during mortality events. The strains virulence was verified through challenge tests with Artemia franciscana nauplii and P. vannamei postlarvae. Through 16S rRNA sequence analysis, strains showed a great similarity to the Vibrio sequences reported as pathogens, with 95% belonging to the Harveyi clade. Through antibiograms and minimal inhibitory concentration (MIC) in vitro tests we found that furazolidone, ciprofloxacin, chloramphenicol, norfloxacin, nalidixic acid, florfenicol, fosfomycin and enrofloxacin inhibited the growth of all or most of the strains. Less efficient antibiotics were penicillin, oxytetracycline and tetracycline. A multiple antibiotic resistance (MAR) index of 0.23 showed some level of resistance to antibiotics, with two MAR prevalent patterns (Penicillin-Oxytetracycline and Penicillin-Oxytetracycline-Tetracycline). From a total of 16 natural products (five probiotics, nine organic acids and two essential oils), only three (one probiotic, one organic acid and one essential oil) were effective to control most of the strains. Shrimp producers can apply relatively simple in vitro analyses, such as those employed in this study, to help take adequate management decisions to reduce the impact of bacterial diseases and increase profit.
Introduction
The high demand of postlarvae to support the cultured shrimp industry and consequently the intensification at hatchery level, together with the trade of aquatic animals and their associated products, has increased the occurrence of infectious pathogens in this production stage [1]. One of the main concerns in shrimp hatcheries are the bacterial pathogens [2]. Vibrio spp, such as Vibrio harveyi [3–8], Vibrio alginolyticus [9, 10] and Vibrio campbellii [7, 11, 12] are recurrent pathogens in shrimp hatcheries in America and Asia. In Ecuador, shrimp hatcheries have suffered from some bacterial diseases caused by pathogens of the Vibrio genus, such as Bolitas nigricans syndrome, caused by V. harveyi [3], and Zoea 2 syndrome, caused by V. harveyi and V. alginolyticus [4]. Therefore, the efficiency of therapeutic products is of vital importance for the control of aquaculture diseases.
Antibiotics are extensively used as prophylactics against bacterial pathogens [13]. However, the use of antibiotics carries important disadvantages, these being residues in aquaculture products [14–16], development and propagation of resistance between pathogens, including human pathogens [17]. For these reasons, the regulation of antibiotics is rigorously controlled, resulting in few antibiotics authorized for use in aquaculture. In this context, alternative strategies of disease control are necessary to replace antibiotics for use in animal production, which has led to consider the use of natural products to control the growth of pathogens in shrimp hatcheries. The administration of probiotics is one of the alternative strategies that may be used in aquaculture [13]; their benefits include the potential for colonization in the gastrointestinal tract, selective antagonism against bacterial pathogens, improvement of the shrimp immune system, enhanced shrimp growth and survival, degradation of detritus and maintenance of water quality [18–20]. The use of organic acids, produced by organisms and used as preservatives and bacterial control in food, agriculture, and animal production, is another potential strategy to control bacterial diseases in animal production [21–23]. They inhibit the growth of pathogenic V. harveyi, Vibrio cholera, V. alginolyticus, Vibrio parahaemolyticus and V. campbellii [24–26], exhibit immunostimulant properties [24, 27], and improve the nutritional and health state of shrimp [28, 29]. Similarly, essential oils have shown to have antimicrobial [30], antioxidant [31] and antifungal [32] properties, which can be an alternative to the use of additives and drugs in shrimp production [33]. Although the use of organic acids and essential oils in Ecuadorian hatcheries is relatively new, there is an increasing demand for their application as control strategies of bacterial diseases in shrimp hatcheries. In general, there are a huge number of products marketed as therapeutic products for shrimp hatcheries worldwide, therefore producers should take suitable decisions as to which products are effective based on technical information and further tests in their own facilities.
The main objective of this study was to determine through in vitro analyses the antimicrobial effectiveness of antibiotics and some commercial products used in Ecuador as therapeutic agents for shrimp larviculture. To determine this, we first performed a survey to identify the pathogenic bacterial strains circulating in Ecuadorian shrimp hatcheries, confirming their virulence through challenge tests and verifying their molecular similarity with previously reported pathogenic Vibrio. Then, we tested the antimicrobial effectivity of some antibiotics and commercial products used in Ecuador as therapeutics against the pathogenic circulating strains through in vitro tests.
Material and methods
Sample collection and processing
In 2015, 31 samples of Penaeus (Litopenaeus) vannamei larvae [from Nauplii 5 (N5) to 13 days postlarvae (PL13)] were collected from tanks of 10 shrimp hatcheries (Santa Elena, Ecuador) during mortality events. Samples were sent by farmers to the Centro Nacional de Acuicultura e Investigaciones Marinas (CENAIM, Santa Elena, Ecuador) for the quantification of shrimp bacterial load (microbiologic analysis services performed by CENAIM). Bacterial strains isolated from these samples were used for this study. Larvae presented clinical signs of abnormal swimming behavior, empty digestive tract, low activity and retardation of larval development. Samples were transported to the research facilities of CENAIM taking a maximum time of two hours from sampling at the hatcheries to processing at the laboratory. At the laboratory, they were rinsed with 2% sterile NaCl solution and each sample (1 g of larvae) was macerated to homogenize the bacterial load associated with the animals.
Isolation and preservation of bacterial strains
Aliquots (100 μL) of serial 10-fold dilutions (10−3 to 10−5) of larvae were macerated in duplicate in 2% sterile NaCl solution and plated on marine agar 2216 (MA, Difco). The same procedure was performed with serial 10-fold dilutions (10−1 to 10−3) on thiosulfate citrate bile salt sucrose agar (TCBS, Difco). All plates were incubated at 30°C. After one to two days of growth, bacterial counts were performed from plates containing 30 to 300 colonies. Bacterial counts were expressed as colony-forming units (CFU) per gram. Presumptive pathogenic strains were selected from colonies of samples with high bacterial counts on MA (>106 CFU g-1) or TCBS (>105 CFU g-1) agars. The criteria of selection were: (1) all different bacterial strains by morphological criterion and (2) luminescent strains. All selected strains were coded and frozen at -80°C after addition of trypticase soy broth (TSB, Difco) supplemented with 2.0% (w/v) NaCl and 20% (v/v) glycerol.
Challenge tests
The pathogenicity of the presumptive pathogenic strains was first evaluated in brine shrimp Artemia franciscana nauplii, following the procedures described by [34], with few modifications. Briefly, 1 g cysts of A. franciscana (Batch 02143, INVE Aquaculture, Belgium) were hydrated under continuous aeration in 100 mL of filtered and autoclaved distilled water for 1 h, and then transferred to a mixture of 10 mL of sodium hypochlorite (10%) and 15 mL of sodium hydroxide (40%), until the cysts changed color from brown to orange. The decapsulated cysts were washed with filtered and autoclaved sea water and transferred to an imhoff funnel with 500 mL of filtered and autoclaved sea water, providing continuous aeration and illumination with a white lamp, remaining at 28°C in a sterile environment for 24 h. Nauplii were harvested under sterile conditions with an autoclaved mesh of 100 μm. Eighty-four groups, each composed of 30 Artemia nauplii, were transferred to 50 mL sterile tubes containing 30 mL of filtered and autoclaved sea water. Eighty tubes were used to test the pathogenicity of the 20 bacterial isolates (four replicates per strain) and four tubes were used for the negative control (without bacteria, four replicates). Bacteria were activated on trypticase soy agar (TSA, Difco) and a colony from each strain was transferred to 150 mL of TSB and incubated for seven hours at 30°C. Bacterial cells, at a density of 106 cell mL-1, were added to the corresponding sterile tubes immediately after the transfer of the Artemia nauplii. Nauplii from all treatments and control were fed once with an inactivated V. alginolyticus commercial probiotic (107) [35] four hours after infection. For this, the probiotic was cultured in liquid medium for six hours, inactivated by heat (autoclaved at 121°C for 20 min), centrifuged at 10000 rpm for 10 min and resuspended with autoclaved seawater. Water exchange (50%) was performed 24 hours after infection and the mortality of Artemia was quantified 48 h post-infection. The whole challenge was performed in a Class II Biological Security Cabinet (CSB-180 A). To verify the asepsis condition of the negative control, water samples were collected at the end of the challenge, thus verifying the absence of Vibrio growth in TCBS agar.
Bacterial strains causing higher mortalities in the Artemia challenge test were selected and their pathogenicity was again verified by a challenge test using healthy Penaeus vannamei postlarvae, following the procedures described by [36]. Bacteria were activated following the procedure described in the Artemia challenge test. Thirty shrimp postlarvae (PL2) per replicate were distributed in sterile petri dishes and exposed to the corresponding bacterial treatment, at a concentration of 108 bacteria mL-1 for 6 min. Larvae were then transferred to 500 mL plastic containers containing 300 mL of sterile seawater and 106 bacteria mL-1. Larvae were fed during the challenge with a pure culture of Thalassiosira weissflogii (913 cell mL-1) every 2 h after exposure to the bacteria. Survival was determined by counting the larvae every 4 h until 38 hours after infection. A negative control was also included in the challenge test (shrimp postlarvae PL2 without bacterial treatment). All treatments including the control had four replicates. The whole challenge was performed in a Class II Biological Security Cabinet (CSB-180 A), with constant aeration. To verify the asepsis condition of the negative control, water samples were collected at the end of the challenge, verifying the absence of Vibrio growth in TCBS agar.
Bacterial characterization by 16S rRNA sequence analysis
Identification of the presumptive pathogenic strains was performed by 16S rRNA sequence analysis. Total genomic DNA was extracted from pure cultures of the bacterial strains after they were cultured on TSA and incubated for 24 h at 30°C. Bacteria were lysed by incubation at 55°C for 1 h in 200 μL of STE buffer (10 mM Tris-HCl, 1 mM EDTA and 100 mM NaCl, pH 8), followed by purification, adding an equal volume of phenol-chloroform-isoamylalcohol (25:24:1) and continuing with a chloroform-isoamylalcohol extraction (24:1). DNA was recovered by adding ethanol (100%) followed by centrifugation at 13000 rpm for 10 min. The pellet was washed with 70% ethanol, dried and resuspended in 50 μL of ultrapure water (pH 7.0). DNA was preserved at -20°C for further use. DNA concentration and purity were estimated with a Varioskan LUX multimode microplate reader (Thermo Fisher Scientific). The 16S rRNA complete gene was amplified using primers suggested by [37] (27F: 5'-AGAGTTTGATCMTGGCTCAG-3', 1492R: 5'-TACGGYTACCTTGTTACGACTT-3'). PCR was performed in a 30 μL reaction mixture containing 1X Buffer NH4 (Bioline, Sydney, Australia), 2.5 mM MgCl2 (Invitrogen, Carlsbad, CA), 2 mM of each dNTP, 0.3 μM of each primer, 0.5 units of Taq DNA polymerase and 2 μL of DNA. PCR cycling conditions were: DNA denatured for 5 min at 94°C, 35 cycles at 94°C for denaturation, 1 min at 52°C for annealing step and 1 min at 72°C for elongation; and a last cycle of 10 min at 72°C to complete the elongation. Amplicons were separated by 1.5% (w/v) agarose gel electrophoresis, stained with SYBR Safe DNA gel stain (Thermo Fisher Scientific), and illuminated under UV light. Images were captured with an E-Gel Imager System (Thermo Fisher Scientific). PCR products were purified and dissolved in 30 μL of ultrapure water for direct sequencing (Macrogen, Korea). BigDye Terminator Cycle sequencing kit (Perkin Elmer) was used for the sequencing. The sequencing products were analyzed with the ABI 3000 sequencer (Applied Biosystems, Foster City, CA, USA). A phylogenetic analysis was carried out with the complete 16S rRNA sequences (1465 bp) from the bacterial isolates, together with 16S rRNA sequences (n = 362) of different Vibrio (pathogens and non-pathogens) obtained from GenBank. The amino acid sequence alignments were generated with ClustalW and the specific region of 16S rRNA was identified using Bioedit 7.0.0. [38]. Phylogenetic trees were built using Maximum Parsimony (MP), Maximum Likelihood (ML), Neighbor Joining (NJ) and Bayesian Inference (BI). The molecular evolution model was selected through a wide range of phylogenetic and evolutionary tools using a dataset composed only of the unique haplotypes and sequences obtained in the present investigation. JModeltest 2.0 was used to test the evolution models based on the hierarchical likelihood ratio test [39]. Values of amino acid substitutions per site for the gene were calculated with MEGA 6.0 (Molecular Evolutionary Genetics Analysis). The 16S rRNA sequences were deposited in GenBank under accession numbers MH997724 to MH997742.
Antimicrobial effectiveness
The antimicrobial effectivity of 16 natural products (five probiotics, nine organic acids and two essential oils, Table 1) used in Ecuador as therapeutic agents against shrimp bacterial diseases and eleven antibiotics was screened in terms of the susceptibility of the pathogenic circulating strains to these products through antibiogram and minimal inhibitory concentration tests (Table 1). We denominated as pathogenic circulating strains those strains that caused high mortality (> 50%) in the Artemia and shrimp postlarvae challenge tests and presented molecular similarity to species previously reported as Vibrio pathogens. The product details are provided in S1 Table.
Table 1. Description of the products marketed in Ecuador as therapeutic agents against shrimp bacterial disease.
| Product code | Declared composition | Declared dosage / dosage used by producers | Presentation | Country manufacturer |
|---|---|---|---|---|
| P1 | Probiotic microorganisms: total aerobesa. Concentration: > 4 × 109 CFU g-1 | 2–10 μg mL-1 | Powder | USA |
| P2 | Probiotic microorganisms: total aerobesa. Concentration: ≥ 2 × 109 CFU g-1 | 5 μg mL-1 | Powder | USA |
| P3 | Strains of Bacillus subtilis, Bacillus licheniformis and Bacillus pumilus. Concentration: minimum 2 × 1010 CFU g-1 | 1 to 5 g kg-1 | Powder | USA |
| P4 | Mixture of strains of Bacillus spp. Concentration: 5 × 1010 CFU g-1 | 100–200 g ha-1 | Powder | USA |
| P5 | Vibrio alginolyticus. Concentration: 1 × 108 CFU mL-1 | 10 mL t-1 | Liquid | Ecuador |
| OA1 | Calcium formate, calcium propionate, premix carvacrol and thymol, premix allicin, yeast cell wall, calcium lactate, nucleotide, vitamin C 35%, fumaric acid, calcium citrate, organic zinc, organic manganese, inositol, vitamin E, sodium acetate, benzoic acid, BHT, betaglucans, organic iron, citric acid, niacin, vitamin A, zinc, calcium pantothenate, vitamin B6, potassium sorbate, magnesium chloride, copper sulfate, organic copper, monosodium phosphate, vitamin D3, organic selenium and sodium selenium | 1–7 kg t-1 of feed | Powder | Ecuador |
| OA2 | Formic acid, propionic acid, ammonium formate, acetic acid, silic acid and vermiculite | 0.6 kg t-1 of feed | Powder | Austria |
| OA3 | Calcium propionate 16%, calcium formate 18% and calcium carbonate 66% | 1–2 kg t-1 | Powder | Ecuador |
| OA4 | Propionic acid 25%, formic acid 25% and formaldehyde 15% | 1–3 kg ha-1 | Powder | Ecuador |
| OA5 | Formic acid, and its salts, mixture of flavors (essences and plant extracts: Allium sativum, Origanum vulgare, Cinnamomum zeylanicum, Eugenia caryophyllata), propionic acid and its salts, citric acid, malic acid, anti-caking agent | 2–3 kg t -1 of feed | Powder | Spain |
| OA6 | Lactic acid 23%, fumaric acid 20%, citric acid 20%, malic acid 25% and succinic acid 10% | 2–4 μg mL-1 | Powder | Ecuador |
| OA7 | Acid formic 35.4%, formate 34.6% and potassium 30.0% | 2–5 kg t -1 of feed | Powder | Germany |
| OA8 | Formaldehyde 35%: 28.6%, propionic acid 10%, bentonite 39% and silicic acid 22.4% | 1 kg t -1 of feed | Powder | Spain |
| OA9 | Mixture of short chain organic acids, acetic acid, propionic acid, formic acid and formaldehyde | 0.5–2 kg t-1 of feed | Powder | Spain |
| EO1 | Oregano oil extract | 1–5 mL t-1 | Liquid | USA |
| EO2 | Highly concentrated mix of essential oils | 1–10 mL t-1 | Liquid | Spain |
Probiotics: P1, P2, P3, P4 and P5; Organic acids: OA1, OA2, OA3, OA4, OA5, OA6, OA7, OA8 and OA9; Essential oils: EO1 and EO2.
a Specific bacterial strain are not declared in the product
Susceptibility of pathogenic Vibrio strains to antibiotics
The susceptibility of the pathogenic circulating strains to antibiotics was determined by the in vitro agar diffusion method [40, 41] using antimicrobial susceptibility test discs (diameter 6 mm, Bioanalyse and Oxoid). For this study, eleven different antimicrobial discs were used per duplicate [chloramphenicol (C, 30 μg), ciprofloxacin (CIP, 5 μg), fosfomycin (FF, 10 μg), furazolidone (FUR, 100 μg), norfloxacin (NOR, 10 μg), oxytetracycline (T, 30 μg), penicillin (P, 10 U), tetracycline (TE, 30 μg), nalidixic acid (NAL, 30 μg), enrofloxacin (E, 5 μg) and florfenicol (F, 30 μg)]. Isolates were activated on TSA and incubated at 30°C for 24 h. Afterwards, a colony from each isolate was transferred to TSB liquid medium (2% NaCl) and incubated again at 30°C for 4 h. The bacterial suspensions were standardized with McFarland´s 0.5 Barium Sulfate Standard Solution (1.5 × 108 CFU mL-1) and diluted to 106. Standardized bacterial suspensions (100 μL) were inoculated onto Mueller-Hinton plates using sterile cotton buds. Plates were incubated agar down between 24 and 48 h at 30°C. Diameters of inhibition halos surrounding the discs were measured and expressed in millimeters. Results were interpreted as sensitive, intermediate or resistant, following the guidelines of the Clinical and Laboratory Standards Institute [42, 43].
The minimal inhibitory concentration (MIC) for the antibiotics approved for use in aquaculture (oxytetracycline and florfenicol, 99% purity, Zhejiang Medicines and Health Company, China) on the growth of the bacterial strains was also determined. The bacterial isolates were activated in TSB liquid medium and incubated at 30°C for 4 h. The bacterial suspensions were diluted with TSB liquid medium to an approximate density of 106 CFU mL-1 by using McFarland´s 0.5 Barium Sulfate Standard Solution. Ten grams of each antibiotic were diluted in culture broth TSB with 2% NaCl solution. Sixteen concentrations of oxytetracycline, ranging from 1 to 3500 μg mL-1 were distributed into wells of round-bottom 96-well microplates. Each well was inoculated with 20 μl of bacterial suspension, including the positive control (bacterial growth at TSB culture, without any antibiotic). The microplates were incubated at 30°C between 24 and 48 h. All measurements were performed in triplicate, including those of the controls. Bacterial growth was detected by optical density at 620 nm (enzyme-linked immunosorbent assay ELISA reader, Varioskan Lux). MIC values were obtained as the lowest concentrations of each antibiotic that completely inhibited the bacterial growth. Absence of bacterial growth during the MIC process was confirmed on TSA agar. The same methodology was performed with the MIC tests for florfenicol, testing 15 concentrations of this antibiotic, ranging from 0.1 to 1000 μg mL-1. Finally, the patterns of multiple antibiotic resistance were analyzed to determine common patterns of resistance between the bacterial strains.
Susceptibility of pathogenic Vibrio strains to probiotics
The susceptibility of the pathogenic circulating strains to five commercial shrimp probiotics (Table 1) was determined by the agar plug diffusion method [40]. Briefly, the probiotics were cultured on Mueller-Hinton agar at 30°C for 24 h (≈ 107 CFU mL-1). In parallel, the pathogenic bacteria were cultured under the same conditions as the probiotics. After incubation, an agar-plot from the probiotic culture was aseptically cut and deposited on the agar surface of the plate inoculated with the pathogenic bacteria. Diameters of inhibition halos surrounding the agar-plots were measured and expressed in millimeters 24 and 48 h after the agar-plots were transferred. The strains were considered as: sensitive, intermediate and resistant when the diameters of the inhibition halos were ≥ 10 mm, between 4 and 9 mm and ≤ 3 mm, respectively.
Susceptibilities of pathogenic Vibrio strains to organic acids and essential oils
The susceptibility of the pathogenic circulating strains to nine organic acids and two essential oils (Table 1) was determined in a similar way to that implemented for the MIC determination for antibiotics. The evaluated concentrations of organic acids and essential oils ranged between 100 to 3500 μg mL-1 and 100 to 3000 μg mL-1, respectively.
Cell toxicity of selected products
The toxicity of the most efficient natural products, as well as the antibiotics authorized for use in aquaculture, was evaluated through the in vitro assay of cell viability of shrimp haemocytes [44]. Briefly, the assay was based on the ability of the mitochondria to convert yellow colored 3- (4,5-dimethylthiazol-2-yl) -2-5-diphenyltetrazole bromide (MTT) in purple colored formazan through the enzyme succinate dehydrogenase. The primary culture of shrimp haemocytes was activated in Hank’s salts for 75 min and exposed to each product for 2 h at different concentrations and then incubated for 2 h with 5 mg mL-1 of MTT in Hank’cs salts. The supernatant and the formazan crystals were diluted with isopropanol mixed with 0.04 N hydrochloric acid. The colorimetric reaction was read at 620 nm. The results were transformed to percent of cell viability, considering the primary culture of haemocytes without chemical exposure as the positive response of optimal cellular respiration (maximum cell viability). The concentrations evaluated of antibiotics were 100, 200, 400, 500, 1000, 2000, 2500, 3000, 4000, 5000, 6000 and 7000 μg mL-1. The concentrations evaluated for the natural products were below the corresponding minimum inhibitory concentrations.
Data analysis
The index of multiple antibiotic resistance (MAR) was calculated according to [45] and calculated as the number of antibiotics to which the isolate was resistant divided by the total number of antibiotics tested.
Differences in cumulative mortality among treatments (presumptive pathogenic strains) were analyzed by one-way analysis of variance (ANOVA) at the end of each challenge test (48 and 38 h after infection for A. franciscana nauplli and P. vannamei postlarvae, respectively). The null hypothesis (no treatment effect) was rejected with a P-value ≤ 0.05. Variance homogeneity of all treatments was examined using the Bartlett test. Assumption of normality was examined through the Shapiro-Wilk normality test. Tukey’s Honest Significant Difference test was used to compare treatment means. The effect of differences between treatments was considered significant at P-value ≤ 0.05. The same statistical analysis was performed to evaluate differences of percent of cell viability (cell toxicity) for each one of selected products. All statistical tests were carried out with the R statistical software [46].
Results
Isolation and preservation of bacterial strains
A total of 20 different bacterial strains were selected by morphological and luminescence criteria from 121 colonies from 31 samples of P. vannamei larvae with high bacterial counts on MA (>106 CFU g-1) or TCBS (>105 CFU g-1) agars from 10 hatcheries collected during mortality events.
Challenge tests
To estimate the virulence of the presumptive pathogenic strains, a first screening was performed challenging A. franciscana nauplii with all bacterial strains. All bacterial strains presented variable and high mortality in the challenge test (>74.2%, Table 2). A group of 11 strains caused the highest mortality in A. franciscana (> 95.0%, Table 2). The strain L15.19.1, belonging to this group caused 100% mortality in all replicates (Table 2). No significant mortality differences were observed among the Artemia nauplii challenged with the remaining ten strains of this group (P > 0.995, Table 2). These strains were used for a second challenge test with shrimp P. vannamei postlarvae (PL2). No significant mortality differences were observed among the postlarvae challenged with these strains (P = 0.148, Table 2).
Table 2. Mortality (average ± standard deviation) of Artemia franciscana nauplii and Penaeus vannamei postlarvae (PL2) after 48 and 38 h of challenge, respectively, with presumptive pathogenic bacterial strains.
| Bacterial strain | Mortality (%) | ||
|---|---|---|---|
| A. franciscana nauplii | P. vannamei postlarvae (PL2) | ||
| L15.12.2 | 74.2 ± 5.7 a | NI | |
| L15.13.2 | 77.5 ± 3.2 ab | NI | |
| L15.31.1 | 79.2 ± 3.2 ab | NI | |
| L15.29.2 | 80.8 ± 4.2 ab | NI | |
| L15.23.3 | 83.3 ± 5.4 ab | NI | |
| L15.26.1 | 83.3 ± 7.2 ab | NI | |
| L15.26.3 | 85.0 ± 5.8 abc | NI | |
| L15.5.2 | 87.5 ± 3.2 bd | NI | |
| L15.19.2 | 87.5 ± 6.9 bd | NI | |
| L15.25.3 | 95.0 ± 4.3 cde | 66.7 ± 13.0 a | |
| L15.29.1 | 95.8 ± 4.2 cde | 64.2 ± 12.6 a | |
| L15.23.1 | 96.7 ± 2.7 de | 61.7 ± 23.5 a | |
| L15.25.1 | 96.7 ± 2.7 de | 72.5 ± 17.9 a | |
| L15.21.1 | 97.5 ± 1.7 de | NI | |
| L15.21.2 | 97.5 ± 3.2 de | 70.9 ± 18.3 a | |
| L15.11.2 | 97.5 ± 3.2 de | 77.5 ± 3.2 a | |
| L15.10.3 | 98.3 ± 1.9 de | 86.6 ± 22.5 a | |
| L15.12.1 | 98.3 ± 3.3 de | 51.7 ± 10.4 a | |
| L15.10.4 | 99.2 ± 1.7 e | 83.4 ± 4.7 a | |
| L15.19.1 | 100.0 ± 0 | 71.7 ± 20.8 a | |
| Negative control | 0 ± 0 | 5.8 ± 9.6 | |
NI: strain not included in the challenge test. Means with different letters within a column indicate significant differences at P ≤ 0.05 by ANOVA and Tukey’s Honest Significant Difference tests.
Bacterial characterization by 16S rRNA sequence analysis
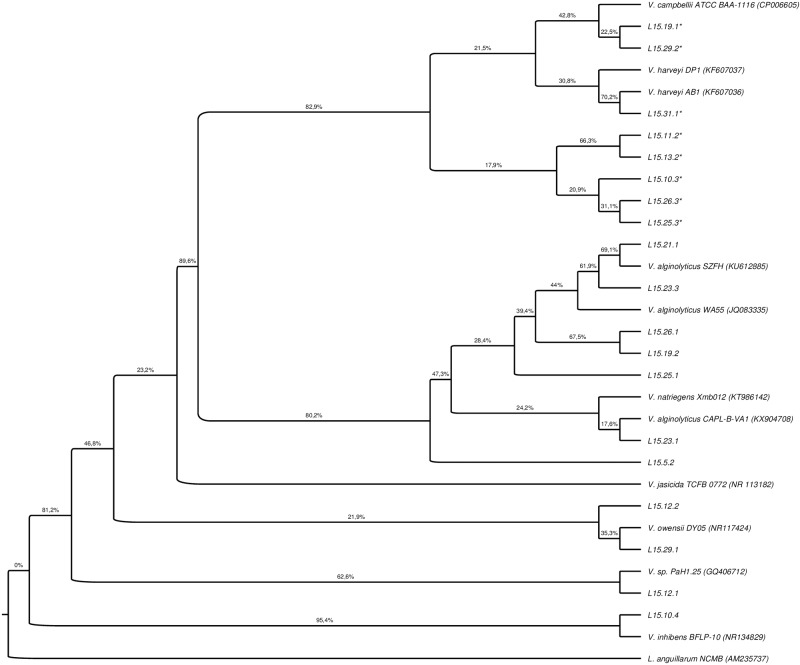
An initial screening of phylogenetic analysis was performed with the 16S rRNA gene sequences of the pathogenic circulating strains and 362 Vibrio sequences reported in GenBank as pathogens and non-pathogens. The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories, +G, parameter = 0.1000). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 9.76% sites). The pathogenic circulating strains of this study showed a greater similarity to the sequences of Vibrio reported as pathogens. To identify the isolates, a last analysis was performed using exclusively the 16S rRNA sequences obtained from GenBank that were more related to our sequences (Fig 1). Two groups of strains with high values of bootstrap support were identified (Fig 1). The first group contained eight sequences that presented similarity levels with sequences identified as V. campbellii and V. harveyi (Fig 1). All strains characterized in this study as luminescent belong to this group (Fig 1). A ninth luminescent strain was not included in the phylogenetic analysis, as the molecular identification was not possible with the obtained sequences (Fig 1). The second group included seven sequences of bacterial isolates with a high similarity level with identified sequences of V. alginolyticus and Vibrio natriegens (Fig 1). In addition, three different groups with low bootstrap values were also identified, which contained four sequences with similarities to Vibrio inhibens, Vibrio owensii and Vibrio sp. Strain PaH1.25 (Fig 1). In total, ninety-five percent of the isolated strains (19/20) belong to the Harveyi clade. Most of the strains were identified as V. harveyi and V. alginolyticus (12/19 strains). The least frequent strains were V. campbelli, V. owensii, V. inhibens and V. natriegens (Fig 1).
Fig 1. Phylogenetic tree of complete 16S rRNA sequences of the pathogenic circulating strains.
The analysis included 11 related Vibrio pathogen sequences reported in GenBank. Listonella anguillarum was used as outgroup (GenBank accession no. AM235737). The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories, +G, parameter = 0.1000). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 9.76% sites). Bootstrap values (percentage of 1000 replicates) appear next to each corresponding branch. The tree was built with the Maximum Likelihood inference method, which was supported by the trees built with Maximum Parsimony, Neighbor Joining and Bayesian inference methods. * indicates luminescent strain.
Susceptibility of pathogenic Vibrio strains to antibiotics
All pathogenic strains were sensitive to furazolidone, ciprofloxacin, norfloxacin, nalidixic acid, chloramphenicol and florfenicol (Table 3). A total of 60, 95 and 90% of the strains were sensitive to tetracycline, fosfomycin and enrofloxacin (Table 3). All strains showed resistance or intermediate sensitivity to penicillin and oxytetracycline (Table 3). Few strains exhibited intermediate sensitivity to enrofloxacin (10%), oxytetracycline (15%) and tetracycline (10%) (Table 3). The complete list of diameters of the inhibition halos is provided in S2 Table.
Table 3. Multiple antibiotic resistance (MAR) and susceptibility of pathogenic circulating strains to antibiotics and commercial probiotics, through the results of the antibiogram tests.
| Bacterial strain | Antibiotic susceptibility | Probiotic susceptibility | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | T | TE | FF | FUR | CIP | NOR | NAL | E | C | F | MAR index | P1 | P2 | P3 | P4 | P5 | |
| L15.25.1 | R | R | R | S | S | S | S | S | S | S | S | 0.27 | R | R | S | R | I |
| L15.13.2 | R | I | R | S | S | S | S | S | S | S | S | 0.27 | I | S | S | S | S |
| L15.31.1 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | R | R | S | S | S |
| L15.19.2 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | R | R | R | R | S |
| L15.26.1 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | S | S | R | R | S |
| L15.10.3 | R | R | R | S | S | S | S | S | S | S | S | 0.27 | R | R | R | R | I |
| L15.29.1 | R | R | R | R | S | S | S | S | S | S | S | 0.36 | R | R | R | R | S |
| L15.21.2 | R | I | S | S | S | S | S | S | S | S | S | 0.18 | R | R | R | R | S |
| L15.5.2 | R | R | R | S | S | S | S | S | S | S | S | 0.27 | S | R | R | R | S |
| L15.12.2 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | R | R | R | R | S |
| L15.19.1 | R | R | I | S | S | S | S | S | S | S | S | 0.27 | R | R | R | R | S |
| L15.25.3 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | R | S | S | I | S |
| L15.26.3 | R | R | R | S | S | S | S | S | S | S | S | 0.27 | I | R | R | R | S |
| L15.21.1 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | S | R | R | R | S |
| L15.11.2 | R | R | S | S | S | S | S | S | I | S | S | 0.27 | R | R | I | I | S |
| L15.23.3 | R | R | S | S | S | S | S | S | I | S | S | 0.27 | S | I | S | S | S |
| L15.23.1 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | R | R | R | R | S |
| L15.12.1 | R | R | S | S | S | S | S | S | S | S | S | 0.18 | S | R | R | R | S |
| L15.10.4 | R | R | I | S | S | S | S | S | S | S | S | 0.27 | R | R | R | R | I |
| L15.29.2 | R | I | S | S | S | S | S | S | S | S | S | 0.18 | R | R | S | R | S |
Antibiotics: penicillin (P), oxytetracycline (T), tetracycline (TE), fosfomycin (FF), furazolidone (FUR), ciprofloxacin (CIP), norfloxacin (NOR), nalidixic acid (NAL), enrofloxacin (E), chloramphenicol (C) and florfenicol (F). Probiotics: P1, P2, P3, P4 and P5. Susceptibility of the pathogenic circulating strains to antibiotics and probiotics are expressed as sensitive (S), intermediate (I) and resistant (R).
All strains presented resistance to at least two antibiotics at the same time, being 50%, 45% and 5% of the strains resistant to 2, 3 and 4 antibiotics, respectively (Tables 3 and 4). The MAR index on average was 0.23 (Tables 3 and 4). The most prevalent pattern of multiple resistance was P-T (10/20 strains = prevalence of 50%, Table 4). The other most prevalent pattern was P-T-TE (7/20 strains = prevalence of 35%, Table 4). The other two MAR patterns were: P-T-Te (2/20 strains = prevalence of 10%, Table 4) and P-T-TE-FF (1/20 strains = prevalence of 5%, Table 4). All strains were resistant at the same time to penicillin and oxytetracycline (Tables 3 and 4). Forty percent (8/20) of the strains were resistant to both antibiotics of the tetracycline group (Tables 3 and 4).
Table 4. Patterns of multiple antibiotic resistance (MAR) and MAR index.
| Number of bacterial strains | MAR pattern | MAR index |
|---|---|---|
| 10 | P-T | 0.18 |
| 7 | P-T-TE | 0.27 |
| 2 | P-T-E | 0.27 |
| 1 | P-T-TE-FF | 0.36 |
Penicillin (P), oxytetracycline (T), tetracycline (TE), enrofloxacin (E) and fosfomycin (FF).
Forty-five percent of the strains exhibited MIC values for oxytetracycline of less than 100 μg mL-1 (Table 5). Forty-five percent of the strains presented MIC values between 200–500 μg mL-1 (Table 5). Ten percent of the strains presented a high MIC value for oxytetracycline (> 3500 μg mL-1, Table 5). All strains presented sensitivity to florfenicol at low concentrations (≤ 40 μg mL-1, Table 5).
Table 5. Results of minimal inhibitory concentration (MIC) tests for authorized antibiotics for use in aquaculture, organic acids and essential oils on the growth of pathogenic circulating strains.
| Bacterial strain | Minimal inhibitory concentration (μg mL-1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | Organic acid | Essential oil | |||||||||||
| T | F | OA1 | 0A2 | 0A3 | 0A4 | 0A5 | 0A6 | 0A7 | 0A8 | OA9 | EO1 | EO2 | |
| L15.12.1 | 500 | 5 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 100 | 1000 | 3000 |
| L15.12.2 | 500 | 8 | >3500 | >3500 | >3500 | 200 | >3500 | 2500 | 2500 | >3500 | 300 | 1000 | >3000 |
| L15.5.2 | >3500 | 2 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 2000 | >3000 |
| L15.21.1 | 100 | 4 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 2000 | 3000 |
| L15.21.2 | 5 | 8 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 900 | 3000 |
| L15.23.1 | 20 | 10 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 2000 | >3000 |
| L15.23.3 | 100 | 40 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 2000 | >3000 |
| L15.25.1 | >3500 | 4 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 500 | 200 | >3000 |
| L15.25.3 | 250 | 40 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 400 | 2000 | >3000 |
| L15.26.1 | 100 | 5 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 200 | 2000 | 3000 |
| L15.26.3 | 50 | 20 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 100 | 2000 | 3000 |
| L15.13.2 | 500 | 20 | >3500 | >3500 | >3500 | 2500 | >3500 | 1500 | 300 | >3500 | 500 | 2000 | >3000 |
| L15.11.2 | 10 | 8 | >3500 | >3500 | >3500 | 2500 | >3500 | 200 | 2500 | >3500 | 200 | 2000 | >3000 |
| L15.29.1 | 200 | 5 | >3500 | >3500 | >3500 | 2500 | >3500 | 500 | 2500 | >3500 | 200 | 1000 | >3000 |
| L15.29.2 | 200 | 8 | >3500 | >3500 | >3500 | 2500 | >3500 | 1000 | 2500 | >3500 | 2500 | 2000 | >3000 |
| L15.10.3 | 500 | 8 | >3500 | >3500 | >3500 | 600 | >3500 | 1000 | 300 | >3500 | 300 | 2000 | >3000 |
| L15.10.4 | 5 | 2 | >3500 | >3500 | >3500 | 1500 | >3500 | 2500 | 2500 | >3500 | 200 | 2000 | >3000 |
| L15.19.1 | 500 | 8 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 200 | 2000 | 3000 |
| L15.19.2 | 200 | 8 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 200 | 3000 | 3000 |
| L15.31.1 | 5 | 5 | >3500 | >3500 | >3500 | 2500 | >3500 | 2500 | 2500 | >3500 | 100 | 3000 | 3000 |
Antibiotics: oxytetracycline (T) and florfenicol (F); Organic acids: OA1, OA2, OA3, OA4, OA5, OA6, OA7, OA8 and OA9; Essential oils: EO1 and EO2. Maximum concentration analyzed: 3500 (oxytetracycline and organic acids) and 3000 (essential oils) μg mL-1.
Susceptibility of pathogenic Vibrio strains to probiotics
In general, the pathogenic strains exhibited low sensitivity to the tested probiotics, except to P5 (85% of sensitivity); 25, 15, 30, 5 and 15% of the strains were sensitive to P1, P2, P3 and P4, respectively (Table 3). Intermediate sensitivity was observed for 10, 5, 5, 10 and 15% of the strains to the probiotics P1, P2, P3, P4 and P5, respectively (Table 3). In terms of resistance, 65, 80, 65, 75 and 0% of the pathogenic strains were resistant to P1, P2, P3, P4 and P5, respectively (Table 3). Two isolates (L15.10.3 and L.15.10.4) were resistant or intermediate resistant to all tested probiotics (Table 3); strains L15.10.3 and L15.10.4 also presented the highest mortalities in the P. vannamei challenge test. The complete list of diameters of the inhibition halos is provided in S2 Table.
Susceptibilities of pathogenic Vibrio strains to organic acids and essential oils
The MIC analyses showed that 100% of the strains were resistant to 5 products (OA1, OA2, OA3, OA5 and OA8) up to 3500 μg mL-1 of each product (Table 5), and 15, 25 and 10% of the strains were sensitive to OA4, OA6 and OA7, respectively, at concentrations equal to or lower than 1500 μg mL-1 (Table 5). In general, most of the strains showed sensitivity to product (OA9, Table 5). Product EO1 controlled 100% of the strains between 100 to 3000 μg mL-1. Forty percent of the strains (8/20) were sensitive up to 3000 μg mL-1 against EO2 (Table 5).
Cell toxicity of selected products
The most efficient products in terms of bacterial sensitivity were organic acids OA6 and OA9, but OA9 was cytotoxic at all assayed concentrations, not showing significant differences of cell viability between the evaluated concentrations of OA9 (P = 0.802, Table 6). However, OA6 was not toxic for shrimp haemocytes between 100 and 400 μg mL-1, exhibiting similar values of cell viability at those levels (P ≥ 0.848, Table 6). Significantly lower levels of cell viability were reported for concentrations of OA6 from 1000 to 3000 μg mL-1 compared with concentrations from 100 to 400 μg mL-1 (P < 0.001, Table 6). No significant differences of cell viability were found at concentrations of OA6 from 1000 to 3000 μg mL-1 (P ≥ 0.129, Table 6). The haemocytes cell viability was not affected by florfenicol and oxytetracycline up to 2000 and 4000 μg mL-1, respectively (Table 6). Cell viability of all four control replicates was 100%.
Table 6. Cell viability (%) of P. vannamei shrimp haemocytes after exposure at varied concentrations of authorized antibiotics for use in aquaculture and the two most effective natural products against pathogenic circulating bacterial strains.
| Concentration of products (μg mL-1) | Product | |||
|---|---|---|---|---|
| F | T | OA6 | OA9 | |
| 100 | 98.6 ± 4.7 d | 97.2 ± 2.7 b | 99.0 ± 1.5 c | 23.6 ± 2.4 a |
| 200 | 98.6 ± 5.0 d | 99.2 ± 0.9 b | 99.6 ± 0.6 c | 24.2 ± 3.4 a |
| 400 | 98.6 ± 3.4 d | 97.7 ± 2.7 b | 97.5 ± 2.5 c | 23.4 ± 3.1 a |
| 500 | 98.6 ± 5.3 d | 98.5 ± 2.4 b | 89.2 ± 2.6 b | 22.5 ± 1.5 a |
| 1000 | 96.1 ± 4.7 d | 98.7 ± 1.2 b | 26.4 ± 1.5 a | 21.9 ± 1.0 a |
| 2000 | 90.0 ± 3.1 d | 97.7 ± 4.3 b | 24.3 ± 2.0 a | - |
| 2500 | 72.0 ± 6.8 c | 97.7 ± 2.0 b | 22.1 ± 1.8 a | - |
| 3000 | 62.0 ± 3.7 bc | 92.7 ± 4.0 b | 22.7 ± 0.9 a | - |
| 4000 | 58.7 ± 4.7 b | 92.7 ± 3.1 b | - | - |
| 5000 | 44.3 ± 2.0 a | 84.3 ± 1.5 a | - | - |
| 6000 | 37.7 ± 1.5 a | 79.0 ± 1.0 a | - | - |
| 7000 | 37.3 ± 1.2 a | 76.3 ± 4.0 a | - | - |
Organic acids: OA6 and OA9; Antibiotics: florfenicol (F) and oxytetracycline (T). Means with different letters within a column indicate significant differences at P ≤ 0.05 by ANOVA and Tukey´s Honest Significant Difference tests.
Discussion
The efficiency of therapeutic products is of vital importance for the control of aquaculture diseases. In the present study, we investigate through some in vitro analyses the antimicrobial effectiveness of antibiotics and commercially available therapeutic products used in Ecuador to control pathogenic bacterial strains of shrimp larvae. To perform these analyses, we isolated the strains that were circulating in the shrimp hatcheries, verified their virulence through challenge tests and identified their molecular similarity with previously reported pathogenic Vibrio. By doing this, we confirmed that we were working with the circulating strains that cause real problems at the production level. The results were dependent on the product, concentration of the product and bacterial strain.
Antibiotics were the most efficient therapeutic agents against the growth of pathogenic bacteria. Eight of the antibiotics inhibited the growth of all or most of the pathogenic bacterial strains, but most of these products are not authorized for use in aquaculture. We included several antibiotics in our evaluation because we wanted to investigate if the pathogenic circulating strains exhibited patterns of multiple antibiotic resistance, which could be associated with antimicrobial use [47]. In our study, the MAR index was on average 0.23, showing some level of resistance to antibiotics. High antibiotic resistance has been found in hatcheries worldwide, as well as higher MAR indexes in hatcheries rather than in shrimp farms [48, 49]. For instance, MAR index ranges from 0.21 to 0.38 have been reported for bacteria isolated from shrimp hatcheries [50, 49], whereas, MAR indexes in shrimp farms range from 0.11 and 0.32 [49–52]. The average MAR index determined in this study is low compared to values reported in other shrimp hatcheries. However, all strains were resistant at the same time to penicillin and oxytetracycline, both antibiotics used in human medicine. Similar observations of isolates with higher resistance to antibiotics used in human medicine than those used in aquaculture have been reported by several authors [53, 48]. Most of the sampled hatcheries are in a region of multiple anthropogenic activities, without wastewater treatment, which could be a source of antibiotic pollution.
In our screening, in addition to penicillin and tetracycline, oxytetracycline was a less efficient antibiotic, which is one of the two authorized antibiotics for use in aquaculture, and the antimicrobial most commonly used in Ecuador for shrimp larval stages, although now it is in decline. The MIC values found in our study are high and similar to those reported by the shrimp culture. For instance, an average MIC of 304 μg mL-1 has been reported for Vibrio isolated in Mexico [53], values up to 400 mg mL-1 for Vibrio isolated from water in hatcheries and farms in Brazil [50] and up to 512 μg mL-1 for Vibrio isolated from Penaeus monodon and P. vannamei shrimp in Thailand [54]. Nevertheless, oxytetracycline is toxic for Penaeus stylirostris larvae at concentrations from 135.5 to 238 μg mL-1 [55]. Fifty-five percent of our screened strains (11/20) presented MIC values for oxytetracycline in this toxic range or above, indicating that the application of this antibiotic is not suitable for most of the pathogenic circulating strains. Considering this observation, it cannot be discounted that the prolonged use of oxytetracycline may explain the observed resistance.
Florfenicol is the other authorized antibiotic for use in aquaculture and was highly efficient in controlling bacterial growth at low concentrations, with a MIC of 8 μg mL-1 for 75% of the strains. Our results are in accordance with the MIC values from 0.5 to 4 μg mL-1 found for Vibrio spp. isolated in Ecuador, USA, Japan and Thailand [56, 57] and values from 0.25 to 8 μg mL-1 for Vibrio spp. isolated in Mexico [53]. Florfenicol is a broad-spectrum antibiotic, with the same mechanism of action to that of chloramphenicol (inhibition of protein synthesis), and commonly used for the treatment of bacterial diseases in shrimp, such as necrotizing hepatopancreatitis [58]. Although it has been reported that florfenicol is toxic for P. vannamei larvae (Zoea 1) at concentrations higher than 20 μg mL-1 [59], we found that haemocytes could thrive up to 2000 ug mL-1 without any problem of cell unviability. Possibly, these seemingly contradictory results could be explained by the fact that MTT analyses were performed using haemocytes of juvenile shrimp, and at this stage, shrimp could handle a high concentration of this antibiotic. Sixteen of our screened strains (80%) showed MIC values equal to or higher than 5 μg mL-1, indicating a loss of bacterial sensitivity and a development of resistance to this antibiotic, and therefore not recommended as a control strategy at production level.
Given these conditions, it is therefore necessary for producers to consider alternative strategies for the control of pathogenic bacteria. In our study, only one commercial probiotic (P5) exhibited a high antagonistic capacity against the bacterial strains (85% of the strains). P5, whose declared composition is V. alginolyticus, has been employed as a probiotic in Ecuadorian hatcheries since 1992 [60, 61] and it has been observed that it enhances postlarvae survival and immune response when shrimp get to the juvenile stages [35]. The rest of the probiotics could inhibit the growth of 15–30% of the strains, which showed intermediate effects, and therefore could be considered as functional for the growth control of some pathogenic bacterial strains. Of the two strains that were not completely inhibited by probiotic P5, neither inhibited the other probiotics. The administration of either multiple or single probiotics remains controversial [62, 63], and, as regards this, although we did not test many probiotics, P5, a single strain, was the most efficient probiotic, whereas, P1, P2, P3 and P4 declared as mixture strains were not particularly effective.
Only one organic acid (OA9) showed inhibition of growth of most of the strains, at low concentrations. This product is a mixture of acetic acid, propionic acid and formic acid. These acids, as well as butyric acid, are efficient for the control of aquatic and shrimp pathogenic Vibrio [26, 28, 29]. OA6 was the second most efficient organic acid, and contains lactic, fumaric, citric, malic and succinic acids. Lactic and citric acids seemed to be the best organic acids to control pathogenic V. harveyi in Macrobrachium rosenbergii [64], but lactic acid can inhibit the pathogenic microbiota of fishes [65]. OA4 contains propionic acid and formic acid, whereas OA7 contains formic acid. Three of these four organic acids contain formic acid, which is considered to be particularly effective against pathogenic Vibrio [26]. In addition, OA9 contains three of the four organic acids reported as good bacterial inhibiters, including acetic acid, which is a good disinfectant of V. parahaemolyticus [66]. Possibly the five organic acids whose MIC were not determined, could be effective at higher concentrations than tested in this study, which makes them inefficient products.
Essential oils are effective for inhibiting bacterial growth; in our study the essential oil EO1, whose declared composition includes extract of oregano oil, efficiently inhibited the growth of all evaluated bacterial strains, with MIC values equal to or lower than 3 mg L-1. Similar results were observed by Teixeira et al. 2013 for other bacterial genera, where the Origanum vulgare essential oil was effective at MIC values lower than 5 mg mL-1 [67]. Possibly, the efficacy for the bacterial inhibition of EO1 might be related to the presence of thymol and carvacrol, two of the compounds of oregano essential oil [68, 69] that decrease the bacterial counts of V. vulnificus, V. parahaemolyticus and V. cholera in muscle and hepatopancreas of juvenile P. vannamei [70]. Carvacrol also increases the survival of A. franciscana larvae challenged with V. harveyi [71]. However, despite the high potential of the essential oil against bacterial growth, it could be toxic at high concentrations. Concentrations higher than 14.9 mg L-1 of carvacrol turned out to be toxic for A. franciscana [71], whereas concentrations of O. vulgare leaf crude extract and essential oil were toxic for both A. salina [72] and P. vannamei shrimp [44] at concentrations higher than 2 mg L-1 and 10 mg L-1, respectively. Other authors have mentioned the ability of essential oils to interrupt bacterial communication, decreasing bacterial virulence and pathogenicity [73, 74]. Therefore, it would be advisable to evaluate these properties of oregano essential oils and at the same time its toxic effect.
The tests performed in this work are designed to analyze whether the commercial products inhibit the growth or kill the pathogenic bacteria, but the natural products evaluated in this work could exhibit other modes of action not studied in this work. Therefore, further studies will be necessary to evaluate their efficacies, in terms of others mode of action, such as: capacity to disrupt bacterial communication, improvement of the shrimp immune system, colonization of the gastrointestinal tract, enhanced shrimp growth and survival, among others.
Ninety-five percent of the isolated strains (19/20) belong to the Harveyi clade, known to be the pathogenic clade for shrimp [75]. This was consistent with the results of the challenge tests, thus verifying that the isolated strains were pathogenic. Most of the strains were identified as V. harveyi and V. alginolyticus (12/20 strains), which have been a continual problem for the Ecuadorian hatcheries since 1988–1989 [2–4]. This study, however, has identified the fact that new pathogenic strains have appeared (V. campbellii, V. owensii, V. inhibens and V. natriegens) in the Ecuadorian hatcheries, diversifying the circulating bacteria and making it crucial to study the effectiveness of treatments for each pathogenic strain. Periodic surveys at regional level and further challenge tests could be performed to identify the pathogenic circulating bacterial strains and to focus on the bacteria of concern. At the same time, shrimp producers can apply relatively simple in vitro and in vivo analyses, such as those employed in this study and take adequate management decisions based on these results, which in turn could reduce the impact of bacterial diseases and increase profits.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Martha Borbor, Ramiro Solórzano, Juan Rodríguez, Rosa Malavé and Rubén Román for their technical assistance in this study. We would like to thank Dr. Stanislaus Sonnenholzner for their kind reviewing of the manuscript and suggestions.
Data Availability
All relevant data are within the paper and supporting table. The 16S rRna sequences of bacterial strains are available from GenBank (accession numbers MH997724 to MH997742).
Funding Statement
This study was funded by the Secretaria de Educación Superior, Ciencia, Tecnología e Innovación, SENESCYT (www.educacionsuperior.gob.ec) in the framework of the PIC-14-CENAIM-003 Project Desarrollo e implementación de métodos de control y prevención de enfermedades en especies acuáticas de uso comercial y uso potencial en maricultura y repoblación (Grant to BB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rodgers CJ, Mohan CV, Peeler EJ. The spread of pathogens through trade in aquatic animals and their products. Rev Sci Tech. 2011; 30(1): 241–256. [DOI] [PubMed] [Google Scholar]
- 2.Le Groumellec ML, Haffner P, Martin B, Martin C. Comparative study of bacterial infections responsible for mass mortality in penaeid shrimp hatcheries of the Pacific zone In: Shariff M, Arthur JR, Subasinhge RP. Diseases in Asian Aquaculture II. Manila: Fish Health Section, Asian Fisheries Society; 1995. pp. 163–173. [Google Scholar]
- 3.Robertson PA, Calderon J, Carrera L, Stark JR, Zherdmant M, Austin B. Experimental Vibrio harveyi infections in Penaeus vannamei larvae. Dis Aquat Organ. 1998; 32 (2): 151–155. [Google Scholar]
- 4.Vandenberghe J, Verdonck L, Robles-Arozarena R, Rivera G, Bolland A, Balladares M, et al. Vibrios Associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts. J Appl Environ Microbiol. 1999; 65 (6): 2592–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandenberghe J, Li Y, Verdonck L, Li J, Sorgeloos P, Xu HS, et al. Vibrios associated with Penaeus chinensis (Crustacea: Decapoda) larvae in Chinese shrimp hatcheries. Aquaculture. 1998; 169: 121–132. [Google Scholar]
- 6.Karunasagar I, Pai R, Malathi GR, Karunasagar I. Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection. Aquaculture. 1994; 128 (3–4): 203–209. [Google Scholar]
- 7.Haldar S, Chatterjee S, Sugimoto N, Das S, Chowdhury N, Hinenoya A, et al. Identification of Vibrio campbellii isolated from diseased farm-shrimps from south India and establishment of its pathogenic portencial in an Artemia model. Microbiology. 2011; 157 (1): 179–188. [DOI] [PubMed] [Google Scholar]
- 8.Mirbakhsh M, Akhavan S, Afsharnasab M, Khanafari A, Razavi MR. Molecular identification of Vibrio harveyi from larval stage of Pacific white shrimp (Litopenaeus vannamei) Boone (Crustacea: Decapoda) by polimerase chain reaction and 16S rDNA sequencing. Iran J Fish Sci. 2014; 13 (2): 384–393. [Google Scholar]
- 9.Karunasagar I, Otta SK, Karunasagar I. Disease problems affecting cultured penaeid shrimp in India. Fish Pathol. 1998; 33(4): 413–419. [Google Scholar]
- 10.Hasan MA, Siddique MA, Hasan M, Hossain MA, Rahman MS. 16S rRNA gene sequence based identification of Vibrio spp. in shrimp and tilapia hatcheries of Bangladesh. Dhaka Univ. J Biol Sci. 2017; 26(1): 45‐58. [Google Scholar]
- 11.Soto-Rodríguez SA, Simoes N, Roque A, Gómez-Gil B. Pathogenicity and colonization of Litopenaeus vannamei larvae by luminescent vibrios. Aquaculture. 2006; 258 (1–4):109–115. [Google Scholar]
- 12.Hameed AS. Susceptibility of three Penaeus species to a Vibrio campbellii‐like bacterium. J World Aquac Soc. 1995; 26 (3): 315–319. [Google Scholar]
- 13.Moriarty DJ. Control of luminous Vibrio species in penaeid aquaculture ponds. Aquaculture. 1998; 164 (1–4): 351–358. [Google Scholar]
- 14.Chen H, Liu S, Xu XR, Diao ZH, Sun KF, Hao QW, et al. Tissue distribution, bioaccumulation characteristics and health risk of antibiotics in cultured fish from a typical aquaculture area. J. Hazard Mater. 2018; 343: 140–148. 10.1016/j.jhazmat.2017.09.017 [DOI] [PubMed] [Google Scholar]
- 15.FAO. Antibiotic residues in aquaculture products In: The state of world fisheries and aquaculture. Rome: Food and Agriculture Organization of the United Nations; 2002. pp. 74–83. [Google Scholar]
- 16.Liu X, Steele JC, Meng XZ. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: a review. Environ Pollut. 2017; 223: 161–169. 10.1016/j.envpol.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 17.Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol. 2006; 8 (7): 1137–1144. 10.1111/j.1462-2920.2006.01054.x [DOI] [PubMed] [Google Scholar]
- 18.Gullian M, Thompson F, Rodriguez J. Selection of probiotic bacteria and study of their immunostimulatory effect in Penaeus vannamei. Aquaculture. 2004; 233 (1–4): 1–14. [Google Scholar]
- 19.Balcázar JL, Rojas-Luna T, Cunningham DP. Effect of the addition of four potential probiotic strains on the survival of pacific white shrimp (Litopenaeus vannamei) following immersion challenge with Vibrio parahaemolyticus. J Invertebr Pathol. 2007; 96 (2): 147–150. 10.1016/j.jip.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 20.Verschuere L, Rombaut G, Sorgeloos P, Verstraete W. Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev. 2000; 64 (4): 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Short-chain fatty acids and poly-β-hydroxyalkanoates: (New) Biocontrol agents for a sustainable animal production. Biotechnol Adv. 2009; 27 (6): 680–685. 10.1016/j.biotechadv.2009.04.026 [DOI] [PubMed] [Google Scholar]
- 22.Kara K, Özkaya S, Erbaş S, Baytok E. Effect of dietary formic acid on the in vitro ruminal fermentation parameters of barley-based concentrated mix feed of beef cattle. J Appl Anim Res. 2018; 46 (1): 178–183. [Google Scholar]
- 23.Hassaan MS, Soltan MA, Jarmołowicz S, Abdo HS. Combined effects of dietary malic acid and Bacillus subtilis on growth, gut microbiota and blood parameters of Nile tilapia (Oreochromis niloticus). Aquac Nutr. 2018; 24(1): 83–93. [Google Scholar]
- 24.Ng WK, Koh CB, Teoh CY, Romano N. Farm-raised tiger shrimp, Penaeus monodon, fed commercial feeds with added organic acids showed enhanced nutrient utilization, immune response and resistance to Vibrio harveyi challenge. Aquaculture. 2015; 449 (1): 69–77. [Google Scholar]
- 25.Mine S, Boopathy R. Effect of organic acids on shrimp pathogen, Vibrio harveyi. Curr Microbiol. 2011; 63 (1): 1–7. 10.1007/s00284-011-9932-2 [DOI] [PubMed] [Google Scholar]
- 26.Adams D, Boopathy R. Use of formic acid to control vibriosis in shrimp aquaculture. Biologia. 2013; 68: 1017–1021. [Google Scholar]
- 27.Anuta JD, Buentello A, Patnaik S, Lawrence AL, Mustafa A, Hume ME, et al. Effect of dietary supplementation of acidic calcium sulfate (Vitoxal) on growth, survival, immune response and gut microbiota of the Pacific white shrimp, Litopenaeus vannamei. J World Aquac Soc. 2011; 42 (6): 834–844. [Google Scholar]
- 28.Da Silva BC, Vieira FDN, Mouriño JLP, Ferreira GS, Seiffert WQ. Salts of organic acids selection by multiple characteristics for marine shrimp nutrition. Aquaculture. 2013; 384–387: 104–110. [Google Scholar]
- 29.Defoirdt T, Halet D, Sorgeloos P, Bossier P, Verstraete W. Short-chain fatty acids protect gnotobiotic Artemia franciscana from pathogenic Vibrio campbellii. Aquaculture. 2006; 261 (2): 804–808. [Google Scholar]
- 30.Lambert RJ, Skandamis PN, Coote PJ, Nychas GJ. A study of the minimum inhibitory concentration and mode of action of oregano essential oil, thymol and carvacrol. J Appl Microbiol. 2001; 91(3): 453–462. [DOI] [PubMed] [Google Scholar]
- 31.Al‐Sagheer AA, Mahmoud HK, Reda FM, Mahgoub SA, Ayyat MS. Supplementation of diets for Oreochromis niloticus with essential oil extracts from lemongrass (Cymbopogon citratus) and geranium (Pelargonium graveolens) and effects on growth, intestinal microbiota, antioxidant and immune activities. Aquac Nutr. 2018; 24 (3): 1006–1014. [Google Scholar]
- 32.Kordali S, Cakir A, Ozer H, Cakmakci R, Kesdek M, Mete E. Antifungal, phytotoxic and insecticidal properties of essential oil isolated from Turkish Origanum acutidens and its three components, carvacrol, thymol and p-cymene. Bioresour Technol. 2008; 99: 8788–8795. 10.1016/j.biortech.2008.04.048 [DOI] [PubMed] [Google Scholar]
- 33.Sarter S, Randrianarivelo R, Ruez P, Raherimandimby M, Danthu P. Antimicrobial effects of essential oils of Cinnamosma fragrans on the bacterial communities in the rearing water of Penaeus monodon larvae. Vector Borne Zoonotic Dis. 2011; 1(4): 433–437. [DOI] [PubMed] [Google Scholar]
- 34.Defoirdt T, Bossier P, Sorgeelos P, Verstraete W. The impact of mutations in the quorum sensing systems of Aeromonas hydrophila, Vibrio anguillarum and Vibrio harveyi on their virulence towards gnotobiotically cultured Artemia franciscana. Environ Microbiol. 2005; 7 (8): 1239–1247. 10.1111/j.1462-2920.2005.00807.x [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez J, Espinosa Y, Echeverria F, Cárdenas G, Román R, Stern S. Exposure to probiotics and β-1,3/1,6-glucans in larviculture modifies the immune response of Penaeus vannamei juveniles and both the survival to white spot syndrome virus challenge and pond culture. Aquaculture. 2007; 273 (4): 405–415. [Google Scholar]
- 36.Tran L, Nunan L, Redman RM, Mohney LL, Pantoja CR, Fitzsimmons K, et al. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Organ. 2013; 105 (1): 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- 37.Avannis-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. Biotechniques. 1994; 17: 144–149. [PubMed] [Google Scholar]
- 38.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999; 41: 95–98. Available from: http://www.mbio.ncsu.edu/BioEdit/bioedit.html [Google Scholar]
- 39.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and and parallel computing. Nat Methods. 2012; 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal. 2016; 6 (2): 71–79. 10.1016/j.jpha.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zidour M, Chevalier M, Belguesmia Y, Cudennec B, Grard T, Drider D, et al. Isolation and characterization of bacteria colonizing Arcatia tonsa copepod eggs and displaying antagonist effects agains Vibrio anguillarum, Vibrio alginolyticus and other pathogenic strains. Front Microbiol. 2017; 8 (1919): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CLSI. Performance standards for antimicrobial susceptibility testing; Twenty-Fifth informational supplement CLSI document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 43.NCCLS. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Approved standard-second edition. NCCLS document M31-A2. Wayne, PA: National Committee for Clinical Laboratory Standards; 2002. [Google Scholar]
- 44.Domínguez-Borbor C, Chalén-Alvarado B, Rodríguez JA. A simple in vitro method to evaluate the toxicity of functional additives used in shrimp aquaculture. MethodsX. 2018; 5: 90–95. 10.1016/j.mex.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster LF, Thompson BC, Fulton MH, Chestnut DE, Van Dolah RF, Leight AK, et al. Identification of sources of Escherichia coli in South Carolina estuaries using antibiotic resistance analysis. J Exp Mar Bio Ecol. 2004; 298 (2): 179–195. [Google Scholar]
- 46.R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. http://www.R-project.org/. [Google Scholar]
- 47.Tendencia EA, De la Peña LD. Antibiotic resistance of bacteria from shrimp ponds. Aquaculture. 2001; 195 (3–4): 193–204. [Google Scholar]
- 48.Vaseeharan B, Ramasamy P, Murugan T, Chen JC. In vitro susceptibility of antibiotics against Vibrio spp. and Aeromonas spp. isolated from Penaeus monodon hatcheries and ponds. Int J Antimicrob Agents. 2005; 26 (4): 285–291. 10.1016/j.ijantimicag.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 49.Zhang YB, Li Y, Sun XL. Antibiotic resistance of bacteria isolated from shrimp hatcheries and cultural ponds on Donghai Island, China. Mar Pollut Bull. 2011; 62 (11): 2299–2307. 10.1016/j.marpolbul.2011.08.048 [DOI] [PubMed] [Google Scholar]
- 50.Reboucas RH, de Sousa OV, Lima AS, Vasconcelos FR, de Carvalho PB, Vieria RH. Antimicrobial resistance profile of Vibrio species isolated from marine shrimp farming enviroments (Litopenaeus vannamei) at Ceará, Brazil. Environ Res J. 2011; 111; 21–24 [DOI] [PubMed] [Google Scholar]
- 51.de Menezes FG, da Silva S, de Carvalho EM, de Sousa OV, dos Fernandes Vieira RH. Multiresistance in vibrios isolated from tissues and hemolynph of Litopenaeus vannamei farmed in northeastern Brazil. Arquivos de Ciencias do March 2013; 46 (1); 84–89. [Google Scholar]
- 52.Silvester R, Alexander D, George M, Hatha AA. Prevalence and multiple antibiotic resistance of Vibrio coralliilyticus, along the southwest coast of India. Curr Sci. 2017; 112 (8): 1749–1755. [Google Scholar]
- 53.Roque A, Molina-Aja A, Bolán-Mejía C, Gómez-Gil B. In vitro susceptibility to 15 antibiotics of vibrios isolated from penaeid shrimps in Northwestern Mexico. Int J Antimicrob Agents. 2001; 17 (5): 383–387. [DOI] [PubMed] [Google Scholar]
- 54.Maisak H, Tipmongkolsilp N, Wongtavatchai J. Minimum inhibitory concentrations of antimicrobials against clinical Vibrio and Streptococcus isolated from aquaculture In: Bondad-Reantaso MG, Jones JB, Corsin F, Aoki T. Diseases in Asian Aquaculture II. Selangor, Malaysia: Fish Health Section, Asian Fisheries Society; 1995. pp. 309–316. [Google Scholar]
- 55.Williams RR, Bell TA, Lightner DV. Shrimp antimicrobial Testing. II. Toxicity testing and safety determination for twelve antimicrobials with penaeid shrimp larvae. J Aquat Anim Health. 1992; 4 (4): 262–270. [Google Scholar]
- 56.Mohney LL, Bell TA, Lightner DV. Shrimp Antimicrobial Testing. I. In Vitro susceptibility of thirteen Gram-Negative bacteria to twelve antimicrobials. J Aquat Anim Health. 1992; 4 (4): 257–261. [Google Scholar]
- 57.Tipmongkolsilp N, Limpanon Y, Patamalai B, Lusanandana P, Wongtavatchai J. Oral medication with florfenicol for black tiger shrimps Penaeus monodon. Tha Thai Journal of Veterinary Medicine 2006; 36 (2): 39–47. [Google Scholar]
- 58.Morales-Covarrubias MS, Tlahuel-Vargas L, Martinez-Rodriguez IE, Lozano-Olvera R, Palacios-Arriaga JM. Necrotising hepatobacterium (NHPB) infection in Penaeus vannamei with florfenicol and oxytetracycline: a comparative experimental study. Rev Cient (Maracaibo). 2012; 22 (1): 72–80. [Google Scholar]
- 59.Soto-Rodríguez S, Armenta M, Gómez-Gil B. Effects of enrofloxacin and florfenicol on survival and bacterial population in an experimental infection with luminescent Vibrio campbellii in shrimp larvae of Litopenaeus vannamei. Aquaculture. 2006; 255: 48–54. [Google Scholar]
- 60.Garriques D, Arevalo G. An evaluation of the production and use off five bacterial isolates to manipulate production of P. vannamei post larvae in Ecuador. In World Aquaculture Meeting. 1995; 1–14. [Google Scholar]
- 61.Zherdmant MT, San Miguel L, Serrano J, Donoso E, Miahle E. Estudio y utilización de probióticos en el Ecuador. Panorama Acuícola. 1997; 2: 28. [Google Scholar]
- 62.Barman P, Raut S, Sen SK, Shaikh U, Bandyopadhyay P, Mahopatra PK. Effect of a three-component bacterial consortium in white shrimp farming for growth, survival and water quality management. Acta Biologica Szegediensis. 2017; 61 (1): 35–44. [Google Scholar]
- 63.Newaj-Fyzul A, Al-Harbi AH, Austin B. Developments in the use of probiotics for disease control in aquaculture. Aquaculture. 2014; 431: 1–11. [Google Scholar]
- 64.Ng WK, Lim CL, Romano N, Kua BC. Dietary short-chain organic acids enhanced resistance to bacterial infection and hepatopancreatic structural integrity of the giant freshwater prawn, Macrobrachium rosenbergii. Int Aquat Res. 2017; 9 (4): 293–302. [Google Scholar]
- 65.Vázquez JA, González MP, Murado MA. Effects of lactic acid bacteria cultures on pathogenic microbiota from fishes. Aquaculture. 2005; 245 (1–4): 149–161. [Google Scholar]
- 66.Salem AM, Amin RA. Evaluation of some organic acids as potential decontaminants of Vibrio parahaemolyticus in fresh shrimp. World Journal of Dairy & Food Sciences. 2012; 7 (1): 41–48. [Google Scholar]
- 67.Teixeira B, Marques A, Ramos C, Serrano C, Matos O, Neng NR, et al. Chemical composition and bioactivity of different oregano (Origanum vulgare) extracts and essential oil. J Sci Food Agric. 2013; 93 (11): 2707–2714. 10.1002/jsfa.6089 [DOI] [PubMed] [Google Scholar]
- 68.Russo M, Galletti GC, Bocchini P, Carnacini A. Essential oil chemical composition of wild populations of Italian Oregano Spice (Origanum vulgare ssp. hirtum (Link) Ietswaart): A preliminary evaluation of their use in chemotaxonomy by cluster analysis. 1. Inflorescences. J Agric Food Chem. 1998; 46 (9): 3741–3746. [Google Scholar]
- 69.Figiel A, Szumny A, Gutiérrez-Ortíz A, Carbonell-Barrachina ÁA. Composition of oregano essential oil (Origanum vulgare) as affected by drying method. J Food Eng. 2010; 98 (2): 240–247. [Google Scholar]
- 70.Gracia-Valenzuela MH, Vergara-Jimenez MJ, Baez-Flores ME, Cabrera-Chavez F. Antimicrobial effect of dietary oregano essential oil against Vibrio bacteria in shrimps. Arch Biol Sci. 2014; 66 (4): 1367–1370. [Google Scholar]
- 71.Baruah K, Norouzitallab P, Phong HP, Smagghe G, Bossier P. Enhanced resistance against Vibrio harveyi infection by carvacrol and its association with the induction of heat shock protein 72 in gnotobiotic Artemia franciscana. Cell Stress Chaperones. 2017; 22 (3): 377–387. 10.1007/s12192-017-0775-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manzanares DL, Morilla LJ, Malawani AD, Lagare NJ, Abrenica-Adamat L. Effects of oregano (Origanum vulgare) leaf extract on early life stages of Artemia salina. AES Bioflux. 2015; 7 (3): 468–474. [Google Scholar]
- 73.Szabó MÁ, Varga GZ, Hohmann J, Schelz Z, Szegedi E, Amaral L, et al. Inhibition of quorum-sensing signals by essential oils. Phytother Res. 2010; 24(5): 782–786. 10.1002/ptr.3010 [DOI] [PubMed] [Google Scholar]
- 74.Kerekes EB, Deák É, Takó M, Tserennadmid R, Petkovits T, Vágvölgyi C, et al. Anti-biofilm forming and anti-quorum sensing activity of selected essential oils and their main components on food-related microorganisms. J Appl Microbiol. 2013; 115(4): 933–942. 10.1111/jam.12289 [DOI] [PubMed] [Google Scholar]
- 75.Ruwandeepika HA, Jayaweera TS, Bhowmick PP, Karunasagar I, Bossier P, Defoirdt T. Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the Harveyi clade. Rev Aquac. 2012; 4(2): 59–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and supporting table. The 16S rRna sequences of bacterial strains are available from GenBank (accession numbers MH997724 to MH997742).